Key Points
Question
How much do genetic factors contribute to the variation in lifetime risk of developing amyotrophic lateral sclerosis?
Findings
This population-based parent-offspring heritability study found that genetic factors account for half of the variance in the risk of developing amyotrophic lateral sclerosis overall and close to 40% in a population devoid of known gene mutations. Heritability estimates are higher in female-specific pairings, suggesting that the C9orf72 repeat expansion may be transmitted in a sex-dependent manner.
Meaning
Inherited and noninherited factors contribute approximately equally toward amyotrophic lateral sclerosis; even in a population devoid of known gene mutations, amyotrophic lateral sclerosis heritability remains high, supporting ongoing efforts to identify causative genes.
Abstract
Importance
Heritability describes the proportion of variance in the risk of developing a condition that is explained by genetic factors. Although amyotrophic lateral sclerosis (ALS) is known to have a complex genetic origin, disease heritability remains unclear.
Objectives
To determine the extent of ALS heritability and assess the association of sex with disease transmission.
Design, Setting, and Participants
A prospective population-based parent-offspring heritability study was conducted from January 1, 2008, to December 31, 2017 to assess ALS heritability, and was the first study to assess heritability in the context of known gene mutations of large effect. A total of 1123 incident cases of ALS, diagnosed according to the El Escorial criteria and recorded on the Irish ALS register, were identified. Ninety-two individuals were excluded (non-Irish parental origin [n = 86] and familial ALS [n = 6]), and 1117 patients were included in the final analysis.
Main Outcomes and Measures
Annual age-specific and sex-specific standardized ALS incidence and mortality-adjusted lifetime risk were determined. Sex-specific heritability estimates were calculated for the overall study cohort, for those known to carry the C9orf72 (OMIM 614260) variant, and for those with no known genetic risk.
Results
A total of 32 parent-child ALS dyads were identified during the study period. Affected offspring were younger at the onset of disease (mean age, 52.0 years; 95% CI, 48.8-55.3 years) compared with their parents (mean age, 69.6 years; 95% CI, 62.4-76.9 years; P = .008). Lifetime risk of developing ALS in first-degree relatives of individuals with ALS was increased compared with the general population (1.4% [32 of 2234] vs 0.3% [2.6 of 1000]; P < .001). Mean lifetime heritability of ALS for the overall study cohort was 52.3% (95% CI, 42.9%-61.7%) and 36.9% (95% CI, 19.8%-53.9%) for those with no known genetic risk. Heritability estimates were highest in mother-daughter pairings (66.2%; 95% CI, 58.5%-73.9%).
Conclusions and Relevance
This population-based study confirms that up to 50% of variance in ALS has a genetic basis, and that the presence of the C9orf72 variant is an important determinant of heritability. First-degree relatives of individuals with ALS without a known genetic basis remain at increased risk of developing ALS compared with the general population. A higher heritability estimate in mother-daughter pairings points to a sex-mediated effect that has been previously unrecognized.
This population-based parent-offspring heritability study examines the extent of amyotrophic lateral sclerosis heritability and assesses the association of sex with disease transmission
Introduction
Amyotrophic lateral sclerosis (ALS), a multisystem neurodegenerative disease, results from a combination of environmental factors and time that interact with preexisting genetic characteristics.1 Evidence in support of a genetic origin of the disease includes familial aggregation of the disorder and the description of 30 or more genetic mutations that cosegregate with the clinical phenotype. Of these, 4 gene variants account for 55% of familial ALS cases in European populations.2
Although familial clustering of the disease implies the possibility of common genetic causality, it does not indicate the extent to which inherited genetic factors contribute to the phenotype. An alternative measure is heritability, which reflects the proportion of the variation in the risk of developing disease (liability) that is attributable to genetic factors. Viewing the problem in this manner requires an alternative approach to the more familiar binary model of diagnostics (ie, that individuals either have a disease or they do not). The liability model assumes an underlying normally distributed susceptibility for the disease within a population, where an individual’s total risk reflects the combination of genetic and nongenetic factors acting to alter an individual’s chance of developing a disease. In this model, the disease manifests when the “threshold of liability,” a fixed point on the scale of overall risk, has been exceeded (Table 1).3,4,5
Table 1. Description of Key Concepts Discussed With Explanation and Justification of Methodological Approaches Used in This Study.
| Step | Description | Formula | Data Required | Alternative Approaches | Justification for Approach Chosen |
|---|---|---|---|---|---|
| Step 3 | |||||
| Heritabilitya | Proportion of variance in liability that is attributable to additive genetic factors | Falconer’s method3 | (1) No. of affected individuals in sample; (2) total No. of individuals in sample; (3) lifetime risk; (4) sex-specific parent-offspring concordance rates |
|
|
| Liability model3 | Liability model assumes underlying normally distributed susceptibility for disease within population, where an individual’s liability reflects the combination of genetic and nongenetic factors acting to alter an individual’s risk of developing disease | Modeled as standard normal distribution with mean 0 and variance 1 | NA |
|
Liability model allows for an estimation of variance in liability within a population, which is required to estimate heritability using a pedigree approach; requires no prior knowledge about underlying genetic risk factors |
| Threshold of liability3 | Point on scale of liability above which all individuals are affected and below which all are unaffected | Standard normal deviate exceeded by sex-specific lifetime risk in population under study | (1) Sex-specific lifetime risk | NA | Prerequisite for calculating heritability of a binary trait using Falconer’s method3 |
| Concordance and covariance | Concordant parent-offspring pairs consist of proband with ALS whose parent also has ALS4 | No. of concordant pairs divided by total No. of pairs, where pairs mean sex-specific parent-offspring pairsb | (1) No. of concordant parent-offspring pairsb; (2) total No. of parent-offspring pairsb | NA | Prerequisite for calculating heritability of a binary trait using Falconer’s method3 |
| Step 2 | |||||
| Lifetime risk | Likelihood that a person will develop a certain disease during his or her lifetime | Current probability method5 | (1) Age-specific and sex-specific ALS incidence rates; (2) age structure of population; (3) birth cohort specific life-expectancies |
|
|
| Birth cohort–specific life expectancies | Mean number of years a cohort is expected to live, based on year of birth, age, and sex | Data extracted from population-specific period life expectancy tables for specific birth cohorts | (1) Mean year of birth for patients and their parents; (2) period life expectancy at various ages for population under study | NA | Prerequisite for calculating lifetime risk. |
| Step 1 | |||||
| Incidence rate | Rate of new cases per population at risk, during a specified time period | No. of disease onsets divided by sum of person years at risk | (1) No. of ALS cases diagnosed annually, considering 5-y age groups 15 to ≥85 y; (2) size of population at risk (census data); (3) standardized to US 2010 population | NA | Prerequisite for calculating lifetime risk; general population sampled must be representative of population from which individuals with ALS and their parents were drawn; to examine this, it is also necessary to determine whether annual incidence of ALS varied significantly with time |
Abbreviations: ALS, amyotrophic lateral sclerosis; NA, not applicable.
Narrow-sense heritability reflects the proportion of variance in liability attributable to additive genetic variance.
Sex-specific pairs (daughter-mother, son-mother, daughter-father, and son-father).
The classic approach toward assessment of heritability examines the difference in disease concordance between monozygotic and dizygotic twins, with higher concordance in the former implying a strong genetic influence. The challenge with this approach for rare conditions lies in the available numbers of twins. Furthermore, in late-onset diseases such as ALS, the covariance among twins may vary with time, as an unaffected twin may develop the disease at a later stage. By contrast, pedigree studies can achieve greater numbers by assessing differences in phenotype presentation across parent-offspring trios to determine narrow-sense heritability, which indicates the extent to which an exhibited phenotype will be transmitted from parent to offspring. This method is of greater practical use for predictive purposes such as those required for genetic counseling. Estimates of ALS heritability also provide guidance as to the value of the ongoing search for new ALS-associated genes.
Amyotrophic lateral sclerosis heritability has been estimated using twin,6,7 pedigree,4 and population-level genome-wide single-nucleotide polymorphism (SNP) data.8,9,10,11 Twin studies6,7 using monozygotic and dizygotic twin pairs drawn from overlapping clinic and population-based ALS cohorts provided estimates of heritability of between 38% and 85%. A pedigree study4 using data ascertained from a clinic-based registry estimated heritability to be between 40% and 60%. However, heritability estimates could only be calculated using prevalence data taken from the relevant literature, rather than from an equivalent population-based data set.
Studies using genome-wide SNP data in large case-control cohorts with the largest data sets to assess ALS heritability have yielded estimates in the range of 7.2% to 9.5%.10,11 These estimates are limited to additive genetic variance that can be captured by common genetic variants genotyped on an SNP array. These lower heritability estimates from SNP data likely reflect a predominantly rare variant-mediated genetic architecture in ALS,12 a hypothesis that is supported by enrichment of heritability in lower-frequency common SNPs.10
In this study, we have used a pedigree approach to estimate heritability based on parent-offspring concordance with reference to a liability threshold for disease, determined from incident data. More important, the general population sampled to determine incidence must be representative of the population from which individuals with ALS and their parents were drawn. To examine heritability, it is also necessary to include an assessment as to whether the annual incidence of ALS varied significantly with time. This assessment has been possible, as the Irish ALS Register has been in operation for more than 25 years. The purpose of this study was to assess for evidence of temporal change in annual ALS incidence, to determine the current lifetime risk of developing ALS in a well-characterized population, to estimate the heritability of ALS in the same population, and to determine whether heritability is modulated by sex.
Methods
Data Collection
The Irish ALS register, established in 1994, has assembled demographic and clinical data on all individuals who received a diagnosis of ALS in Ireland since its foundation, as described previously.13 In brief, individuals with a confirmed diagnosis of definite, probable, possible, or laboratory-supported probable ALS according to the El Escorial criteria14 complete a structured telephone interview in which demographic and clinical details, including familial status, are gathered. For those unable to complete the interview or those identified posthumously, the data are collected from a nominated family member. Since 2008, detailed family aggregation studies15,16 examining associated neurodegenerative and neuropsychiatric disorders among relatives of individuals with ALS have provided additional family history information. All participants provided informed written consent for the study. This study was approved by the Beaumont Hospital Research Ethics Committee (15/40).
Inclusion and Exclusion Criteria
All incident cases of ALS cases recorded in the Irish ALS register between January 1, 1995, and December 31, 2017, were included in the study for the purpose of assessing ALS incidence and lifetime risk. Only those diagnosed between January 1, 2008, and December 31, 2017, were included in deriving estimates of heritability, to optimize quality of family history information gathered and minimize potential biases associated with long-running registers.17 Furthermore, if more than 1 member of the same family received a diagnosis of ALS during this period, the individual with the most recent diagnosis was included in heritability calculations. Individuals with non-Irish parental origin were excluded.
Determining the Heritability of ALS
To estimate the heritability of ALS within a population (step 3), it is necessary first to determine the current lifetime risk of developing ALS within this population (step 2). Prior to determining the current lifetime risk of developing ALS, the annual incidence if ALS within this population must be determined (step 1) (Table 1).
Step 1: Annual ALS Incidence and Temporal Trends
Annual age-specific and sex-specific standardized ALS incidence rates were calculated using Irish population data averaged for 5 census years (1996, 2002, 2006, 2011, and 2016) and considering 5-year age groups from 15 to 85 years or older. The US 2010 population was used for standardization purposes, as it is the most frequently used standard population in ALS.18 To estimate temporal trends in ALS incidence, a simple linear regression line was fitted using calendar year as the independent variable, assessing both overall and sex-specific incidence rates. Model assumptions were assessed using a normal P-P plot, which showed no deviations, and a residual scatterplot, which demonstrated no systematic variation (standard residual range, −1.4 to 2.2).
Step 2: Lifetime Risk of ALS
The current probability method provides the criterion standard estimate of lifetime risk, as it considers competing mortality risks.5 Using this method, estimations of the number of cases of ALS that would develop in specific birth cohorts were calculated on the basis of the person-years at risk, drawn from the Irish life table from 2010-2012 and the age-specific and sex-specific ALS incidence rates, calculated above.
Potential birth cohorts were identified for patients with ALS and their parents. The mean year of birth for patients was 1946 (men, 1947; and women, 1944) and the mean life expectancy for those surviving past childhood for the patient cohort was 71 years for men and 74 years for women. The mean year of birth for patients’ parents was 1923 (men, 1920; and women, 1925) and the mean life expectancy for those surviving childhood was 66 years for men and 67 years for women. Sex-specific lifetime risk estimates of developing ALS were calculated using these life expectancy estimates and a combined mean calculated.
Risk of ALS in Relatives of Patients
All parents of patients with ALS who had also received a diagnosis of ALS were identified. Where possible, affected parents were examined at the Irish national ALS clinic in Beaumont Hospital. For affected parents identified posthumously, diagnosis was confirmed using death certificates. The sex of affected parents was identified and matched with the sex of their affected offspring. Concordance rates were calculated by dividing the number of sex-specific ALS concordant pairs by the total number of sex-specific pairs (daughter-mother, son-mother, daughter-father, and son-father).
Step 3: Heritability of ALS
Point estimates of heritability and their SEs were derived using formulas proposed by Falconer.3 The threshold of liability was set using lifetime risk as calculated, for both patients and their parent’s generations. The parent-offspring regression coefficient is the difference in the sex-specific deviation of the mean liability of both parents and offspring from the threshold (xgr − xr) as a proportion of the mean deviation of affected individuals from the population mean (ag).3 Subscripts denote the sex-specific lifetime risk in the general population (g) and in relatives (r). Narrow-sense heritability is equal to twice the estimate of the parent-offspring regression coefficient. Calculations were performed for each sex-specific parent-offspring pairing independently, with mean heritability and SE estimates calculated weighted by the reciprocal of the sampling variance for each sex-specific parent-offspring pairing.4
Genetic Screening
The Irish ALS DNA biobank has been in operation since 1999. To date, only the C9orf72 (OMIM 614260) hexanucleotide repeat expansion has been observed to be a significant genetically identifiable cause of ALS in the Irish population,19 accounting for 33% of all known familial cases of ALS.20 C9orf72 repeat expansion testing is performed using repeat-primed polymerase chain reaction plus amplicon length analysis as previously described,21 with expansions of 30 or more repeats deemed positive. Sex-specific ALS concordance rates were calculated as above including only those in whom C9orf72 status was established and used to determine heritability estimates for the C9orf72-negative population. Five previously identified patients with sporadic ALS who carried other putative ALS-causative gene mutations (TARDBP [OMIM 605078; n = 1], FUS [OMIM 137070; n = 2], SOD1 [OMIM 147450; n = 1], and SQSTM1 [OMIM 601530; n = 1])20 were excluded from analysis.
Statistical Analysis
The frequency and percentage were calculated for normally distributed nominal variables and the mean value and SD were calculated for normally distributed continuous variables. A χ2 test was used to determine the degree of association between 2 nominal variables. The probability of difference between 2 variables was tested using independent-samples t tests and probability of difference between 3 variables was tested using one-way analysis of variance with Bonferroni correction. P < .05 was considered statistically significant. All analyses were conducted using IBM SPSS Statistics, version 24 (IBM Corp).
Results
Clinical Characteristics
Comparison of demographics for both the overall incidence cohort (1995-2017) and the heritability subcohort (2008-2017) are presented in Table 2. The higher incidence of cognitive-onset disease observed in the heritability subcohort likely reflects recent improved recognition of this phenomenon. Of those with a parental history of ALS, 17 of 32 (53.1%) were female and 7 of 32 (21.9%) received a diagnosis of cognitive-onset ALS. Individuals with a parental history of ALS had a younger mean age at onset (57.9 years; 95% CI, 55.3-60.6 years) than the remaining heritability subcohort (65.1 years; 95% CI, 64.4-65.9 years; P < .001). For 9 cases, in which age at onset was available for both the affected parent and offspring, the affected offspring were younger at time of onset (mean age, 52.0 years; 95% CI, 48.8-55.3 years) compared with their parents (mean age, 69.6 years; 95% CI, 62.4-76.9 years; P = .008), despite no difference in time from onset to diagnosis between the groups (10.1 vs 13. 7 months; P = .41).
Table 2. Comparison of Demographic and Clinical Characteristics Between Overall Study Cohort (1995-2017) and Heritability Subcohort (2008-2017).
| Characteristic | No./Total No. (%) | P Value | |
|---|---|---|---|
| Incident Study: Patient Cohort 1995-2017 (n = 2128) | Heritability Study: Patient Cohort 2008-2017 (n = 1117) | ||
| Male sex | 1207 (56.7) | 626 (56.0) | .71 |
| Age, mean (SD), y | |||
| Onset | 63.9 (11.6) | 64.9 (11.3) | .03 |
| Diagnosis | 65.3 (11.6) | 66.3 (11.3) | .42 |
| Onset to diagnosis, mean (SD), mo | 14.2 (15.8) | 14.0 (14.2) | .78 |
| Site of onset | |||
| Bulbar | 734/2017 (36.4) | 382/1090 (35.0) | .46 |
| Spinal | 1194/2017 (59.2) | 626/1090 (57.4) | .33 |
| Cognitive | 50/2017 (2.5) | 45/1090 (4.1) | .01 |
| El Escorial criteria at diagnosis | |||
| Definite | 1002/1761 (56.9) | 510/823 (62.0) | .02 |
| Probable | 510/1761 (29.0) | 181/823 (22.0) | <.001 |
Incidence and Lifetime Risk of ALS
A total of 2128 patients received a diagnosis of ALS between 1995 and 2017. The annual age-standardized and sex-standardized incidence of ALS did not change with time (0.019; 95% CI, –0.004 to 0.042 per 100 000 persons; P = .14). The overall mean incidence rate was 3.1 (95% CI, 2.9-3.2) per 100 000 persons. The mean male-specific annual incidence rate was 1.8 (95% CI, 1.7-1.9) per 100 000 persons and the mean female-specific annual incidence rate was 1.3 (95% CI, 1.2-1.4) per 100 000 persons.
The current lifetime risk of developing ALS, adjusted for other-cause mortality, was 2.9 per 1000 men and 2.3 per 1000 women, corresponding to 1 in 347 men and 1 in 436 women. The mortality-adjusted lifetime risk of developing ALS during the patient’s mean life expectancy was 1.8 per 1000 men and 1.5 per 1000 women. The mortality-adjusted lifetime risk of developing ALS during the patients’ parent’s mean life expectancy was 1.2 per 1000 men and 0.9 per 1000 women. The combined mortality-adjusted lifetime risk for both patients and their parents’ lifespan was 1.5 per 1000 men and 1.2 per 1000 women.
Risk in Relatives
Between 2008 and 2017, 1123 incident cases of ALS were identified. A total of 92 individuals were excluded (non-Irish parental origin [n = 86] and familial ALS [n = 6]); 1117 patients were included in final analysis. Complete family history was available for both parents of all patients (n = 2234). A total of 32 parents had a diagnosis of ALS. Total and sex-specific concordance rates are reported in Table 3. Concordance was highest in female-female parent-offspring pairs (13 of 491 [2.6%]). Concordance was similar for female-male parent-offspring pairs (4 of 491 [0.8%]) and male-female parent-offspring pairs (5 of 626 [0.8%]). Overall, we observed that in a population devoid of known mendelian-inherited genes, the total lifetime risk of developing ALS was 0.7% (9 of 1210) in any first-degree relative of an individual with ALS. The lifetime risk of developing ALS in first-degree relatives of individuals with ALS whose genetic status is unknown is 1.4% (32 of 2234) (Table 3).
Table 3. Parent-Offspring Amyotrophic Lateral Sclerosis Concordance Rates by Overall Heritability Cohort and C9orf72-Negative Subcohort.
| Proband | Parent | Total Concordance, No./Total No. (%) | |
|---|---|---|---|
| Heritability Cohort (n = 1117) | C9orf72-Negative Cohort (n = 605) | ||
| Female | Female | 13/491 (2.6) | 3/232 (1.3) |
| Male | Female | 5/626 (0.8) | 1/373 (0.3) |
| Female | Male | 4/491 (0.8) | 2/232 (0.9) |
| Male | Male | 10/626 (1.6) | 3/373 (0.8) |
| Total | 32/2234 (1.4) | 9/1210 (0.7) | |
C9orf72
C9orf72 status was available for 674 patients diagnosed between 2008 and 2017. A total of 69 patients were C9orf72 positive; 14 of all C9orf72-positive patients (20.3%) reported a parental history of ALS.
A total of 32 patients had a parental history of ALS; 23 of these patients had C9orf72 testing performed, of whom 14 (60.9%) were C9orf72 positive. No other definitive ALS-causing mutation has been identified in these patients.19,22 Four of 9 patients (44.4%) whose fathers had ALS carried the repeat expansion. By contrast, 10 of 14 patients (71.4%) whose mothers had ALS carried the C9orf72 repeat expansion.
Heritability
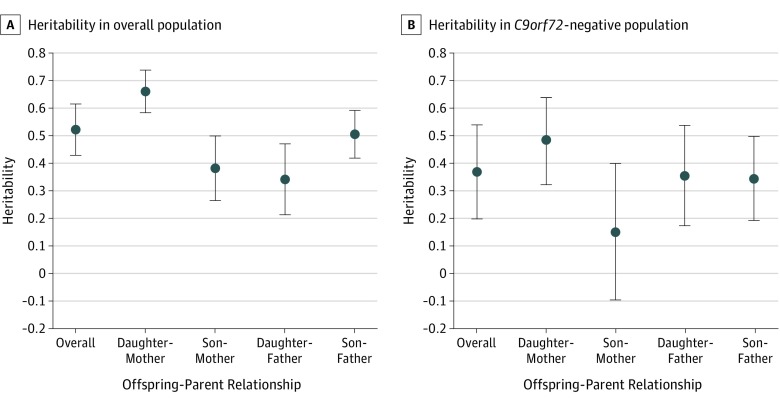
The overall mean lifetime heritability of ALS for the study cohort was 52.3% (95% CI, 42.9%-61.7%). Lifetime heritability estimates by sex pairings and genetic status are displayed in the Figure and Table 4. Unlike-sexed relative pairings, that are sex equivalent, provided the most similar heritability estimates (mother-son, 38.2%; 95% CI, 26.5%-49.9%; and father-daughter 34.2%; 95% CI, 21.4%-47.2%). If ALS is considered as a polygenic disease, heritability should be equal for all parent-offspring pairings. As such, weighing the unlike-sexed relatives’ heritability estimates by the reciprocal of their variance provides us a mean heritability estimate of 36.4% (95% CI, 24.2%-48.9%). By contrast, heritability was significantly higher in sex-specific pairings (combined mother-daughter and father-son pairings, 59.3%; 95% CI, 51.2%-67.5%; P = .002). Overall heritability estimates were highest in mother-daughter pairings (66.2%; 95% CI, 58.5%-73.9%).
Figure. Sex-Specific Heritability Estimates by Overall Heritability Cohort and C9orf72-Negative Subcohort.
A, Mean heritability estimates for overall patient cohort, with lifetime risk of developing amyotrophic lateral sclerosis of 1.5 per 1000 males and 1.2 per 1000 females. B, Mean heritability estimates for C9orf72-negative patient subcohort, with lifetime risk of developing amyotrophic lateral sclerosis of 1.5 per 1000 males and 1.2 per 1000 females. Vertical lines indicate 95% CIs.
Table 4. Exact Heritability Estimates per Study Cohort and Offspring-Parent Relationship.
| Characteristic | Heritability, % (95% CI) | |
|---|---|---|
| Overall Population | C9orf72-Negative Population | |
| Overall | 52.3 (42.9 to 61.7) | 36.9 (19.8 to 53.9) |
| Daughter-mother | 66.2 (58.5 to 73.9) | 48.5 (33.2 to 63.8) |
| Son-mother | 38.2 (26.5 to 49.9) | 15.2 (–9.5 to 40.0) |
| Daughter-father | 34.3 (21.4 to 47.2) | 35.5 (17.3 to 53.8) |
| Son-father | 50.1 (42.0 to 59.4) | 34.5 (19.3 to 49.6) |
In C9orf72-negative patients, the overall mean lifetime heritability of ALS was 36.9% (95% CI, 19.8%-53.9%) (Table 4). Female-specific heritability was 48.5% (95% CI, 33.2%-63.8%). Heritability estimates between mother-son pairings were nonsignificant, which may be partially attributable to the low number of concordant ALS pairs in this group.
Discussion
We have conducted the largest population-based pedigree study to date, to our knowledge, to determine ALS heritability. We observed a mean annual age-standardized and sex-standardized ALS incidence and current lifetime risk of developing ALS, similar to those observed by other methods. Our population-based heritability estimates of between 43% and 62% are similar to those calculated by Wingo et al4 (40%-60%) using a US clinic-based population. However, in our study, ALS heritability was assessed for the first time in a population in whom known genetic mutations have been excluded. We found that heritability estimates were not significantly different in C9orf72-negative patients when compared with the overall heritability study cohort. Although the lifetime risk of developing ALS in first-degree relatives of individuals with ALS whose genetic status is unknown is 1.4% (32 of 2234), first-degree relatives of individuals with ALS who are not known to carry any ALS-associated genetic mutations, remain at increased risk of developing ALS (lifetime risk, 0.7%) compared with the general population (lifetime risk, 0.3%).
A total of 14 of 23 (60.9%) concordant ALS pairs in whom DNA was available carried the C9orf72 repeat expansion. By contrast, only 20.3% of C9orf72-positive patients reported a parental history of ALS, suggesting that genetic anticipation or pleiotropic effects may have masked the clinical phenotype in their parents. Moreover, the affected offspring of affected parents were, on average, 17 years younger at age at onset of ALS than their parents.
Across all estimates calculated, mother-daughter pairings had the highest heritability, strongly suggesting a sex-mediated liability in this cohort. Although female sex hormones may modulate risk,23,24 it is not possible to determine how these factors may be associated with sex-specific heritability differences. A previous study has shown that both clinical phenotype and survival probability of C9orf72 expansion carriers are also influenced by sex,25 suggesting a complex interaction between sex, disease phenotype, and the repeat expansion.
Parental sex has been observed to play a role in the stability of other diseases associated with nucleotide expansions. For example, in Huntington disease, an increase in the CAG trinucleotide expansion length occurs almost exclusively through paternal transmission. By contrast, in myotonic dystrophy 1, CTG trinucleotide expansions are more likely to be transmitted maternally. Our observation of higher ALS heritability in females may be in part be driven by C9orf72 repeat expansion, as 71.4% of patients whose mothers had ALS carried the repeat expansion. That female-specific heritability remains elevated in C9orf72-negative populations, suggesting the possibility of additional uncharacterized repeat expansions associated with ALS.
Limitations
This study has several limitations. Although we found no temporal change in the incidence of ALS in Ireland during the most recent 23 years, it was not possible to determine whether incidence has changed during the entire period that patients and their parents were alive. Nonetheless, as ALS is a late-onset disease, the period assessed was that of most interest for these cohorts. Moreover, while the current probable lifetime risk describes the current risk of developing ALS for individuals of a certain age, it does not directly reflect the risk of either patients or parents at the time of their birth, nor is it associated with future risk.
Second, the association of environmental variables with risk of ALS was not directly assessed during this study. Heritability estimates are specific to the population and environment in which they are estimated. Of proposed environmental risk factors associated with ALS1,26 other than smoking,27 no clear reported change in these variables has occurred over time. Although it is possible that a changing environmental background during the last century may have been associated with heritability estimates, it seems unlikely that this would be the case given the stability of the incidence of ALS within the population.
Finally, while we have accounted for all known genes of large effect in the Irish population, it is not possible to control for unknown genes of large effect. Evidence from genome-wide association studies would suggest that a large proportion of genetic risk is driven by rare variants. Whether these are mutations within the population or isolated occurrences arising de novo in large kindreds is as yet unknown. Nonetheless, the heritability estimates generated by our study support the ongoing search for such mutations to help us explain further the pathogenic mechanisms driving the disease.
Conclusions
To our knowledge, this is the largest population-based pedigree study assessing ALS heritability to date and the first study to assess heritability in the absence of known gene mutations of large effect. We have shown that the overall heritability of ALS in this population is 52.3%. We have found that, in the absence of known ALS-associated genetic mutations in the proband, first-degree relatives of individuals with ALS remain at increased risk of developing ALS compared with the general population. Finally, we consistently observed that the highest heritability estimates occur in mother-daughter pairings, pointing to a previously unrecognized sex-mediated association with disease heritability.
References
- 1.Al-Chalabi A, Hardiman O. The epidemiology of ALS: a conspiracy of genes, environment and time. Nat Rev Neurol. 2013;9(11):617-628. doi: 10.1038/nrneurol.2013.203 [DOI] [PubMed] [Google Scholar]
- 2.Zou ZY, Zhou ZR, Che CH, Liu CY, He RL, Huang HP. Genetic epidemiology of amyotrophic lateral sclerosis: a systematic review and meta-analysis. J Neurol Neurosurg Psychiatry. 2017;88(7):540-549. doi: 10.1136/jnnp-2016-315018 [DOI] [PubMed] [Google Scholar]
- 3.Falconer DS. The inheritance of liability to certain diseases, estimated from the incidence among relatives. Ann Hum Genet. 1965;29(1):51-76. doi: 10.1111/j.1469-1809.1965.tb00500.x [DOI] [Google Scholar]
- 4.Wingo TS, Cutler DJ, Yarab N, Kelly CM, Glass JD. The heritability of amyotrophic lateral sclerosis in a clinically ascertained United States research registry. PLoS One. 2011;6(11):e27985. doi: 10.1371/journal.pone.0027985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Estève J, Benhamou E, Raymond L. Statistical methods in cancer research, volume IV: descriptive epidemiology. IARC Sci Publ. 1994;(128):1-302. [PubMed] [Google Scholar]
- 6.Graham AJ, Macdonald AM, Hawkes CH. British motor neuron disease twin study. J Neurol Neurosurg Psychiatry. 1997;62(6):562-569. doi: 10.1136/jnnp.62.6.562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Chalabi A, Fang F, Hanby MF, et al. An estimate of amyotrophic lateral sclerosis heritability using twin data. J Neurol Neurosurg Psychiatry. 2010;81(12):1324-1326. doi: 10.1136/jnnp.2010.207464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keller MF, Ferrucci L, Singleton AB, et al. Genome-wide analysis of the heritability of amyotrophic lateral sclerosis. JAMA Neurol. 2014;71(9):1123-1134. doi: 10.1001/jamaneurol.2014.1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fogh I, Ratti A, Gellera C, et al. ; SLAGEN Consortium and Collaborators . A genome-wide association meta-analysis identifies a novel locus at 17q11.2 associated with sporadic amyotrophic lateral sclerosis. Hum Mol Genet. 2014;23(8):2220-2231. doi: 10.1093/hmg/ddt587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Rheenen W, Shatunov A, Dekker AM, et al. ; PARALS Registry; SLALOM Group; SLAP Registry; FALS Sequencing Consortium; SLAGEN Consortium; NNIPPS Study Group . Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat Genet. 2016;48(9):1043-1048. doi: 10.1038/ng.3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLaughlin RL, Schijven D, van Rheenen W, et al. ; Project MinE GWAS Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium . Genetic correlation between amyotrophic lateral sclerosis and schizophrenia. Nat Commun. 2017;8:14774. doi: 10.1038/ncomms14774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLaughlin RL, Vajda A, Hardiman O. Heritability of amyotrophic lateral sclerosis: insights from disparate numbers. JAMA Neurol. 2015;72(8):857-858. doi: 10.1001/jamaneurol.2014.4049 [DOI] [PubMed] [Google Scholar]
- 13.Traynor BJ, Codd MB, Corr B, Forde C, Frost E, Hardiman O. Incidence and prevalence of ALS in Ireland, 1995-1997: a population-based study. Neurology. 1999;52(3):504-509. doi: 10.1212/WNL.52.3.504 [DOI] [PubMed] [Google Scholar]
- 14.Brooks BR, Miller RG, Swash M, Munsat TL; World Federation of Neurology Research Group on Motor Neuron Diseases . El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1(5):293-299. doi: 10.1080/146608200300079536 [DOI] [PubMed] [Google Scholar]
- 15.Byrne S, Heverin M, Elamin M, et al. Aggregation of neurologic and neuropsychiatric disease in amyotrophic lateral sclerosis kindreds: a population-based case-control cohort study of familial and sporadic amyotrophic lateral sclerosis. Ann Neurol. 2013;74(5):699-708. doi: 10.1002/ana.23969 [DOI] [PubMed] [Google Scholar]
- 16.O’Brien M, Burke T, Heverin M, et al. Clustering of neuropsychiatric disease in first-degree and second-degree relatives of patients with amyotrophic lateral sclerosis. JAMA Neurol. 2017;74(12):1425-1430. doi: 10.1001/jamaneurol.2017.2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rooney JPK, Brayne C, Tobin K, Logroscino G, Glymour MM, Hardiman O. Benefits, pitfalls, and future design of population-based registers in neurodegenerative disease. Neurology. 2017;88(24):2321-2329. doi: 10.1212/WNL.0000000000004038 [DOI] [PubMed] [Google Scholar]
- 18.Marin B, Boumédiene F, Logroscino G, et al. Variation in worldwide incidence of amyotrophic lateral sclerosis: a meta-analysis. Int J Epidemiol. 2017;46(1):57-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenna KP, McLaughlin RL, Byrne S, et al. Delineating the genetic heterogeneity of ALS using targeted high-throughput sequencing. J Med Genet. 2013;50(11):776-783. doi: 10.1136/jmedgenet-2013-101795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryan M, Heverin M, Doherty MA, et al. Determining the incidence of familiality in ALS: a study of temporal trends in Ireland from 1994 to 2016. Neurol Genet. 2018;4(3):e239. doi: 10.1212/NXG.0000000000000239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renton AE, Majounie E, Waite A, et al. ; ITALSGEN Consortium . A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257-268. doi: 10.1016/j.neuron.2011.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Project MinE ALS Sequencing Consortium Project MinE: study design and pilot analyses of a large-scale whole-genome sequencing study in amyotrophic lateral sclerosis. Eur J Hum Genet. 2018;26(10):1537-1546. doi: 10.1038/s41431-018-0177-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Jong S, Huisman M, Sutedja N, et al. Endogenous female reproductive hormones and the risk of amyotrophic lateral sclerosis. J Neurol. 2013;260(2):507-512. doi: 10.1007/s00415-012-6665-5 [DOI] [PubMed] [Google Scholar]
- 24.Rooney JPK, Visser AE, D’Ovidio F, et al. ; Euro-MOTOR Consortium . A case-control study of hormonal exposures as etiologic factors for ALS in women: Euro-MOTOR. Neurology. 2017;89(12):1283-1290. doi: 10.1212/WNL.0000000000004390 [DOI] [PubMed] [Google Scholar]
- 25.Rooney J, Fogh I, Westeneng HJ, et al. C9orf72 expansion differentially affects males with spinal onset amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2017;88(4):281. doi: 10.1136/jnnp-2016-314093 [DOI] [PubMed] [Google Scholar]
- 26.Zufiría M, Gil-Bea FJ, Fernández-Torrón R, et al. ALS: A bucket of genes, environment, metabolism and unknown ingredients. Prog Neurobiol. 2016;142:104-129. doi: 10.1016/j.pneurobio.2016.05.004 [DOI] [PubMed] [Google Scholar]
- 27.Forey BHJ, Hamling J, Thornton A, Lee P International smoking statistics (web edition): a collection of worldwide historical data: Ireland. http://www.pnlee.co.uk/Downloads/ISS/ISS-Ireland_131105.pdf. Published November 5, 2013. Accessed October 31, 2018.