Abstract
Granulins (GRNs 1–7) are cysteine-rich proteolytic products of progranulin (PGRN) that have recently been implicated in neurodegenerative diseases including frontotemporal dementia (FTD) and Alzheimer’s disease (AD). Their precise mechanism in these pathologies remains uncertain, but both inflammatory and lysosomal roles have been observed for GRNs. Among the seven GRNs, GRN-3 is well characterized and is implicated within the context of FTD. However, the relationship between GRN-3 and amyloid-β (Aβ), a protein relevant in AD pathology, has not yet been explored. To gain insight into this mechanism, we investigated the effect of both oxidized and reduced GRN-3 on Aβ aggregation and found that both GRN-3 (oxidized) and rGRN-3 (reduced) bind to monomeric and oligomeric Aβ42 to promote rapid fibril formation with subtle rate differences. As low molecular weight oligomers of Aβ are well-established neurotoxins, rapid promotion of fibrils by GRN-3 mitigates Aβ42-induced cellular apoptosis. These data provide valuable insights in understanding GRN-3’s ability to modulate Aβ-induced toxicity under redox control and presents a new perspective toward AD pathology. These results also prompt further investigation into the role(s) of other GRNs in AD pathogenesis.
Introduction
Granulins (GRNs 1–7) are a family of small unique cysteine-rich proteins that are proteolytically cleaved from the precursor protein, progranulin (PGRN) (Supplementary Figure S1A) [1]. All seven GRNs (GRNs 1–7) are ∼6 kDa in size and all, but GRN-1, are characterized by the presence of 12 conserved cysteines that form six intramolecular disulfide bonds (Supplementary Figure S1B) [1–4]. GRNs are known to play role in several physiological processes such as wound healing, tumorigenesis, etc. [5–8]. During the last decade, PGRNs and GRNs have also been implicated in neurodegenerative diseases such as frontotemporal dementia (FTD) [9], Alzheimer’s disease (AD) and other tauopathies [10]. Null mutations in PGRN were shown to be one of the main causes of familial FTD [8,10,11]. In addition, a few missense mutations, mapped to be present in GRN-3 sequence of PGRN, have also been implicated in FTD [12,13]. Furthermore, several polymorphisms of PGRN have been linked to idiopathic AD and haplotypes have been identified that contribute to the increased risk of AD [14,15]. Furthermore, GRNs have also been found colocalized with Aβ plaques in brains of AD patients [16] and transgenic AD mice [17], which prompt investigation into the potential interactions between Aβ and GRNs.
GRNs are unique proteins that contain a high percentage of cysteines (∼17%). Among the seven GRNs, the structure of GRN-2, solved by NMR spectroscopy, shows a folded N-terminal region with the stacked β-sheet arrangement and a disordered C-terminus [18]. Structures of other GRNs remain unsolved; however, all are thought to form a ladder-like putative disulfide bond pattern (Supplementary Figure S1B) [19]. Previously, we discovered that complete abrogation of disulfide bonds in GRN-3 (rGRN-3) renders the protein disordered that is also able to activate moderate levels of NF-κB in neuroblastoma cells [20]. We also discovered that although fully oxidized GRN-3 lacks defined secondary structure it exhibits an ordered structure overall based on NMR spectral dispersion. Despite high-temperature stability, homology modeling showed a structure that is dominated by loops, which indicates the significance of disulfide bonds in the biophysical and biochemical properties of the protein [21].
Since activated microglial cells overexpress PGRN as well as the enzymes that cleave PGRN into GRNs [17,22,23], we hypothesize that GRNs could interact directly with Aβ expressed in neuronal cells and modulate the latter’s aggregation and toxicity. Alternatively, increasing evidence suggests that both PGRN and GRNs play a role in the regulation of lysosomal function and trafficking [24–28]. Furthermore, with the evidence of transport, localization [29] and even production of Aβ in lysosomes [30], which support the autophagic processes in Aβ toxicity [31], we questioned whether GRN-3, both in its oxidized (denoted henceforth as GRN-3) and fully reduced rGRN-3 forms, interacts with Aβ42. In this report, we present a biophysical and biochemical basis for the interaction of Aβ42 with both redox forms of GRN-3. These interactions result in a rapid conversion of both monomers and oligomers of Aβ42 into high molecular mass fibrils. While GRN-3 interacts with Aβ42 monomers more cooperatively and strongly than rGRN-3 to promote fibril formation, rGRN-3 induces chaotropic or coacervation-type effects on Aβ42 to promote fibrils more rapidly than GRN-3. Furthermore, Aβ42–GRN-3 interactions diminish the activation of caspases-3 and −7 in neuroblastoma cells, which are involved in apoptosis. These results provide insights into the potentially significant event(s) in AD that could facilitate understanding the mechanisms of the pathology from an entirely new perspective involving GRNs’ role in the pathology.
Experimental
Cloning and purification of unlabeled and uniformly 15N-labeled GRN-3
Unlabeled and 15N-labeled GRN-3 was expressed and purified from E. coli SHuffle™ cells (New England Biolabs, MA, U.S.A.) as described previously [21]. Briefly, the protein was expressed as a GRN-3:trxA fusion protein and purified using nickel affinity chromatography. The fusion protein was then cleaved by the addition of restriction grade thrombin (EMD Millipore Sigma, MA, U.S.A.) at 1 U per 2 mg of the protein to remove both trxA and the His-tag. The reaction was incubated in a 37°C water bath for 22–24 h. The protein was then fractionated on a semi-preparative Jupiter® 5 μm 10 × 250 mm C18 reverse-phase HPLC column (Phenomenex, CA, U.S.A.), using a gradient elution of 60–80% acetonitrile containing 0.1% TFA as previously described [21]. The concentration of the protein was estimated spectrophotometrically using the molar extinction coefficient of 6250 M−1 cm−1 at 280 nm. Only the freshly purified monomeric GRN-3 was used for every experiment as the oxidized GRN-3. The amount of free cysteines was calculated from Ellman’s assay and by iodoacetamide conjugation as previously reported [21]. rGRN-3 was generated by the addition of 1 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) to the HPLC-fractionated GRN-3 for 1 h at room temperature and was used as such. 15N-labeled GRN-3 and rGRN-3 were generated by growing the cells in M9 minimal media enriched with 15NH4Cl.
Preparation of Aβ42 monomers
Aβ42 monomers were purified from the lyophilized synthetic Aβ42 as described previously [32]. Briefly, 0.8–1.5 mg of peptide was solubilized in 0.5 ml of 35 mM NaOH at 25°C for 30 min prior to fractionation on a Superdex-75 HR 10/30 SEC column using an AKTA FPLC system (GE Healthcare, Buckinghamshire). The column was pre-equilibrated in 20 mM Tris–HCl (pH 8.0). The monomer was purified at a flow rate of 0.5 ml/min, and 1-min fractions were collected. The concentrations of monomeric Aβ42 fractions were determined spectrophotometrically using a molar extinction coefficient of 1450 M−1 cm−1 at 276 nm (expasy.org). The purity of the fractionated monomer was confirmed by MALDI-ToF mass spectrometry, which revealed a monoisotopic molecular mass of 4515 Da. Aβ42 monomers were stored at 4°C and used within 24 h of purification.
Preparation of Aβ42 oligomers
Aβ42 oligomers called large fatty acid-derived oligomers (LFAOs) were prepared and purified as described previously [32]. Briefly, freshly purified Aβ42 monomer (50 μM) was incubated with 50 mM NaCl and 5 mM C12:0 fatty acid at 37°C in quiescent conditions for 48 h. After 48-h incubation, the sample was subjected to centrifugation at 18 000×g for 20 min before SEC purification. Fractions corresponding to the peak near the void volume (V0) were collected and concentration was determined using UV absorbance. Aβ42 LFAOs were stored at 4°C and used within 72 h of purification.
Fluorescence spectroscopy
The effect of GRN-3 on Aβ42 aggregation was monitored by Thioflavin-T (ThT) fluorescence using a BioTek Synergy H1 microplate reader. Freshly purified, aggregate-free Aβ42 monomers were incubated with GRN-3 or rGRN-3 in 1:2 stoichiometric ratio at 37°C quiescent for 24 h along with Aβ42 controls in the absence of GRN-3 and rGRN-3. Binding interactions between GRN-3, rGRN-3, and Aβ42 were studied on a Cary Eclipse spectrometer (Agilent Inc., U.S.A.). In this assay, increasing concentrations of GRN-3 or rGRN-3 were titrated on 5 μM tetra-methyl rhodamine-labeled Aβ42 (TMR-Aβ). The samples were excited at 550 nm (excitation λmax for rhodamine) and the changes in the fluorescence emission were monitored at 580 nm. The normalized intrinsic fluorescence intensities were plotted against the concentration of the titrant. Fluorescence anisotropy was measured simultaneously with intrinsic fluorescence by using the polarization accessory. Anisotropy (r) was measured with the excitation and emission wavelengths fixed at 550 and 580 nm, respectively, corresponding to TMR with a spectral bandwidth of 20 nm (10 nm Ex and 10 nm Em). The ADL program provided by the manufacturer was used and the G factor was calculated for each titration with the same fluorophore. Each titration point was measured in quadruplicate after a brief equilibration time of 1 min, and the data points were averaged. The anisotropy data for rGRN-3 and normalized fluorescence intensities for the three proteins were fit to the following non-cooperative single site binding equation using Origin 8.5 software.
| (1) |
where r0 and rs are anisotropy values in the absence and saturated levels of the ligand, respectively, while Lt and Pt are the respective total ligand (GRN-3 or rGRN-3) and TMR-Aβ concentrations. The ThT kinetic data were fit using the following single-order exponential growth equation corresponding to a seeded aggregation growth.
| (2) |
Heteronuclear multiple quantum coherence spectroscopy
The heteronuclear multiple quantum coherence (HMQC) NMR spectra for 15N GRN-3 or rGRN-3 resuspended in 20 mM Tris–HCl (pH 8.0) with 10% D2O was acquired following incubation with and without Aβ42 (in a stoichiometric ratio of 1:2 Aβ42:GRN-3/rGRN-3) for 24 h inside a Shigemi NMR tube. The data were acquired on a Bruker Advance — III-HD 850 MHz NMR spectrometer equipped with a Bruker TCI cryoprobe at the high-field NMR facility of University of Alabama, Birmingham as described previously [21].
Differential interference contrast microscopy
The microscopy images were obtained after incubating Aβ42 with and without GRN-3 or rGRN-3 for 24 h. Samples were then loaded on a fabricated microscopic glass slide apparatus. Briefly, a double-sided tape was placed onto the edges of the slide followed by loading of a drop of the determined volume of each sample onto the center of the slide. A glass coverslip was then adhered with its edges to double-sided tape, layering the sample and enabling visualization. The samples were observed and imaged on a Nikon Eclipse 80i under 20× magnification with a differential interference contrast (DIC)-polarizing filter.
MALDI-ToF mass spectrometry
Pellet characterization of the co-incubated protein complexes was performed on a Bruker Datonics Microflex LT/SH ToF-MS system. Samples were prepared by mixing the supernatant or pellet aliquots from respective reactions with 12.33 ng of insulin (used as an internal standard). The pellet samples were dissolved with formic acid in a 1:1 ratio to allow disaggregation of fibrils and then spotted onto a Bruker MSP 96 MicroScout Target with 1:1 ratio of sample:sinapinic acid matrix in saturated acetonitrile and water. The laser intensity was kept constant at 75% with 3.5× detector gain. Instrument calibration was performed using Bruker Protein Calibration Standard I (Bruker Daltonics).
Turbidity assays
Turbidity assay to monitor fibril formation was performed using a BioTek Synergy H1 microplate reader. Freshly purified GRN-3 or rGRN-3 were incubated with monomeric Aβ42 in 96-microwell plate at 37°C for a period of 2 h, before one oxidized GRN-3 reaction was reduced with 0.5 mM TCEP and the subsequent change in turbidity was monitored at 360 nm. The data were processed using Origin 8.5.
Gel electrophoresis and immunoblotting
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE) was carried out using 4–20% Mini-Protean pre-cast gels (Bio-Rad). For immunoblotting, the proteins were transferred on a 0.45 μm Amersham Protran Premium nitrocellulose membrane (GE Life Sciences) and blocked with 5% non-fat dry milk in 1× PBS with 0.1% Tween 20. The blots were probed overnight with an Aβ42-specific antibody, Ab5 [33]. Blots were then incubated with a HRP-conjugated anti-mouse anti-Fc secondary antibody (Sigma) for 1–2 h and developed with ECL substrate for 2 min (Thermo Scientific). Images were obtained using a GelDoc molecular imager (Bio-Rad).
Cell preparation and treatment
SH-SY5Y human neuroblastoma cells (American Type Culture Collection, Manassas, VA), maintained at 37°C in a humidified atmosphere of 5% CO2/95% air, were cultured using a 1 : 1 mixture of Ham’s F12K medium and DMEM (F12K/DMEM) containing 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. Prior to experimentation, cells were seeded onto 22 × 22 mm glass coverslips (VWR, Radnor, PA) at a density of 5 × 105 cells/ml and maintained for 24 h. In parallel, oligomerization reactions were prepared in 20 mM Tris–HCl (pH 6.5) containing 50 mM NaCl for final concentrations of 0 (negative controls) or 10 μM Aβ42 and 0 (positive control), 10, or 20 μM GRN-3. Aggregation was initiated through immediate incubation at 37°C. Aggregation was halted after 24 h via dilution in F12K/DMEM supplemented with 1% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin for a final concentration of 50 nM Aβ42 and used immediately for cell treatment. Cells treated with media containing an equivalent dilution of buffer served as vehicle control (VEH). Cells were incubated for 24 h (37°C/5% CO2/95% air) prior to assessment of caspase activity.
Detection of active caspase
The Image-iT LIVE Green Caspase-3, −7 kit (Molecular Probes, Inc., Eugene, OR) was used to assess caspase activation in treated SH-SY5Y cells. This kit utilizes a FLuorescent Inhibitor of CAspases (FLICA™) to specific-ally label active forms of caspase-3 and caspase-7, the ‘executioner caspases’ intrinsic to apoptosis [34]. After treatment removal, cells were briefly rinsed with phenol red-free F12K/DMEM media supplemented with 1% FBS (used for all subsequent rinses and incubations unless otherwise specified). Washed cells were incubated for 1 h with FLICA™ reagent (1 : 150). The cells were then rinsed twice prior to incubation for 10 min with 1 μM Hoechst 33342 to enable visualization of nuclei. After two successive rinses with the kit-included wash buffer, the coverslips were mounted to slides using the kit-included fixative solution. A Nikon Eclipse Ti-e fluorescent microscope was used to image Hoechst- and FLICA-labeled cells at 40×. All slides were imaged within 24 h of fixation; images of five different fields were acquired for each slide. Image acquisition settings remained uniform within each experiment.
Images were analyzed to quantify caspase-3, −7 activities of each individual cell via a custom subroutine written in MATLAB (MathWorks, Natick, MA). This subroutine first converts the Hoechst channel into binary 8-bit images through comparison of individual monochromatic pixels to a common threshold. Pixels representative of nuclear boundaries are identified, and successive ‘layers’ of pixels are iteratively distinguished through the progression from these boundaries to the interior of the cell until convergence is reached. Upon convergence, a radius of exclusivity is established relative to the pixel of convergence to prohibit additional cell detection within this area of exclusivity, and the cell is counted. This component of the subroutine algorithm was calibrated to statistically mimic manual cell counts. The subroutine then assesses and quantifies from the FLICA™ channel the fluorescence within each relative cell boundary to generate an average pixel intensity representative of the activated caspase within each cell. The relative cell boundary is defined as the nuclear area plus an additional specified region beyond the nucleus to encompass the total area of the cell. Cells that demonstrate an average activated caspase pixel intensity >5 are classified as caspase-active. Subroutine output is expressed as the percentage of caspase-active cells; results from each experiment are reported as the fraction of the vehicle (for assessment of the positive control) or a fraction of the positive control (for comparison among samples and controls). Normalized results from each experiment were assessed for significance using GraphPad Prism 7.0c software (La Jolla, CA). A one-way analysis of variance (ANOVA) was used to compare all treatments to the controls, and a Tukey’s multiple comparisons test was used to assess significance between samples.
Results
GRN-3 binds to Aβ42 monomers in both redox states
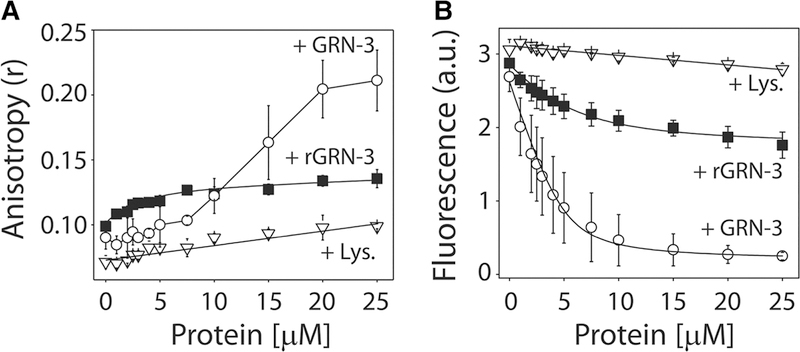
The interaction between Aβ42 monomers and GRN-3 or rGRN-3 was investigated using intrinsic fluorescence, fluorescence anisotropy, and NMR spectroscopy. The titration of GRN-3 on N-terminal-labeled, TMR-Aβ maintained at 5 μM showed a negligible increase in anisotropy below ∼1 : 1 stoichiometry (Figure 1A, ○). Beyond the stoichiometric equivalence, exponential increase in anisotropy was observed that saturates above five-fold stoichiometric excess, exhibiting an overall sigmoidal isotherm (Figure 1A, ○). Upon fitting the data to a Hill equation, the data indicate that the binding of GRN-3 to Aβ involves positive cooperativity (Supplementary Figure S2). Similar titration of rGRN-3 on TMR-Aβ resulted in an exponential increase in anisotropy that saturated at an anisotropy value of 0.14, suggesting a weaker binding (Figure 1A, ■). In comparison, the negative control, BSA, did not show any increase in anisotropy, suggesting no interaction with Aβ42 (Figure 1A, ∇). The binding affinity was also confirmed by intrinsic fluorescence changes in TMR-Aβ. Titration of both GRN-3 and rGRN-3 resulted in an exponential decrease in the intensity (Figure 1B, ○ and ■, respectively). However, GRN-3 induced a more exponential decrease than rGRN-3. Fitting the data using a simple, one-site binding equation (eqn 1) yielded apparent dissociation constants of 0.55 ± 0.1 and 2.55 ± 0.9 μM for GRN-3 and rGRN-3 binding to Aβ42, respectively. As expected, the negative control hen egg lysosome (Amresco Inc.) showed no discernible binding (Figure 1B, ∇). These experiments indicate that both redox states of GRN-3 bind to monomeric Aβ42 with a five-fold difference in binding affinities.
Figure 1. Binding interactions between GRN-3 and Aβ42.
(A) Changes in fluorescence anisotropy (r) of TMR-Aβ upon increasing concentrations of GRN-3 (○), rGRN-3(■), or BSA (∇; negative control). (B) Changes in the normalized intrinsic fluorescence intensities of TMR-Aβ, upon titration with increasing concentrations of GRN-3 (○), rGRN-3(■), or hen egg lysozyme (∇). The data were fit (black line) using a single ligand binding model as described in the Experimental section. The results represented are an average of n = 3 independent datasets.
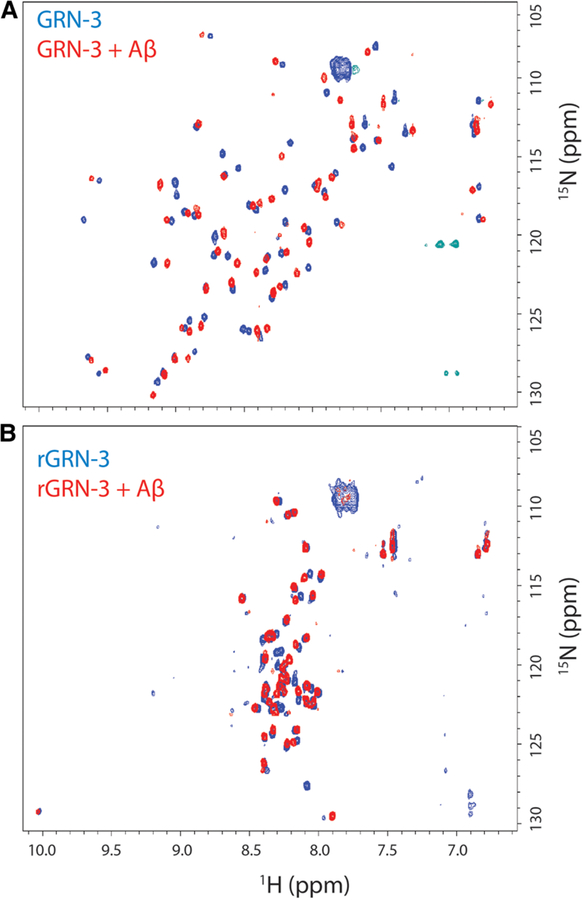
To further identify the binding interactions between GRN-3 or rGRN-3 and Aβ42, 1H–15N HMQC NMR spectroscopy was performed using uniformly 15N-labeled GRN-3. The data obtained showed good chemical shift dispersion on both 1H and 15N dimensions for GRN-3 as reported previously [21] (Figure 2A, blue). In this experiment, we maintained 1:2 stoichiometry but with 20 and 40 μM of Aβ42 and GRN-3, respectively, as the generation of high amounts of 15N-labeled samples became a limiting factor. Approximately, 45% of the peaks were shifted and ∼8% of the resonances could not be observed as previously reported [21]. When GRN-3 was mixed with unlabeled monomeric Aβ42, widespread chemical shift perturbations were observed in the HMQC spectra in both the 1H and 15N dimensions (Figure 2A, red). These observations indicate that the addition of Aβ42 caused a distinct change in the chemical environment of the GRN-3 backbone, and thereby confirming a direct interaction between the two proteins. On the other hand, the HMQC spectrum of rGRN-3 alone showed a markedly lesser number of resonances (50% not observed), and a lower spectral resolution than GRN-3, indicating that the protein is highly disordered (Figure 2B, red), as previously observed [21]. The addition of monomeric Aβ42 showed subtle, yet distinct changes in the chemical shifts suggesting interactions between the two proteins (Figure 2B, blue). Based on the spectra, the addition of Aβ42 did not result in an increase in order within the rGRN-3 structure. However, the widespread chemical shift changes observed suggest that the interactions with Aβ could be multivalent, something which needs further experimental validation. From the extent of chemical shift changes, it would also be inconclusive to infer that the interaction of GRN-3 with Aβ42 may be more or less specific than that of rGRN-3. Further detailed NMR analysis on these aspects will be published at a later time.
Figure 2. NMR analysis of GRN-3 and Aβ42 interactions.
1H–15N HMQC spectra of the amide backbone region for 40 mM, uniformly 15N-labeled GRN-3 (A) or rGRN-3 (B) in the presence (red) or absence (blue) of 20 mM of Aβ42 monomers incubated for 24 h.
Both GRN-3 and rGRN-3 rapidly promote fibril formation upon interacting with monomeric Aβ42
The effect of GRN-3 or rGRN-3 on the aggregation of monomeric Aβ42 was monitored using ThT fluorescence. ThT is an extrinsic fluorescent dye that shows an increase in its fluorescence intensities upon binding to the β-sheet-rich Aβ42 aggregates and therefore is routinely used to monitor Aβ42 aggregation [35]. Freshly purified, aggregate-free Aβ42 monomers were incubated with GRN-3 or rGRN-3 quiescently in 20 mM Tris– HCl (pH 6.5) and 50 mM NaCl at 37°C. To ensure adequate interaction, a two-fold molar excess of both
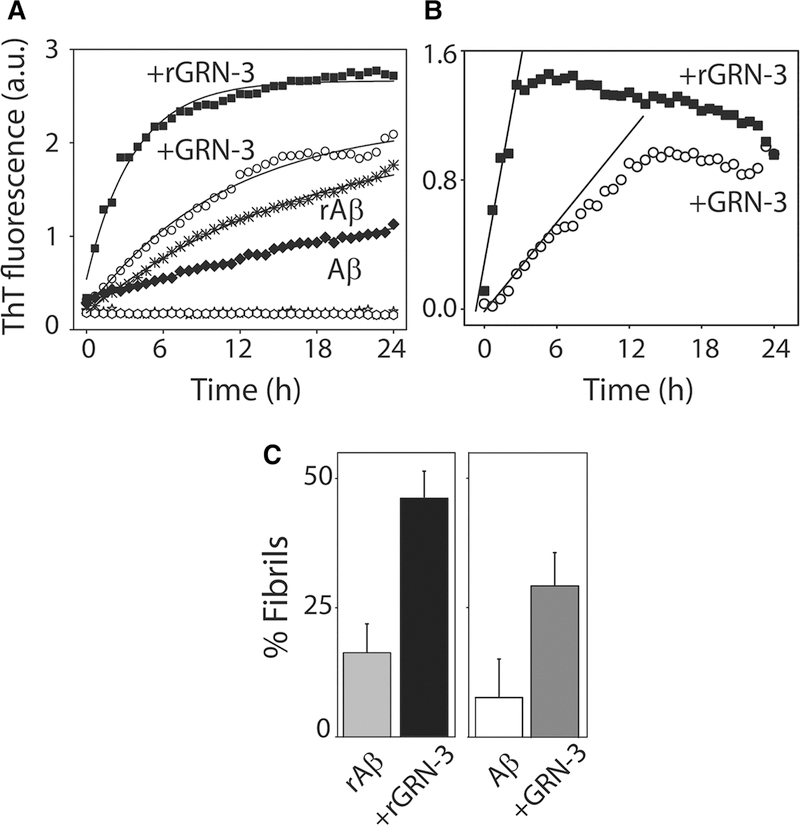
GRN-3 and rGRN-3 over Aβ42 was used. The co-incubated samples showed increased aggregation rates as indicated by the increase in the fluorescent intensity (Figure 3A, ○ and ■), as opposed to the Aβ42 controls (Aβ42 monomers alone or in the presence of TCEP; rAβ) incubated and monitored under identical conditions (Figure 3A, ★ and ♦). Both GRN-3 and rGRN-3 controls did not show any increase in ThT fluorescence (Figure 3A; ■ and ○). Appropriate Aβ42 control-subtracted, net aggregation kinetics of the reactions indicate a faster rate was induced by rGRN-3 (■) than GRN-3 (○) in Figure 3B with first-order rate constants calculated as 68.25 ± 2.48 and 15.25 ± 6.96 h−1, respectively, using the initial rate method (Figure 3B). To identify whether the aggregates formed after 24 h were sedimentable high molecular mass aggregates, the samples were subjected to centrifugation at 19 000×g and the supernatant was subjected to ThT assay. The intensities were then used to quantitate the percentage of sedimentable fibrils formed. The data showed that rGRN-3 induced ∼48% of fibrils as compared with 18% in the absence of GRN-3 (Aβ42 in the presence of TCEP), while GRN-3 induced ∼30% of fibrils as compared with 9% in the absence of GRN-3 (Figure 3C). Collectively, these results suggest that while both rGRN-3 and GRN-3 accelerate Aβ42 aggregation towards the formation of high molecular mass fibrils, the former does so almost 10 times more rapidly than the latter.
Figure 3. Aβ42 fibril formation induced by GRN-3 and rGRN-3.
(A) Aβ aggregation monitored by ThT fluorescence in the presence of rGRN-3 (■) or GRN-3 (○) with respective Aβ42 controls (Aβ42 + TCEP (rAβ); ⬡ and Aβ42 alone; ♦), and GRN-3 (○) or rGRN-3 (■) alone for 24 h. Briefly, 20 μM monomeric Aβ42 was incubated with 40 μM GRN-3 or rGRN-3 in a 1 : 2 stoichiometry. For the controls, 20 μM monomeric Aβ42 was incubated alone or in the presence of TCEP. (B) Net ThT kinetics of reactions from (A) with rGRN-3 (■) and GRN-3 (○) obtained by subtracting corresponding Aβ42 controls. The data were fit (black lines) using the initial rate method to obtain rate constants for the growth kinetics from the slope. (C) Quantification of the fibrils formed after 24 h in the presence or absence of GRN-3 or rGRN-3 determined via the relative difference between the ThT intensities of the samples and their respective supernatants obtained after centrifuging at 18 000×g for 20 min. The results represented are an average of n = 3 independent data sets.
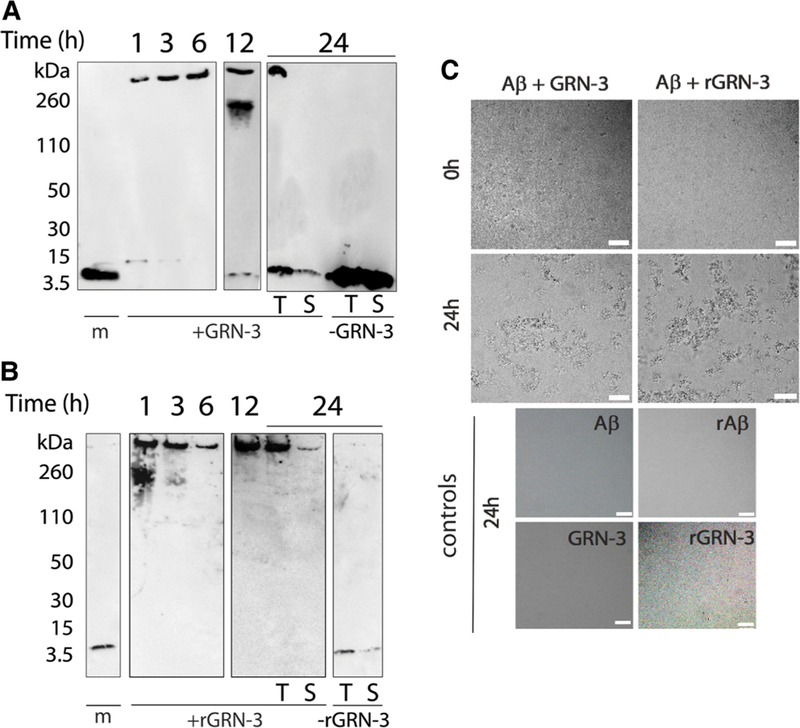
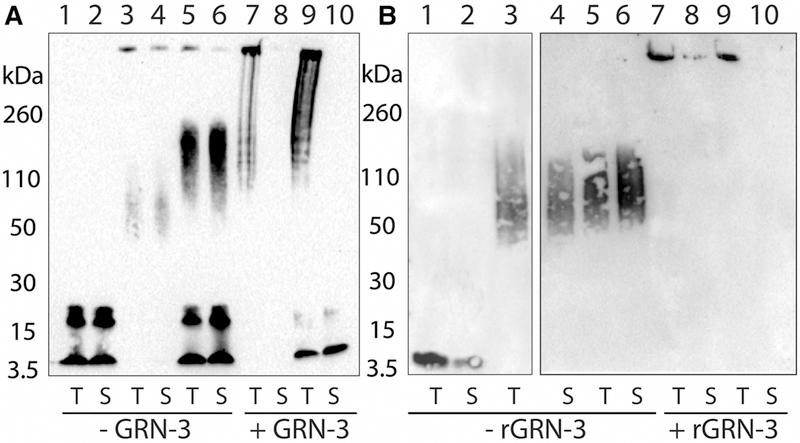
In parallel, aliquots from the co-incubated samples were electrophoresed and immunoblotted using a pan-Aβ42 monoclonal antibody, Ab5 [33], to investigate the nature of the aggregates formed in the co-incubated samples. A high molecular band (>260 kDa) was observed for Aβ42 co-incubated with GRN-3 with an increasing intensity with the incubation time (Figure 4A, lanes 1, 3, 6, 12, and 24 h, +GRN-3). In contrast, Aβ42 control (−GRN-3) incubated under similar conditions showed no high molecular mass band even after 24 h of incubation (Figure 4A, lanes m and T; −GRN-3). The supernatant of the co-incubated samples subjected to centrifugation after 24 h showed no high molecular mass band suggesting the presence of sedimentable fibrils (Figure 4A, T and S; +GRN-3). Similarly, co-incubation of Aβ42 with rGRN-3 also showed the formation of high molecular mass aggregates within 1 h that was sedimentable after 24 h (Figure 4B, lanes 1, 3, 6, 12, and 24 h, +rGRN-3) as opposed to rAβ control (Figure 4B, lanes m and T; −rGRN-3). These results indicate that the increase in the ThT fluorescence for Aβ42 co-incubated with GRN-3 or rGRN-3 is indeed due to the formation of Aβ42 aggregates. To unambiguously confirm the fibrillar nature of the high molecular mass species formed in the co-incubated samples, aliquots from the co-incubated samples were imaged at 0 h and 24 h using DIC microscopy (Figure 4C). Aliquots from control reactions were imaged at 24 h only (Figure 4C). The images demonstrate large fibrillar structures after 24-h co-incubation of Aβ42 with both GRN-3 and rGRN-3, and these fibrils were absent from the controls. These data complement ThT fluorescence results by identifying the aggregates of Aβ42 promoted by both forms of GRN-3 as fibrils.
Figure 4. High molecular mass Aβ42 fibrils detected by immunoblots and differential interference contrast light microscopy (DIC).
(A,B) Aliquots of the co-incubated reactions of GRN-3 (A) and rGRN-3 (B), similar to reactions described in Figure 3, were subjected to electrophoresis and immunoblotting at the indicated time points of 1, 3, 6, 12, and 24 h, respectively. Lane ‘m’ represents Aβ42 monomer control. After 24 h, the samples were centrifuged at 18 000×g and both the sample before (total; T) and after centrifugation (supernatant; S) were electrophoresed along with respective controls. (C) DIC was used to observe the fibrillar structures formed by the interaction of Aβ42 with GRN-3. Scale bar denotes 50 μm.
Characterization of sedimented aggregates suggest potential mechanisms for GRN-3
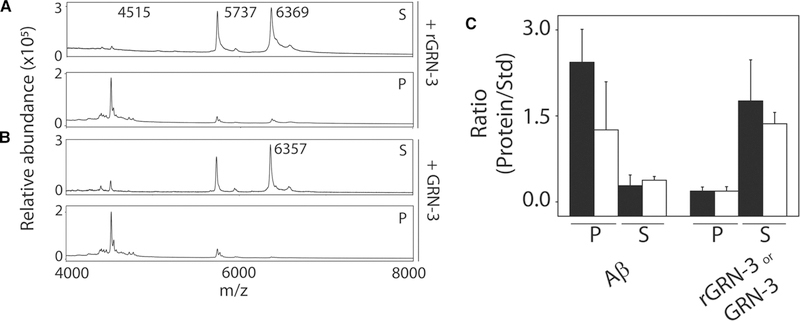
To obtain more insights into the mechanisms of how oxidized and reduced forms of GRN-3 are able to promote fibrils, supernatant and pellet fractions obtained after the sedimentation of 24-h aggregates were examined using MALDI-ToF mass spectrometry. A known amount of insulin (12.33 ng/μl) was used as an internal standard for quantitation. As expected, a significant amount of Aβ42 was observed in the pellet fraction as compared with the supernatant fraction for both GRN-3 and rGRN-3 incubations supporting the data on fibril formation (Figure 5A–C). On the contrary, the amount of both GRN-3 and rGRN-3 were insignificant in the pellet fractions while they were abundant in the supernatant fractions (Figure 5A–C). Overall, these results indicate that both GRN-3 and rGRN-3 promote Aβ42 aggregation rapidly without co-aggregating with Aβ42. While GRN-3 appears to form a co-complex with Aβ42 based on NMR spectra, mass spectrometry data do not indicate the presence of GRN-3 in the fibril pellets in equimolar quantities. Together with binding, mass spectrometric and NMR data, this suggests that both GRN-3 and rGRN-3 interact with Aβ42 monomers in such a way that they form a co-complex which serves as a nucleus for subsequent rapid aggregation and promotion of fibrils. It is likely that there are subtle yet distinct differences in which GRN-3 and rGRN-3 interact to provide the nucleus for Aβ aggregation, but more detailed atomistic data are needed to decipher such differences.
Figure 5. Characterization of fibrils formed upon co-incubation of GRN-3 or rGRN-3 and Aβ42.
MALDI-ToF mass spectra were obtained from the reactions in Figure 4 from both the sedimented pellet and the supernatant after 24 h. Insulin (12.33 μg) was used as an internal standard (Std). (A) MS spectra for the pellet (P) and supernatant (S) from the reaction of Aβ42 (4515 amu) with GRN-3 (6357 amu). (B) MS spectra for the pellet (P) and supernatant (S) from the reaction of Aβ42 (4515 amu) with rGRN-3 (6369 amu). (C) The relative intensities of the proteins in the respective P and S fractions are expressed as a ratio with respect to that of the insulin standard. The closed and open bars represent the reactions with rGRN-3 and GRN-3, respectively.
GRN-3 and rGRN-3 also promote the formation of high molecular mass aggregates by Aβ42 oligomers
Our data demonstrate that GRN-3 is able to interact with Aβ42 monomers and rapidly promote fibril formation. We further questioned whether the protein could interact with oligomers, which are known to be the primary neurotoxic species. Therefore, the interaction between GRN-3 and 12–24mer Aβ42 oligomers called LFAOs was investigated. LFAOs are known to self-propagate in the presence of monomers [32], and, specifically, the effect of GRN-3 in this process was explored. Freshly purified LFAOs (3.0 μM) were incubated with aggregate-free Aβ42 monomers (20 μM) in the presence and absence of GRN-3 and rGRN-3 (40 μM) in 20 mM Tris–HCl (pH 8.0) for 72 h at room temperature. The samples were subjected to SDS–PAGE and immunoblotted using pan anti-Aβ42 monoclonal antibody, Ab5. After 72 h of incubation, Aβ42 monomers remained monomeric with some tetramers (Figure 6A,B, lanes 1 and 2). Similarly, LFAOs also show a band between 50 and 110 kDa as expected [36] (Figure 6A,B, lanes 3 and 4). The addition of LFAO seeds to Aβ42 monomers results in the enhancement in oligomer formation (between 110 and 260 kDa) along with some fibrils (Figure 6A,B, lanes 5 and 6). In all of these samples, centrifugation did not lead to the sedimentation of high molecular mass species as evidenced by the identical banding pattern in total (T) and supernatant (S) lanes. In contrast, both LFAOs and Aβ42 monomers incubated with GRN-3 and rGRN-3 exhibited the formation of fibril-like, high molecular mass oligomers (Figure 6A,B, lanes 7 and 9). Centrifugation of these samples resulted in a decrease in the high molecular mass bands suggesting the formation of sedimentable fibrils in these samples (Figure 6A,B, lanes 8 and 10). These results suggest that GRN-3, in both of its redox states, is capable of promoting Aβ42 fibril formation irrespective of latter’s oligomeric state.
Figure 6. GRN-3 promotes aggregation of oligomeric Aβ42.
Western blot analysis of the effect of GRN-3 (A) or rGRN-3 (B) on Aβ42 oligomers (LFAOs). Aβ42 monomer control (20 μM; lanes 1 and 2). LFAOs alone (3.0 μM; lanes 3 and 4) or after incubation with Aβ42 monomer for 72 h at 25°C (20 μM; lanes 5 and 6). The co-incubation of LFAOs (3.0 μM) and GRN-3 or rGRN-3 (40 μM) in the absence of Aβ42 monomer (lanes 7 and 8), and co-incubation of similar concentrations of LFAOs and GRN-3 or rGRN-3 in the presence of Aβ42 monomer (20 μM; lanes 9 and 10). For lanes 9 and10, GRN-3 (or rGRN-3) and LFAOs were incubated for 1 h prior to the addition of Aβ42 monomers. In all lanes, T represents the total sample while S denotes the supernatant after centrifugation for 20 min at 18 000×g.
Fibril promotion is enhanced in reducing conditions
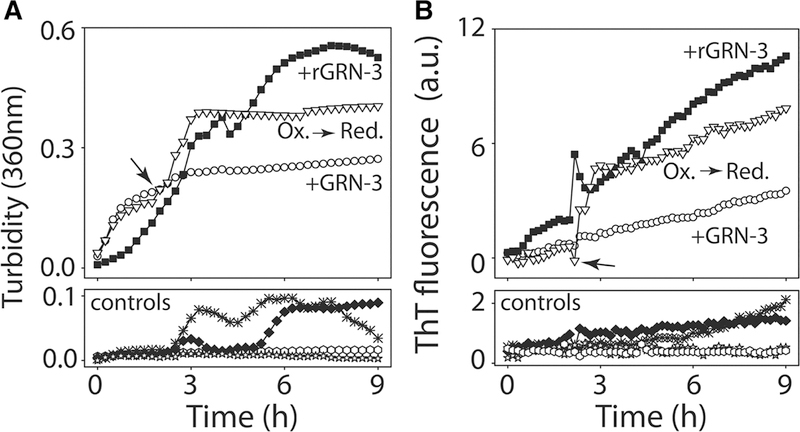
To identify whether redox changes affect the aggregation of Aβ42 in the presence of GRNs, Aβ42, co-incubated with GRN-3, was reduced by the addition of TCEP after 2 h and turbidity and ThT fluorescence were monitored. Turbidity for Aβ42 co-incubated with GRN-3 showed an exponential increase before plateauing after 3 h (Figure 7A, ○). Interestingly, the reduction in GRN-3 co-incubated with Aβ42 by the addition of TCEP at 2 h (indicated by an arrow and hashed line in Figure 7A) resulted in further increase in turbidity (Figure 7A, ∇) that was comparable to the turbidity observed for Aβ42 co-incubated with rGRN-3 (Figure 7A, ■). In contrast, the increase in turbidity for corresponding controls (Figure 7A, controls) was negligible. In parallel, the same reactions were also monitored using ThT fluorescence. A similar trend was observed for ThT fluorescence as that for turbidity measurements. The increase in the ThT fluorescence for Aβ42 co-incubated with GRN-3 that was reduced after 2 h (Figure 7B, ∇) was higher as compared with Aβ42 co-incubated with GRN-3 that was not reduced (Figure 7B, ○). This increase was again comparable to that of Aβ42 co-incubated with rGRN-3 (Figure 7B, ■). The ThT measurements for rGRN-3 and GRN-3 corroborate with our previous data (Figure 3). The controls showed expected behavior with a negligible increase in ThT fluorescence (Figure 7A: Aβ42 monomers alone; ★, or in the presence of TCEP; rAβ ♦ and Figure 7B: GRN-3 and rGRN-3 controls ♦ and ✳). These results suggest that in physiological states with dynamic redox fluctuations, Aβ42 aggregation could be enhanced by the reduced form of GRN-3.
Figure 7. Reduction in GRN-3 induces augmentation in fibril formation.
Incubation of 40 μM GRN-3 with 20 μM Aβ42 at 37°C was monitored using turbidity measurements at 360 nm (A) or by ThT assay (B). After 2 h of incubation, the reaction was reduced by the addition of 500 μM TCEP (arrow) to observe the subsequent kinetics (∇; Ox → Red). Reactions with rGRN-3 (■) and GRN-3 (○) were used as reaction controls along with others such as Aβ (♦), rAβ (⬡), rGRN-3 (✳), and GRN-3 (♦).
Fibril promoting effect of GRN-3 results in attenuation of Aβ42-induced caspase activation in neuronal cells
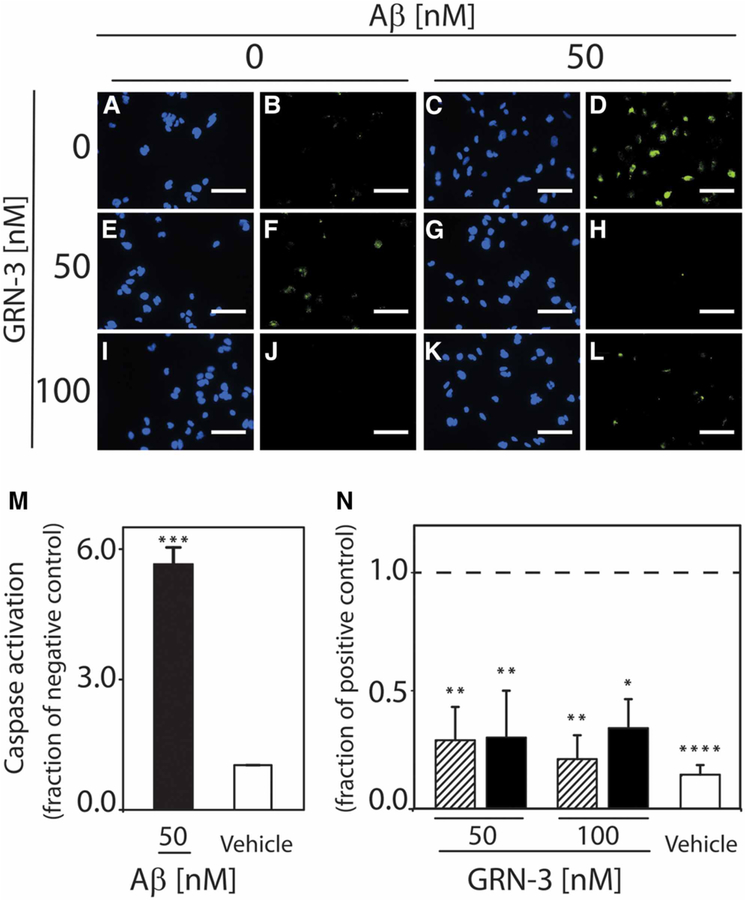
Large Aβ aggregates, including fibrils, have been observed to exhibit lower levels of neurotoxicity than small aggregate species, such as oligomers [37,38]. Thus, the ability of GRN-3 to promote fibril formation could reduce the ability of Aβ to elicit neuronal responses associated with apoptosis. To evaluate the physiological influence that arises from the direct interaction of GRN-3 with Aβ42, Aβ aggregates formed in the absence or presence of GRN-3 were examined for their ability to activate caspase-3 and −7 in SH-SY5Y human neuroblastoma cells. rGRN-3 was not used in these experiments due to the uncertainty of the oxidation state of the protein following introduction into cell culture. In the presence of oxidizing agents, present in cell culture media designed to mimic the in vivo redox environment, reduced proteins will spontaneously regenerate disulfide bonds [39], and this process can occur on the order of hours [40]. Thus, the oxidation state of rGRN exposed to cell culture conditions for 24 h could not be ascertained. Cells treated for 24 h with Aβ42 aggregated alone exhibit significantly increased activation of these ‘executioner’ caspases relative to the vehicle (P < 0.001) (Figure 8A–D,M). In contrast, GRN-3, introduced at the onset of aggregation in equimolar (Figure 8G,H) or two-fold excess (Figure 8K,L) quantities relative to Aβ42, can attenuate Aβ42-induced caspase activation. Cells receiving these treatments exhibit a 60–70% decrease in caspase activation relative to cells treated with Aβ42 aggregated alone (1 : 1 GRN-3:Aβ42, P < 0.01; 2 : 1 GRN-3:Aβ42, P < 0.05) (Figure 8N). Moreover, these cells exhibited no difference in caspase activation when compared with cells treated with GRN-3 alone (Figure 8E,F, I,J,N) or with the vehicle (Figure 8A,B,N). Together, these results demonstrate that GRN-3 exerts a significant protective effect from Aβ42 toxicity.
Figure 8. Direct interaction between GRN-3 and Aβ42 modulates Aβ-induced caspase activation.
SH-SY5Y human neuroblastoma cells were incubated for 24 h with the vehicle (A,B), 50 nM Aβ42 aggregated alone (C,D), 50 nM and 100 nM GRN-3 alone (E,F,I,J), and 50 nM Aβ42 aggregated in the presence of 50 nM GRN-3 (G,H) or 100 nM GRN-3 (K,L). To assess activation of caspases, cells were stained for nuclear markers (Hoechst 33342, blue) and activated forms of caspase-3 and −7 (FLICA, green) and imaged at 40×. Images are representative of three to four independent experiments. Scale bar indicates 50 μm. Image analysis was performed using a custom MATLAB subroutine to determine the percentage of caspase-active cells. (M) The percentage of caspase-active cells following treatment with 50 nM Aβ42 aggregated alone (black bar) or an equivalent dilution of buffer (vehicle, white bar) is shown normalized to the vehicle. ***P < 0.001. Error bars indicate SEM, n = 3–4. (N) The percentage of caspase-active cells following treatment with GRN-3 alone (lined bars) or with 50 nM Aβ42 aggregated in the presence of GRN-3 at a 1:1 or 2:1 molar ratio (GRN-3:Aβ42) (black bars) is shown normalized to the positive control (50 nM Aβ42 aggregated alone). The positive control is indicated by a dashed line at 1. *P < 0.05, **P < 0.01 Error bars indicate SEM, n = 3–4.
Discussion
Emerging evidence suggests that neuroinflammation is not just a passive event activated as a downstream effect of accumulating Aβ plaques and neurofibrillary tau tangles in AD pathology but plays an active role in the disease onset and progression [41–45]. Neuropathological and genetic studies indicate at the onset of AD a surge in the release of proinflammatory markers, such as cytokines like IL-1, IL-6, and TNFα and other inflammatory proteins [46]. In addition, microglial as well as astroglial activation is observed in patients with mild cognitive dementia [41,47]. Chronic activation of the microglia in AD causes these cells to be arrested in a perpetual inflammatory state [48], which can lead to improper clearance of amyloid plaques [49,50]. Therefore, rigorous investigations of these molecular processes, especially the roles of proinflammatory markers and their effect on the onset and progression of AD seem imperative to understand the pathology in greater detail. In this context, the roles of PGRN and GRNs, which have long been known to be involved in multiple biological functions, have captured attention lately in relation to neurodegenerative disorders. Although one report suggests proinflammatory roles for GRNs [6], involvement in lysosomal function and dysfunction is beginning to emerge for both PGRN and GRNs [25,26,28].
Despite their involvement in a multitude of biological functions, the precise structure–function relationship of GRNs is far from clear. One of the intriguing aspects of GRNs is the degree of cysteine content in their sequence (∼17% by weight), which is the highest among mammalian proteins [51]. We recently demonstrated that the disulfide bonds govern the structure and stability of GRN-3, and abrogation of these bonds renders the protein fully disordered [20,21]. Furthermore, even the fully reduced GRN-3 activates NF-κB, suggesting the potential roles of both oxidized and reduced protein in cellular functions [20]. Here, we asked the question of whether GRNs can directly interact with Aβ and modulate neuronal toxicity. As the results indicate, both GRN-3 and rGRN-3 accelerate Aβ42 fibril formation. However, two proteins show subtle differences in the mechanisms by which they augment fibrillation. While GRN-3 binds to Aβ42 monomers more strongly than rGRN-3, the latter promotes fibril formation more rapidly than the former. Although the detailed structural investigation is warranted to understand the precise mechanisms, at this point, one could speculate that while Aβ42–GRN-3 complex nucleates subsequent Aβ42 aggregation, rGRN-3 augments Aβ aggregation by a templating or coacervation-type interaction between the two disordered proteins. It is noteworthy that both redox forms of GRN-3 are not observed in the Aβ42 aggregate pellets, which suggests that they act as nucleating agents. Similarly, both redox forms of GRN-3 are able to promote fibrils from stable oligomers also. Together, the data indicate that both rGRN-3 and GRN-3 promote fibrils at the cost of toxic oligomer formation. This mechanism seems to rescue Aβ42-induced neurotoxicity in neuroblastoma cells at least by oxidized GRN-3. Similar caspase assay could not be performed with rGRN-3 for obvious uncertainty in the precise oxidation state of the protein during the assay. Nevertheless, the mechanisms of interaction between GRN-3 and Aβ42 and their biophysical and cellular consequence are currently being investigated in greater detail and will be reported at a later date. However, our preliminarily NMR results suggest that the interaction between GRN-3 and Aβ42 invokes chemical shifts that involve a wide range of residues in GRN-3 (Figure 2A), indicating the possibility of coacervation between the two proteins. Indeed, further investigation is warranted to ascertain these mechanisms unambiguously, which are currently underway in our laboratory and will be reported at a later date.
Conclusions
The results presented here potentially have far-reaching implications for AD. Although many circumstantial evidences suggest a role of GRNs in AD, a direct interaction between GRNs and Aβ has never been established. Although it remains unclear whether this interaction is extracellular or intracellular, given the localization of GRNs in both places, one could hypothesize two possibilities: extracellular interaction can arise as a consequence of an inflammatory event where GRNs secreted by microglia could interact with Aβ generated from neuronal APP. On the other hand, intracellular interactions can arise from sortillin-based intake of PGRN by the neurons and its eventual cytoplasmic cleavage into GRNs. The interaction of GRNs with intracellular Aβ to promote fibril formation may involve their degradation via lysosomal or autophagic pathways and thereby rescue Aβ-induced toxicity. Nevertheless, it is clear that more questions remain to be answered regarding the precise role of GRNs in neurodegenerative diseases. However, the results presented here do provide intriguing insights into their potential mechanisms and present an entirely new realm of thoughts on how the onset and progression of AD are affected by GRNs.
Supplementary Material
Acknowledgments
Funding
The research was supported by National Center for Research Resources [5P20RR01647–11] and the National Institute of General Medical Sciences [8 P20 GM103476–11] from the National Institutes of Health for funding through INBRE (to V.R.), the National Institute of Aging [R15AG946815, to V.R. and M.A.M.] and the National Institute of General Medical Sciences [R01 GM120634–01A1 to V.R.].
Abbreviations
- Aβ
amyloid-β
- AD
Alzheimer’s disease
- ANOVA
analysis of variance
- DIC
differential interference contrast
- FLICA
FLuorescent Inhibitor of Caspases
- FTD
frontotemporal dementia
- GRN
granulins
- HMQC
heteronuclear multiple quantum coherence
- LFAOs
large fatty acid-derived oligomers
- PGRN
progranulin
- SDS–PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- TCEP
Tris(2-carboxyethyl)phosphine hydrochloride
- ThT
Thioflavin-T
- TMR-Aβ
tetra-methyl rhodamine-labeled Aβ42
- VEH
vehicle control
Footnotes
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Bateman A, Belcourt D, Bennett H, Lazure C and Solomon S (1990) Granulins, a novel class of peptide from leukocytes. Biochem. Biophys. Res. Commun 173, 1161–1168 10.1016/S0006-291X(05)80908-8 [DOI] [PubMed] [Google Scholar]
- 2.Bhandari V, Palfree RG, and Bateman A (1992) Isolation and sequence of the granulin precursor cDNA from human bone marrow reveals tandem cysteine-rich granulin domains. Proc. Natl Acad. Sci. U.S.A 89, 1715–1719 10.1073/pnas.89.5.1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plowman GD, Green JM, Neubauer MG, Buckley SD, McDonald VL, Todaro GJ et al. (1992) The epithelin precursor encodes two proteins with opposing activities on epithelial cell growth. J. Biol. Chem 267, 13073–13078 [PubMed] [Google Scholar]
- 4.Bateman A and Bennett HP (1998) Granulins: the structure and function of an emerging family of growth factors. J. Endocrinol 158, 145–151 10.1677/joe.0.1580145 [DOI] [PubMed] [Google Scholar]
- 5.Liau LM, Lallone RL, Seitz RS, Buznikov A, Gregg JP, Kornblum HI et al. (2000) Identification of a human glioma-associated growth factor gene, granulin, using differential immuno-absorption. Cancer Res 60, 1353–1360 [PubMed] [Google Scholar]
- 6.Zhu J, Nathan C, Jin W, Sim D, Ashcroft GS, Wahl SM et al. (2002) Conversion of proepithelin to epithelins: roles of SLPI and elastase in host defense and wound repair. Cell 111, 867–878 10.1016/S0092-8674(02)01141-8 [DOI] [PubMed] [Google Scholar]
- 7.He Z and Bateman A (2003) Progranulin (granulin-epithelin precursor, PC-cell-derived growth factor, acrogranin) mediates tissue repair and tumorigenesis. J. Mol. Med 81, 600–612 10.1007/s00109-003-0474-3 [DOI] [PubMed] [Google Scholar]
- 8.Gass J, Lee WC, Cook C, Finch N, Stetler C, Jansen-West K et al. (2012) Progranulin regulates neuronal outgrowth independent of sortilin. Mol. Neurodegener 7, 33 10.1186/1750-1326-7-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C et al. (2006) Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 442, 916–919 10.1038/nature05016 [DOI] [PubMed] [Google Scholar]
- 10.Pickering-Brown SM, Baker M, Gass J, Boeve BF, Loy CT, Brooks WS et al. (2006) Mutations in progranulin explain atypical phenotypes with variants in MAPT. Brain 129, 3124–3126 10.1093/brain/awl289 [DOI] [PubMed] [Google Scholar]
- 11.Le Ber I, van der Zee J, Hannequin D, Gijselinck I, Campion D, Puel M et al. (2007) Progranulin null mutations in both sporadic and familial frontotemporal dementia. Hum. Mutat 28, 846–855 10.1002/humu.20520 [DOI] [PubMed] [Google Scholar]
- 12.van der Zee J, Le Ber I, Maurer-Stroh S, Engelborghs S, Gijselinck I, Camuzat A et al. (2007) Mutations other than null mutations producing a pathogenic loss of progranulin in frontotemporal dementia. Hum. Mutat 28, 416 10.1002/humu.9484 [DOI] [PubMed] [Google Scholar]
- 13.Shankaran SS, Capell A, Hruscha AT, Fellerer K, Neumann M, Schmid B et al. (2008) Missense mutations in the progranulin gene linked to frontotemporal lobar degeneration with ubiquitin-immunoreactive inclusions reduce progranulin production and secretion. J. Biol. Chem 283, 1744–1753 10.1074/jbc.M705115200 [DOI] [PubMed] [Google Scholar]
- 14.Brouwers N, Nuytemans K, van der Zee J, Gijselinck I, Engelborghs S, Theuns J et al. (2007) Alzheimer and Parkinson diagnoses in progranulin null mutation carriers in an extended founder family. Arch. Neurol 64, 1436–1446 10.1001/archneur.64.10.1436 [DOI] [PubMed] [Google Scholar]
- 15.Cortini F, Fenoglio C, Guidi I, Venturelli E, Pomati S, Marcone A et al. (2008) Novel exon 1 progranulin gene variant in Alzheimer’s disease. Eur. J. Neurol 15, 1111–1117 10.1111/j.1468-1331.2008.02266.x [DOI] [PubMed] [Google Scholar]
- 16.Pickford F, Marcus J, Camargo LM, Xiao Q, Graham D, Mo J-R et al. (2011) Progranulin is a chemoattractant for microglia and stimulates their endocytic activity. Am. J. Pathol 178, 284–295 10.1016/j.ajpath.2010.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pereson S, Wils H, Kleinberger G, McGowan E, Vandewoestyne M, Van Broeck B et al. (2009) Progranulin expression correlates with dense-core amyloid plaque burden in Alzheimer disease mouse models. J. Pathol 219, 173–181 10.1002/path.2580 [DOI] [PubMed] [Google Scholar]
- 18.Tolkatchev D, Malik S, Vinogradova A, Wang P, Chen Z, Xu P et al. (2008) Structure dissection of human progranulin identifies well-folded granulin/epithelin modules with unique functional activities. Protein Sci 17, 711–724 10.1110/ps.073295308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hrabal R, Chen Z, James S, Bennett H and Ni F (1996) The hairpin stack fold, a novel protein architecture for a new family of protein growth factors. Nat. Struct. Biol 3, 747–752 10.1038/nsb0996-747 [DOI] [PubMed] [Google Scholar]
- 20.Ghag G, Wolf LM, Reed RG, Van Der Munnik NP, Mundoma C, Moss MA et al. (2016) Fully reduced granulin-B is intrinsically disordered and displays concentration-dependent dynamics. Prot. Eng. Des. Sel 29, 177–186 10.1093/protein/gzw005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghag G, Holler CJ, Taylor G, Kukar TL, Uversky VN and Rangachari V (2017) Disulfide bonds and disorder in granulin-3: an unusual handshake between structural stability and plasticity. Protein. Sci 26, 1759–1772 10.1002/pro.3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eriksen JL and Mackenzie IR (2007) Progranulin: normal function and role in neurodegeneration. J. Neurochem 104, 287–297 10.1111/j.1471-4159.2007.04968.x [DOI] [PubMed] [Google Scholar]
- 23.Kessenbrock K, Fröhlich L, Sixt M, Lämmermann T, Pfister H, Bateman A et al. (2008) Proteinase 3 and neutrophil elastase enhance inflammation in mice by inactivating antiinflammatory progranulin. J. Clin. Invest 118, 2438–2447 10.1172/JCI34694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jian J, Hettinghouse A and Liu CJ (2017) Progranulin acts as a shared chaperone and regulates multiple lysosomal enzymes. Genes Dis 4, 125–126 10.1016/j.gendis.2017.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka Y, Suzuki G, Matsuwaki T, Hosokawa M, Serrano G, Beach TG et al. (2017) Progranulin regulates lysosomal function and biogenesis through acidification of lysosomes. Hum. Mol. Genet 26, 969–988 10.1093/hmg/ddx011 [DOI] [PubMed] [Google Scholar]
- 26.Kao AW, McKay A, Singh PP, Brunet A and Huang EJ (2017) Progranulin, lysosomal regulation and neurodegenerative disease. Nat. Rev. Neurosci 18, 325 10.1038/nrn.2017.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou X, Paushter DH, Feng T, Sun L, Reinheckel T and Hu F (2017) Lysosomal processing of progranulin. Mol. Neurodegener 12, 62 10.1186/s13024-017-0205-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holler CJ, Taylor G, Deng Q and Kukar T (2017) Intracellular proteolysis of progranulin generates stable, lysosomal granulins that are haploinsufficient in patients with frontotemporal dementia caused by GRN mutations. eNeuro 4, ENEURO.0100–17.201 10.1523/ENEURO.0100-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pacheco-Quinto J and Eckman EA (2013) Endothelin-converting enzymes degrade intracellular β-amyloid produced within the endosomal/lysosomal pathway and autophagosomes. J. Biol. Chem 288, 5606–5615 10.1074/jbc.M112.422964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tam JH, Seah C and Pasternak SH (2014) The Amyloid Precursor Protein is rapidly transported from the Golgi apparatus to the lysosome and where it is processed into beta-amyloid. Mol. Brain 7, 54 10.1186/s13041-014-0054-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oku Y, Murakami K, Irie K, Hoseki J and Sakai Y (2017) Synthesized Aβ42 caused intracellular oxidative damage, leading to cell death, via lysosome rupture. Cell Struct. Funct 42, 71–79 10.1247/csf.17006 [DOI] [PubMed] [Google Scholar]
- 32.Kumar A, Paslay LC, Lyons D, Morgan SE, Correia JJ and Rangachari V (2012) Specific soluble oligomers of amyloid-β peptide undergo replication and form non-fibrillar aggregates in interfacial environments. J. Biol. Chem 287, 21253–21264 10.1074/jbc.M112.355156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levites Y, Das P, Price RW, Rochette MJ, Kostura LA, McGowan EM et al. (2006) Anti-Aβ42- and anti-Aβ40-specific mAbs attenuate amyloid deposition in an Alzheimer disease mouse model. J. Clin. Invest 116, 193–201 10.1172/JCI25410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walsh JG, Cullen SP, Sheridan C, Luthi AU, Gerner C and Martin SJ (2008) Executioner caspase-3 and caspase-7 are functionally distinct proteases. Proc. Natl Acad. Sci. U.S.A 105, 12815–12819 10.1073/pnas.0707715105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levine H (1993) Thioflavine T interaction with synthetic Alzheimer’s disease β-amyloid peptides: detection of amyloid aggregation in solution. Protein Sci 2, 404–410 10.1002/pro.5560020312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar A, Pate KM, Moss MA, Dean DN and Rangachari V (2014) Self-propagative replication of Aβ oligomers suggests potential transmissibility in Alzheimer disease. PLoS ONE 9, e111492 10.1371/journal.pone.0111492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irie K, Murakami K, Masuda Y, Morimoto A, Ohigashi H, Ohashi R et al. (2005) Structure of β-amyloid fibrils and its relevance to their neurotoxicity: implications for the pathogenesis of Alzheimer’s disease. J. Biosci. Bioeng 99, 437–447 10.1263/jbb.99.437 [DOI] [PubMed] [Google Scholar]
- 38.Bieschke J, Herbst M, Wiglenda T, Friedrich RP, Boeddrich A, Schiele F et al. (2011) Small-molecule conversion of toxic oligomers to nontoxic β-sheet-rich amyloid fibrils. Nat. Chem. Biol 8, 93–101 10.1038/nchembio.719 [DOI] [PubMed] [Google Scholar]
- 39.Rehder DS and Borges CR (2010) Cysteine sulfenic acid as an intermediate in disulfide bond formation and nonenzymatic protein folding. Biochemistry 49, 7748–7755 10.1021/bi1008694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anfinsen CB and Haber E (1961) Studies on the reduction and re-formation of protein disulfide bonds. J. Biol. Chem 236, 1361–1363 [PubMed] [Google Scholar]
- 41.Eikelenboom P, Veerhuis R, Scheper W, Rozemuller AJM, van Gool WA and Hoozemans JJM (2006) The significance of neuroinflammation in understanding Alzheimer’s disease. J. Neural Transm 113, 1685–1695 10.1007/s00702-006-0575-6 [DOI] [PubMed] [Google Scholar]
- 42.Eikelenboom P, van Exel E, Hoozemans JJ, Veerhuis R, Rozemuller AJ and van Gool WA (2010) Neuroinflammation – an early event in both the history and pathogenesis of Alzheimer’s disease. Neurodegener. Dis 7, 38–41 10.1159/000283480 [DOI] [PubMed] [Google Scholar]
- 43.Heneka MT, Carson MJ, Khoury JE, Landreth GE, Brosseron F, Feinstein DL et al. (2015) Neuroinflammation in Alzheimer’s disease. Lancet Neurol 14, 388–405 10.1016/S1474-4422(15)70016-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang B, Gaiteri C, Bodea L-G, Wang Z, McElwee J, Podtelezhnikov AA et al. (2013) Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 153, 707–720 10.1016/j.cell.2013.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wyss-Coray T and Mucke L (2002) Inflammation in neurodegenerative disease—a double-edged sword. Neuron 35, 419–432 10.1016/S0896-6273(02)00794-8 [DOI] [PubMed] [Google Scholar]
- 46.Engelhart M, Geerlings M, Meijer J, Kiliaan A, Ruitenberg A, van Swieten J et al. (2004) Inflammatory proteins in plasma and the risk of dementia: the Rotterdam study. Arch. Neurol 61, 668–672 10.1001/archneur.61.5.668 [DOI] [PubMed] [Google Scholar]
- 47.Webster B, Hansen L, Adame A, Crews L, Torrance M, Thal L et al. (2006) Astroglial activation of extracellular-regulated kinase in early stages of Alzheimer disease. J. Neuropathol. Exp. Neurol 65, 142–151 10.1097/01.jnen.0000199599.63204.6f [DOI] [PubMed] [Google Scholar]
- 48.Morgan D, Gordon MN, Tan J, Wilcock D and Rojiani A (2005) Dynamic complexity of the microglial activation response in transgenic models of amyloid deposition: implications for Alzheimer therapeutics. J. Neuropathol. Exp. Neurol 64, 743–753 10.1097/01.jnen.0000178444.33972.e0 [DOI] [PubMed] [Google Scholar]
- 49.Hickman SE, Allison EK and El Khoury J (2008) Microglial dysfunction and defective β-amyloid clearance pathways in aging Alzheimer’s disease mice. J. Neurosci 28, 8354–8360 10.1523/JNEUROSCI.0616-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El Khoury J, Toft M, Hickman SE, Means TK, Terada K, Geula C et al. (2007) Ccr2 deficiency impairs microglial accumulation and accelerates progression of Alzheimer-like disease. Nat. Med 13, 432–438 10.1038/nm1555 [DOI] [PubMed] [Google Scholar]
- 51.Miseta A and Csutora P (2000) Relationship between the occurrence of cysteine in proteins and the complexity of organisms. Mol. Biol. Evol 17, 1232–1239 10.1093/oxfordjournals.molbev.a026406 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.