Abstract
Background:
The N-myc downstream-regulated gene (NDRG) family, NDRG1-4, has been involved in a wide spectrum of biological functions in multiple cancers. However, their prognostic values remain sparse in gastric cancer (GC). Therefore, it is crucial to systematically investigate the prognostic values of the NDRG family in GC.
Methods:
The prognostic values of the NDRG family were evaluated by Kaplan–Meier Plotter and SurvExpress. The mRNA of the NDRG family was investigated in The Cancer Genome Atlas (TCGA). Transcription factors (TFs) and miRNAs associated with the NDRG family were predicted by NetworkAnalysis. The prognostic values of DNA methylation levels were analyzed by MethSurv. The correlation between immune cells and the NDRG family was evaluated by the Tumor Immune Estimation Resource (TIMER) database.
Results:
High levels of mRNA expression of NDRG2 and NDRG3 were associated with a favorable prognosis in all GCs. In HER2− GC, NDRG1 was significantly associated with a poor prognosis of GC [hazard ratio (HR) = 1.65, 95% confidence interval (CI) = 1.16–2.33, p = 0.0046]. In HER2+ GC, NDRG4 showed a poor prognosis (HR = 1.4, 95% CI: 1.06–1.85, p = 0.017). NDRG4 was an independent prognostic factor in recurrence-free survival by TCGA cohort. The low-risk NDRG-signature group displayed a significantly favorable survival outcome than the high-risk group (HR = 1.76, 95% CI: 1.2–2.59, p = 0.00385). The phosphorylated protein NDRG1 (NDRG1_pT346) displayed a favorable overall survival and was significantly associated with HER2 and phosphorylated HER2. Epidermis development was the top biological process (BP) for coexpressed genes associated with NDRG1 and NDRG4, while mitotic nuclear division and mitotic cell processes were the top BPs for NDRG2 and NDRG3, respectively. Overall, 6 CpGs of NDRG1, 4 CpGs of NDRG2, 3 CpGs of NDRG3 and 24 CpGs of NDRG4 were associated with significant prognosis. CD4+ T-cells showed the highest correlation with NDRG4 (correlation = 0.341, p = 2.14e−11). Furthermore, BCL6 in follicular helper T-cells (Tfh) cells showed the highest association with NDRG4 (correlation = 0.438, p = 00e+00).
Conclusions:
This study analyzed the multilevel prognostic values and biological roles of the NDRG family in GC.
Keywords: gastric cancer, HER2, methylation, NDRG family, prognosis, TCGA
Introduction
Gastric cancer (GC) is one of the leading death-causing, malignant diseases in eastern Asia.1–3 Although improved dietary habits, solid diagnostic screening systems, multiprincipled therapeutic regimes and updated surgical techniques have reduced both the incidence and mortality rates of GC,4–6 the prognosis of GC remains unsatisfactory.3 Thus, identification of reliable biomarkers for the prognostic prediction of GC could facilitate individualized clinical management.
The N-myc downstream-regulated gene (NDRG) family consists of four members, NDRG1, NDRG2, NDRG3 and NDRG4, located on chromosomes 8q24.3, 14q11.2, 20q11.21-23 and 16q21-q22.1 respectively.7–8 Although the four members share 57–65% of amino acid sequences with an alpha/beta hydrolase-fold and an NDR region, they lack catalytic motifs and therefore do not have a hydrolase function.8 NDRG1–4 have been found to be widely expressed in human organs and multiple biological functions have been recently discovered.9 The molecular functions of the NDRG family cover a wide spectrum of biological processes, including cell development and differentiation, stress responses and proliferation, tumor progression and metastasis.7,10–19 NDRG1 has been implicated in embryonic placentation, organ development and cellular skeleton modification,7,10,11 and is induced by hypoxia and DNA damage.12 Global gene expression analysis of breast epithelial cells indicates that NDRG1 is closely associated with cellular vesicle transport and regulation of membrane proteins, such as low-density lipoprotein and E-cadherin endosomal trafficking.13–15 The prognostic values of NDRG1 in solid tumors have been intensively investigated. In esophageal cancer, low NDRG1 mRNA expression indicates a worse prognosis.20 It is also negatively correlated with tumor progression and metastasis in colorectal, breast and prostate cancers,12,21 while associated with an unfavorable prognosis for hepatocellular carcinoma.22
NDRG2, regulated by maturation-associated stimuli, is strongly expressed in dendritic cells16 and is able to maintain activated leukocyte cell adhesion during the entire differentiation progress of dendritic cells.17 NDRG2 expression is found significantly reduced in pancreatic, breast and hepatocellular carcinomas compared with normal counterparts.23–25 Specifically, reduced expression of NDRG2 is correlated with aggressive tumor behavior, higher recurrence and distant metastasis ratio in hepatocellular carcinoma.24 Of note, NDRG2 expression has been found to be negatively associated with a worse prognosis in GC and prostate cancer.26,27
NDRG3 promotes angiogenesis and cell growth and is also involved in the lactate-dependent hypoxia signaling pathway.18 High levels of NDRG3 are associated with shorter overall survival (OS) and relapse-free survival (RFS) in advanced prostate cancer.27
NDRG4 is exclusively expressed in the central nervous system and heart in the embryonic stage, highlighting its essential role of regulating growth and proliferation.19 NDRG4 is reduced in both mRNA and protein expression in colorectal cancer tissues and functionally suppressed in tumor invasion and cell proliferation,28 and is associated with a favorable survival.29
Collectively, the prognostic values of the NDRG family have been noticed in various types of cancers. However, the whole picture of the prognostic value of the entire NDRG family remains poorly investigated in GC. Hereby, based on updated public resources and integrative bioinformatics analysis, the prognostic value of the NDRG family was comprehensively assessed.
Methods
Survival analysis in Kaplan–Meier plotter
The prognostic values of mRNA expression of each NDRG family member to OS were analyzed based on Kaplan–Meier (KM) plotter, a website database based on resources from the Gene Expression Omnibus, including GSE14210, GSE15459, GSE22377, GSE29272, GSE51105 and GSE62254. In fact, GSE62254 was excluded from the total sample survival analysis given its markedly different clinical and genomic data, as suggested by KM plotter. Survival data in each subgroup, including pathological stage, Lauren classification, histological differentiation and human epidermal growth factor receptor 2 (HER2) status, were collected respectively. All four members of the NDRG family were analyzed with various parameters in KM plotter (http://kmplot.com/analysis/index.php?p=service&cancer=gastric).30 The best cutoff values were determined by algorithms embedded in KM plotter.30 The final prognostic KM plots were presented with a hazard ratio (HR), 95% confidence interval (CI) and log-rank p value. A p value <0.05 was considered statistically significant.
Prognosis analysis of NDRG signature via SurvExpress platform
The prognostic value of the NDRG family signature was analyzed via SurvExpress (http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp), which is a platform for integrating public available resources for survival assessment.31 The Stomach Adenocarcinoma (STAD) data of TCGA were selected as the input resource (n = 352). High/low-risk groups were determined by the algorithm of the prognostic risk score. Risk score = expmRNA of NDRG1 × betamRNA of NDRG1 + expmRNA of NDRG2 × betamRNA of NDRG2 + expmRNA of NDRG3 × betamRNA of NDRG3 + expmRNA of NDRG4 × betamRNA of NDRG4, where ‘exp’ indicates the standardized mRNA expression of each selected gene, and ‘beta’ was obtained from the Cox multivariate regression analysis.31 Moreover, the receiver operating characteristics (ROC) curve was used to evaluate the survival curves of the NDRG signature over different event times using the R package, survivalROC.31
Analysis of the mRNA expression of the NDRG family in TCGA
The mRNA expression of the NDRG family was explored in the pathological stage-specific pattern (one-way analysis of variance, violin plots) and among the tumor and normal tissues in the STAD data of TCGA in the Gene Expression Profiling Interactive Analysis platform (GEPIA; http://gepia.cancer-pku.cn/index.html). This web-based tool was established for customized investigation of genomic functionalities based on the resources provided by TCGA and the genotype-tissue expression (GTEx) projects.32 Furthermore, the mRNA expression of the NDRG family, along with other clinic-pathological data, was downloaded from the Xena system (University of California, Santa Cruz, CA, USA), for statistical analysis.33
Protein expression of NDRG1–4 in the Human Protein Atlas
Protein expression of NDRG1–4 in both GC and normal tissues was retrieved from the Human Protein Atlas (www.proteinatlas.org).34
Analysis of the reverse-phase protein array data of The Cancer Proteome Atlas
The Cancer Proteome Atlas (TCPA) dataset (http://tcpaportal.org/tcpa/index.html) mainly provides a comprehensive resource for the assessment, visualization and analysis of cancer proteomic data based on TCGA tumor tissue sample sets. Reverse-phase protein array (RPPA) data are used as a high-throughput antibody-dependent experimental procedure with increased quality and robust quantification.35 The phosphorylated NDRG1 (NDRG1_pT364) and epidermal growth factor receptor (EGFR), HER2, as well as corresponding phosphorylated data (EGFR_pY1068, EGFR_pY1173, HER2_pY1248) were extracted for correlation.35
Prognostic value of NDRG1_pT346 via TRGAted platform
The TRGAted platform (https://nborcherding.shinyapps.io/TRGAted/), is a web tool for survival analysis based on RPPA data retrieved from TCGA.36 Given only NDRG1_pT346 was available in the RPPA of STAD, we only accessed the prognostic value of NDRG1_pT346, including OS and disease-free survival (DFS), between high and low expression groups. The optimal cutoff was determined based on the surv-cutpoint function in the survminer package via TRGAted.36 HR was determined by the Cox proportional hazard regression model.36
Gene ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analysis of the coexpressed genes of NDRG1–4
The genomic alterations of NDRG1–4 were analyzed by cBioPortal, an integrative analytic platform of TCGA.37,38 The coexpressed genes of NDRG1–4 with a Pearson correlation (⩾0.3 or ⩽−0.3) were subject to gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis in the Database for Annotation, Visualization and Integrated Discovery (DAVID).39–41 The cutoff value was a false discover rate (FDR) <0.05 for significant GO and KEGG data.
Prediction of transcription factors and miRNAs for NDRG1–4
Potential transcription factors (TFs) and miRNAs of NDRG1–4 were predicted by NetworkAnalysis (http://www.networkanalyst.ca).42,43 The prediction of TFs for NDRG1–4 was based on the ENCODE database with ChIP-seq data. Only the data with a peak intensity signal value <500 and a potential score value <1 was screened for further analysis. The miRNA-gene interaction data were retrieved from TarBase and miRTarBase via the NetworkAnalysis platform.
DNA methylation data of NDRG1–4 in MethSurv
The DNA methylation of NDRG1–4 in TCGA was analyzed by MethSurv (https://biit.cs.ut.ee/methsurv/).44 The prognostic values and expression levels of CpG methylation in NDRG1–4 were explored.
Tumor-immune infiltrating cells associated with the NDRG family via the Tumor Immune Estimation Resource database
Correlations between all tumor-immune infiltrating cells (TIICs) and the NDRG family were analyzed via the Tumor Immune Estimation Resource (TIMER) platform (https://cistrome.shinyapps.io/timer/), a web tool for gene-specific correlational analysis with TIICs. TIICs included B-cells, CD4+T-cells, CD8+T-cells, dendritic cells, macrophages and neutrophils.45 Tumor purity was used for the correction of Spearman-based correlation analysis.45 Moreover, TIICs with the highest correlation to the NDRG family were selected for further subtype-based biomarker analysis.46,47 Corresponding markers included TBX21 (T-bet), STAT4, STAT1, IFNG (interferon gamma) and TNF (tumor necrosis factor) for T helper (Th)1 cells; BCL6, interleukin (IL)21 for Tfh; GATA3, STAT6, STAT5A and IL13 for Th2; FOXP3, CCR8, STAT5B and TGFB1 (transforming growth factor beta) for T regulatory (Treg) cells; PDCD1 (programmed cell death 1), CTLA4, LAG3, HAVCR2 (TIM-3), GZMB for T-cell exhaustion; STAT3 and IL17A for Th17 cells.46,47
Statistically analysis
SPSS 17.0 (Chicago, IL, USA) and Graphpad Prism 5.0 software (GraphPad Software, San Diego, CA, USA) were used for statistical analysis and illustration. A student’s t test and Pearson correlation test were used for comparison between groups and correlation analysis. Cox regression was used for univariate and multivariate survival analysis. A p value <0.05 was considered significant in all circumstances.
Results
Prognostic values of NDRG members in the whole group of patients with GC
The prognostic values of NDRG mRNA expression in the whole group of patients with GC from KM plotter were collected [Figure 1(a–e)]. NDRG2 and NDRG3 were significantly associated with a better OS prognosis [Figure 1(a, c and d), HR = 0.64, 95% CI: 0.52–0.80, p < 0.0001 and HR = 0.78, 95% CI: 0.63–0.96, p = 0.021] while NDRG1 and NDRG4 showed a modest association with a worse prognosis for the OS [Figure 1(a, b and e), HR = 1.21, 95% CI: 0.98–1.49, p = 0.072 and HR = 1.2, 95% CI: 099–1.45, p = 0.068].
Figure 1.
The prognostic value of the NDRG family mRNA expression in the KM plotter database. (a). Forest plot of the prognostic HRs of NDRG family members in total GC patients. (b–e). Survival curves of NDRG1 (Affymetrix IDs: 200632_s_at), NDRG2 (Affymetrix IDs: 206453_s_at), NDRG3 (Affymetrix IDs: 217286_s_at), NDRG4 (Affymetrix IDs: 209159_s_at) for all GC patients (n = 593).
Red: high expression level; black: low expression level.
GC, gastric cancer; HR: hazard ratio; KM, Kaplan–Meier.
Prognostic values of NDRG members in HER2+/− GC patients
Next, the prognostic values of NDRG family members in HER2+/− GC were assessed [Figure 2(a–i)]. Of note, high mRNA expression of NDRG1 was correlated with a favorable prognosis of HER2+ GC patients, but was not statistically significant [Figure 2(a, i), HR = 0.78, 95% CI = 0.58–1.06, p = 0.11]. Of note, NDRG1 displayed a significantly unfavorable prognosis in HER2− GC [Figure 2(e, i), HR = 1.65, 95% CI = 1.16–2.33, p = 0.0046]. Similarly, NDRG4 showed an inverse prognosis between HER2+/− groups. However, the prognostic value of NDRG4 in HER2− was not significant. In addition, NDRG2 showed a favorable outcome in both HER2+/− groups [Figure 2(b, f)]. NDRG3 only showed a favorable outcome in the HER2+ group [Figure 2(c)].
Figure 2.
Survival curves of NDRG family members in HER2+/− subgroups. (a–h). Survival curves of NDRG1 (Affymetrix IDs: 200632_s_at), NDRG2 (Affymetrix IDs: 206453_s_at), NDRG3 (Affymetrix IDs: 217286_s_at), NDRG4 (Affymetrix IDs: 209159_s_at) are plotted for patients with HER2+/−; (i) Forest plot of the prognostic HRs of NDRG family members in HER2+/− GC.
Red: high expression level; black: low expression level.
GC, gastric cancer; HER2, human epidermal growth factor receptor 2; HR: hazard ratio.
Prognostic values of the NDRG family with different clinicopathological features
In the Lauren classification, a high mRNA expression of NDRG2 was correlated with a worse prognosis in mixed types [Figure 3(a), HR = 5.07, 95% CI: 1.1–23.28, p = 0.021]. NDRG3 [Figure 3(B), HR = 0.58, 95% CI: 0.34–0.99, p = 0.045] was correlated with a favorable prognosis in diffuse types. High mRNA expression of NDRG4 was correlated with a worse prognosis in intestinal types [Figure 3(c), HR = 2.02, 95% CI: 1.33–3.06, p = 0.00079]. In histological differentiation, high mRNA expression of NDRG1 was correlated with a worse prognosis in poor differentiation types [Figure 3(d), HR = 1.84, 95% CI: 1.08–3.15, p = 0.023], and high mRNA expressions of NDRG2 and NDRG4 were correlated with worse prognosis in well differentiated types [Figure 3(e), HR = 3.32, 95% CI: 1.36–8.15, p = 0.0056; Figure 3(f), HR = 11.61, 95% CI: 1.55–87.17, p = 0.0027]. The rest of the NDRG members showed no significant prognostic correlation in both Lauren and histological subtypes.
Figure 3.
Survival curves of the NDRG family with different clinicopathological features. (a) Survival curve of NDRG2 (Affymetrix IDs: 206453_s_at) with mixed type in Lauren classification; (b) survival curve of NDRG3 (Affymetrix IDs: 217286_s_at) with diffuse type in the Lauren classification; (c) survival curve of NDRG4 (Affymetrix IDs: 209159_s_at) with intestinal type in Lauren classification; (d) survival curve of NDRG1 (Affymetrix IDs: 200632_s_at) with poor histological differentiation; (e) survival curve of NDRG2 with well histological differentiation; (f) survival curve of NDRG4 with well histological differentiation; (g) forest plots of the prognostic HR of NDRG1–4 in GC with distant metastasis status; (h) forest plots of the prognostic HR of NDRG1-4 with lymph node status; (i) forest plots of the prognostic HR of NDRG1–4 with pathological staging. Red: high expression level; black: low expression level.
GC, gastric cancer; HR, hazard ratio.
We next evaluated the prognostic values of NDRG family members on distant metastasis status, lymph node status and pathological stages. High mRNA expression of NDRG2 was correlated with better prognosis in the distant metastasis negative group [Figure 3(g), HR = 0.51, 95% CI: 0.32–0.81, p = 0.0041]. Furthermore, high mRNA expressions of NDRG1 and NDRG2 were found to be correlated with better prognosis in lymph node-negative and positive subgroups respectively [Figure 3(h), HR = 0.25, 95% CI: 0.08–0.73, p = 0.0069; HR = 0.65, 95% CI: 0.42–1.00, p = 0.05]. In pathological stages, high mRNA expression of NDRG4 was correlated with a worse prognosis in stage II and III while NDRG3 was correlated with a better prognosis [Figure 3(i), HR = 0.48, 95% CI: 0.25–0.89, p = 0.018].
NDRG4 was validated as an independent prognostic factor
The prognostic values of the NDRG family had been studied in KM plotter. Furthermore, they were further validated in the TCGA database (STAD). The Cox regression was analyzed for both univariate and multivariate process, including sex, age, TNM stage and mRNA expression of the NDRG family (Table 1 and 2). The results indicated that only NDRG4 was determined as an independent prognostic factor for GC in recurrence-free survival results (HR = 1.247, 95% CI: 1.057–1.470, p = 0.009; Table 2).
Table 1.
The univariate and multivariate analysis of overall survival of NDRG family and clinical-pathological data from TCGA.
| Characteristics | Univariate Cox |
Multivariate Cox |
||||
|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | p value | Hazard ratio | 95% CI | p value | |
| Sex | 1.315 | 0.919–1.880 | 0.134 | - | - | - |
| Age | 1.615 | 1.105–2.360 | 0.013 | 1.967 | 1.333–2.903 | 0.001 |
| T | 1.769 | 1.154–2.713 | 0.009 | 1.364 | 0.839–2.218 | 0.211 |
| N | 2.076 | 1.365–3.157 | 0.001 | 1.53 | 0.88–2.660 | 0.132 |
| Metastasis | 2.194 | 1.261–3.817 | 0.005 | 1.851 | 1.033–3.315 | 0.038 |
| Stage | 2.025 | 1.420–2.889 | <0.0001 | 1.388 | 0.830–2.322 | 0.211 |
| NDRG1 | 1.015 | 0.853–1.206 | 0.868 | – | – | – |
| NDRG2 | 1.09 | 0.932–1.276 | 0.281 | – | – | – |
| NDRG3 | 0.895 | 0.645–1.243 | 0.51 | – | – | – |
| NDRG4 | 1.12 | 0.998–1.258 | 0.055 | – | – | – |
CI, confidence interval; STAD, stomach adenocarcinoma; TCGA, The Cancer Genome Atlas.
Table 2.
The univariate and multivariate analysis of recurrence-free survival of the NDRG family and clinical-pathological data from TCGA.
| Characteristics | Univariate Cox |
Multivariate Cox |
||||
|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | p value | Hazard ratio | 95% CI | p value | |
| Sex | 2.182 | 1.224–3.889 | 0.008 | 2.199 | 1.234–3.919 | 0.008 |
| Age | 0.869 | 0.527–1.433 | 0.582 | – | – | – |
| T | 0.87 | 0.512–1.480 | 0.608 | – | – | – |
| N | 1.246 | 0.730–2.129 | 0.42 | – | – | – |
| Metastasis | 1.259 | 0.457–3.471 | 0.656 | – | – | – |
| Stage | 1.042 | 0.640–1.696 | 0.87 | – | – | – |
| NDRG1 | 1.245 | 0.956–1.621 | 0.104 | – | – | – |
| NDRG2 | 1.067 | 0.851–1.337 | 0.575 | – | – | – |
| NDRG3 | 0.824 | 0.499–1.361 | 0.449 | – | – | – |
| NDRG4 | 1.241 | 1.053–1.462 | 0.01 | 1.247 | 1.057–1.470 | 0.009 |
CI, confidence interval; STAD, stomach adenocarcinoma; TCGA, The Cancer Genome Atlas.
Prognostic value of NDRG family signature
For each individual in the STAD data of TCGA, a prognostic risk score was computed based on the risk score equation. Risk score = expmRNA of NDRG1 × −0.102+ expmRNA of NDRG2 × 0.057+ expmRNA of NDRG3 × 0.046+ expmRNA of NDRG4 × 0.153. All cases were assigned to the high/low-risk groups based on the score value with an optimal cutoff. In fact, distinct expression patterns of NDRG members were noticed between low (n = 132) and high-risk (n = 220) groups, particularly NDRG1 and NDRG4 [Figure 4(a, b)]. The low-risk group displayed a significantly favorable survival outcome than the high-risk group [Figure 4(c), HR = 1.76, 95% CI: 1.2–2.59, p value = 0.00385]. Of note, the ROC value increased to 0.679 as follow-up periods increased [Figure 4(d)].
Figure 4.
The prognostic values of the NDRG family signature. (a) Heat map for the clustered expression of the NDRG family between low (green, n = 132) and high (red, n = 220) risk groups; (b) Comparison of expression between low and high-risk groups for each NDRG member; (c) Survival curves of low and high-risk groups of the NDRG signature; (d) the ROC of survival curves over different times.
AUC, area under curve; CI, confidence interval; KM, Kaplan–Meier; Prog.Idx, prognostic index; ROC receiver operating characteristics.
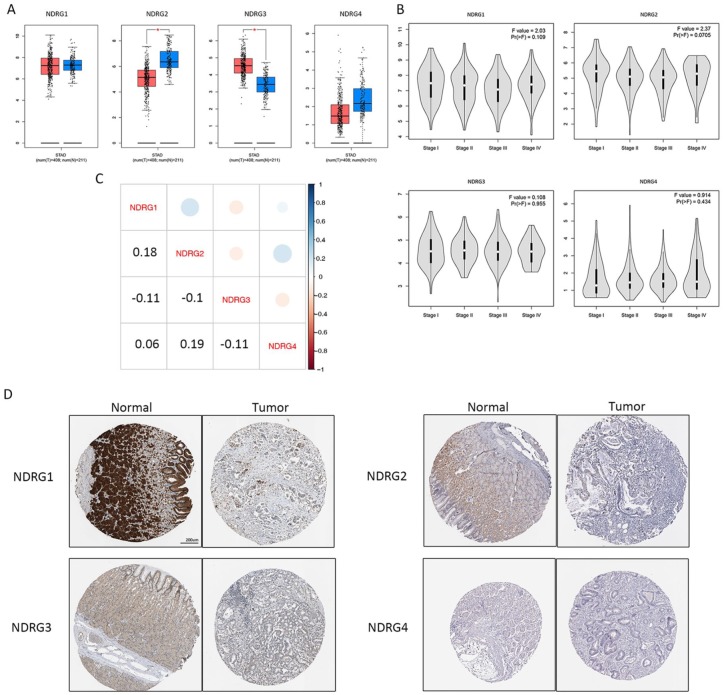
The mRNA and protein expression of NDRG family
Next, we explored the mRNA and protein expression of the NDRG family between tumor and normal tissues. The mRNA expression of NDRG2 was significantly reduced in tumor while the level of NDRG3 was significantly elevated in tumor [Figure 5(a)]. Noteworthy, the entire NDRG family did not show diverse expression in stage-specific manner [Figure 5(b)]. The mRNA expression correlation among each NDRG member was comparably low, excluding potential direct correlational analysis [Figure 5(c)]. Moreover, the protein expression of NDRG members was also displayed [Figure 5(d)].
Figure 5.
The mRNA and protein expression of NDRG family members. (a) The mRNA expression of NDRG family in tumor versus normal in STAD; red: tumor; blue: normal; (b) the mRNA expression of the NDRG family in different pathological stages; (c) the mRNA expression correlation among each NDRG member; (d) the protein expression of NDRG members in gastric cancer from the Human Protein Atlas.
STAD, stomach adenocarcinoma.
The mRNA and protein correlation between HER2/EGFR and NDRG1
Given the fact from Figure 2 that NDRG1, and NDRG4 may feature inverse prognostic values in HER2+/− groups, we further explored the mRNA expression correlation between NDRG1, NDGR4 and HER2. Moreover, given the close functional relationship between HER2 and EGFR, EGFR was also included for correlational analysis. Interestingly, no significant result was identified [Figure 6(a–d)]. Next, the protein expression correlation was investigated in the RPPA data of TCGA. Of note, only NDRG1_pT346 was available. In fact, NDRG1_pT346 was significantly associated with EGFR (r = −0.117, p = 0.02), EGFR_pY1068 (r = 0.218, p < 0.001), EGFR_pY1173 (r = −0.228, p < 0.001), HER2 (r = 0.114, p = 0.024) and HER2_pY1248 [r = 0.135, p = 0.008; Figure 6(e–i)]. Moreover, the prognostic value of NDRG1_pT346 was analyzed. In fact, only high expression of NDRG1_pT346 showed a favorable OS (p = 0.0014).
Figure 6.
The mRNA and protein correlation between HER2/EGFR and NDRG1 and the prognostic value assessment of NDRG1_pT346. (a–d) The mRNA correlation between NDRG1/NDRG4 and HER2/EGFR in the STAD data of TCGA; (e–i) Correlations between NDRG1_pT346 and HER2, HER2_pY1248, EGFR, EGFR_pY1068 and EGFR_pY1173 in STAD of TCGA; (j) OS of prognostic value for NDRG1_pT346 in STAD of TCGA; (k) DFS of prognostic value for NDRG1_pT346 in STAD of TCGA.
Red: high expression; blue: low expression.
DFS, disease-free survival; EGFR, epidermal growth factor receptor; HR: hazard ratio; OS, overall survival; STAD, stomach adenocarcinoma; TCGA, The Cancer Genome Atlas.
GO enrichment analysis and genomic alterations of NDRG1-4
All the coexpressed genes of NDRG1–4 (Pearson correlation ⩾0.3 or ⩽−0.3) were annotated by GO and the KEGG pathway [Figure 7(a–d)]. In fact, epidermis development, extracellular exosome was the top significant biological processes (BP) and cellular components (CC) terms in NDRG1 with no significant terms in molecular functions (MF) and KEGG [Figure 7(e)]. Mitotic nuclear division was the top BP of NDRG2 with no significant results in CC, MF and KEGG [Figure 7(f)]. Mitotic cell process, nucleoplasm and adenyl nucleotide binding were the top significant BP, CC and MF terms in NDRG3 with no significant term in KEGG [Figure 7(g)]. Epidermis development, cornified envelope and structural molecule activity were the top significant terms in BP, CC and MF in NDRG4 with no significant term in KEGG [Figure 7(h)]. The genomic alterations of NDRG1–4 included missense mutation, truncating mutation, amplification, deep deletion and mRNA upregulation [Figure 7(i)]. Moreover, given the relative weak mRNA correlation among each NDRG member, this study further explored potential TFs and miRNA that predicted to be connected with NDRG members. However, no miRNA or TF was predicted to synchronously correlate with all NDRG members or at least three of them [Figure 7(j, k)].
Figure 7.
GO enrichment, genomic alterations and miRNA/TFs prediction of NDRG1–4. (a–d) Coexpressed genes associated with NDRG1–4 (Pearson correlation ⩾0.3 or ⩽−0.3); the chromosomal positions of all genes coexpressed with NDRG1–4 were displayed using various colorful lines; (e–h) GO enrichment for coexpressed genes associated with NDRG1–4; (e) GO enrichment for coexpressed genes with NDRG1; red: BP terms; green: CC terms; (f) GO enrichment for coexpressed genes with NDRG2; red: BP terms; (g) GO enrichment for coexpressed genes with NDRG3; red: BP terms; green: CC terms; blue: MF; (h) GO enrichment for coexpressed genes with NDRG4; BP terms; green: CC terms; blue: MF terms; (i) the genomic alterations of NDRG1–4 in STAD of TCGA; green in grey: missense mutation (unknown significance); darker grey in grey: truncating mutation (unknown significance); red: amplification; blue: deep deletion; pink circle in grey: mRNA upregulation; grey only: no alteration; (j) The predicted networks of TFs and NDRG1–4; red: NDRG family; light blue: predicted TFs; line: predicted interactions; (k) the predicted networks of miRNAs and NDRG1–4, red, NDRG family; violet, predicted miRNA; line, predicted interactions.
BP, biological process; CC, cellular components; GO, gene ontology; MF, molecular functions; STAD, stomach adenocarcinoma; TCGA, The Cancer Genome Atlas; TF, transcription factor.
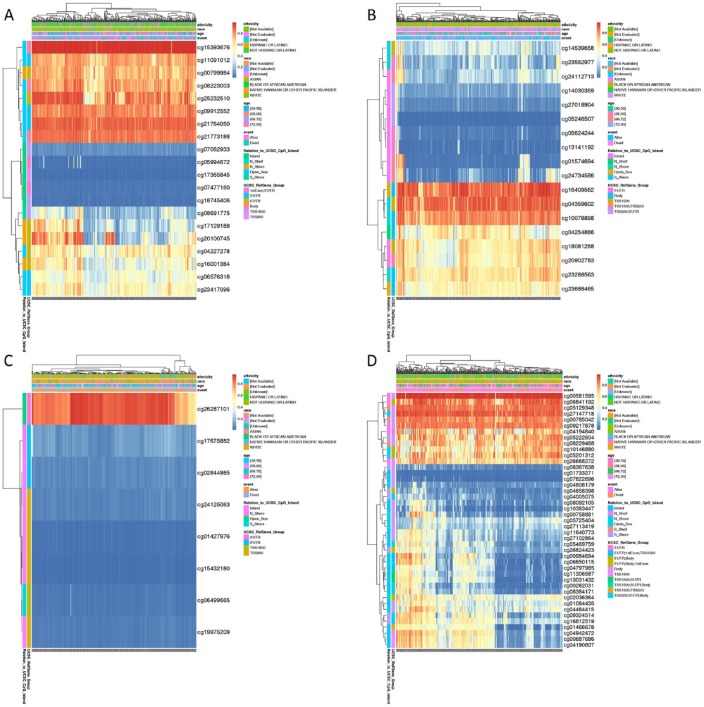
Prognostic values of NDRG1–4 DNA methylation in MethSurv
The DNA methylation levels of NDRG1–4 with the prognostic values of each single CpG in TCGA were analyzed by MethSurv [Figure 8(a–d), Table 1]. In fact, cg15393676 of NDRG1, cg16409562 of NDRG2, cg26287101 of NDRG3 and cg00581595 of NDRG4 showed the highest DNA methylation [Figure 8(A–D)]. Overall, 6 CpGs of NDRG1, 4 CpGs of NDRG2, 3 CpGs of NDRG3 and 24 CpGs of NDRG4 were associated with significant prognosis (Table 3).
Figure 8.
DNA methylation of NDRG1–4 in MethSurv. (a–d) The DNA methylation levels of NDRG1–4; (a) the DNA methylation clustered expression of NDRG1; (b) The DNA methylation clustered expression of NDRG2; (c) The DNA methylation clustered expression of NDRG3; (d) The DNA methylation clustered expression of NDRG4; Red to blue: high expression to low expression. Various colorful side boxes were used to characterize the ethnicity, race, age, event, relation to UCSC_CpG_island, UCSC_refGene_Group.
Table 3.
The significantly prognostic values of CpG in the NDRG family.
The prognostic values of CpG in the NDRG family by MethSurv (p < 0.05).
| Gene-CpG | HR | LR test p value |
|---|---|---|
| NDRG1 − 1stExon;5’UTR−Island−cg05994672 | 1.392 | 0.048 |
| NDRG1 − 3’UTR−Open_Sea−cg09912552 | 0.629 | 0.0052 |
| NDRG1 − 3’UTR−Open_Sea−cg21764050 | 0.687 | 0.042 |
| NDRG1 − 5’UTR−N_Shore−cg16001384 | 0.68 | 0.028 |
| NDRG1 − TSS1500−Island−cg07062933 | 1.622 | 0.014 |
| NDRG1 − TSS200−Island−cg17365845 | 1.433 | 0.038 |
| NDRG2 − Body−N_Shelf−cg04254886 | 1.715 | 0.0051 |
| NDRG2 − TSS1500−S_Shore−cg04359602 | 0.719 | 0.045 |
| NDRG2 − TSS200;5’UTR−Island−cg05246507 | 0.672 | 0.02 |
| NDRG2 − 5’UTR.Island.cg13141192 | 0.647 | 0.014 |
| NDRG3 − 3’UTR−Open_Sea−cg26287101 | 0.675 | 0.029 |
| NDRG3 − 5’UTR−Island−cg02844985 | 1.519 | 0.033 |
| NDRG3 − 5’UTR−N_Shore−cg17675882 | 1.49 | 0.029 |
| NDRG4 − 5’UTR;1stExon; TSS1500−Island−cg00984694 | 0.618 | 0.019 |
| NDRG4 − 5’UTR;1stExon; TSS1500−Island−cg04797985 | 0.603 | 0.015 |
| NDRG4 − 5’UTR; Body;1stExon−S_Shore−cg26824423 | 0.67 | 0.026 |
| NDRG4 − 5’UTR; Body−S_Shore−cg04858398 | 0.653 | 0.034 |
| NDRG4 − Body−Island−cg01084435 | 0.595 | 0.0023 |
| NDRG4 − Body−Island−cg08092105 | 0.644 | 0.026 |
| NDRG4 − Body−Island−cg10383447 | 0.596 | 0.0061 |
| NDRG4 − Body−Island−cg11640773 | 0.594 | 0.0024 |
| NDRG4 − Body−Island−cg27102864 | 0.648 | 0.0098 |
| NDRG4 − Body−N_Shore−cg04484415 | 0.642 | 0.017 |
| NDRG4 − Body−S_Shelf−cg00785042 | 0.591 | 0.0084 |
| NDRG4 − Body−S_Shelf−cg05129348 | 0.585 | 0.0019 |
| NDRG4 − TSS1500;5’UTR; Body−Island−cg05469759 | 0.637 | 0.0079 |
| NDRG4 − TSS1500;5’UTR; Body−Island−cg08384171 | 0.659 | 0.02 |
| NDRG4 − TSS1500; 5’UTR−Island−cg13031432 | 0.632 | 0.024 |
| NDRG4 − TSS1500−Island−cg00687686 | 0.698 | 0.029 |
| NDRG4 − TSS1500−Island−cg04190807 | 0.647 | 0.0084 |
| NDRG4 − TSS1500−Island−cg04942472 | 0.641 | 0.0066 |
| NDRG4 − TSS1500−N_Shore−cg27147718 | 0.558 | 0.0047 |
| NDRG4 − TSS200;5’UTR; Body−Island−cg00262031 | 0.582 | 0.0012 |
| NDRG4 − TSS200;5’UTR; Body−Island−cg06650115 | 0.689 | 0.025 |
| NDRG4 − TSS200;5’UTR; Body−S_Shore−cg04005075 | 0.676 | 0.018 |
| NDRG4 − TSS200;5’UTR; Body−S_Shore−cg09324514 | 0.538 | 0.00065 |
| NDRG4 − TSS200;5’UTR; Body−S_Shore−cg16812519 | 0.662 | 0.013 |
HR, hazard ratio; LR, log-rank.
Correlation between TIICs and NDRG members
Given the increasing association between immunological feature and prognosis in cancer, we further explored the correlation between TIICs and NDRG members. In fact, only CD4+ T-cells showed the highest correlation with NDRG4 [correlation = 0.341, p = 2.14e−11; Figure 9(a)]. Given a variety of immune cells were defined by CD4+ T-cells, we further examined the gene markers in each subtype [Figure 9(b–g)]. Of note NDRG4 was highly associated with BCL6 in Tfh cells [correlation = 0.438, p = 00e+00; Figure 9(c)].
Figure 9.
Correlation analysis between NDRG members and TIICs. (a) The correlation between each type of TIICs (B-cells, CD4+ T-cells, CD8+ T-cells, neutrophils, macrophages and dendritic cells) and NDRG family; (b) The correlation between the gene markers [TBX21 (T-bet), STAT4, STAT1, IFNG (interferon gamma) and TNF (tumor necrosis factor alpha)] of Th1 and NDRG4; (c) the correlation between the genes markers (BCL6, IL21) of Tfh cells and NDRG4; (d) the correlation between the genes markers (GATA3, STAT6, STAT5A and IL13) of Th2 cells and NDRG4; (e) the correlation between the genes markers [FOXP3, CCR8, STAT5B and TGFB1 (transforming growth factor beta)] of Treg cells and NDRG4; (f) the correlation between gene markers [PDCD1 (programmed cell death 1), CTLA4, LAG3, HAVCR2 (TIM-3) and GZMB)] of T-cell exhaustion and NDRG4; (g) the correlation between gene markers (STAT3 and IL17A) of Th17 cells and NDRG4.
IL, interleukin; TIICs, tumor infiltrating immune cells; STAT4, signal transducer and activator of transcription 4; BCL6, B-Cell Lymphoma 6; IL21, interleukin 21; GATA3, GATA binding protein 3; FOXP3, forkhead box P3; CCR8, C-C Motif Chemokine Receptor 8; TGFB1, Transforming Growth Factor Beta 1; CTLA4, cluster of differentiation 152; LAG3, Lymphocyte activation gene 3; HAVCR2. TIM-3, Hepatitis A Virus Cellular Receptor 2; GZMB, Granzyme B..
Discussion
The increasing availability of published mRNA data, clinical outcomes and standardized analysis platforms has provided the opportunities for exploring the correlation between gene expressions and type-specific cancer prognosis. This in silico study demonstrated distinct prognostic and biological values of NDRG family members in GC with mRNA expression and DNA methylation based on multiple cohorts from KM plotter and TCGA.
NDRG1 had been implicated in the regulation of embryonic placentation and organ development,7 the cellular vesicle transport system,13 endocytosis and recycling of membrane proteins.14,15 The reduced expression of NDRG1 had been associated with a worse prognosis in esophageal,20 colorectal21 and breast cancers12 while leading to favorable clinical outcomes in hepatocellular carcinoma.22 This paradoxical fact may be tumor type-specific, further highlighting the complicated biological function and processes that NDRG1 is involved with. In fact, NDRG1 was associated with a decrease in the proliferation and induction of apoptosis of cancer cells by the regulation of Bcl-2 and Ca2+-associated protein 43,48,49 and the dysregulation of epithelial-mesenchymal transition (EMT).50–52 Nonetheless, NDRG1 might exert inconsistent effects in GC prognosis.49,50,53,54
In this study, although NDRG1 was not significantly associated with overall prognosis in all cases, 6 CpGs of NDRG1 were associated with significant prognosis. Of note, inverse prognostic values of NDRG1 in HER2+/− groups indicated a potential correlation between NDRG1 and HER2. However, no significant mRNA expression correlation was identified between NDRG1 and HER2/EGFR. Furthermore, the protein expression of NDRG1 phosphorylation level, NDRG1_pT346, was found to be significantly associated with EGFR, EGFR_pY1068, EGFR_pY1173, HER2 and HER2_pY1248. Previous study indicated that, in colon cancer and pancreatic cancer, NDRG1 significantly reduced the expression of HER2 (general expression, heterodimerization and phosphorylation) and the activation of downstream MAPKK in response to the epidermal growth factor ligand.55 For the first time, our study highlighted a potential role of NDRG1 associated with HER2 status in GC. Given the HER2 targeting drug, trastuzumab, has been widely used in GC,56 digging into the NDRG1-related mechanisms may shed light upon further biological and pharmacological values.
Collectively, although NDRG1 was not validated as an independent general prognostic factor in multivariate analysis from TCGA, the prognostic value of NDRG1 was highlighted in GC subsets with significant correlation to HER2.
The NDRG2 expression had been found significantly reduced in pancreatic, breast cancers and hepatocellular carcinoma compared with normal tissues, accompanied by more aggressive features and a high ratio of relapse.23–25 In this study, NDRG2 was associated with favorable prognosis in all and significantly reduced in tumors compared with normal tissues, consistent with previous studies.26 However, no significance was found in the stage-specific pattern. Of note, increased chemo-resistance and decreased Fas-mediated cell death had been validated due to the inhibition of NDRG2.26 Interestingly, the promoter methylation of NDRG2 was frequently hypermethylated, leading to decreasing expression of NDRG2 at both mRNA and protein level and further associated with worse prognosis of GC.57 Similar prognostic role of NDRG2 had been validated in prostate cancer. The downregulation of NDRG2 was associated with advanced pathological stages and identified as an independent prognostic factor for short recurrence-free survival and OS.27 Of note, overexpression of NDRG2 could decrease the radiosensitization of Hela cells by the regulation of Bax signaling.58
NDRG3 was found to be significantly upregulated in prostate cancer, and was associated with advanced pathological stage and a worse prognosis of prostate cancer, contrary to NDRG2.27 Currently the role of NDRG3 had not been fully investigated in GC. Our study had revealed that NDRG3 was significantly associated with a favorable prognosis for the OS of all patients, as well as the HER2+ and diffuse type subgroups. Furthermore, NDRG3 was significantly increased in tumor compared with normal, with no significant distribution in various stages. In fact, although NDRG3 showed a favorable outcome based on the outcome from the KM plotter, a significant upregulation of NDRG3 was found in tumors compared with normal tissues in TCGA. Moreover, upregulation of NDRG3 was also associated with the high-risk group in the NDRG signature. There are a few issues that need to be clarified. First, clinical heterogeneity may account for the controversial outcome between TCGA and KM plotter (GSE14210, GSE15459, GSE22377, GSE29272, GSE51105 and GSE62254). Second, multivariate Cox analysis of the NDRG family using TCGA also eluded the potential independent prognostic value of NDRG3, both in OS and RFS. However, given the current studies remained sparse, the biological and prognostic values of NDRG3 warrant further intensive investigation. It may be insightful to systematically explore the prognostic value of NDRG3 using meta-analysis.
Previously, NDRG4 was found reduced in both mRNA and protein expression in colorectal cancer tissues compared with normal counterparts, and significantly suppressed tumor invasion and proliferation.28 However, it was not significantly reduced in tumors compared with normal tissues in GC from this study. Similar to NDRG3, the role of NDRG4 had not been clear. Moreover, 24 CpGs of NDRG4 exhibited significant prognostic values. Despite the correlation between NDRG4 and HER2, mRNA expression was not significant based on TCGA data, it was perceived that NDRG4 could exert effects on the downstream signaling components of HER2, such as RAS/RAF/MAPK/ERK. Interestingly, only NDRG4 was validated as an independent prognostic factor in TCGA dataset, further indicating a possible association between NDRG4 and recurrence in GC. For the rest of the NDRG family, it remained far from conclusive due to possible race diversity and other confounding factors such as radiochemotherapy.
Interestingly, although NDRG1 and NDRG4 did not show significantly differential expression between tumor and normal tissues in STAD using the GEPIA platform, the high/low-risk groups exhibited distinct expression patterns of NDRG1 and NDRG4 using the same dataset [Figure 4(a, b)]. In fact, the NDRG member signature may provide insightful clues on the prognostic values of combinational analysis, rather than a single gene.
The current network regulation of the NDRG family associated with GC was summarized (Figure 10).26,50,53–55,57,59–64 NDRG1, 2 and 4 have been reported to feature aberrant methylation in GC compared with normal tissues.57,63,64 Reduced expression of NDRG1 was associated with enhanced migration, invasion and metastasis via several mechanisms, including EMT, MMP-2 and MMP-9.50,53,54,59,60 Although NDRG2 was not an independent prognostic indicator in this manuscript, it was determined as an independent risk factor by Choi and colleagues.26 Moreover, silencing of NDRG2 increased the proliferation and resistance of cisplatin in GC cell lines.26 Up to now, studies focusing on the association between NDRG3 and 4 and GC remain limited. Interestingly, in this manuscript, NDRG4 was highly associated with BCL6 in Tfh cells. Up to now, this is the first study reporting the correlation between BCL6 and NDRG4 in GC, indicating a potential role of NDRG4 in follicular helper CD4+ T-cells. Moreover, potential inverse prognostic values of NDRG1 between HER2+/− GC and the significant association between protein expression of NDRG1 (NDRG1_pT346) and HER2/HER2_pY1248 indicated possible connection as well. However, in silico findings warrant further experimental validation.
Figure 10.
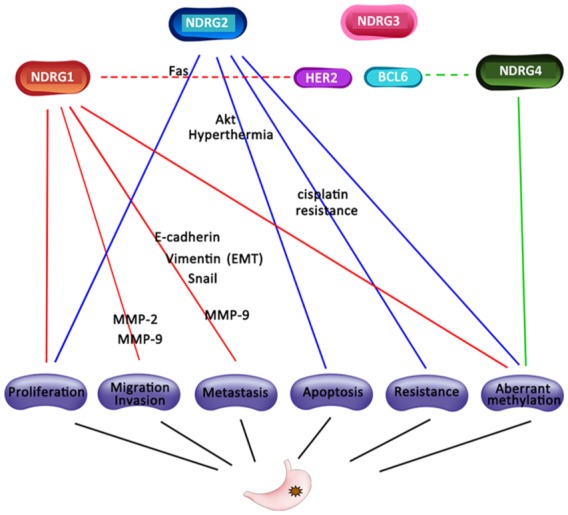
Regulation network of NDRG family in GC. Red: NDRG1-associated mechanism; blue: NDRG2-associated mechanism; pink: NDRG3-associated mechanism; green: NDRG4-associated mechanism; dash line indicated predicted correlation.
EMT, epithelial-mesenchymal transition; GC, gastric cancer.
Up to now, methylation-related study of the NDRG family remains limited. High levels of NDRG1 promoter methylation in the CpG islands were found in both GC cell lines and tissues.64 Interestingly, no mutation of NDRG1 was detected in this study.64 Consistently, our finding also indicated rare cases of NDRG1 mutation. For NDRG2, hypermethylation status was detected in the NDRG2 promoter both in GC cell lines and tissues.62 In fact, the reduced expression of NDRG2 in GC compared with normal tissues was highly correlated with the promoter hypermethylation.62 For NDRG4, both promoter and gene body methylation levels were increased in GC tissues.63 Interestingly, opposite clinical results of NDRG4 were found between the Chinese samples from Chen and colleagues and TCGA data, highlighting the race difference beneath the prognostic values of NDRG4.63
The limitation of this study was the lack of experimental validation and externally clinical cohort validation. The limited number of some subgroups of KM plotter for prognostic analysis and potential sample heterogeneity could bias the results. Further validation on a larger sample size is also required.
Conclusions
This in silico study investigated the biological and prognostic values of the NDRG family in GC based on KM plotter and TCGA, providing insights for further investigation of NDRG family as potential targets in GC.
Acknowledgments
We appreciate the academic support from Dr Mary Goldman from UC Santa Cruz Genomics Institute. Chaoran Yu and Xiaohui Hao contributed as co-first authors. Minhua Zheng and Jing Sun contributed equally as corresponding authors. The contributions of authors are as follows:
CY, XH, SZ, WH and JS carried out data analysis.
CY, XH, JL and MZ drafted the manuscript
MZ, JS and CY participated in study design, data collection and analysis.
MZ, JS and CY revised the manuscript
All authors read and approved the final manuscript.
Footnotes
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The study is financially supported by National Natural Science Foundation of China (NSFC; 81402423, 81572818, 81871984), Shanghai Municipal Commission of Health and Family Planning (2017YQ062), as well as the Shanghai Science and Technology Committee (18695841400).
Conflict of interest statement: The authors declare that there is no conflict of interest.
ORCID iD: Chaoran Yu  https://orcid.org/0000-0003-4657-7975
https://orcid.org/0000-0003-4657-7975
Contributor Information
Chaoran Yu, Department of Gastrointestinal Surgery, Ruijin Hospital, Shanghai Jiao Tong University, School of Medicine, Shanghai, China; Shanghai Minimally Invasive Surgery Center, Ruijin Hospital, Shanghai Jiao Tong University, School of Medicine, Shanghai, China.
Xiaohui Hao, Department of Gastrointestinal Surgery, Ruijin Hospital, Shanghai Jiao Tong University, School of Medicine, Shanghai, China; Shanghai Minimally Invasive Surgery Center, Ruijin Hospital, Shanghai Jiao Tong University, School of Medicine, Shanghai, China.
Sen Zhang, Department of Gastrointestinal Surgery, Ruijin Hospital, Shanghai Jiao Tong University, School of Medicine, Shanghai, China; Shanghai Minimally Invasive Surgery Center, Ruijin Hospital, Shanghai Jiao Tong University, School of Medicine, Shanghai, China.
Wenjun Hu, Department of Gastrointestinal Surgery, Ruijin Hospital, Shanghai Jiao Tong University, School of Medicine, Shanghai, China; Shanghai Minimally Invasive Surgery Center, Ruijin Hospital, Shanghai Jiao Tong University, School of Medicine, Shanghai, China.
Jianwen Li, Department of Gastrointestinal Surgery, Ruijin Hospital, Shanghai Jiao Tong University, School of Medicine, Shanghai, China; Shanghai Minimally Invasive Surgery Center, Ruijin Hospital, Shanghai Jiao Tong University, School of Medicine, Shanghai, China.
Jing Sun, Department of Gastrointestinal Surgery, Ruijin Hospital, Shanghai Jiao Tong University, School of Medicine, Shanghai, China; Shanghai Minimally Invasive Surgery Center, Ruijin Hospital, Shanghai Jiao Tong University, School of Medicine, Shanghai, China.
Minhua Zheng, Department of Gastrointestinal Surgery, Shanghai Minimally Invasive Surgery Center, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, P.R. China.
References
- 1. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016; 66: 115–132. [DOI] [PubMed] [Google Scholar]
- 2. Zheng R, Zeng H, Zhang S, et al. Estimates of cancer incidence and mortality in China, 2013. Chin J Cancer 2017; 36: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ajani JA, D’Amico TA, Almhanna K, et al. Gastric cancer, version 3.2016, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 2016; 14: 1286–1312. [DOI] [PubMed] [Google Scholar]
- 4. Tsugane S, Sasazuki S. Diet and the risk of gastric cancer: review of epidemiological evidence. Gastric Cancer 2007; 10: 75–83. [DOI] [PubMed] [Google Scholar]
- 5. Wilke H, Muro K, Van Cutsem E, et al. Ramucirumab plus paclitaxel versus placebo plus paclitaxel in patients with previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (RAINBOW): a double-blind, randomised phase 3 trial. Lancet Oncol 2014; 15: 1224–1235. [DOI] [PubMed] [Google Scholar]
- 6. Songun I, Putter H, Kranenbarg EMK, et al. Surgical treatment of gastric cancer: 15-year follow-up results of the randomised nationwide Dutch D1D2 trial. Lancet Oncol 2010; 11: 439–449. [DOI] [PubMed] [Google Scholar]
- 7. Melotte V, Qu X, Ongenaert M, et al. The N-myc downstream regulated gene (NDRG) family: diverse functions, multiple applications. FASEB J 2010; 24: 4153–4166. [DOI] [PubMed] [Google Scholar]
- 8. Shaw E, McCue LA, Lawrence CE, et al. Identification of a novel class in the α/β hydrolase fold superfamily: the N-myc differentiation-related proteins. Proteins 2002; 47: 163–168. [DOI] [PubMed] [Google Scholar]
- 9. Ellen TP, Ke Q, Zhang P, et al. NDRG1, a growth and cancer related gene: regulation of gene expression and function in normal and disease states. Carcinogenesis 2007; 29: 2–8. [DOI] [PubMed] [Google Scholar]
- 10. Sun J, Zhang D, Bae DH, et al. Metastasis suppressor, NDRG1, mediates its activity through signaling pathways and molecular motors. Carcinogenesis 2013; 34: 1943–1954. [DOI] [PubMed] [Google Scholar]
- 11. Sun J, Zhang D, Zheng Y, et al. Targeting the metastasis suppressor, NDRG1, using novel iron chelators: regulation of stress fiber-mediated tumor cell migration via modulation of the ROCK1/pMLC2 signaling pathway. Mol Pharmacol 2013; 83: 454–469. [DOI] [PubMed] [Google Scholar]
- 12. Kovacevic Z, Richardson DR. The metastasis suppressor, Ndrg-1: a new ally in the fight against cancer. Carcinogenesis 2006; 27: 2355–2366. [DOI] [PubMed] [Google Scholar]
- 13. Askautrud HA, Gjernes E, Gunnes G, et al. Global gene expression analysis reveals a link between NDRG1 and vesicle transport. PloS One 2014; 9: e87268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pietiäinen V, Vassilev B, Blom T, et al. NDRG1 functions in LDL receptor trafficking by regulating endosomal recycling and degradation. J Cell Sci 2013; 126: 3961–3971. [DOI] [PubMed] [Google Scholar]
- 15. Kachhap SK, Faith D, Qian DZ, et al. The N-Myc down regulated Gene1 (NDRG1) Is a Rab4a effector involved in vesicular recycling of E-cadherin. PloS One 2007; 2: e844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Choi SC, Kim KD, Kim JT, et al. Expression and regulation of NDRG2 (N-myc downstream regulated gene 2) during the differentiation of dendritic cells. FEBS Lett 2003; 553: 413–418. [DOI] [PubMed] [Google Scholar]
- 17. Choi SC, Kim KD, Kim JT, et al. Expression of human NDRG2 by myeloid dendritic cells inhibits down-regulation of activated leukocyte cell adhesion molecule (ALCAM) and contributes to maintenance of T cell stimulatory activity. J Leukoc Biol 2008; 83: 89–98. [DOI] [PubMed] [Google Scholar]
- 18. Park KC, Lee DC, Yeom YI. NDRG3-mediated lactate signaling in hypoxia. BMB Rep 2015; 48: 301–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qu X, Jia H, Garrity DM, et al. Ndrg4 is required for normal myocyte proliferation during early cardiac development in zebrafish. Dev Biol 2008; 317: 486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ando T, Ishiguro H, Kimura M, et al. Decreased expression of NDRG1 is correlated with tumor progression and poor prognosis in patients with esophageal squamous cell carcinoma. Dis Esophagus 2006; 19: 454–458. [DOI] [PubMed] [Google Scholar]
- 21. Mao Z, Sun J, Feng B, et al. The metastasis suppressor, N-myc downregulated gene 1 (NDRG1), is a prognostic biomarker for human colorectal cancer. PLoS One 2013; 8: e68206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cheng J, Xie HY, Xu X, et al. NDRG1 as a biomarker for metastasis, recurrence and of poor prognosis in hepatocellular carcinoma. Cancer Lett 2011; 310: 35–45. [DOI] [PubMed] [Google Scholar]
- 23. Hu XL, Liu XP, Lin SX, et al. NDRG2 expression and mutation in human liver and pancreatic cancers. World J Gastroenterol 2004; 10: 3518–3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee DC, Kang YK, Kim WH, et al. Functional and clinical evidence for NDRG2 as a candidate suppressor of liver cancer metastasis. Cancer Res 2008; 68: 4210–4220. [DOI] [PubMed] [Google Scholar]
- 25. Lorentzen A, Lewinsky RH, Bornholdt J, et al. Expression profile of the N-myc Downstream Regulated Gene 2 (NDRG2) in human cancers with focus on breast cancer. BMC Cancer 2011; 11: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Choi SC, Yoon SR, Park YP, et al. Expression of NDRG2 is related to tumor progression and survival of gastric cancer patients through Fas-mediated cell death. Exp Mol Med 2007; 39: 705–714. [DOI] [PubMed] [Google Scholar]
- 27. Ren GF, Tang L, Yang AQ, et al. Prognostic impact of NDRG2 and NDRG3 in prostate cancer patients undergoing radical prostatectomy. Histol Histopathol 2014; 29: 535–542. [DOI] [PubMed] [Google Scholar]
- 28. Melotte V, Lentjes MHFM, Van den Bosch SM, et al. N-Myc downstream-regulated gene 4 (NDRG4): a candidate tumor suppressor gene and potential biomarker for colorectal cancer. J Natl Cancer Inst 2009; 101: 916–927. [DOI] [PubMed] [Google Scholar]
- 29. Chu D, Zhang Z, Zhou Y, et al. NDRG4, a novel candidate tumor suppressor, is a predictor of overall survival of colorectal cancer patients. Oncotarget 2015; 6: 7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Szász AM, Lánczky A, Nagy Á, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 2016; 7: 49322–49333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aguirre-Gamboa R, Gomez-Rueda H, Martínez-Ledesma E, et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PloS One 2013; 8: e74250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017; 45(W1): W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goldman M, Craft B, Zhu J, et al. The UCSC Xena system for cancer genomics data visualization and interpretation. Cancer Res 2017; 77: abstract 2584. [Google Scholar]
- 34. Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 2010; 28: 1248–1250. [DOI] [PubMed] [Google Scholar]
- 35. Jun Li, Yiling Lu, Akbani R, et al. TCPA: a resource for cancer functional proteomics data. Nat Methods 2013; 10: 1046–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Borcherding N, Bormann N, Voigt A, et al. TRGAted: a web tool for survival analysis using protein data in the Cancer Genome Atlas. F1000Res 2018; 7: 1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. Nat Genet 2000; 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000; 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 42. Lachmann A, Xu H, Krishnan J, et al. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 2010; 26: 2438–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xia J, Gill EE, Hancock RE. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protoc 2015; 10: 823–844. [DOI] [PubMed] [Google Scholar]
- 44. Modhukur V, Iljasenko T, Metsalu T, et al. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics 2018; 10: 277–288. [DOI] [PubMed] [Google Scholar]
- 45. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res 2017; 77: e108–e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Siemers NO, Holloway JL, Chang H, et al. Genome-wide association analysis identifies genetic correlates of immune infiltrates in solid tumors. PLoS One 2017; 12: e0179726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Danaher P, Warren S, Dennis L, et al. Gene expression markers of Tumor Infiltrating Leukocytes. J Immunother Cancer 2017; 5: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang X, Zhu F, Yu C, et al. N-myc downstream-regulated gene 1 promotes oxaliplatin-triggered apoptosis in colorectal cancer cells via enhancing the ubiquitination of Bcl-2. Oncotarget 2017; 8: 47709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kawahara A, Akiba J, Hattori S, et al. Nuclear expression of N-myc downstream regulated gene 1/Ca(2+)-associated protein 43 is closely correlated with tumor angiogenesis and poor survival in patients with gastric cancer. Exp Ther Med 2011; 2: 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ureshino H, Murakami Y, Watari K, et al. N-myc downstream regulated gene 1 (NDRG1) promotes metastasis of human scirrhous gastric cancer cells through epithelial mesenchymal transition. PloS One 2012; 7: e41312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen Z, Zhang D, Yue F, et al. The iron chelators Dp44mT and DFO inhibit TGF-beta-induced epithelial-mesenchymal transition via up-regulation of N-Myc downstream-regulated gene 1 (NDRG1). J Biol Chem 2012; 287: 17016–17028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mi L, Zhu F, Yang X, et al. The metastatic suppressor NDRG1 inhibits EMT, migration and invasion through interaction and promotion of caveolin-1 ubiquitylation in human colorectal cancer cells. Oncogene 2017; 36: 4323–4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jiang K, Shen Z, Ye Y, et al. A novel molecular marker for early detection and evaluating prognosis of gastric cancer: N-myc downstream regulated gene-1 (NDRG1). Scand J Gastroenterol 2010; 45: 898–908. [DOI] [PubMed] [Google Scholar]
- 54. Chang X, Xu X, Ma J, et al. NDRG1 expression is related to the progression and prognosis of gastric cancer patients through modulating proliferation, invasion and cell cycle of gastric cancer cells. Mol Biol Rep 2014; 41: 6215–6223. [DOI] [PubMed] [Google Scholar]
- 55. Kovacevic Z, Menezes SV, Sahni S, et al. The metastasis suppressor, N-Myc downstream-regulated gene-1 (NDRG1), down-regulates the ErbB family of receptors to inhibit downstream oncogenic signaling pathways. J Biol Chem 2016; 291: 1029–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010; 376: 687–697. [DOI] [PubMed] [Google Scholar]
- 57. Ling ZQ, Ge MH, Lu XX, et al. Ndrg2 promoter hypermethylation triggered by helicobacter pylori infection correlates with poor patients survival in human gastric carcinoma. Oncotarget 2015; 6: 8210–8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu J, Zhang J, Wang X, et al. HIF-1 and NDRG2 contribute to hypoxia-induced radioresistance of cervical cancer Hela cells. Exp Cell Res 2010; 316: 1985–1993. [DOI] [PubMed] [Google Scholar]
- 59. Chang X, Xu X, Xue X, et al. NDRG1 controls gastric cancer migration and invasion through regulating MMP-9. Pathol Oncol Res 2016; 22: 789–796. [DOI] [PubMed] [Google Scholar]
- 60. Liu Y, Bai W, Luo W, et al. Downregulation of NDRG1 promotes invasion of human gastric cancer AGS cells through MMP-2. Tumour Biol 2011; 32: 99–105. [DOI] [PubMed] [Google Scholar]
- 61. Tao Y, Guo Y, Liu W, et al. AKT inhibitor suppresses hyperthermia-induced Ndrg2 phosphorylation in gastric cancer cells. Braz J Med Biol Res 2013; 46: 394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chang X, Li Z, Ma J, et al. DNA methylation of NDRG2 in gastric cancer and its clinical significance. Dig Dis Sci 2013; 58: 715–723. [DOI] [PubMed] [Google Scholar]
- 63. Chen X, Yang Y, Liu J, et al. NDRG4 hypermethylation is a potential biomarker for diagnosis and prognosis of gastric cancer in Chinese population. Oncotarget 2017; 8: 8105–8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chang X, Zhang S, Ma J, et al. Association of NDRG1 gene promoter methylation with reduced NDRG1 expression in gastric cancer cells and tissue specimens. Cell Biochem Biophys 2013; 66: 93–101. [DOI] [PubMed] [Google Scholar]