Abstract
Fibroblast growth factor receptor 1 (FGFR1) has been implicated in numerous cancer types including squamous cell lung cancer, a subset of non-small cell lung cancer (NSCLC) with a dismal 5-year survival rate. Small molecule inhibitors targeting FGFR1 are currently in clinical trials, with AZD4547 being one of the furthest along; however, the development of drug resistance is a major challenge for targeted therapies. A prevalent mechanism of drug resistance in kinases occurs through mutation of the gatekeeper residue, V561M in FGFR1; however, mechanisms underlying V561M resistance to AZD4547 are not fully understood. Here, the cellular consequences of the V561M gatekeeper mutation were characterized and it was found that although AZD4547 maintains nanomolar affinity for V561M FGFR1, based on in vitro binding assays, cells expressing V561M demonstrate dramatic resistance to AZD4547 driven by increased STAT3 activation downstream of V561M FGFR1. The data reveal that the V561M mutation biases cells towards a more mesenchymal phenotype, including increased levels of proliferation, migration, invasion and anchorage-independent growth, which was confirmed using CyTOF, a novel single cell analysis tool. Using shRNA knockdown, loss of STAT3 restored sensitivity of cancer cells expressing V561M FGFR1 to AZD4547. Thus, the data demonstrate that combination therapies including FGFR and STAT3 may overcome V561M FGFR1 driven drug resistance in the clinic.
Keywords: FGFR, Lung Cancer, Drug Resistance, Cell Signaling, EMT, STAT3
INTRODUCTION
Fibroblast growth factor receptors (FGFRs) are a family of receptor tyrosine kinases (RTKs) normally involved in tissue repair, hematopoiesis, angiogenesis, and embryonic development (1,2). However, FGFR amplification and/or mutation drives a number of cancer types, including non-small cell lung cancer (NSCLC), breast cancer, gastric cancer and multiple myeloma (3-5) due to their roles in regulating cell growth and survival. FGFRs drive tumorigenesis by constitutively activating signaling pathways involved in cell proliferation and survival, which eventually promotes epithelial-mesenchymal transition (EMT), driving invasion and tumor angiogenesis (6,7).
Gene amplification of FGFR1 resulting in overexpression has been implicated in 20% of squamous cell lung cancer, a type of NSCLC with a 5-year survival rate of only 15% (3,8-10). FGFR1 is activated upon FGF binding to its extracellular domain, resulting in protein dimerization and trans-autophosphorylation of the intracellular tyrosine kinase domains. Such phosphorylation events at multiple tyrosine residues subsequently serve as recruitment sites for binding partners that lead to the activation of downstream signaling pathways (11,12). PLCγ, FRS2, and CrkII directly bind to FGFR1 and lead to activation of protein kinase C (PKC), the MAPK and PI3K/Akt signaling pathways, and JNK, respectively (13-15). Upon FGFR1 amplification, Y677 can also become phosphorylated and serve as a recruitment site for STAT3 signaling (16). FGFR family proteins are also important drivers of the epithelial-mesenchymal transition (EMT), a process important in embryonic development as well as cancer metastasis (17-19).
Due to their implication in numerous cancer types, clinical trials are underway for a number of small molecule inhibitors targeting FGFRs (20,21). However, resistance is a major problem with any targeted therapy using small molecule inhibitors, with one common mechanism of resistance in kinases occurring via the gatekeeper mutation (22,23). Because many RTK small molecule inhibitors compete with ATP to bind in the active site, interaction with a hydrophobic region adjacent to the active site is required to improve specificity for a particular target. The accessibility of this hydrophobic pocket is determined by the gatekeeper residue (24) which is normally a relatively small, hydrophobic amino acid, often valine. Upon treatment with various RTK inhibitors, this residue has been shown to mutate to a larger residue (e.g. methionine, leucine, isoleucine), thus blocking the access of the inhibitor to the pocket from which it gains its affinity and specificity for the protein (25). This mutation accommodates ATP binding, but precludes binding of inhibitors that require access to the hydrophobic pocket. FGFR1 has been shown to gain resistance to a number of inhibitors including AZD4547 upon mutation of valine 561 to a methionine (V561M) (26,27).
In the cases of EGFR, ABL, PDGFR, and Src kinases, it has been shown that the gatekeeper mutation has an activating effect on the kinase domain in addition to conferring resistance (23). The gatekeeper mutants of EGFR, PDGFR, c-ABL, and Src were shown to have activated kinase activity and an increased rate of cellular transformation (16). In addition, the EGFR T790M gatekeeper mutation has been shown to promote EMT (28-30). Previous studies in our lab have kinetically characterized V561M FGFR1 and demonstrated an increase in the kcat of V561M FGFR1 relative to WT. We also identified a slight decrease in AZD4547 affinity for V561M FGFR1, although the Kd was still in the nM, clinically relevant range (22). To confirm the efficacy of AZD4547 in a cellular context, we treated cells expressing full length WT and V561M FGFR1 with AZD4547. To our surprise, although WT cells maintained low nanomolar sensitivity to AZD4547, V561M cells were highly resistant. In this study we utilize a series of cell-based assays including immunoblotting, proliferation, transwell migration and invasion, and anchorage-independent growth assays to characterize the differences between cells expressing WT and V561M FGFR1. To investigate expression changes with single cell resolution, we applied the newly emerging technique Mass Cytometry/Cytometry Time-of-Flight (CyTOF). CyTOF is a novel technology similar to flow cytometry in which antibodies are tagged with heavy metal isotopes rather than fluorophores. This enables simultaneous detection of up to 100 parameters per cell, with minimal overlap between channels. We utilized CyTOF to study a NSCLC cell line known to be heterogeneous with a suspected stem cell population (31). Using this technique, we were able to corroborate our immunoblot findings with single cell data and identify a small percentage of cancer stem cells. This technique provides detailed expression data for multiple signaling partners and markers in a single experiment, enabling profiling of drug resistant cell lines in a higher throughput, multiplexed manner. Other groups have utilized CyTOF to study tumor samples to better understand drug responses (32,33), the roles of stem cell populations (34), and in the study of combination therapy (35). CyTOF has also been used to study drug resistance in Acute Myeloid Leukemia (AML) (36) and glioblastoma (37). In the current study, we demonstrate that this technique can be a valuable tool to investigate drug resistance mechanisms in NSCLC.
It is imperative to fully understand the mechanistic details of common drug resistance mutations prior to inhibitors reaching the clinic. This study elucidates the mechanisms underlying V561M resistance to AZD4547, demonstrating that STAT3 is required for survival in the presence of inhibitor. It also reveals the metastatic consequences of acquiring the gatekeeper mutation, due to an enhanced EMT and increased invasion in cells expressing V561M FGFR1. These results are critical to guide future targeted and combination therapies including FGFR family proteins, providing insight to pathways and processes activated downstream of V561M FGFR1, as well as suggesting methodology for characterizing additional mechanisms of drug resistance.
MATERIALS AND METHODS
Cell lines and culture conditions
The parental L6 cells used in this study were kindly provided by Dr. Joseph Schlessinger at Yale University. L6 cells express no endogenous FGFR or FGF proteins, making them an ideal system with which to compare WT and V561M FGFR1. To generate stable, isogenic cell lines overexpressing WT or V561M FGFR1, the following protocol was utilized: GPG cells were grown to 80% confluence, transfected with a pBABE vector containing WT or V561M FGFR1, and grown in viral production media (DMEM supplemented with 10% FBS) until virus was harvested (4 days). Harvested virus was used to infect L6 cells and cells incorporating the vector were selected using 2.5 μg/mL puromycin for 3-4 weeks. These stable cell lines (L6-WT, L6-V561M) were maintained in DMEM supplemented with 10% heat inactivated FBS, 1% antibiotic-antimycotic (A/A) (Gibco #15240062) and 1 μg/mL puromycin. H1581 NSCLC cell lines overexpressing WT or V561M FGFR1 (H1581-WT, H1581-V561M) were a generous gift from Dr. Roman K. Thomas at the University of Cologne (Köln, Germany). H1581 derived cell lines were grown in RPMI-1640 supplemented with 10% FBS and 1% A/A. All cells were grown at 37°C in a humidified atmosphere with 5% CO2. Cells were tested for mycoplasma and only used up to passage 25.
Reagents and antibodies
Recombinant FGF2 was purchased from Gibco (#PHG0024). Heparin sulfate was purchased from StemCell Technologies (#07980). The following antibodies were used (all primary antibodies are rabbit derived unless otherwise noted): anti-FGFR1 (#9740), phospho-FGFR (Y653/654, #3476, mouse derived), phospho-FRS2 (Y436, #3861; Y196, #3864), PLCγ (#5690), phospho-PLCγ (Y783, #2821), ERK (#4695), phospho-ERK (T202/Y204 #4370), Akt (#4691), phospho-Akt (S473 #4060), STAT3 (#4904), phospho-STAT3 (Y705, #9145), E-cadherin (#3195), Vimentin (#5741), ZEB1 (#3396), N-cadherin (#13116), HIF1α (#14179), Bcl-XL (#2764), Hsp90 (#4877), FGF2 (#61977), TGFβ (#3711), Cyclin D1 (#2978), GAPDH (#2118), horseradish peroxidase (HRP)-linked rabbit IgG secondary antibody (#7074) and HRP-linked mouse IgG secondary antibody (#7076), from Cell Signaling Technology. Anti-FRS2 (#10425, mouse derived) was purchased from Abcam. AZD4547 was purchased from Selleck Chemistry. The following CyTOF antibodies were purchased from Fluidigm Inc.: STAT3 (#3173003A), pSTAT3 (#3158005A), Vimentin (#3154014A), TGFβ (#3163010B), SOX2 (#3150019B), and Nanog (#3169014A). Custom antibody-metal conjugations were performed using kits purchased from Fluidigm, Inc. (PRD002) and carrier-free antibodies targeting FGFR1 (#9740), pFGFR1 (#3476), pFRS2 Y196 (#3864), and ZEB1 (#3396) were purchased from Cell Signaling Technology.
Cell-based assays
L6-WT, L6-V561M, H1581 WT, and H1581 V561M stable cell lines were serum starved for 12 h (RPMI 1640 – glutamine, supplemented with 1% A/A). For AZD4547 treatment experiments, 1000-5000 cells were plated in full growth media (RPMI 1640 + glutamine, supplemented with 1% A/A and 10% FBS). Varying concentrations of AZD4547 (2.5 nM-10 μM) were added after 24 h, and cells were incubated for 2-4 days before quantification using an MTT assay as described previously (38). For signaling experiments, cells were serum starved for 12 h and stimulated with 50 ng/mL FGF2 and 10 μM heparin for 0, 1, 5, and 15 minutes. Timepoints were quenched using ice-cold PBS and immediate addition of lysis buffer (RIPA Lysis Buffer (Sigma #20-188) supplemented with phosphatase (25 mM NaF and 100 mM Sodium orthovanadate) and protease inhibitors (cOmplete, mini, EDTA-free (Sigma #11836170001)). For basal lysates, cells were left untreated, and lysates were collected using ice-cold PBS and immediate addition of lysis buffer. Lysates were cleared by centrifugation, collected, and total protein content was quantified using the Pierce BCA protein assay kit (ThermoFisher #23225). Phosphorylation and/or total expression of FGFR1, downstream signaling partners, STAT3 gene targets, and EMT marker analysis were detected using immunoblotting.
Immunoblot analyses
Equal amounts of protein extracts (5 μg) were resolved by SDS-PAGE (BioRad, 4-20% gradient #4561096) and then transferred to a nitrocellulose membrane using the iBlot® machine from ThermoFisher. The membrane was blocked using 3% BSA in TBST (Tris-buffered saline with 0.1% Tween-20), then incubated with primary antibodies overnight at 4°C, washed, and incubated at room temperature with secondary antibody. Blots were developed using enhanced chemiluminescence and exposed to X-ray film. The level of protein phosphorylation or marker expression was quantified using ImageJ (NIH, Bethesda, MD). Signal density was normalized to GAPDH loading control and total protein expression for phosphorylation analyses. Every blot included in this study has been normalized to a loading control from the same gel. Representative GAPDH blots are displayed in Figures 2, 3, and 4. Data were plotted using GraphPad Prism (GraphPad, San Diego, CA).
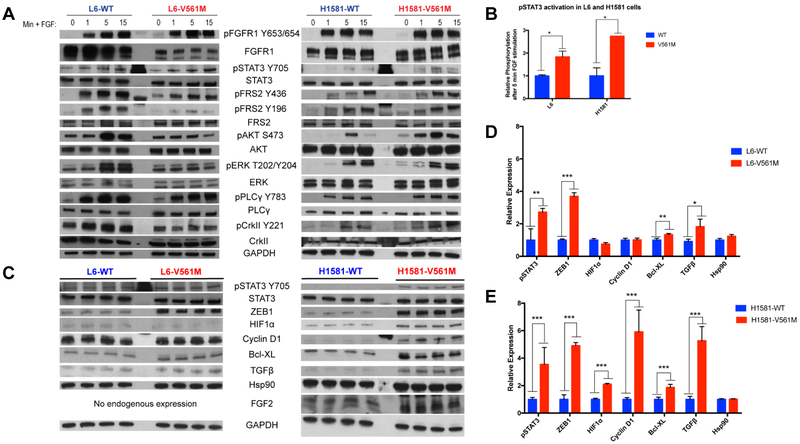
Figure 2: STAT3 is more highly activated downstream of V561M FGFR1 relative to WT.
A L6-WT, L6-V561M, H1581-WT, and H1581-V561M cells were starved for 12 h, then stimulated with 50 ng/mL FGF2 and 10 μg/mL heparin for 0, 1, 5, or 15 minutes before lysis and harvesting. 5 μg of protein was loaded onto an SDS-PAGE gel, transferred to a nitrocellulose membrane, then blotted for total and phospho-protein levels of FGFR and various signaling partners. B pSTAT3 levels for three replicate immunoblots of L6 and H1581 WT and V561M cells stimulated with 50 ng/mL FGF2 and 10 μg/mL heparin for 5 minutes were quantified using ImageJ and plotted using GraphPad Prism. C Basal L6-WT, L6-V561M, H1581-WT and H1581-V561M cells were harvested and lysed, and immunoblots were performed as described for 2A. D pSTAT3 and STAT3 gene target expression in L6- and E H1581- WT and V561M cell lines were normalized against loading controls and quantified using ImageJ. Data were plotted using GraphPad Prism. Significance values: *<0.05; **<0.01, ***<0.001.
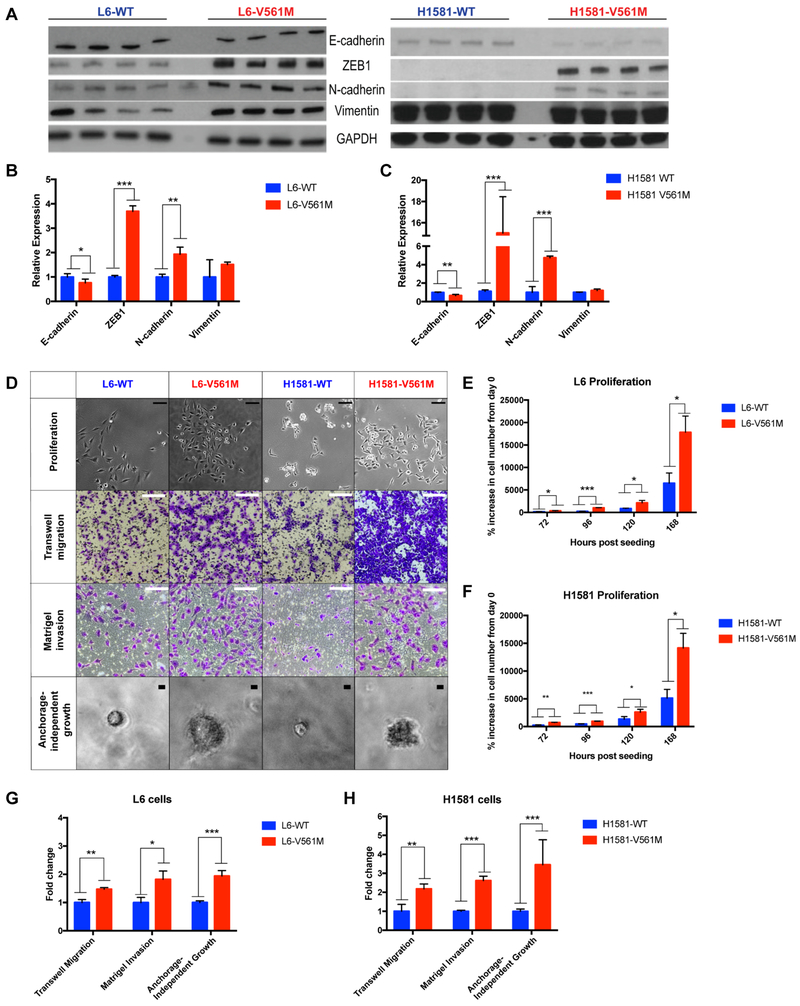
Figure 3: EMT is enhanced in V561M cells relative to WT.
A Basal L6-WT, L6-V561M, H1581-WT and H1581-V561M cells were harvested and lysed, and immunoblots of four replicate lysates were performed as described for 2C. B EMT marker expression in L6-WT, L6-V561M, and C H1581-WT and H1581-V561M cell lines were normalized against loading controls and quantified using ImageJ. Data were plotted using GraphPad Prism. D Representative images of proliferation, transwell migration, Matrigel invasion and anchorage-independent growth in L6 and H1581 cell lines are shown. Scale bars in proliferation images correspond to 100 μm; migration and invasion: 200 μm; anchorage-independent growth: 10 μm E, F Quantification of proliferation of L6 and H1581 cells after 3, 4, 5, and 7 days of proliferation are shown as % cell growth relative to the initial cell count. G, H Transwell migration, Matrigel invasion, and anchorage-independent growth in L6-WT, L6-V561M, H1581-WT, and H1581-V561M were displayed as fold-change of V561M cells relative to WT using GraphPad Prism. Significance values: *<0.05; **<0.01, ***<0.001.
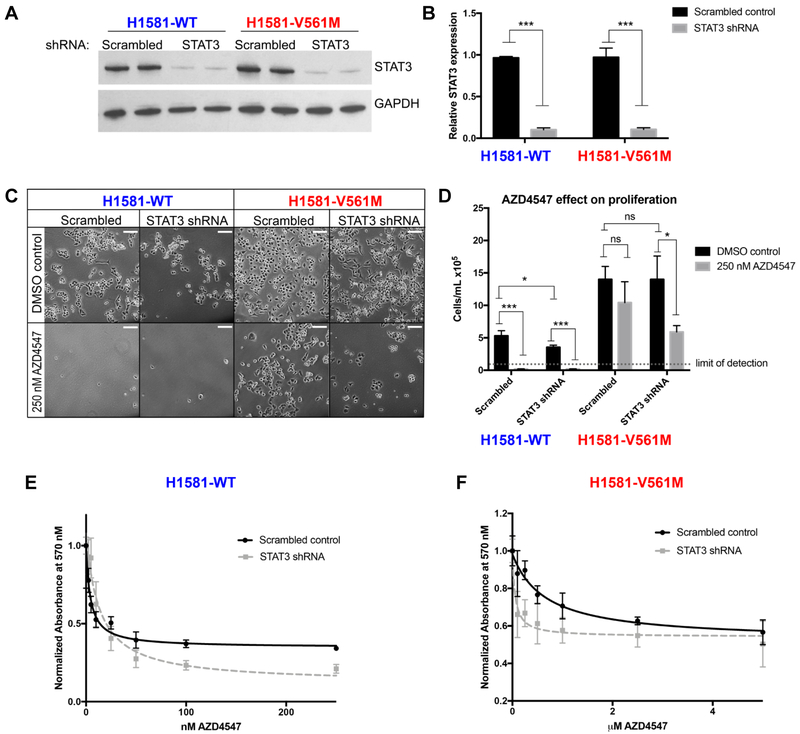
Figure 4: STAT3 knockdown sensitizes H1581-V561M cells to AZD4547 treatment.
A H1581-WT and H1581-V561M cells were infected with virus containing shRNA targeting STAT3 or a non-targeting scrambled control. Lysates were harvested and immunoblots of two replicate lysates were performed as described in 2A. B STAT3 expression levels were quantified using ImageJ and normalized to a GAPDH loading control and plotted using GraphPad Prism. C 5×104 cells were plated in full growth media containing 250 nM AZD4547 or a DMSO control and incubated for 72 h. Cells were imaged using a Nikon Eclipse Ti-S fluorescence microscope. Scale bars correspond to 100 μm. Cells were then trypsinized and quantified using the Countess automated cell counter. D Data were plotted using GraphPad Prism. E, F IC50 curves for AZD4547 treatment of H1581-WT and H1581-V561M cells treated with scrambled control or STAT3 targeting shRNA were generated as described for Figure 1. Significance values: *<0.05; **<0.01, ***<0.001.
Proliferation assay
L6-WT, L6-V561M, H1581-WT, and H1581-V561M cells were serum starved for 12 h. 5×103-5×104 cells were plated in 6-well plates in DMEM or RPMI-1640 (L6 or H1581 cells, respectively) supplemented with 10% FBS and 1% A/A, and appropriate concentration of AZD4547 if applicable. Initial cell counts were confirmed, and cells were then incubated for 48-168 h, trypsinized, and counted using the Countess automated cell counter (Invitrogen #C10227). Data were plotted as % increase in cell number using GraphPad Prism. Cells were imaged using a Nikon Eclipse Ti-S fluorescence microscope and images taken with μManager software.
Transwell migration assay
L6-WT, L6-V561M, H1581-WT, and H1581-V561M cells were serum starved for 12 h. 1×105 cells were then plated onto 8 μm-pore transwell migration filters in 24-well plates (Corning #CLS3464) in starvation media. Growth media (DMEM or RPMI-1640 with 10% FBS) was added to the bottom of the transwell and cells were incubated for either 8 h (L6 cells) or 24 h (H1581 cells) at 37°C under an atmosphere with 5% CO2. Unmigrated cells on the inner surface of the transwell chamber were removed, and migrated cells were stained with crystal violet. Assays were imaged using the Echo Revolve R4 microscope (San Diego, CA). Migrated cells were quantified using ImageJ and data were plotted using GraphPad Prism.
Matrigel invasion assay
L6-WT, L6-V561M, H1581-WT, and H1581-V561M cells were serum starved for 12 h. 2.5×104 (L6) or 5×104 (H1581) cells were then plated onto Matrigel coated 8 μm-pore transwell filters in 24-well plates (Corning #354483) in starvation media. Growth media (DMEM or RPMI-1640 with 10% FBS) was added to the bottom of the transwell and cells were incubated for 24 h (L6 cells) or 72 h (H1581 cells) at 37°C under an atmosphere with 5% CO2. Unmigrated cells on the inner surface of the transwell chamber were removed, and the transwell chambers were stained with crystal violet, then imaged using Echo Revolve R4 microscope. Invasive cells were quantified using ImageJ and data were plotted using GraphPad Prism.
Soft agar anchorage-independent growth assay
The soft agar anchorage-independent growth assay was performed using a modified protocol based on the CytoSelect™ 96-well cell transformation assay kit purchased from CellBio Labs (#CBA130). L6-WT, L6-V561M, H1581-WT, and H1581-V561M cells were serum-starved for 12 h. A bottom layer of agar was plated in a 96-well plate. 1×105 cells were then seeded in agar containing 2x DMEM supplemented with 25% FBS. Finally, complete growth media was plated on top. Plates were incubated for 6-8 days, then quantified using the MTT cell viability assay and a Spectramax M5 plate reader. Data were plotted using GraphPad Prism.
CyTOF Analysis
CyTOF analyses were performed on the CyTOF 2 instrument (Fluidigm, Inc) at the Yale University Core Facility. For CyTOF experiments, cells were grown to 80% confluence, stimulated, treated with cisplatin (live/dead stain) (Fluidigm, Inc. #201064), and trypsinized prior to fixation. Different cell types and treatment conditions were treated with Cell-ID 20-plex Pd Barcodes (#201060), combined into one sample, and stained using heavy metal-tagged surface antibodies. Cells were then permeabilized using methanol and stained using heavy metal-tagged intracellular antibodies. Surface- and intracellular- stained cells were then incubated with an Iridium-tagged intercalation solution (Fluidigm, Inc. #201192B), then run on the CyTOF 2 instrument. Analysis was performed using FlowJo software (FlowJo, LLC).
shRNA knockdown procedures
We obtained 5 different shRNA constructs targeting various parts of the STAT3 gene, as well as a scrambled, non-targeting control as a generous gift from Dr. Ben Turk and Dr. David Calderwood (Yale University) (39,40). HEK293T cells were cotransfected with a pLKO.1 puro vector (Sigma SHC002) containing the shRNA of interest or the scrambled, non-targeting control, vsvG, and dR8.91 packaging vectors using polyethylenimine (PEI). L6 and H1581 cells were subjected to multiple rounds of infection with lentivirus harvested from these cells. The shRNA with the most effective knockdown was selected (shRNA 2: CCTGAGTTGAATTATCAGCTT, scrambled, non-targeting control: CAACAAGATGAAGAGCACCAA). Knockdown efficiency was measured by quantifying immunoblot data using ImageJ. Data were plotted using GraphPad Prism.
RESULTS
V561M FGFR1 maintains nM binding affinity for AZD4547 but confers dramatic resistance in cells
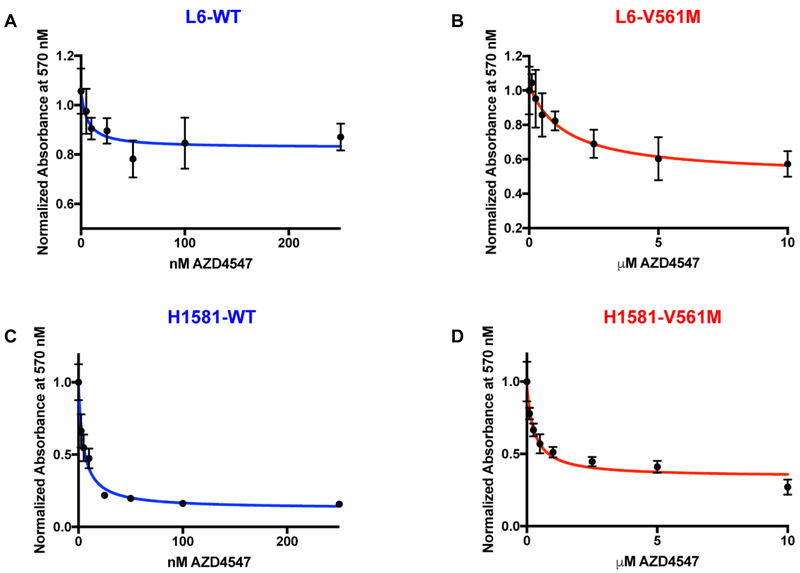
Our previous fluorescence binding studies (22) demonstrated that AZD4547 maintains nM binding affinity for V561M FGFR1 (Table 1). To further investigate the potency of AZD4547 against WT vs. V561M FGFR1, we performed cell-based assays to observe the effects of AZD4547 on cell viability in two cell lines. We utilized L6 cells, rat myoblasts with no endogenous FGFR expression, to ensure an isogenic background, and validated our findings using a non-small cell lung cancer cell line, H1581, engineered to overexpress WT or V561M FGFR1. We calculated IC50 values for AZD4547 treatment of WT and V561M FGFR1 in both L6 and H1581 cells and obtained values of 6 ± 4 nM for L6-WT cells, 1400 ± 700 nM for L6-V561M cells, 4.8 ± 0.8 nM for H1581-WT cells, and 510 ± 90 nM for H1581-V561M cells (Table 1, Figure 1A-D). Thus, while our previously obtained in vitro data demonstrated a 32-fold, modest decrease in binding affinity of AZD4547 for V561M FGFR1 relative to WT, cell-based assays display a 100-200 fold increase in IC50 for cells expressing V561M relative to those expressing WT FGFR1. To understand the mechanism behind V561M driven resistance, we further characterized the FGFR1 V561M mutation, studying differences in downstream signaling and cellular tumorigenicity between cells expressing WT and V561M FGFR1.
Table 1:
Kd and IC50 values for WT and V561M FGFR1
| WT | V561M | Fold-change | |
|---|---|---|---|
| Kd (nM)a | 2±1 | 64±11 | 32 |
| IC50 in L6 cells (nM) | 6±4 | 1400±700 (1.4 μM) | 230 |
| IC50 in H1581 cells (nM) | 4.8±0.8 | 510±90 (0.51 μM) | 100 |
Data are from fluorescence titration experiments performed by Sohl, et al 2015 (22)
Figure 1: V561M FGFR1 displays dramatic AZD4547 resistance in cell-based assays.
A, B L6 and C, D H1581 cells overexpressing WT or V561M FGFR1 were starved for 12 h and 1000-5000 cells were plated in 96 well plates. After 24 h cells were treated with AZD4547, and after 2-4 days cell viability was quantified using an MTT assay. Cell viability was plotted against [AZD4547] using GraphPad prism before fitting to an inhibition curve to obtain IC50 values.
STAT3 is more highly activated downstream of V561M FGFR1 relative to WT FGFR1
To compare the signaling downstream of WT and V561M FGFR1, we starved cells for 12 h, which was the amount of time required to ablate WT FGFR1 phosphorylation. We stimulated cells with FGF and heparin, and measured activation of downstream signaling partners after a series of timepoints using immunoblotting. We found significantly and consistently increased phosphorylation of STAT3 in L6-V561M and H1581-V561M cells (2-fold and 3-fold, respectively; p= 0.03 and 0.01, Figure 2A,B). PLCγ and CrkII were activated to similar levels between WT and V561M in both cell lines. In L6-V561M cells, we observed minimal phosphorylation of Akt and ERK upon FGF stimulation. However, Akt and ERK phosphorylation levels are similar between H1581-WT and H1581-V561M cells, suggesting that this is a cell-line specific phenomenon (Figure 2A). Because STAT3 was consistently upregulated in both L6-V561M and H1581-V561M cells, we chose to further investigate the activation of STAT3 signaling in cells expressing V561M FGFR1. We performed immunoblots probing for known STAT3 gene targets, including ZEB1, HIF1α, Cyclin D1, Bcl-XL, TGFβ, Hsp90, and FGF2 (Figure 2C-E). In H1581-V561M cells, we observed significantly higher levels of pSTAT3, ZEB1, HIF1α, Cyclin D1, and Bcl-XL relative to H1581-WT cells. L6-V561M cells expressed higher levels of pSTAT3, ZEB1, Bcl-XL, and TGFβ relative to H1581-WT cells, but we observed no obvious difference in HIF1α or Cyclin D1 expression. Neither cell line displayed differences in Hsp90 expression. Interestingly, we observed that FGF2 was upregulated in H1581-V561M cells relative to WT (Figure 2C). We confirmed FGF2 expression in the H1581-V561M cells, and demonstrated that the FGF2 levels in L6 cells are extremely low, using qPCR (Supplementary Figure S1). The STAT3 marker upregulation was much more pronounced in the H1581 cells, suggesting that an autocrine loop may be present in the V561M H1581 cells, enhancing the effects of the V561M mutation. Co-amplification of FGFR1 and FGF2 resulting in a similar autocrine signaling loop has been identified as a driver of resistance to EGFR-targeted therapies (41).
The V561M gatekeeper mutation biases cells towards a more mesenchymal phenotype
Among the upregulated STAT3 targets observed in cells expressing V561M FGFR1 are multiple EMT regulators, including TGFβ and ZEB1 (42,43). This led us to investigate whether L6 and H1581 cells expressing V561M FGFR1 display more mesenchymal characteristics relative to WT cells. We performed immunoblotting experiments probing for epithelial and mesenchymal markers (Figure 3A-C) and found that L6-V561M and H1581-V561M cells express consistently higher levels of mesenchymal markers relative to L6-WT and H1581-WT (ZEB1; 3.7-fold and 15-fold, respectively, N-cadherin; 2-fold and 5-fold, respectively; Vimentin, not significant but moderately increased), as well as consistently lower levels of epithelial marker E-cadherin (1.3-fold and 1.5-fold, respectively) (Figure 3A-C).
We then examined the effects of this mutation on cellular behaviors associated with EMT: proliferation, migration, invasion and anchorage-independent growth (Figure 3D-H). L6-V561M and H1581-V561M cells demonstrated significantly enhanced levels of each of these processes relative to their WT counterparts. We observed a 2-4-fold increase in proliferation in V561M cells relative to WT in both L6 and H1581 cell lines by validating initial cell counts and quantifying the % increase in cell number after 72, 96, 144 and 168 h of cell growth (See Supplementary Tables S1 and S2 for cell counts and p-values). Through Boyden chamber transwell migration assays, we found a 2-fold increase in migration in both L6-V561M and H1581-V561M cells relative to WT (p=0.04, 0.01, respectively) by quantifying DAPI stained migrated cells on the bottom layer of the Boyden chamber. Using transwell Matrigel invasion assays, we observed a 1.5-fold increase in invasion in L6 V561M cells relative to WT (p=0.01) and a 2-fold increase in H1581-V561M cells relative to WT (p=0.0004). We observed a 2-fold increase in anchorage-independent growth in L6-V561M cells relative to WT (p=0.001) and a 4-fold increase in H1581-V561M cells (p<0.0001) using an MTT assay. Representative images of proliferation, transwell migration, Matrigel invasion, and anchorage-independent growth experiments are shown in Figure 3D and quantifications are shown in 3E-H. These findings, in combination with the consistently higher expression of mesenchymal markers and lower expression of epithelial markers, demonstrate that the V561M mutation biases cells towards a more mesenchymal phenotype relative to cells expressing WT FGFR1.
STAT3 knockdown restores nM AZD4547 sensitivity in H1581-V561M cells
To investigate the role of STAT3 in the enhanced EMT and AZD4547 resistance observed in H1581-V561M cells relative to WT, we used shRNA to knock down STAT3. We obtained a 10-fold knockdown in H1581-WT and H1581-V561M cells (H1581-WT-STAT3-KD, H1581-VM-STAT3-KD) (Figure 4A,B), and performed cell proliferation assays using H1581-WT-STAT3-KD and H1581-V561M-STAT3-KD cells and control cells treated with a scrambled, non-targeting shRNA. H1581-WT-STAT3-KD cell lines displayed slightly decreased cell proliferation relative to those treated with scrambled shRNA, and proliferation was abolished in both cases with 250 nM AZD4547 (Figure 4C,D). Interestingly, H1581-V561M-STAT3-KD cells did not display a decreased level of proliferation relative to those treated with scrambled shRNA, suggesting that STAT3 alone is not responsible for the increased EMT in H1581-V561M cells. However, H1581-V561M-STAT3-KD cells are significantly sensitized to AZD4547 treatment (p=0.03, Figure 4C,D), demonstrating that they are dependent on STAT3 for survival in the presence of AZD4547. We confirmed this sensitization using cell viability assays, and found IC50 values of 50 ± 30 nM for H1581-V561M-STAT3-KD cells and 700 ± 300 nM for V561M cells treated with the non targeting scrambled control (Table 2, Figure 4E-F). The sensitivity of H1581-WT cells to AZD4547 was largely unaffected by STAT3 knockdown, with IC50s of 4 ± 1 nM for cells treated with the scrambled control and 13 ± 4 nM in H1581-WT-STAT3-KD cells.
Table 2:
AZD4547 sensitivity of H1581 shRNA-treated cells
| IC50 [scrambled] | IC50 [shRNA STAT3] | Fold-change | |
|---|---|---|---|
| H1581-WT | 4 ± 1 nM | 13 ± 4 nM | ↑3 |
| H1581-V561M | 700 ± 300 nM | 50 ± 30 nM | ↓14 |
| Fold-change | 175 | 1.7 | --- |
To confirm that this effect was not cell-line specific, we repeated the STAT3 knockdown experiments in L6-WT and L6-V561M cells and saw a similar sensitization to AZD4547 in L6-V561M cells. We obtained IC50 values of 140 ± 50 nM for L6-V561M-STAT3-KD cells and 700 ± 400 for L6-V561M cells treated with the non targeting scrambled control. As observed with H1581-WT cells, sensitivity of L6-WT cells was unaffected by STAT3 knockdown, with IC50s of 1.4 ± 0.6 nM and 1.2 ± 0.7 nM for L6-WT cells treated with the scrambled, non-targeting control and STAT3 targeting shRNA, respectively (Supplementary Figure S2, Supplementary Table S3). Together, this data suggests that STAT3 plays a critical role in the survival of cells expressing V561M FGFR1.
CyTOF provides a multiplexed, single-cell-resolution approach to characterize drug resistant cell lines
Immunoblot experiments probing for signaling pathway activation downstream of WT and V561M FGFR1 identified STAT3 as a driver of AZD4547 resistance. The value of this type of analysis in guiding combination therapies inspired us to search for a method to expand this type of analysis to characterize drug resistant cell lines in a more high-throughput context. To address this, we utilized CyTOF, a novel technology that enables single cell analysis of up to 20 sample conditions and 38 markers in one experiment (44). Using palladium barcoding (45) and metal-tagged antibodies, we were able to simultaneously probe multiple sample conditions for activation of FGFR1 and its downstream signaling partners, as well as EMT and stem cell marker expression. The power of CyTOF to identify small subpopulations of cells led us to focus our CyTOF studies on H1581 cells due to their previously suggested cancer stem cell population and clinical relevance (31, 46). We stained unstimulated and stimulated (5 or 15 min + FGF2 and heparin) cells with the antibodies listed in Table 3.
Table 3:
CyTOF antibodies and heavy metal conjugates
| Antibody Target | Heavy Metal Isotope |
|---|---|
| FGFR1 | 164Dy |
| pFGFR1 | 165Gd |
| pTyrosine | 144Nd |
| pFRS2 Y196 | 145Nd |
| STAT3 | 173Yb |
| pSTAT3 | 158Gd |
| pERK T202/Y204 | 167Er |
| pAKT S473 | 152Sm |
| TGFβ | 163Dy |
| SOX2 | 150Nd |
| Vimentin | 154Sm |
| Nanog | 169Tm |
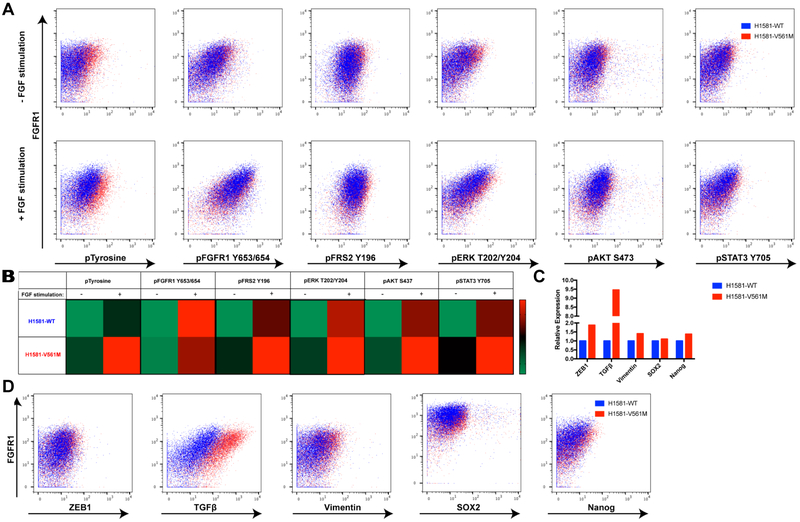
CyTOF data were largely consistent with immunoblots, showing significantly increased levels of TGFβ, ZEB1 and Vimentin expression in V561M H1581 cells relative to WT (Figure 5C,D). Consistent with data demonstrating the enhanced kcat of V561M FGFR1, unstimulated H1581-V561M cells demonstrated higher levels of total phosphotyrosine relative to WT, as well as a corresponding increase with FGF stimulation. We observed slightly increased basal levels of pSTAT3 in H1581-V561M cells relative to WT, as well as a corresponding increase upon FGF stimulation. We also observed similarly increased levels of pFRS2, pERK, and pAKT (Figure 5A,B). We postulate that the increase in STAT3 phosphorylation may have been underestimated in our CyTOF analysis due to limitations with the antibody. Phospho-antibodies often have an appreciable amount of nonspecific binding, which can be accounted for in immunoblot experiments by confirming the correct molecular weight. In CyTOF analysis, this is not possible; since the pSTAT3 signal was already low, the nonspecific background may have masked the increase in pSTAT3. Additional optimization studies are necessary to generate phospho-specific antbodies with reduced background for CyTOF experiments. Notably, we observed a small population of highly SOX2 positive cells in H1581-WT and H1581-V561M cell lines, consistent with studies that have demonstrated cancer stem cell activity in H1581 cells (31). H1581-V561M cells displayed slightly higher overall SOX2 and Nanog levels relative to H1581-WT cells, suggesting that cancer stem cells could play a role in the enhanced EMT observed in H1581-V561M cells, although further studies are required to confirm this possibility.
Figure 5: STAT3 upregulation in H1581-V561M cells and mesenchymal marker expression were confirmed using CyTOF.
A 2×106 cells were barcoded before combining unstimulated and stimulated conditions of H1581-WT and H1581-V561M cells together for both surface and intracellular staining. Cells were surface stained, methanol permeabilized, and stained for intracellular antibodies. Pairwise plots showing marker expression vs FGFR1 in each treatment condition were generated using FlowJo software. B Representative heatmaps showing average activation of each signaling partner before and after FGF stimulation in H1581-V561M and H1581-WT were generated using GraphPad Prism. C Representative bar graphs demonstrating EMT and stem cell marker expression were generated using GraphPad Prism. D Pairwise plots showing marker expression vs FGFR1 in each treatment condition were generated using FlowJo software.
CyTOF is an exciting new technology being used by many labs to obtain single cell expression data to understand cancer biology (34-36,44,45). In this study we have utilized this powerful technique to obtain single-cell data confirming our findings regarding pathway activation, epithelial and mesenchymal marker expression, and STAT3 gene target expression. In addition, CyTOF enabled us to observe a potential cancer stem cell population that may play a role in drug resistance. Our findings, combined with impressive studies from other labs (34-36,44,45) demonstrate the power of CyTOF as a method to profile drug resistant tumors. Through the generation of an antibody panel containing signaling proteins, EMT and stem cell markers, drug resistant tumors can be profiled efficiently, providing an initial snapshot of signaling pathway activation. Subsequent validation using immunoblots can then guide application of combination therapies in the clinic.
DISCUSSION
Targeted therapies are a promising option for cancer treatments, offering the ability to specifically target cancer cells through inhibition of a signaling molecule responsible for oncogenesis. The major drawback of this type of treatment is the development of drug resistance, which can occur through activation of a bypass pathway, or by mutation of the targeted protein (47,48). The gatekeeper mutation is a devastating mechanism of the latter form of resistance in established kinase targets such as BCR-ABL, EGFR, PDGFR, and Src (23). Thus, identifying corresponding gatekeeper mutations in novel targets allows an understanding of their consequences before they reach the clinic. FGFRs are a relatively recent family of proteins to be addressed through targeted therapy, with multiple inhibitors in clinical trials for the treatment cancers such as NSCLC and breast cancer (20,49). Additionally, the V561M mutation of FGFR1 has been observed in humans (50) and requires only a single nucleotide substitution to occur. FGFR gatekeeper mutations have been identified in the literature through acquired resistance experiments using multiple TKIs (26,51), demonstrating the importance of further characterizing this mutation.
AZD4547 is currently in clinical trials for NSCLC (52), and our lab recently found that this inhibitor is able to maintain nM binding affinity for V561M FGFR1 in in vitro binding experiments (22). We determined the IC50 of AZD4547 in L6 and H1581 cells expressing WT or V561M FGFR1 and found that cells expressing V561M FGFR1 are highly resistant to AZD4547, demonstrating that an alternative mechanism is responsible for driving V561M resistance to AZD4547 in addition to the modest decrease in binding affinity. We performed an in-depth analysis of signaling downstream of WT and V561M and identified STAT3 as the driver of AZD4547 resistance through shRNA knockdown. Although STAT3 has been a notoriously undruggable target in past years, recent advances using antisense oligonucleotides (ASOs) have been successful in attenuating cancers driven by STAT3 (53). AZD9150 is an ASO with nanomolar potency for STAT3 that is currently in phase I and II clinical trials for numerous cancer types, including NSCLC, lymphoma and neuroblastoma (53,54).
Another vitally important characteristic we identified in cells expressing V561M FGFR1 is an enhanced EMT relative to cells expressing WT FGFR1. The increased proliferation, migration, invasion and transformation observed in L6-V561M and H1581-V561M suggest that tumors that have acquired this resistance mutation will be more likely to metastasize to surrounding tissues (19). This demonstrates a major pitfall of utilizing single targeted therapies in the clinic; the mechanism of drug resistance has a secondary effect of increasing the metastatic potential of the tumor. EMT has also been identified as a mechanism of drug resistance in EGFR T790M driven cancers (28,29,55), suggesting that this may be a common phenomenon among RTK gatekeeper mutations.
We confirmed that the V561M mutation imparts a growth advantage to both L6 and H1581 cells relative to WT FGFR1, leading to the question of whether V561M could be a driver of tumorigenesis independently of FGFR1 amplification in a clinical setting. Because FGFR family proteins are more recent cancer targets, inhibitors are still progressing through clinical trials, and there are not yet any FDA-approved FGFR-specific inhibitors. We postulate that the V561M likely occurs in cancer patients but has not yet been reported, and we turn to the long-established target EGFR to speculate on what may occur with FGFR1 V561M as inhibitors become more commonly used in the clinic. A study by Bemanian, et. al. has demonstrated that the EGFR T790M mutation is present at low abundance in early breast cancer patients prior to treatment with kinase inhibitors (56). Furthermore, Bell, et. al. have identified EGFR T790M as a germline mutation that increases inherited susceptibility to lung cancer in a European family (57). In addition, the EGFR T790M mutation in parallel with the L858R activating mutation has been shown to drive more aggressive tumors in mice relative to those expressing the T790M mutation alone (58). This indicates that the activating effects of gatekeeper mutations are context-dependent, and that the V561M mutation may only incur a significant growth advantage when it occurs in addition to FGFR1 amplification. The example of the EGFR T790M mutation demonstrates that the gatekeeper mutation can be present at a low abundance independently of kinase inhibition, and become more tumorigenic when it occurs in the context of an additional activating event. Similarly to the EGFR T790M mutation, the V561M FGFR1 mutation may be present at low levels that are difficult to detect, and if patients are treated with inhibitors that are ineffective against V561M FGFR1, this may unintentionally select for more aggressive tumor cells. Additionally, the FGFR4 V550L and V550E gatekeeper mutations have been observed in rhabdomyosarcoma tumors (59), and the FGFR2 V564F gatekeeper mutation has arisen as a mechanism of resistance in cholangiocarcinoma patients expressing FGFR2 fusion proteins treated with the FGFR-specific inhibitor BGJ398 in the clinic (60). Taken together, these studies demonstrate a precedent for FGFR gatekeeper mutations occurring clinically, both before and after treatment with targeted therapies.
Receptor tyrosine kinases signal through many pathways, and drug resistance can occur through activation of any of these signaling partners. We identified STAT3 activation downstream of V561M FGFR1 through immunoblot experiments, and recognized the potential of the novel technique CyTOF to extend the utility of this type of analysis to a higher throughput method that can be completed at a timescale applicable to clinical studies. As a proof-of-concept, we confirmed the upregulation of STAT3 gene target and mesenchymal marker expression in H1581-V561M cells relative to WT cells using CyTOF. We also analyzed signaling pathway activation downstream of FGFR1 in H1581-WT and H1581-V561M cells and observed a modest increase in STAT3 phosphorylation in H1581-V561M cells. However, we also observed similarly increased levels of FRS2, AKT and ERK phosphorylation. We hypothesize that the limitations associated with phospho-specific antibodies, including low signal strength and decreased specificity, mask the differences between STAT3 phosphorylation in H1581-WT and H1581-V561M cells. Our data suggest that CyTOF is a valuable technique to identify subpopulations of cells and to obtain an overall idea of pathway activation downstream of mutant receptors, but that parallel immunoblot analysis is still imperative to confirm CyTOF data. This study shows that CyTOF analysis provides an invaluable set of data that can identify signaling pathway activation, epithelial or mesenchymal characteristics of cells, and even isolate small populations of cancer stem cells that are undetectable with immunoblot approaches.
In this study we have demonstrated that cells expressing the V561M drug resistance mutation of FGFR1 display increased mesenchymal characteristics and depend on STAT3 activity for survival in the presence of AZD4547. Our findings are relevant for clinicians treating NSCLC patients with FGFR1 amplification; while previous data suggested that AZD4547 would be effective against V561M FGFR1 (22), this study confirms the importance of using second-generation FGFR inhibitors that are effective towards the gatekeeper mutant, such as the covalent inhibitors FIIN-1 and 2 (61), or combination therapies including drugs targeting STAT3 such as AZD9150 (53). In addition, the enhanced EMT, including increased invasion, observed in cells expressing V561M FGFR1 reveals the consequences of acquiring this mutation on cancer metastasis. We suggest that patients be screened for FGFR1 mutational status prior to treatment with FGFR targeted inhibitors, and when possible, an inhibitor that maintains efficacy against V561M be utilized as part of a combination therapy. We demonstrate the strength of combining immunoblotting and CyTOF analysis to obtain a thorough understanding of signaling pathway activation in drug resistant cell lines and identify small cellular populations, and affirm CyTOF as a promising technology for rapid cellular characterization in a clinical setting. Our data also suggests that STAT3 is a promising option for targeting FGFR1-driven cancers that have acquired the V561M drug resistance mutation.
Supplementary Material
Implications: The V561M FGFR1 gatekeeper mutation leads to devastating drug resistance through activation of STAT3 and the epithelial-mesenchymal transition; this study demonstrates that FGFR1 inhibitor sensitivity can be restored upon STAT3 knockdown.
Acknowledgements
The authors thank Dr. Joseph Schlessinger for kindly providing L6 parental cell lines. The authors also thank Dr. Roman K. Thomas, Dr. Florian Malchers, Dr. Graziella Bosco, and Ms. Ilona Dahmen for generously providing H1581-WT and H1581-V561M cell lines. We appreciate the help of Dr. Ruth Montgomery and Ms. Shelly Ren for access to the Yale CyTOF Core Facility.
Grant Support
This research was supported by National Institutes of Health Grant NCI F31 CA216915-02 to Molly R. Ryan.
Financial Support: This research was supported by National Institutes of Health Grant NCI F31 CA216915-02 to Molly R. Ryan.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors declare no conflicts of interest.
REFERENCES
- 1.Wesche J, Haglund K, Haugsten EM. Fibroblast growth factors and their receptors in cancer. Biochem J 2011;437(2):199–213 doi 10.1042/BJ20101603. [DOI] [PubMed] [Google Scholar]
- 2.Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov 2009;8(3):235–53 doi 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med 2010;2(62):62ra93 doi 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chioni AM, Grose R. FGFR1 cleavage and nuclear translocation regulates breast cancer cell behavior. The Journal of cell biology 2012;197(6):801–17 doi 10.1083/jcb.201108077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt K, Moser C, Hellerbrand C, Zieker D, Wagner C, Redekopf J, et al. Targeting Fibroblast Growth Factor Receptor (FGFR) with BGJ398 in a Gastric Cancer Model. Anticancer Res 2015;35(12):6655–65. [PubMed] [Google Scholar]
- 6.Greulich H, Pollock PM. Targeting mutant fibroblast growth factor receptors in cancer. Trends Mol Med 2011;17(5):283–92 doi 10.1016/j.molmed.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown WS, Akhand SS, Wendt MK. FGFR signaling maintains a drug persistent cell population following epithelial-mesenchymal transition. Oncotarget 2016. doi 10.18632/oncotarget.13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helsten T, Elkin S, Arthur E, Tomson BN, Carter J, Kurzrock R. The FGFR Landscape in Cancer: Analysis of 4,853 Tumors by Next-Generation Sequencing. Clin Cancer Res 2016;22(1):259–67 doi 10.1158/1078-0432.CCR-14-3212. [DOI] [PubMed] [Google Scholar]
- 9.Jiang T, Gao G, Fan G, Li M, Zhou C. FGFR1 amplification in lung squamous cell carcinoma: a systematic review with meta-analysis. Lung Cancer 2015;87(1):1–7 doi 10.1016/j.lungcan.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 10.Dutt A, Ramos AH, Hammerman PS, Mermel C, Cho J, Sharifnia T, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One 2011;6(6):e20351 doi 10.1371/journal.pone.0020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohammadi M, Dikic I, Sorokin A, Burgess WH, Jaye M, Schlessinger J. Identification of six novel autophosphorylation sites on fibroblast growth factor receptor 1 and elucidation of their importance in receptor activation and signal transduction. Mol Cell Biol 1996;16(3):977–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev 2005;16(2):139–49 doi 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Mohammadi M, Honegger AM, Rotin D, Fischer R, Bellot F, Li W, et al. A tyrosine-phosphorylated carboxy-terminal peptide of the fibroblast growth factor receptor (Flg) is a binding site for the SH2 domain of phospholipase C-gamma 1. Mol Cell Biol 1991;11(10):5068–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ong SH, Guy GR, Hadari YR, Laks S, Gotoh N, Schlessinger J, et al. FRS2 proteins recruit intracellular signaling pathways by binding to diverse targets on fibroblast growth factor and nerve growth factor receptors. Mol Cell Biol 2000;20(3):979–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsson H, Klint P, Landgren E, Claesson-Welsh L. Fibroblast growth factor receptor-1-mediated endothelial cell proliferation is dependent on the Src homology (SH) 2/SH3 domain-containing adaptor protein Crk. J Biol Chem 1999;274(36):25726–34. [DOI] [PubMed] [Google Scholar]
- 16.Dudka AA, Sweet SM, Heath JK. Signal transducers and activators of transcription-3 binding to the fibroblast growth factor receptor is activated by receptor amplification. Cancer Res 2010;70(8):3391–401 doi 10.1158/0008-5472.CAN-09-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gros J, Tabin CJ. Vertebrate limb bud formation is initiated by localized epithelial-to-mesenchymal transition. Science 2014;343(6176):1253–6 doi 10.1126/science.1248228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiao J, Zhao X, Liang Y, Tang D, Pan C. FGF1-FGFR1 axis promotes tongue squamous cell carcinoma (TSCC) metastasis through epithelial-mesenchymal transition (EMT). Biochem Biophys Res Commun 2015;466(3):327–32 doi 10.1016/j.bbrc.2015.09.021. [DOI] [PubMed] [Google Scholar]
- 19.Geiger TR, Peeper DS. Metastasis mechanisms. Biochim Biophys Acta 2009;1796(2):293–308 doi 10.1016/j.bbcan.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 20.Gavine PR, Mooney L, Kilgour E, Thomas AP, Al-Kadhimi K, Beck S, et al. AZD4547: an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res 2012;72(8):2045–56 doi 10.1158/0008-5472.CAN-11-3034. [DOI] [PubMed] [Google Scholar]
- 21.Guffanti F, Chila R, Bello E, Zucchetti M, Zangarini M, Ceriani L, et al. In Vitro and In Vivo Activity of Lucitanib in FGFR1/2 Amplified or Mutated Cancer Models. Neoplasia 2017;19(1):35–42 doi 10.1016/j.neo.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sohl CD, Ryan MR, Luo B, Frey KM, Anderson KS. Illuminating the molecular mechanisms of tyrosine kinase inhibitor resistance for the FGFR1 gatekeeper mutation: the Achilles’ heel of targeted therapy. ACS Chem Biol 2015;10(5):1319–29 doi 10.1021/acschembio.5b00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azam M, Seeliger MA, Gray NS, Kuriyan J, Daley GQ. Activation of tyrosine kinases by mutation of the gatekeeper threonine. Nat Struct Mol Biol 2008;15(10):1109–18 doi 10.1038/nsmb.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Shah K, Yang F, Witucki L, Shokat KM. A molecular gate which controls unnatural ATP analogue recognition by the tyrosine kinase v-Src. Bioorg Med Chem 1998;6(8):1219–26. [DOI] [PubMed] [Google Scholar]
- 25.Bunney TD, Wan S, Thiyagarajan N, Sutto L, Williams SV, Ashford P, et al. The Effect of Mutations on Drug Sensitivity and Kinase Activity of Fibroblast Growth Factor Receptors: A Combined Experimental and Theoretical Study. EBioMedicine 2015;2(3):194–204 doi 10.1016/j.ebiom.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cowell JK, Qin H, Hu T, Wu Q, Bhole A, Ren M. Mutation in the FGFR1 tyrosine kinase domain or inactivation of PTEN is associated with acquired resistance to FGFR inhibitors in FGFR1-driven leukemia/lymphomas. Int J Cancer 2017;141(9):1822–9 doi 10.1002/ijc.30848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang D, Chen Q, Guo Y, Zhang T, Guo W. Insight into resistance mechanisms of AZD4547 and E3810 to FGFR1 gatekeeper mutation via theoretical study. Drug Des Devel Ther 2017;11:451–61 doi 10.2147/DDDT.S129991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rastogi I, Rajanna S, Webb A, Chhabra G, Foster B, Webb B, et al. Mechanism of c-Met and EGFR tyrosine kinase inhibitor resistance through epithelial mesenchymal transition in non-small cell lung cancer. Biochem Biophys Res Commun 2016;477(4):937–44 doi 10.1016/j.bbrc.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang N, Wang D, Li X, Yang Z, Zhang G, Wang Y, et al. A case report of EGFR mutant lung adenocarcinoma that acquired resistance to EGFR-tyrosine kinase inhibitors with T790M mutation and epithelial-to-mesenchymal transition. Respir Med Case Rep 2017;22:183–6 doi 10.1016/j.rmcr.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iderzorig T, Kellen J, Osude C, Singh S, Woodman JA, Garcia C, et al. Comparison of EMT mediated tyrosine kinase inhibitor resistance in NSCLC. Biochem Biophys Res Commun 2018;496(2):770–7 doi 10.1016/j.bbrc.2018.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ji W, Yu Y, Li Z, Wang G, Li F, Xia W, et al. FGFR1 promotes the stem cell-like phenotype of FGFR1-amplified non-small cell lung cancer cells through the Hedgehog pathway. Oncotarget 2016;7(12):15118–34 doi 10.18632/oncotarget.7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subrahmanyam PB, Dong Z, Gusenleitner D, Giobbie-Hurder A, Severgnini M, Zhou J, et al. Distinct predictive biomarker candidates for response to anti-CTLA-4 and anti-PD-1 immunotherapy in melanoma patients. J Immunother Cancer 2018;6(1):18 doi 10.1186/s40425-018-0328-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chew V, Lee YH, Pan L, Nasir NJM, Lim CJ, Chua C, et al. Immune activation underlies a sustained clinical response to Yttrium-90 radioembolisation in hepatocellular carcinoma. Gut 2018. doi 10.1136/gutjnl-2017-315485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng Z, Konopleva M, Andreeff M. Single-Cell Mass Cytometry of Acute Myeloid Leukemia and Leukemia Stem/Progenitor Cells. Methods Mol Biol 2017;1633:75–86 doi 10.1007/978-1-4939-7142-8_5. [DOI] [PubMed] [Google Scholar]
- 35.Anchang B, Davis KL, Fienberg HG, Williamson BD, Bendall SC, Karacosta LG, et al. DRUG-NEM: Optimizing drug combinations using single-cell perturbation response to account for intratumoral heterogeneity. Proc Natl Acad Sci U S A 2018. doi 10.1073/pnas.1711365115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang X, Mak PY, Mu H, Tao W, Mak DH, Kornblau S, et al. Disruption of Wnt/beta-Catenin Exerts Antileukemia Activity and Synergizes with FLT3 Inhibition in FLT3-Mutant Acute Myeloid Leukemia. Clin Cancer Res 2018. doi 10.1158/1078-0432.CCR-17-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qazi MA, Vora P, Venugopal C, Sidhu SS, Moffat J, Swanton C, et al. Intratumoral heterogeneity: pathways to treatment resistance and relapse in human glioblastoma. Ann Oncol 2017;28(7):1448–56 doi 10.1093/annonc/mdx169. [DOI] [PubMed] [Google Scholar]
- 38.Pannecouque C, Daelemans D, De Clercq E. Tetrazolium-based colorimetric assay for the detection of HIV replication inhibitors: revisited 20 years later. Nat Protoc 2008;3(3):427–34 doi 10.1038/nprot.2007.517. [DOI] [PubMed] [Google Scholar]
- 39.Shi X, Mihaylova VT, Kuruvilla L, Chen F, Viviano S, Baldassarre M, et al. Loss of TRIM33 causes resistance to BET bromodomain inhibitors through MYC- and TGF-beta-dependent mechanisms. Proc Natl Acad Sci U S A 2016;113(31):E4558–66 doi 10.1073/pnas.1608319113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A 2008;105(51):20380–5 doi 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terai H, Soejima K, Yasuda H, Nakayama S, Hamamoto J, Arai D, et al. Activation of the FGF2-FGFR1 autocrine pathway: a novel mechanism of acquired resistance to gefitinib in NSCLC. Mol Cancer Res 2013;11(7):759–67 doi 10.1158/1541-7786.MCR-12-0652. [DOI] [PubMed] [Google Scholar]
- 42.Katsuno Y, Lamouille S, Derynck R. TGF-beta signaling and epithelial-mesenchymal transition in cancer progression. Curr Opin Oncol 2013;25(1):76–84 doi 10.1097/CCO.0b013e32835b6371. [DOI] [PubMed] [Google Scholar]
- 43.Takeyama Y, Sato M, Horio M, Hase T, Yoshida K, Yokoyama T, et al. Knockdown of ZEB1, a master epithelial-to-mesenchymal transition (EMT) gene, suppresses anchorage-independent cell growth of lung cancer cells. Cancer Lett 2010;296(2):216–24 doi 10.1016/j.canlet.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roussel M, Ferrell PB Jr., Greenplate AR, Lhomme F, Le Gallou S, Diggins KE, et al. Mass cytometry deep phenotyping of human mononuclear phagocytes and myeloid-derived suppressor cells from human blood and bone marrow. J Leukoc Biol 2017;102(2):437–47 doi 10.1189/jlb.5MA1116-457R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Atkuri KR, Stevens JC, Neubert H. Mass cytometry: a highly multiplexed single-cell technology for advancing drug development. Drug Metab Dispos 2015;43(2):227–33 doi 10.1124/dmd.114.060798. [DOI] [PubMed] [Google Scholar]
- 46.Malchers F, Dietlein F, Schottle J, Lu X, Nogova L, Albus K, et al. Cell-autonomous and non-cell-autonomous mechanisms of transformation by amplified FGFR1 in lung cancer. Cancer Discov 2014;4(2):246–57 doi 10.1158/2159-8290.CD-13-0323. [DOI] [PubMed] [Google Scholar]
- 47.Barouch-Bentov R, Sauer K. Mechanisms of drug resistance in kinases. Expert Opin Investig Drugs 2011;20(2):153–208 doi 10.1517/13543784.2011.546344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engelman JA, Settleman J. Acquired resistance to tyrosine kinase inhibitors during cancer therapy. Curr Opin Genet Dev 2008;18(1):73–9 doi 10.1016/j.gde.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 49.Bello E, Colella G, Scarlato V, Oliva P, Berndt A, Valbusa G, et al. E-3810 is a potent dual inhibitor of VEGFR and FGFR that exerts antitumor activity in multiple preclinical models. Cancer Res 2011;71(4):1396–405 doi 10.1158/0008-5472.CAN-10-2700. [DOI] [PubMed] [Google Scholar]
- 50.Bennett JT, Tan TY, Alcantara D, Tetrault M, Timms AE, Jensen D, et al. Mosaic Activating Mutations in FGFR1 Cause Encephalocraniocutaneous Lipomatosis. Am J Hum Genet 2016;98(3):579–87 doi 10.1016/j.ajhg.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chell V, Balmanno K, Little AS, Wilson M, Andrews S, Blockley L, et al. Tumour cell responses to new fibroblast growth factor receptor tyrosine kinase inhibitors and identification of a gatekeeper mutation in FGFR3 as a mechanism of acquired resistance. Oncogene 2013;32(25):3059–70 doi 10.1038/onc.2012.319. [DOI] [PubMed] [Google Scholar]
- 52.Paik PK, Shen R, Berger MF, Ferry D, Soria JC, Mathewson A, et al. A Phase Ib Open-Label Multicenter Study of AZD4547 in Patients with Advanced Squamous Cell Lung Cancers. Clin Cancer Res 2017;23(18):5366–73 doi 10.1158/1078-0432.CCR-17-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hong D, Kurzrock R, Kim Y, Woessner R, Younes A, Nemunaitis J, et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci Transl Med 2015;7(314):314ra185 doi 10.1126/scitranslmed.aac5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Odate S, Veschi V, Yan S, Lam N, Woessner R, Thiele CJ. Inhibition of STAT3 with the Generation 2.5 Antisense Oligonucleotide, AZD9150, Decreases Neuroblastoma Tumorigenicity and Increases Chemosensitivity. Clin Cancer Res 2017;23(7):1771–84 doi 10.1158/1078-0432.CCR-16-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou J, Hu Q, Zhang X, Zheng J, Xie B, Xu Z, et al. Sensitivity to chemotherapeutics of NSCLC cells with acquired resistance to EGFR-TKIs is mediated by T790M mutation or epithelial-mesenchymal transition. Oncol Rep 2018;39(4):1783–92 doi 10.3892/or.2018.6242. [DOI] [PubMed] [Google Scholar]
- 56.Bemanian V, Sauer T, Touma J, Lindstedt BA, Chen Y, Odegard HP, et al. The epidermal growth factor receptor (EGFR / HER-1) gatekeeper mutation T790M is present in European patients with early breast cancer. PLoS One 2015;10(8):e0134398 doi 10.1371/journal.pone.0134398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bell DW, Gore I, Okimoto RA, Godin-Heymann N, Sordella R, Mulloy R, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet 2005;37(12):1315–6 doi 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- 58.Regales L, Balak MN, Gong Y, Politi K, Sawai A, Le C, et al. Development of new mouse lung tumor models expressing EGFR T790M mutants associated with clinical resistance to kinase inhibitors. PLoS One 2007;2(8):e810 doi 10.1371/journal.pone.0000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taylor JGt, Cheuk AT, Tsang PS, Chung JY, Song YK, Desai K, et al. Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J Clin Invest 2009;119(11):3395–407 doi 10.1172/JCI39703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goyal L, Saha SK, Liu LY, Siravegna G, Leshchiner I, Ahronian LG, et al. Polyclonal Secondary FGFR2 Mutations Drive Acquired Resistance to FGFR Inhibition in Patients with FGFR2 Fusion-Positive Cholangiocarcinoma. Cancer Discov 2017;7(3):252–63 doi 10.1158/2159-8290.CD-16-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan L, Wang J, Tanizaki J, Huang Z, Aref AR, Rusan M, et al. Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors. Proc Natl Acad Sci U S A 2014;111(45):E4869–77 doi 10.1073/pnas.1403438111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.