Abstract
The double-membrane cellular envelope of Gram-negative bacteria enables them to endure harsh environments and represents a barrier to many clinically available antibiotics. The outer membrane (OM) is exposed to the environment and is the first point of contact involved in bacterial processes such as signaling, pathogenesis, and motility. As in the cytoplasmic membrane, the OM in Gram-negative bacteria has a phospholipid-rich inner leaflet and an outer leaflet that is predominantly composed of lipopolysaccharide (LPS). We report on a microfluidic technique for fabricating monodisperse asymmetric giant unilamellar vesicles (GUVs) possessing the Gram-negative bacterial OM lipid composition. Our continuous microfluidic fabrication technique generates 50 to 150 µm diameter water-in-oil-in-water double emulsions at high-throughput. The water-oil and oil-water interfaces facilitate the self-assembly of phospholipid and LPS molecules to create the inner and outer leaflets of the lipid bilayer, respectively. The double emulsions have ultrathin oil shells, which minimizes the amount of residual organic solvent that remains trapped between the leaflets of the GUV membrane. An extraction process by ethanol and micropipette aspiration of the ultrathin oil shells triggers an adhesive interaction between the two lipid monolayers assembled on the water-oil and oil-water interfaces (i.e., dewetting transition), forcing them to contact and form a lipid bilayer membrane. The effect of different inner-leaflet lipid compositions on the emulsion/vesicle stability and the dewetting transition is investigated. We also report on the values for bending and area expansion moduli of synthetic asymmetric model membranes with lipid composition/architecture that is physiologically relevant to the OM in Pseudomonas aeruginosa bacteria.
Introduction
Giant unilamellar vesicles (GUVs) are of great interest as versatile model systems to emulate the function and fundamental properties of the plasma membrane in living cells.1–4 However, it remains a challenge to build physiologically relevant synthetic systems (e.g. vesicles) from scratch using elemental organic and inorganic components and materials. While GUV model systems are too simple to represent the complexity of the entire biological membrane, they are suitable candidates to isolate and study the roles of individual components involved in cellular function.5,6 Synthetic and natural vesicles consist of one or multiple lipid bilayer(s) surrounding an aqueous core that allows encapsulation and protection of active hydrophilic molecules such as nucleic acids. Over the last few decades, researchers have utilized vesicles as models to study biological membrane mechanics and signaling,7–11 lipid and protein distribution,12–16 and as delivery vehicles for DNA, virulence factors, drug delivery, etc.17–20
Gram-negative bacteria are unique because they are characterized by a cell envelope that consists of a relatively thin layer of peptidoglycan between two bilayers of a dissimilar molecular profile.5,21 The cell envelope has proved to be an excellent target for antimicrobial therapy.18,22 The existence of two membranes in Gram-negative bacteria offers two concentric barriers to the external environment with different physical properties (i.e. bending and area expansion moduli).21 The double-membrane cellular envelope in Gram-negative bacteria (e.g. Pseudomonas aeruginosa) enables them to endure harsh environments and represents an important permeability barrier to many clinically available antibiotics. Pseudomonas aeruginosa, a common environmental Gram-negative bacillus, is an opportunistic human pathogen that causes acute or chronic infection in the lung, urinary tract, burns, and blood stream.23 P. aeruginosa is found in 18% of cases of nosocomial pneumonia24 and it is also the major pathogen in cystic fibrosis.25
The outer membrane (OM) in P. aeruginosa is composed of phospholipids in the inner leaflet (mainly phosphoethanolamine (PE), phosphatidylglycerol (PG), and phosphatidylcholine (PC)5) and lipopolysaccharide (LPS) in the outer leaflet.26 Tashiro et. al reported the normalized percentage of PE, PG, and PC headgroups to be 60%, 27%, and 13% for P. aeruginosa OM, respectively.27,28 In addition, the fatty acid compositions of phospholipids in the cellular OM showed a 1:1 molar ratio of saturated to unsaturated aliphatic chains. However, LPS contains three distinct components: a hydrophobic lipid A anchored in the OM, a hydrophilic core polysaccharide chain linked to Lipid A, and a hydrophilic O-antigen side chain.13,29 The outer membrane of gram-negative bacteria is decorated with a potent endotoxin (i.e. Lipid A) which plays a significant role in bacterial survival, pathogenicity, and immune evasion.30–33 It also acts as a physical barrier protecting the cell from chemical attack and represents a significant obstacle for the effective delivery of numerous antimicrobial agents.23
The ability to build and customize biologically relevant vesicles is a sophisticated technology that can advance biological science. Over the last few decades, synthetic biology research has focused on two approaches for building engineered biological cells and vesicles with specialized functions. The first is a top-down methodology to redesign existing biological systems by manipulating the properties of natural cells to achieve or investigate a desired function. The second is a bottom-up approach to build customized cellular systems that may or may not already exist in nature.34,35 With the bottom-up approach, the objective is to: (i) develop a better understanding that would not be possible through the analysis and observation of existing biological systems alone, or (ii) achieve a specialized function that can only be accomplished by creating new systems. Fabrication of synthetic vesicles was first introduced by Angelova and Dimitrov in 1986 using electroformation.36 In this technique, a uniform thin layer of lipids is deposited onto an indium tin oxide (ITO)-coated glass slide and is subjected to an electric field in an aqueous bath.36,37 Electroformation offers limited control over the vesicle size and lipid assembly configuration (i.e. bilayer asymmetry, unilamellarity, and lipid composition). Kubiak et al. developed a protocol using electroformation to create GUVs containing LPS and used them as templates to investigate how these molecules affect the lateral assembly in bacterial model membranes.1 However, GUVs fabricated using their method lack lipid asymmetry and could not fully represent a strong model for Gram-negative bacterial outer membrane model system. In addition, lipid exposure to air throughout the GUVs fabrication process increases the risk of lipid oxidation that constantly needs to be monitored.
Other methods such as gentle hydration,38 micropipette jetting,39,40 and GUV budding assisted by swelling hydrogel films41–43 have been used to build GUVs. Gentle hydration involves the simple rehydration of lipid films with an aqueous solution; however, vesicles formed using this method may display undesirable defects such as vesicles within vesicles and/or lipid aggregates on bilayer leaflets that may hinder their utilization. Micropipette jetting requires the use of high frequency fluid pulses onto a lipid bilayer from an inkjet device to promote the formation of a GUV. In hydrogel-assisted approach, a film of hydrogel, water-swollen networks of cross-linked hydrophilic polymers, is formed on a coverslip followed by a thin film of lipid. This system is rehydrated with physiologically relevant buffers and GUVs with diameters from 10 to 100 μm are readily formed driven by coalescence of sub-μm-sized vesicles into larger vesicles.41 These traditional fabrication techniques allow for relatively facile production of membrane material. However, they provide weak control over membrane lipid composition and architecture. In addition, these methods often lead to non-uniform vesicles in terms of size and lamellarity.
An alternative method for producing GUVs is using microfluidic technology. Hydrodynamic flow focusing,44 pulsed jetting,45 and double emulsion templates with organic phase removal46,47 enable the fabrication of more uniform emulsions at any size range while maintaining unilamellarity and customized lumen content. In addition, these techniques provide the opportunity to deliver functional particles to the emulsion interface at high-throughput.48–51 Microfluidic-based methods exploit oil-water interfaces in droplets/double emulsions to spontaneously assemble lipid bilayers one leaflet at a time.52 This offers a better control over the transbilayer lipid architecture in double emulsion templates which results in acquiring an asymmetric assembly in GUVs (i.e. the composition of lipids in one leaflet of the membrane differs from that of the other leaflet). Our team previously developed a continuous microfluidic fabrication technique for generating 10 to 50 μm diameter water-in-oil-in-water double emulsions at high-throughput for building asymmetric vesicles (Fig. 1).47
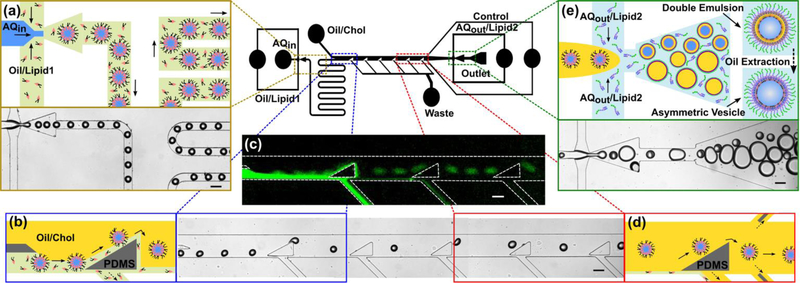
Fig. 1.
Schematics of our microfluidic technique that is capable of generating 50 to 150 μm diameter water-in-oil-in-water double emulsions at high-throughput. (a) Initially, uniform water-in-oil emulsions are made at the first flow focusing region. The phospholipid molecules (i.e. lipid-1) that are present in the oil phase (i.e. green domain) spontaneously assemble on the water-oil interface and proceed along the serpentine channel. (b) After the serpentine channel, a series of triangular posts force the emulsions to move across the two oil phases (i.e. green and yellow domains) and continue along the streamlines in the OL2. (c) The fluorescent image confirms the assembly of NBD-tagged phospholipids on the emulsions and their separation from the OL1 stream (i.e. green flow). The OL1 stream goes into the waste channels located under the triangular posts. (d) Finally, the water-in-oil emulsions, covered with a monolayer of lipid-1 molecules, approach the second flow focusing region. (e) The emulsions are encapsulated in an ultrathin OL2 layer containing cholesterol and form water-in-oil-in-water (w/o/w) double emulsions dispersed in the OA domain in which the LPS molecules are dissolved. Ethanol was introduced to the OA domain in order to extract the intermediate oil layer from the double emulsions and to bring the two lipid monolayers together. Flow rates of the IA, OL1, OL2, and OA streams were set at 12, 22, 400, and 380 μL h−1, respectively. All scale bars denote 100 μm.
In this work, we study the formation of asymmetric model membrane systems (i.e. GUVs) that resemble the outer membrane lipid composition and transbilayer architecture of the Gram-negative bacillus, P. aeruginosa, more accurately. The lipid bilayer architecture and composition of the fabricated model membranes make them suitable candidates to study the bacterial function in antimicrobial studies. We use our microfluidic-based technique to make asymmetric vesicles possessing different compositions of phospholipids in the inner leaflet and LPS in the outer leaflet. The water-oil and oil-water interfaces facilitate the spontaneous self-assembly of phospholipid and LPS molecules to create the inner and outer leaflets of the lipid bilayer, respectively. We also investigate the separation of the organic phase (i.e. dewetting transition) using a chemical oil extraction process and mechanical perturbation. Values for area expansion and bending moduli of asymmetric model membrane systems are reported.
Experimental procedures
Materials
1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dioleoyl-sn-glycero-3-phospho-(1’-rac-glycerol) (DOPG), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), and 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) were purchased from Avanti Polar Lipids. The wild type lipopolysaccharide from P. aeruginosa obtained by phenol extraction (Sigma-Aldrich) was used without further purification. Two fluorescently labeled tracer lipids were used to individually indicate the assembly of the inner and outer leaflets. (N-(7-Nitrobenz-2-Oxa-1,3-Diazol-4-yl)-1,2-Dihexadecanoyl-sn-Glycero-3-Phosphoethanolamine (NBD-DHPE) was purchased from Thermo Fisher Scientific and stocked in chloroform. We used the lipopolysaccharide from Escherichia coli serotype 055:B5, Alexa Flour™ 568 conjugate (Thermo Fisher Scientific) as the tracer to confirm the assembly of LPS in the outer leaflet. Dow SYLGARD™ 184 Silicone Elastomer Clear kit was purchased from Ellsworth. All other chemicals were purchased from VWR.
Preparation of solutions
All phospholipids were stored in chloroform at −20 °C. To prepare a dry lipid film, we evaporated the chloroform under a gentle nitrogen stream for 30 min at 40 °C followed by vacuum exposure for at least 2 h at room temperature. The dried film was dissolved in oleic acid by ultra-sonication for 3 h at 40 °C. Wild type lipopolysaccharide and its fluorescent conjugate were dissolved in aqueous solutions in three cycles of heating to 75 °C and vortex mixing. To prevent the coalescence of double emulsions and GUVs, a nonionic Pluronic surfactant F-68 and polyvinyl alcohol were added to both the inner and outer aqueous phases. In addition, the outer aqueous phase contained 14 vol% glycerol to increase the interfacial viscous stress and facilitate the emulsion formation inside the microfluidic device. This would consequently minimize the resultant oil layer thickness in encapsulating the double emulsions. After adding other constituents (i.e. sugar, surfactants and glycerol) to the aqueous solutions we sonicated them for 1 h at 40 °C. The osmolality of the inner and outer aqueous solutions was measured and matched using a freezing point osmometer (Osmomat 3000, Gonotec). The compositions of the inner aqueous (IA), oil/lipid1 (OL1), oil/Chol (OL2), and outer aqueous (OA) phases are stated below.
IA phase (all solutions were prepared in Milli-Q water): 1 vol% PVA (M.W. ∼ 57,000−66,000 86–89% hydrolyzed), 2 vol% Pluronic F-68, (Fig. 2 and 3; 0.5 M sucrose 99%), (Fig. 4; 1.5 M sucrose 99%), (Fig. 5; 1.2 M sucrose 99%), (Fig. 2f; 1 mM MgSo4.7H2O), and (Fig. 4 and 5; 10 mM MgSo4.7H2O).
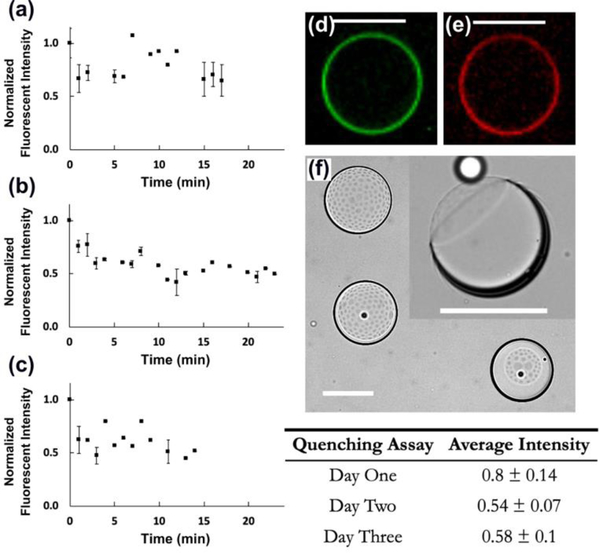
Fig. 2.
The asymmetry assay conducted on the model membrane (i) possessing DOPE/NBD-DHPE (200:1 mol%) in the inner leaflet and LPS/Alexa Flour™ 568 (200:1 mol%) in the outer leaflet (n=3). GUVs were stored in aqueous solution for one, two, and three days in (a), (b), and (c), respectively, followed by the oil extraction process for each. The error bars are ± one standard deviation. The table demonstrates the average normalized steady intensity for each assay ± one standard deviation. The two florescent images, (d) and (e), confirmed the assembly of DOPE and LPS on the inner and outer interfaces, respectively. (f) The top-view of three dewetted oil shells upon adding the ethanol solution. The inset illustrates the side-view of a lipid bilayer membrane with an oil lens attached to it (i.e. the dark ring). All scale bars denote 100 μm.
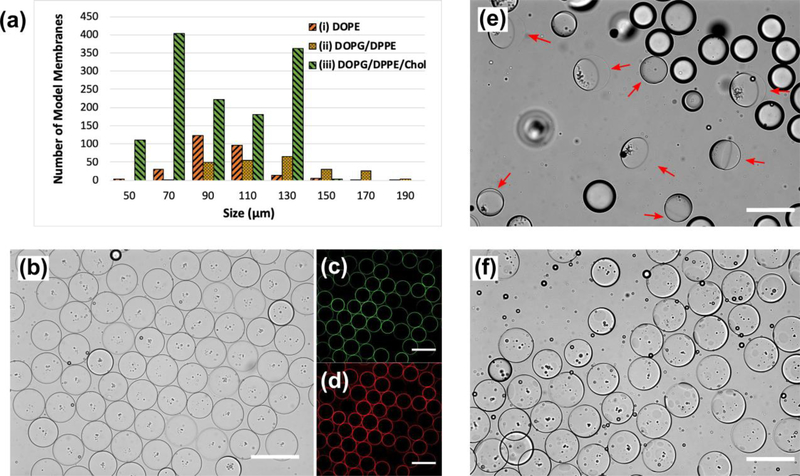
Fig. 3.
All panels illustrate the fabricated bacterial model membrane GUVs possessing phospholipids and LPS in the inner and outer leaflets, respectively. (a) The longevity and size distribution of our three model membrane designs for the oil extraction process. (b) Uniform vesicles containing 40 mol% cholesterol embedded in the lipid bilayer. (c) and (d) are fluorescent images of NBD-labeled DOPG/DPPE (1:1 mol%) and Alexa Flour™ 568-labeled LPS molecules assembly on the inner and outer leaflets, respectively. (e) The ultrathin double emulsion templates (ii) undergoing the partial dewetting transition after adding the ethanol solution to the extracellular domain. Red arrows point at the fully packed lipid bilayer membrane side of the double emulsions. The other droplets were excess oil drops present in the domain. (f) After introducing ethanol to the OA solution, the oil shell in model membrane (iii) ruptured into micro domains protruding outward from the surface. All scale bars denote 200 μm.
Fig. 4.
The dewetting transition of a model membrane composed of DOPC/DPPC/Chol (3:3:4 mol%) and LPS in the inner and outer leaflets, respectively. (a) Illustrating a dewetting event triggered by a mechanical perturbation (i.e. tapping the double emulsion with a micropipette capillary). (b) A slight suction was introduced to the tip of the micropipette capillary that initiated a local dewetting. The lipid bilayer continued to grow throughout the entire surface and resulted in the accumulation of the trapped oil into a droplet. The oil droplet looked blurry since it was out of the focal plane. (c) The schematic of enforced dewetting initiated at the interface of micropipette tip and the oil shell. (d) and (e) confirmed a fully packed assembly of DOPC/DPPC/NBD-DHPE (100:100:1 mol%) and LPS/Alexa Flour™ 568 (100:1 mol%) at the point of contact. All scale bars denote 50 μm
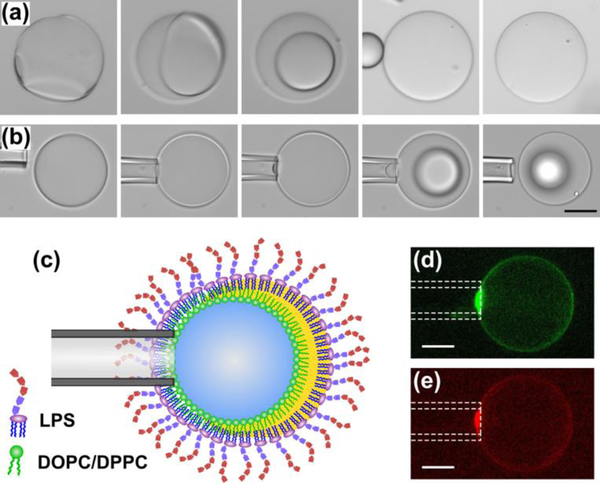
Fig. 5.
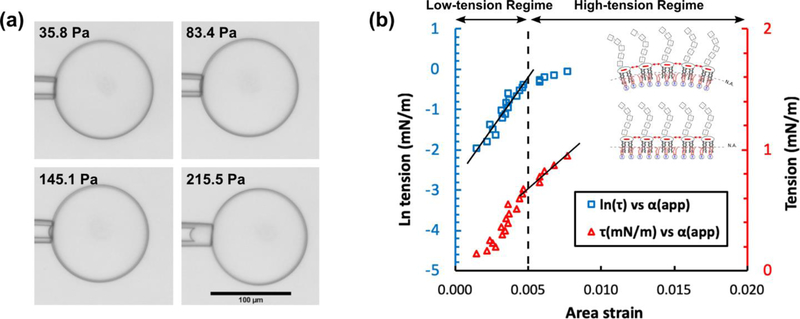
The micropipette aspiration method tested on GUVs possessing DOPC/DPPC/Chol (3:3:4 mol%) and LPS in the inner and outer leaflets, respectively. (a) As is depicted in the four panels, by increasing the withdrawing hydrostatic pressure (i.e. from 36 to 215 Pa) a portion of the lipid bilayer membrane was drawn into the micropipette aspirator. The scale bar denotes 100 μm and is the same in all panels. (b) The plot shows the logarithm of tension (i.e. blue squares) and tension (i.e. red triangles) versus the apparent area strain. The lines are fitted to the blue squares and red triangles in the low- and high-tension regimes, respectively. The R-squared values are larger than 90%. The schematics in the inset illustrate our hypothesis on the effect of repulsive electrostatic stress between the lipid A section of LPS molecules on the bending and area expansion moduli (see text).
OL1 phase: all phospholipid solutions were at 6.25 mg mL−1 in oleic acid (Fig. 2 and 3; DOPE/NBD-DHPE 250:1 mol%), (Fig. 3; DOPG/DPPE/NBD-DHPE 125:125:1 mol%), (Fig. 4 and 5; DOPC/DPPC/NBD-DHPE 100:100:1 mol%).
OL2 phase: (Fig. 2 and 3e; oleic acid) and (Fig. 3, 4, and 5; cholesterol at 2 mg mL−1 in oleic acid).
OA phase (all solutions were prepared in Milli-Q water): 1 vol% PVA (M.W. ∼ 57,000−66,000 86–89% hydrolyzed), 2 vol% Pluronic F-68, 0.5 M D-(+)-Glucose, (Fig. 2, 3, and 4; 14 vol% glycerol), (Fig. 5; 8 vol% glycerol), (Fig. 2f; 1 mM MgSo4.7H2O), (Fig. 4 and 5; 10 mM MgSo4.7H2O), (Fig. 2 and 3; 100 μg mL−1 LPS), (Fig. 4 and 5; 300 μg mL−1 LPS), (Fig. 3 and 5; 1.5 μg mL−1 Alexa Flour™ 568 LPS conjugate), and (Fig. 4; 3 μg mL−1 Alexa Flour™ 568 LPS conjugate).
Microfluidic device fabrication and operation
We built our PDMS device by utilizing the conventional soft lithography process (Fig. 1 illustrates the design).53–55 A mold was first prepared by spin coating a 90 μm thick layer of SU-8 2050 on a four inch silicon wafer. The SU-8 layer was soft baked and patterned by exposure to UV light at 55 mJ cm–2 through a polyester photomask (Fineline Imaging, Inc.). The silicon wafer was post baked (i.e. at 65 °C for 3 min, followed by 95 °C for 9 min, and back at 65 °C for 3 min) and developed in SU-8 developer for 7 min. PDMS was mixed at a ratio of 9:1 of pre-polymer base to curing agent. The PDMS replica was cured at 70 °C for 4 h, peeled from the mold, and diced into individual devices. Connection holes were punched with a blunt dispensing needle. The device was bonded under oxygen plasma to a glass slide covered with a layer of 100 μm spin-coated PDMS. The oxygen plasma treatment generates radical surface silanol groups (Si–OH), alcoholic hydroxyls (C–OH), and carboxylic acids (COOH) on the PDMS surfaces.56 This covalently bonded the two surfaces and created a sealed microfluidic device. Spin-coating PDMS onto the glass slide ensured material consistency inside the channel network since the PDMS was present on all four sides of the channels.
One day after the bonding, all PDMS channels recovered their hydrophobic nature at room temperature.56 However, this can be accelerated at higher temperatures. The microfluidic channels carrying the OA solution (i.e. the blue stream depicted in Fig. 1e) were locally treated with a 2 wt% solution of PVA (M.W. ∼ 57,000−66,000 86–89% hydrolyzed) in DI water to render the surfaces hydrophilic. The device was left under vacuum for 30 min and baked at 120 °C for 12 min. All solutions were loaded in 2.5 mL glass syringes (Hamilton) and were supplied to the inlets of the device through translucent PTFE tubing (1/32” ID, Cole-Parmer). We used syringe pumps (Chemyx) to generate flow in the channels.
Micropipette aspiration operation and theory
The micropipette capillaries were custom-made by pulling 1mm OD, 0.58 mm ID borosilicate glass capillaries (World Precision Instruments, Inc.) using a glass microelectrode puller (Narishige, Japan). The glass capillary tips were cut and bent using microforge MF-900 (Narishige, Japan) to obtain micropipettes with inner diameter of 20 μm at angle of 30°. This guarantees that the tip of micropipette is perfectly parallel to the microscope focal plane. A new micropipette was used for every set of experiments.
The micropipette capillary was precisely located inside a custom-built PDMS observation chamber containing OA solution, using a manual micromanipulator (Sutter Instrument, USA). In the beginning of every measurement, the micropipette tip was located at the same height as the two water reservoirs. The difference between hydrostatic pressures in the water reservoirs was used to apply a suction pressure to a portion of the membrane. The initial zero pressure state inside the micropipette was attained and calibrated by observing the flow of small particles at the tip. The aspiration pressure was controlled by adjusting the relative height of water reservoirs mounted vertically on a linear translational stage (Velmex, Inc.). The acquired hydrostatic pressures between the two reservoirs were measured by an ultra-low range differential pressure transducer (DP 103 Validyne Eng.). The output signal from the transducer was detected by a single-channel USB carrier demodulator (CD 17, Validyne Eng.). In order to ensure the system is at equilibrium, measurements were recorded after 3 min of pressure adjustment. Further details of operating the micropipette aspiration was provided by Lu et al.57
The bending and area expansion moduli were calculated based on the equilibrium theory that divides the stress-strain relationship into low-tension and high-tension regimes.58 In the high-tension regime (τ > 0.5 mN m−1), Evans and Rawicz confirmed that the apparent area strain varied linearly with tension due to the direct expansion of the area per molecule:58
| (1) |
where τ is the applied tension, Kapp is the apparent area expansion modulus, and αapp is the apparent area strain. The isotropic membrane tension is related to the applied withdrawal pressure (ΔP) as:
| (2) |
where RC and RV are the micropipette capillary and vesicle radius, respectively. Also, we can calculate the apparent area strain as:
| (3) |
where, ΔL is the aspirated length of the membrane into the micropipette capillary, A is the membrane area after aspiration in the high-tension regime, and A0 is the membrane area in the initial low-tension regime. In addition, the logarithm of tension varies linearly with area strain in the low-tension regime (τ < 0.5 mN m−1):
| (4) |
where, τ0 is the initial applied tension, κ is the bending modulus, kB is the Boltzmann constant, and T is the absolute temperature.58
Image processing
Bright field images illustrating the GUVs and the flows in the channels were captured using an inverted microscope (Leica) and a CCD camera (Qimaging). The excitation/emission wavelengths for NBD and Alexa Flour™ 568 conjugates were 463/536 nm and 578/603 nm, respectively. We used ImageJ for image processing and analysis.
Results and discussions
Our microfluidic technique generates 50 to 150 μm diameter water-in-oil-in-water ultrathin double emulsions at high-throughput, which are used for building the asymmetric vesicles. Fig. 1 summarizes the entire process on the microfluidic-based chip. Initially, uniform water-in-oil emulsions are made at the first flow focusing region (Fig. 1a). The phospholipid molecules (i.e. lipid-1) that are present in the oil phase spontaneously assemble on the water-oil interface and proceed along the serpentine channel. This provides enough time for the lipid molecules to spontaneously assemble on the water-oil interface and eventually form the inner leaflet of the lipid bilayer membrane. Another advantage of the serpentine channel is to keep the two flow focusing regions apart from each other (i.e. Fig. 1a and 1e) and to dampen the induced perturbations along the microfluidic channels. Downstream from the serpentine channel (Fig. 1b), a series of triangular posts forces the emulsions to deflect across the streamlines (i.e. from the green stream into the yellow domain) and continue along the flow into the OL2 phase containing cholesterol. A comprehensive description of the oil separation performance of the triangular posts was discussed by Lu et al.59 Finally, the water in oil emulsions, covered with a monolayer of lipid-1 molecules (Fig. 1c), approach the second flow focusing region (Fig. 1d). The emulsions are enclosed within an ultrathin shell of OL2 phase containing cholesterol, and form water-in-oil-in-water (w/o/w) double emulsions (Fig. 1e). Through the outlet port, the w/o/w double emulsions and the excess OL2 oil droplets are collected from the chip into Eppendorf centrifuge tubes. The double emulsions encapsulating sucrose in the lumen sink to the bottom of the centrifuge tube in the OA solution with dissolved LPS molecules. This provides enough time for the LPS molecules to spontaneously assemble on the outer oil-water interface due to their amphipathic nature.13,60 LPS molecules can also form micelles above the CMC in the OA solution. It is reported that the CMC of the wild type LPS from E. Coli O55:B5 and O26:B6 are 10 μg mL−1 and 14 μg mL−1, respectively.61,62 We assumed the same properties for the wild type LPS from P. aeruginosa and set the LPS concentrations above CMC to assure enough supply to the interface.
After 30 min, centrifuge tubes were poked from the bottom and the OA phase drained into a custom-built observation chamber. This acted as a filtration method such that the excess OL2 oil droplets remained in the centrifuge tube. A solution of OA phase containing 14 vol% ethanol was gradually added to the OA domain in order to extract the intermediate oil layer from the double emulsions and to bring the two leaflets together. We observed the formation of lipid bilayers with and without cholesterol molecules embedded in the hydrophobic region of the membranes. This was likely due to the Van der Waals interactions between the fatty acid chains of the phospholipids and the lipid A section of the LPS molecules. The oil extraction process took up to 18 hours to complete and was confirmed by the reduction in the thickness of double emulsions.
Asymmetric architecture across the lipid bilayer membrane
The lipid bilayer membrane asymmetry was qualitatively evaluated using a fluorescence-quenching assay. Since membrane-impermeable dithionite (NBD quencher, Na2S2O4) is unable to diffuse across the lipid bilayer, it cannot quench the fluorophore when NBD is deposited in the inner leaflet.63–68 Fig. 1c illustrates a fluorescent image of the water emulsions coated with a layer of NBD-tagged lipid 1 molecules. As mentioned above, a series of triangular posts deflect the water emulsions from oil/lipid 1 stream into the oil/Chol that eventually results an asymmetric architecture across the lipid bilayer membrane. The two fluorescent images, Fig. 2d and 2e, illustrate the assembly of DOPE and LPS on the inner and outer interfaces, respectively. The asymmetrical assembly in synthetic phospholipid-LPS model membranes make them more relevant candidates to study bacterial function than conventional phospholipid-phospholipid membranes. In order to confirm the asymmetry, the fabricated double emulsions were stored in the aqueous solution containing LPS for one, two, and three days (Fig. 2a, 2b, and 2c, respectively) followed by the oil extraction process that took about 18 h (n = 3). The quencher was added in portions of 10 μL every two minutes throughout the assay. As dithionite quenched the fluorophore, the fluorescent intensity was recorded over time and normalized by its maximum value at time zero (i.e. prior to adding the quencher to the extracellular domain). In all the plots, the normalized fluorescent intensity of the GUVs decreased in the first 5 minutes and stayed steady while adding more quencher to the outer aqueous domain. The steady state values of the normalized fluorescent intensity are reported in the table (Fig. 2).
Vesicles that were stored for one day after fabrication showed a 20% drop in intensity. This may be due to the outward translocation (i.e. flip-flop) of the phospholipids across the lipid bilayer membrane.69–72 However, after two and three days, the intensity dropped by 50%. This means the degree of asymmetry across the membrane decreased in time while the ultrathin oil shells (i.e. vesicles with trapped oil layer) were stored in the OA solution73. We propose this was due to lipid flip-flop or lipid transfer across the oil layer. Nevertheless, vesicles used one day after fabrication showed strong asymmetry (i.e. greater than 80%) across the lipid bilayer. This assay confirms that our technique is capable of building stronger synthetic models of the OM in P. aeruginosa with an asymmetric lipid bilayer architecture.
Effect of the inner-leaflet lipid composition on lipid bilayer membrane stability
We built three asymmetric model membranes to investigate the stability of the double emulsions from fabrication throughout the oil extraction process (Fig. 3a). As reported by Tashiro et. al, the normalized percentage of PE, PG, and PC headgroups in P. aeruginosa OM was 60%, 27%, and 13%.27 Also, the fatty acid compositions of phospholipids in the cellular OM showed a 1:1 molar ratio of saturated to unsaturated aliphatic hydrocarbon chains. Therefore, we designed our lipid compositions as follows: (i) DOPE, (ii) DOPG/DPPE (1:1 mol%), and (iii) DOPG/DPPE/Chol (3:3:4 mol%) in the inner leaflet and LPS in the outer leaflet (Fig. 2 and 3). To probe the role of individual components and to make the fabrication process tractable, we did not build model membranes composed of more than two phospholipids. Model (i) is a lipid bilayer membrane with the phosphoethanolamine headgroups and dioleoyl chains (the majority of lipid constituents in the wild type P. aeruginosa OM, e.g. DOPE) in the inner leaflet and LPS in the outer leaflet. In model (ii), membranes were composed of a simplified, yet improved mimic of the OM in P. aeruginosa, lipid composition (i.e. DOPG/DPPE, 1:1 mol%) with a 1:1 molar ratio of saturated to unsaturated acyl chains (i.e. dipalmitoyl to dioleoyl). It has been reported that cholesterol and hopanoids exhibit similar ordering properties in eukaryotic and bacterial membranes, respectively, since they have similar molecular structures.74 Therefore, in model (iii), we added cholesterol to the same binary mixture of the phospholipids as in model (ii) at a total ratio of 3:2 (phospholipid:Chol). This concentration lies in the range of physiological cholesterol content in eukaryotic cells that can be as high as 50 mol%.75–78
As shown in Fig. 3a, the ultrathin double emulsions in model (i) had a normal distribution in size with an average diameter of 98 ± 20 μm. We believe the reason for the polydispersity in size and relatively low longevity was due to the fact that DOPE is more suitable for forming inverted micellar structures than vesicles and spherical, ellipsoidal, or cylindrical micelles.79 The value of packing factor (i.e. v/a0lc, where v and lc are the volume and critical length of the hydrocarbon chain, respectively, and a0 is the optimal surface area per molecule on the interface) for lipids with very small optimal headgroup areas or with bulky polyunsaturated chains (e.g. unsaturated phosphatidylethanolamines, negatively charged lipids in the presence of Ca2+ ions, and cholesterol) is larger than one. Hence, it is energetically unfavourable for them to form lipid bilayers in GUVs. Also, the mismatch between the unsaturated fatty acid chains of DOPE and saturated chains in the Lipid A section of wild type LPS molecules can inhibit Van der Waals interactions. In order to resolve this and tune the lipid composition to a physiologically more relevant condition,27 we used a binary mixture of two phospholipids with higher phase transition temperatures (i.e. –18 °C for DOPG and 63 °C for DPPE). As is shown in Fig. 3a, the size distribution (i.e. an average diameter of 124 ± 25 μm) and longevity did not change significantly in model (ii), compared to model (i). However, adding cholesterol at a total molar ratio of 3:2 (i.e. 40 mol%) drastically enhanced the longevity of model membrane (iii) throughout the oil extraction process (Fig. 3b, 3c, and 3d). We propose that the presence of cholesterol in the ultrathin oil shell increases the affinity of saturated dipalmitoyl in DPPE and the 12 to 16 carbon chains in the lipid A section of wild type LPS80 for oleic acid (i.e. cholesterol acts as a coupling agent and makes them more soluble in oleic acid). This enhances the assembly of the LPS molecules and hence increases the surface density on the outer oil-water interface to stabilize the oil shell.81 Fig. 3b shows the fabricated GUVs after they were transferred to the OA phase containing LPS molecules. GUVs containing cholesterol remained stable and did not merge (Fig. 3b). Also, Fig. 3c and 3d confirmed the success in making stable GUVs possessing the ternary mixture of lipids (i.e. DOPG/DPPE/Chol, 3:3:4 mol%) and LPS, in the inner and outer leaflets, respectively. As was previously reported, cholesterol can assemble in the lipid bilayer and fill the gaps in between the kinks in unsaturated cis fatty acid chains. We found that a 40 mol% cholesterol was necessary to form stable and uniform asymmetric model membrane (iii) of the OM in P. aeruginosa.
The double emulsion templates with ultrathin oil shells were used to form the lipid bilayer membranes. Unlike an aqueous solution, oleic acid is a good solvent for the phospholipids and cholesterol, enabling them to remain fully dissolved during the double emulsion formation. Also, the inner water-oil and outer oil-water interfaces are energetically favorable for the phospholipids and LPS molecules to spontaneously assemble due to their amphipathic nature.13,60 Since oleic acid is insoluble in water but is soluble in ethanol, we introduced an OA solution containing 14 vol% ethanol to extract oleic acid (i.e. Fig. 3e and 3f show model membranes (ii) and (iii), respectively). In both cases, partial spontaneous dewetting events were initiated upon adding the ethanol solution to the extracellular domain. In model membrane (ii), as the ethanol solution was introduced to the extracellular domain, the ultrathin shells divided into two domains: (a) a fully packed lipid bilayer membrane indicated by the red arrows, and (b) the organic phase formed into one oil lens on the opposite side (Fig. 3e). However, model membrane (iii) showed a different spontaneous dewetting event (Fig. 3f). The ultrathin oil shell ruptured into micro domains (i.e. dark circular spots that were visible due to the difference in the refractive index of organic and aqueous phases) that looked like lenses bulging out from the lipid bilayer membrane surface. The observation of partial dewetting implies the existence of an adhesive interaction between the aqueous phases that is comparable to the interfacial tension of the lipid monolayers, as previously observed in both oil-in-water, and water-in-oil single emulsion systems.49,50,81–85 We propose that the dewetting transition happens in cases where the two lipid monolayers, assembled on the inner and outer water-oil interfaces, have a greater affinity for each other than to remain dissolved in the organic phase. Therefore, as the two monolayers come into contact, hydrophobic interactions between the lipid tails initiates the dewetting and stabilizes the inner droplet against coalescence with the exterior aqueous phase.
We also observed a similar partial dewetting transition in model (i) upon adding the ethanol solution (Fig. 2f). As is shown, the organic phase was accumulated to one side forming an oil lens attached to the lipid bilayer membrane (i.e. the black ring). However, during the dewetting transition, some oil was left behind in the membrane in the shape of darker micro domains (i.e. the homogeneous hexagonal pattern in three double emulsions). This is opposed to Fig. 3e in which the dewetting transition left a clean lipid bilayer membrane behind (identified by the red arrows in Fig. 3e). We propose that in model membrane (ii), a stronger attractive interaction between the acyl chains in DPPE, DOPG, and wild type LPS dominates the interfacial tension. Therefore, no oil remains in the lipid bilayer membrane as it assembles. However, in case (i), a weaker interaction between DOPE and LPS leaves some oil behind. This may be similar to model membrane (iii) that possesses 40 mol% cholesterol. It has been reported that cholesterol increases the bending and area expansion moduli of lipid membranes but also, allows them to have a high fluidity.3 Therefore, in the presence of cholesterol, the oil shell ruptures into micro domains forming lenses on the membrane (Fig. 3f). In addition, a crucial aspect of this phenomenon is the choice of the organic solvent as the lipids must be highly soluble in it.83 It is noteworthy to mention that upon adding the ethanol solution, ethanol diffuses into the oleic acid (since they are miscible) and this can effectively lower the affinity of saturated and unsaturated acyl chains for oleic acid to remain fully dissolved in the organic phase. This would initiate the dewetting transition and form the lipid bilayer membrane. We propose that introducing ethanol to the extracellular domain can initiate a dewetting transition on the ultrathin oil shells, and results in forming lipid bilayer membranes with one or multiple oil lenses attached to them.
To further understand the driving mechanism behind the dewetting transition, we introduced a mechanical perturbation to the ultrathin double emulsions to trigger the dewetting event without ethanol. We found that the concentration of 100 μg mL−1 LPS in the OA solution was sufficient to stabilize the ultrathin oil shells initially in Fig. 2 and 3, but too low to result in a complete dewetting transition. Thus, our results suggest that obtaining a complete dewetting transition requires a greater concentration of 300 μg mL−1. Also, to be able to form stable membranes, we picked a different lipid composition that maintains the 1:1 molar ratio of saturated to unsaturated fatty acid chains, but possesses one zwitterionic headgroup (i.e. DOPC/DPPC/Chol 3:3:4 mol%). Fig. 4a shows a series of time lapse images from triggering the dewetting (i.e. a slight impact by the micropipette) until a complete detachment of an oil droplet from the lipid bilayer membrane. In the first panel, the dewetting starts in three separate areas, forming the lipid bilayer membrane and consequently pushing the trapped oil to the opposite side. Ultimately, the excess oil droplet detaches from the lipid bilayer membrane, which contains no oil residuals in between the two leaflets. In addition, Fig. 4b illustrates another dewetting event that was induced by drawing a portion of the oil shell into a micropipette capillary. In the first three panels, the double emulsion was able to withstand the induced tension and remain stable. However, by increasing the tension, the dewetting started from the tip of micropipette capillary and grew across the entire surface, forming an oil droplet located on top of the lipid bilayer membrane. Fig. 4c illustrates a schematic representation of the dewetting event induced by the micropipette aspirator. We propose that this arises from a depletion effect due to the lateral migration of the assembled lipids towards the micropipette tip (Fig. 4d and 4e). As is shown, a brighter fluorescent intensity implies a fully packed assembly of the lipids close to the capillary tip.
Measuring the mechanical properties of lipid bilayer membranes
Biological membranes have a stiffness that is determined by the composition of their lipid bilayer. Therefore, the knowledge of mechanical properties (i.e. bending and area expansion moduli) is necessary to clarify the role of each constituent, and thus to understand the behavior of the membrane. As mentioned above, the assembly of cholesterol in the lipid bilayer membrane (iii) enhanced the stability of the GUVs (Fig. 3a and 3b). In a similar fashion, we observed enhancement in the stability of asymmetric GUVs composed of DOPC/DPPC/Chol in the inner leaflet without any imperfections (i.e. without oil lenses attached to them). The area expansion modulus of GUVs possessing a ternary mixture of DOPC/DPPC/Chol (3:3:4 mol%) in the inner leaflet and LPS in the outer leaflet was measured using micropipette aspiration. By applying the suction pressure in the high-tension regime (τ > 0.5 mN m−1), Evans and Rawicz confirmed the prediction of equilibrium theory that the apparent area strain varied linearly with tension due to the direct expansion of the area per molecule.58 This was observed in our experiments, shown by the red triangles in Fig. 5b. As depicted in the plot in Fig. 5b, by fitting a linear curve onto the data in the high-tension regime, we measured the value for the apparent area expansion modulus (Table 1). GUVs composed of DOPC/DPPC/Chol (3:3:4 mol%) and LPS in the inner and outer leaflets, respectively, showed an apparent area expansion modulus of 119 ± 26 mN m−1 (n = 5). This is lower than the values reported for asymmetric phospholipid GUVs (inner leaflet: DOPC–outer leaflet: DMPC), 224 ± 15 mN m−1.57 We propose this is due to: (i) the excess repulsive electrostatic lateral tension due to the presence of LPS molecules (schematic illustration in Fig. 5b), and (ii) the shorter fatty acid carbon chains for Lipid A compared to DMPC (i.e. 10 to 12 carbons in Lipid A, and 14 in DMPC). The former reason acts in favor of isotropic area expansion and the later provides lesser hydrophobic interactions in between the fatty acids in the lipid bilayer. Consequently, lipid bilayer membranes composed of an asymmetrical assembly of lipids (i.e. a ternary mixture of DOPC/DPPC/Chol (3:3:4 mol%) in the inner leaflet and LPS in the outer leaflet) showed lower resistance to expansion that resulted in smaller values for the apparent area expansion modulus.
Table 1.
Experimental values for bending κ and apparent area expansion Kapp moduli for synthetic lipid bilayer membranes ± one standard deviation. Measurements were obtained using micropipette aspiration at T = 22.5 °C. Values for the phospholipid:phospholipid membranes are from our previous work. n determines the number of membranes that were tested.
In addition, the apparent area strain increases logarithmically with tension in the low-tension regime (τ < 0.5 mN m−1).58 Therefore, the slope of the fitted line on ln(τ) versus αapp in the low-tension regime (i.e. the blue square data points) determines the value of the membrane bending modulus. We measured the bending modulus for the asymmetric GUVs (i.e. a ternary mixture of DOPC/DPPC/Chol (3:3:4 mol%) in the inner leaflet and LPS in the outer leaflet) at T = 22.5°C to be (11.1 ± 3.7) × 10−20 J (n = 3). This is lower than the values reported for asymmetric phospholipid GUVs (inner leaflet: DOPC–outer leaflet: DMPC), (16.8 ± 2.1) × 10−20 J.57 We propose that the two aforementioned reasons for a lower area expansion modulus also result in a lower bending modulus (schematic illustration in Fig. 5b). However, symmetric lipid bilayer membranes (inner leaflet: DOPC/DMPC–outer leaflet: DOPC/DMPC) with a similar value for the bending modulus ((11.2 ± 1.4) × 10−20 J) showed greater resistance against expansion (159 ± 13 mN m−1), compared to asymmetric ones with LPS.57 This suggests that the electrostatic repulsions between the lipid A section of LPS play a significant role in lowering the area expansion modulus.
Note that the apparent area expansion and bending moduli were measured with Mg2+ ions present in the IA and OA solutions. It is noteworthy that a comprehensive understanding of the effect of salt concentration on membrane rigidity is lacking. This knowledge could be used to build membranes with tailored lipid composition (in the presence of an associated salt concentration) to achieve different rigidities for various biological functions. It is believed that Mg2+ ions provide stability by building cross bridges in between the negative phosphate groups in anionic phospholipids and LPS.86,87 The presence of Mg2+ close to the lipid bilayer should suppress the repulsive electrostatic lateral tension and consequently influence the mechanical properties of the membrane (i.e. bending and area expansion moduli).88 Therefore, in the presence of Mg2+, asymmetric phospholipid-LPS GUVs possessing either anionic or zwitterionic phospholipids in the inner leaflet should show an increase in the magnitude of the area expansion modulus and be able to resist strain.
Conclusions
In this work, we reported on the formation of asymmetric model membrane systems (i.e. GUVs) that resembled the outer membrane lipid composition and transbilayer architecture of the Gram-negative bacillus, P. aeruginosa, more accurately. The lipid bilayer architecture and composition of the fabricated model membranes make them suitable candidates to study the bacterial function in antimicrobial studies. Our microfluidic technique generates 50 to 150 μm diameter water-in-oil-in-water ultrathin double emulsions at high-throughput, which are used for building the asymmetric vesicles. We used a microfluidic-based technique to make asymmetric vesicles possessing four different phospholipid compositions in the inner leaflet and LPS in the outer leaflet. The water-oil and oil-water interfaces facilitate the spontaneous self-assembly of phospholipid and LPS molecules to create the inner and outer leaflets of the lipid bilayer, respectively. Membranes that were stored for one day showed a 20% drop in asymmetry across the lipid bilayer compared to a 50% drop for 2 and 3 days of storage. We also investigated the separation of the organic phase mechanism (i.e. dewetting) throughout the oil extraction process and by implementing mechanical aspiration. Changes in the inner-leaflet lipid compositions showed different behavior in dewetting. Model membranes composed of DOPE and DOPG/DPPE (1:1 mol%) showed a lower stability compared to the ones possessing 40 mol% cholesterol. Also, cholesterol had a significant impact on the dewetting transition in the model membranes that were physiologically more relevant to the P. aeruginosa OM. Values for the bending and area expansion moduli of asymmetric model membrane systems were reported. We conclude that the membranes containing LPS comply with the assumption of isotropic area expansion, show lower resistance to lateral expansion, and are more flexible than the ones that are purely composed of phospholipids.
Acknowledgements
This research was supported by the National Science Foundation (Award #1429448) and the National Institute of Allergy and Infectious Diseases (Award #R21AI121848). The authors would like to thank Dr. Noah Malmstadt, Dr. Kaixuan Ren, Lu Wang, Nareh Movsesian, and Ahmed ElBaradei from University of Southern California for their great support and enlightening discussions.
Footnotes
Conflicts of interest
There are no conflicts to declare.
References
- 1.Kubiak J, Brewer J, Hansen S and Bagatolli LA, Biophys J, 2011, 100, 978–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Howse JR, Jones RAL, Battaglia G, Ducker RE, Leggett GJ and Ryan AJ, Nature Materials, 2009, 8, 507–511. [DOI] [PubMed] [Google Scholar]
- 3.Arriaga LR, Rodriguez-Garcia R, Moleiro LH, Prevost S, Lopez-Montero I, Hellweg T and Monroy F, Adv Colloid Interface Sci, 2017, 247, 514–520. [DOI] [PubMed] [Google Scholar]
- 4.Heberle FA and Pabst G, Biophys Rev, 2017, 9, 353–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sohlenkamp C and Geiger O, FEMS Microbiology Reviews, 2016, 40, 133–159. [DOI] [PubMed] [Google Scholar]
- 6.Kim JH, Lee J, Park J and Gho YS, Semin Cell Dev Biol, 2015, 40, 97–104. [DOI] [PubMed] [Google Scholar]
- 7.Schertzer JW and Whiteley M, Journal of Molecular Microbiology and Biotechnology, 2013, 23, 118–130. [DOI] [PubMed] [Google Scholar]
- 8.Florez C, Raab JE, Cooke AC, and Schertzer JW, mBio, 2017, 8, e01034. (doi: 10.1128/mBio.01034-17). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li A, Schertzer JW, and Yong X, J. Biol. Chem, 2019, 294, 1089–1094. (doi: 10.1074/jbc.AC118.006844). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li A, Schertzer JW and Yong X, Physical Chemistry Chemical Physics, 2018, 20, 23635–23648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schertzer JW and Whiteley M, mBio, 2012, 3, e00297–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sperandeo P and Polissi A, Structure, 2016, 24, 847–849. [DOI] [PubMed] [Google Scholar]
- 13.Sperandeo P, Martorana AM and Polissi A, Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids, 2017, 1862, 1451–1460. [DOI] [PubMed] [Google Scholar]
- 14.Choi D-S, Kim D-K, Kim Y-K and Gho YS, Mass Spectrometry Reviews, 2015, 34, 474–490. [DOI] [PubMed] [Google Scholar]
- 15.Lee J, Kim OY and Gho YS, PROTEOMICS - Clinical Applications, 2016, 10, 897–909. [DOI] [PubMed] [Google Scholar]
- 16.Demchenko AP, Biopolymers and Cell, 2012, 28, 24–38. [Google Scholar]
- 17.Toyofuku M, Tashiro Y, Hasegawa Y, Kurosawa M and Nomura N, Advances in Colloid and Interface Science, 2015, 226, 65–77. [DOI] [PubMed] [Google Scholar]
- 18.Kadurugamuwa JL and Beveridge TJ, Journal of Bacteriology, 1995, 177, 3998–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mashburn-Warren LM and Whiteley M, Mol Microbiol, 2006, 61, 839–46. [DOI] [PubMed] [Google Scholar]
- 20.Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, Colas E, Cordeiro-da Silva A, Fais S, Falcon-Perez JM, Ghobrial IM, Giebel B, Gimona M, Graner M, Gursel I, Gursel M, Heegaard NH, Hendrix A, Kierulf P, Kokubun K, Kosanovic M, Kralj-Iglic V, Kramer-Albers EM, Laitinen S, Lasser C, Lener T, Ligeti E, Line A, Lipps G, Llorente A, Lotvall J, Mancek-Keber M, Marcilla A, Mittelbrunn M, Nazarenko I, Nolte-’t Hoen EN, Nyman TA, O’Driscoll L, Olivan M, Oliveira C, Pallinger E, Del Portillo HA, Reventos J, Rigau M, Rohde E, Sammar M, Sanchez-Madrid F, Santarem N, Schallmoser K, Ostenfeld MS, Stoorvogel W, Stukelj R, Van der Grein SG, Vasconcelos MH, Wauben MH and De Wever O, J Extracell Vesicles, 2015, 4, 27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furse S and Scott DJ, Biochemistry, 2016, 55, 4742–4747. [DOI] [PubMed] [Google Scholar]
- 22.MacDonald IA and Kuehn MJ, Journal of Bacteriology, 2013, 195, 2971–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Döring G and Pier GB, Vaccine, 2008, 26, 1011–1024. [DOI] [PubMed] [Google Scholar]
- 24.Van Eldere J, Journal of Antimicrobial Chemotherapy, 2003, 51, 347–352. [DOI] [PubMed] [Google Scholar]
- 25.Lyczak JB, Cannon CL and Pier GB, Microbes and Infection, 2000, 10. [DOI] [PubMed] [Google Scholar]
- 26.Clifton LA, Skoda MWA, Daulton EL, Hughes AV, Le Brun AP, Lakey JH and Holt SA, Journal of The Royal Society Interface, 2013, 10, 20130810–20130810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tashiro Y, Inagaki A, Shimizu M, Ichikawa S, Takaya N, Nakajima-Kambe T, Uchiyama H and Nomura N, Bioscience, Biotechnology, and Biochemistry, 2011, 75, 605–607. [DOI] [PubMed] [Google Scholar]
- 28.Tashiro Y, Uchiyama H and Nomura N, Environmental Microbiology, 2012, 14, 1349–1362. [DOI] [PubMed] [Google Scholar]
- 29.Rocchetta HL, Burrows LL and Lam JS, MICROBIOL. MOL. BIOL. REV, 1999, 63, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cigana C, Curcurù L, Leone MR, Ieranò T, Lorè NI, Bianconi I, Silipo A, Cozzolino F, Lanzetta R, Molinaro A, Bernardini ML and Bragonzi A, PLoS ONE, 2009, 4, e8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahenthiralingam E, Campbell ME, and Speert DP, Infection and Immunity, 1994, 62, 596–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kronborg G, JOURNAL OF CLINICAL MICROBIOLOGY, 1992, 30, 1848–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finlay BB and McFadden G, Cell, 2006, 124, 767–782. [DOI] [PubMed] [Google Scholar]
- 34.Adams PG, Lamoureux L, Swingle KL, Mukundan H and Montaño GA, Biophysical Journal, 2014, 106, 2395–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng H-T, Megha and London E, Journal of Biological Chemistry, 2009, 284, 6079–6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Angelova MI and Dimitrov DS, Faraday discussions of the Chemical Society, 1986, 81, 303–311. [Google Scholar]
- 37.Angelova MI, Soléau S, Méléard P, Faucon F and Bothorel P, in Trends in Colloid and Interface Science VI, eds. Helm C, Lösche M and Möhwald H, Steinkopff, Darmstadt, 1992, vol. 89, pp. 127–131. [Google Scholar]
- 38.Hishida M, Seto H, Yamada NL and Yoshikawa K, Chemical Physics Letters, 2008, 455, 297–302. [Google Scholar]
- 39.Richmond DL, Schmid EM, Martens S, Stachowiak JC, Liska N and Fletcher DA, Proceedings of the National Academy of Sciences, 2011, 108, 9431–9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stachowiak JC, Richmond DL, Li TH, Liu AP, Parekh SH and Fletcher DA, Proceedings of the National Academy of Sciences, 2008, 105, 4697–4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horger KS, Estes DJ, Capone R and Mayer M, Journal of the American Chemical Society, 2009, 131, 1810–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peruzzi J, Gutierrez MG, Mansfield K and Malmstadt N, Langmuir, 2016, 32, 12702–12709. [DOI] [PubMed] [Google Scholar]
- 43.Movsesian N, Tittensor M, Dianat G, Gupta M and Malmstadt N, Langmuir, 2018, 34, 9025–9035. [DOI] [PubMed] [Google Scholar]
- 44.Jahn A, Vreeland WN, Gaitan M and Locascio LE, Journal of the American Chemical Society, 2004, 126, 2674–2675. [DOI] [PubMed] [Google Scholar]
- 45.Funakoshi K, Suzuki H and Takeuchi S, Journal of the American Chemical Society, 2007, 129, 12608–12609. [DOI] [PubMed] [Google Scholar]
- 46.Teh S-Y, Khnouf R, Fan H and Lee AP, Biomicrofluidics, 2011, 5, 044113–044113–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu L, Schertzer JW and Chiarot PR, Lab on a Chip, 2015, 15, 3591–3599. [DOI] [PubMed] [Google Scholar]
- 48.Zhou S, Fan J, Datta SS, Guo M, Guo X and Weitz DA, Advanced Functional Materials, 2013, 23, 5925–5929. [Google Scholar]
- 49.Shum HC, Kim J-W and Weitz DA, Journal of the American Chemical Society, 2008, 130, 9543–9549. [DOI] [PubMed] [Google Scholar]
- 50.Shum HC, Santanach-Carreras E, Kim J-W, Ehrlicher A, Bibette J and Weitz DA, Journal of the American Chemical Society, 2011, 133, 4420–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhargava KC, Ermagan R, Thompson B, Friedman A and Malmstadt N, Micromachines, 2017, 8, 137. [Google Scholar]
- 52.Deng NN, Yelleswarapu M, Zheng L and Huck WT, J Am Chem Soc, 2017, 139, 587–590. [DOI] [PubMed] [Google Scholar]
- 53.McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu H, Schueller OJA and Whitesides GM, ELECTROPHORESIS, 2000, 21, 27–40. [DOI] [PubMed] [Google Scholar]
- 54.Sia SK and Whitesides GM, ELECTROPHORESIS, 2003, 24, 3563–3576. [DOI] [PubMed] [Google Scholar]
- 55.Qin D, Xia Y and Whitesides GM, Nature Protocols, 2010, 5, 491–502. [DOI] [PubMed] [Google Scholar]
- 56.Trantidou T, Elani Y, Parsons E and Ces O, Microsystems & Nanoengineering, 2017, 3, 16091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu L, Doak WJ, Schertzer JW and Chiarot PR, Soft Matter, 2016, 12, 7521–7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Evans E and Rawicz W, Physical Review Letters, 1990, 64, 2094–2097. [DOI] [PubMed] [Google Scholar]
- 59.Lu L, Irwin RM, Coloma MA, Schertzer JW and Chiarot PR, Microfluidics and Nanofluidics, 2015, 18, 1233–1246. [Google Scholar]
- 60.Alam JM and Yamazaki M, Chemistry and Physics of Lipids, 2011, 164, 166–174. [DOI] [PubMed] [Google Scholar]
- 61.Santos NC, Silva AC, Castanho MARB, Martins‐Silva J and Saldanha C, ChemBioChem, 2003, 4, 96–100. [DOI] [PubMed] [Google Scholar]
- 62.Bergstrand A, Svanberg C, Langton M and Nydén M, Colloids and Surfaces B: Biointerfaces, 2006, 53, 9–14. [DOI] [PubMed] [Google Scholar]
- 63.Huijbregts RPH, de Kroon AIPM and de Kruijff B, Biochimica et Biophysica Acta (BBA) - Biomembranes, 1996, 1280, 41–50. [DOI] [PubMed] [Google Scholar]
- 64.Smith BD and Lambert TN, Chemical Communications, 2003, 2261. [DOI] [PubMed] [Google Scholar]
- 65.Kol MA, de Kroon AIPM, Rijkers DTS, Killian JA, and de Kruijff B, Biochemistry, 2001, 40, 10500–10506. (doi: 10.1021/bi010627+). [DOI] [PubMed] [Google Scholar]
- 66.Halevy R, Rozek A, Kolusheva S, Hancock REW and Jelinek R, Peptides, 2003, 24, 1753–1761. [DOI] [PubMed] [Google Scholar]
- 67.McIntyre JC and Sleight RG, Biochemistry, 1991, 30, 11819–11827. [DOI] [PubMed] [Google Scholar]
- 68.Angeletti C and Nichols JW, Biochemistry, 1998, 37, 15114–15119. [DOI] [PubMed] [Google Scholar]
- 69.Son M and London E, J Lipid Res, 2013, 54, 3385–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Devaux PF, Biochemistry, 1991, 30, 1163–1173. [DOI] [PubMed] [Google Scholar]
- 71.Devaux PF, Fellmann P and Hervé P, Chemistry and Physics of Lipids, 2002, 116, 115–134. [DOI] [PubMed] [Google Scholar]
- 72.Liu J and Conboy JC, Biophys J, 2005, 89, 2522–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arriaga LR, Huang Y, Kim S-H, Aragones JL, Ziblat R, Koehler SA, and Weitz DA, Lab Chip, 2019, 19, 749–756. (doi: 10.1039/c8lc00882e). [DOI] [PubMed] [Google Scholar]
- 74.Sáenz JP, Grosser D, Bradley AS, Lagny TJ, Lavrynenko O, Broda M and Simons K, Proceedings of the National Academy of Sciences, 2015, 112, 11971–11976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Meer G, Voelker DR and Feigenson GW, Nature Reviews Molecular Cell Biology, 2008, 9, 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bernardino J de la Serna, Perez-Gil J, Simonsen AC and Bagatolli LA, Journal of Biological Chemistry, 2004, 279, 40715–40722. [DOI] [PubMed] [Google Scholar]
- 77.Saenz JP, Sezgin E, Schwille P and Simons K, Proceedings of the National Academy of Sciences, 2012, 109, 14236–14240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bramkamp M and Lopez D, Microbiology and Molecular Biology Reviews, 2015, 79, 81–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Israelachvili JN, Intermolecular and Surface Forces, Academic Press, 2011. [Google Scholar]
- 80.Pier G, International Journal of Medical Microbiology, 2007, 297, 277–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hayward RC, Utada AS, Dan N and Weitz DA, Langmuir, 2006, 22, 4457–4461. [DOI] [PubMed] [Google Scholar]
- 82.Kim S-H, Kim JW, Kim D-H, Han S-H and Weitz DA, Small, 2013, 9, 124–131. [DOI] [PubMed] [Google Scholar]
- 83.Deshpande S, Caspi Y, Meijering AEC and Dekker C, Nature Communications, 2016, 7, 10447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee MH, Hribar KC, Brugarolas T, Kamat NP, Burdick JA and Lee D, Advanced Functional Materials, 2012, 22, 131–138. [Google Scholar]
- 85.Shum HC, Lee D, Yoon I, Kodger T and Weitz DA, Langmuir, 2008, 24, 7651–7653. [DOI] [PubMed] [Google Scholar]
- 86.Kučerka N, Papp-Szabo E, Nieh M-P, Harroun TA, Schooling SR, Pencer J, Nicholson EA, Beveridge TJ and Katsaras J, The Journal of Physical Chemistry B, 2008, 112, 8057–8062. [DOI] [PubMed] [Google Scholar]
- 87.Akashi K, Miyata H, Itoh H and Kinosita K, Biophysical Journal, 1998, 74, 2973–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Claessens MM, van Oort BF, Leermakers FA, Hoekstra FA and Cohen Stuart MA, Biophys J, 2004, 87, 3882–93. [DOI] [PMC free article] [PubMed] [Google Scholar]