Abstract
Background: Familial non-medullary thyroid cancer (NMTC) accounts for a relatively small proportion of thyroid cancer cases, but it displays strong genetic predisposition. So far, only a few NMTC susceptible genes and low-penetrance variants contributing to NMTC have been described. This study aimed to identify rare germline variants that may predispose individuals to NMTC by sequencing a cohort of 17 NMTC families.
Methods: Whole-genome sequencing and genome-wide linkage analysis were performed in 17 NMTC families. MendelScan and BasePlayer were applied to screen germline variants followed by customized filtering. The remaining candidate variants were subsequently validated by Sanger sequencing. A panel of 277 known cancer predisposition genes was also screened in these families.
Results: A total of 41 rare coding candidate variants in 40 genes identified by whole-genome sequencing are reported, including 24 missense, five frameshift, five splice change, and seven nonsense variants. Sanger sequencing confirmed all 41 rare variants and proved their co-segregation with NMTC in the extended pedigrees. In silico functional analysis of the candidate genes using Ingenuity Pathway Analysis showed that cancer was the top category of “Diseases and Disorders.” Additionally, a targeted search displayed six variants in known cancer predisposition genes, including one frameshift variant and five missense variants.
Conclusions: The data identify rare germline variants that may play important roles in NMTC predisposition. It is proposed that in future research including functional characterization, these variants and genes be considered primary candidates for thyroid cancer predisposition.
Keywords: rare variants, familial non-medullary thyroid carcinoma, whole-genome sequencing, germline predisposition variants, candidate genes
Introduction
Thyroid cancer is recognized as the most common malignancy of the endocrine system. The risk of developing thyroid cancer is influenced by hereditary factors, environmental factors, and somatic replication errors (1). According to a report by the National Cancer Institute for 2018, the estimated number of new thyroid cancer cases in the United States was 53,990 (http://seer.cancer.gov/statfacts/html/thyro.html). More than 90% of all thyroid cancer is non-medullary thyroid carcinoma (NMTC), which includes epithelial tumors that originate from thyroid follicular cells: papillary thyroid carcinoma (PTC), follicular thyroid carcinoma (FTC), Hürthle cell carcinoma (HTC), poorly differentiated thyroid carcinoma (PDTC), and anaplastic thyroid carcinoma (ATC) (2). NMTC is mostly sporadic, but familial aggregation (two or more affected family members) has been reported in 3–9% of all thyroid cancers (3). While some reports conclude that familial cases demonstrate more aggressive features than sporadic NMTC (4–6), other studies dispute such differences (7,8).
Thyroid cancer can also be classified into two categories: high penetrance, often occurring in families, and low penetrance, mostly occurring in sporadic cases. Regarding high-penetrance thyroid cancers, <5% display syndromes, including Cowden syndrome, familial adenomatous polyposis (FAP) of the colorectum, Gardner syndrome (a subtype of FAP), Carney complex, and Werner syndrome (3). In the remaining 95%, a small number of genes have been detected or proposed (9–15). Given the high heritability of thyroid cancer determined by case/control studies (16,17), it is likely that low-penetrance variants contribute to a substantial but as of yet undetermined proportion of all thyroid cancers. Recent studies of sizeable cohorts of patients using genome-wide association study (GWAS) techniques have indeed confirmed that such low-penetrance predisposition variants play important roles (18–21).
The present study aimed to identify and characterize the traditional class of predisposing factors, namely rare high-penetrance gene variants. As mentioned above, several such variants have been identified and annotated. It is obvious that many more such variants must exist. However, it is notoriously difficult to prove the pathogenic nature of such variants, all of which are rare and some of which have been detected in just a single family. The existence of thyroid cancer in large families encompassing three or more affected individuals suggests regular Mendelian inheritance of the predisposition. These types of families fulfill the traditional requirement for successful linkage analysis and are amenable to the now available next-generation sequencing (NGS) approaches. This study identified 41 variants in 40 genes qualifying as primary candidates for high-risk thyroid cancer predisposition using whole-genome sequencing (WGS) and whole-genome linkage analysis. The results highlight several new potential predisposition genes for future study.
Methods
The study was approved by the Institutional Review Board at the Ohio State University, and all subjects gave written informed consent before participation.
Genomic DNA preparation
Genomic DNA was extracted from patient blood samples following standard inorganic isolation procedures (22). To remove RNA contamination, DNA was purified by the standard phenol-chloroform extraction procedure after RNase treatment using PureLink™ RNase A (Thermo Fisher Scientific). DNA concentration was determined with a Qubit DNA Assay Kit (Life Technologies) with a Gemini XPS Microplate Reader (Molecular Devices). Fragment distribution of the DNA library was measured using the Agilent Bioanalyzer 2100 system (Agilent Technologies).
WGS and variant calling
WGS (2 × 150 bp paired-end reads) of the 56 NMTC patient DNA samples was performed using the HiSeq X (Illumina) platform at Novogene Co. Ltd. Primary genome sequencing data analysis was performed using Churchill (23), which implements the GATK “best practices” workflow for alignment, variant discovery, and genotyping. Reads were mapped to the GRCh37 reference sequence (human_g1k_v37_decoy) using BWA v0.7.15. Duplicate reads were identified and removed using samblaster v.0.1.22. SNV/indel discovery and genotyping were performed on a per-family basis (joint calling) using GATK v3.7. SNPeff, ANNOVAR, and custom in-house scripts were used to annotate single nucleotide polymorphisms (SNPs)/insertions and deletions (INDELs) with gene, transcript, function class, damaging scores, and population allele frequencies.
Variant filtering and annotation for the identification of rare variants
Variants were subsequently analyzed using MendelScan v1.2.3 under a dominant disease model. The resulting variants were filtered to obtain those that segregate with disease (segregation score = 1.0), which means all the variants were shared by every WGS tested individual within each family. First, candidate variants were selected at <0.001 maximum allele frequency in any population of the Genome Aggregation Database (gnomAD) data. Customized cutoff thresholds were further applied to two different variant categories. Missense variants present in four or more individuals in gnomAD and truncating variants (nonsense, splice change, and frameshift) present in ≥30 individuals in gnomAD were removed. Variants were annotated using Variant Effect Predictor (VEP, Ensembl release 50) to isolate variants likely to be pathogenic. Truncating variants (nonsense, splice change, and frameshift) were automatically considered pathogenic. Missense variants were scored using 10 prediction software programs offered through VarSome (DANN, MutationTaster, FATHMM, FATHMM-MKL, MetaSVM, MetalR, LRT, MutationAssessor, SIFT, and Provean) and considered as pathogenic if at least 3/10 pathogenic scores were obtained. Missense variants in genes that (i) appear to be tolerant of missense variation (constraint Z-score <1.0), (ii) exhibit low thyroid expression in the GTEx data set, or (iii) encode proteins with unknown functions were excluded.
Additionally, to avoid missing important truncating variants caused by algorithm bias, variants (nonsense, splice change, and frameshift) from the same VCF files were further analyzed using BasePlayer v1.0.0. All the selected variants fit the same population frequency criteria (<30 individuals in gnomAD).
The variants with positive linkage scores (Z-score >0) were further selected, and they were confirmed by Sanger sequencing in all members with DNA available (including the samples used in WGS). These selected variants were evaluated for co-segregation with disease, that is, the variant must be carried by all the affected individuals and not be carried by any of the unaffected individuals. Only the individuals with diagnosed thyroid cancer were considered as affected. For the individuals with benign thyroid diseases, their genotypes were not considered in the selection criteria. The list of 41 rare variants was generated including the variants identified by both methods.
Genome-wide linkage analysis
Genome-wide linkage analysis was performed using genotypes obtained with HumanCytoSNP-12 BeadChip (Illumina) in a total of 107 samples, including all the 56 affected individuals used in WGS and other family members. Nonparametric linkage analysis was applied with MERLIN v1.1.2 (24), and Z-scores and LOD-scores were obtained. The Z-scores of the selected variants were approximated by the average value of Z-scores of two adjacent markers used in linkage analysis.
Polymerase chain reaction and Sanger sequencing validation
To validate the finding of the WGS variants after filtering, Sanger sequencing was performed on the candidate variants in all the family members with DNA available. Polymerase chain reaction (PCR) primers of the 41 variants with co-segregation are shown in Supplementary Table S1. PCR assays were performed using either an AmpliTaq Gold DNA polymerase kit (Thermo Fisher Scientific) or a HotStarTaq DNA Polymerase kit (Qiagen). PCR products were purified using ExoSAP-IT™ PCR Product Cleanup Reagent (Thermo Fisher Scientific) before being sent for sequencing using an ABI3730 DNA Analyzer at the Genomics Shared Facility, OSU.
Variant filtering and annotation in the 277 known cancer predisposition genes
Variants in the coding regions of 277 known cancer predisposition genes were annotated by ANNOVAR (25). Only the variants present in all the WGS patient samples of at least one NMTC family were selected. A cutoff value of 0.05 was used for the maximum allele frequency in any population of the gnomAD, Exome Sequencing Project v. 6500 (ESP6500), Exome Aggregation Consortium (ExAC), and 1000 Genome databases. After filtering, all the variants annotated as “synonymous” or “unknown” were removed. Predicted functional consequences of the missense variants were annotated using PredictSNP2 (26). Only the variants predicted as “deleterious” were kept. One variant each in AGK and RNF213 were excluded, since they had been identified in the 41 rare variants list. Only the variants with positive linkage Z-score were chosen (Z > 0). As a result, a total of six variants, including five missense variants and one frameshift variant, were obtained.
Databases and web resources
gnomAD: http://gnomad.broadinstitute.org/
VEP: https://useast.ensembl.org/info/docs/tools/vep/index.html
Varsome: https://varsome.com/
Hereditary Thyroid Cancer Panel of the University of Chicago: http://dnatesting.uchicago.edu/tests/hereditary-thyroid-cancer-panel
PredictSNP2: https://loschmidt.chemi.muni.cz/predictsnp2/
Data availability statement
WGS data of the 10 NMTC patients in this study are available at dbGAP (accession number phs001758.v1.p1). The sequence data of the rest 46 patients are not publicly available because the contain information that could compromise research privacy/consent.
Results
NMTC patient sample selection for WGS and linkage analysis
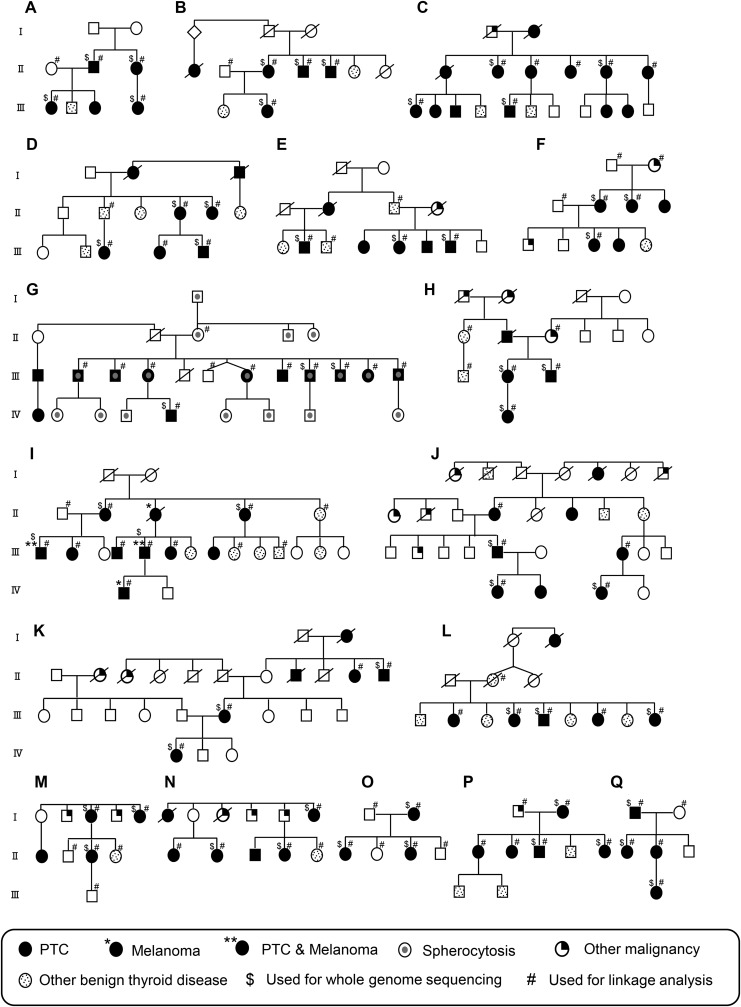
The study focused on NMTC families displaying a Mendelian-like mode of inheritance. The 17 families chosen for this study were of European descent living in the United States comprising three or more NMTC individuals from at least two generations. The number of affected individuals within each family ranged from 3 to 13 (Fig. 1). Individuals diagnosed with papillary thyroid carcinoma were classified as NMTC patients. Individuals with nodules, goiter, and hypothyroidism were classified as having benign thyroid disease. Individuals with other malignancies were not considered affected for the purpose of this study. A total of 56 samples, including at least three samples from affected individuals within each family, were selected for WGS. The main selection criteria for WGS were availability and quality of DNA, the inclusion of family members from different generations, and male sex. The ratio of female to male patients was 2.1:1 (38:18). This deviates from the female-to-male ratio of ∼3:1 in the entire series because affected males were preferentially chosen for WGS when available. This is done because in Mendelian disorders, the individuals belonging to the rarer sex are most likely to carry the sought-after variant. Notably, 60% (34/56) of the patients in this study were diagnosed with NMTC at relatively young age (<50 years old). Of the cases with pathology reports available for review, 23 were microcarcinomas (<1 cm; Supplementary Table S2). Additionally, genome-wide linkage analysis was performed within each family using a total of 107 samples, including 80 affected individuals. These 107 samples included all 56 WGS samples (Fig. 1).
FIG. 1.
Pedigrees of the 17 non-medullary thyroid cancer (NMTC) families used for whole-genome sequencing (WGS). Family structures are indicated by the pedigree maps. Males are indicated by squares, and females are indicated by circles. Generation is labeled using Roman numerals (I, II, III, etc.). Each alphabetic letter represents one family (A–Q). Deceased individuals are displayed as a symbol with a diagonal line. WGS was done on the individuals as indicated.
WGS uncovers rare germline variants in NMTC families
To identify disease predisposition variants from these 17 NMTC families, WGS was conducted on 56 selected patient samples. The study aim to identify missense, splice change, nonsense, and frameshift variants caused by single nucleotide variation (SNV) or insertion/deletion events (INDELs).
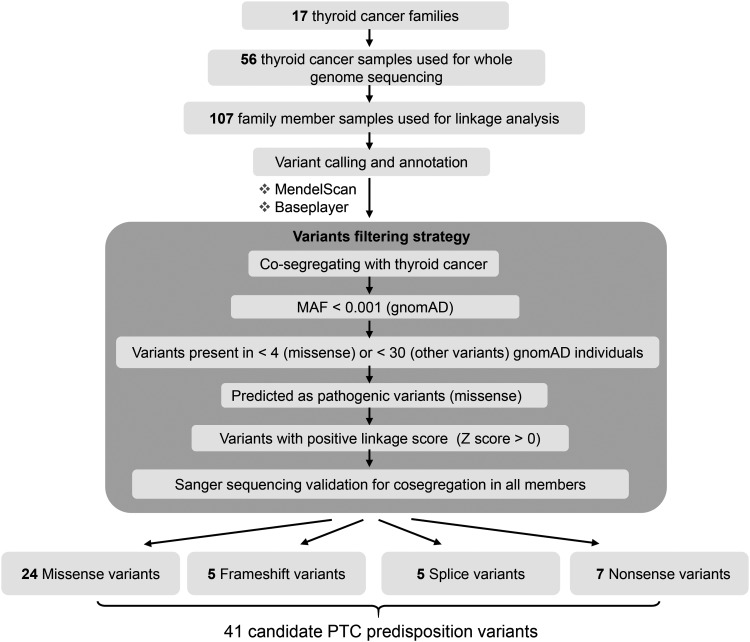
The study generated ∼125 Gbp of uniquely mapped sequence data per sample (range 66–173 Gbp), yielding an average depth of coverage of 32.11 × (range 16.56–40.40 × ) for each individual. GATK v3.7 was used to perform SNV/INDEL detection on a per-family basis (joint calling). On average, WGS uncovered ∼7.64 million variants per family (range 7.10–8.11 million), of which 757,570 segregated with NMTC and 93,024 were also below the MAF threshold. A decision was made to focus on coding variants (∼82 per family), since variants in annotated regulatory non-coding regions (7874 per family on average) yield far too many candidates for further interpretation (Supplementary Table S3). Two different programs, MendelScan (27) and BasePlayer (28), were applied for the variant filtering and annotation steps after processing with GATK (Fig. 2).
FIG. 2.
Overview of the variant filtering scheme for the rare predisposing variant calls in NMTC families. To identify the rare NMTC predisposition variants, variants were called using both MendelScan and BasePlayer as indicated. Variants were then filtered and annotated. Ten programs were used for the missense variant prediction and ranking as described in the Methods. All the remaining variants were validated with Sanger sequencing in all the family individuals with DNA available. MAF, minor allele frequency.
A total of 41 candidate NMTC predisposition variants in 40 genes were identified, including 24 missense, five frameshift, five splice change, and seven nonsense variants. The 41 variants were from 65% (11/17) of the tested families. Thus, in six families, no candidate coding variant was discovered. Notably, 54% (22/41) of the selected variants have not been reported in the dbSNP database (Table 1 and Supplementary Table S4). In silico functional analysis of the 40 identified genes using Ingenuity Pathway Analysis showed that cancer was in the top category of “Diseases and Disorders” with the lowest p-value, which suggested the possible correlation between the candidate genes and NMTC carcinogenesis (Supplementary Table S5). Gene network analysis was also performed. Of the 40 candidate genes, 16 showed direct or indirect interactions within another 19 genes in the top network, and they are involved in cell death and survival, lipid metabolism, and molecular transport (Supplementary Fig. S1).
Table 1.
Details of the 41 Rare Candidate NMTC Predisposing Variants with Co-Segregation in NMTC Families
| Family ID | Position (Hg19) | dbSNP ID | Gene | Variant type | Effect | Z-scorea |
|---|---|---|---|---|---|---|
| B | chr6:46655960 | rs1177620240 | TDRD6 | frameshift_variant | p.Leu32fs | 1.387 |
| A | chr10:94267395 | rs749353444 | IDE | frameshift_variant | p.Met394fs | 1.378 |
| I | chr14:24710238 | N/A | TINF2 | frameshift_variant | p.Gly197fs | 4.583 |
| O | chr17:78318734 | N/A | RNF213 | frameshift_variant | p.His2200fs | 0.456 |
| H | chr7:141321533 | rs1228071168 | AGK | frameshift_variant & splice_region_variant | p.His174fs | 1.382 |
| M | chr1:160340766 | N/A | NHLH1 | missense_variant | p.Thr82Lys | 0.628 |
| O | chr3:129390026 | N/A | TMCC1 | missense_variant | p.Ser41Ala | 0.447 |
| H | chr4:74270076 | N/A | ALB | missense_variant | p.Phe11Tyr | 1.242 |
| B | chr5:79368179 | N/A | THBS4 | missense_variant | p.Asp600Gly | 1.342 |
| B | chr5:133295603 | rs769385290 | C5orf15 | missense_variant | p.Gln83Arg | 2.218 |
| O | chr5:136997689 | rs1234320567 | KLHL3 | missense_variant | p.Tyr223Cys | 0.447 |
| B | chr5:176520192 | N/A | FGFR4 | missense_variant | p.Ile371Val | 1.37 |
| A | chr7:150938678 | rs139896646 | SMARCD3 | missense_variant | p.Arg280His | 1.639 |
| O | chr9:132845876 | N/A | GPR107 | missense_variant | p.Ser187Pro | 0.244 |
| B | chr9:140352197 | N/A | NSMF | missense_variant | p.Pro97 his | 2.198 |
| B | chr10:29782280 | N/A | SVIL | missense_variant | p.Lys86Asn | 1.389 |
| N | chr10:120832522 | rs1241413917 | EIF3A | missense_variant | p.Thr141Ala | 3.331 |
| L | chr11:74459941 | rs749118015 | RNF169 | missense_variant | p.Pro6Ser | 2.813 |
| A | chr11:129744416 | rs200192480 | NFRKB | missense_variant | p.Pro680Ser | 1.682 |
| O | chr12:7171685 | N/A | C1S | missense_variant | p.Lys169Thr | 0.446 |
| Q | chr16:65005565 | rs776739314 | CDH11 | missense_variant | p.Asp520Gly | 1.755 |
| A | chr16:67916998 | N/A | EDC4 | missense_variant | p.Ser1256Cys | 1.367 |
| M | chr19:46376146 | rs372848865 | FOXA3 | missense_variant | p.Asp295His | 1.264 |
| L | chr20:5155891 | N/A | CDS2 | missense_variant | p.Ile86Ser | 2.78 |
| O | chr20:23358093 | N/A | NAPB | missense_variant | p.Asp273Asn | 1.342 |
| O | chr20:50407707 | N/A | SALL4 | missense_variant | p.Pro439Thr | 1.342 |
| H | chr14:55836494 | N/A | ATG14 | missense_variant | p.Pro441Leu | 1.381 |
| P | chr14:94088825 | N/A | UNC79 | missense_variant | p.Leu1572His | 1.94 |
| A | chr22:21351254 | N/A | LZTR1 | missense_variant and splice_region_variant | p.Lys802Arg | 1.228 |
| A | chr1:17316456 | rs866035312 | ATP13A2 | stop_gained | p.Arg819Ter | 1.382 |
| B | chr2:219267800 | N/A | CTDSP1 | stop_gained | p.Arg141Ter | 1.388 |
| H | chr3:50684626 | rs768275251 | MAPKAPK3 | stop_gained | p.Glu330Ter | 1.382 |
| Q | chr16:70304179 | rs756337758 | AARS | stop_gained | p.Arg246Ter | 1.755 |
| B | chr18:61006104 | rs377122315 | KDSR | stop_gained | p.Arg172Ter | 2.172 |
| F | chr19:35175846 | rs775988983 | ZNF302 | stop_gained | p.Gln302Ter | 0.632 |
| H | chr19:57932175 | rs375034689 | ZNF17 | stop_gained | p.Glu441Ter | 1.737 |
| Q | chr16:31434433 | rs769152136 | ITGAD | splice_acceptor_variant | c.2781-2A>C | 1.382 |
| Q | chr16:31434434 | rs759944116 | ITGAD | splice_acceptor_variant | c.2781-1G>A | 1.382 |
| F | chr12:95486473 | N/A | FGD6 | splice_donor_variant | c.3747 + 2A>G | 0.632 |
| Q | chr16:70161297 | N/A | PDPR | splice_donor_variant | c.361 + 1G>C | 1.755 |
| O | chr20:31466614 | N/A | EFCAB8 | splice_donor_variant | c.431 + 1G>C | 1.341 |
Value represents the average Z-score of the two adjacent single nucleotide polymorphism (SNP) markers in linkage analysis within each family.
Identification of candidate NMTC-associated germline variants in known cancer predisposition genes
In addition to the identification of the 41 rare variants, WGS variants were analyzed in known cancer predisposition genes. A total of 277 cancer predisposition genes identified in thyroid cancer and other cancers by functional studies, GWAS, and a commercially available hereditary thyroid cancer panel were selected (Supplementary Table S6) (9,11,13–15,18–21,29–39). For this analysis, the variants were selected in the coding regions of the known cancer genes sharing co-segregation within all WGS members per family. The filter applied with respect to allele frequency was <0.05 and linkage Z-score >0. A total of six candidate variants were identified, including one frameshift variant and five missense variants in the known cancer predisposition genes (Table 2 and Supplementary Table S7). All the missense variants were predicted as deleterious variants by PredictSNP2 (26). The six variants were identified from five families. One variant in the ATM gene was identified in more than one family (families M and N; Table 2). In these families, there are individuals with other malignancies, including kidney, lung, stomach, and prostate cancer (Supplementary Fig. S2).
Table 2.
Candidate Variants of the 277 Known Cancer Susceptibility Genes Identified in the 17 NMTC Families
| Family ID | Position (Hg19) | dbSNP ID | Gene | Variant type | Effect | Average Z-scorea |
|---|---|---|---|---|---|---|
| G | chr22:29091856 | rs555607708 | CHEK2 | frameshift | p.Thr367fs | 2.792 |
| G | chr10:72360387 | rs35947132 | PRF1 | missense | p.Ala91Val | 0.474 |
| N | chr11:108143456 | rs1800057 | ATM | missense | p.Pro1054Arg | 0.456 |
| M | chr11:108143456 | rs1800057 | ATM | missense | p.Pro1054Arg | 0.632 |
| M | chr11:108155132 | rs149711770 | ATM | missense | p.Ala1309Thr | 0.632 |
| H | chr15:86087095 | rs74502151 | AKAP13 | missense | p.Gly191Arg | 1.755 |
| O | chr17:78225167 | rs140131205 | SLC26A11 | missense | p.Gly566Arg | 0.456 |
Value represents the average Z-score of the two adjacent SNP markers in linkage analysis within each family.
Discussion
This study focused on NMTC, which accounts for ∼90% of all thyroid cancers. The aim was to use WGS to identify rare predisposing germline variants in NMTC families.
Genes that confer a risk of human disease when mutated are commonly designated as either rare with high penetrance or common with low penetrance. In reality, much of the genetic predisposition to disease is from genes that fall between the two extremes. The parameter commonly used to categorize variants by frequency is the minor allele frequency (MAF) that is derived from controls (“healthy individuals”) and varies between populations. A commonly used arbitrary cutoff between rare and common alleles is a MAF of 0.005 (5/1000) (40).
Research into rare high-penetrance loci has been possible (linkage, sequencing) for several decades and has uncovered many highly important disease-related loci in various cancers. Nevertheless, the proportion of all heritability caused by these loci is generally low. This has triggered increased research into the common low-penetrance loci, which has become accessible thanks to improved sequencing and automated genotyping, mainly in the past decade. However, importantly, even though the number of risk variants has skyrocketed, the proportion of all heritability caused by presently known genes is low (41,42). There is a clear need for more research to detect and categorize both rare and common variants. In the quest to find additional loci contributing to cancer risk in PTC, 0.001 was chosen as the MAF cutoff between common and rare in this study. This stringent MAF value was chosen because the strategy is based on studying families with several affected individuals. Such families typically harbor rare dominant risk variants that should be readily detectable not only by NGS but also by linkage. The advantage of focusing only on very rare alleles (MAF <0.001) is that “noise” from more common alleles that segregate with the phenotype by chance is reduced. It follows that the chance of missing risk alleles is increased.
In recent years, many rare disease-causing variants for recessively or dominantly inherited diseases have been reported using NGS platforms (43,44). WGS was chosen as the primary method in this study, and a number of rare germline variants in 11/17 NMTC families were identified. The criteria of “co-segregation” and “lack of co-segregation” are challenging in the context of high-penetrance NMTC variants with low-allele frequency. Importantly penetrance is age dependent and not usually complete. Thus, some unaffected individuals especially at a young age may carry the variant and thereby appear as not showing co-segregation with the disease. Collaborative genetic changes (e.g., in a low-penetrance common variant) leading to tumor formation may confound the traditional concept of co-segregation.
Besides the advantage of discovering rare variants, there are limitations to the current method in researching germline variants. In the present results, the 41 rare variants were from 11/17 NMTC families, and there was more than one candidate variant discovered in 8/17 families. Assuming there is one unique causative variant for each family, it is not easy to prioritize any variant based on functional prediction in families with multiple candidates. Functional validation and screening may be necessary to identify the causal variant(s) in the future. Of the 17 families, six did not appear to carry any risk variant. This could be because variants in non-coding genomic regions such as gene regulatory regions and long intergenic noncoding RNA genes were not studied. In addition, germline DNA copy number variations (CNVs) and genomic structural variations such as duplications, deletions, and inversions were not considered. CNVs have been reported to target genes that act in cancer-related pathways, resulting in a high cancer risk (45). Furthermore, a family may not have displayed a candidate variant in cases with imperfect segregation due to phenocopies.
Considering the six variants in known cancer genes identified in NMTC families and the important functions of these genes in tumorigenesis, the variants that were predicted as pathogenic or were able to alter gene structure may play a role in the causation of NMTC. For example, CHEK2 encodes a checkpoint kinase that interacts with cell-cycle regulators and DNA repair proteins. Multiple CHEK2 variants have been reported to be associated with increased risk in various cancer types such as breast cancer (46), colorectal cancer (47), and thyroid cancer (48). In one family, rs555607708 (CHEK2*1100delC) was identified, which is a well-established breast cancer risk variant that is most prevalent in European populations. Its estimated odds ratio (OR) for invasive breast cancer is 2.26 (49). Such an association of CHEK2*1100delC was also confirmed for prostate cancer (50). While it is not possible to explain the events underlying the multicancer involvement of CHEK2 variants, the thyroid is being adding here to the list of affected sites.
In conclusion, a practical method using WGS in searching for rare germline variants in known and previously undescribed cancer genes among NMTC families was applied. Using family-based analysis, a number of rare germline variants that may contribute to NMTC predisposition were identified. Future work including functional annotation and understanding of the molecular mechanisms of these candidate variants will hopefully improve early detection and management of NMTC.
Supplementary Material
Acknowledgments
We thank Isabella Hendrickson for help with PCR and Sanger sequencing. We also thank Jan Lockman and Barbara Fersch for administrative help. The analysis was done in part by an allocation of computing time from the Ohio Supercomputer Center. This work was supported by National Cancer Institute Grants P01CA124570 and P30CA16058, and the Jane and Aatos Erkko Foundation.
Author Disclosure Statement
The authors declare that no competing financial interests exist.
Supplementary Material
References
- 1. Tomasetti C, Li L, Vogelstein B. 2017. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science 355:1330–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xing MZ. 2013. Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer 13:184–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peiling Yang S, Ngeow J. 2016. Familial non-medullary thyroid cancer: unraveling the genetic maze. Endocr Relat Cancer 23:R577–R595 [DOI] [PubMed] [Google Scholar]
- 4. Alsanea O, Clark OH. 2001. Familial thyroid cancer. Curr Opin Oncol 13:44–51 [DOI] [PubMed] [Google Scholar]
- 5. Alsanea O, Wada N, Ain K, Wong M, Taylor K, Ituarte PHG, Treseler PA, Weier HU, Freimer N, Siperstein AE, Duh QY, Takami H, Clark OH. 2000. Is familial non-medullary thyroid carcinoma more aggressive than sporadic thyroid cancer? A multicenter series. Surgery 128:1043–1050 [DOI] [PubMed] [Google Scholar]
- 6. McDonald TJ, Driedger AA, Garcia BM, Van Uum SH, Rachinsky I, Chevendra V, Breadner D, Feinn R, Walsh SJ, Malchoff CD. 2011. Familial papillary thyroid carcinoma: a retrospective analysis. J Oncol 2011:948786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pinto AE, Silva GL, Henrique R, Menezes FD, Teixeira MR, Leite V, Cavaco BM. 2014. Familial vs sporadic papillary thyroid carcinoma: a matched-case comparative study showing similar clinical/prognostic behaviour. Eur J Endocrinol 170:321–327 [DOI] [PubMed] [Google Scholar]
- 8. Vidinov K, Nikolova D. 2017. Familial papillary thyroid carcinoma (FPTC): a retrospective analysis in a sample of the Bulgarian population for a 10-year period. Endocr Pathol 28:54–59 [DOI] [PubMed] [Google Scholar]
- 9. He HL, Bronisz A, Liyanarachchi S, Nagy R, Li W, Huang YG, Akagi K, Saji M, Kula D, Wojcicka A, Sebastian N, Wen B, Puch Z, Kalemba M, Stachlewska E, Czetwertynska M, Dlugosinska J, Dymecka K, Ploski R, Krawczyk M, Morrison PJ, Ringel MD, Kloos RT, Jazdzewski K, Symer DE, Vieland VJ, Ostrowski M, Jarzab B, de la Chapelle A. 2013. SRGAP1 is a candidate gene for papillary thyroid carcinoma susceptibility. J Clin Endocr Metab 98:E973–E980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ngan ESW, Lang BHH, Liu TT, Shum CKY, So MT, Lau DKC, Leon TYY, Cherny SS, Tsai SY, Lo CY, Khoo US, Tam PKH, Garcia-Barcelo MM. 2009. A germline mutation (A339V) in thyroid transcription factor-1 (TITF-1/NKX2.1) in patients with multinodular goiter and papillary thyroid carcinoma. J Natl Cancer Inst 101:162–175 [DOI] [PubMed] [Google Scholar]
- 11. Pereira JS, da Silva JG, Tomaz RA, Pinto AE, Bugalho MJ, Leite V, Cavaco BM. 2015. Identification of a novel germline FOXE1 variant in patients with familial non-medullary thyroid carcinoma (FNMTC). Endocrine 49:204–214 [DOI] [PubMed] [Google Scholar]
- 12. Diquigiovanni C, Bergamini C, Evangelisti C, Isidori F, Vettori A, Tiso N, Argenton F, Costanzini A, Iommarini L, Anbunathan H, Pagotto U, Repaci A, Babbi G, Casadio R, Lenaz G, Rhoden KJ, Porcelli AM, Fato R, Bowcock A, Seri M, Romeo G, Bonora E. 2018. Mutant MYO1F alters the mitochondrial network and induces tumor proliferation in thyroid cancer. Int J Cancer 143:1706–1719 [DOI] [PubMed] [Google Scholar]
- 13. Ni Y, Seballos S, Fletcher B, Romigh T, Yehia L, Mester J, Senter L, Niazi F, Saji M, Ringel MD, LaFramboise T, Eng C. 2017. Germline compound heterozygous poly-glutamine deletion in USF3 may be involved in predisposition to heritable and sporadic epithelial thyroid carcinoma. Hum Mol Genet 26:243–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tomsic J, He HL, Akagi K, Liyanarachchi S, Pan Q, Bertani B, Nagy R, Symer DE, Blencowe BJ, de la Chapelle A. 2015. A germline mutation in SRRM2, a splicing factor gene, is implicated in papillary thyroid carcinoma predisposition. Sci Rep 5:10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yehia L, Niazi F, Ni Y, Ngeow J, Sankunny M, Liu ZG, Wei W, Mester JL, Keri RA, Zhang B, Eng C. 2015. Germline heterozygous variants in SEC23B are associated with Cowden Syndrome and enriched in apparently sporadic thyroid cancer. Am J Hum Genet 97:661–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goldgar DE, Easton DF, Cannonalbright LA, Skolnick MH. 1994. Systematic population-based assessment of cancer risk in first-degree relatives of cancer probands. J Natl Cancer Inst 86:1600–1608 [DOI] [PubMed] [Google Scholar]
- 17. Dong CH, Hemminki K. 2001. Modification of cancer risks in offspring by sibling and parental cancers from 2,112,616 nuclear families. Int J Cancer 92:144–150 [PubMed] [Google Scholar]
- 18. Gudmundsson J, Thorleifsson G, Sigurdsson JK, Stefansdottir L, Jonasson JG, Gudjonsson SA, Gudbjartsson DF, Masson G, Johannsdottir H, Halldorsson GH, Stacey SN, Helgason H, Sulem P, Senter L, He HL, Liyanarachchi S, Ringel MD, Aguillo E, Panadero A, Prats E, Garcia-Castano A, De Juan A, Rivera F, Xu L, Kiemeney LA, Eyjolfsson GI, Sigurdardottir O, Olafsson I, Kristvinsson H, Netea-Maier RT, Jonsson T, Mayordomo JI, Plantinga TS, Hjartarson H, Hrafnkelsson J, Sturgis EM, Thorsteinsdottir U, Rafnar T, de la Chapelle A, Stefansson K. 2017. A genome-wide association study yields five novel thyroid cancer risk loci. Nat Commun 8:14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Son HY, Hwangbo Y, Yoo SK, Im SW, Yang SD, Kwak SJ, Park MS, Kwak SH, Cho SW, Ryu JS, Kim J, Jung YS, Kim TH, Kim SJ, Lee KE, Park DJ, Cho NH, Sung J, Seo JS, Lee EK, Park YJ, Kim JI. 2017. Genome-wide association and expression quantitative trait loci studies identify multiple susceptibility loci for thyroid cancer. Nat Commun 8:15966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gudmundsson J, Sulem P, Gudbjartsson DF, Jonasson JG, Sigurdsson A, Bergthorsson JT, He HL, Blondal T, Geller F, Jakobsdottir M, Magnusdottir DN, Matthiasdottir S, Stacey SN, Skarphedinsson OB, Helgadottir H, Li W, Nagy R, Aguillo E, Faure E, Prats E, Saez B, Martinez M, Eyjolfsson GI, Bjornsdottir US, Holm H, Kristjansson K, Frigge ML, Kristvinsson H, Gulcher JR, Jonsson T, Rafnar T, Hjartarsson H, Mayordomo JI, de la Chapelle A, Hrafnkelsson J, Thorsteinsdottir U, Kong A, Stefansson K. 2009. Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat Genet 41:460–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gudmundsson J, Sulem P, Gudbjartsson DF, Jonasson JG, Masson G, He HL, Jonasdottir A, Sigurdsson A, Stacey SN, Johannsdottir H, Helgadottir HT, Li W, Nagy R, Ringel MD, Kloos RT, de Visser MCH, Plantinga TS, den Heijer M, Aguillo E, Panadero A, Prats E, Garcia-Castano A, De Juan A, Rivera F, Walters GB, Bjarnason H, Tryggvadottir L, Eyjolfsson GI, Bjornsdottir US, Holm H, Olafsson I, Kristjansson K, Kristvinsson H, Magnusson OT, Thorleifsson G, Gulcher JR, Kong A, Kiemeney LALM, Jonsson T, Hjartarson H, Mayordomo JI, Netea-Maier RT, de la Chapelle A, Hrafnkelsson J, Thorsteinsdottir U, Rafnar T, Stefansson K. 2012. Discovery of common variants associated with low TSH levels and thyroid cancer risk. Nat Genet 44:319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Buckingham L, Flaws ML. 2007. Molecular Diagnostics: Fundamentals Methods, and Clinical Applications. F.A. Davis, Philadelphia, PA [Google Scholar]
- 23. Kelly BJ, Fitch JR, Hu YQ, Corsmeier DJ, Zhong HC, Wetzel AN, Nordquist RD, Newsom DL, White P. 2015. Churchill: an ultra-fast, deterministic, highly scalable and balanced parallelization strategy for the discovery of human genetic variation in clinical and population-scale genomics. Genome Biol 16:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Abecasis GR, Cherny SS, Cookson WO, Cardon LR. 2002. Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 [DOI] [PubMed] [Google Scholar]
- 25. Wang K, Li MY, Hakonarson H. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bendl J, Musil M, Stourac J, Zendulka J, Damborsky J, Brezovsky J. 2016. PredictSNP2: a unified platform for accurately evaluating SNP effects by exploiting the different characteristics of variants in distinct genomic regions. PLoS Comput Biol 12:e1004962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koboldt DC, Larson DE, Sullivan LS, Bowne SJ, Steinberg KM, Churchill JD, Buhr AC, Nutter N, Pierce EA, Blanton SH, Weinstock GM, Wilson RK, Daiger SP. 2014. Exome-based mapping and variant prioritization for inherited Mendelian disorders. Am J Hum Genet 94:373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Katainen R, Donner I, Cajuso T, Kaasinen E, Palin K, Makinen V, Aaltonen LA, Pitkanen E. 2018. Discovery of potential causative mutations in human coding and noncoding genome with the interactive software BasePlayer. Nat Protoc 13:2580–2600 [DOI] [PubMed] [Google Scholar]
- 29. Huang KL, Mashl RJ, Wu YG, Ritter DI, Wang JY, Oh C, Paczkowska M, Reynolds S, Wyczalkowski MA, Oak N, Scott AD, Krassowski M, Cherniack AD, Houlahan KE, Jayasinghe R, Wang LB, Zhou DC, Liu D, Cao S, Kim YW, Koire A, McMichael JF, Hucthagowder V, Kim TB, Hahn A, Wang C, McLellan MD, Al-Mulla F, Johnson KJ, Lichtarge O, Boutros PC, Raphael B, Lazar AJ, Zhang W, Wendl MC, Govindan R, Jain S, Wheeler D, Kulkarni S, Dipersio JF, Reimand J, Meric-Bernstam F, Chen K, Shmulevich I, Plon SE, Chen F, Ding L; Cancer Genome Atlas Research Network. 2018. Pathogenic germline variants in 10,389 adult cancers. Cell 173:355–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nikiforova MN, Mercurio S, Wald AI, de Moura MB, Callenberg K, Santana-Santos L, Gooding WE, Yip L, Ferris RL, Nikiforov YE. 2018. Analytical performance of the ThyroSeq v3 genomic classifier for cancer diagnosis in thyroid nodules. Cancer 124:1682–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Siraj AK, Masoodi T, Bu R, Parvathareddy SK, Al-Badawi IA, Al-Sanea N, Ashari LH, Abduljabbar A, Alhomoud S, Al-Sobhi SS, Tulbah A, Ajarim D, Alzoman K, Aljuboury M, Bin Yousef H, Al-Dawish M, Al-Dayel F, Alkuraya FS, Al-Kuraya KS. 2017. Expanding the spectrum of germline variants in cancer. Hum Genet 136:1431–1444 [DOI] [PubMed] [Google Scholar]
- 32. Figlioli G, Chen BW, Elisei R, Romei C, Campo C, Cipollini M, Cristaudo A, Bambi F, Paolicchi E, Hoffmann P, Herms S, Kalemba M, Kula D, Pastor S, Marcos R, Velazquez A, Jarzab B, Landi S, Hemminki K, Gemignani F, Forsti A. 2015. Novel genetic variants in differentiated thyroid cancer and assessment of the cumulative risk. Sci Rep 5:8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mancikova V, Cruz R, Inglada-Perez L, Fernandez-Rozadilla C, Landa I, Cameselle-Teijeiro J, Celeiro C, Pastor S, Velazquez A, Marcos R, Andia V, Alvarez-Escola C, Meoro A, Schiavi F, Opocher G, Quintela I, Ansede-Bermejo J, Ruiz-Ponte C, Santisteban P, Robledo M, Carracedo A. 2015. Thyroid cancer GWAS identifies 10q26.12 and 6q14.1 as novel susceptibility loci and reveals genetic heterogeneity among populations. Int J Cancer 137:1870–1878 [DOI] [PubMed] [Google Scholar]
- 34. Landa I, Ruiz-Llorente S, Montero-Conde C, Inglada-Perez L, Schiavi F, Leskela S, Pita G, Milne R, Maravall J, Ramos I, Andia V, Rodriguez-Poyo P, Jara-Albarran A, Meoro A, del Peso C, Arribas L, Iglesias P, Caballero J, Serrano J, Pico A, Pomares F, Gimenez G, Lopez-Mondejar P, Castello R, Merante-Boschin I, Pelizzo MR, Mauricio D, Opocher G, Rodriguez-Antona C, Gonzalez-Neira A, Matias-Guiu X, Santisteban P, Robledo M. 2009. The variant rs1867277 in FOXE1 gene confers thyroid cancer susceptibility through the recruitment of USF1/USF2 transcription factors. PLoS Genet 5:e1000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Diquigiovanni C, Bergamini C, Evangelisti C, Isidori F, Vettori A, Tiso N, Argenton F, Costanzini A, Iommarini L, Anbunathan H, Pagotto U, Repaci A, Babbi G, Casadio R, Lenaz G, Rhoden KJ, Porcelli AM, Fato R, Bowcock A, Seri M, Romeo G, Bonora E. 2018. Mutant MYO1F alters the mitochondrial network and induces tumor proliferation in thyroid cancer. Int J Cancer 2018. April 19 [Epub ahead of print]; DOI: 10.1002/ijc.31548 [DOI] [PubMed] [Google Scholar]
- 36. Ngan ES, Lang BH, Liu T, Shum CK, So MT, Lau DK, Leon TY, Cherny SS, Tsai SY, Lo CY, Khoo US, Tam PK, Garcia-Barcelo MM. 2009. A germline mutation (A339V) in thyroid transcription factor-1 (TITF-1/NKX2.1) in patients with multinodular goiter and papillary thyroid carcinoma. J Natl Cancer Inst 101:162–175 [DOI] [PubMed] [Google Scholar]
- 37. Liu DX, Yang CF, Bojdani E, Murugan AK, Xing MZ. 2013. Identification of RASAL1 as a major tumor suppressor gene in thyroid cancer. J Natl Cancer Inst 105:1617–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Y, He H, Liyanarachchi S, Genutis LK, Li W, Yu L, Phay JE, Shen R, Brock P, de la Chapelle A. 2018. The role of SMAD3 in the genetic predisposition to papillary thyroid carcinoma. Genet Med 20:927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. He HL, Li W, Liyanarachchi S, Wang YQ, Yu LB, Genutis LK, Maharry S, Phay JE, Shen RL, Brock P, de la Chapelle A. 2018. The role of NRG1 in the predisposition to papillary thyroid carcinoma. J Clin Endocr Metab 103:1369–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bodmer W, Bonilla C. 2008. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet 40:695–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zuk O, Hechter E, Sunyaev SR, Lander ES. 2012. The mystery of missing heritability: genetic interactions create phantom heritability. Proc Natl Acad Sci U S A 109:1193–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. 2009. Finding the missing heritability of complex diseases. Nature 461:747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Boycott KM, Vanstone MR, Bulman DE, MacKenzie AE. 2013. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat Rev Genet 14:681–691 [DOI] [PubMed] [Google Scholar]
- 44. Bahlo M, Tankard R, Lukic V, Oliver KL, Smith KR. 2014. Using familial information for variant filtering in high-throughput sequencing studies. Hum Genet 133:1331–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kuiper RP, Ligtenberg MJ, Hoogerbrugge N, Geurts van Kessel A. 2010. Germline copy number variation and cancer risk. Curr Opin Genet Dev 20:282–289 [DOI] [PubMed] [Google Scholar]
- 46. Apostolou P, Papasotiriou I. 2017. Current perspectives on CHEK2 mutations in breast cancer. Breast Cancer 9:331–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cybulski C, Wokolorczyk D, Kladny J, Kurzawski G, Suchy J, Grabowska E, Gronwald J, Huzarski T, Byrski T, Gorski B T D.EB, Narod SA, Lubinski J. 2007. Germline CHEK2 mutations and colorectal cancer risk: different effects of a missense and truncating mutations? Eur J Hum Genet 15:237–241 [DOI] [PubMed] [Google Scholar]
- 48. Siolek M, Cybulski C, Gasior-Perczak D, Kowalik A, Kozak-Klonowska B, Kowalska A, Chlopek M, Kluzniak W, Wokolorczyk D, Palyga I, Walczyk A, Lizis-Kolus K, Sun P, Lubinski J, Narod SA, Gozdz S. 2015. CHEK2 mutations and the risk of papillary thyroid cancer. Int J Cancer 137:548–552 [DOI] [PubMed] [Google Scholar]
- 49. Schmidt MK, Hogervorst F, van Hien R, Cornelissen S, Broeks A, Adank MA, Meijers H, Waisfisz Q, Hollestelle A, Schutte M, van den Ouweland A, Hooning M, Andrulis IL, Anton-Culver H, Antonenkova NN, Antoniou AC, Arndt V, Bermisheva M, Bogdanova NV, Bolla MK, Brauch H, Brenner H, Bruning T, Burwinkel B, Chang-Claude J, Chenevix-Trench G, Couch FJ, Cox A, Cross SS, Czene K, Dunning AM, Fasching PA, Figueroa J, Fletcher O, Flyger H, Galle E, Garcia-Closas M, Giles GG, Haeberle L, Hall P, Hillemanns P, Hopper JL, Jakubowska A, John EM, Jones M, Khusnutdinova E, Knight JA, Kosma VM, Kristensen V, Lee A, Lindblom A, Lubinski J, Mannermaa A, Margolin S, Meindl A, Milne RL, Muranen TA, Newcomb PA, Offit K, Park-Simon TW, Peto J, Pharoah PD, Robson M, Rudolph A, Sawyer EJ, Schmutzler RK, Seynaeve C, Soens J, Southey MC, Spurdle AB, Surowy H, Swerdlow A, Tollenaar RA, Tomlinson I, Trentham-Dietz A, Vachon C, Wang Q, Whittemore AS, Ziogas A, van der Kolk L, Nevanlinna H, Dork T, Bojesen S, Easton DF. 2016. Age- and tumor subtype-specific breast cancer risk estimates for CHEK2*1100delC Carriers. J Clin Oncol 34:2750–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Seppala EH, Ikonen T, Mononen N, Autio V, Rokman A, Matikainen MP, Tammela TL, Schleutker J. 2003. CHEK2 variants associate with hereditary prostate cancer. Br J Cancer 89:1966–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
WGS data of the 10 NMTC patients in this study are available at dbGAP (accession number phs001758.v1.p1). The sequence data of the rest 46 patients are not publicly available because the contain information that could compromise research privacy/consent.