Abstract
It is hypothesized that retrotransposons have played a fundamental role in primate evolution and that enhanced neurologic retrotransposon activity in humans may underlie the origin of higher cognitive function. As a potential consequence of this enhanced activity, it is likely that neurons are susceptible to deleterious retrotransposon pathways that can disrupt mitochondrial function. An example is observed in the TOMM40 gene, encoding a β-barrel protein critical for mitochondrial preprotein transport. Primate-specific Alu retrotransposons have repeatedly inserted into TOMM40 introns, and at least one variant associated with late-onset Alzheimer’s disease originated from an Alu insertion event. We provide evidence of enriched Alu content in mitochondrial genes and postulate that Alus can disrupt mitochondrial populations in neurons, thereby setting the stage for progressive neurologic dysfunction. This Alu neurodegeneration hypothesis is compatible with decades of research and offers a plausible mechanism for the disruption of neuronal mitochondrial homeostasis, ultimately cascading into neurodegenerative disease.
Keywords: Alternative splicing, Alzheimer’s disease, Epigenetics, H3K9, Inflammation, LINE, Neuroepigenetics, Nonsense-mediated decay, Parkinson’s disease, Retrotransposon, SINE, Somatic mutation, Somatic mosaicism, Spliceosome, A-to-I editing
1. Introduction
The molecular mechanisms underlying sporadic neurodegenerative disorders such as late-onset Alzheimer’s disease (LOAD) and Parkinson’s disease (PD) remain unclear. Although traditional genome-wide association studies (GWASs) have identified numerous candidate genes associated with both LOAD and PD, the explanatory power of these genes is low (approximately 3%−4% per locus in LOAD cases), and effective therapies that disrupt the progression of idiopathic neurodegenerative diseases have yet to be developed [1–4]. Considering this disparity, a growing number of researchers are hypothesizing a link between non-Mendelian mechanisms and sporadic neurodegenerative disease. Functional hypotheses for such mechanisms include epigenetic effects, novel structurai variants influencing alternative gene splicing and gene expression, maternal inheritance of mitochondrial DNA mutations, and microbial infection [5–10]. A common thread across decades of sporadic neurodegenerative disease research is the hypothesis that mitochondrial dysfunction contributes to neuron stress and neuron degeneration, ultimately leading to the diseased state [11–19].
Pathologies associated with age-related neurodegenerative diseases (e.g., senile Aβ plaques, tau aggregates, cerebral atrophy, and age-related cognitive impairment) are not restricted to humans, having been identified in a number of nonhuman primates (e.g., chimpanzee, gorilla, orangutan, rhesus macaque, tamarin, and gray mouse lemur) that collectively span at least 65 million years of primate evolution [20–24]. Of these species, the one that is most distantly related to human, the gray mouse lemur (Microcebus murinus), routinely develops age-related pathologies (within captive individuals in established colonies) that are similar to both Alzheimer’s disease (AD) and PD [25]. The evolutionary perspective that can be gleaned from the spectrum of ~ 65 million years of primate evolution is critically important for understanding the origin of neurodegenerative disease in humans.
Despite the fact that primates share similar age-related disease pathologies, the manifestation of devastating human-specific symptoms associated with sporadic neurodegenerative diseases across the global distribution of our species is suggestive of a common neurologic mechanism that evolved in humans [23,24,26,27]. Following this logic, identification of the genetic factors that contribute to sporadic neurodegenerative disease in humans requires an understanding of the origin of primates and the genetic mechanisms underlying the evolution of enhanced neurologic function that separates humans from our closest primate relatives. Therefore, central to this perspective are the following observations: (1) although primates share common age-related neurodegenerative pathologies, humans display a spectrum of neurologic disorders that are uniquely human; (2) a growing body of evidence supports the hypothesis that non-Mendelian mechanisms contribute to the manifestation of neurodegenerative disease; and (3) mitochondrial dysfunction is consistently hypothesized to be associated with sporadic neurodegenerative diseases such as LOAD, PD, Huntington’s disease (HD), and amyotrophic lateral sclerosis (ALS).
Here we propose a hypothesis, informed by primate evolution, for an age-related genetic mechanism that can contribute to tissue-specific mitochondrial dysfunction eventually resulting in neuronal death. It is important to note that our hypothesis centers on molecular mechanisms underlying cellular stress at the very initial stages of neurologic disease, therefore preceding macroscopic pathologies (e.g., pervasive plaque formation) that are frequently diagnostic of the disease state. We begin with the observation that structural variants of primate-specific retrotransposons (Alu elements) within the translocase of outer mitochondrial membrane 40 (TOMM40) gene are statistically associated with LOAD and that these transposable elements can influence gene function through non-Mendelian pathways. Retrotransposons are mobile elements that can replicate by reverse transcription of an RNA intermediate and then insert themselves into new locations across the genome [28]. Broadly defined, retrotransposons include long terminal repeats, long interspersed elements (LINEs), and short interspersed elements (SINEs). Of these, Alu elements are a highly successful primate-specific SINE and Alus are the most abundant mobile elements in the human genome having more than a million copies that comprise ~ 11% of genomic DNA.
Although traditionally viewed as “junk DNA,” a number of discoveries have shown that retrotransposons have played a fundamental role in primate evolution, including the evolution of our own species, having contributed to the formation of novel genes and gene transcription networks as well as having a role in human disease [29–35]. Moreover, retrotransposons (including Alu) remain active in the human central nervous system throughout life, and it is hypothesized that this activity underlies the origin of higher brain function [32,33,36,37]. We postulate that enhanced somatic retrotransposon activity in human neurologic networks is accompanied by tissue-specific mitochondrial vulnerability that increases with time and/or fluctuating epigenetic landscapes, and can thus be a contributing mechanism to sporadic neurodegeneration. This in turn leads to the specific hypothesis that retrotransposons, operating through primate or human-specific pathways, are a plausible source for environment or age-induced mitochondrial dysfunction that can ultimately contribute to neuron atrophy and death.
2. Mitochondrial integrity and neurodegenerative disease
The human brain has exceptionally high energetic demands, and metabolically active neurons depend on healthy mitochondrial populations for their survival and function. Disrupting mitochondrial homeostasis in neurons can have devastating neurologic consequences, and therefore mitochondrial dysfunction has long been hypothesized to be associated with neurologic diseases (reviewed in [38] and [39]). First proposed in 2004 by Swerdlow and Khan, the “mitochondrial cascade hypothesis” provides the framework by which mitochondrial dysfunction can contribute to the development of sporadic neurodegenerative disease [13]. Although not without controversy, the hypothesis that dysfunctional mitochondria play a role in LOAD, PD, and other neurodegenerative conditions is a consistent theme across decades of research. The mitochondrial genome encodes only 13 proteins, yet mitochondria depend on an estimated 1500 nuclear-encoded proteins for their functionality. Thus, genetic mechanisms that contribute to genomic instability of the nuclear genome, including deleterious retrotransposon-mediated pathways, can directly impact mitochondrial function and contribute to neurologic disease. A clear example involves the relationship between genetic variation of the TOMM40 gene and neurodegenerative diseases including LOAD, PD, HD, and ALS [40].
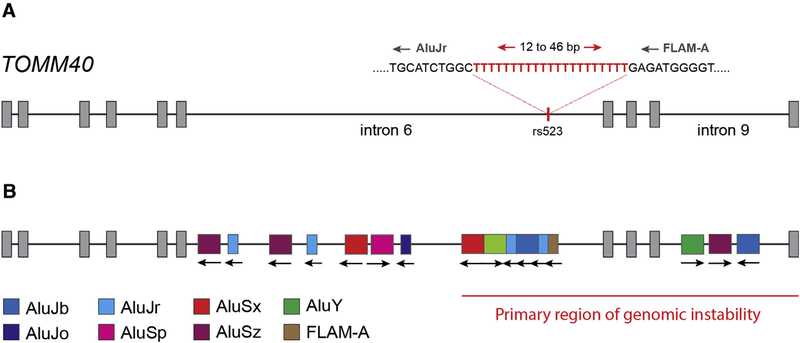
2.1. Insights from TOMM40
The translocase of the outer membrane (TOM) complex is responsible for importing more than 90% of all preproteins into the mitochondrion, and the TOM complex comprises seven subunits (Tom5, Tom6, Tom7, Tom20, Tom22, Tom40, and Tom70). Of these, Tom40 forms a β-barrel protein that is the primary channel through which mitochondrial preproteins pass, and the fully assembled TOM complex contains three Tom40 β-barrel channels arranged in a triangular pattern [41]. The TOMM40 gene is located on human chromosome 19 and encodes 10 exons and nine introns (Fig. 1). A poorly understood yet functional and potentially important paralog of TOMM40, TOMM40L, is located on chromosome 1. Both single-nucleotide polymorphisms and short structural variants within TOMM40 have been implicated in a number of neurologic disorders, ranging from mild cognitive impairment to major neurodegenerative diseases including LOAD and PD (reviewed in [40]) [42–44]. Amyloid precursor protein accumulates at Tom40 in AD brains, clogging the TOM complex and contributing to mitochondrial dysfunction [16]. Moreover, with respect to PD, decreasing Tom40 expression results in increasing α-synuclein accumulation and is directly correlated with an increase in reactive oxygen species, inflammation, oxidative damage, reduced adenosine triphosphate production, and an overall decrease in mitochondrial integrity [45]. Disrupting the TOM complex effectively triggers mitochondrial stress response and can ultimately lead to mitophagy, a process that has especially devastating consequences across neurologic networks [38,46,47].
Fig. 1.
Diagram of human TOMM40 showing location of rs523 structural variant (A) and Alu elements (B). The primary region of genomic instability is identified as the approximate region spanning bases 44,898,490 to 44,903,725 on chromosome 19 (human genome build GRCh38/hg38). This region is hypothesized to be vulnerable to Alu-related mechanisms that contribute to genomic instability including alternative splicing and modification of pre-mRNA transcripts.
Despite numerous lines of evidence that show that the TOM complex plays an essential role in mitochondrial stability, there remains an ongoing debate regarding the significance of TOMM40 sequence variation with respect to mitochondrial dysfunction and potential consequences for neurologic disease. Much of this debate centers on the observation that TOMM40 is in tight linkage disequilibrium with the APOE gene, and thus, questions have been posed as to whether there is an APOE-independent effect of variation within TOMM40 and AD risk (reviewed in [40,48]). In particular, the statistical association between variable deoxythymidine homopolymer repeats within TOMM40 intron 6 (the rs10524523 poly-T; i.e., rs523) and LOAD risk and time of onset has gained much attention. Three categorical length classes of rs523 are identified, short (S; ≤18 bp), long (L; 19–29 bp), and very long (VL; ≥30 bp) based on distributions of rs523 lengths in individuals of Caucasian ancestry [49]. The distribution of categorical length classes observed in Caucasians was also observed in individuals of African, African American, and Asian ancestry. Cis-haplotypes of APOE and rs523 are clearly delineated and are specific to ethnicity. For example, in Caucasians, the APOE ε4 allele is usually linked to an rs523L allele, whereas in West Africans, the APOE ε4 allele can be linked to either an rs523L allele or an rs523 S allele. African American APOE-rs523 haplotype frequencies differ from both West Africans and Caucasians and represent admixture of distinct West African and Caucasian haplotypes [50]. Several studies have reported on statistically significant associations of rs523 genotype and APOE-rs523 haplotypes with the risk and age of onset of AD and with cognitive decline [40,48,49,51–55]. Other meta-analyses of GWAS data have not identified a statistically significant association of rs523 with the risk or age of onset of AD independent of APOE genotype [56,57].
A key observation regarding the origin of rs523 that has been largely overlooked, is that rs523 is part of an Alu mobile element monomer that has inserted into TOMM40 intron 6 in the antisense orientation (Fig. 1). To date, only a single study has noted the relationship between rs523 and Alu elements that are enriched in TOMM40 intron 6 [58]. Indeed, Payton et al. [58] provide critical insight with respect to TOMM40 gene expression and the presence of Alus within TOMM40 introns. We expand on this observation and highlight how Alu retrotransposons in TOMM40 introns 6 and 9 could contribute to transcriptional noise through enhanced nonsense-mediated decay and/or the production of alternative TOMM40 isoforms.
2.2. Alu elements and TOMM40 stability
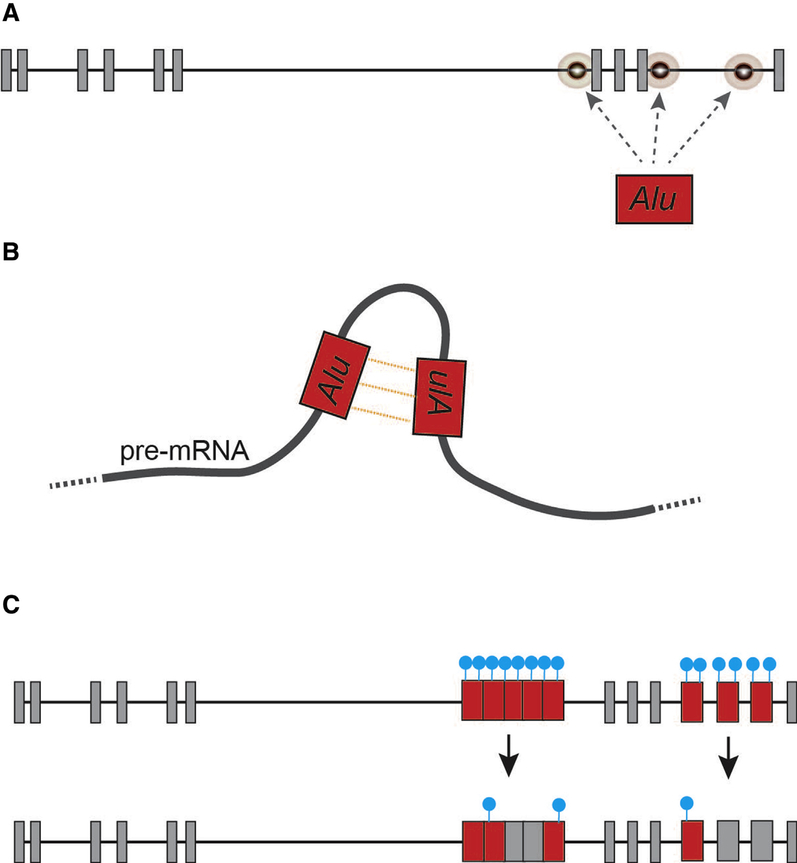
Retrotransposons, including Alu elements, can have a profound influence on messenger RNA (mRNA) stability and gene transcription networks [31]. Sixteen Alu elements have inserted themselves across TOMM40 introns 6 and 9, and the rs523 poly-T is part of an Alu element in antisense orientation. The 3′ end of TOMM40 is particularly rich with Alu elements and, over evolutionary time, these have inserted themselves into the germline in both sense and antisense orientations (Fig. 1B). The relative age of each Alu element and the orientation of Alu insertion events play an essential role with respect to potential Alu exonization and deleterious recombination events that can disrupt TOMM40 mRNA transcripts (Fig. 2) [29,31,58]. With respect to rs523, both poly-T and poly-A rich regions associated with intronic Alu elements can interrupt efficient processing of pre-mRNA transcripts by spliceosome machinery through the activation of premature polyadenylation sites and/or antagonistic binding of AU-rich binding proteins such as hnRNP C, PABP, PTB, and U2AF65 [6,31,59]. Alu-associated poly-T regions resulting from antisense insertion events are known to destabilize gene transcription and contribute to increasing levels of mRNA degradation [60].
Fig. 2.
Select mechanisms by which retrotransposon can influence gene splicing in somatic tissues (also see [31]). (A) de novo Alu retrotransposition events; (B) formation of inverted-repeat Alu duplexes within pre-mRNA transcripts. Such Alu duplexes are the primary target for tissue-specific A-to-I RNA editing path-ways that can destabilize pre-mRNA transcripts [31]. (C) Hypomethylation of Alu contributing to exonization (blue dots represent both DNA and histone methylation landscapes).
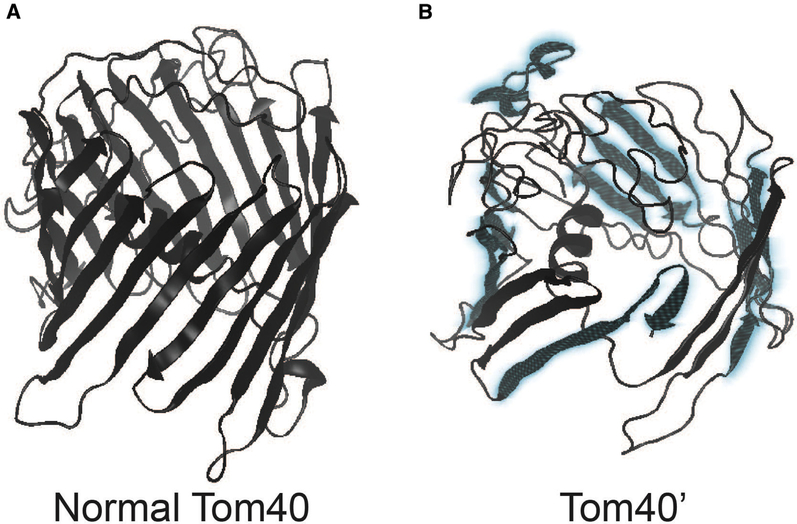
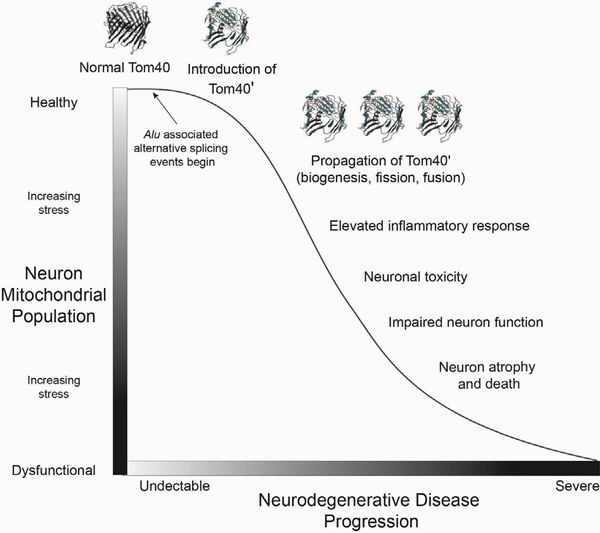
Collectively, these observations identify a region of enhanced genomic instability in TOMM40 that is vulnerable to several Alu-associated pathways proven to alter gene expression and implicated in a growing list of human diseases (Fig. 2) [31]. Inverted repeat Alus, such as those distributed across TOMM40 introns 6 and 9, can disrupt mRNA stability by facilitating premature transcription termination and altering adenosine-to-inosine (A-to-I) RNA editing. These pathways can also contribute to the production of altered protein conformations [31], and alternative TOMM40 isoforms with premature termination coinciding with Alus (i.e., Tom40’) are identified in National Center for Biotechnology Information (NCBI) and Human Protein Atlas databases. If Alu elements enriched across the 3’ end of TOMM40 are contributing to the production of modified yet functional transcripts that escape nonsense-mediated mRNA decay, then it is likely that peptides encoded by these transcripts would still be localized to the TOM complex [61]. Predictive modeling [62] of one potential alternative splicing event that coincides with Alus in TOMM40 intron 9 (Tom40′; NCBI XP_005258468) suggests that a β-barrel protein conformation may still form. If so, Tom40’ would likely display functional differences from normal Tom40, perhaps serving to restrict the passage of preproteins and/or destabilize the TOM complex (Fig. 3) [61]. Moreover, an intriguing possibility with respect to the paralog TOMM40L is that Alu-associated disruption of normal mRNA processing of Tom40 could result in increased localization of Tom40L to the mitochondrial outer membrane. This conformation could alter the efficient processing of mitochondrial preproteins or otherwise destabilize the TOM complex. If accurate, such a mechanism could result in the propagation of inefficient TOM channels through mitochondrial biogenesis, fusion, and fission within individual neurons over variable time-scales (Fig. 4).
Fig. 3.
Predictive model showing the potential influence of premature termination of TOMM40 gene transcripts on Tom40 protein structure. Protein models were generated using normal (A) TOMM40 (NCBI NP_006105; 361 amino acid sequences in length) and truncated (B) TOMM40 (NCBI XP_005258468; 335 aa) alternative gene transcripts. The 3′ end of the truncated 335 aa transcript coincides with the AluY mobile element within intron 9 of TOMM40 (a potential region of enhanced A-to-I editing; Figs. 1 and 2). Regions shaded in blue identify major conformational changes to the p-barrel protein.
Fig. 4.
Model showing the propagation of Alu-induced alternative conformations of Tom40, ultimately resulting in increasing levels of inflammation, neuron toxicity, and neuron death resulting in neurodegenerative disease. The model is applicable to any number of nuclear-encoded mitochondrial proteins critical to mitochondrial stability within neurons and is consistent with the broader “mitochondrial cascade hypothesis” [13]. Transcriptional noise and alternative splicing events associated with increased retrotransposon activity in neurons can contribute to mitochondrial stress over time, ultimately resulting in increasing mitochondrial stress (Y-axis) and progressive neurodegenerative disease resulting in neuron atrophy and death (X-axis).
2.3. Retrotransposons, mitochondrial gene vulnerability, and neurodegenerative disease
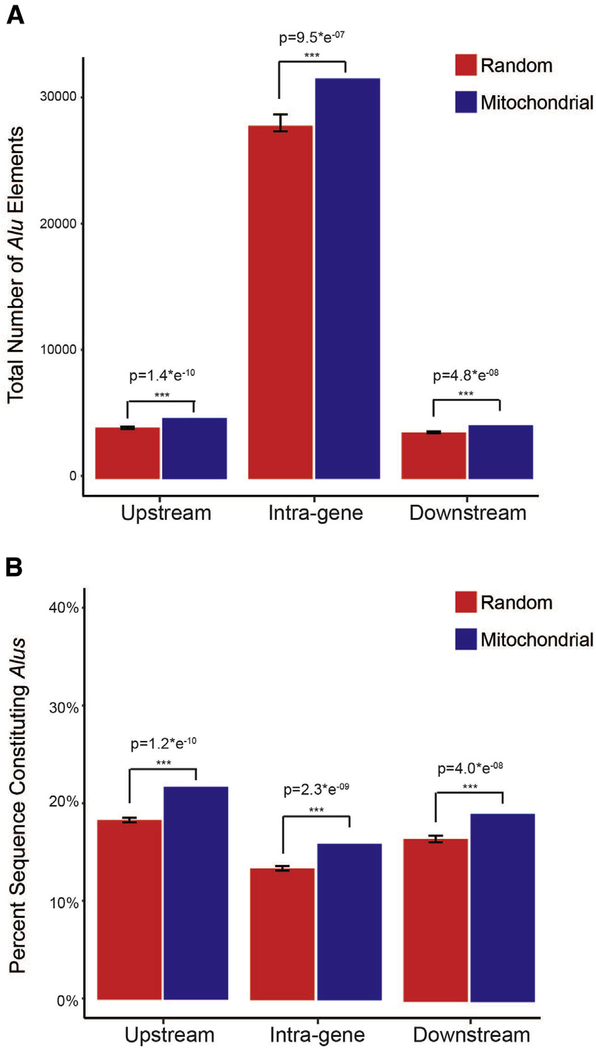
From a broader perspective, Alu exonization and somatic retrotransposition events of both LINEs and Alu have been identified in multiple TOM genes including TOMM5, TOMM7, TOMM22, TOMM40, and TOMM40L [36,63,64]. These patterns indicate TOM genes are actively influenced by and are vulnerable to retrotransposons, perhaps owing to their high transcription rates and open chromatin status as hypothesized by de Andrade et al. [63]. If true, it can be predicted that nuclear-encoded mitochondrial genes would display an enrichment of mobile elements with respect to other nuclear genes. To test this prediction, we examined the mobile element content of 1145 genes that encode mitochondrial proteins [65] and an additional 8973 randomly selected protein-coding genes throughout the human genome (Supplementary Material). Our results provide statistical support for an enrichment of Alu mobile elements within and adjacent to mitochondrial genes (Fig. 5; Supplementary Material), consistent with previous analyses identifying enriched Alu content at transcriptionally active regions of the genome [66]. Thus, transcriptionally active genes essential for mitochondrial function are potentially more vulnerable to deleterious retrotransposon-related mechanisms known to disrupt gene expression pathways (reviewed in [31,35,67]).
Fig. 5.
Alu content measured across 1145 mitochondrial genes and 8973 randomly selected (non-mitochondrial) genes, as well as flanking genomic regions (5 kb), sampled from the human genome (Ensembl build GRCh38). Panel (A) shows raw number of Alus annotated within, and adjacent to, the sampled gene sets. Panel (B) shows the percent of bases constituting Alu elements within each sample. Statistical support was measured using two-tailed t tests and detailed methods are provided in the Supplementary Material. *** Identifies significance at P = .05 and error bars identify 95% confidence intervals.
If operating within energetically demanding neurons, retrotransposon-related destabilization of efficient transcription and translation of mitochondrial genes would likely contribute to the activation of inflammatory response pathways that can cascade to neuronal tissue damage and neurodegenerative disease [47]. It would therefore be anticipated that deleterious retrotransposon activity in nuclear-encoded mitochondrial genes that encode peptides occupying key functional roles would contribute to a variety of neurologic diseases. In addition to the connections observed between TOMM40 Alu-rs523 and LOAD [40,58], there are several striking examples that support this observation. A notable example is found with adrenoleukodystrophy, where multiple Alu insertion events within the ABCD1 gene (a nuclear-encoded mitochondrial gene) contribute to deleterious nonhomologous recombination events, disrupting functional ABCD1 peptides [68]. ABCD1 encodes a protein of the adenosine triphosphate-binding cassette transporter family, the disruption of which contributes to functional and structural destabilization of mitochondria and is hypothesized to result in the accumulation of very long chain fatty acids throughout the nervous system [68,69]. Adrenoleukodystrophy patients suffer from progressive axonal degeneration contributing to a range of neurologic impairments, including progressive memory loss. Another example involves the OPA1 gene where an antisense Alu insertion event is hypothesized to contribute to alternative OPA1 splicing events [70]. OPA1 peptides play a key role in mitochondrial fusion and mitochondrial cristae morphology, and the Alw-disruption of OPA1 expression is linked to autosomal dominant optic atrophy, a disease stemming from progressive degeneration of retinal ganglion cells ultimately resulting in optic nerve atrophy [70,71].
Alu exonization, Alw-mediated copy number variants, and Alw-mediated nonhomologous end joining have also been detected in mitochondrial genes (including mitochondria quality control genes) associated with Leigh syndrome, pyruvate dehydrogenase deficiency, presenile dementia, and PD (e.g., NDUFS2, SLC30A6, PDHA1, and PARK2) [34,64,72,73]. Collectively, these examples demonstrate that primate-specific Alu mobile elements can disrupt the translation and peptide formation of nuclear-encoded mitochondrial genes, potentially contributing to the manifestation of multiple neurodegenerative disorders.
3. Mechanisms for altered gene expression and neurologic disease: an epigenetic connection
Alu elements are believed to have played a critical role in primate evolution, especially in our own species wherein increased Alu insertion events, human-specific SINE-Variable Number of Tandem repeat-Alus, human-specific Alu-derived exons, and enhanced neurologic Alu activity separate us from nonhuman primates [36,64,72,74]. Indeed, an accumulating amount of evidence indicates retrotransposons (including both LINEs and SINEs) are actively influencing human neural stem cells and somatic tissues of the brain, resulting in human-specific neurologic transcription networks [31–33,36,37,75,76]. Advanced genomic approaches, such as single-cell genome sequencing, further support this fascinating possibility and reveal that somatic retrotransposition activity in the brain contributes to the establishment of mosaic genomes of individual neurons [37,75–78]. Although potentially advantageous for higher-order cognition, this enhanced retrotransposon activity in the human central nervous system is likely accompanied by vulnerability in somatic tissue that increases with age [76].
Host genomes have evolved a number of mechanisms to defend against deleterious retrotransposon activity, including DNA and histone methylation and RNA degradation using microRNA-processing enzymes (reviewed in [31]). Theepigenetic silencing of Alu elements is mediated by both DNA and histone H3K9 methylation to suppress transcription and retrotransposition [79,80]. Hypomethylation of Alu elements contributes to enhanced retrotransposon activity, which in turn can increase transcriptional noise by disrupting gene expression pathways, inducing alternative splicing events, and reducing mRNA stability [79]. Genome-wide Alu hypomethylation is part of the aging process and global hypomethylation of Alu is statistically associated with AD [81,82]. More broadly, emerging neuroepigenetic research is establishing a link between age and environment-associated epigenetic modifications and a range of neurologic disorders, from autism and schizophrenia to LOAD and PD [83]. A growing body of evidence suggests that increased retrotransposon activity in the human central nervous system, mediated by epigenetic regulation for beneficial neurologic function and the reduction of deleterious events, is accompanied by enhanced vulnerability in neurons resulting in neurologic disease [31,35,37,76].
With respect to heightened neuronal retrotransposon activity, the potential impact on neuron mitochondrial function remains unexplored. Hypomethylation of Alu elements and/or de novo Alu insertions within, or near, genes that are essential to mitochondrial function could contribute to mRNA instability, ultimately leading to mitochondrial dysfunction. Furthermore, an intriguing observation with respect to histone H3K9 methylation of Alu elements is that H3K9 also regulates APOE transcription [80]. Additional research is required to determine how the epigenetic interplay between the Alu-rich regions within and flanking TOMM40 (Fig. 1; immediately upstream of APOE on human chromosome 19) and APOE can influence either TOMM40 or APOE gene expression, or both. Operating within an epigenetic framework, Alu-related transcriptional noise of nuclear-encoded mitochondrial genes would potentially correlate with senescence and environmental exposures [84]. Tissue-specific methylation patterns of nuclear-encoded mitochondrial genes are observed in mammals and indicate specialized epigenetic modulation for the maintenance of tissue-dependent mitochondrial functional pathways, including specialized pathways in the brain [85]. Collectively, these data provide a putative epigenetic link between time-dependent mitochondrial dysfunction (both slowly accumulating or accelerated) and tissue-specific idiopathic neurodegenerative disease.
Regarding the coevolution of mitochondrial organelles and eukaryotes, the origin of primate-specific retrotransposons is a notably recent event. This is especially interesting when considering the enhanced neurologic retrotransposon activity that separates humans from other primates [33,36,76]. In concert with the increased longevity of modern humans, human-specific age-related neurologic diseases are perhaps indicative of increasing neurologic transcriptional noise because of a relaxation of retrotransposon control mechanisms. It is within this framework that we propose the Alu neurodegeneration hypothesis.
4. The Alu neurodegeneration hypothesis
Of all tissues in the human body, the brain has one of the highest rates of transposable element activity [32,36,75]. This phenomenon is one of the most striking characteristics that separates human neurologic gene networks from our closest primate relatives, leading Friedli and Trono (2015) to conclude that “.the endovirome [the collection of all transposable elements within the genome] and its controllers played a fundamental role in the expansion of higher brain functions that was key to the emergence of modern humans.” Managing this enhanced retrotransposon activity requires exquisite control of the molecular pathways that work together to reap the evolutionary benefits of novel gene function while simultaneously preventing catastrophic events [31,32,76]. When considering the Friedli and Trono (2015) hypothesis, it is conceivable that gain-of-function associated with enhanced transposable element activity and the evolution of higher cognitive ability of modern Homo sapiens may be accompanied by neurologic vulnerability. In light of recent discoveries regarding the epigenetic regulation of transposable elements, such neurologic vulnerability may well correlate with age. Thus, humans, with our steadily increasing life expectancy rates, would be especially vulnerable [86].
We hypothesize that retrotransposons, operating through human-specific neurologic pathways [32,33,74,87–89], contribute to environment and/or age-related neurodegeneration by disrupting functional mitochondrial populations within neurons. This mitochondrial disruption can occur through several retrotransposon-induced mechanisms that can influence the efficient and accurate transcription and/or translation of mitochondrial genes encoded in the nuclear genome, ultimately resulting in depauperate neuron mitochondrial populations. Considering TOMM40, it is plausible that Alu-related conformational changes (both subtle and major) of the outer and inner mitochondrial membrane pores could restrict or prevent the normal translocation of proteins, ultimately contributing to mitochondrial stress, inflammation, and mitophagy (Figs. 1–4). Importantly, the disruption of mitochondrial protein trafficking across both TOMM and translocase of inner mitochondrial membrane complexes has been implicated in several neurodegenerative diseases (AD, PD, HD, and ALS) [47]. An age-related and/or environmental-related pathway for such disruption is found in the epigenetic regulation of retrotransposons in the central nervous system. Both tissue and cell-specific hypomethylation of Alu elements, resulting from fluctuating epigenetic landscapes, can facilitate retrotransposon-induced mitochondrial stress.
The mechanisms by which symbiotic mitochondrial organelles coevolved with eukaryotic genomes provide a potential vulnerability with respect to recently evolved primate and or human-specific genetic mechanisms that disrupt gene stability. This vulnerability can be amplified through mitochondrial biogenesis and downstream mitochondrial fission and fusion events, thus contributing to the initial establishment of inefficient mitochondria that increase mitochondrial stress over time and limit neuron functionality, ultimately leading to a diseased state (Fig. 4) [13,15,18,19]. Under this framework, it would be expected that retrotransposon-mediated dysfunctional mitochondrial events would manifest in different neurologic tissues and in a seemingly temporally sporadic nature that is difficult to predict using traditional approaches such as GWAS. In light of emerging data regarding retrotransposon-induced mosaic genomes of individual neurons, it is likely that only advanced single-cell genomic techniques will provide the appropriate resolution for detecting retrotransposon-related mitochondrial dysfunction, as such events may be restricted to specific neuron populations arising from individual progenitor cells influenced by somatic retrotransposition during brain development [36,37,76,78].
The initiation of tissue-specific retrotransposon-induced dysfunctional mitochondrial cascade events, operating through variable intercellular and intracellular processes and occurring at different life stages, would ultimately result in diseased states that share similar underlying pathologies, such as inflammatory response activation, protein aggregation, and neurodegeneration [47,90,91], although with a spectrum of phenotypic neurologic impairments. Future studies focused on the epigenetic regulation of retrotransposons in individual neurons and across neurologic networks may elucidate the origin of a range of neurologic disorders and serve as the foundation for novel therapeutic approaches. We recommend that the “Alu-neurodegeneration hypothesis” be considered for sporadic tissue-specific neurodegenerative diseases wherein mitochondrial dysfunction has been identified, including AD, PD, HD, and ALS.
Supplementary Material
RESEARCH IN CONTEXT.
Systematic review: Despite enormous research effort the molecular mechanisms underlying sporadic neurodegenerative disease remain elusive. Moreover, although mitochondrial dysfunction is hypothesized to be an early indicator of multiple neurodegenerative diseases, the source of this dysfunction also remains unclear. We postulate that neurological mitochondrial populations are vulnerable to deleterious retrotransposon activity operating on nuclear-encoded mitochondrial genes.
Interpretation: Primate-specific Alu elements are enriched within nuclear-encoded mitochondrial genes, and these genes are subject to Alu-mediated mechanisms that contribute to transcriptional noise. It is likely that Alu-induced transcriptional noise of mitochondrial genes correlates with fluctuating epigenetic landscapes associated with aging and/or environmental stress.
Future directions: Our hypothesis can account for incipient mitochondrial dysfunction observed in sporadic neurodegenerative disease. We recommend future studies focused on the interplay between retrotransposons and nuclear-encoded mitochondrial gene expression and protein formation. Such research may provide new therapeutic approaches that could alleviate the earliest stages of mitochondrial and cellular stress within neurological networks, thereby preventing neurodegenerative cascades.
Acknowledgments
The authors thank W.K. Gottschalk, S.S. Sundeth, O. Chiba-Falek, B.A. Sullivan, A.D. Brown, C.R. Campbell, R.J. Larsen, and L. Pinto for comments and discussion that helped to improve this manuscript. We are grateful for the support of Duke Research Computing and the Duke Data Commons (NIH 1S10OD018164–01). This is Duke Lemur Center publication number 1340.
Footnotes
Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jalz.2017.01.017.
References
- [1].Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 2013; 45:1452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lord J, Cruchaga C. The epigenetic landscape of Alzheimer’s disease. Nat Neurosci 2014;17:1138–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mitsui J, Tsuji S. Genomic aspects of sporadic neurodegenerative diseases. Biochem Biophys Res Commun 2014;452:221–5. [DOI] [PubMed] [Google Scholar]
- [4].Ertekin-Taner N Genetics of Alzheimer disease in the pre-and post-GWAS era. Alzheimers Res Ther 2010;2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].De Jager PL, Srivastava G, Lunnon K, Burgess J, Schalkwyk LC, Yu L, et al. Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. NatNeurosci 2014;17:1156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Roses AD, Akkari PA, Chiba-Falek O, Lutz MW, Gottschalk WK, Saunders AM, et al. Structural variants can be more informative for disease diagnostics, prognostics and translation than current SNP mapping and exon sequencing. Expert Opin Drug Metab Toxicol 2016; 12:135–47. [DOI] [PubMed] [Google Scholar]
- [7].Itzhaki R, Lathe R, Balin BJ, Ball MJ, Bearer EL, Braak H, et al. Microbes and Alzheimer’s Disease. J Alzheimers Dis 2016;51:979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic mechanisms in Alzheimer’s disease. Neurobiol Aging 2011;32:1161–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Linnertz C, Anderson L, Gottschalk W, Crenshaw D, Lutz MW, Allen J, et al. The cis-regulatory effect of an Alzheimer’s disease-associated poly-T locus on expression of TOMM40 and apolipoprotein E genes. Alzheimers Dement 2014;10:541–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Carvalho CM, Lupski JR. Mechanisms underlying structural variant formation in genomic disorders. Nat Rev Genet 2016;17:224–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Castellani R, Hirai K, Aliev G, Drew KL, Nunomura A, Takeda A, et al. Role of mitochondrial dysfunction in Alzheimer’s disease. J Neurosci Res 2002;70:357–60. [DOI] [PubMed] [Google Scholar]
- [12].Leuner K, Müller WE, Reichert AS. From mitochondrial dysfunction to amyloid beta formation: novel insights into the pathogenesis of Alzheimer’s disease. Mol Neurobiol 2012;46:186–93. [DOI] [PubMed] [Google Scholar]
- [13].Swerdlow RH, Khan SM. A “mitochondrial cascade hypothesis” for sporadic Alzheimer’s disease. Med Hypotheses 2004;63:8–20. [DOI] [PubMed] [Google Scholar]
- [14].Hashimoto M, Rockenstein E, Crews L, Masliah E. Role of protein aggregation in mitochondrial dysfunction and neurodegeneration in Alzheimer’s and Parkinson’s diseases. Neuromolecular Med 2003;4:21–35. [DOI] [PubMed] [Google Scholar]
- [15].Sheng B, Wang X, Su B, Hg L, Casadesus G, Perry G, et al. Impaired mitochondrial biogenesis contributes to mitochondrial dysfunction in Alzheimer’s disease. J Neurochem 2012;120:419–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Devi L, Prabhu BM, Galati DF, Avadhani NG, Anandatheerthavarada HK. Accumulation of amyloid precursor protein in the mitochondrial import channels of human Alzheimer’s disease brain is associated with mitochondrial dysfunction. J Neurosci 2006;26:9057–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cha MY, Han SH, Son SM, Hong HS, Choi YJ, Byun J, et al. Mitochondria-specific accumulation of amyloid β induces mitochondrial dysfunction leading to apoptotic cell death. PLoS One 2012;7:e34929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hirai K, Aliev G, Nunomura A, Fujioka H, Russell RL, Atwood CS, et al. Mitochondrial abnormalities in Alzheimer’s disease. J Neurosci 2001;21:3017–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chen H, Chan DC. Mitochondrial dynamics—fusion, fission, movement, and mitophagy—in neurodegenerative diseases. Hum Mol Genet 2009;18:R169–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Verdier JM, Acquatella I, Lautier C, Devau G, Trouche S, Lasbleiz C, et al. Lessons from the analysis of nonhuman primates for understanding human aging and neurodegenerative diseases. Front Neurosci 2015;9:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Perez SE, Sherwood CC, Cranfield MR, Erwin JM, Mudakikwa A, Hof PR, et al. Early Alzheimer’s disease-type pathology in the frontal cortex of wild mountain gorillas (Gorillaberingei beringei). Neurobiol Aging 2016;39:195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gearing M, Tigges J, Mori H, Mirra S. β-Amyloid (Ap) deposition in the brains of aged orangutans. Neurobiol Aging 1997;18:139–46. [DOI] [PubMed] [Google Scholar]
- [23].Collier TJ, Kanaan NM, Kordower JH. Ageing as a primary risk factor for Parkinson’s disease: evidence from studies of non-humanprimates. Nat Rev Neurosci 2011;12:359–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Heuer E, Rosen RF, Cintron A, Walker LC. Nonhuman primate models of Alzheimer-like cerebral proteopathy. Curr Pharm Des 2012; 18:1159–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Languille S, Blanc S, Blin O, Canale C, Dal-Pan A, Devau G, et al. The grey mouse lemur: a non-human primate model for ageing studies. Ageing Res Rev 2012;11:150–62. [DOI] [PubMed] [Google Scholar]
- [26].Rapoport S Hypothesis: Alzheimer’s disease is a phylogenetic disease. Med Hypotheses 1989;29:147–50. [DOI] [PubMed] [Google Scholar]
- [27].Finch CE, Austad SN. Commentary: is Alzheimer’s disease uniquely human? Neurobiol Aging 2015;36:553–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Deininger PL, Moran JV, Batzer MA, Kazazian HH. Mobile elements and mammalian genome evolution. Curr Opin Genet Dev 2003;13:651–8. [DOI] [PubMed] [Google Scholar]
- [29].Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet 2002;3:370–9. [DOI] [PubMed] [Google Scholar]
- [30].Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, et al. SVA elements: a hominid-specific retroposon family. J Mol Biol 2005; 354:994–1007. [DOI] [PubMed] [Google Scholar]
- [31].Elbarbary RA, Lucas BA, Maquat LE. Retrotransposons as regulators of gene expression. Science 2016;351:aac7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Friedli M, Trono D. The developmental control of transposable elements and the evolution of higher species. Annu Rev Cell Dev Biol 2015;31:429–51. [DOI] [PubMed] [Google Scholar]
- [33].Trono D Transposable elements, polydactyl proteins, and the genesis of human-specific transcription networks. Cold Spring Harb Symp Quant Biol 2015;80:281–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Boone PM, Yuan B, Campbell IM, Scull JC, Withers MA, Baggett BC, et al. The Alu-rich genomic architecture of SPAST predisposes to diverse and functionally distinct disease-associated CNV alleles. Am J Hum Genet 2014;95:143–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hancks DC, Kazazian HH. Roles for retrotransposon insertions in human disease. Mob DNA 2016;7:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature 2011;479:534–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Erwin JA, Paquola AC, Singer T, Gallina I, Novotny M, Quayle C, et al. L1-associated genomic regions are deleted in somatic cells of the healthy human brain. Nat Neurosci 2016;19:1583–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Rugarli EI, Langer T. Mitochondrial quality control: a matter of life and death for neurons. EMBO J 2012;31:1336–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chaturvedi RK, Beal MF. Mitochondrial diseases of the brain. Free Radic Biol Med 2013;63:1–29. [DOI] [PubMed] [Google Scholar]
- [40].Gottschalk WK, Lutz MW, He YT, Saunders AM, Burns DK, Roses AD, et al. The broad impact of TOM40 on neurodegenerative diseases in aging. J Parkinsons Dis Alzheimers Dis 2014;1:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Shiota T, Imai K, Qiu J, Hewitt VL, Tan K, Shen HH, et al. Molecular architecture of the active mitochondrial protein gate. Science 2015; 349:1544–8. [DOI] [PubMed] [Google Scholar]
- [42].Lu F, Guan H, Gong B, Liu X, Zhu R, Wang Y, et al. Genetic variants in PVRL2-TOMM40-APOE region are associated with human longevity in a Han Chinese population. PLoS One 2014;9:e99580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Liu X, Bai F, Yue C, Shi Y, Yu H, Luo B, et al. The association between TOMM40 gene polymorphism and spontaneous brain activity in amnestic mild cognitive impairment. J Neurol 2014;261:1499–507. [DOI] [PubMed] [Google Scholar]
- [44].Yu L, Lutz MW, Wilson RS, Burns DK, Roses AD, Saunders AM, et al. TOMM40’ 523 variant and cognitive decline in older persons with APOE e3/3 genotype. Neurology 2017;88:661–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bender A, Desplats P, Spencer B, Rockenstein E, Adame A, Elstner M, et al. TOM40 mediates mitochondrial dysfunction induced by α-synuclein accumulation in Parkinson’s disease. PLoS One 2013;8:e62277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Harbauer AB, Zahedi RP, Sickmann A, Pfanner N, Meisinger C. The protein import machinery of mitochondria—a regulatory hub in metabolism, stress, and disease. Cell Metab 2014;19:357–72. [DOI] [PubMed] [Google Scholar]
- [47].Richards RI, Robertson SA, O’Keefe LV, Fornarino D, Scott A, Lardelli M, et al. The enemy within: innate surveillance-mediated cell death, the common mechanism of neurodegenerative disease. Front Neurosci 2016;10:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lutz MW, Crenshaw D, Welsh-Bohmer KA, Burns DK, Roses AD. New genetic approaches to AD: lessons from APOE-TOMM40 phylogenetics. Curr Neurol Neurosci Rep 2016;16:1–9. [DOI] [PubMed] [Google Scholar]
- [49].Crenshaw D, Gottschalk W, Lutz M, Grossman I, Saunders A, Burke J, et al. Using genetics to enable studies on the prevention of Alzheimer’s disease. Clin Pharmacol Ther 2013;93:177–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Roses AD, Lutz MW, Saunders AM, Goldgaber D, Saul R, Sundseth SS, et al. African-American TOMM40’523-APOE haplotypes are admixture of West African and Caucasian alleles. Alzheimers Dement 2014;10:592–601.e2. [DOI] [PubMed] [Google Scholar]
- [51].Caselli RJ, Dueck AC, Huentelman MJ, Lutz MW, Saunders AM, Reiman EM, et al. Longitudinal modeling of cognitive aging and the TOMM40 effect. Alzheimers Dement 2012;8:490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Greenbaum L, Springer RR, Lutz MW, Heymann A, Lubitz I, Cooper I, et al. The TOMM40 poly-T rs10524523 variant is associated with cognitive performance among non-demented elderly with type 2 diabetes. Eur Neuropsychopharmacol 2014;24:1492–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Hayden KM, McEvoy JM, Linnertz C, Attix D, Kuchibhatla M, Saunders AM, et al. A homopolymer polymorphism in the TOMM40 gene contributes to cognitive performance in aging. Alzheimers Dement 2012;8:381–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Johnson SC, La Rue A, Hermann BP, Xu G, Koscik RL, Jonaitis EM, et al. The effect of TOMM40 poly-T length on gray matter volume and cognition in middle-aged persons with APOEε3/ε3 genotype. Alzheimers Dement 2011;7:456–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Roses A, Lutz M, Amrine-Madsen H, Saunders A, Crenshaw D, Sundseth S, et al. ATOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer’s disease. Pharmacogenomics J 2010; 10:375–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Jun G, Vardarajan BN, Buros J, Yu CE, Hawk MV, Dombroski BA, et al. Comprehensive search for Alzheimer disease susceptibility loci in the APOE region. Arch Neurol 2012;69:1270–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Cruchaga C, Nowotny P, Kauwe JS, Ridge PG, Mayo K, Bertelsen S, et al. Association and expression analyses with single-nucleotide polymorphisms in TOMM40 in Alzheimer disease. Arch Neurol 2011; 68:1013–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Payton A, Sindrewicz P, Pessoa V, Platt H, Horan M, Ollier W,et al. A TOMM40 poly-T variant modulates gene expression and is associated with vocabulary ability and decline in non-pathological ageing. Neurobiol Aging 2016;39:217.e1–7. [DOI] [PubMed] [Google Scholar]
- [59].Zarnack K, König J, Tajnik M, Martincorena I, Eustermann S, Stévant I, et al. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell 2013;152:453–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].An HJ, Lee D, Lee KH, Bhak J. The association of Alu repeats with the generation of potential AU-rich elements (ARE) at 3’untranslated regions. BMC Genomics 2004;5:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kuszak AJ, Jacobs D, Gurnev PA, Shiota T, Louis JM, Lithgow T, et al. Evidence of distinct channel conformations and substrate binding affinities for the mitochondrial outer membrane protein translocase pore Tom40. J Biol Chem 2015;290:26204–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Zhang Y I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 2008;9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].de Andrade A, Wang M, Bonaldo MF, Xie H, Soares MB. Genetic and epigenetic variations contributed by Alu retrotransposition. BMC Genomics 2011;12:617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lin L, Jiang P, Park JW, Wang J, Lu ZX, Lam MP, et al. The contribution of Alu exons to the human proteome. Genome Biol 2016;17:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Calvo SE, Clauser KR, Mootha VK. MitoCarta2. 0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res 2016; 44:D1251–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Ganapathi M, Srivastava P, Sutar SK, Kumar K, Dasgupta D, Singh GP, et al. Comparative analysis of chromatin landscape in regulatory regions of human housekeeping and tissue specific genes. BMC Bioinformatics 2005;6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Deininger PL, Batzer MA. Alu repeats and human disease. Mol Genet Metab 1999;67:183–93. [DOI] [PubMed] [Google Scholar]
- [68].Kutsche K, Ressler B, Katzera HG, Orth U, Gillessen-Kaesbach G, Morlot S, et al. Characterization ofbreakpoint sequences of five rearrangements in L1CAM and ABCD1 (ALD) genes. Hum Mutat 2002;19:526–35. [DOI] [PubMed] [Google Scholar]
- [69].McGuinness M, Lu JF, Zhang HP, Dong GX, Heinzer A, Watkins P, et al. Role of ALDP (ABCD1) and mitochondria in X-linked adreno-leukodystrophy. Mol Cell Biol 2003;23:744–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Gallus GN, Cardaioli E, Rufa A, Da Pozzo P, Bianchi S, D’Eramo C, et al. Alu-element insertion in an OPA1 intron sequence associated with autosomal dominant optic atrophy. Mol Vis 2010;16:178–83. [PMC free article] [PubMed] [Google Scholar]
- [71].Varanita T, Soriano ME, Romanello V, Zaglia T, Quintana-Cabrera R, Semenzato M, et al. The OPA1-dependent mitochondrial cristae remodeling pathway controls atrophic, apoptotic, and ischemic tissue damage. Cell Metab 2015;21:834–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Shen S, Lin L, Cai JJ, Jiang P, Kenkel EJ, Stroik MR, et al. Widespread establishment and regulatory impact of Alu exons in human genes. Proc Natl Acad Sci U S A 2011;108:2837–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Morais S, Bastos-Ferreira R, Sequeiros J, Alonso I. Genomic mechanisms underlying PARK2 large deletions identified in a cohort of patients with PD. Neurol Genet 2016;2:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Hedges DJ, Callinan PA, Cordaux R, Xing J, Barnes E, Batzer MA. Differential Alu mobilization and polymorphism among the human and chimpanzee lineages. Genome Res 2004;14:1068–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, et al. L1 retrotransposition in human neural progenitor cells. Nature 2009;460:1127–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Erwin JA, Marchetto MC, Gage FH. Mobile DNA elements in the generation of diversity and complexity in the brain. Nat Rev Neurosci 2014;15:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Upton KR, Gerhardt DJ, Jesuadian JS, Richardson SR, Sanchez-Luque FJ, Bodea GO, et al. Ubiquitous L1 mosaicism in hippocampal neurons. Cell 2015;161:228–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Evrony GD. One brain, many genomes. Science 2016;354:557–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Bakshi A, Herke SW, Batzer MA, Kim J. DNA methylation variation of human-specific Alu repeats. Epigenetics 2016;11:163–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Varshney D, Vavrova-Anderson J, Oler AJ, Cowling VH, Cairns BR, White RJ. SINE transcription by RNA polymerase III is suppressed by histone methylation but not by DNA methylation. Nat Commun 2015;6:6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Jintaridth P, Mutirangura A. Distinctive patterns of age-dependent hypomethylation in interspersed repetitive sequences. Physiol Genomics 2010;41:194–200. [DOI] [PubMed] [Google Scholar]
- [82].Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, et al. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev 2009;130:234–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Jakovcevski M, Akbarian S. Epigenetic mechanisms in neurological disease. Nat Med 2012;18:1194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet 2009;5:e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Takasugi M, Yagi S, Hirabayashi K, Shiota K. DNA methylation status of nuclear-encoded mitochondrial genes underlies the tissue-dependent mitochondrial functions. BMC Genomics 2010;11:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Finch CE. Evolution of the human lifespan and diseases of aging: roles of infection, inflammation, and nutrition. Proc Natl Acad Sci U S A 2010;107:1718–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Gianfrancesco O, Bubb VJ, Quinn JP. SVA retrotransposons as potential modulators of neuropeptide gene expression. Neuropeptides 2016. 10.1016/j.npep.2016.09.006. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Lee J, Kim YJ, Mun S, Kim HS, Han K. Identification of human-specific AluS elements through comparative genomics. Gene 2015;555:208–16. [DOI] [PubMed] [Google Scholar]
- [89].Robbez-Masson L, Rowe HM. Retrotransposons shape species-specific embryonic stem cell gene expression. Retrovirology 2015;12:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Strittmatter WJ, Weisgraber KH, Goedert M, Saunders AM, Huang D, Corder EH, et al. Hypothesis: microtubule instability and paired helical filament formation in the Alzheimer disease brain are related to apolipoprotein E genotype. Exp Neurol 1994;125:163–71. discussion 72–4. [DOI] [PubMed] [Google Scholar]
- [91].Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med 2004;10 Suppl:S10–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.