Abstract
There is potential value in incorporating biomedical big data (BBD)—observational real-world patient-level genomic and clinical data in multiple sub-populations—into economic evaluations of precision medicine. However, health economists face practical and methodological challenges when using BBD in this context. We conducted a literature review to identify and summarise these challenges. Relevant articles were identified in MEDLINE, EMBASE, EconLit, University of York Centre for Reviews and Dissemination and Cochrane Library from 2000 to 2018. Articles were included if they studied issues relevant to the interconnectedness of biomedical big data, precision medicine, and health economic evaluation. Nineteen articles were included in the review. Challenges identified related to data management, data quality and data analysis. The availability of large volumes of data from multiple sources, the need to conduct data linkages within an environment of opaque data access and sharing procedures, and other data management challenges are primarily practical and may not be long-term obstacles if procedures for data sharing and access are improved. However, the existence of missing data across linked datasets, the need to accommodate dynamic data, and other data quality and analysis challenges may require an evolution in economic evaluation methods. Health economists face challenges when using BBD in economic evaluations of technologies that facilitate precision medicine. Potential solutions to some of these challenges do, however, exist. Going forward, health economists who present work that uses BBD should document challenges and the solutions they have applied to the challenges to support future researcher endeavours.
Electronic supplementary material
The online version of this article (10.1007/s40258-019-00474-7) contains supplementary material, which is available to authorized users.
Key Points for Decision Makers
| We find that challenges of using biomedical big data (BBD) for economic evaluations of precision medicine relate to data management, data quality and data analysis. |
| While data management challenges are primarily practical and may not be long-term obstacles if procedures for data sharing and access are improved, data quality and analysis challenges may require an evolution in economic evaluation methods. |
| Health economists who present work that uses BBD should document challenges and the solutions they have applied to the challenges to support future researcher endeavours. |
Introduction
Advances in genomic technologies have led to rapid growth in the availability of genomic data [1] and a move towards so-called precision medicine, which is often described as the tailoring of health interventions based on patients’ individual characteristics [2]. At the same time, clinical information from electronic health records (EHRs) and healthcare claims data have added to the production of large data volumes [3]. The linkage of genomic and clinical data sources has created “biomedical big data” (BBD), defined as: “the emerging technologically-driven phenomena focusing on analysis of aggregated datasets to improve medical knowledge and clinical care” [4]. As such, BBD can be seen as a separate type of data with distinctive characteristics compared to big data, health records data, and genomic data.
In a genomic context, BBD primarily enables research into genotype-phenotype associations. However, it can also support the assessment of the economic impact of implementing technologies that facilitate precision medicine into routine clinical care. These technologies include next-generation sequencing (NGS) approaches such as whole-genome sequencing. The integration of these technologies into routine clinical care has been widely discussed [5, 6]. In the UK, the National Health Service (NHS) in England recently announced the implementation of a national Genomic Medicine Service to provide genomic precision medicine based on NGS [7]. In the USA, the Centers for Medicare and Medicaid Services (CMS) have recommended covering cancer sequencing nationally [8].
Ideally, technologies that facilitate precision medicine should only be implemented into routine clinical care if evidence indicates their clinical and cost-effectiveness [7]. For instance, while NGS technologies are driving the movement towards precision medicine, and evidence on their diagnostic yield seems to favour implementation, evidence from economic evaluations on their cost-effectiveness is less clear [9]. Economic evaluations that make use of rich BBD on patient resource use and health outcomes over time could contribute to the evidence base on whether precision medicine approaches based on technologies such as NGS are cost-effective. A study by Lorgelly et al. is a good example of the use of BBD within a health economic context, presenting potential benefits and challenges of working with high-volume and high-variety data. This study linked data from the Australian prospective genomic cohort study (Cancer 2015) with healthcare reimbursement data to explain variations in health expenditure, for example assessing the presence of particular mutations and their effect on healthcare costs [10]. The advent of large-scale national genomic sequencing projects such as the 100,000 Genomes Project in the UK could lead to similar studies being undertaken to eventually inform economic evaluations [11].
Although there is potential value in incorporating BBD into economic evaluations of technologies that facilitate precision medicine, there are also practical and methodological challenges. A small but growing literature on the issues surrounding the use of BBD in health economics is emerging, with some researchers tentatively concluding that novel approaches may be required with respect to the management, analysis and interpretation of BBD for use in economic evaluations [12, 13]. However, most of these articles are commentary or perspective articles, while no review has so far been conducted. A review of the challenges of using BBD for health economic evaluations of precision medicine is therefore urgently required. The launch of the Genomic Medicine Service in the UK NHS in October 2018 serves as an example of why urgency is warranted [14]. The decision to widen access to whole-genome sequencing for patients with rare diseases and cancer has been taken within the internal policy-making environment of the NHS, but no health economic evaluations have been published that support sequencing at scale. Studies that use linked BBD to provide decision-makers with information on the cost-effectiveness of genomic precision medicine would therefore be informative. However, when conducting such studies, challenges may arise. The aim of this literature review is to summarise these challenges. Given the interdisciplinary nature of the research conducted on this topic, we also summarise the links between the challenges identified in different disciplines.
Methods
Literature Search Strategy
The following five databases were searched: Medline, Embase, EconLit, The Cochrane Library, and the Centre for Reviews and Dissemination. The development of the search syntax consisted of three steps. First, a preliminary sample of relevant articles [10, 13, 15–18] from the health economics field that were known to the authors were analysed based on their indexed medical subject headings (MeSH), using the Yale MeSH Analyser [19] (a description of these articles is provided in Appendix 1). Second, based on identified keywords and MeSH terms, six broader concepts were defined (Table 1). Third, each of the six broader concepts was defined by various index-terms and free-text words, then combined using “AND” and “OR” operators (the full search syntax is provided in Appendix 2).
Table 1.
Literature search concepts and terms
| Concept | Example of search termsa |
|---|---|
| Data linkage | Record linkage, linked data, linked records, joined data, joint records, medical record linkage, etc. |
| Electronic health records | Clinical data, biomedical data, patient records, phenotype data, etc. |
| Big data | Big data, -omics |
| Genomics | Genomics, genetics, pharmacogenomics, pharmacogenetics, whole exome sequencing, whole genome sequencing, etc. |
| Precision medicine | Precision medicine, personalised medicine, individualised medicine, stratified medicine, etc. |
| Health economics | Health expenditure, health care costs, economic evaluations, cost-effectiveness, cost-benefit, cost-utility, cost-minimisation, cost-consequence, pharmacoeconomics, cost analysis, health technology assessment, etc. |
aThis is a sample of search terms used in the literature search
Article Selection Criteria
Articles were included if they studied issues relevant to the interconnectedness of biomedical big data, precision medicine, and health economic evaluation. The review covered articles published from January 2000 to January 2018 in English, German or Spanish (according to the language proficiencies of the authors). Articles were excluded if they did not meet all of the inclusion criteria, for example discussing precision medicine but not biomedical big data. Non-human-based articles were also excluded.
PF screened all titles and abstracts and excluded irrelevant articles. Full-text articles were then independently assessed by both PF and JB, with any disagreements resolved through discussion. Subsequent to the strategy-driven literature search, two further search methodologies were applied. First, articles that were known to the authors (including those in the grey literature) and that met the inclusion criteria, but that were not identified by the electronic database search, were added to the review. Second, snowballing (the recursive process of gathering, searching and aggregating references) was used to screen the references of included articles for relevant publications [20].
Data Extraction and Analysis
Data extraction was undertaken by PF. Qualitative information related to challenges associated with using BBD to conduct health economic evaluations of precision medicine were extracted from each paper. The identified challenges were grouped into categories that were established retrospectively based on the study findings.
Results
Article Selection
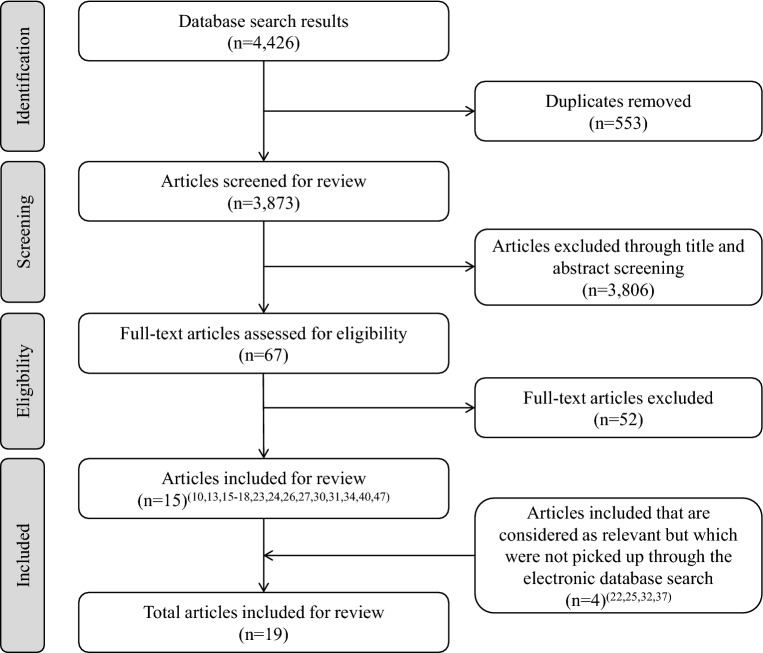
Figure 1 summarises the literature search results. 4426 articles were identified, of which 553 were duplicates. The 3873 remaining articles were screened by title and abstract, and 67 underwent full-text screening. Fifteen articles were eligible for inclusion in the review. Four additional references not identified in the database search but which were known to the authors were then added. Snowballing did not lead to the inclusion of any references. A total of 19 articles were included in the final review.
Fig. 1.
Literature search results
Study Characteristics
Of the 19 articles included in the review, ten were perspective pieces, five were reviews, three were methods papers, and one was a cost analysis. All of the publications were published between 2013 and 2017, with most (n = 13) published in 2016.
Challenges
The challenges identified within the articles were grouped into three categories: data management, data quality and data analysis. These challenges are described below. Table 2 presents a summary of the challenges falling into each category.
Table 2.
Summary of challenges of using biomedical big data (BBD) for health economic evaluations
| Challenge | Description |
| Data management | |
| Data storage and computation |
Increasing demand for data storage capacity and computational processing power [15, 22, 23] Costs associated with data storage and computational demands [13, 15, 23–27] |
| Data integration and linkage |
Presence of semi-structured and unstructured data; lack of adequate and standardised data integration [22, 30] Linking multiple datasets can be administratively complex and time-consuming [10] |
| Data access and sharing | |
| Data quality | |
| General quality issues |
Linking heterogeneous datasets into clinically useful information [34] Uncertainty around the regulatory acceptance of BBD [30] Quality issues of linked datasets due to missing data [32] Lack of contemporaneous data (data velocity) [10] |
| Secondary data |
Presence of bias and unmeasured confounding in observational data and the need for advanced statistical technique to account for this [16, 32, 37, 40] Lack of health outcomes data linked to claims data [26] Clinical miscoding [15, 34, 40] Presence of heterogeneous and complex data formats (e.g. speech recording, medical imaging, semi-structured text) [30] |
| EHRs |
High variability in the implementation and quality of EHRs within and across countries [16, 22, 30, 34] Lack of advanced EHR data cleaning procedures [15] |
| Data analysis | |
| Data heterogeneity | Increasing heterogeneity of both diagnosis and clinical management due to stratification of patients; need for multidimensional data analysis methods [34, 40] |
| Decision-modelling |
Departure from classical decision-modelling towards dynamic analysis models and machine learning techniques [10, 13, 15, 26] Modelling of -omics profiles [18] |
| Clinical trials | Potential need for n-of-1 trials [18, 31, 47] |
EHR electronic health record
Data Management
Data management refers to the administrative procedures undertaken prior to data analysis. Several issues emerged in the literature related to data storage, integration, linkage, sharing and access. These issues can present obstacles to the use of BBD for health economic evaluations.
Data Storage and Computation
Demand for storage capacity for genomic and clinical data is constantly rising [21]. The use of BBD requires extensive storage capacity and high computational processing power [15, 22, 23]. Both requirements involve significant costs, which could present an obstacle for some research institutions, limiting their ability to conduct health economic evaluations in this context [13, 15, 23–27]. The use of cloud storage and computing may alleviate this challenge. However, this will likely introduce issues related to data security and privacy, as an increasing amount of personal data originating from different data sources will be stored centrally, increasing the scale of potential data misuse [28, 29].
Data Integration and Linkage
Data integration refers to the process of combining heterogeneous data sources. An example is the integration of genomic data into EHRs. Genomic-integrated EHRs are a promising source of BBD for economic evaluations as a high quantity of data required by health economists would be available within a single system. However, challenges exist regarding their use. One major challenge is the structure of clinical and genotypic information in EHRs [22, 30]. For example, clinical information is often presented in a semi-structured or unstructured format. This limits the use of such information in economic evaluations unless it is further processed and standardised in a structural manner.
The data linkage environment and the linkage process pose additional challenges for the use of BBD in economic evaluations. Linking multiple datasets can be administratively complex and time-consuming, involving research groups, third-party linkage providers and data custodians. In the Cancer 2015 study, researchers performed part of the data linkage themselves, linking patient records on prescription medicines and medical services with cohort and genomic panel data. However, a trusted third party (data linkage unit) was required to link an additional dataset on secondary care resource use, as this required that a de-identified master dataset was returned to the researchers post-linkage [10].
Data Access and Sharing
Data sharing of EHRs may be necessary to increase study sample sizes, in particular in low-prevalence clinical areas such as rare diseases, where patients access a wide range of clinical services. However, the ease of sharing such components varies and data-sharing procedures are often unavailable [30, 31]. A second challenge relates to data access. Accessing the various datasets that are combined to form BBD can be very costly [15, 32]. In the UK, these costs can quickly add up to several thousand pounds [33]. Both of these challenges will likely prevent health economists from accessing all of the data they need to conduct economic evaluations.
Data Quality
Data quality is defined by the appropriateness of a dataset in light of its intended use. The presence of high-quality data is a precondition for successful data analysis. This section describes quality issues related to BBD that could be an obstacle to health economic evaluations. This includes general quality issues, and issues related to the use of secondary data (e.g. claims data) and EHRs.
General Quality Issues
In health economic evaluations, health outcomes and cost data from a variety of sources are often needed. Some of these sources, such as EHRs or claims data, have been used in economic evaluations for decades and are often referred to as “big data”. What is different with respect to BBD, however, is the linkage component with genomic and/or other-omics data sources. With respect to this, four general data quality challenges should be noted.
First, it will be a challenge to convert linked datasets that are heterogeneous (e.g. genomic data alongside covariates such as a patients treatment history), of complex format (e.g. medical images, text formats and bioinformatics-specific data formats), and dissimilar in quality (e.g. accuracy of semi-structured clinical data in EHRs compared to endpoint-specific data collected in a randomised controlled trial (RCT) research environment), into clinically useful information for health economic evaluations [34, 35]. The volume of information within BBD may become so complex that it will be increasingly difficult to judge the clinical relevance of such information.
Second, there is some uncertainty regarding regulatory acceptance of BBD [30]. A recent European Medicines Agency (EMA) report concluded that BBD is a valuable resource to support medicines assessment, but emphasised the issue of evidence validity, concluding that big data may support but not replace randomised controlled trials [36]. This could have a knock-on effect on the use of BBD within economic evaluations.
Third, missing data may present a particular problem for BBD, as the proportion of missing data points will likely increase with the number of datasets linked [32]. Economic evaluations in stratified patient groups with already small sample sizes may therefore become more challenging due to loss of information and a decrease in statistical power.
Fourth, contemporaneous BBD may not be available for data analysis. A key quality indicator for big data is data velocity (the speed at which data are available for analysis); however, velocity might be lower in datasets used for health economic evaluations due to delays in the linkage process [10].
Secondary Data
Secondary data such as claims data are a key component of BBD [26, 37]. Although the methodological challenges associated with using such observational data for health economic evaluations are well established [38, 39], these challenges may be more difficult to resolve in the context of BBD and precision medicine. There are three specific challenges. The first concerns the presence of bias and confounding in observational studies [32, 37, 40]. Any conclusion on the causality of observed associations in BBD will depend on the control of confounding effects by advanced statistical analysis (e.g. propensity score models, instrumental variable analysis or Bayesian modelling with non-informative priors) [32, 37, 41]. For instance, Dixon et al. used Mendelian randomization to support the estimation of causal effects of health conditions on costs by using genetic variants as instrumental variables [16]. Furthermore, a classic example of a confounding variable is the “physician’s past experience” with regard to subsequent clinical decision-making and consequential effects on health outcomes [42]. A physician’s diagnostic strategy or treatment decision, indicated by the available data on healthcare service utilisation, could suffer from such confounding, especially in the context of an increasing granularity of care options, or in rare and complex genetic diseases. Additionally, when using claims data in the context of rare and complex genetic diseases, healthcare service utilisation might be biased depending on how likely a patient is to access highly specialised care centres.
The second challenge relates to the lack of health outcomes data linked to claims data [26] or EHRs. In economic evaluations, health outcomes data is often used to construct QALYs, which measure the effectiveness of health interventions in the form of aggregated health state utility values (HSUVs). The absence of health outcomes information to construct QALYs is not per se a problem, as HSUVs from other studies with similar patient groups can be used. However, in cases where BBD enables the analysis of specific population sub-groups, e.g. patients with rare diseases, the use of HSUVs from other studies may be inappropriate, and of little value for informing economic evaluations.
A third challenge relates to the handling of missing data [15, 17, 32] and the clinical miscoding of diseases and procedures [15, 34, 40]. The coding and classification system based on the International Classification of Diseases (ICD) is said to have limited value in the context of rare diseases [43], and the presence of clinical miscoding can have serious consequences for data analysis [44], such as the wrongful representation of a patient’s care episode, affecting estimates of both clinical events and resource use/costs. This in turn will impact estimates of cost-effectiveness and increase uncertainty in economic models, and may increase the probability of decision-makers making incorrect decisions. In the context of rare diseases, the challenges associated with miscoding may be exacerbated as coding accuracy in the context of the currently used ICD system is limited [43]. Health economists could potentially make assumptions to resolve miscoding issues, guided by expert opinion. However, as data volumes increase and patient stratification advances, this might be less feasible due to the significant increase in cases experts would have to review.
Electronic Health Records
The availability of EHRs and the breadth of clinical information contained within them have increased considerably in recent years [22, 26, 45]. However, the use of information from EHRs for health economic evaluations can be challenging. One challenge relates to the use of data from EHRs for computation-analytic exercises. This can be technically challenging due to the presence of various data formats (e.g. speech recording, medical imaging, and semi-structured text) [30] and variability in the implementation and quality of EHRs within and across institutions and countries [16, 22, 30, 34, 46]. Moreover, the lack of advanced data-cleaning procedures for EHRs may be an issue for data quality [15].
Data Analysis
The use of BBD for health economic evaluations introduces several challenges for data analysis, including issues related to data heterogeneity, decision-modelling and the conduct of clinical trials.
Data Heterogeneity
Some of the challenges associated with the heterogeneity of BBD have already been noted in the context of data management and data quality. However, the presence of heterogeneous data is equally important with regard to data analysis. The move away from population models towards the stratification of patients into clusters will increase the heterogeneity of both diagnosis and clinical management. Patients may be clustered based on either their specific phenotypic or molecular disease profiles, or their response to treatment [40]. The use of BBD for economic evaluations of precision medicine will therefore require the development of novel and multidimensional methods of data analysis that account for increasing patient heterogeneity [34]. In the long term, the results of such methods could potentially challenge the current approach to health technology assessment decision-making.
Decision-Modelling
Economic decision models (e.g. decision trees or Markov models) that bring together information on costs and health outcomes are used frequently in health economic evaluations. The use of such models may, however, be less feasible in the context of BBD [13]. For example, the modelling of -omics data with underlying biological interactions, multiple -omics profiles, and various time dimensions (e.g. a change in the -omics profile over time) will likely require both novel advanced modelling skills and clinically valid evidence on deep systems biology processes [18, 26]. Dynamic simulation models, in which various parameters and their time-varying behaviour and complexity are modelled, could be one solution to this problem [15]. Additionally, predictive analytic models may increasingly be used for the analysis of big data [10, 15]. Such models are often based on machine-learning techniques and go beyond standard economic probabilistic models.
Clinical Trials
Health economic evaluations alongside clinical trials are key sources of evidence for health intervention implementation. However, trial-based economic evaluations will likely become more complex with the arrival of genomic medicine due to the increasing molecular stratification of trial participants [47]. This complexity will be amplified by the use of BBD to further stratify patients based on data from large observational data sources. Ultimately, such stratification may result in the need for n-of-1 trials [18, 31, 47], in which individual patients are regarded as the single case of observation [48]. A number of challenges have been noted concerning the use of n-of-1 trials to inform economic evaluations. For instance, health outcome measures may be biased due to carryover effects in between two subsequent but different treatments. In addition, questions remain regarding the evaluation of such trials in the context of reimbursement [18]. However, BBD may allow such n-of-1 cases to be modelled on retrospective observational data, which is in line with the idea of utilising real-world data beyond the clinical trial environment. The validity of such real-world evidence will, however, depend on the quality of data used.
Discussion
There is potentially value in incorporating BBD—observational real-world patient-level genomic and clinical data in multiple sub-populations—into economic evaluations of technologies that facilitate precision medicine. However, health economists face practical and methodological challenges when using such data in economic evaluations. The aim of this review was to identify and summarise these challenges. Several challenges were identified related to data management, data quality and data analysis. The availability of large volumes of data from multiple sources and the need to conduct linkages within an environment of opaque data access and unclear sharing procedures can lead to issues in data management. However, these challenges are primarily practical and may not be a long-term obstacle if key stakeholders invest time and effort to improve procedures for data sharing and access. The challenges related to data quality and data analysis may be more difficult to resolve. The use of complex and aggregated heterogeneous BBD sources, of which observational data make up a significant proportion, results in challenges related to data quality, such as missing data or the presence of unobserved confounding. Data quality issues also arise due to the increased potential for missing data when multiple linkages are preformed using datasets developed or maintained by several different stakeholders. Finally, the dynamic nature of genomics and the increase in information provided by BBD will likely require health economists to develop dynamic and potentially more complex economic evaluation methods going forward.
This is the first literature review that has considered the challenges of using BBD for economic evaluations of precision medicine. Whilst other articles have analysed potential issues arising in economic evaluations of precision medicine (e.g. in the context of predictive biomarkers, companion diagnostics and/or digital health applications) [49–51], their focus has not been on BBD. The economic case for NGS technologies in the context of precision medicine has recently been discussed in a special report [52], but the use of linked observational data (e.g. BBD) within economic evaluations of these technologies was not considered, even though some of the existing economic evaluations of NGS technologies use such data [53, 54]. In light of the various national genomic sequencing initiatives, observational real-world patient-level genomic and clinical data will likely be used to an increasing extent. The challenges synthesised as part of this review, including articles from outside the discipline of health economics, add valuable insight on the use of biomedical big data in the context of precision medicine.
The following limitations should, however, be noted. Issues related to the use of BBD are by their nature multidisciplinary, hence the literature search strategy may have missed relevant articles. Given this, our search strategy was designed to minimise false negatives, which meant that included articles were a small proportion of all screened articles. The search syntax and the results were discussed with a librarian specialised in reviews and were considered to be appropriate. A further limitation is that study quality was not assessed. However, few empirical articles on this topic were identified, so it would not have been possible to apply a quality checklist.
Within clinical contexts such as precision cancer genomics and the development of companion diagnostics, genomic data is commonly used in the form of specific genotype variables that support precision treatments based on the molecular profile of a patient [55]. The challenges that we have identified may be less relevant in this context when conducting health economic evaluations. However, these challenges are particularly relevant in the context of rare diseases. Few interventions are available in this context and the value of undergoing genomic sequencing may be justified by receiving a diagnosis and avoiding a diagnostic odyssey. The use of retrospective real-world observational data linked with genomic data is vital when assessing value in this rare disease context (e.g. when establishing the optimal time point for the use of whole-genome sequencing in a diagnostic pathway), but presents many of the challenges documented in this paper.
The lack of economic evaluations of technologies that facilitate precision medicine using BBD supports the view that incorporating such information into economic evaluations in this context is challenging. However, this is an area of research that is still in its infancy. Going forward, machine learning techniques might be able to resolve some of these issues; however, it will be challenging to prove dominance of novel big data analytic techniques over commonly applied statistical methods [40]. For the foreseeable future, health economists who present work that uses BBD should comprehensively document challenges and potential solutions to support future researchers in this field.
Conclusion
Recent years have seen a massive increase in the production of big biomedical data sets with the potential to inform health economic evaluations of precision medicine. Large-scale BBD linkage initiatives will drive the discipline forward, offering health economists opportunities to analyse individuals and patient subgroups on a micro-level using real-world evidence. Although health economists face various challenges at present when using BBD in economic evaluations, potential solutions to some of these challenges do exist [56], and these should be thoroughly tested in the coming years. Solving some of these challenges will likely require effective collaboration between highly skilled individuals from different disciplines (including bio-informatics, statistics and medicine, as well as health economics). The challenge of keeping up with the dynamic developments in the field of big data analytics will have to be overcome if the full potential of BBD is to be realised from a health economic perspective [22, 23, 27].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author Contributions
PF initiated and led the study, designed the literature review, extracted and tabulated data, interpreted the results and drafted the manuscript. JB assisted with the review of the literature, the interpretation of the results, and reviewed and modified the manuscript for important intellectual content. SW assisted with the interpretation of the results, and reviewed and modified the manuscript for important intellectual content. All authors approved the final manuscript. PF acts as the overall guarantor for this work.
Compliance with Ethical Standards
P. F., J. B. and S. W. report no conflicts of interest. P. F. is funded by the German Academic Scholarship Foundation. J. B. and S. W. are funded by the National Institute for Health Research Oxford Biomedical Research Centre.
References
- 1.Stephens ZD, Lee SY, Faghri F, Campbell RH, Zhai C, Efron MJ, et al. Big data: astronomical or genomical? PLoS Biol. 2015;13(7):e1002195. doi: 10.1371/journal.pbio.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372(9):793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raghupathi W, Raghupathi V. Big data analytics in healthcare: promise and potential. Health Inf Sci Syst. 2014;2(1):3. doi: 10.1186/2047-2501-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mittelstadt BD, Floridi L. Introduction. The ethics of biomedical big data. New York: Springer; 2016. p. 3. [Google Scholar]
- 5.Marino P, Touzani R, Perrier L, Rouleau E, Kossi DS, Zhaomin Z, et al. Cost of cancer diagnosis using next-generation sequencing targeted gene panels in routine practice: a nationwide French study. Eur J Hum Genet. 2018;26(3):314–323. doi: 10.1038/s41431-017-0081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alam K, Schofield D. Economic evaluation of genomic sequencing in the paediatric population: a critical review. Eur J Hum Genet. 2018 doi: 10.1038/s41431-018-0175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.NHS England. Creating a genomic medicine service to lay the foundation to deliver personalised interventions and treatments. 2017. https://www.england.nhs.uk/wp-content/uploads/2017/03/board-paper-300317-item-6.pdf. Accessed 17 Dec 2017.
- 8.Centers for Medicare & Medicaid Services. CMS finalizes coverage of Next Generation Sequencing tests, ensuring enhanced access for cancer patients. 2018. https://www.cms.gov/Newsroom/MediaReleaseDatabase/Press-releases/2018-Press-releases-items/2018-03-16.html. Accessed 24 Mar 2018.
- 9.Schwarze K, Buchanan J, Taylor JC, Wordsworth S. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet Med. 2018 doi: 10.1038/gim.2017.247. [DOI] [PubMed] [Google Scholar]
- 10.Lorgelly PK, Doble B, Knott RJ, Investigators C. Realising the value of linked data to health economic analyses of cancer care: a case study of cancer 2015. Pharmacoeconomics. 2016;34(2):139–154. doi: 10.1007/s40273-015-0343-2. [DOI] [PubMed] [Google Scholar]
- 11.Genomics England. The 100,000 Genomes Project. 2018. https://www.genomicsengland.co.uk/the-100000-genomes-project/. Accessed 01 May 2018.
- 12.Onukwugha E. Big data and its role in health economics and outcomes research: a collection of perspectives on data sources, measurement, and analysis. Pharmacoeconomics. 2016;34(2):91–93. doi: 10.1007/s40273-015-0378-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins B. Big data and health economics: strengths, weaknesses, opportunities and threats. Pharmacoeconomics. 2016;34(2):101–106. doi: 10.1007/s40273-015-0306-7. [DOI] [PubMed] [Google Scholar]
- 14.NHS England. National Genomic Test Directories. 2018. https://www.england.nhs.uk/publication/national-genomic-test-directories/. Accessed 16 Oct 2018.
- 15.Marshall DA, Burgos-Liz L, Pasupathy KS, Padula WV, IJzerman MJ, Wong PK, et al. Transforming healthcare delivery: integrating dynamic simulation modelling and big data in health economics and outcomes research. Pharmacoeconomics. 2016;34(2):115–126. doi: 10.1007/s40273-015-0330-7. [DOI] [PubMed] [Google Scholar]
- 16.Dixon P, Davey Smith G, von Hinke S, Davies NM, Hollingworth W. Estimating marginal healthcare costs using genetic variants as instrumental variables: Mendelian randomization in economic evaluation. Pharmacoeconomics. 2016;34(11):1075–1086. doi: 10.1007/s40273-016-0432-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Payakachat N, Tilford JM, Ungar WJ. National Database for Autism Research (NDAR): big data opportunities for health services research and health technology assessment. Pharmacoeconomics. 2016;34(2):127–138. doi: 10.1007/s40273-015-0331-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doble B, Harris A, Thomas DM, Fox S, Lorgelly P. Multiomics medicine in oncology: assessing effectiveness, cost-effectiveness and future research priorities for the molecularly unique individual. Pharmacogenomics. 2013;14(12):1405–1417. doi: 10.2217/pgs.13.142. [DOI] [PubMed] [Google Scholar]
- 19.Grossetta Nardini HK, Wang, L. The Yale MeSH Analyzer. 2018. http://mesh.med.yale.edu/.
- 20.Greenhalgh T, Peacock R. Effectiveness and efficiency of search methods in systematic reviews of complex evidence: audit of primary sources. Bmj. 2005;331(7524):1064–1065. doi: 10.1136/bmj.38636.593461.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Check Hayden E. Genome researchers raise alarm over big data. Nature News. 2015. doi:10.1038/nature.2015.17912.
- 22.He KY, Ge D, He MM. Big data analytics for genomic medicine. Int J Mol Sci. 2017 doi: 10.3390/ijms18020412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alessandrini M, Chaudhry M, Dodgen TM, Pepper MS. Pharmacogenomics and global precision medicine in the context of adverse drug reactions: Top 10 opportunities and challenges for the next decade. OMICS J Integr Biol. 2016;20(10):593–603. doi: 10.1089/omi.2016.0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costa FF. Big data in biomedicine. Drug Discov Today. 2014;19(4):433–440. doi: 10.1016/j.drudis.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 25.Kruse CS, Goswamy R, Raval Y, Marawi S. Challenges and opportunities of big data in health care: a systematic review. JMIR Med Inf. 2016;4(4):e38. doi: 10.2196/medinform.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Guzauskas GF, Gu C, Wang BC, Furnback WE, Xie G, et al. Precision health economics and outcomes research to support precision medicine: big data meets patient heterogeneity on the road to value. J Pers Med. 2016;6(4):20. doi: 10.3390/jpm6040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alyass A, Turcotte M, Meyre D. From big data analysis to personalized medicine for all: challenges and opportunities. BMC Med Genom [Electronic Resource]. 2015;8:33. doi: 10.1186/s12920-015-0108-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patil HK, Seshadri R. Big data security and privacy issues in healthcare. Int Congr Big Data. 2014 [Google Scholar]
- 29.Abouelmehdi K, Beni-Hessane A, Khaloufi H. Big healthcare data: preserving security and privacy. J Big Data. 2018;5(1):1. [Google Scholar]
- 30.Auffray C, Balling R, Barroso I, Bencze L, Benson M, Bergeron J, et al. Making sense of big data in health research: towards an EU action plan. Genome Med. 2016;8(1):71. doi: 10.1186/s13073-016-0323-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertier G, Carrot-Zhang J, Ragoussis V, Joly Y. Integrating precision cancer medicine into healthcare-policy, practice, and research challenges. Genome Med. 2016;8(1):108. doi: 10.1186/s13073-016-0362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asche CV, Seal B, Kahler KH, Oehrlein EM, Baumgartner MG. Evaluation of healthcare interventions and big data: review of associated data issues. Pharmacoeconomics. 2017;35(8):759–765. doi: 10.1007/s40273-017-0513-5. [DOI] [PubMed] [Google Scholar]
- 33.NHS Digital. Data Access Request Service (DARS) charges 2018/2019. 2018. https://digital.nhs.uk/services/data-access-request-service-dars/data-access-request-service-dars-charges-2018-19. Accessed 14 Aug 2018.
- 34.Beckmann JS, Lew D. Reconciling evidence-based medicine and precision medicine in the era of big data: challenges and opportunities. Genome Med. 2016;8(1):134. doi: 10.1186/s13073-016-0388-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiskopf NG, Weng C. Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. J Am Med Inf Assoc. 2013;20(1):144–151. doi: 10.1136/amiajnl-2011-000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.European Medicines Agency. Identifying opportunities for ‘big data’ in medicines development and regulatory science. 2016. http://www.ema.europa.eu/docs/en_GB/document_library/Report/2017/02/WC500221938.pdf. Accessed 20 Mar 2018.
- 37.Lee CH, Yoon HJ. Medical big data: promise and challenges. Kidney Res Clin Pract. 2017;36(1):3–11. doi: 10.23876/j.krcp.2017.36.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berger ML, Mamdani M, Atkins D, Johnson ML. Good research practices for comparative effectiveness research: defining, reporting and interpreting nonrandomized studies of treatment effects using secondary data sources: the ISPOR Good Research Practices for Retrospective Database Analysis Task Force Report—Part I. Value Health. 2009;12(8):1044–1052. doi: 10.1111/j.1524-4733.2009.00600.x. [DOI] [PubMed] [Google Scholar]
- 39.Cox E, Martin BC, Van Staa T, Garbe E, Siebert U, Johnson ML. Good research practices for comparative effectiveness research: approaches to mitigate bias and confounding in the design of nonrandomized studies of treatment effects using secondary data sources: the International Society for Pharmacoeconomics and Outcomes Research Good Research Practices for Retrospective Database Analysis Task Force Report—Part II. Value Health. 2009;12(8):1053–1061. doi: 10.1111/j.1524-4733.2009.00601.x. [DOI] [PubMed] [Google Scholar]
- 40.Rumsfeld JS, Joynt KE, Maddox TM. Big data analytics to improve cardiovascular care: promise and challenges. Nat Rev Cardiol. 2016;13(6):350–359. doi: 10.1038/nrcardio.2016.42. [DOI] [PubMed] [Google Scholar]
- 41.Johnson ML, Crown W, Martin BC, Dormuth CR, Siebert U. Good research practices for comparative effectiveness research: analytic methods to improve causal inference from nonrandomized studies of treatment effects using secondary data sources: the ISPOR Good Research Practices for Retrospective Database Analysis Task Force Report—Part III. Value Health. 2009;12(8):1062–1073. doi: 10.1111/j.1524-4733.2009.00602.x. [DOI] [PubMed] [Google Scholar]
- 42.Brookhart MA, Stürmer T, Glynn RJ, Rassen J, Schneeweiss S. Confounding control in healthcare database research: challenges and potential approaches. Medical care. 2010;48(6 0):S114. [DOI] [PMC free article] [PubMed]
- 43.Department of Health. The UK Strategy for Rare Diseases. 2013. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/260562/UK_Strategy_for_Rare_Diseases.pdf. Accessed 22 Nov 2017.
- 44.Cheng P, Gilchrist A, Robinson KM, Paul L. The risk and consequences of clinical miscoding due to inadequate medical documentation: a case study of the impact on health services funding. Health Inf Manag J. 2009;38(1):35–46. doi: 10.1177/183335830903800105. [DOI] [PubMed] [Google Scholar]
- 45.Murdoch TB, Detsky AS. The inevitable application of big data to health care. Jama. 2013;309(13):1351–1352. doi: 10.1001/jama.2013.393. [DOI] [PubMed] [Google Scholar]
- 46.Sullivan T. Why EHR data interoperability is such a mess in 3 charts. Healthcare IT News. 2018. https://www.healthcareitnews.com/news/why-ehr-data-interoperability-such-mess-3-charts. Accessed 14 Jan 2019.
- 47.Barker R. Precision medicine: what’s all the fuss about? Scand J Clin Lab Investig Suppl. 2016;245:S2–S5. doi: 10.1080/00365513.2016.1206434. [DOI] [PubMed] [Google Scholar]
- 48.Lillie EO, Patay B, Diamant J, Issell B, Topol EJ, Schork NJ. The n-of-1 clinical trial: the ultimate strategy for individualizing medicine? Pers Med. 2011;8(2):161–173. doi: 10.2217/pme.11.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garattini L, Curto A, Freemantle N. Personalized medicine and economic evaluation in oncology: all theory and no practice? Expert Rev Pharmacoecon Outcomes Res. 2015;15(5):733–738. doi: 10.1586/14737167.2015.1078239. [DOI] [PubMed] [Google Scholar]
- 50.IJzerman MJ, Manca A, Keizer J, Ramsey SD. Implementation of comparative effectiveness research in personalized medicine applications in oncology: current and future perspectives. Comp Effect Res. 2015;5:65–72. [Google Scholar]
- 51.Love-Koh J, Peel A, Rejon-Parrilla JC, Ennis K, Lovett R, Manca A, et al. The future of precision medicine: potential impacts for health technology assessment. PharmacoEconomics. 2018;36(12):1439–1451. doi: 10.1007/s40273-018-0686-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gavan SP, Thompson AJ, Payne K. The economic case for precision medicine. Expert Rev Precis Med Drug Dev. 2018;3(1):1–9. doi: 10.1080/23808993.2018.1421858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palmer EE, Schofield D, Shrestha R, Kandula T, Macintosh R, Lawson JA, et al. Integrating exome sequencing into a diagnostic pathway for epileptic encephalopathy: evidence of clinical utility and cost effectiveness. Mol Genet Genom Med. 2018;6(2):186–199. doi: 10.1002/mgg3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schofield D, Alam K, Douglas L, Shrestha R, MacArthur DG, Davis M, et al. Cost-effectiveness of massively parallel sequencing for diagnosis of paediatric muscle diseases. NPJ Genom Med. 2017;2(1):4. doi: 10.1038/s41525-017-0006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daemen A, Griffith OL, Heiser LM, Wang NJ, Enache OM, Sanborn Z, et al. Modeling precision treatment of breast cancer. Genome Biol. 2013;14(10):R110. doi: 10.1186/gb-2013-14-10-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wordsworth S, Doble B, Payne K, Buchanan J, Marshall DA, McCabe C, et al. Using “big data” in the cost-effectiveness analysis of next-generation sequencing technologies: challenges and potential solutions. Value Health. 2018;21(9):1048–1053. doi: 10.1016/j.jval.2018.06.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.