Abstract
In MRI scans of patients with anorexia nervosa (AN), reductions in brain volume are often apparent. However, it is unknown whether such brain abnormalities are influenced by genetic determinants that partially overlap with those underlying AN. Here, we used a battery of methods (LD score regression, genetic risk scores, sign test, SNP effect concordance analysis, and Mendelian randomization) to investigate the genetic covariation between subcortical brain volumes and risk for AN based on summary measures retrieved from genome-wide association studies of regional brain volumes (ENIGMA consortium, n = 13,170) and genetic risk for AN (PGC-ED consortium, n = 14,477). Genetic correlations ranged from − 0.10 to 0.23 (all p > 0.05). There were some signs of an inverse concordance between greater thalamus volume and risk for AN (permuted p = 0.009, 95% CI: [0.005, 0.017]). A genetic variant in the vicinity of ZW10, a gene involved in cell division, and neurotransmitter and immune system relevant genes, in particular DRD2, was significantly associated with AN only after conditioning on its association with caudate volume (pFDR = 0.025). Another genetic variant linked to LRRC4C, important in axonal and synaptic development, reached significance after conditioning on hippocampal volume (pFDR = 0.021). In this comprehensive set of analyses and based on the largest available sample sizes to date, there was weak evidence for associations between risk for AN and risk for abnormal subcortical brain volumes at a global level (that is, common variant genetic architecture), but suggestive evidence for effects of single genetic markers. Highly powered multimodal brain- and disorder-related genome-wide studies are needed to further dissect the shared genetic influences on brain structure and risk for AN.
Electronic supplementary material
The online version of this article (10.1007/s12035-018-1439-4) contains supplementary material, which is available to authorized users.
Keywords: Anorexia nervosa, Brain structure, Genetic correlation
Introduction
Anorexia nervosa (AN) is an often life-threatening, adolescent-onset eating disorder characterized by severe emaciation, and typically by severe food restriction. AN mortality rates are the highest in psychiatry [1].
Despite high twin-based heritability estimates of around 50–80% for AN [2], we do not fully understand the pathophysiology of the disorder. So far only one common variant on chromosome 12 and no rare variants above genome-wide thresholds have been detected in four studies based on up to 3500 AN cases and 15,000 controls [3–6].
In acutely ill patients with AN, reduction of brain tissue is often readily visible in individual patients’ brain scans. A number of structural MRI studies (albeit with small sample sizes) have documented decreases in both gray and white matter volumes (for review, see ref. [7, 8]). More recent, larger studies and a meta-analysis have confirmed widespread gray matter volume reductions in AN, with regional effects especially in reward-related and somatosensory areas [9–11]. In weight-recovered patients, subcortical and cortical gray matter deficits seemed to normalize, but several studies have reported small group differences even after recovery [9, 12–14].
In light of these structural abnormalities, one question is whether certain structural brain differences share a similar genetic architecture with AN and may possibly even reflect a predisposition for AN. Magnetic resonance imaging (MRI) measures of brain structure are moderately heritable [15] and several volumetric measures have high reproducibility and low measurement error [16]. Ideally, an answer to the above question might require longitudinal, prospective population-based studies with brain scans in healthy individuals who will go on to develop AN later in life. Considering the high cost, low power, and logistical challenges of such a study, a more feasible alternative approach would be to examine the genetic overlap between AN and brain structure. Unfortunately, the number of existing genetic and neuroimaging studies in AN is relatively small compared to other major neuropsychiatric disorders. To date, only two imaging genetics studies have been published [17, 18], suggesting that COMT and 5-HTTLPR genotype may modulate functional connectivity in AN patients.
We are now able to leverage results from large-scale, genome-wide association studies (GWAS) based on tens of thousands of individuals. Data from GWAS on brain structure and on genetic risk for AN allows us to investigate the genetic covariation between brain structure and disease risk. Investigating this genetic correlation should inform us about shared genetic influences between brain structure and AN. A large degree of genetic overlap could indicate potential pleiotropic effects, where the same genetic variants influence both traits. Results could then help to derive or adapt hypotheses about how brain structure is involved in AN etiology. Several groups have investigated genetic overlap between structural brain measures and risk for schizophrenia as well as other psychiatric disorders [19–22].
In the current study, we followed a roadmap for the analysis of genetic covariation between brain volumes and AN using a battery of complementary methods including those suggested by Franke et al. [20]. To date, well-powered summary estimates are available for eight structural brain measures (intracranial volume (ICV) and seven subcortical regions) from published GWAS [16]. Therefore, our analysis focused on ICV and these regional subcortical volumes. In detail, we investigated the potential for a shared genetic architecture based on common genetic variation as well as of an overlap of individual genetic risk variants between both disorder and brain measures.
Materials and Methods
In this article, we used independent data from separate GWAS on brain structure and on genetic risk for AN to study the genetic covariation between these measures.
Samples
Subcortical Brain Volume GWAS Summary Statistics from the Enhancing NeuroImaging Genetics Through Meta-Analysis Consortium (ENIGMA)
ENIGMA MRI summary measures from genetic association analyses of ICV and seven subcortical volumes [16] were available online at http://enigma.usc.edu/research/download-enigma-gwas-results/. These analyses were based on brain MRI scans and genome-wide genotype data for 13,170 subjects from 28 cohorts Online Resource, section 1.1 and Table S1). All participants in all cohorts in this study gave written informed consent and sites involved obtained approval from local research ethics committees or Institutional Review Boards. In the original analysis, GWAS statistics from each of the 28 sites had been combined using a fixed-effect inverse variance-weighted meta-analysis as implemented in METAL [23].
Anorexia Nervosa GWAS Summary Statistics from the Eating Disorders Working Group of the Psychiatric Genomics Consortium (PGC-ED)
Anorexia nervosa (AN) cases met DSM-IV criteria for either lifetime AN (restricting or binge-purge subtype) or lifetime eating disorders ‘not otherwise specified’ AN-subtype (i.e., exhibiting the core features of AN) [24]. Summary measures were available on genetic association analyses on AN diagnosis (https://www.med.unc.edu/pgc/results-and-downloads) [4]. These analyses were based on AN phenotype and genome-wide genotype data for 3495 AN cases and 10,982 control subjects from 12 cohorts [3, 6] (Table S2). To our knowledge, PGC-ED AN cohorts did not overlap with cohorts in the current ENIGMA GWAS. Participants clustered with subjects of known European ancestry. Genomic data were imputed to a reference panel (1000 Genomes, phase 3), using SHAPEIT [25] for phasing and IMPUTE2 for imputation [26]. Tests of AN association within datasets were performed using an additive model in PLINK [27] with the first ten principal components as covariates. Fixed-effects meta-analysis across the 12 datasets was carried out using METAL [23] with inverse variance weighting. For more information, see Online Resource section 1.2 and reference [4].
Statistical Analysis
Linkage Disequilibrium Score Regression
Linkage disequilibrium score (LDSC) regression [28] was used to assess genome-wide common variant heritability and genetic correlations between AN and subcortical volumes. In detail, LDSC assesses whether inflation in GWAS test statistics is due to polygenicity or other confounding biases such as cryptic relatedness or population stratification. For this analysis, each dataset was filtered to only include markers overlapping with HapMap Project Phase 3 SNPs (Noverlap = 1,161,164), as these tend to be well-imputed across studies and alleles will match those listed in the data used to estimate the LD score. No SNPs had out-of-bound p values or were strand-ambiguous. Because ENIGMA subcortical brain volume and PGC-ED AN measures were based on European samples, we used pre-computed LD scores for European populations, as provided on the LDSC website (https://github.com/bulik/ldsc). Standard errors were estimated using a block jackknife procedure and used to calculate p values. p values were adjusted for seven tests (eight brain regions minus the amygdala; see Franke et al., [20] and Results below) to account for multiple testing.
Genetic Risk Score Analysis
We used the grs.summary function developed by Johnson [29] and implemented in PRSice (version 1.25, ref. [30]), which approximates the regression of a response variable (i.e., risk for AN based on PGC-ED GWAS) onto an additive multi-SNP genetic risk score. Risk score coefficients are weighted by single SNP regression coefficients estimated from one set of GWAS results (here: ENIGMA subcortical brain volume). We investigated the effect at four p value thresholds (1*10−4, 1*10−3, 1*10−2, 5*10−2) and adjusted for 28 tests (eight brain regions minus the amygdala * four thresholds) to account for multiple testing.
Sign Test
We employed a sign test as an additional method to investigate a potential overlap of positive or inverse direction effects of SNPs between both datasets at p value thresholds (1*10−4, 1*10−3, 1*10−2, 5*10−2). Using the binom.test function from the stats package in R, we tested the significance of the number of SNPs with opposite direction effects between datasets at these four thresholds over the total number of SNPs. p values were adjusted for 28 tests (eight brain regions minus the amygdala * four thresholds) to account for multiple testing.
SNP Effect Concordance Analysis
SNP effect concordance analysis (SECA) tests for pleiotropy, concordance, and ‘pleiotropy-informed’ conditional false discovery rate (FDR) results between two sets of GWAS summary results [31]. SECA estimates whether (a) there is an excess of SNPs associated with the respective phenotype in both datasets (pleiotropy); (b) the directions of effect are in agreement across datasets (concordance); and (c) single SNPs in dataset 2 (here: PGC-ED AN) gain in significance after conditioning on their strength of association in dataset 1 (here: ENIGMA subcortical brain volumes; conditional results). Concordance analysis in SECA tests for positive concordance (i.e., whether a larger ORAN relates to a larger BETAsubcortical). However, since we were interested in the opposite relationship between these variables (i.e., whether a ORAN greater than one relates to a negative BETAsubcortical), we derived and used the inverse of the ORAN in the concordance analysis. For all SECA analyses, overlapping SNPs between both datasets (N = 7,868,363) were pruned for LD using a p value informed method, a 1 Mb window and r2 > 0.1 (all default settings in SECA). This resulted in N = 26,558 SNPs, which were entered in the analysis. p values were adjusted for seven tests (eight brain regions minus the amygdala; see Franke et al., [20] and Results below) to account for multiple testing.
Mendelian Randomization
To investigate potentially causal relationships between subcortical brain volumes and AN, we applied a two-sample Mendelian Randomization (MR) approach, which only requires GWAS summary level data. MR is a method of investigating causal relationships by using genetic variants as instrumental variables [32]. The main assumptions and strengths of the technique have been outlined in detail elsewhere [33–35]. Briefly, to select SNPs that are strong instrumental variables (relevance assumption), we investigated only brain volumes and SNPs where the genetic variants associated with brain volume at a genome-wide level significant level (caudate (1 SNP), hippocampus (2 SNPs), putamen (4 SNPs, of which 3 were available in the AN GWAS summary data), and ICV (1 SNP); as reported in Hibar et al. [16]). We were not able to investigate the causal effect of AN on brain volumes, as the AN-linked variant (rs4622308) was not available in the subcortical GWAS summary data. Due to the limited number of strongly associated genetic variants per structure, we used the Wald ratio method and hence were unable to investigate presence of horizontal pleiotropy as a potential violation of the MR exclusion restriction assumption. To limit confounding due to population stratification (a potential violation of the independence assumption), we used GWAS summary data based on largely European populations. The TwoSampleMR package in R (also available as part of the MR-Base (www.mrbase.org) platform [36] was used for all MR analyses.
Results
The following analyses were based on summary statistics for (a) eight brain volume measures of 13,170 participants from the ENIGMA consortium and (b) AN case-control data from 3495 AN patients and 10,982 healthy individuals. We focused on ICV and all subcortical regions (caudate, hippocampus, pallidum, nucleus accumbens, putamen, thalamus, amygdala) that were investigated in [16], and hence had GWAS summary data available.
Linkage Disequilibrium Score Regression
Linkage disequilibrium score (LDSC) regression examines the relationship between two sets of GWAS test statistics using linkage disequilibrium. Restricting our analyses to Hapmap3 SNPs (as recommended, see methods) left 1,161,164 SNPs in the ENIGMA datasets and 1,217,311 in the AN dataset with a total of 1,094,348 overlapping SNPs. SNP-based heritability estimates for the traits were 26% (caudate, 95% CI [18; 34]), 15% (hippocampus [95% CI 7; 23] and pallidum [95% CI 7; 24]), 9% (nucleus accumbens, 95% CI [1; 17]), 31% (putamen; 95% CI [21; 41]), 14% (thalamus; 95% CI [6; 22]), 18% (ICV; 95% CI [9; 27]), and 23% (AN; 95% CI [16; 31]; Table 1). The GWAS-estimated heritability for the amygdala volume was not significantly different from zero (− 2%, 95% CI [− 9; 4]). This structure was therefore omitted from the following analyses. Using LDSC regression, we did not identify a significant genetic correlation between any of the remaining seven brain volumes and AN (Table 1).
Table 1.
Genetic overlap analyses between anorexia nervosa (AN) and seven subcortical brain volumes
| ROI | LDSC | GRSa | Sign testa | SECA | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Volume | h2 in % | 95% CI | rg | se | p(rg) | R2 | p(GRS) | p threshold (brain volume) | Estimateb | 95% CI | p(binomial) | p threshold (brain volume) | p(pleiotropy) | p(concordance) |
| Caudate | 25.65 | [17.61,33.69] | 0.14 | 0.11 | 0.19 | 9.55E-05 | 0.120 | 1E-04 | 0.513 | [0.49,0.535] | 0.268 | 1E-02 | 1.000 | 1.000 |
| Hippocampus | 14.86 | [6.61,23.11] | − 0.06 | 0.15 | 0.66 | 3.42E-04 | 0.013c | 1E-02 | 0.522 | [0.499,0.544] | 0.061 | 1E-02 | 1.000 | 0.138 |
| Nucleus accumbens | 8.93 | [0.76,17.1] | 0.14 | 0.19 | 0.45 | 1.43E-04 | 0.075 | 1E-03 | 0.450 | [0.384,0.518] | 0.159 | 1E-03 | 0.057 | 0.052 |
| Pallidum | 15.25 | [6.84,23.66] | 0.10 | 0.14 | 0.47 | 2.65E-04 | 0.025c | 1E-02 | 0.522 | [0.499,0.545] | 0.065 | 1E-02 | 1.000 | 0.311 |
| Putamen | 30.80 | [20.78,40.82] | 0.05 | 0.10 | 0.61 | 1.26E-04 | 0.089 | 1E-04 | 0.438 | [0.374,0.503] | 0.061 | 1E-03 | 0.153 | 0.170 |
| Thalamus | 13.74 | [5.78,21.7] | − 0.10 | 0.15 | 0.49 | 2.23E-04 | 0.036c | 5E-02 | 0.512 | [0.502,0.523] | 0.019d | 5E-02 | 0.160 | 0.009c,d |
| ICV | 17.96 | [8.59,27.33] | 0.23 | 0.14 | 0.10 | 3.28E-04 | 0.015c | 1E-03 | 0.560 | [0.495,0.623] | 0.072 | 1E-03 | 1.000 | 0.078 |
ROI, region of interest; ICV, intracranial volume; LDSC, LD score regression; GRS, genetic risk score; SECA, SNP effect concordance analysis; amygdala volume was omitted from the analyses because of low heritability; heritability estimates on the observed scale; rg = genetic correlation with AN; p(rg) = p value of the genetic correlation; se = standard error; CI = confidence interval; a tests were carried out on subsets of four different thresholds (1*10−4, 1*10−3, 1*10−2, 5*10−2) in the discovery dataset (i.e., ENIGMA subcortical brain volume), but only the strongest results are reported in the table. For detailed results, see Tables S3 and S4. b An estimate > 0.5 indicates a higher-than-chance proportion of SNPs with a risk effect on brain volume (BETA<0) and on AN (OR > 1); c nominally significant at p < 0.05; d trend significance after Bonferroni-correction for 7 tests (number of brain volumes)
Genetic Risk Score Analysis
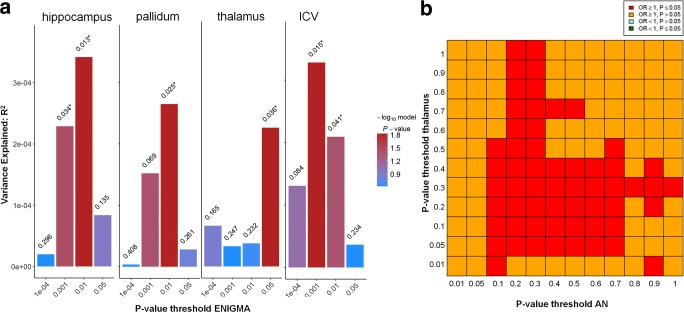
Next, we tested the effect of additive multi-SNP genetic risk scores, weighted by ENIGMA subcortical brain volume betas, onto PGC-ED AN, to see whether risk for altered subcortical brain volume is related to risk for AN. No effects were observed after correcting for 28 tests (seven brain regions * four thresholds). For four brain regions (hippocampus, pallidum, thalamus, and ICV), nominal effects were observed, but the amount of explained variance was negligible (less than 0.034%). See Table 1, Fig. 1a and Table S3 for further details.
Fig. 1.
a Genetic risk score analysis results for four subcortical brain volumes with nominally significant results. p value criteria used to threshold ENIGMA input data are plotted on the x-axis, the amount of variance of AN liability explained (R2) on the y-axis. The color bar indicates the level of significance for a GRS effect on AN. b SECA analysis indicated significant concordance effects between AN (x-axis) and thalamus volume (y-axis). For computational purposes, ORAN was inversely coded, so that red indicates concordance of SNP effects between increased risk for AN and lower thalamus volume
Sign Test
Testing for an accumulation of positive or negative direction effects of SNPs in the PGC-ED AN and ENIGMA datasets at four different ENIGMA p value thresholds (1*10−4, 1*10−3, 1*10−2, 5*10−2), we could not identify any significant effects after adjusting for multiple testing. There was a nominally significant effect (pthreshold < 0.05 = 0.019) for the thalamus, where we found a higher proportion of SNP alleles with a negative effect on brain volume (BETA < 0) and an increased risk for AN (OR > 1) than would be expected by chance. This effect was not found when restricting the analyses to SNPs passing thresholds based on the AN GWAS results instead. See Table 1 and Table S4 for further details.
SNP Effect Concordance Analysis
SNP effect concordance analysis (SECA) extracts subsets of independent SNPs (present in both datasets) to test at 12 different p value thresholds (resulting in 144 subsets) for an excess of SNPs associated in both datasets (pleiotropy), for concordance of effect directions, and for conditional effects. After LD-pruning, 26,558 SNPs were left in the analysis. There was an overall effect for an inverse concordance between the thalamus and AN (i.e., SNP effects related to smaller thalamus volume and larger ORAN), which remained significant only at a trend level after correcting for seven tests (uncorrected pconcordance = 0.009; Table 1 and Fig. 1b). There were no other signs of pleiotropy or concordance between any of the remaining volumes and AN (Table 1). Testing for conditional effects, we identified individual SNPs that gained in association significance with AN after conditioning on their strength of association with subcortical volume for the caudate, hippocampus, nucleus accumbens and the pallidum. In detail, rs3863294 on chromosome 11—located close to the genes ZW10 (involved in cell division) and TMPRSS5 (highly expressed in brain tissue) and also in the vicinity of neurotransmitter and immune system relevant genes such as DRD2, HTR3B, HTR3A, and NCAM (Fig. S1)—was significantly associated with AN only after conditioning on its association with caudate volume (pFDR-noCond = 0.324 to pFDR-caudCond = 0.0246). Follow-up analyses on functional effects of rs3863294 on DNA methylation or gene expression indicated an influence on DNA methylation linked to ZW10 in blood, but not in brain tissue and an effect on gene expression of TMPRSS5 in brain tissue and of DRD2 in peripheral tissue (Online Resource section 2.4).
Similarly, rs7945461—linked to LRRC4C which is important in axonal and synaptic development—gained significance after conditioning on hippocampal volume (pFDR-noCond = 0.272 to pFDR-hipp-Cond = 0.0208). Two SNPs that were conditional on volumes of the nucleus accumbens and the pallidum (rs6708971; pFDR-noCond = 0.127 to pFDR-accumb-Cond = 0.009; and rs873507; pFDR-noCond = 0.093 to pFDR-pall-Cond = 0.034) were located in intergenic regions. For further details on these conditional analyses, see Online Resource section 2.5.
Mendelian Randomization
To assess potential causal effects of brain volume on AN, genetic variants linked to each of the four brain volumes with genome-wide level markers were used as instruments. Wald ratios did not indicate causal effects of any brain volume on risk for AN (Table S5).
Discussion
We evaluated the relationship between common genetic variants implicated in subcortical brain volumes and those associated with a clinical diagnosis of AN. The sample sizes were the largest yet applied to these questions. With a comprehensive set of analyses, we found weak evidence for strong genetic correlations at a global level (that is, common variant genetic architecture) and suggestive evidence for effects of single genetic markers.
There were signs of an inverse concordance between thalamus volume and risk for AN (i.e., SNP effects related to smaller thalamus volume and larger ORAN). For individual genetic variants, we identified a variant in the vicinity of neurotransmitter, development and immune system relevant genes such as ZW10, TMPRSS5 and DRD2, HTR3B, HTR3A, and NCAM to be significantly associated with AN only after conditioning on its association with caudate volume. Another variant linked to LRRC4C, important in axonal and synaptic development, gained significance after conditioning on hippocampal volume.
Several reasons could explain why we did not detect a major genetic overlap between AN and subcortical volumes in the current study. In this study, we focused on volume alterations in subcortical regions which are potentially implicated in AN, but other neuroimaging phenotypes, such as measures of cortical thickness or surface area as well as resting-state functional or anatomical connectivity, may also be informative. Similarly, genetic risk for AN might relate to specific cell types or structures that do not easily relate to those properties detected by the structural imaging approach applied in this study. Second, it is possible that the samples in the PGC-ED GWAS are somewhat heterogeneous with respect to disease severity or subtype. More robust differences might be found when larger sample sizes and deeply phenotyped data allow us to focus on, e.g., subtype effects (restrictive versus binge-purge) or select individuals with more severe and enduring psychopathologies or those with particularly high genetic risk for AN. Third, the data analyzed in this study are largely based on adult samples. Heritability estimates vary with age and the degree of genetic overlap between subcortical volume and AN may be larger in younger populations. Fourth, we evaluated only common genetic variation. It is possible that rare variants play a role in the shared genetic architecture between brain volume and AN. Last but not least, it is indeed possible that there is no link between the genetics of subcortical volumes and AN. However, the results of the conditional analysis draw such a conclusion into question.
Our results are similar to other studies that reported a lack of a shared genetic basis between brain volumes and schizophrenia as well as major depression [20, 22]. However, another recent study reported an enriched contribution of schizophrenia GWAS loci to intracranial volume (a global measure of brain morphology; Lee et al. [21]). The latter study also included several other psychiatric traits as part of an exploratory analysis and found no associations between risk for AN and intracranial volume. It is likely that genetic variants that are linked to complex disorders such as AN may exert their effects through various biological pathways affecting different systems to varying degrees. This high level of heterogeneity might prevent us from identifying more distinct genetic signals on brain volume. Alternative methods such as those described in Smeland et al. [37] and Lee et al. [21], or parallel ICA [38], some of which are able to delineate independent genetic signals on distinct brain networks might provide a potentially promising approach to study the relationship between brain volume and AN.
Despite our negative findings, several studies have reported genetic correlations between AN and different clinical traits. A significant twin-based genetic correlation between AN and OCD has been observed using data from the population-based Swedish Twin Registry (additive genetic correlation = 0.52; ref. [39]) and has since been replicated with a SNP-based genetic correlation by the PGC consortium (rg = 0.53; ref. [40]). A recent study [4] has found positive genetic correlations between AN and a range of psychiatric traits including schizophrenia and neuroticism, perhaps reflecting genetic risk for general psychopathology. Negative genetic associations were observed with several “unfavorable” metabolic phenotypes (such as fasting insulin, fasting glucose or insulin resistance), suggesting that metabolic factors might be involved in dysregulation of weight and appetite in AN.
We detected an inverse concordance between genetic determinants of thalamus volume and AN. Gray matter atrophy in the thalamus in AN has been reported in previous studies [10, 14] and diffusion-based MRI investigations have frequently implicated alterations of white matter tracts connecting the thalamus and especially fronto-parietal regions with AN [41–43]. Studies using functional MRI have also found differences in thalamus functioning in AN [44], indicating that this region might be involved in motivation-related behavior in AN. Moreover, thalamo-frontal circuit abnormalities in AN (measured using an fMRI-based resting-state functional connectivity approach) were linked to cognitive control and working memory performance in patients [45]. Reduced local thalamic network efficiency (indicated through decreased connectivity strength and increased path length) in patients with AN was further supported using an approach that modeled the entire brain as a complex network [46, 47]. These results are in line with a model for human awareness and subjectivity by Craig et al. [48], which postulates that afferent representations of the physiological state of the body ascend from the spinal cord via the brain stem and the thalamus to the insula. Therefore, reduced connectivity in a network including the thalamus might reflect a modified calibration of signals such as body size or hunger and may subsequently contribute to typical AN symptoms. Such abnormalities in signal processing might be genetically determined, arise during neurodevelopment and manifest in altered thalamus volumes and connectivity. However, considering largely trend level effects in our analyses, as well as the fact that we could not investigate causal associations through Mendelian randomization due to the absence of strong genetic loci associated with thalamus volume, we cannot draw any conclusions about the causality or direction of effect (i.e., whether reduced thalamus volume is a risk factor or consequence of AN or whether this association is due to confounding).
We also found that genetic variant rs3863294, located close to the genes ZW10 (involved in cell division) and TMPRSS5 (highly expressed in brain tissue) and also in the vicinity of neurotransmitter and immune system relevant genes such as DRD2, HTR3B, HTR3A, and NCAM, was significantly associated with AN after conditioning on its genetic association with caudate volume. A direct link between rs3863294 and these genes deserves further investigation. However, it is interesting to note that prior candidate gene studies suggest that many genes found in this region could be associated with AN. For instance, genetic variants associated with the serotonergic system (linked to the genes HTR3A and HTR3B) may be associated with the restrictive subtype of AN [49] and genetic variants of genes involved in the dopaminergic system (e.g., DRD2) might play a role in the susceptibility for AN in some populations [50, 51]. Interestingly the caudate is a brain structure that is strongly modulated by ascending dopaminergic projections [52] and implicated in reward processing [53, 54]. A number of PET studies indicate that aberrant striatal dopamine function may contribute to the behavioral phenotype in AN [55], although findings might be specific to the stage of recovery. For instance, the interaction between dopamine receptor and serotonin transporter binding was predictive of harm avoidance in recovered eating disorder patients [56, 57]. AN-related alterations in dopaminergic reward-related brain regions such as the caudate have also been shown using fMRI and monetary rewards, taste- or food-related stimuli [58]. However, the direction of change (increased or decreased activation) varied across studies [59–64].
Last, the genetic variant rs7945461—linked to LRRC4C, important in axonal and synaptic development [65, 66]—gained significance after conditioning on hippocampal volume. LRRC4C mRNA is abundant in hippocampal pyramidal neurons and dentate granule cells [67] and several studies have reported hippocampal volume reduction in acute AN patients relative to controls [7]. There has been no direct link so far between cognitive function and hippocampal alterations in AN [68], but there might be an indirect effect via estrogen-related hormone levels, which have been associated both with cognitive performance in AN [69] and with hippocampal volume regeneration upon weight restoration [14].
Conclusion
In this comprehensive set of analyses, we found weak evidence for a relationship between common genetic variants implicated in AN and those associated with subcortical brain volumes at a high level (that is, common variant genetic architecture), but some suggestive evidence for effects of single genetic markers. Despite the sample sizes being the largest yet applied to these questions, more detailed multimodal brain- and genome-wide studies are needed to dissect the potential impact of genetic risk for AN on brain structure or function.
Electronic Supplementary Material
(DOCX 773 kb)
Acknowledgements
We would like to thank the following members of the ENIGMA Genetics Working Group and PGC-ED consortium who were involved in the respective GWAS analyses:
ENIGMA Genetics Working Group
Derrek P. Hibar, Jason L. Stein, Miguel E. Renteria, Alejandro Arias- Vasquez, Sylvane Desrivières, Neda Jahanshad, Roberto Toro, Katharina Wittfeld, Lucija Abramovic, Micael Andersson, Benjamin S. Aribisala, Nicola J. Armstrong, Manon Bernard, Marc M. Bohlken, Marco P. Boks, Janita Bralten, Andrew A. Brown, M. Mallar Chakravarty, Qiang Chen, Christopher R.K. Ching, Gabriel Cuellar- Partida, Anouk den Braber, Sudheer Giddaluru, Aaron L. Goldman, Oliver Grimm, Tulio Guadalupe, Johanna Hass, Girma Woldehawariat, Avram J. Holmes, Martine Hoogman, Deborah Janowitz, Tianye Jia, Sungeun Kim, Marieke Klein, Bernd Kraemer, Phil H. Lee, Loes M. Olde Loohuis, Michelle Luciano, Christine Macare, Karen A. Mather, Manuel Mattheisen, Yuri Milaneschi, Kwangsik Nho, Martina Papmeyer, Adaikalavan Ramasamy, Shannon L. Risacher, Roberto Roiz-Santiañez, Emma J. Rose, Alireza Salami, Philipp G. Sämann, Lianne Schmaal, Andrew J. Schork, Jean Shin, Lachlan T. Strike, Alexander Teumer, Marjolein M.J. van Donkelaar, Kristel R. van Eijk, Raymond K. Walters, Lars T. Westlye, Christopher D. Whelan, Anderson M. Winkler, Marcel P. Zwiers, Saud Alhusaini, Lavinia Athanasiu, Stefan Ehrlich, Marina M.H. Hakobjan, Cecilie B. Hartberg, Unn K. Haukvik, Angelien J.G.A.M. Heister, David Höhn, Dalia Kasperaviciute, David C.M. Liewald, Lorna M. Lopez, Remco R.R. Makkinje, Mar Matarin, Marlies A.M. Naber, David R. McKay, Margaret Needham, Allison C. Nugent, Benno Pütz, Natalie A. Royle, Li Shen, Emma Sprooten, Daniah Trabzuni, Saskia S.L. van der Marel, Kimm J.E. van Hulzen, Esther Walton, Christiane Wolf, Laura Almasy, David Ames, Sampath Arepalli, Amelia A. Assareh, Mark E. Bastin, Henry Brodaty, Kazima B. Bulayeva, Melanie A. Carless, Sven Cichon, Aiden Corvin, Joanne E. Curran, Michael Czisch, Greig I. de Zubicaray, Allissa Dillman, Ravi Duggirala, Thomas D. Dyer, Susanne Erk, Iryna O. Fedko, Luigi Ferrucci, Tatiana M. Foroud, Peter T. Fox, Masaki Fukunaga, J. Raphael Gibbs, Harald H.H. Göring, Robert C. Green, Sebastian Guelfi, Narelle K. Hansell, Catharina A. Hartman, Katrin Hegenscheid, Andreas Heinz, Dena G. Hernandez, Dirk J. Heslenfeld, Pieter J. Hoekstra, Florian Holsboer, Georg Homuth, Jouke- Jan Hottenga, Masashi Ikeda, Clifford R. Jack Jr., Mark Jenkinson, Robert Johnson, Ryota Kanai, Maria Keil, Jack W. Kent Jr., Peter Kochunov, John B. Kwok, Stephen M. Lawrie, Xinmin Liu, Dan L. Longo, Katie L. McMahon, Eva Meisenzahl, Ingrid Melle, Sebastian Mohnke, Grant W. Montgomery, Jeanette C. Mostert, Thomas W. Mühleisen, Michael A. Nalls, Thomas E. Nichols, Lars G. Nilsson, Markus M. Nöthen, Kazutaka Ohi, Rene L. Olvera, Rocio Perez-Iglesias, G. Bruce Pike, Steven G. Potkin, Ivar Reinvang, Simone Reppermund, Marcella Rietschel, Nina Romanczuk-Seiferth, Glenn D. Rosen, Dan Rujescu, Knut Schnell, Peter R. Schofield, Colin Smith, Vidar M. Steen, Jessika E. Sussmann, Anbupalam Thalamuthu, Arthur W. Toga, Bryan J. Traynor, Juan Troncoso, Jessica A. Turner, Maria C. Valdés Hernández, Dennis van ‘t Ent, Marcel van der Brug, Nic J.A. van der Wee, Marie-Jose van Tol, Dick J. Veltman, Thomas H. Wassink, Eric Westman, Ronald H. Zielke, Alan B. Zonderman, David G. Ashbrook, Reinmar Hager, Lu Lu, Francis J. McMahon, Derek W. Morris, Robert W. Williams, Han G. Brunner, Randy L. Buckner, Jan K. Buitelaar, Wiepke Cahn, Vince D. Calhoun, Gianpiero L. Cavalleri, Benedicto Crespo-Facorro, Anders M. Dale, Gareth E. Davies, Norman Delanty, Chantal Depondt, Srdjan Djurovic, Wayne C. Drevets, Thomas Espeseth, Randy L. Gollub, Beng-Choon Ho, Wolfgang Hoffmann, Norbert Hosten, René S. Kahn, Stephanie Le Hellard, Andreas Meyer-Lindenberg, Bertram Müller-Myhsok, Matthias Nauck, Lars Nyberg, Massimo Pandolfo, Brenda W.J.H. Penninx, Joshua L. Roffman, Sanjay M. Sisodiya, Jordan W. Smoller, Hans van Bokhoven, Neeltje E.M. van Haren, Henry Völzke, Henrik Walter, Michael W. Weiner, Wei Wen, Tonya White, Ingrid Agartz, Ole A. Andreassen, John Blangero, Dorret I. Boomsma, Rachel M. Brouwer, Dara M. Cannon, Mark R. Cookson, Eco J.C. de Geus, Ian J. Deary, Gary Donohoe, Guillén Fernández, Simon E. Fisher, Clyde Francks, David C. Glahn, Hans J. Grabe, Oliver Gruber, John Hardy, Ryota Hashimoto, Hilleke E. Hulshoff Pol, Erik G. Jönsson, Iwona Kloszewska, Simon Lovestone, Venkata S. Mattay, Patrizia Mecocci, Colm McDonald, Andrew M. McIntosh, Roel A. Ophoff, Tomas Paus, Zdenka Pausova, Mina Ryten, Perminder S. Sachdev, Andrew J. Saykin, Andrew Simmons, Andrew Singleton, Hilkka Soininen, Joanna M. Wardlaw, Michael E. Weale, Daniel R. Weinberger, Hieab H.H. Adams, Lenore J. Launer, Stephan Seiler, Reinhold Schmidt, Ganesh Chauhan, Claudia L. Satizabal, James T. Becker, Lisa R. Yanek, Sven J. van der Lee, Maritza Ebling, Bruce Fischl, W.T. Longstreth, Douglas Greve, Helena Schmidt, Paul Nyquist, Louis N. Vinke, Cornelia M. van Duijn, Xue Luting, Bernard Mazoyer, Joshua C. Bis, Vilmundur Gudnason, Sudha Seshadri, M. Arfan Ikram, Nicholas G. Martin, Margaret J. Wright, Gunter Schumann, Barbara Franke, Paul M. Thompson, Sarah E. Medland.
Eating Disorders Working Group of the Psychiatric Genomics Consortium
Laramie Duncan, PhD; Zeynep Yilmaz, PhD; Raymond Walters, PhD; Jackie Goldstein, PhD; Verneri Anttila, PhD; Brendan Bulik-Sullivan, PhD; Stephan Ripke, MD, PhD; Roger Adan, PhD; Lars Alfredsson, PhD; Tetsuya Ando, MD, PhD; Ole Andreassen, MD, PhD; Harald Aschauer, MD; Jessica Baker, PhD; Jeffrey Barrett, PhD; Vladimir Bencko, MD, PhD; Andrew Bergen, PhD; Wade Berrettini, MD, PhD; Andreas Birgegård, PhD; Claudette Boni, PhD; Vesna Boraska Perica, PhD; Harry Brandt, MD; Roland Burghardt, MD; Laura Carlberg, MD; Matteo Cassina, MD; Carolyn Cesta Sven Cichon, PhD; Maurizio Clementi, MD; Sarah Cohen-Woods, PhD; Joni Coleman, MSc; Roger Cone, PhD; Philippe Courtet, MD; Steven Crawford, MD; Scott Crow, MD; Jim Crowley, PhD; Unna Danner, PhD; Oliver Davis, MSc, PhD; Martina de Zwaan, MD; George Dedoussis, PhD; Daniela Degortes, PhD; Janiece DeSocio, PhD;, RN, PMHNP-BC Danielle Dick, PhD; Dimitris Dikeos, MD; Christian Dina, PhD; Bo Ding, PhD; Monika Dmitrzak-Weglarz, PhD; Elisa Docampo, MD, PhD; Karin Egberts, MD; Stefan Ehrlich, MD; Geòrgia Escaramís, PhD; Tõnu Esko, PhD; Thomas Espeseth, PhD; Xavier Estivill, MD, PhD; Angela Favaro, MD, PhD; Fernando Fernández-Aranda, PhD; FAED Manfred Fichter, MD, Dipl-Psych Chris Finan, PhD; Krista Fischer, PhD; James Floyd, PhD; Manuel Föcker, MD; Lenka Foretova, MD, PhD; Monica Forzan, PhD; Caroline Fox, MD; Christopher Franklin, PhD; Valerie Gaborieau Steven Gallinger, MD; Giovanni Gambaro, MD, PhD; Héléna Gaspar, PhD; Ina Giegling, PhD; Fragiskos Gonidakis, MD; Philip Gorwood, MD, PhD; Monica Gratacos, MD, PhD; Sébastien Guillaume, MD, PhD; Yiran Guo, PhD; Hakon Hakonarson, MD, PhD; Katherine Halmi, MD; Rebecca Harrison Konstantinos Hatzikotoulas, MD, PhD; Joanna Hauser, MD, PhD; Johannes Hebebrand, MD; Sietske Helder, PhD; Judith Hendriks, BSc Stefan Herms, PhD; Beate Herpertz-Dahlmann, MD; Wolfgang Herzog, MD; Christopher Hilliard, BS; Laura Huckins, PhD; James Hudson, MD, ScD; Julia Huemer, MD; Hartmut Imgart, MD; Hidetoshi Inoko, PhD; Sigrid Jall Stephane Jamain, PhD; Vladimir Janout, PhD; Susana Jiménez-Murcia, PhD; Craig Johnson, PhD; Jenny Jordan, PhD; Antonio Julià, PhD; Anders Juréus, PhD; Gursharan Kalsi, PhD; Allan Kaplan, MSc, MD, FRCP(C); Jaakko Kaprio, MD, PhD; Leila Karhunen, PhD; Andreas Karwautz, MD, FAED; Martien Kas, PhD; Walter Kaye, MD; Martin Kennedy, PhD; James Kennedy, MD, FRCP(C); Anna Keski-Rahkonen, MD, PhD, MPH; Kirsty Kiezebrink, BSc (Hons), PGDip, PhD, FHEA; RNutr Youl-Ri Kim, MD, PhD; Lars Klareskog, MD; Kelly Klump, PhD; Gun Peggy Knudsen, PhD; Bobby Koeleman, PhD; Doris Koubek, MD; Maria La Via, MD; Mikael Landén, MD, PhD; Stephanie Le Hellard, PhD; Marion Leboyer. MD, PhD; Robert Levitan, MD; Dong Li, PhD; Paul Lichtenstein, PhD; Lisa Lilenfeld, PhD; Jolanta Lissowska, PhD; Astri Lundervold, PhD; Pierre Magistretti, PhD; Mario Maj, MD, PhD; Katrin Mannik, PhD; Sara Marsal, MD, PhD; Debora Kaminska, PhD; Nicholas Martin, PhD; Morten Mattingsdal, PhD; Sara McDevitt, MB, MD, MRCPsych, MMedED; Peter McGuffin, MD; Elisabeth Merl, MD; Andres Metspalu, PhD, MD; Ingrid Meulenbelt, PhD; Nadia Micali, MD, PhD; James Mitchell, MD; Karen Mitchell, PhD; Palmiero Monteleone, MD; Alessio Maria Monteleone, MD; Grant Montgomery, PhD; Preben Mortensen, MD, DrMedSc, Melissa Munn-Chernoff, PhD; Timo Müller, PhD; Benedetta Nacmias, PhD; Marie Navratilova, MUDr., PhD; Ida Nilsson, PhD; Claes Norring, PhD; Ioanna Ntalla, PhD; Roel Ophoff, PhD; Julie O’Toole, MD; Aarno Palotie, MD, PhD; Jacques Pantel, PhD; Hana Papezova, MD, PhD; Richard Parker Dalila Pinto, PhD; Raquel Rabionet, PhD; Anu Raevuori, MD, PhD; Andrzej Rajewski, MD, PhD; Nicolas Ramoz, PhD; N. William Rayner, PhD; Ted Reichborn-Kjennerud, MD; Valdo Ricca, MD; Samuli Ripatti, PhD; Franziska Ritschel, MSc; Marion Roberts, PhD; Alessandro Rotondo, MD; Dan Rujescu, MD; Filip Rybakowski, MD, PhD; Paolo Santonastaso, MD; André Scherag, PhD; Stephen Scherer, PhD, FRSC; Ulrike Schmidt, MD, PhD; Nicholas Schork, PhD; Alexandra Schosser, PhD; Laura Scott, PhD; Jochen Seitz, MD; Lenka Slachtova, PhD; Robert Sladek, MD; P. Eline Slagboom, PhD; Margarita Slof-Op ‘t Landt, PhD; Agnieszka Slopien, MD; Tosha Smith, PhD; Nicole Soranzo, PhD; Sandro Sorbi, MD; Lorraine Southam, BSc Vidar Steen, MD, PhD; Eric Strengman, BS; Michael Strober, PhD; Jin Szatkiewicz, PhD; Neonila Szeszenia-Dabrowska, MD, PhD; Ioanna Tachmazidou, PhD; Elena Tenconi, MD; Alfonso Tortorella, MD; Federica Tozzi, MD; Janet Treasure, PhD, FRCP, FRCPsych; Matthias Tschöp, MD; Artemis Tsitsika, MD, PhD; Konstantinos Tziouvas, MD, MSc Annemarie van Elburg, MD, PhD; Eric van Furth, PhD; Tracey Wade, PhD; Gudrun Wagner, Dr., MSc, DPO; Esther Walton, Dr. rer. nat., PhD; Hunna Watson, PhD; H-Erich Wichmann, PhD; Elisabeth Widen, MD, PhD; D. Blake Woodside, MD; Jack Yanovski, MD, PhD; Shuyang Yao, MSc, BSc; Stephanie Zerwas, PhD; Stephan Zipfel, MD; Laura Thornton, PhD; Anke Hinney, PhD; Gerome Breen, PhD; Cynthia M. Bulik, PhD.
Authors’ Contributions
EW carried out the analyses and wrote the manuscript. DH aided in the statistical work. PT and CB oversaw the ENIGMA and PGC-ED consortium work, while ZY, JC, and NJ were responsible for descriptive, methodological, and analytical questions with respect to the PGC-ED and ENIGMA datasets. VZ helped writing the manuscript and creating the figures and tables. JS, PT, and CB contributed to the interpretation of results. SE supervised the project and the drafting of the manuscript.
Funding
This work was funded through the Collaborative research center grant (DFG, SFB 940/2) and Schweizer Anorexia Nervosa Stiftung (both to SE), the National Institutes of Health K01MH109782 (ZY), NIH Big Data to Knowledge Initiative U54EB020403 and the Kavli Foundation (both to PT). CB acknowledges funding from the Swedish Research Council (VR Dnr: 538-2013-8864).
Compliance with Ethical Standards
All participants in all ENIGMA cohorts in this study gave written informed consent and sites involved obtained approval from local research ethics committees or Institutional Review Boards. All PGC-ED sites had documented permission from local ethical committees and all participants provided informed consent.
Conflict of Interest
CB received travel and research grants and honoraria for speaking and participating in advisory boards from Shire Pharmaceuticals and advances and royalties from Pearson and Walker. DPH is now an employee of Janssen R&D, LLC. All other authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Papadopoulos FC, Ekbom A, Brandt L, Ekselius L. Excess mortality, causes of death and prognostic factors in anorexia nervosa. Br J Psychiatry. 2009;194:10–17. doi: 10.1192/bjp.bp.108.054742. [DOI] [PubMed] [Google Scholar]
- 2.Kaye WH, Fudge JL, Paulus M. New insights into symptoms and neurocircuit function of anorexia nervosa. Nat Rev Neurosci. 2009;10:573–584. doi: 10.1038/nrn2682. [DOI] [PubMed] [Google Scholar]
- 3.Boraska V, Franklin CS, Floyd JA, et al. A genome-wide association study of anorexia nervosa. Mol Psychiatry. 2014;19:1085–1094. doi: 10.1038/mp.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duncan Laramie, Yilmaz Zeynep, Gaspar Helena, Walters Raymond, Goldstein Jackie, Anttila Verneri, Bulik-Sullivan Brendan, Ripke Stephan, Thornton Laura, Hinney Anke, Daly Mark, Sullivan Patrick F., Zeggini Eleftheria, Breen Gerome, Bulik Cynthia M., Duncan Laramie, Yilmaz Zeynep, Gaspar Héléna, Walters Raymond, Goldstein Jackie, Anttila Verneri, Bulik-Sullivan Brendan, Ripke Stephan, Adan Roger, Alfredsson Lars, Ando Tetsuya, Andreassen Ole, Aschauer Harald, Baker Jessica, Barrett Jeffrey, Bencko Vladimir, Bergen Andrew, Berrettini Wade, Birgegård Andreas, Boni Claudette, Perica Vesna Boraska, Brandt Harry, Burghardt Roland, Carlberg Laura, Cassina Matteo, Cesta Carolyn, Cichon Sven, Clementi Maurizio, Cohen-Woods Sarah, Coleman Joni, Cone Roger, Courtet Philippe, Crawford Steven, Crow Scott, Crowley Jim, Danner Unna, Davis Oliver, de Zwaan Martina, Dedoussis George, Degortes Daniela, DeSocio Janiece, Dick Danielle, Dikeos Dimitris, Dina Christian, Ding Bo, Dmitrzak-Weglarz Monika, Docampo Elisa, Egberts Karin, Ehrlich Stefan, Escaramís Geòrgia, Esko Tõnu, Espeseth Thomas, Estivill Xavier, Favaro Angela, Fernández-Aranda Fernando, Fichter Manfred, Finan Chris, Fischer Krista, Floyd James, Föcker Manuel, Foretova Lenka, Forzan Monica, Fox Caroline, Franklin Christopher, Gaborieau Valerie, Gallinger Steven, Gambaro Giovanni, Giegling Ina, Gonidakis Fragiskos, Gorwood Philip, Gratacos Monica, Guillaume Sébastien, Guo Yiran, Hakonarson Hakon, Halmi Katherine, Harrison Rebecca, Hatzikotoulas Konstantinos, Hauser Joanna, Hebebrand Johannes, Helder Sietske, Hendriks Judith, Herms Stefan, Herpertz-Dahlmann Beate, Herzog Wolfgang, Hilliard Christopher, Huckins Laura, Hudson James, Huemer Julia, Imgart Hartmut, Inoko Hidetoshi, Jall Sigrid, Jamain Stephane, Janout Vladimir, Jiménez-Murcia Susana, Johnson Craig, Jordan Jenny, Julià Antonio, Juréus Anders, Kalsi Gursharan, Kaplan Allan, Kaprio Jaakko, Karhunen Leila, Karwautz Andreas, Kas Martien, Kaye Walter, Kennedy Martin, Kennedy James, Keski-Rahkonen Anna, Kiezebrink Kirsty, Kim Youl-Ri, Klareskog Lars, Klump Kelly, Knudsen Gun Peggy, Koeleman Bobby, Koubek Doris, La Via Maria, Landén Mikael, Le Hellard Stephanie, Leboyer Marion, Levitan Robert, Li Dong, Lichtenstein Paul, Lilenfeld Lisa, Lissowska Jolanta, Lundervold Astri, Magistretti Pierre, Maj Mario, Mannik Katrin, Marsal Sara, Kaminska Debora, Martin Nicholas, Mattingsdal Morten, McDevitt Sara, McGuffin Peter, Merl Elisabeth, Metspalu Andres, Meulenbelt Ingrid, Micali Nadia, Mitchell James, Mitchell Karen, Monteleone Palmiero, Monteleone Alessio Maria, Montgomery Grant, Mortensen Preben, Munn-Chernoff Melissa, Müller Timo, Nacmias Benedetta, Navratilova Marie, Nilsson Ida, Norring Claes, Ntalla Ioanna, Ophoff Roel, O’Toole Julie, Palotie Aarno, Pantel Jacques, Papezova Hana, Parker Richard, Pinto Dalila, Rabionet Raquel, Raevuori Anu, Rajewski Andrzej, Ramoz Nicolas, Rayner N. William, Reichborn-Kjennerud Ted, Ricca Valdo, Ripatti Samuli, Ritschel Franziska, Roberts Marion, Rotondo Alessandro, Rujescu Dan, Rybakowski Filip, Santonastaso Paolo, Scherag André, Scherer Stephen, Schmidt Ulrike, Schork Nicholas, Schosser Alexandra, Scott Laura, Seitz Jochen, Slachtova Lenka, Sladek Robert, Slagboom P. Eline, ’t Landt Margarita Slof-Op, Slopien Agnieszka, Smith Tosha, Soranzo Nicole, Sorbi Sandro, Southam Lorraine, Steen Vidar, Strengman Eric, Strober Michael, Szatkiewicz Jin, Szeszenia-Dabrowska Neonila, Tachmazidou Ioanna, Tenconi Elena, Tortorella Alfonso, Tozzi Federica, Treasure Janet, Tschöp Matthias, Tsitsika Artemis, Tziouvas Konstantinos, van Elburg Annemarie, van Furth Eric, Wade Tracey, Wagner Gudrun, Walton Esther, Watson Hunna, Wichmann H-Erich, Widen Elisabeth, Woodside D. Blake, Yanovski Jack, Yao Shuyang, Zerwas Stephanie, Zipfel Stephan, Thornton Laura, Hinney Anke, Daly Mark, Sullivan Patrick F., Zeggini Eleftheria, Breen Gerome, Bulik Cynthia M. Significant Locus and Metabolic Genetic Correlations Revealed in Genome-Wide Association Study of Anorexia Nervosa. American Journal of Psychiatry. 2017;174(9):850–858. doi: 10.1176/appi.ajp.2017.16121402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huckins L M, Hatzikotoulas K, Southam L, Thornton L M, Steinberg J, Aguilera-McKay F, Treasure J, Schmidt U, Gunasinghe C, Romero A, Curtis C, Rhodes D, Moens J, Kalsi G, Dempster D, Leung R, Keohane A, Burghardt R, Ehrlich S, Hebebrand J, Hinney A, Ludolph A, Walton E, Deloukas P, Hofman A, Palotie A, Palta P, van Rooij F J A, Stirrups K, Adan R, Boni C, Cone R, Dedoussis G, van Furth E, Gonidakis F, Gorwood P, Hudson J, Kaprio J, Kas M, Keski-Rahonen A, Kiezebrink K, Knudsen G-P, Slof-Op 't Landt M C T, Maj M, Monteleone A M, Monteleone P, Raevuori A H, Reichborn-Kjennerud T, Tozzi F, Tsitsika A, van Elburg A, Collier D A, Sullivan P F, Breen G, Bulik C M, Zeggini E. Investigation of common, low-frequency and rare genome-wide variation in anorexia nervosa. Molecular Psychiatry. 2017;23(5):1169–1180. doi: 10.1038/mp.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang K, Zhang H, Bloss CS, et al. A genome-wide association study on common SNPs and rare CNVs in anorexia nervosa. Mol Psychiatry. 2011;16:949–959. doi: 10.1038/mp.2010.107. [DOI] [PubMed] [Google Scholar]
- 7.Seitz J, Herpertz-Dahlmann B, Konrad K. Brain morphological changes in adolescent and adult patients with anorexia nervosa. J Neural Transm (Vienna) 2016;123:949–959. doi: 10.1007/s00702-016-1567-9. [DOI] [PubMed] [Google Scholar]
- 8.Van den Eynde F, Suda M, Broadbent H, et al. Structural magnetic resonance imaging in eating disorders: a systematic review of voxel-based morphometry studies. Eur Eat Disord Rev. 2012;20:94–105. doi: 10.1002/erv.1163. [DOI] [PubMed] [Google Scholar]
- 9.King JA, Geisler D, Ritschel F, Boehm I, Seidel M, Roschinski B, Soltwedel L, Zwipp J, Pfuhl G, Marxen M, Roessner V, Ehrlich S. Global cortical thinning in acute anorexia nervosa normalizes following long-term weight restoration. Biol Psychiatry. 2015;77:624–632. doi: 10.1016/j.biopsych.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Seitz J, Walter M, Mainz V, Herpertz-Dahlmann B, Konrad K, von Polier G. Brain volume reduction predicts weight development in adolescent patients with anorexia nervosa. J Psychiatr Res. 2015;68:228–237. doi: 10.1016/j.jpsychires.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 11.Titova OE, Hjorth OC, Schiöth HB, Brooks SJ. Anorexia nervosa is linked to reduced brain structure in reward and somatosensory regions: a meta-analysis of VBM studies. BMC Psychiatry. 2013;13:110. doi: 10.1186/1471-244X-13-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernardoni F, King JA, Geisler D, Stein E, Jaite C, Nätsch D, Tam FI, Boehm I, Seidel M, Roessner V, Ehrlich S. Weight restoration therapy rapidly reverses cortical thinning in anorexia nervosa: a longitudinal study. NeuroImage. 2016;130:214–222. doi: 10.1016/j.neuroimage.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Bomba M, Riva A, Morzenti S, Grimaldi M, Neri F, Nacinovich R. Global and regional brain volumes normalization in weight-recovered adolescents with anorexia nervosa: preliminary findings of a longitudinal voxel-based morphometry study. Neuropsychiatr Dis Treat. 2015;11:637–645. doi: 10.2147/NDT.S73239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mainz V, Schulte-Rüther M, Fink GR, Herpertz-Dahlmann B, Konrad K. Structural brain abnormalities in adolescent anorexia nervosa before and after weight recovery and associated hormonal changes. Psychosom Med. 2012;74:574–582. doi: 10.1097/PSY.0b013e31824ef10e. [DOI] [PubMed] [Google Scholar]
- 15.Blokland GAM, de Zubicaray GI, McMahon KL, Wright MJ. Genetic and environmental influences on neuroimaging phenotypes: a meta-analytical perspective on twin imaging studies. Twin Res Hum Genet. 2012;15:351–371. doi: 10.1017/thg.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hibar DP, Stein JL, Renteria ME, et al. Common genetic variants influence human subcortical brain structures. Nature. 2015;520:224–229. doi: 10.1038/nature14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collantoni E, Michelon S, Tenconi E, Degortes D, Titton F, Manara R, Clementi M, Pinato C, Forzan M, Cassina M, Santonastaso P, Favaro A. Functional connectivity correlates of response inhibition impairment in anorexia nervosa. Psychiatry Res. 2016;247:9–16. doi: 10.1016/j.pscychresns.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Favaro A, Clementi M, Manara R, Bosello R, Forzan M, Bruson A, Tenconi E, Degortes D, Titton F, di Salle F, Santonastaso P. Catechol-O-methyltransferase genotype modifies executive functioning and prefrontal functional connectivity in women with anorexia nervosa. J Psychiatry Neurosci. 2013;38:241–248. doi: 10.1503/jpn.120068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J, Calhoun VD, Pearlson GD, Perrone-Bizzozero N, Sui J, Turner JA, Bustillo JR, Ehrlich S, Sponheim SR, Cañive JM, Ho BC, Liu J. Guided exploration of genomic risk for gray matter abnormalities in schizophrenia using parallel independent component analysis with reference. Neuroimage. 2013;83:384–396. doi: 10.1016/j.neuroimage.2013.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franke B, Stein JL, Ripke S, et al. Genetic influences on schizophrenia and subcortical brain volumes: large-scale proof of concept. Nat Neurosci. 2016;19:420–431. doi: 10.1038/nn.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee PH, Baker JT, Holmes AJ, Jahanshad N, Ge T, Jung JY, Cruz Y, Manoach DS, Hibar DP, Faskowitz J, McMahon KL, de Zubicaray GI, Martin NH, Wright MJ, Öngür D, Buckner R, Roffman J, Thompson PM, Smoller JW. Partitioning heritability analysis reveals a shared genetic basis of brain anatomy and schizophrenia. Mol Psychiatry. 2016;21:1680–1689. doi: 10.1038/mp.2016.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wigmore EM, Clarke T-K, Howard DM, Adams MJ, Hall LS, Zeng Y, Gibson J, Davies G, Fernandez-Pujals AM, Thomson PA, Hayward C, Smith BH, Hocking LJ, Padmanabhan S, Deary IJ, Porteous DJ, Nicodemus KK, McIntosh AM. Do regional brain volumes and major depressive disorder share genetic architecture? A study of Generation Scotland (n=19 762), UK Biobank (n=24 048) and the English Longitudinal Study of Ageing (n=5766) Transl Psychiatry. 2017;7:e1205. doi: 10.1038/tp.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.American Psychiatric Association (2000) Diagnostic and statistical manual of mental disorders, 4th ed. Author, Washington, DC
- 25.Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 26.Howie B, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bulik-Sullivan BK, Loh P-R, Finucane HK, et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson T (2012) Efficient calculation for multi-SNP genetic risk scores. San Francisco
- 30.Euesden J, Lewis CM, O’Reilly PF. PRSice: polygenic risk score software. Bioinformatics. 2015;31:1466–1468. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nyholt DR. SECA: SNP effect concordance analysis using genome-wide association summary results. Bioinformatics. 2014;30:2086–2088. doi: 10.1093/bioinformatics/btu171. [DOI] [PubMed] [Google Scholar]
- 32.Davey Smith G, Ebrahim S. Mendelian randomization: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 33.Burgess S, Scott RA, Timpson NJ, et al. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol. 2015;30:543–552. doi: 10.1007/s10654-015-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith GD, Ebrahim S. Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol. 2004;33:30–42. doi: 10.1093/ije/dyh132. [DOI] [PubMed] [Google Scholar]
- 35.Davies NM, Holmes MV, Smith GD. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. doi: 10.1136/bmj.k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hemani G, Zheng J, Wade KH, et al (2016) MR-Base: a platform for systematic causal inference across the phenome using billions of genetic associations BioRxiv 078972. 10.1101/078972
- 37.Smeland OB, Wang Y, Lo M-T, Li W, Frei O, Witoelar A, Tesli M, Hinds DA, Tung JY, Djurovic S, Chen CH, Dale AM, Andreassen OA. Identification of genetic loci shared between schizophrenia and the big five personality traits. Sci Rep. 2017;7:2222. doi: 10.1038/s41598-017-02346-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pearlson GD, Liu J, Calhoun VD. An introductory review of parallel independent component analysis (p-ICA) and a guide to applying p-ICA to genetic data and imaging phenotypes to identify disease-associated biological pathways and systems in common complex disorders. Front Genet. 2015;6:276. doi: 10.3389/fgene.2015.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cederlöf M, Thornton LM, Baker J, Lichtenstein P, Larsson H, Rück C, Bulik CM, Mataix-Cols D. Etiological overlap between obsessive-compulsive disorder and anorexia nervosa: a longitudinal cohort, multigenerational family and twin study. World Psychiatry. 2015;14:333–338. doi: 10.1002/wps.20251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brainstorm Consortium. Anttila V, Bulik-Sullivan B, et al. Analysis of shared heritability in common disorders of the brain. Science. 2018;360:eaap8757. doi: 10.1126/science.aap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cha J, Ide JS, Bowman FD, Simpson HB, Posner J, Steinglass JE. Abnormal reward circuitry in anorexia nervosa: a longitudinal, multimodal MRI study. Hum Brain Mapp. 2016;37:3835–3846. doi: 10.1002/hbm.23279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayes DJ, Lipsman N, Chen DQ, Woodside DB, Davis KD, Lozano AM, Hodaie M. Subcallosal cingulate connectivity in anorexia nervosa patients differs from healthy controls: a multi-tensor tractography study. Brain Stimul. 2015;8:758–768. doi: 10.1016/j.brs.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 43.Vogel K, Timmers I, Kumar V, Nickl-Jockschat T, Bastiani M, Roebroek A, Herpertz-Dahlmann B, Konrad K, Goebel R, Seitz J. White matter microstructural changes in adolescent anorexia nervosa including an exploratory longitudinal study. Neuroimage Clin. 2016;11:614–621. doi: 10.1016/j.nicl.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zastrow A, Kaiser S, Stippich C, et al. Neural correlates of impaired cognitive-behavioral flexibility in anorexia nervosa. Am J Psychiatry. 2009;166:608–616. doi: 10.1176/appi.ajp.2008.08050775. [DOI] [PubMed] [Google Scholar]
- 45.Biezonski D, Cha J, Steinglass J, Posner J. Evidence for thalamocortical circuit abnormalities and associated cognitive dysfunctions in underweight individuals with anorexia nervosa. Neuropsychopharmacology. 2016;41:1560–1568. doi: 10.1038/npp.2015.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ehrlich S, Lord AR, Geisler D, Borchardt V, Boehm I, Seidel M, Ritschel F, Schulze A, King JA, Weidner K, Roessner V, Walter M. Reduced functional connectivity in the thalamo-insular subnetwork in patients with acute anorexia nervosa. Hum Brain Mapp. 2015;36:1772–1781. doi: 10.1002/hbm.22736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geisler D, Borchardt V, Lord AR, Boehm I, Ritschel F, Zwipp J, Clas S, King JA, Wolff-Stephan S, Roessner V, Walter M, Ehrlich S. Abnormal functional global and local brain connectivity in female patients with anorexia nervosa. J Psychiatry Neurosci. 2016;41:6–15. doi: 10.1503/jpn.140310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Craig ADB. Significance of the insula for the evolution of human awareness of feelings from the body. Ann N Y Acad Sci. 2011;1225:72–82. doi: 10.1111/j.1749-6632.2011.05990.x. [DOI] [PubMed] [Google Scholar]
- 49.Hammer C, Kapeller J, Endele M, Fischer C, Hebebrand J, Hinney A, Friedel S, Gratacòs M, Estivill X, Fichter M, Fernández-Aranda F, Ehrlich S, Rappold G, Niesler B. Functional variants of the serotonin receptor type 3A and B gene are associated with eating disorders. Pharmacogenet Genomics. 2009;19:790–799. doi: 10.1097/FPC.0b013e32833132b3. [DOI] [PubMed] [Google Scholar]
- 50.Bergen AW, Yeager M, Welch RA, Haque K, Ganjei JK, van den Bree MBM, Mazzanti C, Nardi I, Fichter MM, Halmi KA, Kaplan AS, Strober M, Treasure J, Woodside DB, Bulik CM, Bacanu SA, Devlin B, Berrettini WH, Goldman D, Kaye WH. Association of multiple DRD2 polymorphisms with anorexia nervosa. Neuropsychopharmacology. 2005;30:1703–1710. doi: 10.1038/sj.npp.1300719. [DOI] [PubMed] [Google Scholar]
- 51.Peng S, Yu S, Wang Q, Kang Q, Zhang Y, Zhang R, Jiang W, Qian Y, Zhang H, Zhang M, Xiao Z, Chen J. Dopamine receptor D2 and catechol-O-methyltransferase gene polymorphisms associated with anorexia nervosa in Chinese Han population: DRD2 and COMT gene polymorphisms were associated with AN. Neurosci Lett. 2016;616:147–151. doi: 10.1016/j.neulet.2016.01.036. [DOI] [PubMed] [Google Scholar]
- 52.Fallon JH. Topographic organization of ascending dopaminergic projections. Ann N Y Acad Sci. 1988;537:1–9. doi: 10.1111/j.1749-6632.1988.tb42093.x. [DOI] [PubMed] [Google Scholar]
- 53.Harsay HA, Cohen MX, Oosterhof NN, Forstmann BU, Mars RB, Ridderinkhof KR. Functional connectivity of the striatum links motivation to action control in humans. J Neurosci. 2011;31:10701–10711. doi: 10.1523/JNEUROSCI.5415-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ridderinkhof KR, van den Wildenberg WPM, Segalowitz SJ, Carter CS. Neurocognitive mechanisms of cognitive control: the role of prefrontal cortex in action selection, response inhibition, performance monitoring, and reward-based learning. Brain Cogn. 2004;56:129–140. doi: 10.1016/j.bandc.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 55.Kaye WH, Wierenga CE, Bailer UF, Simmons AN, Bischoff-Grethe A. Nothing tastes as good as skinny feels: The neurobiology of anorexia nervosa. Trends Neurosci. 2013;36:110–120. doi: 10.1016/j.tins.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bailer UF, Frank GK, Price JC, Meltzer CC, Becker C, Mathis CA, Wagner A, Barbarich-Marsteller NC, Bloss CS, Putnam K, Schork NJ, Gamst A, Kaye WH. Interaction between serotonin transporter and dopamine D2/D3 receptor radioligand measures is associated with harm avoidant symptoms in anorexia and bulimia nervosa. Psychiatry Res. 2013;211:160–168. doi: 10.1016/j.pscychresns.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frank GK, Bailer UF, Henry SE, Drevets W, Meltzer CC, Price JC, Mathis CA, Wagner A, Hoge J, Ziolko S, Barbarich-Marsteller N, Weissfeld L, Kaye WH. Increased dopamine D2/D3 receptor binding after recovery from anorexia nervosa measured by positron emission tomography and [11c]raclopride. Biol Psychiatry. 2005;58:908–912. doi: 10.1016/j.biopsych.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 58.Wierenga CE, Ely A, Bischoff-Grethe A, Bailer UF, Simmons AN, Kaye WH. Are extremes of consumption in eating disorders related to an altered balance between reward and inhibition? Front Behav Neurosci. 2014;8:410. doi: 10.3389/fnbeh.2014.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Foerde K, Steinglass JE, Shohamy D, Walsh BT. Neural mechanisms supporting maladaptive food choices in anorexia nervosa. Nat Neurosci. 2015;18:1571–1573. doi: 10.1038/nn.4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frank GK, Shott ME, Hagman JO, Mittal VA. Alterations in brain structures related to taste reward circuitry in ill and recovered anorexia nervosa and in bulimia nervosa. Am J Psychiatry. 2013;170:1152–1160. doi: 10.1176/appi.ajp.2013.12101294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frank S, Kullmann S, Veit R. Food related processes in the insular cortex. Front Hum Neurosci. 2013;7:499. doi: 10.3389/fnhum.2013.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Joos AAB, Saum B, van Elst LT, Perlov E, Glauche V, Hartmann A, Freyer T, Tüscher O, Zeeck A. Amygdala hyperreactivity in restrictive anorexia nervosa. Psychiatry Res. 2011;191:189–195. doi: 10.1016/j.pscychresns.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 63.Kim KR, Ku J, Lee J-H, Lee H, Jung YC. Functional and effective connectivity of anterior insula in anorexia nervosa and bulimia nervosa. Neurosci Lett. 2012;521:152–157. doi: 10.1016/j.neulet.2012.05.075. [DOI] [PubMed] [Google Scholar]
- 64.Sanders N, Smeets PAM, van Elburg AA, Danner UN, van Meer F, Hoek HW, Adan RAH (2015) Altered food-cue processing in chronically ill and recovered women with anorexia nervosa. Front Behav Neurosci 9(46). 10.3389/fnbeh.2015.00046 [DOI] [PMC free article] [PubMed]
- 65.Lin JC, Ho W-H, Gurney A, Rosenthal A. The netrin-G1 ligand NGL-1 promotes the outgrowth of thalamocortical axons. Nat Neurosci. 2003;6:1270–1276. doi: 10.1038/nn1148. [DOI] [PubMed] [Google Scholar]
- 66.Song YS, Lee H-J, Prosselkov P, Itohara S, Kim E. Trans-induced cis interaction in the tripartite NGL-1, netrin-G1 and LAR adhesion complex promotes development of excitatory synapses. J Cell Sci. 2013;126:4926–4938. doi: 10.1242/jcs.129718. [DOI] [PubMed] [Google Scholar]
- 67.Nishimura-Akiyoshi S, Niimi K, Nakashiba T, Itohara S. Axonal netrin-Gs transneuronally determine lamina-specific subdendritic segments. Proc Natl Acad Sci U S A. 2007;104:14801–14806. doi: 10.1073/pnas.0706919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Connan F, Murphy F, Connor SEJ, Rich P, Murphy T, Bara-Carill N, Landau S, Krljes S, Ng V, Williams S, Morris RG, Campbell IC, Treasure J. Hippocampal volume and cognitive function in anorexia nervosa. Psychiatry Res. 2006;146:117–125. doi: 10.1016/j.pscychresns.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 69.Buehren K, Konrad K, Schaefer K, Kratzsch J, Kahraman-Lanzerath B, Lente C, Herpertz-Dahlmann B. Association between neuroendocrinological parameters and learning and memory functions in adolescent anorexia nervosa before and after weight recovery. J Neural Transm (Vienna) 2011;118:963–968. doi: 10.1007/s00702-010-0567-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 773 kb)