Abstract
This study has evaluated the clinical applicability of a single‐tube multiplex RT‐ PCR as compared with a two‐step nested RT‐PCR for the diagnosis as well as serotyping of dengue virus in patient's samples. Seventy‐six acute phase blood samples collected from clinically suspected dengue patients during the 2008 outbreak were subjected to two‐step nested RT‐PCR and single‐tube multiplex RT‐PCR for dengue diagnosis and serotyping. Of the 76 samples, 17 (22.4%) were positive for dengue viral RNA. Single dengue virus infection was found in 16 cases and 1 had concurrent infection with two serotypes (3&1). Dengue serotype 3 was the predominant serotype (70.5%), followed by serotype 1 (23.5%). Single‐tube multiplex PCR had concordant result with that of two‐step nested RT‐PCR including the one with concomitant infection. This study reveals the predominance of dengue serotype 3 in North India in addition to the co‐circulation of multiple serotypes and concomitant infection. The rapid and accurate diagnostic capability of single‐tube multiplex RT‐PCR used in the study appears to be promising enough to be commonly used for dengue viral detection as well as serotyping. J. Clin. Lab. Anal. 25:76–78, 2011. © 2011 Wiley‐Liss, Inc.
Keywords: dengue virus, dengue fever, dengue hemorrhagic fever, concurrent infection, single‐tube RT‐PCR
INTRODUCTION
Dengue is the most important arboviral infection with around 2.5 billion people living within dengue endemic areas. The four dengue serotypes (DENV 1–4) are capable of producing diseases ranging from self‐limiting dengue fever (DF) to severe life‐threatening dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) 1, 2, 3. With the changing epidemiology of DF and DHF both globally as well as nationally, more number of DHF cases are being reported 1. DHF is postulated as an outcome of secondary dengue infection with a heterologous serotype. In addition, certain serotypes have been reported to be more commonly associated with severity 4; therefore, rapid and serotype‐specific diagnosis has become paramount importance. Conventionally, dengue serotyping is carried out by virus isolation followed by typing with type‐specific monoclonal antibody by indirect immunofluorescence, which is laborious and time consuming 5. Currently, a two‐step nested reverse transcriptase polymerase chain reaction (RT‐PCR) with multiplex PCR as the second step is widely used for the serotyping of dengue viruses 6, 7, 8. However, this method also suffers from the possibility of carry over contamination and is more time consuming 9. A single‐tube multiplex PCR was used in this study for direct detection of dengue serotype in the clinical samples of 2008 dengue outbreak.
MATERIALS AND METHODS
A total of 76 acute phase blood samples (within 5 days of fever) were collected from clinically suspected dengue patients during the 2008 dengue outbreak. Samples were transported in ice to the laboratory and serum was separated in cold centrifuge. All the samples were stored at −85°C deep freezer till tested.
Detection of Dengue Virus Serotypes in Clinical Samples
All the 76 serum samples were subjected to two‐step nested PCR and single‐tube multiplex PCR for the detection and serotyping of dengue virus.
Extraction of Dengue Viral RNA
One hundred and forty microliters of patient's serum sample was subjected to extraction of viral RNA using QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany), following the manufacturer's protocol. Finally, the RNA was eluted in 60 μl of elution buffer.
Detection and Serotyping of Dengue Virus by Two‐Step Nested RT‐PCR
The extracted viral RNA was reverse transcribed to cDNA using murine Moloney Leukemia Virus (mMULV) reverse transcriptase (MBI Fermentas, Glen Burnie, MD) by dengue virus downstream consensus primer (D2). Subsequent serotype Taq Polymerase amplification of 1:100 diluted cDNA was carried out with the dengue virus consensus forward primer (D1) and four dengue serotype‐specific reverse primers (TS1–TS4). The components of RT‐PCR and the thermal profile were used as described by Lanciotti et al. 10.
Single‐Tube Dengue Multiplex RT‐PCR
Five microliters of extracted RNA of the patient serum was amplified in 50 μl reaction mix with 0.5 μM each of dengue virus D1 upstream primer and four downstream dengue serotype‐specific primers (TS1–TS4) using QIAGEN One‐Step RT‐PCR kit (Qiagen) according to the manufacturer's instructions. The single‐tube reaction mix contained: 1× QI Buffer containing 2.5 mM MgCl2, Tris–Cl, KCl, (NH4)2SO4; 1× Q Solution; 400 μM of each dNTP; 1 U QIAGEN One‐Step RT‐PCR enzyme mix and 5 U of RNase inhibitor. Tubes were spun briefly and reverse transcriptase (RT) reaction was carried out with one cycle at 50°C for 30 min and 35 cycles of PCR amplification using the one‐step program (95°C, 30 sec; 55°C, 45 sec, and 72°C, 2 min). The serotype‐specific bands characteristic of each dengue virus, i.e. 482 bp for Den‐1, 119 bp for Den‐2, 290 bp for Den‐3, and 389 bp for Den‐4, were visualized by 2% gel electrophoresis under a digital gel documentation system (Alpha Innotech, San Leandro, CA).
RESULTS
Of the 76 samples, 17 (22.4%) were positive for dengue viral RNA. Single dengue virus infection was found in 16 cases and 1 had concurrent infection with two DENV serotypes. Dengue serotype 3 was found to be the predominant serotype constituting 70.5% (12/17) of the positive samples, followed by serotype 1 in 23.5% (4/17). The only case of concurrent infection was found to have the co‐infection with dengue virus type1 and type3 (Fig. 1). The dengue virus serotyping detected by the single‐tube multiplex PCR was found to be concordant with the result of two‐step nested RT‐PCR including the one with concomitant infection.
Figure 1.
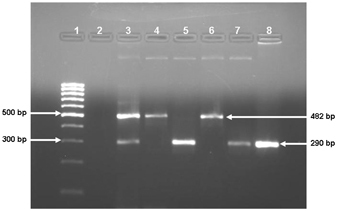
Agarose gel analyses of single‐tube multiplex RT‐PCR. Lane‐1: 100 bp Molecular Marker; Lane‐2: No template control; Lane‐3: Co‐infection of Type‐1 and Type‐3 dengue virus; Lane‐4 and 6: Type‐1 dengue virus infection (482 bp); Lane‐5, 7 and 8: Type‐3 dengue virus infection (290 bp).
No cross‐reaction was observed with Japanese encephalitis virus and Chikungunya virus, suggesting that this method of dengue virus detection is specific. The age of the patients' positive for dengue viral RNA was found to vary from 2 months to 70 years and among these patients' males outnumbered the females with a ratio of 2:1. Of the 17 dengue‐positive patients, 15 had dengue fever and two had dengue hemorrhagic fever.
DISCUSSION
With the changing global epidemiology of dengue fever and dengue hemorrhagic fever, the scenario in India is also becoming critical 6, 11. India is known to be endemic for dengue infection where the disease was mostly occurring in the form of self‐limiting dengue fever; however, since last decade, the number of DHF and DSS cases is being increasingly reported 12. Moreover DHF is known to be caused mostly during secondary dengue infection. Lack of an effective dengue vaccine has also made the need of a rapid and specific diagnosis of dengue serotypes of paramount importance. The widely used two‐step nested RT‐PCR is a rapid method as compared with that of virus isolation and followed by identification; however, it still suffers from the risk of carry over contamination and effectively takes at least two working days for serotype detection 8, 9. Therefore, a diagnostic system that would provide rapid diagnosis along with the serotype detection has become an absolute need for proper management of the cases.
The present method could detect all the dengue virus serotypes as were detected by two‐step nested PCR method including the concurrent infection with more than one dengue viral serotypes. This is in agreement with Saxena et al. 9 where single‐step multiplex PCR has been reported to have equal sensitivity as with two‐step PCR method. This study for the first time reports the successful detection of concurrent infection using the single‐tube multiplex PCR. The serotype determination could be carried out on the same day of sample collection, whereas the two‐step nested PCR effectively takes more than 2 days 9, 13. Second, the cost of the test is reduced almost one‐forth 5, 9, 13.
The city, Chandigarh, is situated at the northwest part of India and is endemoepidemic for dengue viral infection 14. Both the DHF cases had DENV‐3 infection. Several epidemics of DHF caused by DENV serotype 3 (subtype III) have been reported in the last decade from various countries including India 15, 16. However, the finding of concurrent infection along with the co‐circulation of multiple serotypes is unique for this outbreak and reported for the first time from this part of the country. Concurrent infection with different dengue viruses has been reported from areas where multiple serotypes co‐circulate. In India, so far, this has been reported only from Delhi 6. Though a severe clinical outcome is usually expected in patients with concurrent infection, the present case had uneventful recovery. No significant association of concurrent dengue virus infection with clinical outcome has been observed 17. However, due to small number of cases of concurrent dengue infection reported so far, this aspect needs to be further investigated with larger number of cases. More importantly the presence of concomitant infection leads the possibility of occurrence of recombination events 18.
This study reveals the predominance of dengue serotype 3 in this part of the country with circulation of multiple serotypes and occurrence of concomitant infection. The rapid and accurate diagnostic capability of the single‐tube multiplex RT‐PCR used in the study appears to be promising enough to be commonly used for dengue viral detection as well as serotyping.
Acknowledgements
Authors are thankful to Mr. H.C. Gera, Nodal Officer for National Vector Borne Control Programme, UT, Chandigarh and Dr. Abha, Govt. Multi Specialty Hospital, Chandigarh for sample collection.
Contributor Information
Bajayantimala Mishra, Email: bm_mishra@hotmail.com.
Radha Kanta Ratho, Email: rathopgi@yahoo.com.
REFERENCES
- 1. Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev 1998;11:480–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Monath TP, Heinz FX. Flaviviruses In: Fields BN, Knipe DN, Howley PM, et al., editors. Fields Virology. Philadelphia, PA: Lippincott Raven Publishers, 1996. p 961–1034. [Google Scholar]
- 3. World Health Organization . Dengue haemorrhagic fever: Diagnosis, treatment, prevention and control In: Prasittisuk C, Longmire CM, Gubler DJ, editors. Geneva: World Health Organization, 1997, http://who.int/csr/resources/publications/dengue/Dengepublication/en. [Google Scholar]
- 4. Vaughn DW, Green S, Kalayanarooj S, et al. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis 2000;181:2–9. [DOI] [PubMed] [Google Scholar]
- 5. Harris E, Roberts TG, Smith L, et al. Typing of Dengue viruses in clinical specimens and mosquitoes by single‐ tube multiplex reverse transcriptase PCR. J Clin Microbiol 1998;36:2634–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bharaj P, Chahar HS, Pandey A, et al. Concurrent infections by all four dengue virus serotypes during an outbreak of dengue in 2006 in Delhi, India. Virol J 2008;5:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cam BV, Fonsmark L, Hue NB, Phoung NT, Poulesen A, Heegaard ED. Prospective case‐control study of encephalopathy in children with dengue hemorrhagic fever. Am J Trop Med Hyg 2001;65:848–851. [DOI] [PubMed] [Google Scholar]
- 8. Ratho RK, Mishra B, Kaur J, Kakkar N, Sharma K. An outbreak of dengue fever in periurban slums of Chandigarh, India, with special reference to entomological and climatic factors. Indian J Med Sci 2005;59:518–526. [PubMed] [Google Scholar]
- 9. Saxena P, Dash PK, Santhosh SR, Shrivastava A, Parida M, Rao PVL. Development and evaluation of one step single tube multiplex RT‐PCR for rapid detection and typing of dengue viruses. Virol J 2008;5:20–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lanciotti RS, Calisher CH, Gubler DJ, Chang G, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase‐polymerase chain reaction. J Clin Microbiol 1992;30:545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gupta E, Dar L, Kapoor G, Broor S. The changing epidemiology of dengue in Delhi, India. Virol J 2006;3:92–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chaturvedi UC. The curse of Dengue. Indian J Med Res 2006;124:467–470. [PubMed] [Google Scholar]
- 13. Kumaria R, Chakravarti A. Molecular detection and serotypic characterization of dengue viruses by single‐tube multiplex reverse transcriptase–polymerase chain reaction. Diagn Microbiol Infect Dis 2005;52:311–316. [DOI] [PubMed] [Google Scholar]
- 14. Ratho RK, Mishra B, Kumar S, Varma S. Dengue fever/dengue haemorrhagic fever in Chandigarh (North India). Dengue Bulletin 2006;30:278–280. [Google Scholar]
- 15. Dash PK, Parida MM, Saxena P, et al. Reemergence of dengue virus type‐3 (subtype‐III) in India: Implications for increased incidence of DHF & DSS. Virol J 2006;3:55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Messer WB, Gubler DJ, Harris E, Sivananthan K, de Silva AM. Emergence and global spread of dengue serotype3, subtypeIII virus. Emerg Infect Dis 2003;9:800–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Morita K, Tanaka M, Igarashi A. Rapid identification of dengue virus serotypes by using polymerase chain reaction. J Clin Microbiol 1991;29:2107–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Worobey M, Rambaut A, Holmes EC. Widespread intraserotype recombination in natural populations of dengue virus. Proc Natl Acad Sci USA 1999;96:7352–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]


