Abstract
A study of multiple tissues was conducted to identify potential metabolic differences in cattle differing in feed efficiency. Individual feed intake and body weight was measured on 144 steers during 105 days on a high-concentrate ration. Steers were selected according to differences in average daily gain (ADG) with those with the greatest ADG (n = 8; 1.96 ± 0.02 kg/day) and least ADG (n = 8; 1.57 ± 0.02 kg/day), whose dry matter intake was within 0.32 SD of the mean intake (10.10 ± 0.05 kg/day). Duodenum, liver, adipose, and longissimus-dorsi were collected at slaughter, and metabolomics profiles were performed by ultra performance liquid chromatography quadrupole-time of-flight mass spectrometry. Principal components analyses, t-tests, and fold changes in tissues profile were used to identify differential metabolites between ADG groups. These were primarily involved in α-linolenic metabolism, which was downregulated in the greatest ADG as compared to least-ADG group in duodenum, adipose, and longissimus-dorsi. However, taurine and glycerophospholipids metabolisms were both upregulated in the greatest ADG compared with least-ADG group in the liver. The phospholipids and cholesterol were quantified in the tissues. Lipid transport and oxidation were the main common metabolic mechanisms associated with cattle feed efficiency. Combining analyses of multiple tissues may offer a powerful approach for defining the molecular basis of differences in performance among cattle for key production attributes.
Introduction
Increased sustainability of livestock production is needed by the growing human population.1 Conversion of feed into meat is associated, among others, with emission of greenhouse gases and excretion of nitrogen and phosphorus.2 Thus, improving production efficiency by increasing beef produced per amount of feed offered would reduce both operating cost and waste from beef cattle production.
Through more than 50 years of research focused on nutritional management and genetic selection for beef cattle production, feed efficiency conversion rate has been improved on average from 10.2:13,4 to 5.4:1.5,6 Some of the traditional strategies are the use of high-concentrate diets and animal selection by moderately heritable variables such as intake and gain.7,8 However, large phenotypic differences regarding many complex traits of general biological interest in cattle such as growth, fat deposition, and energy partitioning are not completely understood.9 Application of new technologies, including “omics” selections that incorporate molecular breeding values, plays an important role in meeting these challenges.10 The use of genome-wide SNP markers is a promising predictor of the genetic merit of animals, despite genetics limiting to explain the differences of the phenotype.10 Thus, the final “omic” level in a biological system, metabolomics may provide the most “functional” information of the omics technologies, allowing the measure of the end products of the complex genetic epigenetic and environmental interactions.11 This analytical technique is increasingly used in livestock research for providing a distinctive insight into the biochemical activity and robustly correlate with animal production traits.11
For the first time, we previously characterized metabolite changes in the rumen using the distinct ruminal profiling method, identifying 33 biomarkers related to differences in average daily gain (ADG).12 Linoleic/α-linolenic acids (ALA) metabolism and biosynthesis of aromatic amino acids were the most altered functional pathways in the rumen associated with production efficiency of beef steers. In addition, fatty acids were quantified and evaluated as potential biomarkers to predict ADG in cattle.
An identification of a distinctive physiological process across multiple tissues could therefore usefully contribute for understanding the mechanism associated with cattle feed efficiency. We hypothesize that differences in ADG of cattle, with similar intake, are associated with common biochemical pathways at multiple levels of the tissues metabolism. A combination of metabolomics fingerprint and targeted lipidomics analysis was used to identify metabolites from animals that differed with feed efficiency in duodenum, liver, subcutaneous adipose, and longissimus-dorsi.
Results
Tissues Metabolomics Profiling
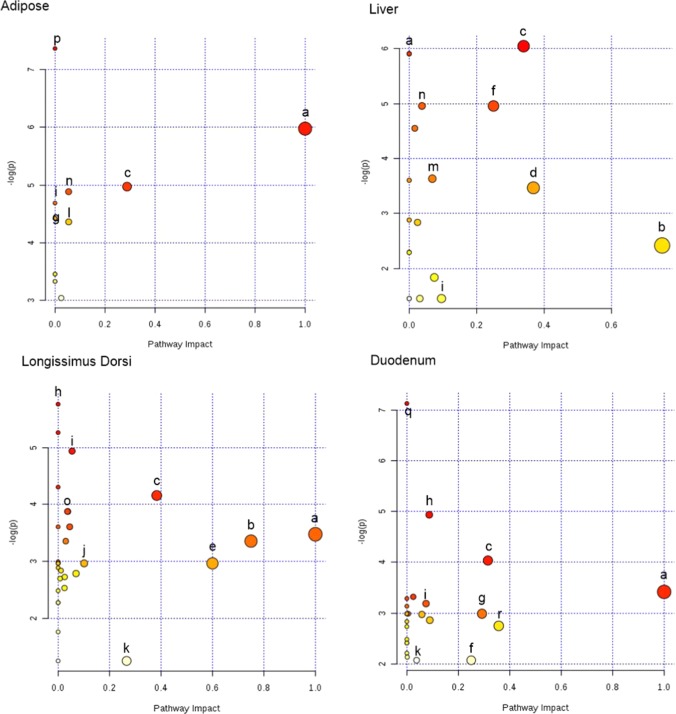
Differences in metabolites were identified between least-ADG and greatest-ADG animals (p < 0.05; see Supporting Information Table S1) in duodenum (n = 87), liver (n = 92); longissimus-dorsi (LD, n = 95), and adipose (n = 52) tissue. The analysis of these metabolites revealed specific metabolic pathways to ADG groups in the duodenum, liver, adipose, and LD (Figures 1 and 2; see Supporting Information Table S2). The α-linolenic acid (ALA) metabolism was the most relevant metabolic pathway downregulated in adipose, duodenum, and LD tissues (impact value = 1 and p < 0.05), whereas taurine and glutathione biosynthesis were upregulated in hepatic tissue (impact value = 0.5 and p < 0.08) in greatest ADG compared to least-ADG animals.
Figure 1.
Tissues metabolomics pathway analysis by MetaboAnalyst 3.0 Software on the steers with the greatest average daily gain compare to the least ADG with similar average dry matter intake according to Bos taurus KEGG pathway database. Ultra performance liquid chromatography quadrupole-time of-flight mass spectrometry (UPLC-q-Tof MS) metabolites identified to differ between ADG by t-test (p < 0.05). (a) α-Linolenic acid metabolism. (b) Taurine and hypotaurine metabolism. (c) Glycerophospholipids (GLP). (d) Glutathione metabolism. (e) Synthesis and degradation of ketone bodies. (f) Primary bile biosynthesis. (g) Glycine, serine, and threonine metabolism. (h) Histidine metabolism. (i) Gluconeogenesis/glucogenolisis metabolisms. (j) Purine metabolism. (k) Steroids metabolism. (l) Pyruvate metabolism. (m) Cholic acid biosynthesis. (n) Cysteine and methione metabolism. (o) Creatinine metabolism. (p) Unsaturated fatty acids. (q) Ether lipids metabolism. (r) Pentose and glucoronate interconversion. The darker the color and larger the size represent higher p-value from enrichment analysis and greater impact from the pathway topology analysis, respectively.
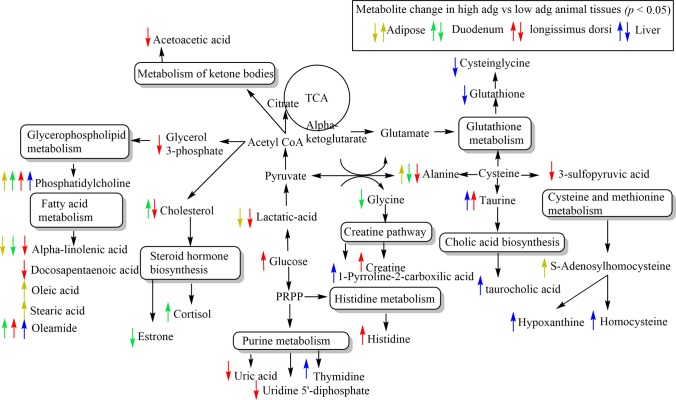
Figure 2.
Integrated pathways metabolomics analysis in adipose, duodenum, longissimus-dorsi, and liver compare high-ADG vs low-ADG beef cattle with similar average dry matter intake according to Bos taurus KEGG pathway database. UPLC-q-Tof MS metabolites identified to differ between ADG groups by t-test (p < 0.05).
No common identified metabolites were found in the four tissues evaluated (Figure 3). However, alanine and ALA were downregulated in duodenum, LD, and adipose tissues, whereas oleamide was upregulated in duodenum, LD, and liver tissues, in greatest ADG compared to least-ADG animals (Figure 3). In addition, phosphatidylcholine (PC) was upregulated in high-ADG animals in the four tissues but differed within fatty acids (Supporting Information Table S1).
Figure 3.
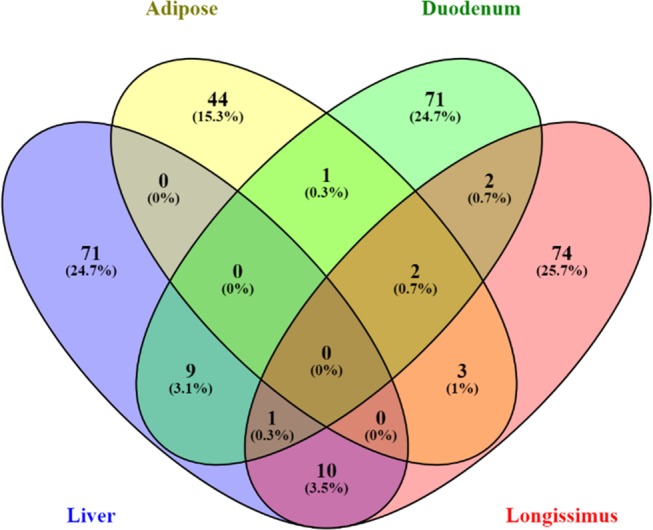
Venn diagram showing the overlap across tissues of the metabolites identified that differ between ADG groups by t-test (p < 0.05).
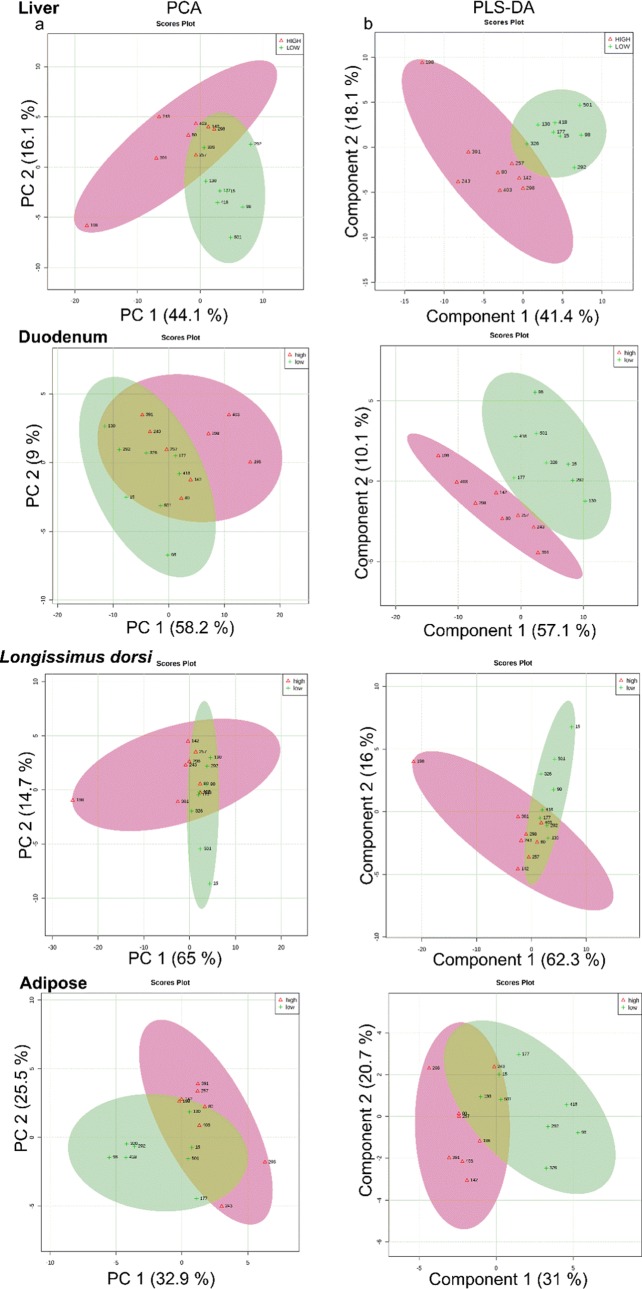
Principal component analysis and score plot from supervised partial least square of discriminant analysis (PLS-DA) were conducted on the metabolites identified in the tissues. The analysis revealed a clear clustering between least-ADG and greatest-ADG animals (Figure 4), which suggest that biochemistry changed in the tissues according to cattle feed efficiency. The p-value for 1000 permutations was p < 0.05 in the tissues, indicating that the PLS-DA model was valid. For the first component of PLS-DA, descriptive statistics from model fitting by accuracy, estimates of the goodness of fit (R2), and estimates of goodness prediction (Q2) by tissue were: accuracy = 0.81, R2 = 0.52, and Q2 = 0.40 for LD; accuracy = 0.75, R2 = 0.64, and Q2 = 0.42 for adipose; accuracy = 0.94, R2 = 0.94, and Q2 = 0.62 for duodenum; accuracy = 0.75, R2 = 0.77, Q2 = 0.39 for liver.
Figure 4.
Tissues metabolomic profile of the steers with the greatest average daily gain (high-ADG; open red triangle) and the least ADG (low-ADG; green plus) with similar average dry matter intake. (A) Principal component analysis for UPLC-q-Tof MS metabolites identified to differ between ADG by t-test (p < 0.05). (B) Partial least square-discriminant analysis for UPLC-q-Tof MS metabolites identified to differ between ADG by t-test (p < 0.1). One data point represents one steer.
Phospholipids Quantification in Tissues
On the basis of the findings of the nontargeted metabolomic analysis, phospholipids were quantified in liver (Table 1), duodenum (Table 2), adipose (Table 3), and LD (Table 4). The performance of phospholipids as biomarkers was assessed using receiver–operator characteristic (ROC) curves. According to the accepted classification of biomarker utility, the area under the curve (AUC) was the metric used to assess the metabolites as candidate markers.17
Table 1. Phospholipid and Cholesterol Concentrations (mg/g) in Longissimus-Dorsi on Steers with the Low and High Average Daily Gain.
| groupa |
||||||
|---|---|---|---|---|---|---|
| phospholipids and cholesterol | high-ADG | low-ADG | SEM | AUCb | p | FDRc |
| phosphatidylcholine | ||||||
| 16:0/16:0 | 8.30 | 7.26 | 0.87 | 0.61 | 0.42 | 0.47 |
| 16:0/18:1 | 7.09 | 5.82 | 0.89 | 0.66 | 0.33 | 0.47 |
| 16:/20:3 | 8.99 | 7.31 | 1.01 | 0.67 | 0.26 | 0.43 |
| 16:0/20:4 | 13.1 | 10.2 | 1.57 | 0.63 | 0.21 | 0.38 |
| 18:0/18:1 | 2.01 | 1.72 | 0.26 | 0.61 | 0.43 | 0.47 |
| 18:0/18:2, 18:1/18:1 | 2.42 | 1.85 | 0.29 | 0.69 | 0.19 | 0.38 |
| 18:0/20:3 | 1.12 | 0.55 | 0.16 | 0.86 | <0.01 | 0.03 |
| 18:0/20:4 | 2.93 | 1.77 | 0.37 | 0.80 | 0.04 | 0.25 |
| 18:0/22:5 | 1.16 | 0.52 | 0.21 | 0.81 | <0.01 | 0.05 |
| 18:0/22:6, 18:1/22:5 | 2.53 | 1.72 | 0.31 | 0.75 | 0.09 | 0.26 |
| 18:1/20:4, 18:0/20:5, 16:0/22:5 | 3.63 | 2.67 | 0.46 | 0.70 | 0.16 | 0.37 |
| phosphatidylethanolamine | ||||||
| 16:0/18:2 | 0.28 | 0.23 | 0.06 | 0.55 | 0.58 | 0.58 |
| 18:0/18:0 | 0.76 | 0.52 | 0.12 | 0.66 | 0.16 | 0.37 |
| 18:0/22:5 | 0.10 | 0.09 | 0.02 | 0.53 | 0.89 | 0.89 |
| 18:0/22:6 | 0.28 | 0.22 | 0.05 | 0.56 | 0.45 | 0.47 |
| lysophosphatidylcholine | ||||||
| 18:0 | 0.25 | 0.37 | 0.09 | 0.63 | 0.37 | 0.47 |
| 18:1 | 0.19 | 0.20 | 0.06 | 0.52 | 0.94 | 0.94 |
| 18:2 | 0.07 | 0.06 | 0.14 | 0.53 | 0.62 | 0.62 |
| lysophosphatidylethanolamine | ||||||
| 16:0 | 0.04 | 0.05 | 0.01 | 0.61 | 0.43 | 0.47 |
| 18:0 | 0.59 | 0.755 | 0.06 | 0.75 | 0.08 | 0.18 |
| cholesterol | 1.25 | 1.71 | 0.08 | 0.78 | 0.07 | 0.18 |
Groups were least-ADG steers (n = 8), with the least average daily gain, and greatest-ADG steers (n = 8), with the greatest average daily gain, with groups having similar dry matter intake.
AUC = area under the curve calculated by receiver–operator characteristic curve analysis.
FDR = false discovery rate p-adjustment.
Table 2. Phospholipid and Cholesterol Concentrations in Liver (mg/g) on Steers with the Low and High Average Daily Gain.
| groupa |
||||||
|---|---|---|---|---|---|---|
| phospholipids and cholesterol | low-ADG | high-ADG | SEM | AUCb | p | FDRc |
| phosphatidylcholine | ||||||
| 16:0/16:0 | 31.9 | 31.5 | 2.62 | 0.59 | 0.69 | 0.68 |
| 16:0/18:1 | 6.19 | 5.01 | 0.13 | 0.67 | 0.14 | 0.13 |
| 16:/20:3 | 16.1 | 12.1 | 1.27 | 0.79 | 0.05 | 0.07 |
| 16:0/20:4 | 16.9 | 14.1 | 1.25 | 0.64 | 0.13 | 0.13 |
| 18:0/18:1 | 17.9 | 14.1 | 1.46 | 0.81 | 0.06 | 0.09 |
| 18:0/18:2, 18:1/18:1 | 5.24 | 4.08 | 0.25 | 0.78 | 0.09 | 0.09 |
| 18:0/20:3 | 7.53 | 6.07 | 0.29 | 0.64 | 0.12 | 0.12 |
| 18:0/20:4 | 10.6 | 8.63 | 0.88 | 0.62 | 0.12 | 0.12 |
| 18:0/22:5 | 15.4 | 12.6 | 1.21 | 0.64 | 0.13 | 0.13 |
| 18:0/22:6, 18:1/22:5 | 12.0 | 9.71 | 0.99 | 0.64 | 0.12 | 0.12 |
| 18:1/20:4, 18:0/20:5, 16:0/22:5 | 13.9 | 11.8 | 0.95 | 0.61 | 0.13 | 0.13 |
| phosphatidylethanolamine | ||||||
| 16:0/18:2 | 0.20 | 0.21 | 0.02 | 0.53 | 0.92 | 0.92 |
| 18:0/18:0 | 0.18 | 0.15 | 0.02 | 0.72 | 0.23 | 0.24 |
| 18:0/22:5 | 0.16 | 0.18 | 0.01 | 0.61 | 0.27 | 0.27 |
| 18:0/22:6 | 0.12 | 0.12 | 0.01 | 0.61 | 0.27 | 0.27 |
| lysophosphatidylcholine | ||||||
| 18:0 | 0.74 | 1.26 | 0.20 | 0.89 | 0.07 | 0.07 |
| 18:1 | 1.55 | 2.47 | 0.17 | 0.98 | <0.01 | 0.01 |
| 18:2 | 0.65 | 0.75 | 0.14 | 0.63 | 0.32 | 0.32 |
| lysophosphatidylethanolamine | ||||||
| 16:0 | 1.54 | 2.14 | 0.29 | 0.70 | 0.16 | 0.16 |
| 18:0 | 0.71 | 1.02 | 0.11 | 0.78 | 0.07 | 0.07 |
| cholesterol | 3.40 | 5.06 | 0.47 | 0.99 | <0.01 | 0.02 |
Groups were least-ADG steers (n = 8), with the least average daily gain, and greatest-ADG steers (n = 8), with the greatest average daily gain, with groups having similar dry matter intake.
AUC = area under the curve calculated by receiver–operator characteristic curve analysis.
FDR = false discovery rate p-adjustment.
Table 3. Phospholipid and Cholesterol Concentrations in Adipose Tissue (mg/g) on Steers with the Low and High Average Daily Gain.
| groupa |
||||||
|---|---|---|---|---|---|---|
| phospholipids and cholesterol | high-ADG | low-ADG | SEM | AUCb | p | FDRc |
| phosphatidylcholine | ||||||
| 16:0/16:0 | 14.1 | 9.6 | 2.08 | 0.70 | 0.14 | 0.59 |
| 16:0/18:1 | 3.77 | 3.2 | 0.64 | 0.62 | 0.55 | 0.67 |
| 16:/20:3 | 6.36 | 5.3 | 1.04 | 0.52 | 0.80 | 0.85 |
| 16:0/20:4 | 8.21 | 6.7 | 1.13 | 0.58 | 0.35 | 0.59 |
| 18:0/18:1 | 1.28 | 1.6 | 0.26 | 0.67 | 0.44 | 0.59 |
| 18:0/18:2, 18:1/18:1 | 1.81 | 1.6 | 0.31 | 0.67 | 0.40 | 0.59 |
| 18:0/20:3 | 2.05 | 1.7 | 0.30 | 0.63 | 0.35 | 0.59 |
| 18:0/20:4 | 3.69 | 2.9 | 0.54 | 0.55 | 0.31 | 0.59 |
| 18:0/22:5 | 1.35 | 0.9 | 0.25 | 0.62 | 0.20 | 0.59 |
| 18:0/22:6, 18:1/22:5 | 2.84 | 2.5 | 0.43 | 0.68 | 0.21 | 0.59 |
| 18:1/20:4, 18:0/20:5, 16:0/22:5 | 3.65 | 2.9 | 0.50 | 0.63 | 0.30 | 0.59 |
| phosphatidylethanolamine | ||||||
| 16:0/18:2 | 0.39 | 0.21 | 0.06 | 0.77 | 0.06 | 0.59 |
| 18:0/18:0 | 0.15 | 0.09 | 0.02 | 0.71 | 0.14 | 0.59 |
| 18:0/22:5 | 0.47 | 0.29 | 0.05 | 0.69 | 0.19 | 0.59 |
| 18:0/22:6 | 0.71 | 0.52 | 0.12 | 0.63 | 0.30 | 0.59 |
| lysophosphatidylcholine | ||||||
| 18:0 | 0.03 | 0.04 | 0.01 | 0.61 | 0.45 | 0.59 |
| 18:1 | 0.06 | 0.07 | 0.02 | 0.52 | 0.89 | 0.89 |
| 18:2 | 0.38 | 0.32 | 0.05 | 0.53 | 0.81 | 0.85 |
| lysophosphatidylethanolamine | ||||||
| 16:0 | 0.03 | 0.02 | 0.01 | 0.55 | 0.45 | 0.59 |
| 18:0 | 0.76 | 0.70 | 0.15 | 0.60 | 0.28 | 0.59 |
| cholesterol | 3.29 | 3.23 | 0.22 | 0.53 | 0.78 | 0.59 |
Groups were least-ADG steers (n = 8), with the least average daily gain, and greatest-ADG steers (n = 8), with the greatest average daily gain, with groups having similar dry matter intake.
AUC = area under the curve calculated by receiver–operator characteristic curve analysis.
FDR = false discovery rate p-adjustment.
Table 4. Phospholipid and Cholesterol Concentrations in Duodenum (mg/g) on Steers with the Low and High Average Daily Gain.
| groupa |
||||||
|---|---|---|---|---|---|---|
| phospholipids and cholesterol | high-ADG | low-ADG | SEM | AUCb | p | FDRc |
| phosphatidylcholine | ||||||
| 16:0/16:0 | 10.3 | 8.84 | 0.65 | 0.72 | 0.13 | 0.24 |
| 16:0/18:1 | 1.04 | 0.87 | 0.07 | 0.75 | 0.10 | 0.21 |
| 16:/20:3 | 2.03 | 1.64 | 0.13 | 0.73 | 0.06 | 0.21 |
| 16:0/20:4 | 2.57 | 2.08 | 0.16 | 0.77 | 0.06 | 0.21 |
| 18:0/18:1 | 0.42 | 0.34 | 0.03 | 0.75 | 0.10 | 0.21 |
| 18:0/18:2, 18:1/18:1 | 0.63 | 0.50 | 0.05 | 0.77 | 0.07 | 0.21 |
| 18:0/20:3 | 0.40 | 0.34 | 0.03 | 0.67 | 0.21 | 0.26 |
| 18:0/20:4 | 0.86 | 0.74 | 0.06 | 0.70 | 0.17 | 0.25 |
| 18:0/22:5 | 1.85 | 1.73 | 0.28 | 0.66 | 0.18 | 0.25 |
| 18:0/22:6, 18:1/22:5 | 1.81 | 1.39 | 0.17 | 0.75 | 0.10 | 0.21 |
| 18:1/20:4, 18:0/20:5, 16:0/22:5 | 1.14 | 0.96 | 0.08 | 0.73 | 0.16 | 0.25 |
| phosphatidylethanolamine | ||||||
| 16:0/18:2 | 0.42 | 0.39 | 0.06 | 0.56 | 0.76 | 0.76 |
| 18:0/18:0 | 0.20 | 0.22 | 0.02 | 0.60 | 0.57 | 0.59 |
| 18:0/22:5 | 0.59 | 0.47 | 0.02 | 0.52 | 0.75 | 0.75 |
| 18:0/22:6 | 0.05 | 0.06 | 0.01 | 0.53 | 0.59 | 0.59 |
| lysophosphatidylcholine | ||||||
| 18:0 | 0.92 | 0.81 | 0.08 | 0.67 | 0.38 | 0.43 |
| 18:1 | 3.70 | 3.41 | 0.53 | 0.57 | 0.70 | 0.70 |
| 18:2 | 21.1 | 21.8 | 4.94 | 0.56 | 0.91 | 0.91 |
| lysophosphatidylethanolamine | ||||||
| 16:0 | 3.73 | 4.27 | 1.06 | 0.52 | 0.72 | 0.72 |
| 18:0 | 1.23 | 1.07 | 0.09 | 0.73 | 0.84 | 0.84 |
| cholesterol | 1.46 | 1.20 | 0.09 | 0.62 | 0.10 | 0.21 |
Groups were least-ADG steers (n = 8), with the least average daily gain, and greatest-ADG steers (n = 8), with the greatest average daily gain, with groups having similar dry matter intake.
AUC = area under the curve calculated by receiver–operator characteristic curve analysis.
FDR = false discovery rate p-adjustment.
The concentrations of compounds with the greatest sensitivity/specificity for ADG differences were cholesterol and LPC-18:1 in liver (AUC = 0.99, AUC = 0.98; FDR < 0.05; respectively) followed by PC-18:0/20:3 in LD (AUC = 0.86; FDR = 0.03). Steers with the greatest ADG had lower concentration of cholesterol and LPC-18:1 in liver and higher concentration of PC-18/20:3 in LD than least-ADG steers.
No significant differences (p > 0.05) in phospholipids concentration were found in the duodenum and adipose tissues across ADG classification. However, liver cholesterol concentration was associated with cholesterol concentration in LD (r = 0.68; p < 0.01) and in duodenum (r = −0.67; p < 0.01), whereas liver cholesterol was not associated with cholesterol concentration in adipose tissue (r = 0.03; p = 0.40).
Discussion
This study represents the first comprehensive metabolome evaluation based on phenotypic differences in cattle efficiency for four tissues, duodenum, liver, adipose, and LD, combining metabolomics and targeted lipidomics approaches. Previously,12 rumen fluid was evaluated by UPLC-q-Tof MS metabolomics analysis associated with univariate and multivariate statistical methods to identify metabolites that differentiate steers with greatest ADG from least-ADG. On the basis of this previous approach, metabolomics analyses in multiple tissues on the same animals were conducted to provide evidence on physiological mechanisms involved in differences in weight-to-gain ratios in cattle with similar DMI. Furthermore, targeted lipidomics analysis was conducted to support the metabolomics results and provided valuable potential biomarkers for feed efficiency.
In the current study, the compounds identified according to differences in ADG, reflected the complexity and diversity of the metabolism in different tissues. For instance, the adipose tissues presented 51 compounds mainly associated with fatty acid metabolism, whereas in the liver 92 compounds were associated mainly with taurine and glutathione metabolism. Indeed, no common identified metabolites were found across the four tissues evaluated. However, ALA metabolism was downregulated in adipose, duodenum, and LD tissues in animals with greatest ADG. Lower level of ALA in more efficient animals might reflect more extensive ruminal biohydrogenation as reported before in ruminal fluid and plasma in the same animals.12 The ALA entering tissues is rapidly accumulated, although a certain percentage of ALA is subjected to desaturation or β-oxidation depending on the tissues needs. For instance, longer n-3 fatty acids, such EPA or DHA or others energetic carbons products, including ATP, monosaturated fatty acids, and phospholipids.18 In vivo, the two basic ALA metabolic fates are associated with the expressions of lipogenic enzymes, substrate level, and product inhibition.19 Therefore, considering that in this study the intake is similar between the two ADG groups, ALA levels in tissues probably reflected a predominant catabolic fate producing more energy in more efficient animals.
On the basis of fingerprint analysis, glycerophospholipid (GLP) compounds were the main lipid category, and their metabolism was displayed in the four tissues evaluated. According to differences on GLP head group, PC was the main phospholipid class in LD, liver, and adipose tissue, whereas the GLP acyl chain length was widely distributed across tissues. The specific functions of these GLP class/species are not full elucidated. However, the fatty acid pattern of GLP membrane together with the class distribution of phospholipids are major lipid-related factors that characterize the properties and functions of the membranes, including the regulation of transport processes, energy homeostasis, and signal transduction.20 Thus, the GLP profile is associated with diverse metabolic states. For instance, the PC acyl chain length was associated with the concentrations of glucose, insulin, leptin, and hepatic cholesterol in humans, and mice fed a high-fat diet..21−23 Furthermore, in our results, a fatty acid amide synthesized from phospholipids membrane, Oleamide, was upregulated in duodenum, liver, LD, and previously, in rumen fluid.12 The fatty acid amines belong to a group of endogenous lipid mediators, endocannabinoids, implicated on feed efficiency and carcass composition of beef cattle.25 Considering GLP is not only an energy-storage compound but also an interactive player in various metabolic processes, we sought to quantify main phospholipids species and cholesterol associated with ADG in cattle.
Phosphatidylcholine is required for many energetic mechanisms such as the efflux of triglycerides and cholesterol by VLDL and HDL, respectively. In the hepatocytes, it also regulates fatty acid synthesis and cholesterol oxidation.24 Indeed, the supplementation of rumen-protected choline (as a precursor of phospholipids) reduced triacylglycerol accumulation in the liver and increased milk yield during the transition period in dairy ruminants.26,27 However, phosphatidylcholine has not been extensively studied in beef cattle. Certainly, dietary supplementation with lecithin improves the feed conversion rate in pigs on a high-fat diet during finishing period,28 and rumen-protected choline supplementation has been shown to promote weight gain and improve meat quality in young lambs.29 In addition, differential gene expression and transcripts already highlighted the importance of phospholipids metabolism in cattle feed efficiency.30,31 Despite dietary phospholipids are extensively degraded by the rumen32 and PC concentration depends almost exclusively upon endogenous synthesis,33 understanding of the requirements of PC is needed to improve feed efficiency in beef cattle.
The concentration of cholesterol is considered a robust biomarker of feed efficiency in cattle34,35 as consistent with our results, lower cholesterol in liver and blood is associated with more efficient beef cattle. Cholesterol concentrations in fact reflect the capacity of synthesis, transport, and oxidation by the liver.36 Particularly in ruminants, cholesterol is derived from hepatic de-novo biosynthesis,37 and its major catabolism represents the bile acid synthesis.38 In our results, cholesterol oxidation products, such as glycine or taurine conjugated, were in turn higher in the liver of animals with greater ADG. Therefore, the excretion and reabsorption of bile acid forms represent the basis of the enterohepatic circulation, an essential mechanism for the digestion and absorption of fatty acids, which likely relates to cattle feed efficiency.
By contrast, the less efficient animal presented higher hepatic concentration of cholesterol with an increase of glutathione metabolism. The liver is the primary organ for the synthesis of glutathione from glutamate, glycine, and cysteine and it is a major antioxidant, which regulates the homeostasis of free radicals.39 Perturbations of glutathione status are usually a consequence of oxidative stress and redox shift that impair liver function.40 Thus, the accumulation of cholesterol in the liver may result in part from a decrease in cholesterol oxidation and transport probably linked with hepatic performance.
In our results, the concentration of cholesterol was positively associated in liver and muscle. Despite the lack of information on body composition or carcass traits in this study, improving cattle feed efficiency potentially has a beneficial effect on carcass composition, particularly for the human health aspect. Furthermore, metabolomics results in LD from animals that are more efficient and presented a higher level of creatinine and glucose and lower levels of lactic acid and acetoacetic acid, which might reflect differences in muscle metabolism. The physiological role of creatine in the skeletal muscle is as an “energy shuttle” transferring ATP energy from mitochondria to the myofibrils, whereas glucose contributes to an intramuscular great proportion of acetyl units for fatty acid biosynthesis.41,42 In this experimental model, where diet and intake have been fixed, group differences according to feed efficiency classification are most likely involved with differences in cattle physiology and metabolism. Hence, elucidating the metabolic mechanisms of muscle growth and development associated with feed efficiency and more importantly developing the ability to advantageously manipulate gain-to-intake ratios would allow significant contributions to meat composition.
Conclusions
Overall, lipid transport and oxidation were the main common metabolic mechanism involved in weight-gain differences at multiple metabolic levels of complexity in beef cattle notwithstanding the uniqueness of the molecular composition of each tissue. Both mechanisms were associated with the levels of ALA, phosphatidylcholine, and cholesterol that could be considered as useful biomarkers (in other sample formats e.g., blood, plasma) for feed efficiency in beef cattle. The integration of the metabolomics profiles of tissues and metabolites points to the hepatic function as an important metabolic crossroad that has the potential to improve the gain-to-feed ratios.
Combining analyses of multiple tissues may offer a powerful approach for defining the molecular basis of differences in performance among cattle for key production attributes
Experimental Procedures
Study Design and Samples Details
This study was approved by the Institutional Animal Care and Use Committee (IACUC) at the U.S. Meat Animal Research Center.
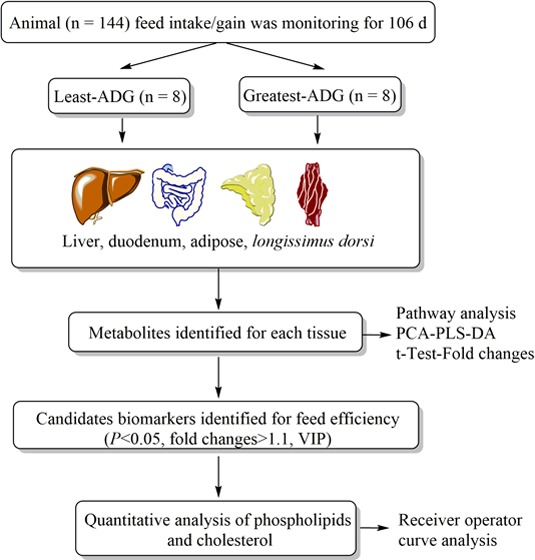
After weaning, steers (n = 144 Angus-sired) were housed in a facility with Calan Broadbent electronic headgates (American Calan, Inc., Northwood, NH) to measure individual feed intake. The diet consists on a DM basis, of 8% chopped alfalfa hay, 20% wet distillers grains with solubles, 67.75% dry-rolled corn, and 4.25% commercial vitamin and mineral supplement; the supplement contained monensin (Rumensin 80; Elanco Animal Health, Greenfield, IN) to meet the nutrient requirements (NRC, 2016) and provide a finished marketed carcass. Further details about the experimental design were provided previously.12 Briefly, feed intake was measured for 105 days in steers, and body weight was measured on days 0, 1, 21, 42, 63, 84, 104, and 105 of the experiment.
After the end of intake study, steers received the same ration ad libitum and remained in the same pen until slaughter (5–8 days). On the basis of the DMI and ADG data, steers were selected according to differences in ADG (p = 0.01) with those with the greatest ADG (n = 8; 1.96 ± 0.02 kg/day) and least ADG (n = 8; 1.57 ± 0.02 kg/day), whose dry matter intake was within 0.32 SD of the mean intake (10.10 ± 0.05 kg/day; p = 0.41). The final body weight of the steers with the greatest ADG was 582.80 ± 52.06 kg and least ADG was 596.80 ± 58.87 kg.
Steers were slaughtered 5 days after the feeding period for 4 consecutive days (4 animals/day). Before slaughter, steers had ad libitum access to feed and water. Tissues were collected immediately after slaughter; the mucosa layer was removed from the wall of duodenum with a sterile scalpel blade from the small intestine from a section removed approximately 5 cm caudal of the cranial duodenal flexure; subcutaneous adipose tissues were collected from the tail–head; liver samples were collected from left lobule, and longissimus-dorsi (LD) from the section adjacent from the 12th rib. All tissue samples were immediately placed in liquid nitrogen, with storage at −80 °C after collection.
Nontargeted Metabolomics Analysis
Duplicate samples of each tissues were extracted for metabolomics analysis. The extraction procedure was adapted from Artegoitia et al.12 Briefly, tissues were individually pulverized with 6875D Freezer/Mill (Spex sample prep; Metuchen, NJ), and 50 mg of each tissue was weighed and diluted with 1 mL of chloroform/methanol/water (1:2:0.8 v/v), vortexed, and centrifuged at 16 000g for 10 min at 4 °C. The supernatant and solid precipitate were separated in different vials for aqueous (supernatant) and organic (precipitate) extractions, respectively. For the aqueous extraction, the supernatant was transferred to a new vial, dried under a nitrogen stream, and resuspended in 500 μL of acetonitrile/water (9:1 v/v). For the organic extraction, the solid precipitate was dissolved in 1 mL of dichlormethane/methanol (3:1 v/v), centrifuged (16 000g, 10 min at 4 °C), dried under a nitrogen stream, and resuspended in 500 μL in acetonitrile/water (9:1 v/v). The ultra-performance liquid-chromatography/mass spectrometry (UPLC/MS) analysis was carried out using a Waters ACQUITY ultra-performance liquid-chromatography (UPLC) system (Waters Corp., Milford, MA) equipped with an autosampler and coupled with a hybrid triple quadrupole-time of-flight mass spectrometry (XEVO-G2-S-qTOF; Waters Corp.). Details of instrument calibration, quality control samples, chromatogram separation, and MS system parameters were carried out as described before.13
Quantitative Lipidomics Analysis
The phospholipids were quantified in duodenum, liver, longissimus-dorsi (LD), and adipose tissue by UPLC-triple quadrupole mass spectrometry using an adapted methodology.13 Briefly, phospholipids were extracted from 50 mg of tissues using the Bligh and Dyer method.14 A mixture (1 μL) of deuterated phospholipids (Avanti Polar 330707, Alabaster, AL) was used as an internal standard. The calibration curves for lysophosphatidylcholine (LPC; Avanti Polar 83007, Alabaster, AL), lysophosphatidylamine (Avanti Polar 840081, Alabaster, AL), phosphatidylethanolamine (Avanti Polar 330707, Alabaster, AL), phosphatidylcholine (PC; Avanti Polar 840055, Alabaster, AL), and cholesterol (Avanti Polar 700000, Alabaster, AL) were prepared by serial dilution (8-fold) in water/methanol (1:9 v/v).
The UPLC/MS analysis was carried out using a Waters ACQUITY ultra-performance liquid-chromatography (UPLC, Acquity BEH HILIC, 2.1 × 100 mm × 1.7 μm; Waters Corp., Milford, MA) equipped with an autosampler and coupled with a triple quadrupole mass spectrometry (XEVO-TQS; Waters Corp.). Details of instrument calibration, chromatogram separation, and MS system parameters were carried out as described before.13
Data and Statistical Analysis
Data processing and analysis were conducted individually for each tissue. Raw data from the fingerprinting analysis was aligned and normalized using total ion intensity using Progenesis QI v1.0 software (Waters Corp.). The bovine Metabolome Database (http://www.cowmetdb.ca/) was used to identify tissue’s compounds by using exact m/z values and retention times.
Data were tested for normality and log transformed and standardized using the Pareto scaling technique. The t-test, principal components analysis, and partial least square of discriminant analysis (PLS-DA) were conducted to identify and visualize differences of the compounds identified between least-ADG and greatest-ADG animals using MetaboAnalyst 3.0 software according to previously published recommended statistical procedure for metabolomics analysis.15 Raw p-values were adjust by PROC MULTTEST procedure of SAS 9.3 (SAS Inst. Inc., Cary, NC) and reported as fold discovered rate (FDR) p-adjusted.
The Venn diagram of the identified metabolites between ADG groups was conducted across the four tissues.16 Pathway analysis was performed using a Bos taurus pathway library, which integrates global pathway enrichment analysis and relative between centrality pathway topology analysis from MetaboAnalyst 3.0 software. The identification and visualization of the top altered pathway were based on KEGG (http://www.genome.jp/kegg/) database sources.
The concentration of phospholipids was evaluated by t-test (p-adjusted), and a receiver–operator characteristic curve (ROC) used to evaluate the sensitivity and specificity of potential metabolic biomarkers.17 In addition, the association of cholesterol concentration among tissues was evaluated by PROC CORR procedure of SAS 9.3 (SAS Inst. Inc., Cary, NC).
Acknowledgments
The authors are grateful to all of the personnel from USMARC for the assistance of the cattle operations and laboratory support during the execution of the study.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b02494.
Identification of longissimus-dorsi, adipose, liver and duodenum metabolites for feed efficiency (Table S1); results from tissues pathway analysis (Table S2) (PDF)
Author Present Address
§ Department of Food Science, Aarhus University, Årslev 05792, Denmark (V.M.A.).
Author Contributions
A.P.F., H.C.F., and V.M.A. designed and planned the experiment. A.P.F. conducted the feeding and growth study. A.P.F and V.M.A. collected tissue samples. V.M.A performed the LC-MS analysis, data analysis, and wrote the manuscript. R.M.L. consulted on data analysis and revised the manuscript.
The authors declare no competing financial interest.
Notes
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendations or endorsement by the USDA. The USDA is an equal opportunity provider and employer.
Supplementary Material
References
- Dijkstra J.; France J.; Ellis J.; Strathe A.; Kebreab E.; Bannink A.. Production Efficiency of Ruminants: Feed, Nitrogen and Methane. In Sustainable Animal Production; Kebreab E., Ed.; AB International: Wallingford, U.K., 2013; pp 10–25. [Google Scholar]
- Tilman D.; Cassman K. G.; Matson P. A.; Naylor R.; Polasky S. Agricultural sustainability and intensive production practices. Nature 2002, 418, 671–677. 10.1038/nature01014. [DOI] [PubMed] [Google Scholar]
- Bailey C. M.; Probert C. L.; Bohman V. R. Growth rate, feed utilization and body composition of young bulls and steers. J. Anim. Sci. 1966, 25, 132–137. 10.2527/jas1966.251132x. [DOI] [PubMed] [Google Scholar]
- Klosterman E. W. Beef cattle size for maximum efficiency. J. Anim. Sci. 1972, 34, 875–880. 10.2527/jas1972.345875x. [DOI] [Google Scholar]
- Rolfe K. M.; Snelling W. M.; Nielsen M. K.; Freetly H. C.; Ferrell C. L.; Jenkins T. G. Genetic and phenotypic parameter estimates for feed intake and other traits in growing beef cattle, and opportunities for selection. J. Anim. Sci. 2011, 89, 3452–3459. 10.2527/jas.2011-3961. [DOI] [PubMed] [Google Scholar]
- Foote A. P.; Hales K. E.; Tait R. G.; Berry E. D.; Lents C. A.; Wells J. E.; Lindholm-Perry A. K.; Freetly H. C. Relationship of glucocorticoids and hematological measures with feed intake, growth, and efficiency of finishing beef cattle. J. Anim. Sci. 2016, 94, 275–283. 10.2527/jas.2015-9407. [DOI] [PubMed] [Google Scholar]
- Asem-Hiablie S.; Rotz C. A.; Stout R.; Stackhouse-Lawson K. Management characteristics of beef cattle production in the Northern Plains and Midwest regions of the United States. Prof. Anim. Sci. 2016, 32, 736–749. 10.15232/pas.2016-01539. [DOI] [Google Scholar]
- Retallick K. J.; Bormann J. M.; Weaber R. L.; MacNeil M. D.; Bradford H. L.; Freetly H. C.; Hales K. E.; Moser D. W.; Snelling W. M.; Thallman R. M.; Kuehn L. A. Genetic variance and covariance and breed differences for feed intake and average daily gain to improve feed efficiency in growing cattle. J. Anim. Sci. 2017, 95, 1444–1450. 10.2527/jas.2016.1260. [DOI] [PubMed] [Google Scholar]
- Nkrumah J.; Okine E.; Mathison G.; Schmid K.; Li C.; Basarab J.; Price M.; Wang Z.; Moore S. Relationships of feedlot feed efficiency, performance, and feeding behavior with metabolic rate, methane production, and energy partitioning in beef cattle. J. Anim. Sci. 2006, 84, 145–153. 10.2527/2006.841145x. [DOI] [PubMed] [Google Scholar]
- Suravajhala P.; Kogelman L. J. A.; Kadarmideen H. N. Multi-omic data integration and analysis using systems genomics approaches: methods and applications in animal production, health and welfare. Genet., Sel., Evol. 2016, 48, 38 10.1186/s12711-016-0217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldansaz S. A.; Guo A. C.; Sajed T.; Steele M. A.; Plastow G. S.; Wishart D. S. Livestock metabolomics and the livestock metabolome: A systematic review. PLoS One 2017, 12, e0177675 10.1371/journal.pone.0177675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artegoitia V. M.; Foote A. P.; Lewis R. M.; Freetly H. C. Rumen fluid metabolomics analysis associated with feed efficiency on crossbred steers. Sci. Rep. 2017, 7, 2864 10.1038/s41598-017-02856-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artegoitia V. M.; Middleton J. L.; Harte F. M.; Campagna S. R.; de Veth M. J. Choline and choline metabolite patterns and associations in blood and milk during lactation in dairy cows. PLoS One 2014, 9, No. e103412 10.1371/journal.pone.0103412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh E.; Dyer W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. 10.1139/y59-099. [DOI] [PubMed] [Google Scholar]
- Xia J.; Sinelnikov I. V.; Han B.; Wishart D. S. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveros J. C.VENNY. An Interactive Tool for Comparing Lists with Venn Diagrams. http://bioinfogp.cnb.csic.es/tools/venny, 2007. (date of access 06/11/2018).
- Xia J.; Broadhurst D. I.; Wilson M.; Wishart D. S. Translational biomarker discovery in clinical metabolomics: an introductory tutorial. Metabolomics. 2013, 9, 280–299. 10.1007/s11306-012-0482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barceló-Coblijn G.; Murphy E. J. Alpha-linolenic acid and its conversion to longer chain n– 3 fatty acids: Benefits for human health and a role in maintaining tissue n– 3 fatty acid levels. Prog. Lipid Res. 2009, 48, 355–374. 10.1016/j.plipres.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Wang S.-h.; Pan Y.; Li J.; Chen H.-q.; Zhang H.; Chen W.; Gu Z.-n.; Chen Y. Q. Endogenous omega-3 long-chain fatty acid biosynthesis from alpha-linolenic acid is affected by substrate levels, gene expression, and product inhibition. RSC Adv. 2017, 7, 40946–40951. 10.1039/C7RA06728C. [DOI] [Google Scholar]
- Ecker J.; Liebisch G. Application of stable isotopes to investigate the metabolism of fatty acids, glycerophospholipid and sphingolipid species. Prog. Lipid Res. 2014, 54, 14–31. 10.1016/j.plipres.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Pietiläinen K. H.; Sysi-Aho M.; Rissanen A.; Seppänen-Laakso T.; Yki-Järvinen H.; Kaprio J.; Orešič M. Acquired obesity is associated with changes in the serum lipidomic profile independent of genetic effects–a monozygotic twin study. PLoS One 2007, 2, e218 10.1371/journal.pone.0000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.-J.; Kim J. H.; Noh S.; Hur H. J.; Sung M. J.; Hwang J.-T.; Park J. H.; Yang H. J.; Kim M.-S.; Kwon D. Y.; Yoon S. K. Metabolomic analysis of livers and serum from high-fat diet induced obese mice. J. Prot. Res. 2011, 10, 722–731. 10.1021/pr100892r. [DOI] [PubMed] [Google Scholar]
- Nam M.; Choi M.-S.; Jung S.; Jung Y.; Choi J.-Y.; Ryu D. H.; Hwang G.-S. Lipidomic profiling of liver tissue from obesity-prone and obesity-resistant mice fed a high fat diet. Sci. Rep. 2015, 5, 16984 10.1038/srep16984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Vance D. E. Thematic review series: glycerolipids. Phosphatidylcholine and choline homeostasis. J. Lipid Res. 2008, 49, 1187–1194. 10.1194/jlr.R700019-JLR200. [DOI] [PubMed] [Google Scholar]
- Artegoitia V. M.; Foote A. P.; Tait R. G.; Kuehn L. A.; Lewis R. M.; Wheeler T. L.; Freetly H. C. Endocannabinoid concentrations in plasma during the finishing period are associated with feed efficiency and carcass composition of beef cattle. J. Anim. Sci. 2017, 95, 4568–4574. 10.2527/jas2017.1629. [DOI] [PubMed] [Google Scholar]
- Leiva T.; Cooke R.; Brandao A.; Marques R.; Vasconcelos J. Effects of rumen-protected choline supplementation on metabolic and performance responses of transition dairy cows. J. Anim. Sci. 2015, 93, 1896–1904. 10.2527/jas.2014-8606. [DOI] [PubMed] [Google Scholar]
- Shahsavari A.; Michael J.; Al Jassim R. The role of rumen-protected choline in hepatic function and performance of transition dairy cows. Br. J. Nutr. 2016, 116, 35–44. 10.1017/S0007114516001641. [DOI] [PubMed] [Google Scholar]
- Akit H.; Collins C.; Fahri F.; Hung A.; D’Souza D.; Leury B.; Dunshea F. Dietary lecithin improves feed efficiency without impacting meat quality in immunocastrated male pigs and gilts fed a summer ration containing added fat. Anim. Nutr. 2018, 4, 203–209. 10.1016/j.aninu.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Wang H.; Yu L.; Wang M.; Liu S.; Sun L.; Chen Q. Effects of supplementation of rumen-protected choline on growth performance, meat quality and gene expression in longissimus dorsi muscle of lambs. Arch. Anim. Nutr. 2015, 69, 340–350. 10.1080/1745039X.2015.1073001. [DOI] [PubMed] [Google Scholar]
- Alexandre P. A.; Kogelman L. J.; Santana M. H.; Passarelli D.; Pulz L. H.; Fantinato-Neto P.; Silva P. L.; Leme P. R.; Strefezzi R. F.; Coutinho L. L. Liver transcriptomic networks reveal main biological processes associated with feed efficiency in beef cattle. BMC Genomics 2015, 16, 1073 10.1186/s12864-015-2292-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida Santana M. H.; Junior G. A. O.; Cesar A. S. M.; Freua M. C.; da Costa Gomes R.; da Luz e Silva S.; Leme P. R.; Fukumasu H.; Carvalho M. E.; Ventura R. V.; et al. Copy number variations and genome-wide associations reveal putative genes and metabolic pathways involved with the feed conversion ratio in beef cattle. J. Appl. Genet. 2016, 57, 495–504. 10.1007/s13353-016-0344-7. [DOI] [PubMed] [Google Scholar]
- Erdman R. A.; Sharma B. Effect of dietary rumen-protected choline in lactating dairy cows. J. Dairy Sci. 1991, 74, 1641–1647. 10.3168/jds.S0022-0302(91)78326-4. [DOI] [PubMed] [Google Scholar]
- Baldi A.; Pinotti L. Choline metabolism in high-producing dairy cows: Metabolic and nutritional basis. Can. J. Anim. Sci. 2006, 86, 207–212. 10.4141/A05-061. [DOI] [Google Scholar]
- Bourgon S.; de Amorim M. D.; Miller S.; Montanholi Y. Associations of blood parameters with age, feed efficiency and sampling routine in young beef bulls. Livest. Sci. 2017, 195, 27–37. 10.1016/j.livsci.2016.11.003. [DOI] [Google Scholar]
- Montanholi Y. R.; Haas L. S.; Swanson K. C.; Coomber B. L.; Yamashiro S.; Miller S. P. Erratum to: Liver morphometrics and metabolic blood profile across divergent phenotypes for feed efficiency in the bovine. Acta Vet. Scand. 2017, 59, 39 10.1186/s13028-017-0308-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wulp M. Y. M.; Verkade H. J.; Groen A. K. Regulation of cholesterol homeostasis. Mol. Cell Endocrinol. 2013, 368, 1–16. 10.1016/j.mce.2012.06.007. [DOI] [PubMed] [Google Scholar]
- Nestel P. J.; Poyser A.; Hood R.; Mills S.; Willis M.; Cook L.; Scott T. The effect of dietary fat supplements on cholesterol metabolism in ruminants. J. Lipid Res. 1978, 19, 899–909. [PubMed] [Google Scholar]
- Li T.; Chiang J. Y. Bile acids as metabolic regulators. Curr. Opin. Gastroenterol. 2015, 31, 159 10.1097/MOG.0000000000000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G.; Fang Y.-Z.; Yang S.; Lupton J. R.; Turner N. D. Glutathione metabolism and its implications for health. J. Nutr. 2004, 134, 489–492. 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]
- Yuan L.; Kaplowitz N. Glutathione in liver diseases and hepatotoxicity. Mol. Aspects Med. 2009, 30, 29–41. 10.1016/j.mam.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Smith S. B.; Crouse J. D. Relative contributions of acetate, lactate and glucose to lipogenesis in bovine intramuscular and subcutaneous adipose tissue. J. Nutr. 1984, 114, 792–800. 10.1093/jn/114.4.792. [DOI] [PubMed] [Google Scholar]
- Ceddia R. B.; Sweeney G. Creatine supplementation increases glucose oxidation and AMPK phosphorylation and reduces lactate production in L6 rat skeletal muscle cells. J. Physiol. 2004, 555, 409–421. 10.1113/jphysiol.2003.056291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.