Abstract
Lesion‐symptom mapping has become a cornerstone of neuroscience research seeking to localize cognitive function in the brain by examining the sequelae of brain lesions. Recently, multivariate lesion‐symptom mapping methods have emerged, such as support vector regression, which simultaneously consider many voxels at once when determining whether damaged regions contribute to behavioral deficits (Zhang, Kimberg, Coslett, Schwartz, & Wang, 2014). Such multivariate approaches are capable of identifying complex dependences that traditional mass‐univariate approach cannot. Here, we provide a new toolbox for support vector regression lesion‐symptom mapping (SVR‐LSM) that provides a graphical interface and enhances the flexibility and rigor of analyses that can be conducted using this method. Specifically, the toolbox provides cluster‐level family‐wise error correction via permutation testing, the capacity to incorporate arbitrary nuisance models for behavioral data and lesion data and makes available a range of lesion volume correction methods including a new approach that regresses lesion volume out of each voxel in the lesion maps. We demonstrate these new tools in a cohort of chronic left‐hemisphere stroke survivors and examine the difference between results achieved with various lesion volume control methods. A strong bias was found toward brain wide lesion‐deficit associations in both SVR‐LSM and traditional mass‐univariate voxel‐based lesion symptom mapping when lesion volume was not adequately controlled. This bias was corrected using three different regression approaches; among these, regressing lesion volume out of both the behavioral score and the lesion maps provided the greatest sensitivity in analyses.
Keywords: Aphasia, lesion‐symptom mapping, lesion volume, support vector regression
1. INTRODUCTION
A central goal of cognitive neuroscience is to understand the relationship between structure and function in the brain. Early insights into this relationship came from linking behavioral deficits exhibited by individuals while they were alive with the general location of a brain lesion discovered on autopsy. Using this logic, 19th century aphasiologists established the foundation of our modern understanding of language in the brain, localizing language function to the left hemisphere, with frontal regions supporting speech production faculties (Broca, 1866) and posterior regions supporting auditory language comprehension (Wernicke, 1874).
The introduction of computed tomography (CT) in the 1970s and the spread of magnetic resonance imaging (MRI) in the 1980s allowed lesions to be considered in vivo with greater spatial resolution, enabling approximately congruous slices to be compared across patients. Through the 1990s, researchers used this logic to study groups of individuals categorized by behavioral syndrome, lesion characteristics, or specific behavioral deficit. This approach is limited because it requires categorization of individuals using arbitrary cutoffs based on behavioral symptoms or lesion distribution. Within the domain of language, this work both supported traditional views of the lesion‐deficit relationship in aphasia (Mazzocchi & Vignolo, 1979; Naeser & Hayward, 1978; Yarnell, Monroe, & Sobel, 1976) and highlighted important exceptions that suggested the traditional views were incomplete (Basso, Lecours, Moraschini, & Vanier, 1985). This work also suggested that critical brain regions extended beyond classical language areas (Alexander, Naeser, & Palumbo, 1990), and indeed that the role of classical language regions like Broca's area must be revisited (Mohr et al., 1978).
Today, lesion‐deficit correlation studies are conducted with millimeter‐scale precision by relying on high‐resolution structural MRI scans. This voxel‐based lesion‐symptom mapping (VLSM) technique produces a statistical map showing the strength of the relationship between damage at any given voxel and performance on a behavioral measure of interest across a group of individuals with brain lesions (Bates et al., 2003). Analyses are performed on lesion maps delineated by hand (Ashton et al., 2003) or through automated methods (Pustina et al., 2016) on anatomical scans which are then warped to a standard space. Statistical analysis conceptually involves conducting a between‐group comparison at each voxel in the sample of spatially normalized lesion maps, with behavioral score binned by lesion status at that voxel. Hot regions in the resulting statistical map correspond to voxels where lesioned status predicts the measured behavior over and above chance alone. These regions are inferred to relate to the cognitive functions relied on by the behavior under investigation. A similar technique, voxel‐based morphometry, uses the same logic except tissue values are continuous rather than binary lesion maps (Ashburner & Friston, 2000; Geva, Baron, Jones, Price, & Warburton, 2012). These procedures not only exploit the full spatial resolution of modern medical imaging but alleviate the need to categorize patients with arbitrary cutoffs, as they model behavioral measures on continuous scales (Bates et al., 2003).
In addition to demonstrating proof of concept, the initial VLSM publication challenged traditional notions of Broca's and Wernicke's areas, finding anterior insula important for fluency and middle temporal gyrus important for auditory comprehension over and above Wernicke's area (Bates et al., 2003). In a relatively short period of time as their introduction, univariate voxelwise lesion‐symptom mapping techniques have become widely used, resulting in important findings on crucial brain structures for language (Mirman et al., 2015), written language processing (Rapcsak et al., 2009), visual attention (Karnath, Fruhmann Berger, Küker, & Rorden, 2004), and cognitive control (Gläscher et al., 2012), among many other functions.
One limitation of traditional VLSM is its reliance on a mass‐univariate analytical approach, where the relationship between lesion status and behavioral performance is considered one voxel at a time. A consequence of this approach is that information concerning the spatial relationship between a specific voxel and neighboring voxels is not incorporated into the analysis. This constraint equates to an assumption of statistical independence across voxels, but such an assumption is unwarranted given both the spatial autocorrelation across voxels inherent to neuroimaging data and the nonrandom distribution of lesions throughout the brain (Husain & Nachev, 2007; Mah, Husain, Rees, & Nachev, 2014). The consequences of assuming voxelwise independence in VLSM includes not only a loss of statistical power but also a potential bias in regional localization of the lesion‐deficit relationship, in particular when a behavior of interest relies on multiple distinct brain regions (Herbet, Lafargue, & Duffau, 2015; Kimberg, Coslett, & Schwartz, 2007; Mah et al., 2014). Indeed, large‐scale simulations have demonstrated that the assumption of statistical independence across voxels can lead traditional mass‐univariate VLSM analyses to produce biased lesion‐symptom maps. The same work has highlighted the potential for multivariate inference methods to reduce or remove this source of error (Mah et al., 2014). By considering the contribution of many voxels simultaneously, these methods enable the detection of multivariate relationships between lesion location and behavioral deficit.
Recently, Zhang et al. (2014) proposed and validated an approach to lesion‐symptom mapping using support vector regression (SVR), an application of support vector machines (SVMs) that permits regression involving continuous variables such as behavioral scores (Cortes & Vapnik, 1995). This new approach, called support vector regression‐based lesion‐symptom mapping (SVR‐LSM), provides a multivariate method that addresses these important limitations of the traditional mass‐univariate approach. In their study, the authors demonstrated that SVR‐LSM has higher sensitivity and specificity compared to VLSM when applied to simulated data, and higher sensitivity to lesion‐behavior relationships in real data.
Zhang et al. have made their implementation of SVR‐LSM publicly available online as a free download. Here we describe a new MATLAB toolbox for SVR‐LSM that provides several upgrades over this prior implementation, including a graphical interface to improve usability and new options that allow more flexible and rigorous analysis designs. First, we implement a permutation‐based cluster‐level correction for multiple comparisons correction. This method addresses limitations of the voxelwise FDR approach implemented previously in SVR‐LSM, which treated each voxel as an independent comparison, resulting in potentially flawed corrections. Permutation‐based multiple‐comparisons correction methods are considered the gold standard for lesion symptom mapping because they account for the lesion autocorrelation structure inherent to the data sets (Kimberg et al., 2007; Mirman et al., 2018; Nichols & Holmes, 2002; Wilson, 2017). Second, we provide the ability to control for multiple covariates in the analysis design and provide more flexibility in how the covariates are handled via nuisance models. Although permutation‐based multiple comparisons correction and support for multiple covariates has been available in mass‐univariate lesion‐symptom mapping tools like vlsm2 (Bates et al., 2003) and npm (Rorden, Karnath, & Bonilha, 2007), these techniques have not yet to our knowledge been implemented in a multivariate environment. Third, we provide new options for lesion volume correction in multivariate lesion‐symptom mapping, including a method to control for lesion volume that removes bias from all voxels in the data.
Lesion volume is an important confounding variable in lesion‐symptom mapping analyses because larger lesions tend to cause more severe deficits regardless of location (Price, Hope, & Seghier, 2017). Further, at any given voxel, the patients with a lesion occupying that voxel are likely to have larger strokes than those without a lesion at that voxel. This imbalance is likely to lead to a bias toward identifying lesion‐behavior associations throughout the brain, even in regions with no role in the behavior under investigation. One approach to control for lesion volume has been to regress it out of behavioral scores, which transforms behavioral scores to remove their relationship with lesion volume prior to lesion‐symptom mapping. These transformed behavioral scores are then used in a subsequent lesion‐symptom mapping analysis (Karnath et al., 2004; Schwartz, Faseyitan, Kim, & Coslett, 2012). This approach is analogous to a semi‐partial correlation, which is an atypical method of covariate handling in general statistical analyses but is advantageous in that any between‐group statistical test can be used for the lesion‐symptom mapping analysis (e.g., t‐test, Brunner‐Munzel test). Software packages that model the lesion‐deficit relationship using multiple regression (e.g., vlsm2) can simply include lesion volume as second predictor along with the behavioral score, thus controlling for the lesion volume confound in both the voxel data and the behavior. This approach limits the statistical test used for lesion‐symptom mapping to voxelwise regressions. Because translation of this approach to SVR‐LSM is not straightforward, Zhang et al. (2014) introduced “direct total lesion volume control” (dTLVC) which places greater weight on the contribution of smaller lesions to control for lesion volume in the lesion map data. Prior to SVR analysis, the voxel values in each lesion map are divided by the square root of the lesion volume for that patient. Although the method addresses bias in lesioned voxels, unlesioned voxels remain uncorrected because zero values are unaffected by the transform. Here, we introduce an additional approach in which lesion volume is regressed out of each voxel in the lesion maps prior to SVR analysis, thus correcting both voxels with and without lesions. In the new SVR‐LSM toolbox, we provide several options for lesion volume correction: dTLVC, regression of lesion volume out of behavioral scores, regression of lesion volume out of lesion maps, and regression of lesion volume out of both behavioral scores and lesion maps. This last approach is conceptually appealing because it is mathematically nearly equivalent to the way in which covariates are typically treated in between‐group statistical comparisons (e.g., ANCOVA), and also the way covariates including lesion volume are handled in regression‐based mass‐univariate lesion‐symptom mapping software (e.g., vlsm2). Below, in addition to describing the features of the new SVR‐LSM toolbox, we test the impact of each of the available correction methods on the results of analyses on a cohort of left hemisphere stroke survivors. Finally, we examine two cases of how controlling for lesion volume affects the results and interpretation of multivariate lesion‐symptom mapping analyses. To demonstrate the generality of our findings about lesion volume correction beyond multivariate lesion‐symptom mapping, we also replicate some analyses using a mass‐univariate VLSM approach (see Supporting Information Figures).
2. METHODS
2.1. Preexisting implementation
The SVM classifier is a type of supervised machine learning model that has become popular for multivariate data analysis. SVMs take labeled training data as input, nonlinearly transform the data into a high‐dimensional feature space, and then in this feature space estimate a linear decision boundary, or hyperplane, based on the input features according to a loss‐function that seeks to correctly categorize as many training points as possible. The subset of training data points that constrain and straddle the estimated hyperplane are referred to as support vectors. A trained SVM model can then be used to classify novel, unobserved data points. Complicated patterns in the data can be learned by applying the so‐called “kernel trick” in which a kernel function assists with an otherwise potentially intractable mapping of the data to feature space. SVR extends the technique to regression (Cortes & Vapnik, 1995) which can operate on continuous variables such as the behavioral measures associated with a lesion‐symptom mapping study. When applied to lesion‐symptom mapping, the voxelwise lesion statuses in a predefined mask region are collapsed into a column vector for each subject and combined so that rows correspond to unique spatial locations across the sample of lesion maps. These data are used as input to train an SVR model to predict the behavioral data under investigation. To recover the relevant anatomical locations, the trained model's predictive hyperplane is back projected into the input data space, which can then be overlaid onto a standard brain image for interpretation. The behavior of the SVR algorithm utilized here (ε‐SVM; epsilon‐insensitive SVM) is determined by two hyperparameters. The first is epsilon, which defines a range around the hyperplane margins within which the model is not penalized for misclassified data points. The second is cost, which specifies the penalty for misclassified data points, where a higher penalty encourages more support vector points and a more complex fit to the data. In addition to these two hyperparameters, the use of a nonlinear radial basis function (Gaussian) kernel introduces a third hyperparameter, kernel scale (also referred to as sigma or gamma) corresponding to the width of the Gaussian kernel function. Hyperparameters are chosen prior to running the SVR, and choices are often optimized based on properties of the data at hand. The mathematical equations underlying the core SVR‐LSM functionality were described by Zhang et al. (2014) and will not be described again here. The original implementation of SVR‐LSM by Zhang et al. (2014) is available for free from the SPM website (http://www.fil.ion.ucl.ac.uk/spm/ext/#SVRLSMtbx) and the author's github website (https://github.com/yongsheng-zhang/SVR-LSM). Procedures are implemented in MATLAB scripts and rely on both SPM (Wellcome Institute of Imaging Neuroscience, London, UK) functions for basic image manipulation and LibSVM (Chang & Lin, 2011), a third‐party package, for SVR functionality. Analyses are configured by editing a MATLAB script file such that relevant variable values reflect a user's analysis choices. The software allows up to one behavioral covariate, and correction for lesion volume may be incorporated into an analysis via dTLVC (Zhang et al., 2014). The software generates a map of SVR‐β values which are then converted to p values via permutation testing at the voxel level, and optionally applies a FDR correction threshold to the permutation analysis results. Although this existing implementation of SVR‐LSM supports a minimum cluster size threshold, this arbitrary value is provided by the user.
The original SVR‐LSM software used fixed hyperparameter values, with a cost of 30 and a gamma of 5. These parameters are recommended by the authors of the initial SVR‐LSM publication (Zhang et al., 2014), in which a metric of prediction accuracy and an index of reproducibility were derived from their large data set to determine optimal parameters for performing lesion‐symptom mapping using SVR. Specifically, they state “…there is no golden standard for choosing the optimal [gamma and cost] values, they were chosen to have a compromised high reproducibility and prediction accuracy. … while the optimal values were determined using the 106 patients’ data, similar values should work for other data too as we empirically tested (data not shown).” Based on this recommendation, the analyses in this manuscript also utilize these recommended hyperparameter values. The default epsilon value in LibSVM (0.1) was used.
2.2. New implementation
We have adapted the preexisting implementation of SVR‐LSM and provide several upgrades to the original software to improve usability and to allow for more flexible analysis designs, as detailed below. The free software may be downloaded at its github repository (http://github.com/atdemarco/svrlsmgui).
2.2.1. Handling of lesion volume
Although it is recognized that correction for lesion volume is important in lesion‐symptom mapping, it is not agreed on how best to perform the correction. We introduce a method of removing correlations between lesion volume and lesion status at each voxel that affects both the one‐ and zero‐valued voxels in the raw lesion data. Specifically, prior to SVR analysis, we perform a linear regression between the lesion status values at each voxel and the lesion volume values, residualizing the voxel values with respect to their independent relationship with lesion volumes. This has the desirable effect of correcting lesion volume biases in both in the lesioned voxels and in unlesioned voxels, whose values are zero, which make up much of each data set. As noted above, transforming either the behavioral data or the lesion data prior to lesion‐symptom mapping analysis is common in current analysis packages. The general approach is analogous to a nuisance model commonly used in resting state fMRI analysis (e.g., Poldrack, Mumford, & Nichols, 2011, p. 133), and also to hierarchical regression, in which one builds an initial model using a set of baseline predictors, and then subsequently adds predictors of interest to determine if they predict the dependent variable above and beyond the variables specified in the original model.
As our primary goal is to make a variety of tools available to investigators and not restrict methodological choice, in the new software we make available this novel method, in addition to previously used methods available for correcting for lesion volume. The five options include: (a) no correction for lesion volume, (b) dTLVC correction (Zhang et al., 2014), (c) regressing lesion volume out of the behavior of interest (Karnath et al., 2004; Schwartz et al., 2012), (d) regressing lesion volume out of the lesion voxel data, and finally (e) regressing lesion volume out of both the behavioral data and lesion data. This last approach is nearly equivalent to a regular partial correlation, ANCOVA model, or including lesion volume as an additional predictor in a multiple regression, such as is utilized in vlsm2 (Bates et al., 2003).
To explore the effect that each of the five approaches to lesion volume correction has on the stringency of results of lesion‐symptom maps, we calculated number of suprathreshold voxels for twenty behaviors in a cohort of individuals with chronic left‐hemisphere stroke (N = 49) using each method. Behaviors investigated included oral and written naming performance on the Philadelphia Naming Test (Roach, Schwartz, Martin, Grewal, & Brecher, 1996), category and letter fluency scores, subtests from the Western Aphasia Battery (Kertesz, 1982) including Spontaneous Speech Content, Spontaneous Speech Fluency, Repetition, Auditory Verbal Word Recognition, Auditory Verbal Yes No, and Sequential Commands, Pseudoword Repetition (in‐house), imageability effect (concrete minus abstract word reading, in‐house), regularity effect (regular minus exception word reading, in‐house), lexicality effect (word minus pseudoword reading, in‐house), Pyramids and Palm Trees (Howard & Patterson, 1992), Digit Span Backward, Picture Pointing, score on Complex Ideational Material from the Boston Diagnostic Aphasia Examination (Goodglass & Kaplan, 2001), and the Attention and Symbol trails subtest scores from the Cognitive Linguistics Quick Test (Helm‐Estabrooks, 2001). These measures were chosen to sample across a range of lesion effect sizes and potential anatomical loci. Participants were drawn from other studies examining lesion‐behavior relationships (Lacey, Skipper‐Kallal, Xing, Fama, & Turkeltaub, 2017; Xing et al., 2016; Xing, Lacey, Skipper‐Kallal, Zeng, & Turkeltaub, 2017). All participants provided written informed consent and all studies were approved by the Georgetown University IRB.
To assess the impact of each method on systematic biases in voxelwise statistics because of lesion volume effects, we also examined the effect of the lesion volume correction methods on the distribution of unthresholded SVR‐β values throughout the brain. All computations were conducted on an Intel Xeon (E5–2680 v4) with CPUs at 2.4 GHz and 58 GB of RAM, running Ubuntu 16.04, parallelized across the 14 system cores.
2.2.2. Handling of covariates (including multiple covariates)
In the new SVR‐LSM toolbox, an arbitrary number of covariates may be incorporated into an analysis, allowing for flexible analysis designs. Nonetheless, as in typical regression models, including too many covariates could result in invalid results. Because the valid upper limit of number of covariates that may be included is not clear, we emphasize that care should be taken in including more than a few. As with controlling for lesion volume, covariates are handled using nuisance models and a partialing out strategy, which residualizes the single behavioral score under investigation with respect to the specified covariates prior to SVR analysis. Behavioral covariates may also be factored out of the raw lesion data, effectively allowing behavioral data to be included in the model in each way the lesion volume may be handled (see above). Behavioral covariates are specified through the graphical interface in an interactive listbox and incorporated into the analysis by checkboxes indicating whether to covary values out of the behavior of interest, the lesion data, or both.
2.2.3. Statistical thresholding
The voxelwise SVR‐β values produced by the SVR analysis must be converted into p values via permutation testing before they can be interpreted. Permutation testing involves randomly reassigning the actual behavioral scores in the data set to the lesion masks in the data set, simulating the null hypothesis that there is only a chance level association between the behavior and the lesion distributions. These permutations are first used to generate voxelwise distributions of SVR‐β values obtained through these chance associations. Importantly, the distribution at each voxel is catalogued separately as the distributional properties depend on the number of patients lesioned at a given voxel, the volume of those lesions, and other characteristics that differ across voxels. This allows a critical SVR‐β value to be determined for each voxel that is associated with the user‐selected uncorrected p value threshold. A map of the critical SVR‐β values at each voxel is then assembled and acts as a voxelwise mask to threshold for voxels which are statistically significant before correction for cluster extent. The new cluster‐level correction procedure then evaluates each of the previous permutations, which were saved to disk, first applying the voxelwise mask and then recording the size of the single largest cluster surviving from each permutation map. The distribution of these cluster sizes provides the null distribution from which the FWE‐corrected cluster threshold is determined. Adapted from functional neuroimaging analysis (Nichols & Holmes, 2002), this procedure has been applied to univariate lesion‐mapping studies (Mirman et al., 2015; Pillay, Stengel, Humphries, Book, & Binder, 2014) and was recently shown to adequately control for false positive rate (Mirman et al., 2018). Although permutation‐based correction for multiple comparisons controls false positives in univariate VLSM, this approach does not control for false negative findings. Mass‐univariate analyses are still susceptible to mislocalization and false negatives when behaviors rely on multiple structures (Mah et al., 2014; Zhang et al., 2014).
2.2.4. Inclusion of MATLAB SVM
The original implementation of SVR‐LSM runs within the MATLAB environment, but relies on a popular open‐source third‐party machine learning library for SVR functionality, LibSVM (Chang & Lin, 2011). Thus, when installing this implementation of SVR‐LSM, a user may be required to compile LibSVM on their system from binaries. Although this process is easy in some computing environments, it can be burdensome in others due to variable support of compilers across operating systems and MATLAB versions. MATLAB (2014) introduced SVR functionality into their Statistics and Machine Learning Toolbox™, including the type utilized by the existing implementation of SVR‐LSM (Zhang et al., 2014). To alleviate the need to install the LibSVM package, we provide the option of using SVR functionality from the Statistics and Machine Learning Toolbox™, configurable through a program menu. We retain functionality of LibSVM and leave the cost and gamma parameters user‐configurable.
2.2.5. Graphical user interface
We wrap these functions in a graphical interface constructed in MATLAB. As shown in Figure 1, the interface consists of a single dialog window launched in MATLAB. Within the Analysis Configuration pane, the user specifies a directory containing lesion images, a design file containing a list of patients and relevant behavioral scores, and an output filename. The directionality of the hypothesis is then specified as well as the lesion volume correction method and minimum lesion overlap cutoff for voxels to be included in an analysis. Covariates may be specified in the covariates pane, and may be regressed out of the lesion data, the behavioral score data, or both. Permutation testing can be configured in the top right pane by indicating the number of permutations desired and critical values at the voxelwise and clusterwise levels. Once these parameters are configured, the user clicks “Run Analysis” and waits for the analysis to conclude. A pane in the bottom right of the interface displays progress to the user as the analysis proceeds.
Figure 1.
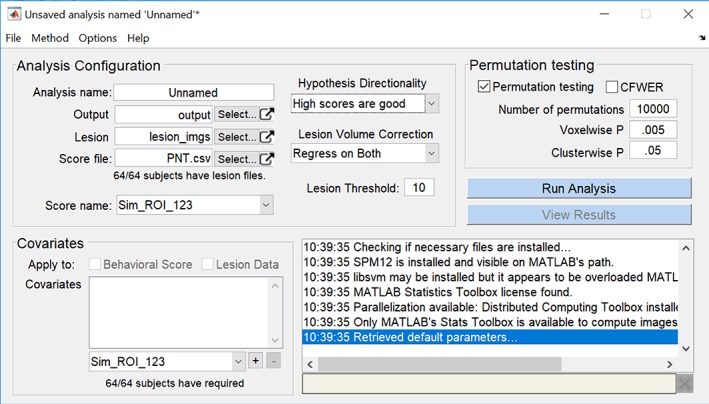
Screenshot of the MATLAB‐based graphical user interface for SVR‐LSM. The single window contains a pane for configuring an analysis, a pane for configuring covariates, a pane for permutation testing, and a feedback pane that reports the progress of an analysis [Color figure can be viewed at http://wileyonlinelibrary.com]
2.2.6. Other new features
In addition to the technical advances outlined above, we include other additions that assist in analysis configuration, execution, diagnostics, and interpretation. An online reference file details all options within the program. A few notable features include:
Analyses configured through the graphical interface may be saved to disk and loaded later or may be executed from the command line without user interaction.
To speed up analyses, an option is provided to parallelize the permutation procedures through the graphical interface. When enabled, this option will utilize the parallel pool with the number of workers specified in the MATLAB Parallel Computing Toolbox preferences. A system that supports parallel processing is necessary for this functionality.
To assist with diagnostics and interpretation of results, each analysis produces an overview file that may be viewed in the browser which contains a short narrative describing the analysis specifications and thumbnails of various output steps. Thumbnails include a lesion overlay image delineating a lesion overlap map and a mask of voxels to which the analysis was restricted based on the minimum lesion overlap threshold, unthresholded SVR‐β maps, voxelwise thresholded SVR‐β maps, and SVR‐β maps thresholded at both the voxel and cluster level, with numerically labeled clusters. To assist with detection of correlation among the main behavioral predictor and covariates specified in the behavioral nuisance model, including lesion volume, output also includes a matrix of pairwise scatter plots to visualize the correlation matrix of those variables. Each plot displays the least squares slope and correlation coefficients, with significant t‐statistics highlighted red. Finally, the output file also plots the largest observed clusters and how they relate to the critical cluster threshold as it changed throughout the permutation testing procedure. This last output can help assess when the critical cluster threshold has reached a point of stability and can give insight into the robustness of observed cluster sizes relative to the permutation testing.
3. RESULTS
3.1. Comparison between results of different SVR‐LSM implementations
To ensure that the new implementation reproduces the results achieved with the prior implementation of SVR‐LSM, we ran an analysis using WAB Repetition as the behavioral score and parameter values cost of 30 and gamma of 5 using both implementations. Because MATLAB utilizes a formulation of the Gaussian radial‐basis function kernel that differs from LibSVM's formulation, we used the algebraic equivalent of a gamma of 5 (sigma = 0.316) for MATLAB SVR analyses. As shown in Figure 2, the two implementations produce SVR‐β maps that are identical (voxelwise r[433] = 1.00, p < .001). Next, to ensure that the MATLAB and LibSVM implementations within the new toolbox produce the same result, we reran the same analysis using the native MATLAB SVR functionality. The results were nearly identical, with negligible differences in exact SVR‐β values (voxelwise r[433] = .94, p < .001). Voxelwise correlations were restricted to voxels that were nonzero in any of the three analyses.
Figure 2.
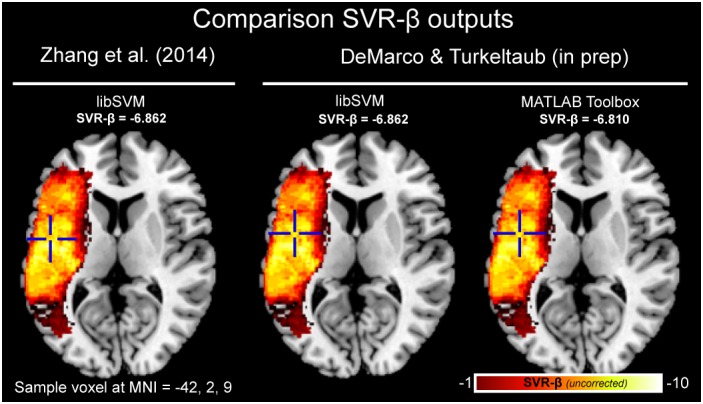
Comparison of uncorrected SVR‐β value output for original SVR‐LSM (left; Zhang et al., 2014), for the new implementation using LibSVM (middle), and MATLAB's SVM functionality (right). For each output, the value is shown at the same sample voxel coordinate of MNI = −42, 2, 9 also indicated by crosshairs. This analysis was conducted on individuals with chronic left‐hemisphere stroke (N = 48) modeling the score for WAB repetition and SVR parameters of cost = 30 and gamma = 5, minimum lesion threshold of 10 [Color figure can be viewed at http://wileyonlinelibrary.com]
3.2. Computing time
The time required to run an analysis will depend on the resolution of the lesion maps, the sample size of individuals in the analysis, the number of permutations used for statistical thresholding, and the specifications of the computer on which the analysis is run. To provide rough estimates of the time required for typical analyses, we timed several analyses varying the parameters of parallelization (on/off), method of correcting for lesion volume (no correction, dTLVC, Regression on both behavior and lesion), number of permutations computed (1,000, 5,000, 10,000), and sample size (50, 100, 150). Analyses were carried out on an Intel Xeon (E5–2680 v4) with CPUs at 2.4 GHz and 58 GB of RAM, running Ubuntu 16.04. Parallelized analyses utilized all 14 system cores. Results for these analyses are listed in Table 1. Execution time grew with sample size, lasting on average 67/16 minutes (unparallelized/parallelized) for a sample size of 50, 217/66 minutes for a sample size of 100, and 445/132 minutes for a sample size of 150. Likewise, execution time grew with the number of permutations, lasting on average 47/14 minutes for 1,000 permutations, 227/68 minutes for 5,000 permutations, and 455/131 minutes for 10,000 permutations. Enabling parallelization resulted in analyses completing on average six times faster, with at least a two‐fold decrease and at most a nine‐fold decrease in execution time. Duration of analyses using dTLVC correction was 131/16 minutes, which was comparable to analyses with no correction applied, 128/16 minutes. Regressing out lesion volume resulted in slower execution time, lasting on average 470/181 minutes. Regressing out lesion volume results is slower analyses because the SVR takes longer to converge, and this extra time accumulates over the permutation portions of the analysis.
Table 1.
Analysis duration for SVR‐LSM with and without parallelization for three sample sizes (50, 100, 150), three numbers of permutations (1,000, 5,000, 10,000) and for three methods of lesion volume correction (no correction, dTLVC, and regression on both behavior and lesion volume)
| Correction method | Sample size | Number of permutations | Analysis duration (minutes) | |
|---|---|---|---|---|
| Unparallelized | Parallelized | |||
| No correction for lesion volume | 50 | 1,000 | 7 | 1 |
| 5,000 | 33 | 5 | ||
| 10,000 | 64 | 11 | ||
| 100 | 1,000 | 22 | 3 | |
| 5,000 | 105 | 14 | ||
| 10,000 | 211 | 27 | ||
| 150 | 1,000 | 46 | 5 | |
| 5,000 | 222 | 26 | ||
| 10,000 | 443 | 51 | ||
| DTLVC | 50 | 1,000 | 7 | 1 |
| 5,000 | 32 | 5 | ||
| 10,000 | 62 | 11 | ||
| 100 | 1,000 | 22 | 3 | |
| 5,000 | 106 | 14 | ||
| 10,000 | 221 | 27 | ||
| 150 | 1,000 | 45 | 5 | |
| 5,000 | 228 | 26 | ||
| 10,000 | 458 | 52 | ||
| Regress on both behavior and lesion | 50 | 1,000 | 27 | 7 |
| 5,000 | 127 | 34 | ||
| 10,000 | 248 | 67 | ||
| 100 | 1,000 | 83 | 33 | |
| 5,000 | 396 | 159 | ||
| 10,000 | 786 | 311 | ||
| 150 | 1,000 | 161 | 72 | |
| 5,000 | 795 | 327 | ||
| 10,000 | 1604 | 621 | ||
3.3. Distribution of bias related to lesion volume
To explore the distribution of bias related to lesion volume in a hypothetical lesion‐symptom mapping analysis, we examined the average lesion volumes of those with and without lesions at each voxel in a cohort of left‐hemisphere stroke survivors (N = 48). These maps revealed that the presence of lesions at most locations was associated with large lesion volume (Figure 3a), while the absence of lesions at the same voxels was associated with small lesion volume (Figure 3b). Larger lesions were particularly associated with lesion status at the border of middle cerebral artery (MCA) territory, and the largest lesions were associated with lesion status in anterior cerebral artery territory. A difference map of average lesion volume in those with and without lesions at every voxel reveals large bias throughout most of the left hemisphere (Figure 3c), a difference that was statistically significant throughout a large portion of MCA territory (Figure 3d).
Figure 3.
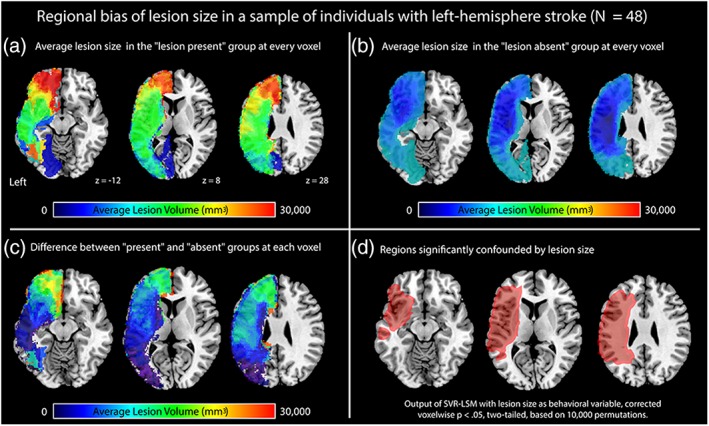
Figure showing the regional bias caused by lesion volume in a cohort of individuals with left‐hemisphere stroke (N = 48). The panels show maps of average lesion volume in (a) the “lesion present” group at each voxel, (b) the “lesion absent” group at each voxel, and (c) a difference between average lesion volume in the “lesion present” and “lesion absent” groups, and (d) regions where lesion volume was significantly different (red outline, red translucent fill). The analysis displayed in panel d was conducted in SVR‐LSM treating lesion volume as the behavioral variable, thresholded voxelwise at p < .05 (two‐tailed) based on 10,000 permutations [Color figure can be viewed at http://wileyonlinelibrary.com]
3.4. Comparison of lesion volume correction methods
To compare the effects of lesion volume correction using the methods available in the new SVR‐LSM toolbox, we conducted analyses on 20 different behaviors using no correction and the four lesion volume correction methods provided. Conducting analyses without lesion volume correction produced the greatest number of suprathreshold voxels after voxelwise and clusterwise correction. Of the 20 behaviors tested, 16 produced significant results (24,887 significant voxels on average). When the dTLVC transform was applied to correct for lesion volume, the same 16 behaviors produced significant results, although the average volume of significant voxels was reduced (10,718 significant voxels on average). With lesion volume regressed out of the behavior, two of 20 analyses produced significant results (3,827 significant voxels on average). With lesion volume regressed out of the raw lesion data, one of the 20 analyses produced significant results (4,095 voxels). Finally, with lesion volume regressed out of both behavioral data and raw lesion data, seven of 20 analyses produced significant results (2,884 voxels on average). A spaghetti plot of the 20 analyses for five correction choices is shown in Figure 4. Table 2 lists the behaviors that were significant for each lesion volume correction method. See Supporting Information Figure for thumbnails of the SVR‐β maps for the 20 behaviors across the five lesion‐volume correction methods. See Supporting Information Figure and Supporting Information Table for a demonstration that this effect also applies to mass‐univariate VLSM (for VLSM methods, see Supporting Information Methods).
Figure 4.
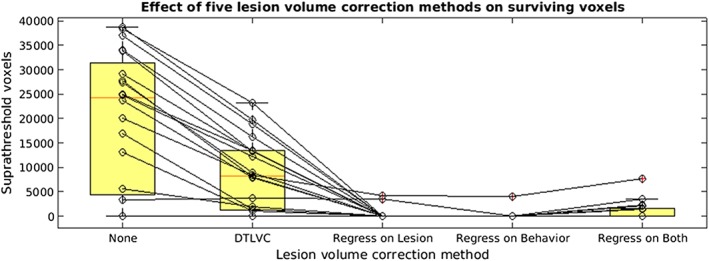
Spaghetti plot showing the effect of five lesion volume correction methods on twenty behaviors in a group of individuals with left‐hemisphere stroke (N = 49). Lesion volume methods include (a) none, (b) dTLVC, (c), regression of lesion volume out of the raw voxel data, (d) regression of lesion volume out of the behavioral data, and (e) regression of lesion volume out of both behavioral data and out of raw voxel data. All analyses were thresholded at voxelwise at p < .005 (one‐tailed) and clusterwise correction at p < .05, based on 10,000 permutations [Color figure can be viewed at http://wileyonlinelibrary.com]
Table 2.
Table listing which of 20 behaviors were significant for each of five lesion volume correction methods using support vector regression lesion‐symptom mapping
| Lesion volume correction method | |||||
|---|---|---|---|---|---|
| Behavior under investigation | No correction | dTLVC | Regress on lesion | Regress on behavior | Regress on both |
| Philadelphia naming test (oral) | • | • | ○ | ○ | |
| Philadelphia naming test (written) | • | • | ○ | ○ | ○ |
| Category fluency | • | • | ○ | ○ | ○ |
| Letter fluency | • | • | ○ | ○ | • |
| Western aphasia battery content | • | • | ○ | ○ | ○ |
| Western aphasia battery fluency | • | • | ○ | ○ | • |
| Backward digit span | • | • | ○ | ○ | ○ |
| Western aphasia battery repetition | • | • | ○ | ○ | ○ |
| Pseudoword repetition | • | • | ○ | ○ | • |
| Imageability effect (concrete minus abstract) | ○ | ○ | ○ | ○ | ○ |
| Regularity effect (regular minus exception) | • | • | • | ○ | • |
| Lexicality effect (word minus pseudoword) | ○ | ○ | ○ | ○ | ○ |
| Pyramids and palm trees | • | • | ○ | ○ | ○ |
| Western aphasia battery word recognition | • | • | ○ | ○ | • |
| Western aphasia battery yes/no | • | • | • | • | • |
| Picture pointing | • | • | ○ | ||
| BDAE complex ideational material | • | • | ○ | ○ | ○ |
| CLQT attention score | ○ | ○ | ○ | ○ | |
| CLQT symbol trails score | ○ | ○ | ○ | ○ | ○ |
| Western aphasia battery sequential commands | • | • | ○ | ○ | • |
A solid bullet (•) indicates that significant findings for a given combination of behavior and lesion volume correction method which survived voxelwise thresholding, p < .005 (one‐tailed) and clusterwise correction, p < .05 based on 10,000 permutations. An empty bullet (○) indicates that the analysis resulted in voxels that survived voxelwise thresholding but did not survive cluster correction.
Because the motivation for performing lesion‐volume correction is to remove bias in the data, we also examined uncorrected SVR‐β maps for evidence of this bias and the extent to which each lesion‐volume correction method addressed it (Figure 5). We examined two behaviors, one with significant findings across all methods (WAB Fluency) and one with significant findings only without lesion volume correction or with dTLVC (Pyramids and Palm Trees). In addition to plotting a histogram of SVR‐β values for each lesion correction method, we also generated distributions of SVR‐β maps resulting from 1,000 permutations for each analysis to examine the null distributions against which the real data would be compared in an SVR‐LSM analysis.
Figure 5.
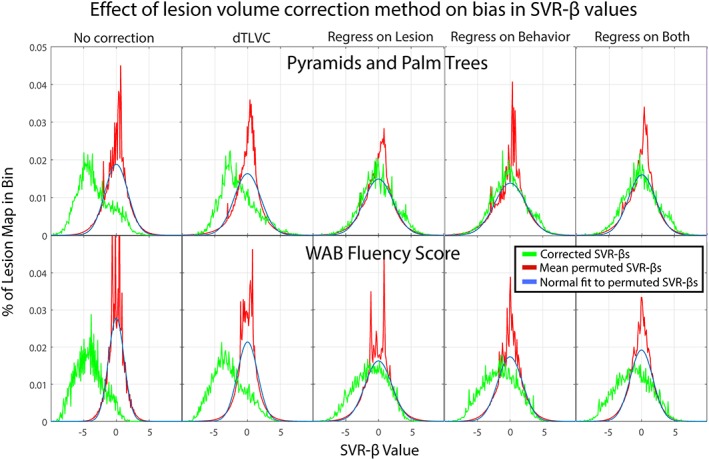
Histograms of uncorrected SVR‐β values after five lesion volume correction methods (green) for two behaviors, including Pyramids and Palm Trees (top row) and WAB Fluency (bottom row). Also shown is the average histogram of 1,000 permuted SVR‐β maps for each behavior and correction method (red) and a normal distribution fit to the permutations, scaled to the histograms [Color figure can be viewed at http://wileyonlinelibrary.com]
The SVR‐β values for the permutation data were normally distributed and centered at 0 for all analyses of both behaviors. The width of these distributions varied somewhat across correction methods with the narrowest distributions (i.e., least variance in SVR‐β values) observed in the permutations without lesion volume correction.
The SVR‐β distributions for both analyses without lesion volume correction were substantially shifted leftward, such that nearly all SVR‐β values in these analyses were negative. This indicates that lesions at most voxels in these analyses were associated with negative effects on behavior. DTLVC had only a marginal effect on this leftward bias, with the distributions slightly shifted rightward for both behaviors compared to the analyses without lesion volume correction. In contrast, all three regression methods qualitatively changed the shape of the distributions compared to no correction or dTLVC. For Pyramids and Palm Trees, which did not yield any significant results with lesion volume regressed out of analyses, all three methods resulted in normal distributions centered on 0, little different than the permutation distributions. For WAB Fluency, which did yield significant results in these analyses, the distributions were wider than the permutation distributions and shifted leftward such that the left (negative) tail extended beyond the null distributions.
3.5. Lesion correction effects on localization of behaviors
To explore whether controlling for lesions size alters localization in lesion symptom mapping analyses, we first examined the SVR‐LSM maps of the 20 behavioral scores (Section 3.4, Supporting Information Figure). Several of the analyses without lesion volume correction produced similar maps despite examining different behaviors, with large areas of significance falling in the region of lesion volume bias shown in Figure 3d. When significant clusters survived in the lesion volume corrected maps, they were substantially smaller, but not spatially displaced from the uncorrected results. The same effects were also observed when the analyses were computed using a mass‐univariate VLSM approach (Supporting Information Figure). This suggests that the data transformations required for lesion volume correction did not cause mislocalization of the behaviors.
To examine more directly the effect of lesion volume control on localization of behaviors strongly or weakly associated with lesion volume, we next performed lesion‐symptom mapping analyses of two simulated behaviors. Based on the earlier result (Figure 3d), one simulated behavior was generated by taking the number of lesioned voxels in a 4 mm sphere surrounding a voxel strongly related to lesion volume (Figure 6a; MNI = −48, −17, 28; SVR‐β = 9.47; p < .001 based on 10,000 permutations, 23 overlapping lesions), and one behavior was generating using the same approach at a voxel less associated with lesion volume (Figure 6b; MNI = −47, −51, 14; SVR‐β = 3.12, p = .07 based on 10,000 permutations, 18 overlapping lesions). We then examined the effect of each lesion correction method on the localization of the simulated behaviors. Lesion‐symptom mapping of these behaviors should ideally produce significant results in a fairly focal area surrounding the seed.
Figure 6.
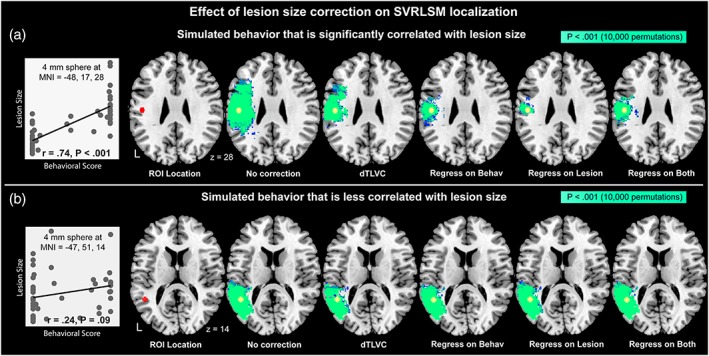
Effect of lesion volume correction on SVR‐LSM localization for (a) a simulated behavior significantly correlated with lesion volume and (b) a simulated behavior less correlated with lesion volume. Behaviors were simulated by summing the number of lesioned voxels within a 4 mm sphere (red) placed based on its correlation with lesion volume. At the left of each pane is shown a scatterplot of lesion volume versus the simulated behavior across the patient sample. Voxels with statistically significant results at voxelwise p < .001 based on 10,000 permutations are shown in bright green [Color figure can be viewed at http://wileyonlinelibrary.com]
As depicted in Figure 6a, lesion symptom mapping of the simulated behavior that was strongly related to lesion volume produced significant results in a large area surrounding the seed from which the behavior was derived (red). When dTLVC was applied, the region that was identified as significantly related to the simulated behavior became less dispersed relative to no correction. When regression methods were applied to correct for lesion volume, the region that was identified as significantly related to the simulated behavior became more focal, and was still centered on the seed location. For the simulated behavior that was less correlated with lesion volume (Figure 6b), all correction methods produced significant results surrounding the seed voxel without a qualitative difference in the number or distribution of significant voxels. See Supporting Information Figure for a demonstration that this effect also applies to mass‐univariate lesion‐symptom mapping.
3.6. Case studies of the effect of lesion‐volume correction on multivariate lesion‐symptom mapping
To illustrate the effects of lesion volume correction on a hypothetical lesion‐symptom mapping analysis, we focus here on SVR‐LSM results for two behaviors, specifically Pseudoword Repetition and WAB Yes/No. Pseudoword repetition tests phonological processing without lexical‐semantic support by asking participants to repeat aloud pronounceable nonwords that are dictated by the examiner. WAB Yes/No tests sentence‐level comprehension by asking participants to respond yes or no to a series of question sentences (e.g., “Will paper burn in fire?”). Each analysis was first conducted without controlling for lesion volume, and then conducted again with lesion volume regressed out of both voxelwise lesion data and behavioral scores.
When lesion volume was not controlled, lesions were associated with impaired Pseudoword Repetition in a single large cluster of regions that included portions of left inferior frontal gyrus, superior temporal gyrus, ventral premotor cortex, and much of the inferior parietal lobe (Figure 7a, left). When lesion volume was regressed out of both behavior and lesion data as a covariate, many of these regions were no longer significant. Rather, regions where lesions resulted in impaired Pseudoword Reading were contained in single smaller cluster, containing posterior superior temporal gyrus, supramarginal gyrus, and ventral post‐central gyrus, roughly surrounding area Spt in the temporoparietal junction (Figure 7a, right).
Figure 7.
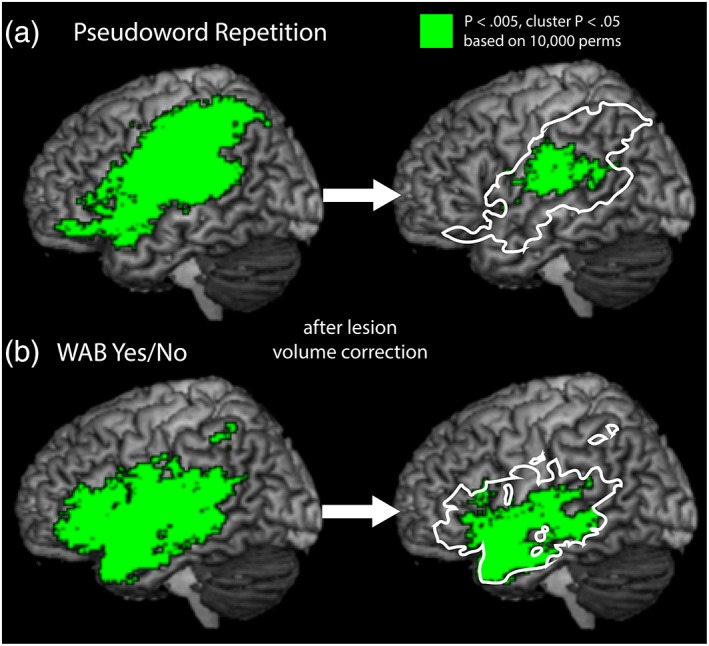
Brain regions where lesions are significantly related to behavioral deficits in (a) Pseudoword Repetition and (b) WAB Yes/No prior to lesion‐volume correction (left) and following lesion volume correction by regressing lesion volume out of both behavior and lesion data prior to SVR (right). Resulting statistical maps are corrected voxelwise at p < .005, and by cluster extent at p < .05 based on 10,000 permutations [Color figure can be viewed at http://wileyonlinelibrary.com]
When lesion volume was not controlled, lesions were associated with impaired WAB Yes/No score in a single large cluster of regions that again included inferior frontal gyrus, ventral premotor cortex, insula, ventral post‐central gyrus, and a large swath of superior and middle temporal gyri, extending subcortically into the parietal lobe (Figure 7b, left). When lesion volume was regressed out of both behavior and lesion data as a covariate, lesions were associated with impaired WAB Yes/No scores in a single smaller cluster primarily consisting of the middle and superior temporal gyri, extending into the insula and posterior inferior frontal gyrus (Figure 6b, right).
4. DISCUSSION
Lesion‐symptom mapping has become a cornerstone of cognitive neuroscience research, and methodological developments have enabled multivariate approaches with benefits over traditional mass‐univariate methods. Despite the availability of multivariate methods for lesion‐symptom mapping, these techniques have yet to be widely adopted, perhaps in part due to limitations of previously available implementations. Here, we have provided new methods for SVR‐LSM that make it easier to implement and expand on the types of analyses that can be conducted. We have also provided tools, such as permutation‐based FWE correction at the cluster level, to improve the rigor of SVR‐LSM analyses.
Lesion volume confounds limit the rigor of lesion symptom mapping analyses, but there is no consensus about how to best control for these effects in practice. We compared several different approaches here, including a new approach as described above. Although our focus was on the comparison using SVR‐LSM, we replicated the analyses using mass‐univariate methods (Supporting Information Figures and Supporting Information Table) to illustrate that the lesion volume problem is similar in univariate VLSM. The results demonstrated that the most liberal approach is to ignore lesion volume as a confound. Doing so resulted in significant lesion‐deficit associations in 16 of 20 behaviors examined, many including large numbers of significant voxels. In a simulated behavior strongly related to lesion volume, lack of correction resulted in identifying significant lesion‐behavior associations in a large area of the brain. Using dTLVC was only modestly more conservative, with significant results again observed for the same 16 behaviors and somewhat fewer suprathreshold voxels. Examination of the SVR‐β distributions for both of these approaches revealed that voxelwise associations with behavior were dramatically skewed toward negative relationships, a bias related to the influence of lesion volume on voxelwise statistical values, as demonstrated by the correction of the bias after regressing out lesion volume. Lesion volume has a large impact on lesion‐symptom maps because the likelihood of any given voxel being lesioned is directly proportionate to the size of the lesion. Thus, at any given voxel, patients with large lesions are more likely to have a lesion at that voxel compared to patients with small lesions. In our cohort, this lesion volume difference was present throughout most of the perisylvian tissue as well as much of the underlying white matter, demonstrating how widespread these effects are in the brain. The voxelwise lesion volume difference means that statistical values at most voxels in an uncorrected map result from comparisons between groups (lesion present and lesion absent) that are mismatched on lesion volume, producing a strong bias toward finding lesion‐deficit associations in large areas of the brain when behaviors are related to lesion volume. This bias was demonstrated here by the similarity of lesion‐symptom maps generated for different behaviors when lesion volume correction was not used, and by the analysis of the simulated behaviors, which yielded a large area of significance for the behavior associated with lesion volume when this relationship was not corrected. DTLVC only partially corrects this bias because it still leaves bias in unlesioned voxels, which make up a majority of most lesion maps.
The decision of how to address the lesion volume bias ultimately should be driven by the research question. Permitting the bias to remain in the analysis may be appropriate if the question is “Where are lesions associated with deficits in behavior X?” as lesions should almost always be associated with negative changes in behavior. This may be reasonable for clinical questions about expected deficits based on lesion distribution, but results should not be taken to indicate that the areas of the brain identified as significant are important for the behavior under investigation.
Allowing the lesion volume bias to impact results is not appropriate if the analysis aims to localize the substrates for a given behavior by asking “Is behavior X more strongly associated with lesions in certain brain areas compared to others?” This is the type of question motivating most lesion‐symptom mapping studies, and in this case the lesion volume bias confounds the association between the behavior and lesion location. As an analogy, imagine a hypothetical experiment aiming to determine if the left inferior frontal gyrus is important for cognitive control. The experimenters compare two groups of patients, one with lesions involving left inferior frontal gyrus and one with lesions elsewhere. The left inferior frontal gyrus lesion group performs worse on the cognitive control tasks as predicted, but also has larger lesions than the control lesion group. One cannot conclude from these results that the left inferior frontal gyrus is associated with cognitive control because the observed behavioral difference may have been driven purely by the difference in lesion volume between the groups. Performing lesion‐symptom mapping without adequate lesion volume correction is like performing similarly biased comparisons at each voxel in the brain.
Our analyses of Pseudoword repetition and WAB Yes/No illustrate this principle. Without controlling for lesion volume, impaired performance on these measures are associated with spatially nonspecific regions of the brain that span frontal, parietal, and temporal lobes. Controlling for lesion volume reveals that pseudoword repetition relates to more specific cortical regions, specifically area Spt and nearby cortical areas. Area Spt is a key component of the dorsal stream of modern neuroanatomical language models (Hickok & Poeppel, 2007) and is believed to perform the sensory‐motor transformations that enable direct mapping from auditory input to motor output, which is required for behaviors like pseudoword repetition. Indeed, this finding is highly congruent with previous functional neuroimaging studies of healthy adults, and this region is implicated in conduction aphasia, a syndrome in which this skill is particularly impaired (Buchsbaum et al., 2011; Hickok et al., 2000; Hickok, Buchsbaum, Humphries, & Muftuler, 2003). Controlling for lesion volume reveals that poor performance on WAB Yes/No score results from lesions to superior and middle temporal gyri, and portions of posterior inferior frontal gyrus. These findings also converge with existing work examining regions relevant to auditory comprehension of single words and sentences, which are the primary linguistic skills required for this task. Many lines evidence from patients with aphasia indicate auditory comprehension is associated with superior temporal cortex (Hillis, Rorden, & Fridriksson, 2017), but there is also evidence for a broadly distributed network supporting sentence comprehension throughout the middle temporal gyrus (Turken & Dronkers, 2011). Recent work on the semantic variant of primary progressive aphasia underlines the additional contributions of the anterior temporal lobe for language comprehension (e.g., Mesulam et al., 2015). These two cases illustrate that without controlling for lesion volume, analysis results are broadly distributed, whereas more specific and informative results that conform to modern neuroanatomical language models are identified with lesion volume control.
For the type of research question described above, statistically controlling for lesion volume is necessary to interpret a significant result as indicating a role for the identified brain structure in the behavior under investigation. DTLVC appears to only partially correct for lesion volume biases, based on the residual negative skew in the SVR‐β histograms and the modest effects of this method on both the analyses of real behaviors and the simulated behavior strongly related to lesion volume. In contrast, all three regression methods provided more conservative correction, completely addressing the skew in SVR‐β histograms and improving the spatial specificity for localizing the behavior strongly related to lesion volume. Importantly, spatial specificity was not affected by lesion volume correction when the behavior was less strongly related to lesion volume, and none of the lesion volume correction methods introduced mislocalization of lesion‐behavior associations. Together these findings suggest that correcting for lesion volume offers a practical benefit of gaining greater spatial specificity in localizing a lesion‐symptom relationship in regions confounded by lesion volume, without negatively influencing localization in regions less confounded with lesion volume.
The cost of rigorously addressing lesion volume is more conservative voxelwise thresholding, as demonstrated by the relative paucity of significant results in the analyses using any of the three regression approaches. Among the three options examined here, regressing lesion volume out of both the behavioral scores and the lesion maps appeared to be the most sensitive method. This may be because true lesion‐deficit relationships are obscured by applying a nuisance model to one variable (i.e., dependent or independent) but not the other. In other words, the lesion‐symptom relationship was more detectable when both the dependent and independent variables were transformed to control for lesion volume prior to the analysis, rather than just one or the other. This method is also appealing because it is mathematically nearly equivalent to how lesion volume (or any other covariate) is dealt with in statistical analyses of studies like the hypothetical one described above. It is notable, that given the relatively small sample size used here, identifying significant associations between lesion location and deficits for seven out of 20 behaviors suggests that this method is conservative, but not overly so.
It should also be noted that correcting lesion volume is likely unnecessary when the behavioral variable has no relationship to lesion volume. This is illustrated by the analyses of the simulated behavior that was not strongly related to lesion volume (Figure 6b). In this case, lesion volume correction did not qualitatively alter the lesion symptom maps. Similarly, if behavioral covariates are included that have a similar relationship to lesion volume as the test behavior, correcting for lesion volume may be redundant. However, even a weak residual relationship between the behavior of interest and lesion volume could induce a small brain wide bias, so erring on the side of lesion volume correction seems appropriately conservative in general.
Although lesion volume was strongly related to lesion location in our patient sample, it is possible that in some cohorts this relationship will be weaker. In these cases, the lesion volume confound would still systematically bias the resulting lesion‐symptom map toward finding large regions of lesion‐symptom association but would cause less dramatic spatial displacement in the location of these effects. Unfortunately, it is often difficult to assess the degree to which a lesion volume confound might influence the results of existing lesion‐symptom mapping literature. For instance, in reviewing the literature for lesion‐symptom mapping studies of our example of pseudoword repetition, we found a number of studies that examined the behavior (Baldo, Katseff, & Dronkers, 2012; Fridriksson et al., 2010), but in only one study (Rogalsky et al., 2015) was it clear that lesion volume was included as a covariate. These studies generally converge on consistent findings regarding the role of temporoparietal cortex in this task, which may suggest that lesion volume confounds cause less severe spatial displacement of results than we observed here.
Multivariate lesion‐symptom mapping is still in its infancy and there were a number of questions about how to optimize the approach that we were not able to address here. First, we did not investigate procedures to optimize our SVR hyperparameters. Rather, we used fixed hyperparameter values that were recommended by the initial SVR‐LSM publication (Zhang et al., 2014). It is likely that optimal hyperparameter values may differ depending on analysis characteristics such as voxel size, and future work should examine this issue. Although our analyses could have slightly different solutions with optimized hyperparameters, importantly, our findings about lesion volume correction held for both multivariate and mass‐univariate lesion‐symptom mapping analyses. Second, in this toolbox we handled covariates by applying nuisance models prior to submitting the data to the SVR. Although it is possible to include an arbitrary number of covariates in this way, it is not clear how many covariates are too many and future work should explore this question. Furthermore, future work should also explore an alternative approach to controlling for lesion volume and behavioral covariates by adding them as additional features directly in the SVR model. Finally, this toolbox will continue to be developed to keep abreast with new developments in the field of lesion‐symptom mapping. For instance, methods we intend to add in future releases include hyperparameter optimization for each analysis and innovative thresholding procedures such as continuous family‐wise error rate correction (Mirman et al., 2018).
In conclusion, we have provided a new software implementation with a graphical interface that may facilitate the adoption of SVR‐LSM as a multivariate alternative to traditional VLSM. The new implementation takes advantage of MATLAB's SVR functionality in its native Statistics Toolbox, addresses prior limitations and adds new functionality, including cluster‐level family‐wise error correction using permutation testing, highly flexible handling of behavioral covariates through nuisance models, and a novel method for lesion volume correction that supplements a number of existing methods. We have also demonstrated the degree of bias lesion volume effects can induce in lesion symptom maps and explored the impact of several different approaches to handling lesion volume, which may aid investigators in selecting the most appropriate approach for their specific research questions.
Supporting information
Supplemental Figure 1 This figure plots the correlation between behavioral performance and lesion volume for 20 behaviors measured in a cohort of left‐hemisphere stroke survivors (N = 48), with significant correlations in red. Below each plotted correlation, thumbnails are shown of SVR‐LSM results using five different lesion volume correction methods. Values displayed are suprathreshold SVR‐β values thresholded voxelwise at p < .005. Columns correspond to behaviors and rows correspond to lesion volume correction methods. MNI coordinate in the z plane is shown in white for each column. The lesion volume correction methods displayed include no correction, dTLVC, regression of lesion volume out of behavior, regression of lesion volume out of voxelwise lesion statuses, and regression of lesion volume out of both behavior and lesion data. The top row of lesion‐symptom maps, which are uncorrected for lesion volume, also depicts regions from Figure 3d for behaviors significantly correlated (p < .05) with lesion volume (red outline) and not significantly correlated with lesion volume (dotted white outline). Behaviors include Philadelphia Naming Test score (PNT), pseudoword repetition, regularity effect, Pyramids and Palm Trees (PPT), backward digit span, Western Aphasia Battery (WAB) Fluency score, category fluency, score on complex ideational material (CIM) from the Boston Diagnostic Aphasia Examination (BDAE), WAB Content score, WAB Sequential Commands score, WAB Yes/No score, letter fluency, imageability effect, picture pointing score, WAB Repetition score, lexicality effect, WAB Word Recognition score, written PNT score, and the Trails and Attention subscores on the Cognitive Linguistics Quick Test (CLQT)
Supplemental Figure 2 This figure replicates the results from Supplemental Figure which used SVR‐LSM, with traditional mass‐univariate lesion‐symptom mapping. This figure demonstrates that the concern about lesion volume as a confound is not limited to multivariate lesion‐symptom mapping. In this figure, P values are derived from beta values from voxelwise linear regression (see Supplemental Methods). Statistical maps are corrected at voxelwise p < .005, based on 10,000 permutations. Each of the 20 columns corresponds to one behavior and each and five rows correspond to a lesion volume correction methods, as in Supplemental Figure
Supplemental Figure 3 This figure replicates the results from Figure 6, which used SVR‐LSM, with traditional mass‐univariate lesion‐symptom mapping methods. Effect of lesion volume correction on mass‐univariate VLSM localization for (A) a simulated behavior significantly correlated with lesion volume and (B) a simulated behavior less correlated with lesion volume. Behaviors were simulated by summing the number of lesioned voxels within a 4 mm sphere (red) placed based on its correlation with lesion volume. At the left of each pane is shown a scatterplot of lesion volume versus the simulated behavior across the patient sample. Voxels with statistically significant results at voxelwise p < .001 based on 10,000 permutations are shown in bright green
Supplemental Table 1 Table listing which of 20 behaviors were significant for each of five lesion volume correction methods using traditional, mass‐univariate lesion‐symptom mapping. A solid bullet (•) indicates that significant findings for a given combination of behavior and lesion volume correction method which survived voxelwise thresholding, p < .005 (one‐tailed) and clusterwise correction, p < .05 based on 10,000 permutations. An empty bullet (○) indicates that the analysis resulted in voxels that survived voxelwise thresholding but did not survive cluster correction
Supplemental Methods
ACKNOWLEDGMENTS
We thank Ze Wang for providing the SVR‐LSM software on which the toolbox we present is based. We also thank Elizabeth Lacey, Mackenzie Fama, Zainab Anbari, and Kate Spiegel for data collection. The research was supported by the Doris Duke Charitable Foundation (grant #2012062), NIH/NCATS via GHUCCTS (KL2TR000102 and TL1TR001431), and by the NIDCD (R01DC014960).
DeMarco AT, Turkeltaub PE. A multivariate lesion symptom mapping toolbox and examination of lesion‐volume biases and correction methods in lesion‐symptom mapping. Hum Brain Mapp. 2018;39:4169–4182. 10.1002/hbm.24289
Funding information National Center for Advancing Translational Sciences, Grant/Award Number: KL2TR000102/TL1TR001431; National Institute on Deafness and Other Communication Disorders, Grant/Award Number: R01DC014960; Doris Duke Charitable Foundation, Grant/Award Number: 2012062
Contributor Information
Andrew T. DeMarco, Email: andrew.demarco@georgetown.edu.
Peter E. Turkeltaub, Email: turkelp@georgetown.edu.
REFERENCES
- Alexander, M. P. , Naeser, M. A. , & Palumbo, C. (1990). Broca's area aphasias: Aphasia after lesions including the frontal operculum. Neurology, 40(2), 353–362. [DOI] [PubMed] [Google Scholar]
- Ashburner, J. , & Friston, K. J. (2000). Voxel‐based morphometry—the methods. NeuroImage, 11(6), 805–821. [DOI] [PubMed] [Google Scholar]
- Ashton, E. A. , Takahashi, C. , Berg, M. J. , Goodman, A. , Totterman, S. , & Ekholm, S. (2003). Accuracy and reproducibility of manual and semiautomated quantification of MS lesions by MRI. Journal of Magnetic Resonance Imaging: JMRI, 17(3), 300–308. 10.1002/jmri.10258 [DOI] [PubMed] [Google Scholar]
- Baldo, J. V. , Katseff, S. , & Dronkers, N. F. (2012). Brain regions underlying repetition and auditory‐verbal short‐term memory deficits in aphasia: Evidence from voxel‐based lesion symptom mapping. Aphasiology, 26(3–4), 338–354. 10.1080/02687038.2011.602391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso, A. , Lecours, A. R. , Moraschini, S. , & Vanier, M. (1985). Anatomoclinical correlations of the aphasias as defined through computerized tomography: Exceptions. Brain and Language, 26(2), 201–229. [DOI] [PubMed] [Google Scholar]
- Bates, E. , Wilson, S. M. , Saygin, A. P. , Dick, F. , Sereno, M. I. , Knight, R. T. , & Dronkers, N. F. (2003). Voxel‐based lesion–symptom mapping. Nature Neuroscience, 6(5), 448–450. 10.1038/nn1050 [DOI] [PubMed] [Google Scholar]
- Broca, P. (1866). Sur la faculté générale du language, dans ses rapports avec la faculté du language articulé. Bulletin de La Société d'Anthropologie Deuxième Série, 1, 377–382. [Google Scholar]
- Buchsbaum, B. R. , Baldo, J. , Okada, K. , Berman, K. F. , Dronkers, N. , D'Esposito, M. , & Hickok, G. (2011). Conduction aphasia, sensory‐motor integration, and phonological short‐term memory ‐ an aggregate analysis of lesion and fMRI data. Brain and Language, 119(3), 119–128. 10.1016/j.bandl.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C.‐C. , & Lin, C.‐J. (2011). LIBSVM: A library for support vector machines. ACM Transactions on Intelligent Systems and Technology (TIST), 2(3), 27. [Google Scholar]
- Cortes, C. , & Vapnik, V. (1995). Support‐Vector Networks. Machine Learning, 20(3), 273–297. 10.1023/A:1022627411411 [DOI] [Google Scholar]
- Fridriksson, J. , Kjartansson, O. , Morgan, P. S. , Hjaltason, H. , Magnusdottir, S. , Bonilha, L. , & Rorden, C. (2010). Impaired speech repetition and left parietal lobe damage., Impaired speech repetition and left parietal lobe damage. The Journal of neuroscience: The official journal of the Society for Neuroscience, 30(33), 11057–11061. 10.1523/JNEUROSCI.1120-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geva, S. , Baron, J.‐C. , Jones, P. S. , Price, C. J. , & Warburton, E. A. (2012). A comparison of VLSM and VBM in a cohort of patients with post‐stroke aphasia. NeuroImage: Clinical, 1(1), 37–47. 10.1016/j.nicl.2012.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gläscher, J. , Adolphs, R. , Damasio, H. , Bechara, A. , Rudrauf, D. , Calamia, M. , … Tranel, D. (2012). Lesion mapping of cognitive control and value‐based decision making in the prefrontal cortex. Proceedings of the National Academy of Sciences, 109(36), 14681–14686. 10.1073/pnas.1206608109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodglass, H. , & Kaplan, E. (2001). The assessment of aphasia and related disorders. Philadelphia, PA: Lippincott Williams & Wilkins. [Google Scholar]
- Helm‐Estabrooks, N. (2001). Cognitive linguistic quick test: CLQT. San Antonio, TX: PsychCorp: Pearson. [Google Scholar]
- Herbet, G. , Lafargue, G. , & Duffau, H. (2015). Rethinking voxel‐wise lesion‐deficit analysis: A new challenge for computational neuropsychology. Cortex; A Journal Devoted to the Study of the Nervous System and Behavior, 64, 413–416. 10.1016/j.cortex.2014.10.021 [DOI] [PubMed] [Google Scholar]
- Hickok, G. , Buchsbaum, B. R. , Humphries, C. , & Muftuler, T. (2003). Auditory–motor interaction revealed by fMRI: Speech, music, and working memory in area Spt. Journal of Cognitive Neuroscience, 15(5), 673–682. [DOI] [PubMed] [Google Scholar]
- Hickok, G. , Erhard, P. , Kassubek, J. , Helms‐Tillery, A. K. , Naeve‐Velguth, S. , Strupp, J. P. , … Ugurbil, K. (2000). A functional magnetic resonance imaging study of the role of left posterior superior temporal gyrus in speech production: Implications for the explanation of conduction aphasia. Neuroscience Letters, 287(2), 156–160. [DOI] [PubMed] [Google Scholar]
- Hickok, G. , & Poeppel, D. (2007). The cortical organization of speech processing. Nature Reviews. Neuroscience, 8(5), 393–402. 10.1038/nrn2113 [DOI] [PubMed] [Google Scholar]
- Hillis, A. E. , Rorden, C. , & Fridriksson, J. (2017). Brain regions essential for word comprehension: Drawing inferences from patients. Annals of Neurology, 81(6), 759–768. 10.1002/ana.24941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, D. , & Patterson, K. E. (1992). The pyramids and palm trees test: A test of semantic access from words and pictures. Bury St Edmunds, England: Thames Valley Test Company; Retrieved from http://trove.nla.gov.au/version/46509214 [Google Scholar]
- Husain, M. , & Nachev, P. (2007). Space and the parietal cortex. Trends in Cognitive Sciences, 11(1), 30–36. 10.1016/j.tics.2006.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnath, H.‐O. , Fruhmann Berger, M. , Küker, W. , & Rorden, C. (2004). The anatomy of spatial neglect based on voxelwise statistical analysis: A study of 140 patients. Cerebral Cortex (New York, N.Y.: 1991), 14(10), 1164–1172. 10.1093/cercor/bhh076 [DOI] [PubMed] [Google Scholar]
- Kertesz, A. (1982). The western aphasia battery. New York, NY: Grune & Stratton. [Google Scholar]
- Kimberg, D. Y. , Coslett, H. B. , & Schwartz, M. F. (2007). Power in voxel‐based lesion‐symptom mapping. Journal of Cognitive Neuroscience, 19(7), 1067–1080. 10.1162/jocn.2007.19.7.1067 [DOI] [PubMed] [Google Scholar]
- Lacey, E. H. , Skipper‐Kallal, L. M. , Xing, S. , Fama, M. E. , & Turkeltaub, P. E. (2017). Mapping common aphasia assessments to underlying cognitive processes and their neural substrates. Neurorehabilitation and Neural Repair, 31(5), 442–450. 10.1177/1545968316688797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah, Y.‐H. , Husain, M. , Rees, G. , & Nachev, P. (2014). Human brain lesion‐deficit inference remapped. Brain: A Journal of Neurology, 137(pt 9), 2522–2531. 10.1093/brain/awu164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzocchi, F. , & Vignolo, L. A. (1979). Localisation of lesions in aphasia: Clinical‐CT scan correlations in stroke patients. Cortex; a Journal Devoted to the Study of the Nervous System and Behavior, 15(4), 627–653. [DOI] [PubMed] [Google Scholar]
- Mesulam, M. M. , Thompson, C. K. , Weintraub, S. , & Rogalski, E. J. (2015). The Wernicke conundrum and the anatomy of language comprehension in primary progressive aphasia. Brain, 138(8), 2423–2437. 10.1093/brain/awv154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirman, D. , Chen, Q. , Zhang, Y. , Wang, Z. , Faseyitan, O. K. , Coslett, H. B. , & Schwartz, M. F. (2015). Neural organization of spoken language revealed by lesion‐symptom mapping. Nature Communications, 6, 6762 10.1038/ncomms7762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirman, D. , Landrigan, J.‐F. , Kokolis, S. , Verillo, S. , Ferrara, C. , & Pustina, D. (2018). Corrections for multiple comparisons in voxel‐based lesion‐symptom mapping. Neuropsychologia, 115, 112–123. 10.1016/j.neuropsychologia.2017.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr, J. P. , Pessin, M. S. , Finkelstein, S. , Funkenstein, H. H. , Duncan, G. W. , & Davis, K. R. (1978). Broca aphasia pathologic and clinical. Neurology, 28(4), 311–311, 324. 10.1212/WNL.28.4.311 [DOI] [PubMed] [Google Scholar]
- Naeser, M. A. , & Hayward, R. W. (1978). Lesion localization in aphasia with cranial computed tomography and the Boston Diagnostic Aphasia Exam. Neurology, 28(6), 545–551. [DOI] [PubMed] [Google Scholar]
- Nichols, T. E. , & Holmes, A. P. (2002). Nonparametric permutation tests for functional neuroimaging: A primer with examples. Human Brain Mapping, 15(1), 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay, S. B. , Stengel, B. C. , Humphries, C. , Book, D. S. , & Binder, J. R. (2014). Cerebral localization of impaired phonological retrieval during rhyme judgment. Annals of Neurology, 76(5), 738–746. 10.1002/ana.24266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack, R. A. , Mumford, J. A. , & Nichols, T. E. (2011). Handbook of Functional MRI Data Analysis (1 edition) Cambridge New York Melbourne Madrid: Cambridge University Press. [Google Scholar]
- Price, C. J. , Hope, T. M. , & Seghier, M. L. (2017). Ten problems and solutions when predicting individual outcome from lesion site after stroke. NeuroImage, 145(pt B), 200–208. 10.1016/j.neuroimage.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pustina, D. , Coslett, H. B. , Turkeltaub, P. E. , Tustison, N. , Schwartz, M. F. , & Avants, B. (2016). Automated segmentation of chronic stroke lesions using LINDA: Lesion identification with neighborhood data analysis. Human Brain Mapping, 37(4), 1405–1421. 10.1002/hbm.23110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapcsak, S. Z. , Beeson, P. M. , Henry, M. L. , Leyden, A. , Kim, E. , Rising, K. , … Cho, H. S. (2009). Phonological dyslexia and dysgraphia: Cognitive mechanisms and neural substrates. Cortex, 45(5), 575–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach, A. , Schwartz, M. F. , Martin, N. , Grewal, R. S. , & Brecher, A. (1996). The Philadelphia naming test: Scoring and rationale. Clinical Aphasiology, 24, 121–133. http://aphasiology.pitt.edu/215/ [Google Scholar]
- Rogalsky, C. , Poppa, T. , Chen, K.‐H. , Anderson, S. W. , Damasio, H. , Love, T. , & Hickok, G. (2015). Speech repetition as a window on the neurobiology of auditory–motor integration for speech: A voxel‐based lesion symptom mapping study. Neuropsychologia, 71, 18–27. 10.1016/j.neuropsychologia.2015.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorden, C. , Karnath, H.‐O. , & Bonilha, L. (2007). Improving lesion‐symptom mapping. Journal of Cognitive Neuroscience, 19(7), 1081–1088. 10.1162/jocn.2007.19.7.1081 [DOI] [PubMed] [Google Scholar]
- Schwartz, M. F. , Faseyitan, O. , Kim, J. , & Coslett, H. B. (2012). The dorsal stream contribution to phonological retrieval in object naming. Brain: A Journal of Neurology, 135(pt 12), 3799–3814. 10.1093/brain/aws300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turken, A. U. , & Dronkers, N. F. (2011). The neural architecture of the language comprehension network: Converging evidence from lesion and connectivity analyses. Frontiers in Systems Neuroscience, 5, 1 10.3389/fnsys.2011.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernicke, C. (1874). Der aphasische Symptomencomplex: Eine psychologische Studie auf anatomischer Basis. Breslau, Poland: Cohn. [Google Scholar]
- Wilson, S. M. (2017). Lesion‐symptom mapping in the study of spoken language understanding. Language, Cognition and Neuroscience, 32(7), 891–899. 10.1080/23273798.2016.1248984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, S. , Lacey, E. H. , Skipper‐Kallal, L. M. , Jiang, X. , Harris‐Love, M. L. , Zeng, J. , & Turkeltaub, P. E. (2016). Right hemisphere grey matter structure and language outcomes in chronic left hemisphere stroke. Brain, 139(1), 227–241. 10.1093/brain/awv323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, S. , Lacey, E. , Skipper‐Kallal, L. M. , Zeng, J. , & Turkeltaub, P. (2017). White matter correlates of auditory comprehension outcomes in chronic post‐stroke aphasia. Frontiers in Neurology, 8, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnell, P. , Monroe, P. , & Sobel, L. (1976). Aphasia outcome in stroke: A clinical neuroradiological correlation. Stroke, 7(5), 516–522. 10.1161/01.STR.7.5.516 [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Kimberg, D. Y. , Coslett, H. B. , Schwartz, M. F. , & Wang, Z. (2014). Multivariate lesion‐symptom mapping using support vector regression. Human Brain Mapping, 35(12), 5861–5876. 10.1002/hbm.22590 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 This figure plots the correlation between behavioral performance and lesion volume for 20 behaviors measured in a cohort of left‐hemisphere stroke survivors (N = 48), with significant correlations in red. Below each plotted correlation, thumbnails are shown of SVR‐LSM results using five different lesion volume correction methods. Values displayed are suprathreshold SVR‐β values thresholded voxelwise at p < .005. Columns correspond to behaviors and rows correspond to lesion volume correction methods. MNI coordinate in the z plane is shown in white for each column. The lesion volume correction methods displayed include no correction, dTLVC, regression of lesion volume out of behavior, regression of lesion volume out of voxelwise lesion statuses, and regression of lesion volume out of both behavior and lesion data. The top row of lesion‐symptom maps, which are uncorrected for lesion volume, also depicts regions from Figure 3d for behaviors significantly correlated (p < .05) with lesion volume (red outline) and not significantly correlated with lesion volume (dotted white outline). Behaviors include Philadelphia Naming Test score (PNT), pseudoword repetition, regularity effect, Pyramids and Palm Trees (PPT), backward digit span, Western Aphasia Battery (WAB) Fluency score, category fluency, score on complex ideational material (CIM) from the Boston Diagnostic Aphasia Examination (BDAE), WAB Content score, WAB Sequential Commands score, WAB Yes/No score, letter fluency, imageability effect, picture pointing score, WAB Repetition score, lexicality effect, WAB Word Recognition score, written PNT score, and the Trails and Attention subscores on the Cognitive Linguistics Quick Test (CLQT)
Supplemental Figure 2 This figure replicates the results from Supplemental Figure which used SVR‐LSM, with traditional mass‐univariate lesion‐symptom mapping. This figure demonstrates that the concern about lesion volume as a confound is not limited to multivariate lesion‐symptom mapping. In this figure, P values are derived from beta values from voxelwise linear regression (see Supplemental Methods). Statistical maps are corrected at voxelwise p < .005, based on 10,000 permutations. Each of the 20 columns corresponds to one behavior and each and five rows correspond to a lesion volume correction methods, as in Supplemental Figure
Supplemental Figure 3 This figure replicates the results from Figure 6, which used SVR‐LSM, with traditional mass‐univariate lesion‐symptom mapping methods. Effect of lesion volume correction on mass‐univariate VLSM localization for (A) a simulated behavior significantly correlated with lesion volume and (B) a simulated behavior less correlated with lesion volume. Behaviors were simulated by summing the number of lesioned voxels within a 4 mm sphere (red) placed based on its correlation with lesion volume. At the left of each pane is shown a scatterplot of lesion volume versus the simulated behavior across the patient sample. Voxels with statistically significant results at voxelwise p < .001 based on 10,000 permutations are shown in bright green
Supplemental Table 1 Table listing which of 20 behaviors were significant for each of five lesion volume correction methods using traditional, mass‐univariate lesion‐symptom mapping. A solid bullet (•) indicates that significant findings for a given combination of behavior and lesion volume correction method which survived voxelwise thresholding, p < .005 (one‐tailed) and clusterwise correction, p < .05 based on 10,000 permutations. An empty bullet (○) indicates that the analysis resulted in voxels that survived voxelwise thresholding but did not survive cluster correction
Supplemental Methods


