Abstract
ATP-binding cassette (ABC) superfamily members have a key role as nutrient importers and exporters in bacteria. However, their role as drug exporters in eukaryotes brought this superfamily member to even greater prominence. The capacity of ABC transporters to efflux a broad spectrum of xenobiotics represents one of the major mechanisms of clinical multidrug resistance in pathogenic fungi including Candida species. Candida auris, a newly emerged multidrug-resistant fungal pathogen of humans, has been responsible for multiple outbreaks of drug-resistant infections in hospitals around the globe. Our study has analyzed the entire complement of ABC superfamily transporters to assess whether these play a major role in drug resistance mechanisms of C. auris. Our bioinformatics analyses identified 28 putative ABC proteins encoded in the genome of the C. auris type-strain CBS 10913T; 20 of which contain transmembrane domains (TMDs). Quantitative real-time PCR confirmed the expression of all 20 TMD transporters, underlining their potential in contributing to the C. auris drug-resistant phenotype. Changes in transcript levels after short-term exposure of drugs and in drug-resistant C. auris isolates suggested their importance in the drug resistance phenotype of this pathogen. CAUR_02725 orthologous to CDR1, a major multidrug exporter in other yeasts, showed consistently higher expression in multidrug-resistant strains of C. auris. Homologs of other ABC transporter genes, such as CDR4, CDR6, and SNQ2, also displayed raised expression in a sub-set of clinical isolates. Together, our analysis supports the involvement of these transporters in multidrug resistance in C. auris.
Keywords: Candida auris, multidrug resistance, ABC proteins, drug efflux pumps, fluconazole
Introduction
The ATP-binding cassette (ABC) proteins are one of the largest superfamily of proteins found in both prokaryotes and higher eukaryotes. The eukaryotic ABC transporters are promiscuous exporters and can extrude a variety of substrates including metals, drugs, xenobiotics, lipids, and other cellular metabolites (Prasad and Goffeau, 2012). A large number of ABC proteins are multidrug transporters and function as efflux pumps. Rapid active drug extrusion from cells represents one of the major mechanisms of multidrug resistance, frequently encountered in organisms ranging from bacteria to mammals (Prasad and Goffeau, 2012). However, the function of ABC proteins is not limited to detoxification or drug expulsion, since many of its members perform diverse cellular functions. Among others, these include roles in vacuole fusion, maintaining mitochondrial integrity, lipid translocation, and pheromone secretion (Khandelwal et al., 2016; Prasad et al., 2016). Notably, the ABC proteins play major roles in various human diseases, such as cystic fibrosis, Tangier’s disease, adrenoleukodystrophy, and cancer (Dean et al., 2001b).
Candida species are the fourth most common cause of fungal blood stream infections, and among them Candida albicans is the most prevalent agent of superficial and systemic disease (Wisplinghoff et al., 2004; Tortorano et al., 2006). However, recent global emergence of multidrug-resistant Candida auris has become a major health concern (Cortegiani et al., 2018). Compared to other pathogenic Candida species, C. auris has unique features, which make it difficult to treat and eradicate from intensive care hospital wards (Forsberg et al., 2019). This fungus is mostly associated with clonal outbreaks that spread rapidly through health care facilities. C. auris is frequently resistant to commonly used frontline antifungals of different classes. An initial study on C. auris showed that it is phylogenetically related to Candida haemulonii and has the largest numbers of orthologs with Candida lusitaniae (Chatterjee et al., 2015).
Whole-genome sequence (WGS) analyses of C. auris isolates from different countries led to a subdivision into four distinct clades specific to a geographic area, thus indicating the simultaneous emergence of C. auris in different continents (Lockhart et al., 2017). Based on the geographically restricted area of isolation, the authors have classified the clades as follows: clade I comprises isolates from India and Pakistan, clade II from Japan and South Korea, clade III from South Africa, and clade IV from Venezuela (Schelenz et al., 2016; Lockhart et al., 2017; Muñoz et al., 2018; Rhodes et al., 2018). As per this classification, the C. auris strain, CBS 10913T belongs to clade II (Japan and South Korea) and the three Indian clinical-resistant isolates used in this study probably belong to clade I (India and Pakistan). The different clades are differentiated by thousands of single-nucleotide polymorphisms (SNPs), whereas genetic differences within each geographic clade are minimal (Lockhart et al., 2017). Importantly, not much is known about phenotypic differences between isolates from different C. auris clades. Initial WGS of a C. auris isolate has revealed that approximately 2.4% of its genes encode ABC and major facilitator superfamily (MFS) transporters along with other transporters like oligopeptide transporters and iron transporters, etc. (Chatterjee et al., 2015). Two recent simultaneously published reports confirmed the role of ABC transporter CDR1 and MFS transporter MDR1 in azole resistance (Kim et al., 2019; Rybak et al., 2019).
Our study presents the identification and expression analysis of ABC proteins on a genome-wide scale in the pathogenic yeast C. auris. For this, we performed WGS of the C. auris type-strain, CBS 10913T (clade II – Japan and South Korea), recovered in 2009 from a Japanese patient with an ear infection (Satoh et al., 2009). Our bioinformatics analysis of the genome of CBS 10913T identified 28 putative ABC proteins. Based on phylogenetic analysis, domain organization, and following the nomenclature adopted by the Human Genome Organization (HUGO) scheme (Vasiliou et al., 2009), these proteins are classified into six subfamilies ABCB/MDR, ABCC/multidrug resistance-associated protein (MRP), ABCD/ALDp, ABCE/RLI, ABCF/YEF3, and ABCG/PDR. Among these, 20 contained transmembrane domains (TMDs). The TMD ABC transporters predominantly belong to the ABCG/PDR, ABCB/MDR, ABCC/MRP, and ABCD/ALDp subfamilies. Comparative phylogenetic analysis of the ABC proteins with C. albicans, Saccharomyces cerevisiae, and Debaryomyces hansenii revealed orthologous relationships and highlighted their conservation. We also demonstrate that the expression of various ABC transporters changes substantially in the presence of different antifungal drugs suggesting a potential role in drug resistance in C. auris. The comparative expression landscape of all transcripts of ABC transporters in the clade I multidrug-resistant hospital isolates of C. auris suggests a possible role of selected ABC proteins in conferring drug resistance.
Materials and Methods
Yeast Strains, Culture Conditions, and Antifungal Drugs
The C. auris type strain CBS 10913T was obtained from the CBS-KNAW fungal culture collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands. Although B11220 and CBS 10913T have apparently been isolated from the same original material they seem not to be completely identical. Additionally, their karyotypes are overall quite similar, but show some differences (Bravo Ruiz et al., 2019).
Three drug-resistant clinical isolates (Isolate 1-NCCPF470150, Isolate 2-NCCPF 470156, and Isolate 3-NCCPF470114) of C. auris were provided by the National Culture Collection of Fungal Pathogens, Postgraduate Institute of Medical Education & Research, Chandigarh, India. All yeast strains used in this study are listed in Supplementary Table S1. Yeasts were grown on yeast extract/peptone/dextrose (YEPD; Difco, Sparks, MD, United States). The antifungals amphotericin B (AMPB), terbinafine (TRB), and fluconazole (FLU) were procured from the Sigma Chemical Co. (St. Louis, MO, United States).
Whole-Genome Sequencing (WGS) and Assembly
Genomic DNA of strain CBS 10913T was extracted from an overnight culture in YEPD broth using an established protocol. Briefly, cells were resuspended in equal volumes of extraction buffer (2% Triton X-100, 1% SDS, 100 mM NaCl, 10 mM Tris pH 8.0, 1mM EDTA) and phenol:chloroform:isoamyl alcohol (25:24:1). Vigorous shaking in the presence of acid-washed glass beads was applied to break the cell wall. After addition of a volume of 1× Tris–EDTA (TE pH 7.5) and centrifugation (5 min, 14,000 × g), the aqueous layer was mixed with 2.5 volumes of 100% ethanol to precipitate the DNA. Following centrifugation (2 min, 14,000 × g) the pelleted genomic DNA was suspended in 1 volume of 1× TE containing 250 μg/ml RNase A and incubated for 20 min at 37°C. DNA was then precipitated by adding 1/20 of a volume of 3 M sodium acetate and 2.5 volumes of 100% ethanol followed by centrifugation (10 min, 14,000 × g), finally DNA was resuspended in sterile water. Concentration and purity of the DNA preparation were determined on a NanoDropTM Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States). A genomic library was prepared and barcoded at the Centre of Genome-Enabled Biology and Medicine (CGEBM, University of Aberdeen) using the Nextera DNA Library Preparation Kit (Illumina, Inc., San Diego, CA, United States), and WGS was performed on an Illumina MiSeq sequencer at CGEBM. Sequence reads were subjected to quality control inspection and trimming using FASTX-toolkit1 and FastQC2, and adaptor sequences were removed using Cutadapt (Martin, 2011). Velvet (Zerbino, 2010) and ABySS (Simpson et al., 2009) assemblers were used in standard and optimized mode with three sets of paired end reads: untrimmed reads, reads trimmed to a base quality score of 36, and to a base quality score of 28. CEGMA was used to conduct assembly quality control checks for genome completeness (Parra et al., 2007). The initial assembly was scaffolded using read alignments with BESST (Sahlin et al., 2014) and extended using IMAGE in PAGIT. The contiguity of the assembly was then improved by additional scaffolding against the published genome of the closely related species C. lusitaniae using Newbler aligner. Resulting contigs were curated to retain only contigs with read coverage >100 and <10,000 and a length of >1,000 bp, gap closing was performed using SSPACE-LongRead (Boetzer and Pirovano, 2014). The raw sequence files are available at https://www.ebi.ac.uk/ena/data/view/PRJEB29190 (ENA Accession No. PRJEB29190).
Identification of ABC Proteins
A total of 5,279 proteins were identified in the genome of C. auris CBS 10913T. The HMM profile of the ABC-tran model (accession PF00005) obtained from the Pfam database3 was used as query against the proteins of C. auris in the HMM search program of HMMER package4 with default parameters (Finn et al., 2016). For further filtering of hits obtained after HMM search cut-off was decided on the basis of plots between domain scores and E-values. Domain (bit) scores above 69.4 and E-values below 1.3e−19 were considered positive for having an NBD domain. Sequences thus selected were taken forward as putative ABC proteins for detailed analysis.
Retrieving of Sequences and Phylogenetic Analysis
For combined phylogenetic analysis S. cerevisiae, C. albicans, and D. hansenii sequences were retrieved from UniProt using previously published information (Wasi et al., 2018), and aligned with C. auris sequences by ClustalΩ using default parameters. The phylogenetic tree was constructed by MEGA 6.06 (Tamura et al., 2013) using the neighbor-joining method (Wasi et al., 2018) for the clustering, and the Poisson substitution for calculating the evolutionary distance. 1,000 bootstrap replicates were utilized to establish the tree topology. To calculate the sequence identities of ABC proteins among different species the web-based BLASTp program from NCBI5 was used.
Subcellular Localization Prediction
To predict the subcellular localization of ABC transporters, we used WoLF PSORT6 and DEEPLOC7 software.
RNA Isolation, cDNA Synthesis, and Quantitative Real-Time PCR
For the total RNA isolation, a primary culture was grown overnight to saturation. From this a secondary culture was inoculated in YEPD broth at an OD600 of 0.2, and grown for 6 h. The cultures were incubated for another 60 min with the indicated concentrations of drugs (Supplementary Table S2), and then collected by centrifugation, and washed with DEPC-treated water. Total RNA was isolated using an RNeasy Mini Kit (Qiagen, Hilden, Germany), following the manufacturer’s specifications. cDNA synthesis was performed using the RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, United States) according to the manufacturer’s instruction. iTaq Universal SYBR green super mix Bio-Rad was used along with the desired gene-specific oligonucleotide primers (Supplementary Table S3) to evaluate the quantitative expression profile.
TDH1 (CAUR_02457), an ortholog of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control for normalization of expression. For the constitutive expression profile the ΔCT of TDH1 was subtracted from the specific gene. The relative expression profiles under drug stresses were calculated using the 2–ΔΔCT method (Livak and Schmittgen, 2001) in comparison to untreated cells. The experiments were performed in biological duplicates and technical triplicates. Statistical significance values were calculated using a two-way ANOVA (uncorrected Fisher’s LSD) in GraphPad Prism 6 software.
MIC50 Determination
Antifungal susceptibility testing was performed according to the broth micro-dilution technique guidelines of the Clinical and Laboratory Standards Institute (CLSI) for AMPB, FLU, voriconazole (VRC), and itraconazole (ITR) (Clinical and Laboratory Standards Institute, 2008). Due to clinical breakpoints not being available for C. auris, the interpretation of the breakpoints suggested for yeast was followed. For AMPB, an MIC50 of 1 mg/l was considered resistant. Candida krusei (ATCC 6258) and Candida parapsilosis (ATCC 22019) were used as control organisms (Supplementary Table S1).
Results and Discussion
Ploidy Determination and Genome-Wide Sequencing of C. auris
A WGS (close to 200× coverage) of the type strain of C. auris, CBS 10913T belonging to the E. Asian geographical clade (Lockhart et al., 2017) was generated using an Illumina MiSeq sequencer (ENA Accession No. PRJEB29190). Final assembly produced a 12.1 Mb genome distributed over 71 contigs with an N50 of 379.6 kb and a GC content of 45%. A total of 5,279 open-reading frames (ORFs) have been identified in the genome of CBS 10913T, which is comparable to published WGSs of other C. auris isolates (Muñoz et al., 2018; Rhodes et al., 2018). The genome ploidy of C. auris CBS 10913T was confirmed as being haploid using flow cytometry by comparison to a bona fide haploid S. cerevisiae strain BY4741 (Supplementary Figure S1). The resulting assembly size of 12.1 Mb and the flow cytometric analysis are in line with previously reported haploid genome sizes of C. auris (Lockhart et al., 2017; Muñoz et al., 2018; Rhodes et al., 2018; Bravo Ruiz et al., 2019).
Identification of Putative ABC Proteins of C. auris
To identify the ABC proteins in C. auris CBS 10913T, we used the hidden Markov models (HMMs) profile of ATP-binding domain alignment Pfam (00005) as a query against the 5,279 protein sequences extracted from the C. auris CBS 10913T WGS. The initial screening of the HMM profile resulted in 51 sequences as proteins of interest (Supplementary Table S4). After applying cut-off values of 69.4 for the bit scores and 1.3e−19 for the E-values, 28 sequences were identified as significant matches and were subjected to further analysis (Table 1 and Supplementary Figure S2). These 28 sequences were confirmed to contain the ABC-specific nucleotide-binding domain (NBD) by ScanProsite8 (Table 1).
TABLE 1.
List of ABC proteins identified in C. auris and their molecular size features.
| Protein name | Number of amino acids | Size in kDa | Subfamily | Orthologs in C. albicans |
| CAUR_02725 | 1, 508 | 169.9196 | PDR/ABCG | CDR1 |
| CAUR_01285 | 1, 452 | 163.474 | PDR/ABCG | SNQ2 |
| CAUR_01276 | 1, 486 | 166.899 | PDR/ABCG | SNQ2 |
| CAUR_05555 | 1, 441 | 163.6461 | PDR/ABCG | CDR4 |
| CAUR_02773 | 1, 469 | 166.9447 | PDR/ABCG | CDR4 |
| CAUR_04233 | 1, 264 | 141.1817 | PDR/ABCG | CDR6(ROA1) |
| CAUR_03774 | 1, 012 | 113.6363 | PDR/ABCG | ADP1 |
| CAUR_01852 | 1, 907 | 213.6577 | MDR/ABCB | HST6 |
| CAUR_03761 | 679 | 75.3979 | MDR/ABCB | MDL1 |
| CAUR_04565 | 1, 432 | 160.1326 | MDR/ABCB | MDL2 |
| CAUR_02994 | 703 | 78.3624 | MDR/ABCB | ATM1 |
| CAUR_01188 | 1, 659 | 185.6801 | MRP/ABCC | YBT1 |
| CAUR_00156 | 1, 469 | 165.7274 | MRP/ABCC | YCF1 |
| CAUR_00964 | 1, 604 | 180.0822 | MRP/ABCC | MLT1 |
| CAUR_01719 | 1, 560 | 176.1635 | MRP/ABCC | YOR1 |
| CAUR_00368 | 1, 424 | 160.218 | MRP/ABCC | YOR1 |
| CAUR_03320 | 1, 566 | 175.1847 | MRP/ABCC | YCF1 |
| CAUR_02951 | 1, 406 | 158.184 | MRP/ABCC | YOR1 |
| CAUR_00862 | 758 | 85.9342 | ALDP/ABCD | PXA2 |
| CAUR_04133 | 810 | 94.1119 | ALDP/ABCD | PXA1 |
| CAUR_02351 | 1, 106 | 124.1289 | ABCF/YEF3 | GCN20 |
| CAUR_03795 | 1, 420 | 157.7549 | ABCF/YEF3 | GCN20 |
| CAUR_04953 | 751 | 84.7145 | ABCF/YEF3 | NEW1 |
| CAUR_00824 | 1, 051 | 116.4815 | ABCF/YEF3 | GCN20 |
| CAUR_04813 | 610 | 68.1726 | ABCF/YEF3 | HEF3 |
| CAUR_04997 | 617 | 68.8987 | RLI/ABCE | RLI1 |
| CAUR_03076 | 322 | 36.9176 | OTHERS | CAF16 |
| CAUR_04824 | 547 | 61.7759 | OTHERS | CAF16 |
To be classified as an ABC transporter protein, at least one NBD and one TMD must be present in the protein sequence. ABC proteins which lack TMDs are considered as soluble ABC proteins (Prasad et al., 2015). To predict the TMDs among all the putative ABC proteins, we used TOPOCONS and TMPred software, which led to the identification of 20 proteins, which harbor at least one TMD and one NBD; we consider these to be localized to membranes (Supplementary Table S5). The remaining eight proteins lacking a TMD likely are soluble ABC proteins (Supplementary Table S5).
Phylogenetic Analysis, Domain Organization, and Subfamily Prediction of C. auris ABC Proteins
We used the characteristic conserved sequence of NBDs to classify C. auris ABC proteins into subfamilies. According to the recent nomenclature suggested by the HUGO gene nomenclature committee9, eukaryotic ABC superfamily proteins can be classified into nine subfamilies from ABCA to ABCI (Dean et al., 2001a). However, not all subfamily members are uniformly present in all organisms. For instance, the ABCA subfamily does not exist in yeast, and their presence is limited to plants and some higher organisms (Kovalchuk and Driessen, 2010). ABCI is the most recent subfamily added to this classification, whose existence is confined only to plant taxa (Wasi et al., 2018). Likewise, members of another subfamily, ABCH, are only found in insects and zebrafish (Wasi et al., 2018).
For subfamily predictions, the NBD sequences of putative ABC proteins of C. auris were extracted using ScanProsite, and were aligned with the NBD sequences of C. albicans. An unrooted phylogenetic tree was constructed using the MEGA6.06 software as described in the section “Materials and Methods.” The well-defined ABC proteins of C. albicans (Gaur et al., 2005) allowed us to assign the subfamilies of C. auris (Figure 1). The NBD-based phylogenetic analysis revealed that the ABC proteins of C. auris clustered into six major subfamilies: (I) pleiotropic drug resistance (PDR)/ABCG, (II) multidrug resistance (MDR)/ABCB, (III) MRP/ABCC, (IV) adrenoleukodystrophy protein (ALDp)/ABCD, (V) RNase-L inhibitor (RLI)/ABCE, and (VI) elongation factor 3 (YEF3)/ABCF. The proteins not segregating with any of these subfamilies were categorized as “Others.” Interestingly, our phylogeny analysis reveals that within the same subfamily N-terminal and C-terminal NBDs of all the proteins form separate clusters. This separation of clusters within a subfamily corroborates previous studies, which suggested that a common ancestor gives rise to each half of the proteins within each subfamily, and also may allocate different functions (Daumke and Knittler, 2001). More recent data also point to this functional asymmetry between N- and C-terminal NBDs (Banerjee et al., 2018). Notably, the presence and the positional order of TMDs and NBDs in each subfamily are specific, and this domain arrangement creates a topological difference among different members of subfamilies. Thus, ABC proteins are clustered into three groups. The first group has a forward topology where the NBD is preceded by the TMD (TMD-NBD); this is observed in the members of the ABCC/MRP, ABCB/MDR, and ABCD/ALDp subfamilies. The second group has a reverse topology where NBD is followed by TMD (NBD–TMD); this is a typical feature of the ABCG/PDR subfamily in fungi and plants. A third group of members of two subfamilies ABCE/RLI and ABCF/YEF3 do not possess any TMD domain and belong to the soluble ABC proteins.
FIGURE 1.
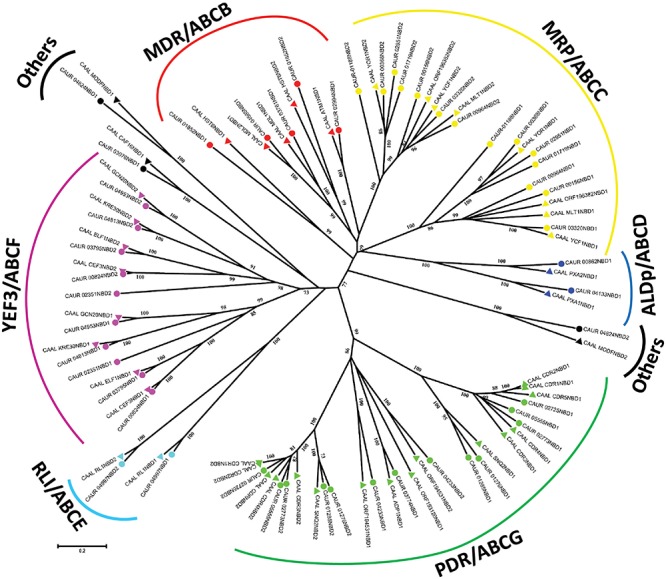
A phylogenetic tree depicting ABC protein subfamilies based on the NBD sequences of C. auris (CBS 10913T) and C. albicans (SC5314). C. auris (circle) and C. albicans (triangle) ABC protein NBD sequences were aligned using ClustalΩ. The evolutionary relationship was inferred using the neighbor-joining method, and the evolutionary distances were computed using the Poisson correction method in units of number of amino acid substitutions per site of the MEGA 6.06 package. Reliability of the tree topology was confirmed by bootstrap analysis employing 1,000 replicates. NBD1 represents the N-terminal NBD and NBD2 the C-terminal NBD.
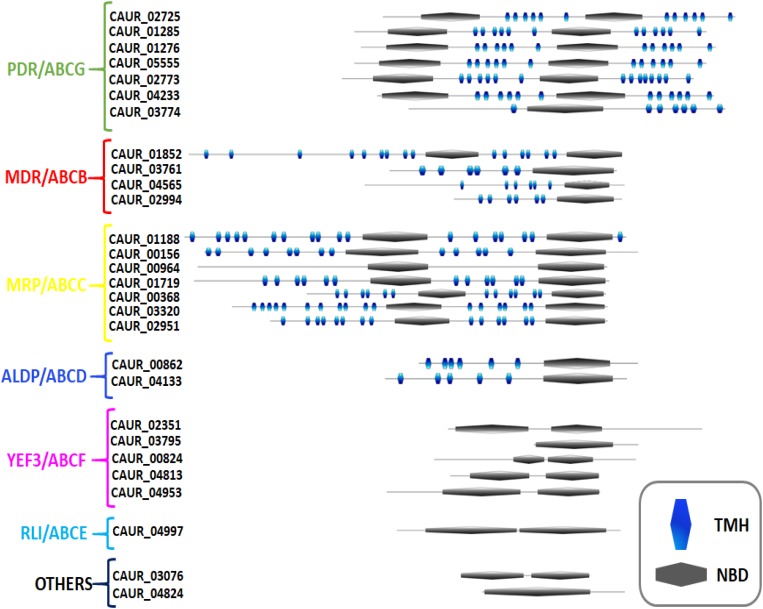
We utilized the topological distinction to validate our phylogeny-based subfamily clustering of C. auris ABC proteins. Using TOPOCONS and TMPRED, the number of transmembrane helices (TMHs) and their arrangement with NBDs were identified (Figure 2 and Supplementary Table S5). Our topology prediction suggests that out of the 20 ABC proteins possessing a TMD, 6 proteins are half-size transporters (containing one NBD and one TMD each), while 14 members can be considered as full transporters because they have 2 TMDs and 2 NBDs each. A further eight proteins do not have a TMD and are thus considered soluble ABC proteins (Figure 2 and Supplementary Table S5).
FIGURE 2.
Predicted topology of ABC proteins of C. auris. The domain arrangement of C. auris ABC proteins was generated using my domain builder tool in PROSITE. The transmembrane helices (TMHs) in transmembrane domain (TMD) were identified with the help of TOPOCONS and nucleotide-binding domain by ScanProsite. The subfamilies were assigned on the guidelines proposed by Human Genome Organization (HUGO).
To get better insight into the evolutionary status of C. auris ABC proteins, we performed a phylogenetic analysis along with the full ABC proteins of S. cerevisiae, C. albicans, and D. hansenii (Figure 3A). Descriptions of each subfamily of C. auris and their comparative analysis are detailed in the following sections.
FIGURE 3.
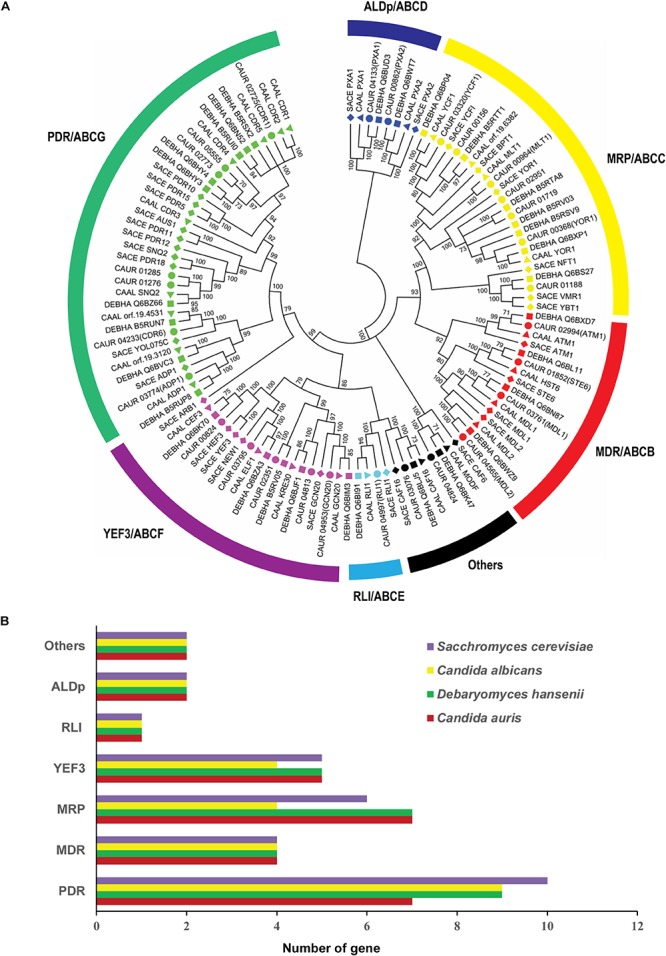
Comparative analysis of C. auris ABC proteins with other yeast. (A) Comparative phylogenetic relationship of C. auris ABC proteins with C. albicans, D. hansenii, and S. cerevisiae. Symbols: circles represent C. auris proteins, squares D. hansenii proteins, triangles C. albicans proteins, and diamonds S. cerevisiae proteins. (B) The number of ABC proteins in each subfamily in C. auris, D. hansenii, C. albicans, and S. cerevisiae. Different subfamilies are plotted on the y-axis and the number of genes is indicated on the x-axis.
ABCG/PDR Subfamily
ABCG/PDR is ubiquitous in eukaryotes and one of the largest subfamily. Based on combined phylogenetic analysis and topology predictions, we observed that C. auris harbors seven ABCG/PDR subfamily members. The number of PDR members varies among different yeasts. While C. glabrata, C. albicans, and D. hansenii have higher numbers, the fission yeast Schizosaccharomyces pombe contains only two PDR genes (Lamping et al., 2010; Kumari et al., 2018; Wasi et al., 2018).
Our topology analysis confirmed that all PDR subfamily members of C. auris follow the reverse topology (NBD–TMD) arrangement. The full transporters contain two NBD–TMD units following each other (NBD–TMD–NBD–TMD), while half-size transporters possess only one of each (NBD–TMD) (Lamping et al., 2010). Out of the seven PDR members, CAUR_02725, CAUR_02773, CAUR_04233, CAUR_01276, CAUR_01285, and CAUR_05555 are full transporters, while CAUR_03774 is a half-size transporter (Figure 2). Each TMD usually possesses six TMHs; however, in the case of CAUR_02773 protein, its C-terminal TMD is predicted to consist of eight TMHs; this is also observed in the PDR-type ABC member DEBHA_Q6BHY3 of D. hansenii (Wasi et al., 2018). Surprisingly, the PDR subfamily in C. auris has fewer members compared to other yeast species, such as S. cerevisiae (10 members), C. albicans (9 members), and D. hansenii (9 members) (Figure 3B). Most PDR subfamily members of other yeasts are multidrug transporters. However, only one of the PDR subfamily members (CDR1) of C. auris has recently been characterized as being directly involved in azole resistance in C. auris (Kim et al., 2019; Rybak et al., 2019). The overexpression of PDR transporters correlates well with the emergence of multidrug resistance in C. albicans, C. glabrata, and other pathogenic fungi (Prasad and Goffeau, 2012). In S. cerevisiae, Pdr5 is one of the well-characterized members of this subfamily. It is capable of extruding a wide range of xenobiotics and functions as a promiscuous drug transporter (Jungwirth and Kuchler, 2006; Lamping et al., 2010). C. albicans PDR subfamily members CDR1 and CDR2 can complement a null mutant of PDR5 in S. cerevisiae, and importantly are also involved in the multidrug resistance phenotype of clinical C. albicans isolates (Prasad et al., 1995; Sanglard et al., 1997). Apart from their role in drug transport, members of the PDR subfamily have also shown to impact lipid homeostasis. For instance, Pdr5 in S. cerevisiae and Cdr1, Cdr2, Cdr3, and Cdr4 of C. albicans are well-characterized lipid translocators (Prasad et al., 2016).
Based on sequence identity with functionally characterized members of other yeasts, the functions of C. auris proteins can be predicted. For instance, CAUR_02725 of C. auris shows 73 and 68% sequence identity with Cdr1 and Cdr2 of C. albicans, respectively, thus forming a subgroup. Notably, our analysis does not reveal an obvious independent homolog of Cdr2 as in C. albicans and C. glabrata. CAUR_02773 and CAUR_05555 of C. auris show 64% and 68% sequence identity, respectively, with Cdr4 of C. albicans, and along with DEBHA_Q6BHY3 and DEBHA_Q6BHY4 of D. hansenii cluster into a separate subgroup. Interestingly, two proteins, CAUR_01276 and CAUR_01285, of C. auris cluster together on a separate branch with Snq2 of C. albicans. However, independent blast searches show that both the proteins display 60% sequence identity with Snq2 of C. albicans, which strongly suggests that C. auris might possess two independent genes coding for Snq2-like proteins, in contrast to C. albicans and D. hansenii which only possess a single ortholog. An additional copy of SNQ2 has also been reported in a C. auris isolate belonging to the S. African geographical clade, B11221 (Muñoz et al., 2018). A closer examination suggests that these two Snq2-like proteins form a separate branch from that formed by S. cerevisiae Snq2 and Pdr18, which perform overlapping functions and are considered paralogs (Godinho et al., 2018). The separate blast searches with these two S. cerevisiae proteins show that C. auris CAUR_01276 and CAUR_01285 have 55 and 54% sequence identity with Pdr18 and Snq2, respectively. S. cerevisiae YOL075C of the PDR subfamily and its homologs C. albicans Cdr6 and D. hansenii DEBHA_B5RUN7 constitute a distinct cluster, to which CAUR_04233 also belongs; CAUR_04233 has 51% sequence identity with Cdr6. The separation of this cluster of proteins from other PDR subfamily members suggests an early origin, as it is among the few ABCG protein members, that are also observed in primitive eukaryotes like Batrachochytrium dendrobatidis and Encephalitozoon cuniculi (Kovalchuk and Driessen, 2010). While YOL075C of S. cerevisiae remains uncharacterized, its homolog in C. albicans Cdr6 has recently been shown to impact drug susceptibility mediated by TOR signaling (Khandelwal et al., 2018).
The only half-size transporter member of the PDR subfamily, CAUR_03774, harbors a TMH1–NBD–TMH6 topology (Figure 2), and shows a close phylogenetic relationship with Adp1 of S. cerevisiae and C. albicans. CAUR_03774 has 54% sequence identity with S. cerevisiae Adp1, and 67% sequence identity with C. albicans Adp1. Similar to Adp1, CAUR_03774 is characterized by the presence of a domain containing epidermal growth factor repeats at its N-terminus, where the first NBD would be situated, if it were a full-size ABC transporter (Figure 2). As with D. hansenii and C. albicans, there is no homolog of S. cerevisiae Aus1 observed in C. auris (Wilcox et al., 2002). Our subcellular predictions show that similar to the most closely related PDR subfamily members in S. cerevisiae, most of the PDR-type ABC proteins in C. auris are probably localized to the plasma membrane (Supplementary Table S6).
ABCB/MDR Subfamily
The ABCB/MDR subfamily is characterized by a forward topology, comprising full and half-size transporters. Our analysis revealed that the MDR subfamily of C. auris has four ABC transporters: CAUR_01852, CAUR_03761, CAUR_04565, and CAUR_02994 (Figure 1). Notably, CAUR_03761, CAUR_04565, and CAUR_02994 are half-size transporters, while CAUR_01852 is predicted to be a full transporter (TMD–NBD–TMD–NBD) (Figure 2). Unlike the PDR subfamily in C. auris, the number of MDR subfamily members is equal to S. cerevisiae, C. albicans, and D. hansenii pointing toward a conserved functional nature across different fungal species (Figure 3). One of the most studied examples of this subfamily is human P-glycoprotein, which is a full transporter, and its overexpression in cancer cells is associated with multidrug resistance (Ambudkar et al., 1992). The only full transporter, CAUR_01852, shows close homology with S. cerevisiae Ste6, which transports pheromones, and with C. albicans Hst6, which functionally complements Ste6 in S. cerevisiae (Raymond et al., 1998). One of the half-size transporters, CAUR_02994, displays high sequence identity 77% and 68% with Atm1 of C. albicans and S. cerevisiae, respectively. In both species Atm1 is a mitochondrial transporter involved in exporting Fe–S clusters from the inner mitochondrial membrane into the cytosol (Leighton and Schatz, 1995). The other two half-size transporters, CAUR_03761 and CAUR_04565, display an orthologous relationship with Mdl1 and Mdl2, respectively, which are mitochondrial peptide transporters in S. cerevisiae (Young et al., 2001; Figure 3A). Our subcellular localization prediction suggests that CAUR_01852 (the Hst6-homolog) is localized to plasma membranes, similar to its homolog in S. cerevisiae (Supplementary Table S6). Notably, the other three members of MDR family, the Mdl1-homolog CAUR_03761, the Mdl2-homolog CAUR_04565, and the Atm1-homolog CAUR_02994, might be present on mitochondrial membranes (Supplementary Table S6).
ABCC/MRP Subfamily
The ABCC/MRP subfamily is the second largest subfamily of the ABC superfamily found in most eukaryotes and contains only full-length transporters. Our analysis showed that C. auris harbors seven putative MRP subfamily members: CAUR_01188, CAUR_00156, CAUR_00368, CAUR_00964, CAUR_01719, CAUR_03320, and CAUR_02951 (Figures 1, 2). Notably, our comparative analysis reveals that the number of MRP subfamily members in C. auris (seven members) is equivalent to D. hansenii (seven members), but expanded compared to C. albicans (four members) and S. cerevisiae (six members) (Figure 3B).
Multidrug resistance-associated protein subfamily members form two major clusters: one with CAUR_01188, which aligns with Ybt1, Vmr1, and Nft1 of S. cerevisiae, and the second major cluster, which branches into three subgroups (Figures 1, 3A). Out of these three, one subgroup is formed by the CAUR_00368, CAUR_01719, and CAUR_02951 of C. auris, and similar to Yor1 of C. albicans and S. cerevisiae; the second consists of CAUR_00964 and C. albicans Mlt1; and the third subgroup contains CAUR_03320 with 67% sequence identity to C. albicans Ycf1 and CAUR_00156, which displays similarity to S. cerevisiae Ybt1 and C. albicans Orf19.6382 (Figure 3A).
This subfamily is also known for the presence of long MRP proteins, which have an extra TMD called the N-terminal extension (NTE) or (TMD)0. Usually the (TMD)0 consists of five TMHs and hence long MRPs have a (TMD)0–(TMD–NBD–TMD–NBD) topology (Figure 2). It is well-documented in S. cerevisiae that the (TMD)0 of Ycf1 is required for proper localization to the vacuolar membrane (Mason, 2002). Our analysis shows that out of the seven predicted members, two members CAUR_01188 and CAUR_03320 possess a (TMD)0–(TMD–NBD–TMD–NBD) topology and are long MRPs (Figure 2). Interestingly, another two MRP members, CAUR_00156 and CAUR_02951, have two additional TMHs at their N-terminus, displaying an unusual TMH2–(TMH6–NBD–TMH6–NBD) arrangement (14 TMHs in total) (Figure 2). However, the phylogenetic analysis clusters CAUR_00156 with D. hansenii DEBHA_B5RTT1 and C. albicans Orf19.6382, which are predicted to have 17 TMHs, a (TMD)0–(TMD–NBD–TMD–NBD) topology (Wasi et al., 2018). CAUR_00368 and CAUR_01719 show a typical (TMD–NBD–TMD–NBD) topology. Notably, no TMDs are predicted for CAUR_00964 by TOPOCONS software, but it appears to have 12 TMDs and a (TMD–NBD–TMD–NBD) according to TMPRED software (Supplementary Table S5).
The members of the MRP subfamily are known to be localized to vacuoles in various yeasts (Snider et al., 2013; Khandelwal et al., 2016) and are well known for their roles in cellular detoxification and sequestration of heavy metal ions in the form of conjugates with glutathione (GSH), glucuronate, or sulfate (Paumi et al., 2007). Recently, our group has shown vacuolar localization for Ybt1 in S. cerevisiae and Mlt1 in C. albicans, which sequester azoles into the vesicle lumen (Khandelwal et al., 2019). The MRP transporters are also involved in the translocation of bile acids and phosphatidylcholine into the vacuole and affect the regulation of vacuolar fusion (Gulshan and Moye-Rowley, 2011; Sasser and Fratti, 2014; Khandelwal et al., 2016). The other member, yeast cadmium factor (Ycf1) of S. cerevisiae helps the cell with detoxification of heavy metal conjugates with GSH, vacuolar fusion, and also affects chronological aging (Paumi et al., 2007; Sasser and Fratti, 2014). Notably, in C. auris out of seven MRP subfamily members, three transporters (CAUR_00368, CAUR_01719, and CAUR_02951) lack the extra five TMHs at the very N-terminus (NTE/TMD0), and hence are predicted to be localized to the plasma membrane (Supplementary Table S6).
ABCD/ALDp Subfamily
Similar to S. cerevisiae, C. albicans, and D. hansenii, the ALDp subfamily of C. auris has two members, namely CAUR_00862 and CAUR_04133 (Figures 1–3). Predominantly, members of the ALDp subfamily are half-size transporters with a forward topology (TMD–NBD); in plants also full-length transporters of this subfamily have been described (Wasi et al., 2018). The half-size ALDp transporters turn into functionally complete transporters after hetero- or homo-dimerization (Morita and Imanaka, 2012).
The members of the ALDp subfamily are also known as the peroxisomal ABC transporters, which are involved in fatty acid regulation (Morita and Imanaka, 2012). Our localization predictions suggest that C. auris members of this subfamily are also peroxisomal transporters (Supplementary Table S6). Phylogenetic analysis indicates that CAUR_04133 is homologous to Pxa1 (44% identity), while CAUR_00862 is 42% identical with Pxa2 of S. cerevisiae (Figure 3A). Pxa1 and Pxa2 are long chain fatty acid transporters in S. cerevisiae (van Roermund et al., 2012). The clear evolutionary orthologous relationship between the ALDp subfamily members in various species strongly points toward conserved functions.
ABCE/RLI Subfamily
This subfamily belongs to the soluble ABC proteins as its members are devoid of TMD regions and have only two NBDs in tandem (NBD–NBD) (Figure 2). ABCE-subfamily proteins are also known as RLIs because of their ability to inhibit the double-stranded RNA nuclease RNase L. However, these proteins are also present in organisms, which do not have RNase L implying their involvement in other important cellular functions (Wasi et al., 2018). The RLI subfamily has the lowest member count among all ABC proteins (Figure 3B). Indeed, C. auris contains a single member, CAUR_04997, belonging to this subfamily. The main distinctive feature of RLI subfamily proteins is the presence of a conserved ferredoxin iron–sulfur cluster (4Fe4S-type) (CX2CX2CX3C) motif (Gaur et al., 2005). RLI-subfamily proteins are implicated in processes such as ribosome biogenesis and translation initiation at the 3′-untranslated region of ORFs (Wasi et al., 2018).
ABCF/YEF3 Subfamily
Similar to RLI-type ABC proteins, ABCF/YEF3 subfamily members characteristically have two NBDs (NBD–NBD) and no TMDs (Figure 2), and are also considered soluble ABC proteins. In C. auris, this subfamily has five protein members: CAUR_04813, CAUR_02351, CAUR_03795, CAUR_04953, and CAUR_00824. The combined phylogenetic analyses showed that CAUR_00824 forms a tight cluster with C. albicans Cef3 and D. hansenii DEBHA_Q6BK70, and a wider cluster with its ortholog in S. cerevisiae, Yef3 and Hef3 (Figure 3A). It is worth pointing out that except for S. cerevisiae with two YEF3-like proteins, most fungi have only a single YEF3 ortholog (Kovalchuk and Driessen, 2010). Yef3 in S. cerevisiae is involved in translation elongation by supporting the binding of aminoacyl-tRNA to the ribosome. CAUR_04953 of C. auris shows high sequence identity with S. cerevisiae Gcn20 (72%) and with C. albicans Gcn20 (87%). Gcn20 is one of the most-studied members of the YEF3 subfamily, and it is involved in amino acid biosynthesis through the positive regulation of Gcn2 kinase activity (Yuan et al., 2017). Similar to the other CTG-clade yeasts C. albicans and D. hansenii, no Arb1-like protein can be detected in C. auris (Figure 3A). Arb1 is majorly responsible for maintaining ribosome biogenesis in S. cerevisiae (Dong et al., 2005).
Others: Unclassified ABC Proteins
This group of ABC proteins comprises members that remain separated from the main subfamilies. It contains soluble ABC proteins, because of the absence of TMDs within their sequences. Our analysis shows that C. auris has two proteins in this group, namely CAUR_3076 and CAUR_04824 (Figure 1). Topology predictions show that CAUR_04824 has NBD-NBD architecture, while CAUR_3076 harbors a single NBD (Figure 2). The phylogenetics points to a close relationship between CAUR_04824 and Modf of C. albicans with a sequence identity of 50%, while CAUR_3076 seems orthologous to S. cerevisiae Caf16, which is part of the CCR4–NOT complex and has a regulatory role in transcription (Liu et al., 2011).
Expression Analysis Confirms Putative ABC Transporter Genes Transcripts
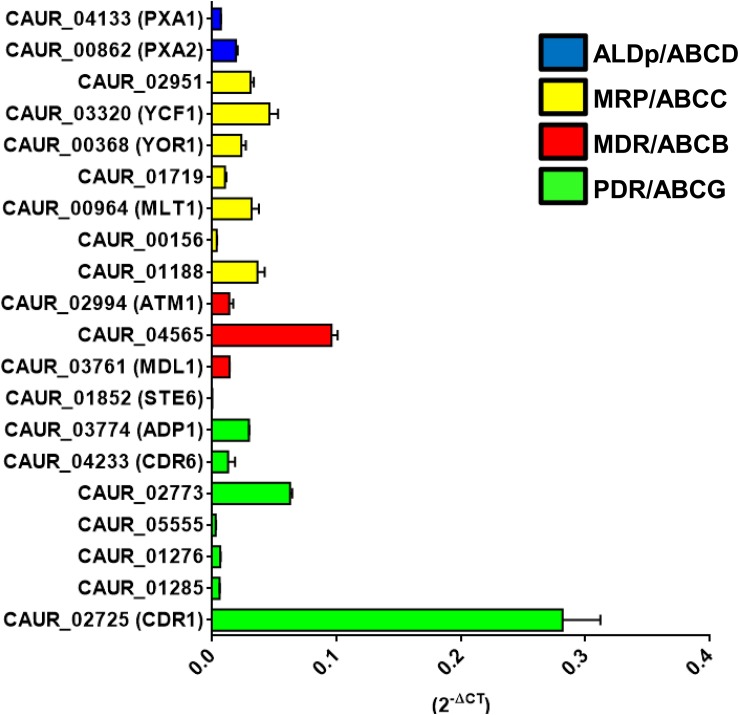
As a goal of evaluating the role of ABC proteins as drug transporters in C. auris (CBS 10913T), we first assessed the basal expression of all 20 genes encoding membrane-bound ABC proteins. The basal expression profile of all the putative ABC transporter genes was quantified and normalized in comparison to TDH1 (CAUR_02457) gene expression. TDH1 is a housekeeping gene, which is an ortholog of GAPDH, a housekeeping gene commonly used as internal control for qRT-PCR (Kumari et al., 2018). The expression profile of the CBS 10913T strain established the presence of transcripts of all the 20 predicted putative ABC transporter genes, and highlighted a variability of basal expression between them (Figure 4). Although all the ABC transporters showed lower expression than housekeeping gene GAPDH, ABC transporters do show constitutive differential expression, which suggests possible different roles. The C. albicans CDR1-ortholog CAUR_02725 showed the highest level of basal expression. CAUR_01852 (the homolog of STE6/HST6) showed the least basal expression among the 20 ABC transporter genes. Notably, similar low expression of C. glabrata CAGL0K00363g, an ortholog of S. cerevisiae STE6 has been reported (Kumari et al., 2018). SNQ2 orthologs, CAUR_01276 and CAUR_01285, showed similar levels of basal expression. Notably, CAUR_01852 lacks two nucleotides at positions 3,309 and 3,310 in clade I strains B8441, 6684, and VPCI 479/P/13, and could be considered a pseudogene in these. However, in isolates from all other clades, CBS 10913T, B11220, B11221, and B11243, these residues are present and the ORFs seem to be intact. Considering this and that CAUR_01852 is transcribed, albeit at low levels, raises the possibility that CAUR_01852 is functional in C. auris isolates from clades II, III, and IV.
FIGURE 4.
Basal expression levels of ABC transporter genes in C auris CBS10913T. The presence of true ABC transporters ORFs was confirmed using qRT-PCR. Expression levels were calculated using the 2–ΔCT methods with the house-keeping TDH1 gene as internal control. Here 2–ΔCT represents the relative expression of ABC transporter in comparison to TDH1 gene and its value 1 meaning that the gene expressing equal to the internal control TDH1. The values are means ± SE (error bars) from triplicates of biological duplicates (n = 6).
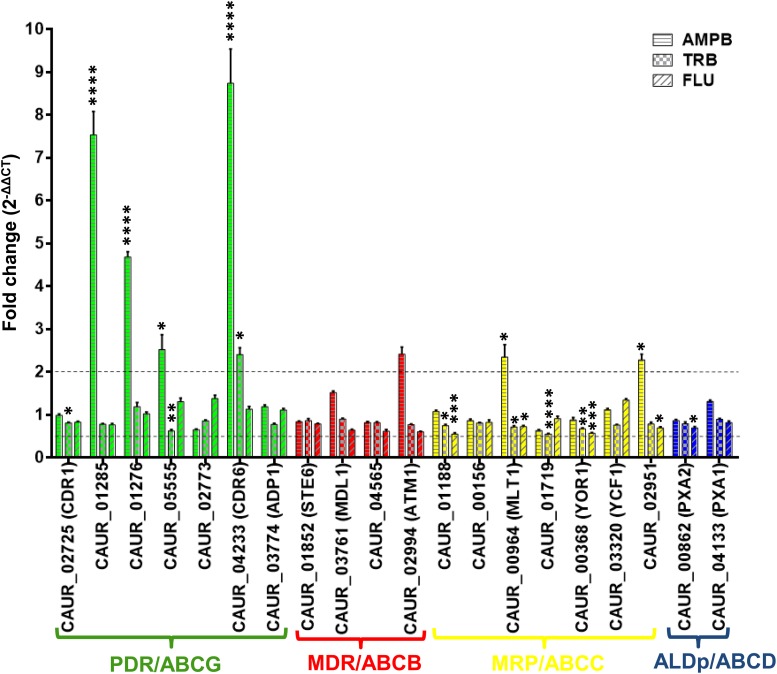
ABC Transporters Display Selective Response to Drug Treatment
To assess the role of ABC transporters in drug resistance, we examined the changes in ABC transporter transcript levels in response to short-term drug exposure by qRT-PCR. For this, we treated C. auris CBS 10913T with different classes of known antifungals, specifically the polyene AMPB, the allylamine TRB, and the triazole FLU. The following MIC50 values were used for each drug in YEPD broth for 60 min treatments of the C. auris CBS 10913T strain: AMPB at 0.7 μg/ml, TRB at 0.75 μg/ml, and FLU at 6 μg/ml (Supplementary Table S2). The qRT-PCR analysis was performed after the drug treatment and compared with untreated cells (Figure 5). The transcripts of subfamily members were differentially regulated in the presence of the antifungals (Figure 5). PDR subfamily members are well-known drug transporters implicated in clinical drug resistance encountered in different fungi. The observed upregulation of PDR subfamily members of C. auris strongly highlights a potential role in drug resistance. Among all the three drugs, AMPB exposure presented maximum response in terms of upregulated expression of selected ABC transporter genes. Particularly, the PDR subfamily genes exhibited higher expression after AMPB exposure, including CAUR_05555 (one of the homologs of CDR4), CAUR_04233 (homolog of CDR6), and both homologs of SNQ2: CAUR_01285 and CAUR_01276 (Figure 5). An elevated expression of the SNQ2 homologs in C. glabrata and S. cerevisiae has also been observed after AMPB treatment (Rogers et al., 2001); this indicates that CAUR_01285 and CAUR_01276 could be involved in drug susceptibility in C. auris as well. Incidentally, the more than eightfold higher expression of the CDR6-homolog CAUR_04233 was highest among all ABC transporter transcripts (Figure 5). Interestingly, CAUR_04233 was also strongly upregulated among transcripts following TRB treatment (Figure 5).
FIGURE 5.
Drug induced expression profiles of various ABC transporter genes in C. auris. The expression levels of different genes were checked by qRT-PCR and data were measured by 2–ΔΔCT method in the presence of amphotericin B (0.7 μg/ml), terbinafine (0.75 μg/ml), and fluconazole (6 μg/ml) in comparison to sample with no drug treatment. TDH1 was used as an internal control. The dotted lines represent the selected biological significant cut off (more than or equal to twofold). 2–ΔΔCT represents the fold change and its values 2, 1, and 0.5 represent twofold up, no change, and twofold down regulation, respectively. Data presented as means ± SE (error bars) from biological replicates with technical triplicates (n = 6). P-values (*p ≤ 0.05, ∗∗p ≤ 0.001, ∗∗∗p ≤ 0.0001, and ****p ≤ 0.00001) were calculated with two-way ANOVA (uncorrected Fisher’s LSD) using GraphPad prism. P-values are for differences from the fold change of 1.
In our present study, ABC genes show maximum transcriptional response to AMPB treatment. This observation also matches the situation in S. cerevisiae, in that transcriptional changes in transporters is a major category (10.3% of total genes) affected by AMPB treatment (Zhang, 2002; Agarwal et al., 2003). Also, in D. hansenii AMPB treatment resulted in a significant increase in the expression of several ABC transporters (Wasi et al., 2018). This suggests that AMPB treatment-dependent change in expression of ABC transporters is a common response observed in various yeasts.
Considering the well-established role of ABC transporters in membrane lipid homeostasis and the fact that the expression level of ABC transporters can impact AMPB sensitivity, the commonly observed up-regulation of ABC transporters could be a crucial defense mechanism (Smriti et al., 2002; Rockwell et al., 2009; Guan et al., 2010; Nagi et al., 2013; Gulati et al., 2015). The TOR1 pathway also governs AMPB persistence in yeast (Bojsen et al., 2016), indeed, we have recently shown that the ABC transporter Cdr6 can impact TOR signaling and its absence induces the TOR signaling cascade (Khandelwal et al., 2018). This suggests that ABC transporters and TOR signaling are linked and over-expression of ABC transporters could also impact the AMPB sensitivity via TOR signaling.
Strikingly, unlike AMPB and TRB exposure, none of the ABC transporter transcripts showed any significant changes in their expression following FLU treatment (Figure 5). A similar observation was recently reported by Cuomo and co-workers, where they show a lack of response after VRC treatment (Muñoz et al., 2018).
PDR Subfamily Genes Predominantly Display Elevated Transcript Levels in Drug-Resistant Clinical Isolates of C. auris
The upregulated expression of certain ABC transporters correlates well with an increased resistance to antifungals in clinical isolates of C. albicans, C. glabrata, and other pathogenic fungi (Sanglard et al., 1999). The upregulation of ABC transporter genes associated with rapid expulsion of antifungal drugs is one of the major strategies adapted by clinical multidrug-resistant strains. To evaluate the relevance of ABC transporter expression levels in multidrug-resistant C. auris, we evaluated their expression in three selected clinical isolates of C. auris in comparison to C. auris (CBS 10913T). These were resistant to AMPB and FLU, and also displayed elevated MIC50s to VRC and ITR (Table 2).
TABLE 2.
In vitro susceptibility profile (MIC50 values in RPMI media) of C. auris strains used in the study to antifungal drugs.
| Strain | AMPB | FLU | VRC | ITR |
| C. auris CBS 10913T | 1 | 1 | 0.03 | 0.12 |
| Isolate-1 | 1 | 1.00 | 0.25 | |
| Isolate-2 | 1 | 0.50 | 0.13 | |
| Isolate-3 | 4 | 64 | 1.00 | 0.25 |
MIC values are given in μg/ml. AMPB, amphotericin B; FLU, fluconazole; VRC, voriconazole; ITR, itraconazole.
The transcript analysis confirmed the involvement of PDR subfamily members in drug resistance of the emerging pathogenic C. auris. Accordingly, the CDR1-homolog CAUR_02725 and the SNQ2-homolog CAUR_01276 displayed higher transcript levels in all tested clinical isolates suggesting a role as a drug exporter in C. auris (Figure 6). A role in multidrug resistance including resistance to azoles is well-established for Cdr1 in C. albicans and Snq2 in C. glabrata (Whaley et al., 2018).
FIGURE 6.
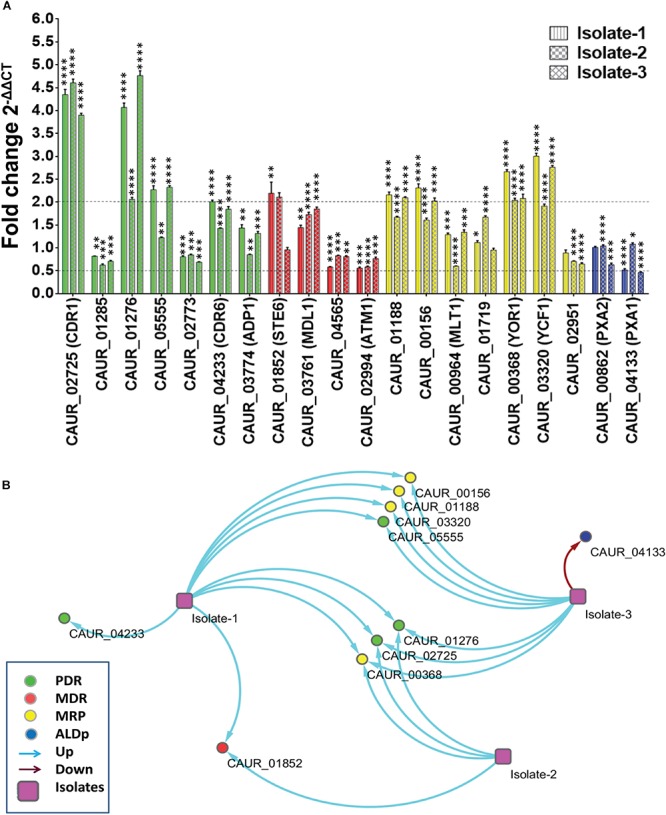
Expression levels of ABC transporter genes in drug-resistant clinical isolates of C. auris. (A) The expression levels of the various ABC transporter genes in three clinical isolates in comparison to the reference strain CBS 10913T was checked by qRT-PCR. Fold change (2–ΔΔCT) measured and presented as means ± SE (error bars) (n = 3). TDH1 was used as an internal control. The dotted lines represent the selected biological significant cut off (more than or equal to twofold). P-values (*p ≤ 0.05, ∗∗p ≤ 0.001, ∗∗∗p ≤ 0.0001, and ****p ≤ 0.00001) were calculated with two-way ANOVA (uncorrected Fisher’s LSD) using GraphPad prism. P-values are for differences from the fold change of 1. (B) Interaction map of ABC transporters in the clinical isolates was made by taking the arbitrary cut off of (more than or equal to twofold) change with the help of Cytoscape software version 3.4.0 (Shannon et al., 2003).
Another PDR subfamily member, CAUR_04233 (CDR6 homolog), is (more than or equal to twofold) upregulated in Isolate-1. It also showed a trend toward higher expression in Isolate-2 and Isolate-3, but it was not considered significant since it was below our selected threshold (more than or equal to twofold) of upregulation (Figure 6A). In C. albicans the Cdr6 transporter is known as an exporter of xenobiotics (Khandelwal et al., 2018). One of the homologs of CDR4 (CAUR_05555) is upregulated in clinical Isolate-1 and Isolate-3 (Figure 6A), interestingly, elevated expression of CDR4 in clinical multidrug-resistant C. albicans and C. glabrata strains has not been reported.
Among the MDR subfamily members, the homolog of STE6 (CAUR_001852) was the only member that exhibited elevated expression in Isolate-1 and Isolate-2 (Figure 6A). Notably, Ste6 is not an established drug transporter, but a dedicated pheromone transporter in S. cerevisiae (Kumari et al., 2018). However, one study has reported that Ste6 could also impart resistance to valinomycin in S. pombe (Christensen et al., 1997). In C. glabrata, the transcripts of a STE6 homolog (CAGL0K00363g) displayed variable response to drug exposure (Kumari et al., 2018), and also, the human Ste6 homolog, ABCB1, has been shown to extrude xenobiotics in various chemotherapy-resistant tumors (He et al., 2011). Notably, none of the MDR transporters showed more than or equal to twofold downregulation. However, CAUR_02994 (ATM1 homolog) of the MDR subfamily showed 1.8- and 1.7-fold downregulation in Isolate-1 and Isolate-2, respectively (Figure 6A). This is in contrast to what was observed after transient exposure of C. auris CBS 10913T to AMPB where a more than twofold increase in CAUR_02994 expression was observed. The homolog of ATM1 in C. glabrata (CAGL0M13739g) displayed raised expression levels following ketoconazole treatment (Kumari et al., 2018).
Recent studies established the roles of MRP subfamily members in fungal drug resistance and virulence (Gulshan and Moye-Rowley, 2011; Khandelwal et al., 2016, 2019). We observed that CAUR_00368, which displays close identity with YOR1, is upregulated in all three clinical isolates (Figure 6B). In C. albicans, Yor1 is involved in expulsion of beauvericin, a natural antimicrobial (Shekhar-Guturja et al., 2016). Upregulation of YOR1 transcription in C. auris indicates a potential role in multidrug resistance of this pathogen. CAUR_01188/YBT1, CAUR_00156/BPT1, and CAUR_03320/YCF1 show upregulation in two isolates (Isolate-1 and -3). These three transporters are known to be involved extruding various xenobiotics in S. cerevisiae (Paumi et al., 2007).
The ALDp subfamily member CAUR_04133/PXA1 transcript is significantly (more than or equal to twofold) down regulated in clinical Isolate-3 (Figure 6B). In clinical Isolate-1, the expression of CAUR_04133 is also decreased (1.93-fold) compared to WT (CBS10913T) (Figure 6A). Coincidently, in C. glabrata its homolog CAGL0M02387 is also downregulated when treated with ketoconazole (Kumari et al., 2018).
Conclusion
The increasing reports of C. auris outbreaks and its emerging resistance to common antifungal pose an alarming clinical situation for hospitals. An established role of ABC transporters in multidrug resistance of different fungal pathogens highlights an essential requirement for understanding the impact these ABC proteins might have in C. auris. Here, we have analyzed the full repertoire of ABC proteins of the C. auris type strain CBS 10913T.
During the preparation of this manuscript two studies were published (Kim et al., 2019; Rybak et al., 2019) which further highlighted the role of some of the ABC transporters (mainly of PDR family) of C. auris. Our study is a comprehensive analysis of total landscape of ABC protein in C. auris. Our evaluation identified 28 putative ABC proteins, which is based on phylogeny and domain organization, demonstrated that these could be clustered into the six major ABC transporter subfamilies. Of these, 20 of the putative ABC proteins contain TMDs and are categorized into the four membrane-localized subfamilies. The constitutive expression of these membrane-localized transporters confirmed their presence as true ORFs, while differential expression of the genes encoding ABC transporters implies different biological and clinical relevance. The selective responses of some of these transporters to antifungal drug treatment possibly provide a clue of their relevance in clinically drug resistance mechanisms. The higher expression (more than or equal to twofold) of the CAUR_02725 (CDR1) and CAUR_01276 (SNQ2) ABC transporters in multidrug-resistance clinical isolates of C. auris also supports that these transporters may be involved in drug resistance. Our study therefore provides the first complete landscape of ABC transporters of this pathogen, which presents a pivotal platform for further exploration of this important class of proteins and their relevance in multidrug resistance in C. auris.
Data Availability
The datasets analyzed for this study can be found at https://www.ebi.ac.uk/ena/data/view/PRJEB29190 (ENA Accession No. PRJEB29190).
Author Contributions
RP, NG, MW, and NK conceived and designed the experiments. MW, NK, AJM, RN, and PV performed the bioinformatics analysis. AJM, GR, ZR, AL, and NG carried out the ploidy determination and genome-wide sequencing of C. auris. MW and NK carried out the constitutive and drug-induced expression profiling. MW performed the expression analysis of the clinical isolates. RP, NK, MW, RN, and AL analyzed the data and wrote the manuscript. SR and AC provided the clinical isolates and performed the drug profiling experiments. AKM, RP, AML, NG, and AL provided the lab space and reagents for the experiments. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer RC declared a past co-authorship with one of the authors RP to the handling editor.
Acknowledgments
We thank the Centre for Genome Enabled Biology and Medicine at the University of Aberdeen (E. Collie-Duguid and S. Shaw) for sequencing and support with genome analysis.
Funding. This work was supported by the ICMR (AMR/149/2018-ECD-II) and DBT (BT/PR14117/BRB/10/1420/2015) to RP. AKM appreciates the support by research grant EMR/2016/001927 and DST PURSE II from the Department of Science and Technology (IN). MW was grateful for a Senior Research Fellowship from the University Grant Commission. NG acknowledges the Wellcome Trust support of a Senior Investigator (101873/Z/13/Z), Collaborative (200208/A/15/Z), and Strategic Awards (097377/Z11/Z), and the MRC Centre for Medical Mycology (MR/N006364/1). AJM was supported by the University of Aberdeen studentship. Work in AL’s laboratory was supported by the Wellcome Trust (212524/Z/18/Z) and the Medical Research Council (MRC) Centre for Medical Mycology at the University of Aberdeen (MR/P501955/1 and MR/N006364/1).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01445/full#supplementary-material
References
- Agarwal A. K., Rogers P. D., Baerson S. R., Jacob M. R., Barker K. S., Cleary J. D., et al. (2003). Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J. Biol. Chem. 278 34998–35015. 10.1074/jbc.M306291200 [DOI] [PubMed] [Google Scholar]
- Ambudkar S. V., Lelong I. H., Zhang J., Cardarelli C. O., Gottesman M. M., Pastan I. (1992). Partial purification and reconstitution of the human multidrug-resistance pump: characterization of the drug-stimulatable ATP hydrolysis. Proc. Natl. Acad. Sci. U.S.A. 89 8472–8476. 10.1073/pnas.89.18.8472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Shah A. H., Redhu A. K., Moreno A., Falson P., Prasad R. (2018). W1038 near D-loop of NBD2 is a focal point for inter-domain communication in multidrug transporter Cdr1 of Candida albicans. Biochim. Biophys. Acta Biomembr. 1860 965–972. 10.1016/j.bbamem.2018.01.022 [DOI] [PubMed] [Google Scholar]
- Boetzer M., Pirovano W. (2014). SSPACE-longread: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics 15:211. 10.1186/1471-2105-15-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojsen R., Regenberg B., Gresham D., Folkesson A. (2016). A common mechanism involving the TORC1 pathway can lead to amphotericin B-persistence in biofilm and planktonic Saccharomyces cerevisiae populations. Sci. Rep. 6:21874. 10.1038/srep21874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo Ruiz B. G., Ross Z. K., Holmes E., Schelenz S., Gow N. A. R., Lorenz A. (2019). Rapid and extensive karyotype diversification in haploid clinical Candida auris isolates. Curr. Genet. 10.1007/s00294-019-00976-w [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S., Alampalli S. V., Nageshan R. K., Chettiar S. T., Joshi S., Tatu U. S. (2015). Draft genome of a commonly misdiagnosed multidrug resistant pathogen Candida auris. BMC Genomics 16:686. 10.1186/s12864-015-1863-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen P. U., Davey J., Nielsen O. (1997). The Schizosaccharomyces pombe mam1 gene encodes an ABC transporter mediating secretion of M-factor. Mol. Gen. Genet. 255 226–236. 10.1007/s004380050493 [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (2008). Reference Method For Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard Clsi Document M27-A3, 3rd Edn Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Cortegiani A., Misseri G., Fasciana T., Giammanco A., Giarratano A., Chowdhary A. (2018). Epidemiology, clinical characteristics, resistance, and treatment of infections by Candida auris. J. Intensive Care 6:69. 10.1186/s40560-018-0342-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daumke O., Knittler M. R. (2001). Functional asymmetry of the ATP-binding-cassettes of the ABC transporter TAP is determined by intrinsic properties of the nucleotide binding domains. Eur. J. Biochem. 268 4776–4786. 10.1046/j.1432-1327.2001.02406.x [DOI] [PubMed] [Google Scholar]
- Dean M., Hamon Y., Chimini G. (2001a). The human ATP-binding cassette transporter superfamily. J. Lipid Res. 42 1007–1017. [PubMed] [Google Scholar]
- Dean M., Rzhetsky A., Allikmets R. (2001b). The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 11 1156–1166. 10.1101/gr.184901 [DOI] [PubMed] [Google Scholar]
- Dong J., Lai R., Jennings J. L., Link A. J., Hinnebusch A. G. (2005). The novel ATP-binding cassette protein ARB1 Is a shuttling factor that stimulates 40S and 60S ribosome biogenesis. Mol. Cell. Biol. 25 9859–9873. 10.1128/MCB.25.22.9859-9873.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J., Mitchell A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44 D279–D285. 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg K., Woodworth K., Walters M., Berkow E. L., Jackson B., Chiller T., et al. (2019). Candida auris: the recent emergence of a multidrug-resistant fungal pathogen. Med. Mycol. 57 1–12. 10.1093/mmy/myy054 [DOI] [PubMed] [Google Scholar]
- Gaur M., Choudhury D., Prasad R. (2005). Complete inventory of ABC proteins in human pathogenic yeast, Candida albicans. J. Mol. Microbiol. Biotechnol. 9 3–15. 10.1159/000088141 [DOI] [PubMed] [Google Scholar]
- Godinho C. P., Dias P. J., Poncot E., Sa-Correia I. (2018). The paralogous genes pdr18 and snq2, encoding multidrug resistance abc transporters, derive from a recent duplication event, pdr18 being specific to the Saccharomyces genus. Front. Genet. 9:476. 10.3389/fgene.2018.00476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W., Jiang H., Guo X., Mancera E., Xu L., Li Y., et al. (2010). Antagonistic changes in sensitivity to antifungal drugs by mutations of an important ABC transporter gene in a fungal pathogen. PLoS One 5:e11309. 10.1371/journal.pone.0011309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati S., Balderes D., Kim C., Guo Z. A., Wilcox L., Area-Gomez E., et al. (2015). ATP-binding cassette transporters and sterol O-acyltransferases interact at membrane microdomains to modulate sterol uptake and esterification. FASEB J. 29 4682–4694. 10.1096/fj.14-264796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulshan K., Moye-Rowley W. S. (2011). Vacuolar import of phosphatidylcholine requires the ATP-binding cassette transporter ybt1. Traffic 12 1257–1268. 10.1111/j.1600-0854.2011.01228.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S.-M., Li R. R., Kanwar J., Zhou S.-F. (2011). Structural and functional properties of human multidrug resistance protein 1 (MRP1/ABCC1). Curr. Med. Chem. 18 439–481. 10.2174/092986711794839197 [DOI] [PubMed] [Google Scholar]
- Jungwirth H., Kuchler K. (2006). Yeast ABC transporters - A tale of sex, stress, drugs and aging. FEBS Lett. 580 1131–1138. 10.1016/j.febslet.2005.12.050 [DOI] [PubMed] [Google Scholar]
- Khandelwal N. K., Chauhan N., Sarkar P., Esquivel B. D., Coccetti P., Singha A., et al. (2018). Azole resistance in a Candida albicans mutant lacking the ABC transporter CDR6/ROA1 depends on TOR signaling. J. Biol. Chem. 293 412–432. 10.1074/jbc.M117.807032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelwal N. K., Kaemmer P., Forster T. M., Singh A., Coste A. T., Andes D. R., et al. (2016). Pleiotropic effects of the vacuolar ABC transporter MLT1 of Candida albicans on cell function and virulence. Biochem. J. 473 1537–1552. 10.1042/BCJ20160024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelwal N. K., Wasi M., Nair R., Gupta M., Kumar M., Mondal A. K., et al. (2019). Vacuolar sequestration of azoles: a novel strategy of azole antifungal resistance conserved across pathogenic and non-pathogenic yeast. Antimicrob. Agents Chemother. 63:e01347-18. 10.1128/AAC.01347-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Iyer K. R., Pardeshi L., Munoz J. F., Robbins N., Cuomo C. A., et al. (2019). Genetic analysis of Candida auris implicates Hsp90 in morphogenesis and azole tolerance and Cdr1 in azole resistance. MBio 10:e00346-19. 10.1128/mBio.02529-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk A., Driessen A. J. M. (2010). Phylogenetic analysis of fungal ABC transporters. BMC Genomics 11:177. 10.1186/1471-2164-11-177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S., Kumar M., Khandelwal N. K., Kumari P., Varma M., Vishwakarma P., et al. (2018). ABC transportome inventory of human pathogenic yeast Candida glabrata: phylogenetic and expression analysis. PLoS One 13:e0202993. 10.1371/journal.pone.0202993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamping E., Baret P. V., Holmes A. R., Monk B. C., Goffeau A., Cannon R. D. (2010). Fungal PDR transporters: phylogeny, topology, motifs and function. Fungal Genet. Biol. 47 127–142. 10.1016/j.fgb.2009.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton J., Schatz G. (1995). An ABC transporter in the mitochondrial inner membrane is required for normal growth of yeast. EMBO J. 14 188–195. 10.1002/j.1460-2075.1995.tb06989.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Zhou S., Tian L., Guo E., Luan Y., Zhang J., et al. (2011). Genome-wide identification and characterization of ATP-binding cassette transporters in the silkworm, Bombyx mori. BMC Genomics 12:491. 10.1186/1471-2164-12-491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lockhart S. R., Etienne K. A., Vallabhaneni S., Farooqi J., Chowdhary A., Govender N. P., et al. (2017). Simultaneous emergence of multidrug-resistant candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 64 134–140. 10.1093/cid/ciw691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Mason D. L. (2002). Requirement of the N-Terminal extension for vacuolar trafficking and transport activity of yeast Ycf1p, an ATP-binding cassette transporter. Mol. Biol. Cell 13 4443–4455. 10.1091/mbc.E02-07-0405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M., Imanaka T. (2012). Peroxisomal ABC transporters: structure, function and role in disease. Biochim. Biophys. Acta 1822 1387–1396. 10.1016/j.bbadis.2012.02.009 [DOI] [PubMed] [Google Scholar]
- Muñoz J. F., Gade L., Chow N. A., Loparev V. N., Juieng P., Berkow E. L., et al. (2018). Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat. Commun. 9:5346. 10.1038/s41467-018-07779-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagi M., Tanabe K., Ueno K., Nakayama H., Aoyama T., Chibana H., et al. (2013). The Candida glabrata sterol scavenging mechanism, mediated by the ATP-binding cassette transporter Aus1p, is regulated by iron limitation. Mol. Microbiol. 88 371–381. 10.1111/mmi.12189 [DOI] [PubMed] [Google Scholar]
- Parra G., Bradnam K., Korf I. (2007). CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23 1061–1067. 10.1093/bioinformatics/btm071 [DOI] [PubMed] [Google Scholar]
- Paumi C. M., Menendez J., Arnoldo A., Engels K., Iyer K. R., Thaminy S., et al. (2007). Mapping protein-protein interactions for the yeast ABC transporter Ycf1p by integrated split-ubiquitin membrane yeast two-hybrid analysis. Mol. Cell 26 15–25. 10.1016/j.molcel.2007.03.011 [DOI] [PubMed] [Google Scholar]
- Prasad R., Banerjee A., Khandelwa N. K., Dhamgaye S. (2015). The ABCs of Candida albicans multidrug transporter Cdr1. Eukaryot. Cell 14 1154–1164. 10.1128/EC.00137-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R., De Wergifosse P., Goffeau A., Balzi E. (1995). Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr. Genet. 27 320–329. 10.1007/BF00352101 [DOI] [PubMed] [Google Scholar]
- Prasad R., Goffeau A. (2012). Yeast ATP-Binding cassette transporters conferring multidrug resistance. Annu. Rev. Microbiol. 66 39–63. 10.1146/annurev-micro-092611-150111 [DOI] [PubMed] [Google Scholar]
- Prasad R., Khandelwal N. K., Banerjee A. (2016). Yeast ABC transporters in lipid trafficking. Fungal Genet. Biol. 93 25–34. 10.1016/j.fgb.2016.05.008 [DOI] [PubMed] [Google Scholar]
- Raymond M., Dignard D., Alarco A. M., Mainville N., Magee B. B., Thomas D. Y. (1998). A Ste6p/P-glycoprotein homologue from the asexual yeast Candida albicans transports the a-factor mating pheromone in Saccharomyces cerevisiae. Mol. Microbiol. 27 587–598. 10.1046/j.1365-2958.1998.00704.x [DOI] [PubMed] [Google Scholar]
- Rhodes J., Abdolrasouli A., Farrer R. A., Cuomo C. A., Aanensen D. M., Armstrong-James D., et al. (2018). Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg. Microbes Infect. 7:43. 10.1038/s41426-018-0045-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell N. C., Wolfger H., Kuchler K., Thorner J. (2009). ABC transporter pdr10 regulates the membrane microenvironment of pdr12 in Saccharomyces cerevisiae. J. Membr. Biol. 229 27–52. 10.1007/s00232-009-9173-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers B., Decottignies A., Kolaczkowski M., Carvajal E., Balzi E., Goffeau A. (2001). The pleitropic drug ABC transporters from Saccharomyces cerevisiae. J. Mol. Microbiol. Biotechnol. 3 207–214. [PubMed] [Google Scholar]
- Rybak J. M., Doorley L. A., Nishimoto A. T., Barker K. S., Palmer G. E., Rogers P. D. (2019). Abrogation of triazole resistance upon deletion of CDR1 in a clinical isolate of Candida auris. Antimicrob. Agents Chemother. 63:e00057-19. 10.1128/AAC.00057-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlin K., Vezzi F., Nystedt B., Lundeberg J., Arvestad L. (2014). BESST - efficient scaffolding of large fragmented assemblies. BMC Bioinformatics 15:281. 10.1186/1471-2105-15-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D., Ischer F., Calabrese D., Majcherczyk P. A., Bille J. (1999). The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob. Agents Chemother. 43 2753–2765. 10.1128/aac.43.11.2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D., Ischer F., Monod M., Bille J. (1997). Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 143 405–416. 10.1099/00221287-143-2-405 [DOI] [PubMed] [Google Scholar]
- Sasser T. L., Fratti R. A. (2014). Class C ABC transporters and Saccharomyces cerevisiae vacuole fusion. Cell. Logist. 4:e943588. 10.4161/21592780.2014.943588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh K., Makimura K., Hasumi Y., Nishiyama Y., Uchida K., Yamaguchi H. (2009). Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 53 41–44. 10.1111/j.1348-0421.2008.00083.x [DOI] [PubMed] [Google Scholar]
- Schelenz S., Hagen F., Rhodes J. L., Abdolrasouli A., Chowdhary A., Hall A., et al. (2016). First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob. Resist. Infect. Control 5:35. 10.1186/s13756-016-0132-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar-Guturja T., Tebung W. A., Mount H., Liu N., Kohler J. R., Whiteway M., et al. (2016). Beauvericin potentiates azole activity via inhibition of multidrug efflux, blocks Candida albicans morphogenesis, and is effluxed via Yor1 and circuitry controlled by Zcf29. Antimicrob. Agents Chemother. 60 7468–7480. 10.1128/AAC.01959-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson J. T., Wong K., Jackman S. D., Schein J. E., Jones S. J. M., Birol I. (2009). ABySS: a parallel assembler for short read sequence data. Genome Res. 19 1117–1123. 10.1101/gr.089532.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smriti X., Krishnamurthy S., Dixit B. L., Gupta C. M., Milewski S., Prasad R. (2002). ABC transporters Cdr1p, Cdr2p and Cdr3p of a human pathogen Candida albicans are general phospholipid translocators. Yeast 19 303–318. 10.1002/yea.818 [DOI] [PubMed] [Google Scholar]
- Snider J., Hanif A., Lee M. E., Jin K., Yu A. R., Graham C., et al. (2013). Mapping the functional yeast ABC transporter interactome. Nat. Chem. Biol. 9 565–574. 10.1038/nchembio.1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorano A. M., Kibbler C., Peman J., Bernhardt H., Klingspor L., Grillot R. (2006). Candidaemia in Europe: epidemiology and resistance. Int. J. Antimicrob. Agents 27 359–366. 10.1016/j.ijantimicag.2006.01.002 [DOI] [PubMed] [Google Scholar]
- van Roermund C. W. T., Ijlst L., Majczak W., Waterham H. R., Folkerts H., Wanders R. J. A., et al. (2012). Peroxisomal fatty acid uptake mechanism in Saccharomyces cerevisiae. J. Biol. Chem. 287 20144–20153. 10.1074/jbc.M111.332833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiliou V., Vasiliou K., Nebert D. W. (2009). Human ATP-binding cassette (ABC) transporter family. Hum. Genomics 3 281–290. 10.1186/1479-7364-3-3-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasi M., Khandelwal N. K., Vishwakarma P., Lynn A. M., Mondal A. K., Prasad R. (2018). Inventory of ABC proteins and their putative role in salt and drug tolerance in Debaryomyces hansenii. Gene 676 227–242. 10.1016/j.gene.2018.07.029 [DOI] [PubMed] [Google Scholar]
- Whaley S. G., Zhang Q., Caudle K. E., Rogers P. D. (2018). Relative Contribution of the ABC transporters Cdr1, Pdh1, and Snq2 to azole resistance in Candida glabrata. Antimicrob. Agents Chemother. 62:e01070-18. 10.1128/AAC.01070-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox L. J., Balderes D. A., Wharton B., Tinkelenberg A. H., Rao G., Sturley S. L. (2002). Transcriptional profiling identifies two members of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast. J. Biol. Chem. 277 32466–32472. 10.1074/jbc.M204707200 [DOI] [PubMed] [Google Scholar]
- Wisplinghoff H., Bischoff T., Tallent S. M., Seifert H., Wenzel R. P., Edmond M. B. (2004). Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39 309–317. 10.1086/421946 [DOI] [PubMed] [Google Scholar]
- Young L., Leonhard K., Tatsuta T., Trowsdale J., Langer T. (2001). Role of the ABC transporter Mdl1 in peptide export from mitochondria. Science 291 2135–2138. 10.1126/science.1056957 [DOI] [PubMed] [Google Scholar]
- Yuan W., Guo S., Gao J., Zhong M., Yan G., Wu W., et al. (2017). General Control nonderepressible 2 (GCN2) kinase inhibits target of rapamycin complex 1 in response to amino acid starvation in Saccharomyces cerevisiae. J. Biol. Chem. 292 2660–2669. 10.1074/jbc.M116.772194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino D. R. (2010). Using the velvet de novo assembler for short-read sequencing technologies. Curr. Protoc. Bioinformatics 31 11.5.1–11.5.12. 10.1002/0471250953.bi1105s31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. (2002). Response of gene expression in Saccharomyces cerevisiae to amphotericin B and nystatin measured by microarrays. J. Antimicrob. Chemother. 49 905–915. 10.1093/jac/dkf001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed for this study can be found at https://www.ebi.ac.uk/ena/data/view/PRJEB29190 (ENA Accession No. PRJEB29190).