Abstract
Hydroxyl amino acids have tremendous potential applications in food and pharmaceutical industries. However, available dioxygenases are limited for selective and efficient hydroxylation of free amino acids. Here, we identified a 2-oxoglutarate-dependent dioxygenase from Kutzneria albida by gene mining and characterized the encoded protein (KaPH1). KaPH1 was estimated to have a molecular weight of 29 kDa. The optimal pH and temperature for its l-proline hydroxylation activity were 6.5 and 30 °C, respectively. The Km and kcat values of KaPH1 were 1.07 mM and 0.54 s–1, respectively, for this reaction by which 120 mM l-proline was converted to trans-4-hydroxy-l-proline with 92.8% yield (3.93 g·L–1·h–1). EDTA, [1,10-phenanthroline], Cu2+, Zn2+, Co2+, and Ni2+ inhibited this reaction. KaPH1 was also active toward l-isoleucine for 4-hydroxyisoleucine synthesis. Additionally, the unique biophysical features of KaPH1 were predicted by molecular modeling whereby this study also contributes to our understanding of the catalytic mechanisms of 2-oxoglutarate-dependent dioxygenases.
Introduction
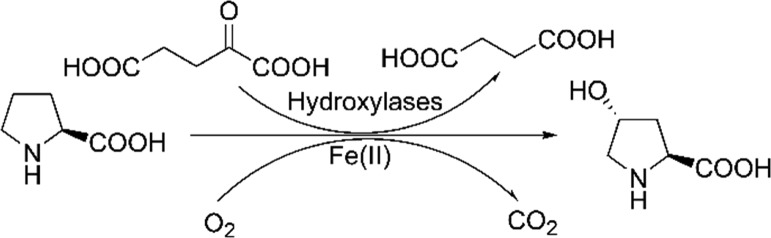
Hydroxyl amino acids are used as nutritional supplements in the food industry and as intermediates in the pharmaceutical industry. For example, trans-4-hydroxy-l-proline (trans-4-Hyp) is a major component of collagen and used as a nutritional supplement in animal diets in lieu of collagen.1trans-4-Hyp has also been reported as a precursor for synthesis of pharmaceuticals and other important compounds, such as (−)-kainic acid,2N-aryl pyrrole,3 and the alkaloid TAN1251A.4 Given that trans-4-Hyp has such a versatile use with high demand, efficient manufacturing methods that are environmental friendly are greatly needed. Similar situations, albeit less drastic, exist also for other hydroxyl amino acids. Compared with conventional methods involving tissue extraction and chemical synthesis,5 biosynthesis has increasingly shown unique advantages of high specificity, low energy consumption, and low environmental pollution. For example, a 2-oxoglutarate (2-OG)-dependent dioxygenase can be used to synthesize trans-4-Hyp via l-proline hydroxylation simply with 2-OG, Fe(II), and dioxygen in the reaction (Scheme 1).
Scheme 1. Hydroxylation Reaction Catalyzed by KaPH1.
2-OG-dependent dioxygenases for l-proline hydroxylation have been discovered in various microorganisms, such as Dactylosporangium sp. RH1,6Streptomyces griseoviridus P8648,7 and Clonostachys cylindrospora SANK 14591.8 Among them, a 2-OG-dependent dioxygenase from Dactylosporangium sp. (P4H), which catalyzes trans-4-Hyp production, has previously been cloned into Escherichia coli (E. coli).6 This P4H-expressing recombinant E. coli strain (W1485/pWFH1) was then cultured for 100 h in a jar fermenter, yielding 41 g·L–1trans-4-Hyp.9 Although various 2-OG-dependent dioxygenases have been identified for biocatalytic conversion of l-proline to trans-4-Hyp, efficient enzymes with high selectivity are still limited, necessitating development or identification of a new dioxygenase.
The members of the 2-OG-dependent dioxygenase superfamily, such as clavaminic acid synthase10 and arginine hydroxylase,11 have similar reaction mechanisms due to characteristic amino acid residues. Thus, here we sought to identify a new 2-OG-dependent dioxygenase for l-proline hydroxylation by gene mining. We thereby identified a new Fe(II)- and 2-OG-dependent dioxygenase KaPH1, which showed high l-proline hydroxylating activity. KaPH1 was then cloned into an E. coli BL21(DE3) strain, expressed, and purified. The purified protein was used to characterize the catalytic activity of KaPH1 for l-proline hydroxylation. Additionally, the unique biophysical features of KaPH1 were predicted by molecular modeling whereby this study also contributes to our understanding of the catalytic mechanisms of 2-oxoglutarate-dependent dioxygenases.
Results and Discussion
Gene Mining-Based Identification of Functional Dioxygenases
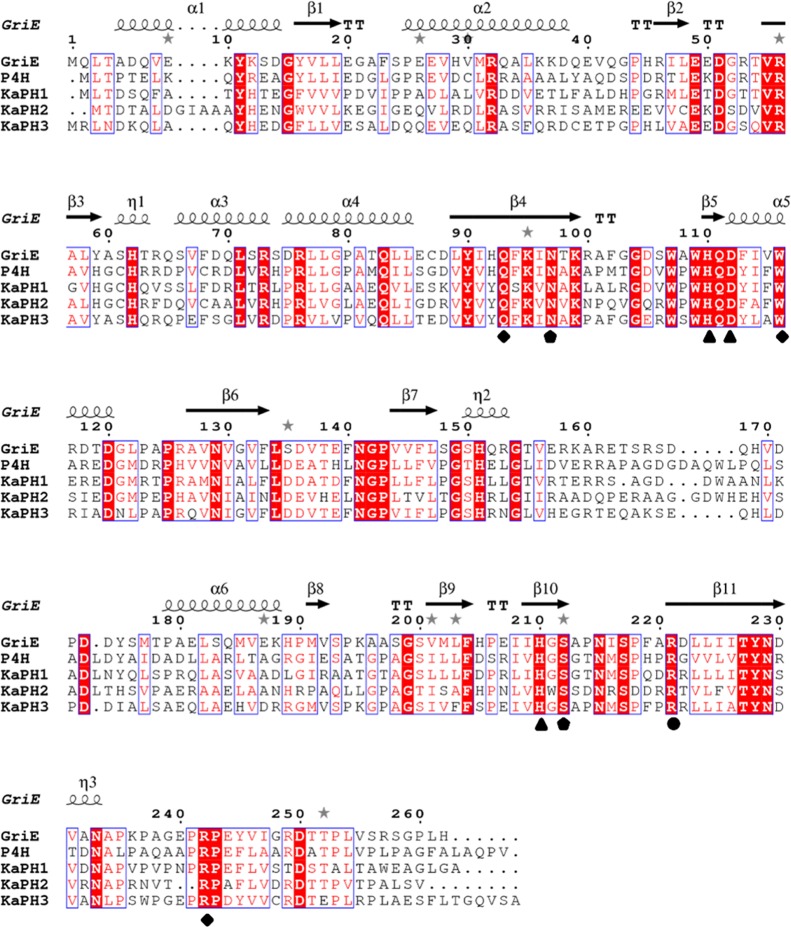
Hydroxylation of nonaromatic amino acids catalyzed by Fe(II)- and 2-OG-dependent dioxygenases, such as clavaminic acid synthase CAS12 and l-arginine dioxygenase VioC,13 occurs through a similar reaction mechanism. These two enzymes exhibit approximately 32% amino acid sequence identity. Thus, gene mining was performed based on sequence similarity. Toward this end, the amino acid sequence of P4H (GenBank: BAA20094.1) from Dactylosporangium sp. was used as the template whereby three putative functional proteins from Kutzneria albida (K. albida) were identified. Among these, KaPH1 (GenBank: WP_025358137.1) exhibited 51% amino acid sequence identity to P4H, KaPH2 (GenBank: WP_030110684.1) exhibited 45% amino acid sequence identity to P4H, and KaPH3 (GenBank: WP_025355730.1) exhibited 42% amino acid sequence identity to P4H (Figure 1).
Figure 1.
Sequence comparison among GriE, P4H, KaPH1, KaPH2, and KaPH3. Gaps in the aligned sequences are indicated with dotted lines. Conserved amino acid residues are indicated in the blue box in which the same amino acid residues are shown with red background. Fe(II)- and 2-OG-binding residues are marked with triangles and circles, respectively. Predicted binding sites for l-proline and 2-OG are marked with rhombi and pentagons, respectively. Secondary structures are indicated for P4H as spirals (helices) and arrows. The multiple sequence alignment was generated by the ESPript program.
Fe(II)/2-OG-dependent dioxygenases have a β-strand “jellyroll” structural fold that contains three metal-binding residues in a His-X-Asp/Glu-Xn-His motif.14 Furthermore, 2-OG chelates Fe(II) with its C-2 keto group and C-1 carboxylate functions as the cofactor, while its C-5 carboxylate mediates an additional interaction with other side chains.15 In consistency with this, His109, Asp111, and His215 of P4H are involved in metal binding. Additionally, an Arg residue residing at the active site of P4H (Arg226) is involved in 2-OG C-5 stabilization. Notably, the “His-X-Asp” carboxylate motif and Arg residue of the active site are conserved in KaPH1, KaPH2, and KaPH3. The structural similarity between the active site of P4H and that of KaPH1, KaPH2, and KaPH3 suggested that the three proteins may catalyze l-proline hydroxylation.
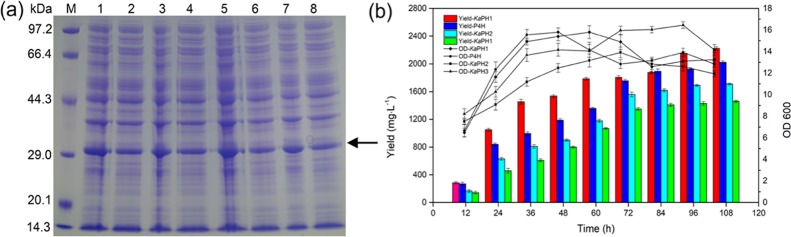
The corresponding genes encoding KaPH1, KaPH2, KaPH3, and P4H were cloned and expressed in recombinant E. coli. The recombinants were cultured in autoinduction media at 20 °C. As shown in Figure 2a, all the enzymes were heterologously expressed in soluble forms. Additionally, they all showed distinct bands in SDS-PAGE in agreement with the predicted molecular weights (P4H, 29.71 kDa; KaPH1, 29.02 kDa; KaPH2, 29.48 kDa; KaPH3, 30.1 kDa).
Figure 2.
Expression of the dioxygenases and their catalytic activities on l-proline. (a) Heterologous expression of recombinant enzymes induced at 20 °C. M: molecular weight standards (Bio-Rad). Lanes 1 and 2: the total protein and soluble fraction of P4H; lanes 3and 4: the total protein and soluble fraction of KaPH1; lanes 5 and 6: the total protein and soluble fraction of KaPH2; lanes 7 and 8: the total protein and soluble fraction of KaPH3. (b) Production of trans-4-Hyp in the growth phase of the recombinant strains. These recombinants were cultivated in autoinduction media containing 20 mM l-proline as the substrate. The trans-4-Hyp content of the fermentation supernatant was measured under standard conditions.
The catalytic activities of the recombinant proteins for l-proline hydroxylation were further evaluated with 20 mM l-proline as the substrate. The products were isolated and subjected to structural analysis as described above. The spectra of 1H NMR (Figure S1), 13C NMR (Figure S2), and MS (Figure S4a) are shown in the Supporting Information. As shown in Figure 2b, 2223 mg·L–1 of trans-4-Hyp was produced in the growth phase of the recombinant strain KaPH1, with a yield of 84.8%, while 2022 mg·L–1trans-4-Hyp was achieved in the growth phase by P4H with a yield of 77.1%. The yields obtained from recombinant strains KaPH2 and KaPH3 were 64.8% (1723 mg·L–1) and 54.7% (1455 mg·L–1), respectively. No significant difference was observed in the OD600 values of the recombinants, suggesting that growth of the recombinants is normal. The results indicated that the recombinant E. coli strain KaPH1 derived from gene mining showed a relatively high l-proline hydroxylating activity compared with P4H. Therefore, KaPH1 was further processed by affinity purification.
Kinetic Parameters of KaPH1
The Michaelis–Menten kinetic parameters of KaPH1 and P4H were measured by varying l-proline and 2-OG concentrations. Comparisons of the kinetic parameters are shown in Table 1. The kcat value of KaPH1 for l-proline was 1.47 times higher than that of P4H, while the Km value of KaPH1 was slightly higher than that of P4H. The kcat value of KaPH1 for 2-OG was higher than that of P4H with a lower Km value. Although P4H had a higher affinity for l-proline than KaPH1, the reaction rate of KaPH1 was 1.47 times higher than that of P4H with a higher affinity and higher reaction rate for 2-OG. The kcat/Km values of KaPH1 for both substrates were higher than those of P4H, indicating that KaPH1 would be favorable for l-proline hydroxylation that involves 2-OG decarboxylation.
Table 1. Kinetics Data Collected for P4H and KaPH1 with l-Proline.
| enzyme | substrate | Km (mM) | Vmax (μM·min–1·mg–1) | kcat (s–1) | kcat/Km (s–1·mM–1) |
|---|---|---|---|---|---|
| P4H | l-proline | 1.00 ± 0.07 | 0.76 ± 0.04 | 0.38 ± 0.02 | 0.37 ± 0.02 |
| 2-OG | 1.35 ± 0.04 | 0.92 ± 0.05 | 0.45 ± 0.01 | 0.34 ± 0.07 | |
| KaPH1 | l-proline | 1.07 ± 0.11 | 1.12 ± 0.06 | 0.54 ± 0.01 | 0.51 ± 0.05 |
| 2-OG | 0.84 ± 0.10 | 1.03 ± 0.09 | 0.50 ± 0.02 | 0.59 ± 0.05 |
To elucidate the biophysical differences between P4H and KaPH1, homology modeling, and molecular docking analyses were performed. The leucine dioxygenase GriE (PDB: 5NCI) as the highest scoring template was submitted for the prediction of potential binding sites of KaPH1 and P4H. It exhibits 41.34 and 41.9% identity to KaPH1 and P4H, respectively. The structural alignment was performed as shown in Figure 3a. The active center of KaPH1 (purple) almost coincides with that of P4H (green). Notably, the residues (Glu150-Asp183) of P4H form a rigid α-helix near the HXD/E···H Fe(II)-binding motif, while the residues (Leu150-Leu183) of KaPH1 form flexible loops. Both components are probably important in the conversion of l-proline, causing the difference in the catalytic performance between P4H and KaPH1. Additionally, the crystal structures of GriE showed conformational changes in the loop regions of which the loop comprising residues 159–176 was assumed to be a lid for the active site.16
Figure 3.
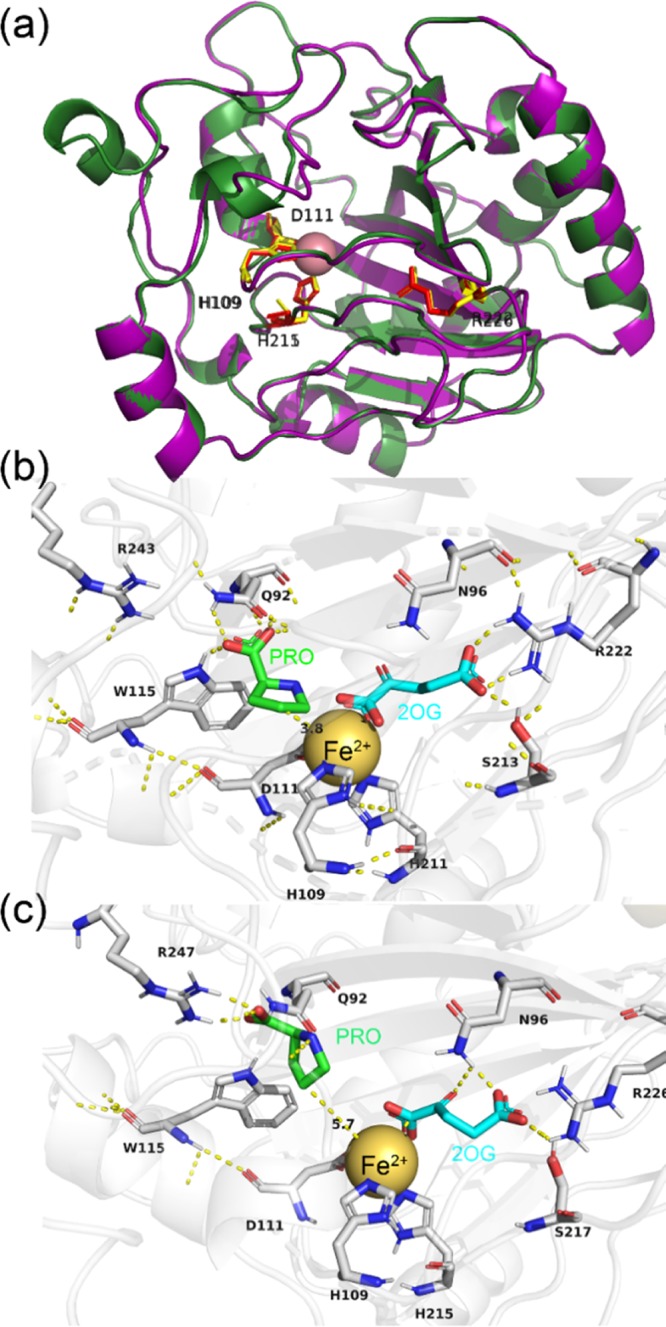
Biophysical analysis of the dioxygenases. (a) Structural alignment of KaPH1 and P4H. The model of KaPH1 and P4H is displayed in purple and green, respectively. Molecular docking results of 2-OG and substrate l-proline in (b) KaPH1 and (c) P4H. The distances between C4 of l-proline/C1 of 2-OG and Fe(II) are illustrated by yellow dashed lines.
Furthermore, Fe(II), 2-OG, and l-proline were docked into the active center of KaPH1 and P4H using AutoDock. The distance between l-proline and Fe(II) as well as 2-OG were calculated and illustrated in Figure 3b,c. The Fe(II) is bound with three residues (His109, Asp111, and His211/215) in the docking models, while 2-OG binds to the Fe(II) with the C-1 carboxylate and C-2 ketone, and its C-5 carboxylate interacts with another residue (Arg222/226) for additional structural stabilization. The side chains of three residues (His109, Asp111, and His211/215), Gln92, and Trp115 are neatly arranged near Fe(II). Among them, Gln92 and Trp115 may interact with the proline carboxylate, properly positioning proline with the C4 bond for oxidation with respect to Fe(II).15 Next, the side chains of three residues (Asn96, Ser 213/217, and Arg222/226) may help the carbonyl group of 2-OG to position toward the Fe(II). Conformation of the active site that contains Fe(II) and l-proline converts to a Fe(IV)-peroxyhemiketal transition state as dioxygen binds to the vacant coordination site and interacts with the carbonyl group of 2-OG.16 Afterward, 2-OG is decarboxylated followed by hydroxylation of l-proline.17 Notably, the distance between C4 of l-proline and Fe(II) is 3.8 Å in the active center of KaPH1, while it is 5.7 Å with P4H. The distance between C1 of 2-OG and Fe(II) in the active center of KaPH1 (2.5 Å) is shorter than that with P4H (2.6 Å). The proximity may lead to a higher catalytic efficiency in KaPH1 toward the substrate/cofactor. Molecular modeling analysis suggested that the structures of KaPH1 and P4H are similar, especially in the conserved motif. The structural differences in flexibility and interaction with metals and substrates are expected to influence the catalytic activities of the enzymes.
Effects of Temperature and pH on KaPH1 Activity
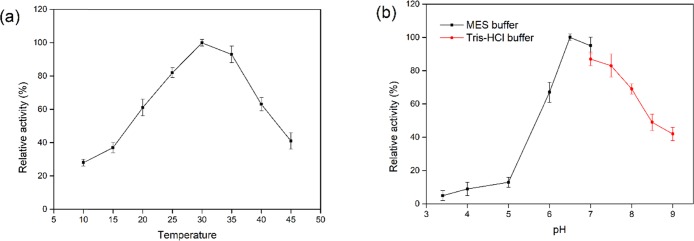
2-OG-dependent dioxygenases specific for l-proline hydroxylation from various microbial strains exhibit an optimum temperature range of 30–40 °C and pH range of 6.0–7.5.18 The effects of temperature and pH on the catalytic activity of recombinant KaPH1 were investigated. The relative activity of recombinant KaPH1 was maintained above 60% at a temperature range of 20–35 °C and reached the maximum activity at 30 °C (Figure 4a). According to the pH profile (Figure 4b), KaPH1 showed the highest activity around pH 6.5 in the MES buffer (0.05 M) with an abrupt increase at pH 6.0, and it was stable under slightly acidic conditions.
Figure 4.
Effects of temperature and pH on the activity of KaPH1. (a) Optimum temperature of l-proline hydroxylation by KaPH1. (b) Optimum pH of l-proline hydroxylation by KaPH1. The following buffers were used: MES–Tris buffer (0.05 M), pH 4–7.0; Tris–HCl buffer (0.05 M), pH 7.0–9.0. The maximum KaPH1 enzymatic activity was set to 100%.
Effects of Common Cofactors and Inhibitors on the Enzymatic Activity
The effects of several common enzymatic cofactors and inhibitors on the enzyme activity were investigated in the activity assay system. As shown in Table 2, the requirement of 2-OG appeared to be very strict, and no trans-4-Hyp was detected without 2-OG. This phenomenon has also been found in other 2-OG-dependent dioxygenases with 2-OG as the decarboxylation cosubstrate.19
Table 2. . Effects of Common Cofactors and Inhibitors on the Enzymatic Activity.
| components | compoundsa | concn (mM) | relative activity (%) |
|---|---|---|---|
| Pro, Fe(II), 2-OG, VC | none | 0 | 100 |
| Pro, Fe(II), VC | none | 0 | 0 |
| Pro, 2-OG, VC | none | 0 | 14 |
| Pro, Fe(II), 2-OG | none | 0 | 69 |
| Pro, Fe(II), 2-OG, VC | EDTA | 4 | 3 |
| Pro, Fe(II), 2-OG, VC | 1,10-phenanthroline | 4 | 2 |
| Pro, Fe(II), 2-OG, VC | diethyl pyrocarbonate | 4 | 0 |
| Pro, Fe(II), 2-OG, VC | MgSO4 | 2 | 82 |
| Pro, Fe(II), 2-OG, VC | MnCl2 | 2 | 79 |
| Pro, Fe(II), 2-OG, VC | CoCl2 | 2 | 22 |
| Pro, Fe(II), 2-OG, VC | ZnSO4 | 2 | 12 |
| Pro, Fe(II), 2-OG, VC | CuSO4 | 2 | 14 |
| Pro, Fe(II), 2-OG, VC | NiSO4 | 2 | 7 |
Effects of chemicals and metal ions were assayed under standard assay conditions. Relative activity was calculated as percentages to the enzymatic activity under the following conditions: l-proline (Pro, 10 mM), 2-oxoglutarate (2-OG, 10 mM), ferrous sulfate (Fe(II), 1.5 mM), l-ascorbic acid (VC, 10 mM).
The 2-OG-dependent dioxygenases are likely to have the relevant structural feature where the Asp and two His residues are conserved.20 As a selective His-modifying reagent,21 the effect of diethyl pyrocarbonate on the enzymatic activity of KaPH1 was investigated. Complete inhibition of the enzymatic activity of KaPH1 was detected with 4 mM diethyl pyrocarbonate, suggesting that the His residues are critical for KaPH1’s enzymatic activity. l-ascorbate was reported to be involved in biochemical reactions catalyzed by dioxygenases that require Fe(II) and 2-OG as cofactors,22 and thus, as expected, l-ascorbate promoted KaPH1 activity. Furthermore, EDTA and 1,10-phenanthroline as the chelating agents strongly inhibited the enzymatic activity, suggesting the requirement for Fe(II). l-proline hydroxylating activity of KaPH1 without FeSO4 in the reaction mixture is presumably due to KaPH1’s interaction with the Fe(II) pool of the E. coli cells before the purification, as reported previously.23
Inhibition of 2-OG dioxygenases by divalent transition metal ions has been extensively observed. Mn2+, Ni2+, Co2+, Cu2+, and Zn2+ were generally found to be the most potent inhibitors.24 The enzymatic activity of KaPH1 declined in the presence of divalent transition metal ions (Table 2), and the strongest inhibition was caused by Zn2+, Ni2+, Co2+, and Cu2+. Inhibition is mostly attributed to competition with Fe(II) for binding to the active site, although such competition was observed at nonactive sites in some cases as well.25
Enzymatic Activity toward l-Isoleucine
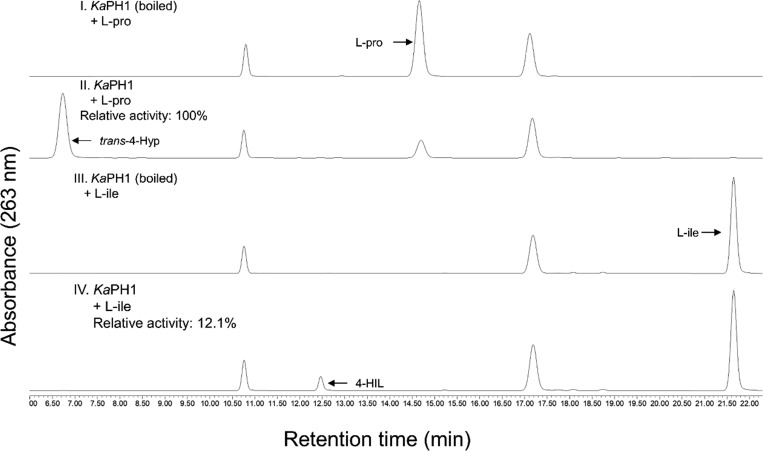
Besides trans-4-Hyp, 4-HIL is another major hydroxyl amino acid constituent of important pharmaceuticals.26,27 It can be synthesized via l-isoleucine hydroxylation by 2-OG-dependent dioxygenases.28l-Isoleucine is an aliphatic amino acid and thus differs from l-proline, which has a heterocyclic structure. Therefore, l-isoleucine hydroxylation activity of KaPH1 was also evaluated under standard conditions. The reaction mixture was subjected to HPLC analysis as described above. As shown in Figure 5, KaPH1 was active toward l-isoleucine. The products were characterized by 1H NMR spectroscopy (Figure S3) and MS (Figure S4b). These results indicate that KaPH1 can hydroxylate different amino acids whereby the most useful intermediates for food and pharmaceutical industries are produced.
Figure 5.
l-Isoleucine hydroxylation by KaPH1. trans-4-Hyp was detected by LC–MS analysis after the incubation of KaPH1 with l-proline (l-Pro) as the substrate. 4-HIL was detected by LC–MS analysis after the incubation of KaPH1 with l-isoleucine (L-Ile) as the substrate. Relative activities are presented with the assumption that the activity on l-Pro is 100%. 4-HIL was further characterized by MS and 1H NMR as described in the ESI.
l-Proline Hydroxylation by KaPH1
Hydroxylation of l-proline, with the concentration of 10–120 mM, was catalyzed by 1 g·L–1 purified KaPH1 in 10–210 min. As shown in Figure 6, KaPH1 showed a robust catalytic activity for l-proline hydroxylation, completely converting 10 mM l-proline within 10 min. Substrate inhibition was not evident during the hydroxylation reaction under any substrate concentration. In addition, KaPH1 generated 92.8% trans-4-Hyp yield with 120 mM l-proline after 4 h (3.93 g·L–1·h–1). P4H-containing recombinant E. coli W1485/pWFH1 yielded 41 g·L–1 of trans-4-Hyp after 100 h of culturing.9 Bontoux et al.100 evaluated five strains of Clonostachys cylindrospora that produce trans-4-Hyp. Among these, SANK 14591 generated trans-4-Hyp with a yield of 13.8 mg·L–1.8 By comparison, KaPH1 would be a highly efficient biocatalyst for in vitro enzymatic generation of trans-4-Hyp.
Figure 6.
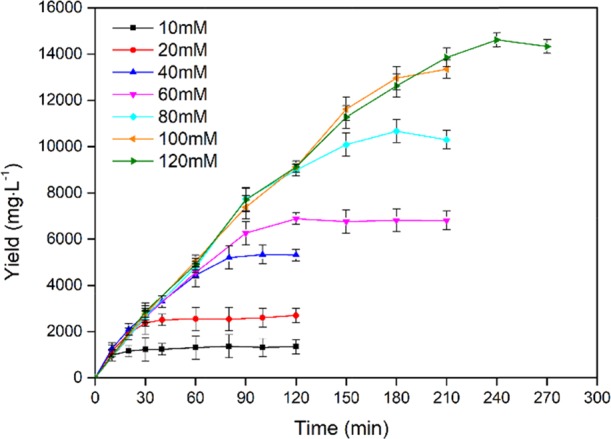
l-Proline hydroxylation catalyzed by the recombinant KaPH1. Samples were taken every 10 min, and the content of trans-4-Hyp was measured under standard conditions.
Conclusions
In this paper, three putative dioxygenases from K. albida were identified by gene mining against P4H. By evaluating the l-proline hydroxylation capacity of the putative enzymes, a new Fe(II)- and 2-OG-dependent dioxygenase KaPH1 from K. albida was identified and characterized for its efficiency in converting free l-proline to trans-4-Hyp. Furthermore, the findings demonstrated the high biotransformation efficiency of KaPH1 (a 92.8% yield when 120 mM l-proline was used as the substrate) as well as its l-isoleucine hydroxylation activity. In conclusion, the newly identified enzyme KaPH1 is a promising biocatalyst for in vitro generation of trans-4-Hyp and other hydroxyl amino acids.
Materials and Methods
Materials
A Kutzneria albida CGMCC 4.1347 strain was obtained from the China General Microbiological Culture Collection Center (CGMCC, Beijing, China). An E. coli BL21(DE3) strain was used for the gene expression. The plasmid pET-28a was obtained from Novagen (U.S.A.) and served as the expression vector. All the enzymes for the genetic manipulations were purchased from Takara Biotechnology Co., Ltd. (Dalian, China). The standard samples of trans-4-Hyp and cis-4-Hyp were purchased from Cambridge Sigma-Aldrich (Munich, Germany). Unless otherwise stated, all the other chemicals were purchased from Sinopharm Chemical Reagent Company (Shanghai, China).
Candidate Gene Screening and Sequence Analysis
To identify potential dioxygenases for l-proline hydroxylation, we searched the National Center for Biotechnology Information database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) using the amino acid sequence of P4H as the query in the protein BLAST algorithm (GenBank accession number: BAA20094.1).29 Multiple sequence alignments with secondary structure elements were generated by ESPript.30
Cloning of the Candidate Dioxygenase Genes
The genes encoding the potential dioxygenases with 42–51% homology to P4H were amplified from the K. albida genome using appropriate primer pairs containing the NdeI/HindIII and XhoI restriction sites (Table 3). As a control, the P4H gene (GenBank accession number: D78338) was synthesized by the Ruidi Biotech Company (Shanghai, China). The purified fragments were digested with the abovementioned restriction enzymes and ligated into the pET-28a expression vector. Next, verified recombinant plasmids were transformed into E. coli BL21 (DE3) competent cells.
Table 3. Strains/Plasmids/Primers Used in this Study.
| strains/plasmids/primers | features | source |
|---|---|---|
| Kutzneria albida CGMCC 4.1347 | origin for potential genes | CGMCC |
| pET-28a-P4H | pET-28a containing P4H | this study |
| pET-28a-KaPH1 | pET-28a containing KaPH1 | this study |
| pET-28a-KaPH2 | pET-28a containing KaPH2 | this study |
| pET-28a-KaPH3 | pET-28a containing KaPH3 | this study |
| F-KaPH1 | 5′-TGCATCATATGCTCACCGATTCGCAGT-3′ | |
| R-KaPH1 | 5′- TTAAGCTTTCATGCGCCCAGGC-3′ | |
| F-KaPH2 | 5′-CAAGCTTGGCTGATGACTGACACCGCACT-3′ | |
| R-KaPH2 | 5′-CCTCGAGGGGGTCAGACGGACAGCGC-3′ | |
| F-KaPH3 | 5′-CAAGCTTGGACCATGCGTTTGAACGACAAGCA-3′ | |
| R-KaPH3 | 5′- CCTCGAGGGCTCATGCCGACACCTGGC-3′ | |
Bacterial Culture and Recombinant Gene Expression
Recombinant E. coli BL21(DE3) strains were grown at 37 °C in LB liquid medium containing 50 μg·mL–1 kanamycin. Cells were inoculated into 30 mL of autoinduction media (tryptone, 10 g·L–1; yeast extract, 5 g·L–1; glycerol, 5 g·L–1; glucose, 0.5 g·L–1; α-lactose, 10 g·L–1; Na2HPO4, 7.1 g·L–1; KH2PO4, 6.8 g·L–1; NH4Cl, 2.68 g·L–1; Na2SO4, 0.71 g·L–1; MgSO4·7H2O, 0.74 g·L–1; CaCl2, 0.015 g·L–1; pH 7.0).31 Cultures were grown in baffled Erlenmeyer flasks on rotary shakers. Cell densities were monitored by measuring the optical density at 600 nm (OD600) using a A380 scanning UV–vis spectrophotometer (AOE Instruments Company, Shanghai, China). Recombinant protein production was analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE).32 Digital images of the gels were captured on a Gel Doc 2000 instrument (Bio-Rad, Shanghai, China).
Identification of Functional Dioxygenases Catalyzing l-Proline Hydroxylation
The recombinant strains were inoculated into the whole cell conversion system where glucose (20 mM), l-proline (20 mM), l-ascorbic acid (5 mM), 2-OG (20 mM), FeSO4·7H2O (0.5 mM), MgSO4 (2 mM), and CaCl2 (0.5 mM) were added into 30 mL of autoinduction media. The recombinant strains were first cultured at 37 °C, 200 rpm for 3 h, and then incubated at 20 °C, 250 rpm for 96 h for whole cell conversion. Samples were taken every 12 h. They were boiled for 10 min, centrifuged at 18,514 × g for 10 min at 4 °C, and the supernatant was analyzed by high-performance liquid chromatography (HPLC)33 whereby trans-4-Hyp was detected.
Protein Purification
The crude enzymes were obtained after disruption of the bacterial pellet and centrifugation, and the supernatant was purified using a HisTrap HP affinity column (GE Healthcare, U.S.A.).34 Then, the purified fractions were applied to disposable PD-10 desalting columns (GE Healthcare, U.S.A.) to remove the high salt content derived from the elution buffer of the affinity columns.35 The protein yields were evaluated by a NanoDrop 8000 microvolume UV–vis spectrophotometer (Thermo Scientific, U.S.A.).
Enzymatic Activity Assays
The concentrations of the purified enzymes were determined by Thermo Scientific NanoDrop 8000 (Thermo Fisher Scientific, U.S.A.). Meanwhile, a mix of 10 mM l-Pro, 1.5 mM FeSO4, 10 mM l-ascorbic acid, 10 mM 2-OG, and 150 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer (pH 6.5) was incubated in a Thermomixer Comfort incubator (Eppendorf, Hamburg, Germany) at 30 °C, 500 rpm. The reaction was started by adding 1 g·L–1 purified enzyme in a final volume of 500 μL.36 Samples were taken 0, 5, and 10 min after adding the enzyme and analyzed by HPLC.33 One unit of enzymatic activity was defined as the amount of enzyme required to catalyze the synthesis of 1 μmol trans-4-Hyp/min under standard conditions.
Determination of Kinetic Parameters
The kinetic parameters (Km and Vmax) were determined in the assay mixture in a final volume of 300 μL. For l-proline, the standard assay conditions were used, with the concentration of l-proline ranging between 0.1 and 5 mM. Likewise, the concentration of 2-OG, where applicable, varied between 0.1 and 5 mM. The assays were carried out in triplicate. KaPH1 activity was measured as described above, and the kinetic parameters were fitted into a Michaelis–Menten model.37
Effects of pH and Temperature on KaPH1 Activity
To determine the effect of pH, we measured KaPH1 activity at a pH range of 4.0–9.0 with the standard reaction mixture. The effect of temperature on KaPH1 activity was determined with the standard reaction mixture incubated for 10 min at different temperatures ranging from 10 to 45 °C.
Effects of Common Enzymatic Cofactors and Inhibitors on KaPH1 Activity
The effects of commonly found enzymatic cofactors (l-ascorbic acid, 2-OG, Mn2+, Mg2+, Co2+, Zn2+, Cu2+, and Ni2+), a divalent cation chelator (EDTA), a metal chelator (1,10-phenanthroline), and a nuclease inhibitor (diethyl pyrocarbonate) on KaPH1 activity were investigated in the enzymatic activity assay system. KaPH1 activity was measured as described above.
l-Proline Hydroxylation by KaPH1
l-proline hydroxylation by the recombinant KaPH1 was investigated under different substrate concentrations. The reaction mixture contained 1.5 mM FeSO4, 10 mM l-ascorbic acid, 10–120 mM l-proline, 10–120 mM 2-OG, 1 g·L–1 purified enzyme, and 150 mM MES buffer (pH 6.5) in a final volume of 500 μL. This mixture was incubated in a Thermomixer Comfort incubator (Eppendorf, Hamburg, Germany) at 30 °C, 500 rpm. Notably, l-proline and 2-OG were used at the same concentration.
Analytical Methods
The analyses of amino acids were carried out by the post column derivatization method with Fmoc-Cl.33 The samples containing hydroxyproline were analyzed by using the Waters 2695 HPLC system equipped with a Diomansil C18 column (250 mm long with 4.6 mm in inner diameter). The chromatographic conditions were as follows: mobile phase A [NaAc–HAc buffer (50 mM, pH 4.2):acetonitrile, 70:30] and mobile phase B [NaAc–HAc buffer (50 mM, pH 4.2):acetonitrile, 30:70] were used with a gradient elution program at a flow rate of 1 mL·min–1, and the column temperature was kept at 25 °C. The injection volume was 10 μL. Fmoc-Cl derivatives of amino acids and hydroxyl amino acids formed in the reaction mixture were detected spectrofluorometrically at 263 nm.
LC–MS analysis was carried out using a Waters ACQUITY UPLC–MS system with a Waters ACQUITY UPLC HSS C18 reversed phase column (inner diameter: 1.8 μm). The inlet, MS transfer line, and ion source temperatures were set at 280, 280, and 230 °C, respectively.
Nuclear Magnetic Resonance (NMR) Analysis
The products derived from l-proline hydroxylation by purified enzyme KaPH1 were isolated from the reaction mixtures by the methods reported previously.38 After the cation exchange chromatography using a strong cation resin (C100E (Na+ form), Purolite, England), the fractions with NH4OH were concentrated under vacuum. The reactions generated 2.13 mM trans-4-Hyp with 69% isolated yield. The concentrated product was dissolved in D2O, and then 1H NMR (400 MHz) and 13C NMR (100 MHz) analyses of the trans-4-Hyp were recorded using an Avance 400 (Bruker, Billerica, MA, U.S.A.). The chemical shifts were provided in parts per million, and coupling constants were reported in Hertz (Hz). Multiplicity was indicated as follows: s (singlet), d (doublet), t (triplet), q (quartet), m (multiplet), dd (doublet of doublet), and br (broad). 1H NMR of trans-4-Hyp: 4.60 (s, 1H), 4.28 (d, J = 2.0 Hz, 1H), 3.40 (d, J = 3.7 Hz, 1H), 3.34–3.21 (m, 1H), 2.35 (d, J = 8.0 Hz, 1H), 2.09 (s, 1H). 13C NMR of trans-4-Hyp: 174.23 (s), 70.07 (s), 59.88 (s), 52.97 (s), 37.44 (s). The spectrum of the prepared trans-4-Hyp is consistent with that reported in the literature.39
4-Hydroxyisoleucine (4-HIL) was generated in vitro by KaPH1-mediated hydroxylation of l-isoleucine. The reactions generated 0.97 mM 4-HIL with 47% isolated yield. The spectrum of 4-HIL was recorded on an Avance 400 (Bruker, Billerica, MA, U.S.A.) at 400 MHz (1H NMR) as described before.281H NMR of 4-HIL: δ 3.84 (d, J = 4.4 Hz, 1H), 3.79 (dd, J = 13.6, 6.8 Hz, 1H), 1.92–1.82 (m, 1H), 1.19 (d, J = 6.3 Hz, 3H), 0.91 (d, J = 7.1 Hz, 3H).
Homology Modeling and Molecular Docking
Homology structures of KaPH1 and P4H were constructed with Phyre 240 (PDB: 5NCI) as a template. All docking calculations of 2-OG and substrate l-proline were achieved with AutoDock 4.2.41 Rigid receptor–flexible ligand docking was carried out with the standard parameters for interactive growing and subsequent scoring.
Funding Resources
Financial support from the National Key R&D Program of China (no. 2018YFC1604100), National Natural Science Foundation of China (NSFC) (nos. 21676120 and 31872891), 111 Project (no. 111-2-06), High-end Foreign Experts Recruitment Program (no. GDT20183200136), Program for Advanced Talents within Six Industries of Jiangsu Province (no. 2015-NY-007), National Program for Support of Top-notch Young Professionals, Fundamental Research Funds for the Central Universities (no. JUSRP51504), the project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, Top-notch Academic Programs Project of Jiangsu Higher Education Institutions, the Jiangsu Province ″Collaborative Innovation Center for Advanced Industrial Fermentation″ Industry Development Program, and the National First-Class Discipline Program of Light Industry Technology and Engineering (no. LITE2018-09) is greatly appreciated.
Acknowledgments
We would like to thank Editage (www.editage.cn) for the English language editing.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.9b00983.
1H and 13C NMR of trans-4-Hyp; 1H NMR of 4-HIL; mass spectrum of Fmoc-trans-4-Hyp and Fmoc-4-HIL (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Li P.; Wu G. Roles of dietary glycine, proline, and hydroxyproline in collagen synthesis and animal growth. Amino Acids 2018, 50, 29–38. 10.1007/s00726-017-2490-6. [DOI] [PubMed] [Google Scholar]
- Poisson J.-F.; Orellana A.; Greene A. E. Stereocontrolled synthesis of (−)-kainic acid from trans-4-hydroxy-L-proline. J. Org. Chem. 2005, 70, 10860–10863. 10.1021/jo051508t. [DOI] [PubMed] [Google Scholar]
- Reddy V. P.; Kumar A. V.; Rao K. R. New strategy for the synthesis of N-aryl pyrroles: Cu-catalyzed C–N cross-coupling reaction of trans-4-hydroxy-L-proline with aryl halides. Tetrahedron Lett. 2011, 52, 777–780. 10.1016/j.tetlet.2010.12.016. [DOI] [Google Scholar]
- Nagumo S.; Matoba A.; Ishii Y.; Yamaguchi S.; Akutsu N.; Nishijima H.; Nishida A.; Kawahara N. Synthesis of (−)-TAN1251A using 4-hydroxy-L-proline as a chiral source. Tetrahedron 2002, 58, 9871–9877. 10.1016/S0040-4020(02)01292-9. [DOI] [Google Scholar]
- Gajare V. S.; Khobare S. R.; Malavika B.; Rajana N.; Venkateswara Rao B.; Syam Kumar U. K. A short diastereoselective synthesis of cis-(2S,4S) and cis-(2R,4R)-4-hydroxyprolines. Tetrahedron Lett. 2015, 56, 3743–3746. 10.1016/j.tetlet.2015.03.119. [DOI] [Google Scholar]
- Shibasaki T.; Mori H.; Chiba S.; Ozaki A. Microbial proline 4-hydroxylase screening and gene cloning. Appl. Environ. Microbiol. 1999, 65, 4028–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. C.; Sobey W. J.; Field R. A.; Baldwin J. E.; Schofield C. J. Purification and initial characterization of proline 4-hydroxylase from Streptomyces griseoviridus P8648: a 2-oxoacid, ferrous-dependent dioxygenase involved in etamycin biosynthesis. Biochem. J. 1996, 313, 185–191. 10.1042/bj3130185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa N.; Matsuoka T.; Hosoya T.; Furuya K. Fermentative production of trans-4-Hydroxy-L-proline by Clonostachys cylindrospora. Biosci., Biotechnol., Biochem. 1995, 59, 555–557. 10.1271/bbb.59.555. [DOI] [Google Scholar]
- Shibasaki T.; Mori H.; Ozaki A. Enzymatic production of trans-4-hydroxy-L-proline by regio- and stereospecific hydroxylation of L-proline. Biosci., Biotechnol., Biochem. 2000, 64, 746–750. 10.1271/bbb.64.746. [DOI] [PubMed] [Google Scholar]
- Salowe S. P.; Marsh E. N.; Townsend C. A. Purification and characterization of clavaminate synthase from Streptomyces clavuligerus: an unusual oxidative enzyme in natural product biosynthesis. Biochemistry 1990, 29, 6499–6508. 10.1021/bi00479a023. [DOI] [PubMed] [Google Scholar]
- Haltli B.; Tan Y.; Magarvey N. A.; Wagenaar M.; Yin X.; Greenstein M.; Hucul J. A.; Zabriskie T. M. Investigating β-hydroxyenduracididine formation in the biosynthesis of the mannopeptimycins. Chem. Biol. 2005, 12, 1163–1168. 10.1016/j.chembiol.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Jensen S. E.; Paradkar A. S. Biosynthesis and molecular genetics of clavulanic acid. Antonie van Leeuwenhoek 1999, 75, 125–133. 10.1023/A:1001755724055. [DOI] [PubMed] [Google Scholar]
- Helmetag V.; Samel S. A.; Thomas M. G.; Marahiel M. A.; Essen L.-O. Structural basis for the erythro-stereospecificity of the L-arginine oxygenase VioC in viomycin biosynthesis. FEBS J. 2009, 276, 3669–3682. 10.1111/j.1742-4658.2009.07085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantri M.; Zhang Z.; Mcdonough M. A.; Schofield C. J. Autocatalysed oxidative modifications to 2-oxoglutarate dependent oxygenases. FEBS J. 2012, 279, 1563–1575. 10.1111/j.1742-4658.2012.08496.x. [DOI] [PubMed] [Google Scholar]
- Hausinger R. P. Fe (II)/α-ketoglutarate-dependent hydroxylases and related enzymes. Crit. Rev. Biochem. Mol. Biol. 2004, 39, 21–68. 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- Lukat P.; Katsuyama Y.; Wenzel S.; Binz T.; König C.; Blankenfeldt W.; Brönstrup M.; Müller R. Biosynthesis of methyl-proline containing griselimycins, natural products with anti-tuberculosis activity. Chem. Sci. 2017, 8, 7521–7527. 10.1039/C7SC02622F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez S.; Hausinger R. P. Catalytic mechanisms of Fe (II)- and 2-oxoglutarate-dependent oxygenases. J. Biol. Chem. 2015, 290, 20702–20711. 10.1074/jbc.R115.648691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y.; Sheng H.; Li Z.; Ye Q. Biosynthesis of trans-4-hydroxyproline by recombinant strains of Corynebacterium glutamicum and Escherichia coli. BMC Biotechnol. 2014, 14, 44. 10.1186/1472-6750-14-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C.; Tian K.; Ban Q.; Wang L.; Sun Q.; He Y.; Yang Y.; Pan Y.; Li Y.; Jiang J.; Jiang C. Genome-wide analysis of the biosynthesis and deactivation of gibberellin-dioxygenases gene family in Camellia sinensis (L.) O. Kuntze. Genes 2017, 8, 235. 10.3390/genes8090235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markolovic S.; Wilkins S. E.; Schofield C. J. Protein hydroxylation catalyzed by 2-oxoglutarate-dependent oxygenases. J. Biol. Chem. 2015, 290, 20712–20722. 10.1074/jbc.R115.662627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.; Rosenberg R. C. Carbethoxylation of coordinated histidine by diethylpyrocarbonate. J. Inorg. Biochem. 1993, 51, 727–735. 10.1016/0162-0134(93)85005-S. [DOI] [PubMed] [Google Scholar]
- Guz J.; Oliński R. The role of vitamin C in epigenetic regulation. Postepy Hig. Med. Dosw. 2017, 71, 747–760. [DOI] [PubMed] [Google Scholar]
- Hara R.; Kino K. Characterization of novel 2-oxoglutarate dependent dioxygenases converting L-proline to cis-4-hydroxy-L-proline. Biochem. Biophys. Res. Commun. 2009, 379, 882–886. 10.1016/j.bbrc.2008.12.158. [DOI] [PubMed] [Google Scholar]
- Rose N. R.; Mcdonough M. A.; King O. N. F.; Kawamura A.; Schofield C. J. Inhibition of 2-oxoglutarate dependent oxygenases. Chem. Soc. Rev. 2011, 40, 4364–4397. 10.1039/c0cs00203h. [DOI] [PubMed] [Google Scholar]
- Tian Y.-M.; Yeoh K. K.; Lee M. K.; Eriksson T.; Kessler B. M.; Kramer H. B.; Edelmann M. J.; Willam C.; Pugh C. W.; Schofield C. J.; Ratcliffe P. J. Differential sensitivity of hypoxia inducible factor hydroxylation sites to hypoxia and hydroxylase inhibitors. J. Biol. Chem. 2011, 286, 13041–13051. 10.1074/jbc.M110.211110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam S.; Ishrat N.; Yadav P.; Singh R.; Narender T.; Srivastava A. K. 4-Hydroxyisoleucine attenuates the inflammation-mediated insulin resistance by the activation of AMPK and suppression of SOCS-3 coimmunoprecipitation with both the IR-β subunit as well as IRS-1. Mol. Cell. Biochem. 2016, 414, 95–104. 10.1007/s11010-016-2662-9. [DOI] [PubMed] [Google Scholar]
- Rawat A. K.; Korthikunta V.; Gautam S.; Pal S.; Tadigoppula N.; Tamrakar A. K.; Srivastava A. K. 4-Hydroxyisoleucine improves insulin resistance by promoting mitochondrial biogenesis and act through AMPK and Akt dependent pathway. Fitoterapia 2014, 99, 307–317. 10.1016/j.fitote.2014.10.006. [DOI] [PubMed] [Google Scholar]
- Kodera T.; Smirnov S. V.; Samsonova N. N.; Kozlov Y. I.; Koyama R.; Hibi M.; Ogawa J.; Yokozeki K.; Shimizu S. A novel L-isoleucine hydroxylating enzyme, L-isoleucine dioxygenase from Bacillus thuringiensis, produces (2S, 3R, 4S)-4-hydroxyisoleucine. Biochem. Biophys. Res. Commun. 2009, 390, 506–510. 10.1016/j.bbrc.2009.09.126. [DOI] [PubMed] [Google Scholar]
- Bontoux M. C.; Gelo-Pujic M. Microbial screening in hydroxylation of L-proline. Tetrahedron Lett. 2006, 47, 9073–9076. 10.1016/j.tetlet.2006.10.094. [DOI] [Google Scholar]
- Matsuda F.; Tsugawa H.; Fukusaki E. Method for assessing the statistical significance of mass spectral similarities using basic local alignment search tool statistics. Anal. Chem. 2013, 85, 8291–8297. 10.1021/ac401564v. [DOI] [PubMed] [Google Scholar]
- Gouet P.; Robert X.; Courcelle E. ESPript/ENDscript: sequence and 3D information from protein structures. Acta Crystallogr. 2005, 61, c42–c43. 10.1107/S0108767305098211. [DOI] [Google Scholar]
- Studier F. W. Protein production by auto-induction in high density shaking cultures. Protein Expression Purif. 2005, 41, 207–234. 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Herbert P.; Santos L.; Alves A. Simultaneous quantification of primary, secondary amino acids, and biogenic amines in musts and wines using OPA/3-MPA/FMOC-CI fluorescent derivatives. J. Food Sci. 2001, 66, 1319–1325. 10.1111/j.1365-2621.2001.tb15208.x. [DOI] [Google Scholar]
- Wang X.; Nie Y.; Xu Y. Improvement of the activity and stability of starch-debranching pullulanase from Bacillus naganoensis via tailoring of the active sites lining the catalytic pocket. J. Agric. Food Chem. 2018, 66, 13236–13242. 10.1021/acs.jafc.8b06002. [DOI] [PubMed] [Google Scholar]
- Xiao R.; Anderson S.; Aramini J.; Belote R.; Buchwald W. A.; Ciccosanti C.; Conover K.; Everett J. K.; Hamilton K.; Huang Y. J.; Janjua H.; Jiang M.; Kornhaber G. J.; Lee D. Y.; Locke J. Y.; Ma L.-C.; Maglaqui M.; Mao L.; Mitra S.; Patel D.; Rossi P.; Sahdev S.; Sharma S.; Shastry R.; Swapna G. V. T.; Tong S. N.; Wang D.; Wang H.; Zhao L.; Montelione G. T.; Acton T. B. The high-throughput protein sample production platform of the northeast structural genomics consortium. J. Struct. Biol. 2010, 172, 21–33. 10.1016/j.jsb.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcioni F.; Blank L. M.; Frick O.; Karau A.; Bühler B.; Schmid A. Proline availability regulates proline-4-hydroxylase synthesis and substrate uptake in proline-hydroxylating recombinant Escherichia coli. Appl. Environ. Microbiol. 2013, 79, 3091–3100. 10.1128/AEM.03640-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheiner L. B.; Beal S. L. Evaluation of methods for estimating population pharmacokinetic parameters. I. Michaelis-menten model: Routine clinical pharmacokinetic data. J. Pharmacokinet. Biopharm. 1980, 8, 553–571. 10.1007/BF01060053. [DOI] [PubMed] [Google Scholar]
- Mori H.; Shibasaki T.; Uozaki Y.; Ochiai K.; Ozaki A. Detection of novel proline 3-hydroxylase activities in Streptomyces and Bacillus spp. by regio- and stereospecific hydroxylation of L-proline. Appl. Environ. Microbiol. 1996, 62, 1903–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier R. J.; Dinesh P. Vibrational spectra, monomer, dimer, NBO, HOMO, LUMO and NMR analyses of trans-4-hydroxy-L-proline. Spectrochim. Acta, Part A 2014, 128, 54–68. 10.1016/j.saa.2014.02.047. [DOI] [PubMed] [Google Scholar]
- Kelley L. A.; Mezulis S.; Yates C. M.; Wass M. N.; Sternberg M. J. E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. M.; Huey R.; Lindstrom W.; Sanner M. F.; Belew R. K.; Goodsell D. S.; Olson A. J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.