Abstract
Enzyme-embedded polymer degradation was reported to be an attractive alternative approach to the conventional surface pouring method for efficient degradation of polymers using fungal-derived enzyme Candida antarctica lipase B. Despite the enormous potential, this approach is still in its infancy. In the present study, a probiotic lipase obtained from Lactobacillus plantarum has been employed for the first time to study the enzyme-embedded polymer degradation approach using poly(ε-caprolactone) (PCL) as the semicrystalline polymer candidate. PCL films embedded with 2 to 8 wt % lipase are studied under static conditions for their enzymatic degradation up to 8 days of incubation. Thermogravimetric analyses (TGA) have shown a clear trend in decreasing thermal stability of the polymer with increasing lipase content and number of incubation days. Differential thermal analyses have revealed that the percentage crystallinity of the leftover PCL films increases with progress in enzymatic degradation because of the efficient action of lipase over the amorphous regions of the films. Thus, the higher lipase loading in the PCL matrix and more number of incubation days have resulted in higher percentage crystallinity in the leftover PCL films, which has further been corroborated by X-ray diffraction analyses. In a similar line, higher percentage mass loss of the PCL films has been observed with increased enzyme loading and number of incubation days. Field emission scanning electron microscopy (FE-SEM) has been employed to follow the surface and cross-sectional morphologies of the polymer films, which has revealed micron-scale pores on the surface as well as a bulk polymer matrix with progress in enzymatic polymer degradation. Additionally, FE-SEM studies have revealed the efficient enzyme-catalyzed hydrolysis of the polymer matrix in a three-dimensional fashion, which is unique to this approach. In addition to the first-time utility of a probiotic lipase for the embedded polymer degradation approach, the present work provides insight into the PCL degradation under static and ambient temperature conditions with no replenishment of enzymes.
Introduction
Polymer biodegradation is of paramount interest for several applications including drug delivery, tissue engineering, and biomedical sutures.1−3 Conventionally, polymer degradation was carried out either through chemical or thermal treatment.4,5 Chemical treatments often involve harsh conditions such as strong acids, bases, or peroxides, which are not eco-friendly. Thermal degradation of polymers generally releases greenhouse gases and at times highly toxic gases such as dioxin.6,7 On the other hand, biodegradation by microbial enzymes has emerged as an attractive alternate route for polymer degradation because of the mild conditions, and biocompatibility.8−13 Due to its room temperature operation, enzymatic polymer degradation is also considered to be energy efficient.14
Enzymes from various sources such as fungi and bacteria have been shown to have great potential for polymer degradation. Several enzymes like amylases, proteases, and lipases are employed for polymer degradation because of their hydrolytic action on the functional groups such as glycosidic, amide, and ester, respectively.15−21 Bacterial lipases are generally considered to be one of the important classes of enzymes that can easily be genetically modified and produced at a mass level. Also, they are promising over fungal lipases owing to their alkaline nature, thermal stability, and tolerance to organic solvents.22 Among the various bacterial strains, lactic acid bacteria such as Lactobacillus sp. are established as probiotics, avirulent, and part of human mucosal surfaces.23
Because many of the preferred biodegradable polymers possess an ester functionality, lipases are majorly used for such hydrolytic degradation.24 Some of the preferred ester-based bioresorbable polymers include poly(ε-caprolactone) (PCL), poly(glycolic acid), poly(l-lactic acid) and poly(lactide-co-glycolide).25 Among these polymers, PCL has been identified as a promising candidate for various biomedical and environmental applications. In particular, PCL has been demonstrated for its enzymatic biodegradation toward controlled drug delivery and tissue engineering applications.26 It is a soft semicrystalline polyester having a melting point at ∼60 °C. It has advantages such as easy processability at low temperatures (∼80 °C) and safe elimination of water-soluble byproducts after hydrolysis.27 Therefore, in this study, lipase from Lactobacillus plantarum has been chosen to degrade PCL because of the enzyme’s probiotic nature and high efficacy toward ester hydrolysis.
Most enzymatic polymer degradation studies utilized the application of lipase over the surface of polyesters to result in surface degradation.28 In this approach, enzymes have to penetrate from the surface into the bulk of the polymer matrix to degrade it efficiently. This approach warrants frequent changing of enzyme-containing buffer solution yet provides only a low degradation rate.29 On the other hand, new embedded enzymatic polymer degradation was reported by Ganesh and Gross that utilized a fungal derived enzyme Candida antarctica lipase B (CALB). They have demonstrated that embedding of the enzyme into the polymer matrix helped in initiating the hydrolysis simultaneously in the surface as well as in the bulk and thus resulting in rapid degradation.29 In their further work, the polymer degradation using CALB-embedded PCL films was optimized under shaking and flow conditions.30 Despite the high potential of this approach, it is still in its infancy, and no further studies were followed. In the current work, we have studied the embedded enzymatic degradation of PCL using lipase obtained from the probiotic source L. plantarum. In addition, we have chosen static conditions without changing the enzyme-containing buffer solution to get insights into the efficacy of the probiotic lipase on polymer degradation in the absence of any additional physical variables. The change in thermal mass loss behavior, percentage crystallinity, and morphology have been studied as a function of enzymatic polymer degradation.
Experimental Section
Materials and Methods
L. plantarum (MTCC 4461) was purchased from IMTECH, Chandigarh, India. PCL pellets (average molecular weight of 45,000 Da), chloroform, and Tween 20 were purchased from Sigma-Aldrich and used as received. Lipase was extracted from L. plantarum and purified using the previously reported literature.31 The change in the percentage crystallinity and thermal mass loss behavior of the samples were studied by differential thermal and thermogravimetric analyses using Shimadzu DTG-60. For this, the enzyme-embedded PCL films were subjected to heating in a nitrogen atmosphere from 35 to 600 °C at a heating rate of 10 °C/min under a steady nitrogen flow rate of 20 mL/min. X-ray diffraction (XRD) analyses over selected PCL films were performed using Rigaku Ultima IV with Cu Kα radiation (λ = 1.5418 A) at a scan rate of 0.3 °/min. The morphology and cross-sectional aspects of the enzyme degraded polymers at various intervals were studied using FEI Apreo field emission scanning electron microscopy (FE-SEM). Gel permeation chromatography (GPC) for molecular weight analyses was performed using a Waters GPC instrument equipped with a 2414 RI detector.
Preparation of Enzyme-Embedded PCL Films
PCL films loaded with different weight percentages such as 2, 4, 6, and 8 wt % lipase were prepared by the following procedure. Initially, PCL pellets were dissolved in chloroform to obtain 1% w/v solution, to which a calculated amount of lyophilized lipase powder was added. To this solution, a nonionic surfactant, Tween 20, was added in a proportion of 1:4 weight ratio to that of lipase to stabilize the enzyme in the polymer matrix. The obtained solutions were casted onto culture Petri dishes and dried overnight to get PCL films having a thickness in the range of ∼100 to 120 μm. The dry films were cut into smaller pieces (10 × 10 mm) and used for further studies.
Embedded Enzymatic PCL Degradation
The polymer degradation was performed by incubating the 10 × 10 mm lipase-embedded PCL film in a Petri dish containing 10 mL of 0.01 M Tris–HCl buffer having pH 8.1. The experimental setup was maintained at ambient temperature throughout the degradation studies. The incubated films were gently removed from the buffer solution at regular intervals, washed with distilled water, and then dried in a vacuum desiccator at room temperature for 24 h. The polymer-incubated buffer solutions were recovered to study the enzyme release kinetics. The leached out lipase was quantified using the standard spectrophotometric protocol using p-nitrophenyl palmitate as the substrate. p-Nitrophenyl palmitate was converted to p-nitrophenol by lipase-mediated hydrolysis, whose absorbance was measured at 410 nm.31 The weight of the enzyme-embedded PCL films was measured before and after degradation to obtain the percentage mass loss. All the experiments were performed in triplicate, and the results are presented with standard deviation.
Results and Discussion
To understand the polymer degradation through this embedded approach, the experiments were performed under static conditions as shaking could assist leaching of partially degraded polymer chains into the aqueous medium. In addition, no further enzyme has been supplemented during the entire process so as to realize the degradation effect solely from the initially loaded enzyme. Thus, we have employed very mild conditions that could mimic a polymer substrate in a static environment.
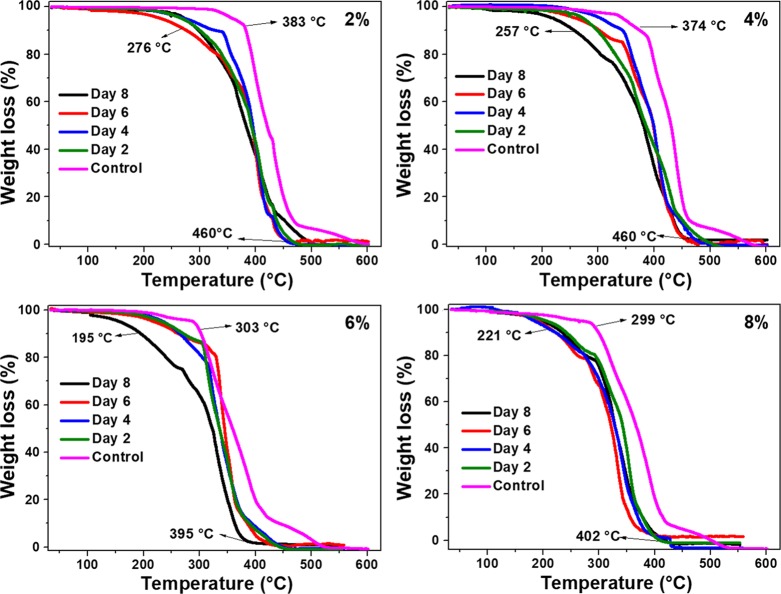
First, to study the efficiency of the enzyme-embedded approach toward polymer degradation, PCL films embedded with lipase of different percentages (2, 4, 6, and 8) were incubated in Tris–HCl buffer solution. TGA and DTA studies were performed on these samples to understand the thermal degradation behavior and crystallinity of the leftover PCL film after degradation. For this, the remaining PCL films were removed at regular time intervals of 2, 4, 6, and 8 days from each incubated plate, rinsed, and dried well. Control experiments were also performed with PCL films that were not subjected to enzymatic degradation but embedded with the corresponding amounts of lipase. Figure 1 shows the TGA analyses of all the samples used in this study. It is expected that the onset mass loss of the PCL films degraded by the enzymes would be less than that of the control ones, as the enzyme degraded films would have been converted into lower-molecular-weight polymers. It can be seen that the 2% control film yielded ∼10% mass loss (considered as onset) at 383 °C and 93% mass loss at 480 °C. After the enzymatic degradation with 2 to 8% of lipase loading, the onset decomposition temperature was found to be in the range of 195 to 300 °C, and the complete decomposition was observed by 395 to 460 °C with a residual mass of <2%. It is also evident from the graph that the thermal stability of the degraded film was less at any point of temperature, when compared to the control. Such a decrease in thermal stability is in line with the expected trend. With 4% lipase embedded films, the onset of the thermal mass loss in 2 to 6 days of enzyme degraded films was in the range of 300 to 330 °C, whereas the 8 day-degraded film started to decompose from 257 °C onward. With these samples, also ∼98% of the thermal decomposition was found to occur by 460 °C. Further increasing the enzyme loading as in 6 and 8% resulted in a further decrease of the onset thermal mass loss of PCL much below 300 °C (∼195 to 250 °C), whereas 98% of thermal decomposition was observed at ∼400 °C. The substantial decrement in the thermal stability of the lipase-embedded PCL films clearly indicated the efficient enzymatic polymer degradation at 6–8% of enzyme loading.
Figure 1.
TGA mass loss analyses of 2, 4, 6, and 8% lipase-embedded PCL films before (control) and after enzymatic polymer degradation.
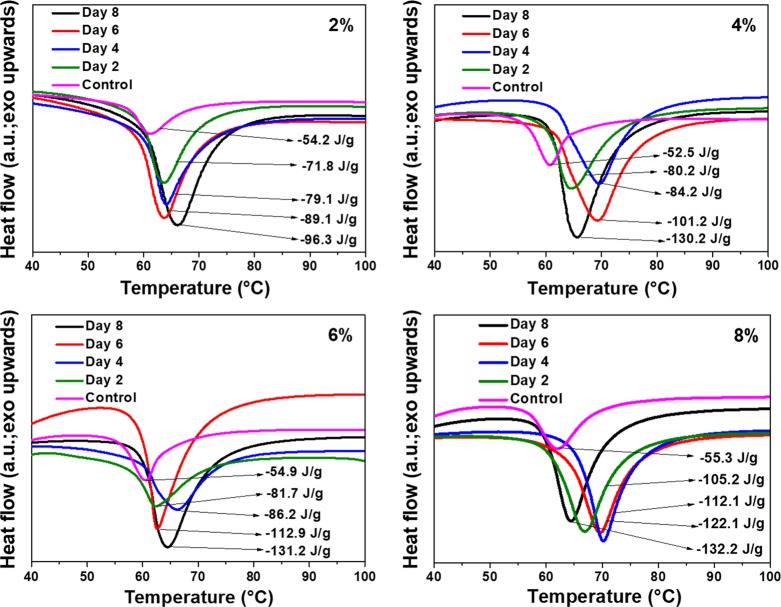
The semicrystalline nature of PCL allows one to probe the change in crystallinity as a function of polymer degradation. DTA analyses on all the PCL samples used in this study were performed before and after enzymatic degradation (Figure 2). The 2 to 8% lipase-embedded PCL films possessed melting points in the range of 60 to 70 °C. In all the samples, the melting point was found to be increased by 5 to 10 °C after the enzymatic degradation due to the increase in crystallinity. Table 1 summarizes the variation in percentage crystallinity of the enzyme-degraded PCL films as a function of time and compared against the corresponding controls. It can be noted that the pristine PCL film without any embedded enzyme was reported to possess a percentage crystallinity of ∼55 %.31,32 The enthalpy of fusion for the 100% crystalline PCL is known to be 139.5 J/g, using which the percentage crystallinity of our samples were calculated from the enthalpy of fusion values obtained from DTA analyses.33 The percentage crystallinity of 2 to 8% lipase-embedded PCL films was found to be in the range of 37 to 39%. Thus, the embedding of enzymes in the PCL matrix caused a significant degree of amorphization that could be due to the incorporation of enzymes into the polymer matrix, thereby disrupting the crystalline regions. After 2 days of enzymatic degradation, the percentage crystallinity of the residual PCL films was found to be significantly increased to the range of 51 to 75%. This substantial increase in the percentage crystallinity indicates the efficient enzymatic degradation of amorphous regions in the PCL matrix even by 2 days. It is known that the enzymatic degradation of semicrystalline polymers mainly occur in amorphous regions.34,35 Further incubation of the PCL films for a longer duration up to 8 days resulted in a gradual increase in the percentage crystallinity. By 8 days of degradation, the percentage crystallinity with varying enzyme-loaded PCL films was found to increase to the range of ∼70 to 95%. These observations clearly indicate that the amorphous regions of the PCL films have been degraded more effectively than the crystalline regions and the 8% lipase-embedded PCL film subjected to 8 days of enzymatic degradation was found to be the most efficient.
Figure 2.
DTA percentage crystallinity analyses of 2, 4, 6, and 8% lipase-embedded PCL films before (control) and after enzymatic polymer degradation.
Table 1. Variation in Percentage Crystallinity Obtained from DTA Measurements with Varying Enzyme Loading against the Number of Incubation Days.
| amount
of enzyme loading |
||||
|---|---|---|---|---|
| number of days | 2% | 4% | 6% | 8% |
| 0 | 39 | 37 | 39 | 39 |
| 2 | 51 | 57 | 58 | 75 |
| 4 | 57 | 60 | 61 | 80 |
| 6 | 64 | 72 | 81 | 87 |
| 8 | 69 | 93 | 94 | 95 |
To further ascertain the enzymatic degradation of the amorphous PCL regions, we have performed XRD analyses of the 8% lipase-embedded polymer film before and after subjecting to 8 days of degradation (Figure 3). As seen from the figure, the pristine PCL film exhibited peaks corresponding to (110) and (200) planes at 21.6 and 23.8° 2θ values, respectively.36,37 In addition, a small shoulder at 2θ = 22.2° corresponding to the (111) plane was observed, proving the characteristic semicrystalline nature of PCL. The crystallite size of the (110) and (200) peaks calculated using Scherrer’s formula was 23.2 and 11.7 nm, respectively. The 8% enzyme-loaded PCL control film, on the other hand, exhibited a gentle decrease in the crystallinity with a crystallite size of 16.8 and 10.8 nm for the (110) and (200) peaks, respectively. The decrease in the percentage crystallinity with enzyme loading corroborated the amorphization of the PCL matrix with the embedding of lipase, as indicated by DTA analyses. However, after 8 days of degradation, the percentage crystallinity was found to be substantially increased. The crystallite sizes corresponding to the respective (110) and (200) peaks in this case were found to be 26.5, and 22.8 nm. These results additionally corroborate the mechanism that the amorphous regions of the polymer films were degraded by the lipase, thereby increasing the percentage crystallinity of the remaining PCL films.
Figure 3.
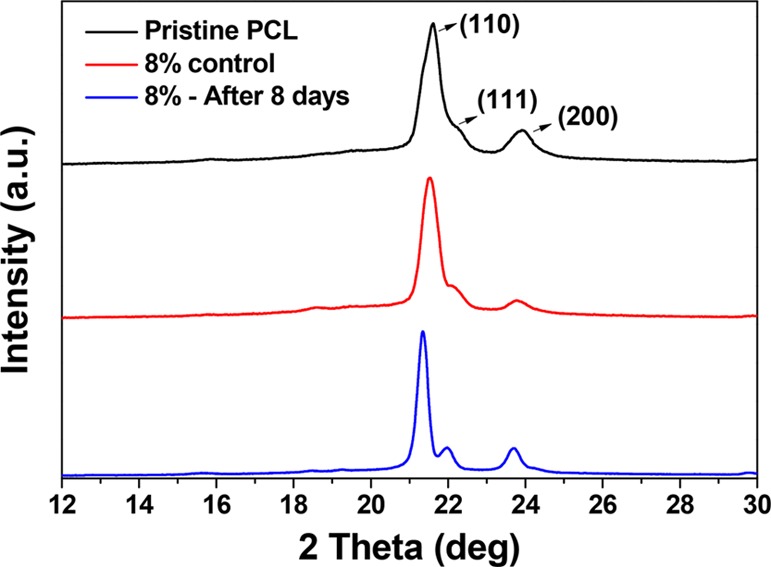
XRD patterns of pristine PCL film, 8% lipase-embedded PCL film (control), and 8% lipase-embedded PCL film after 8 days of incubation.
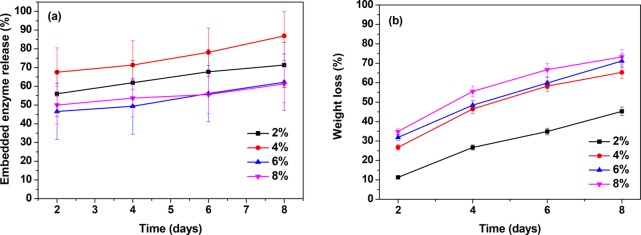
The enzyme release kinetics of the lipase-embedded PCL films was studied to quantify the amount of enzyme leached from the polymer matrix to the buffer solution (Figure 4a). Simultaneously, the percentage mass loss of the lipase-embedded PCL films was studied to monitor the polymer degradation (Figure 4b). For this, PCL films embedded with 2 to 8% L. plantarum lipase were incubated for different time periods of 2 to 8 days in the Tris–HCl buffer. The supernatant solution was collected and analyzed for the amount of enzyme leached. It was observed that 50–65 % of the embedded lipase leached out of the PCL matrix in 2 days. With further incubation of 4 days, additional 2–5% of the lipase leaching to the buffer solution was observed. A similar magnitude of enzyme release from the PCL matrix was observed on days 6 and 8 as well. Thus, a significant portion of the embedded enzyme was released to the buffer solution over a course of time. It is presumed that the enzyme leaching from the PCL matrix was assisted by the rapid polymer degradation within 2 days. The percentage mass loss studies of enzyme-embedded PCL films revealed a clear trend of increasing polymer degradation with time as well as with higher loading of lipase. The lowest mass loss of 11% was observed with 2% lipase-embedded PCL film in 2 days. During the same period of time, the observed mass losses with 4, 6, and 8% lipase-embedded PCL films were 27, 32, and 35%, respectively. This shows that a significant portion of the polymer matrix got degraded within 2 days, which could also have assisted in the leaching of the embedded lipase. Among all the enzyme loadings, 2 to 6% yielded a steady increase in the percentage degradation. Between 6 and 8% loadings, the latter showed only a marginal increment in the percentage mass loss. The percentage mass loss after 8 days was observed to be 71 and 73% with 6 and 8% lipase loadings, respectively. Correlating with the DTA studies, it is obvious that the enzymes efficiently hydrolyzed the amorphous regions of the PCL film. These results show the high efficacy of the L. plantarum lipase in degrading the PCL films, when embedded in the matrix.
Figure 4.
(a) Lipase release kinetics and (b) polymer mass loss analyses of 2, 4, 6, and 8% lipase-embedded PCL films after 2, 4, 6, and 8 days of incubation.
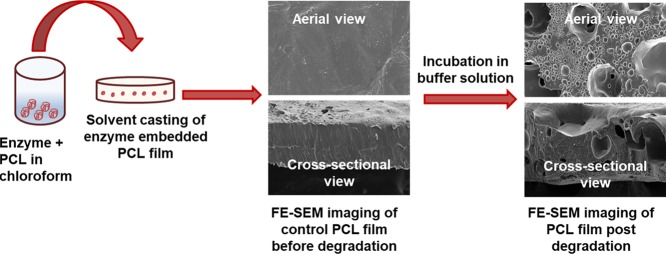
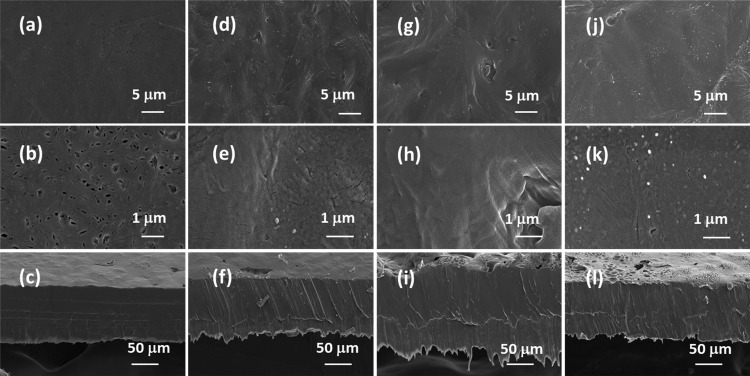
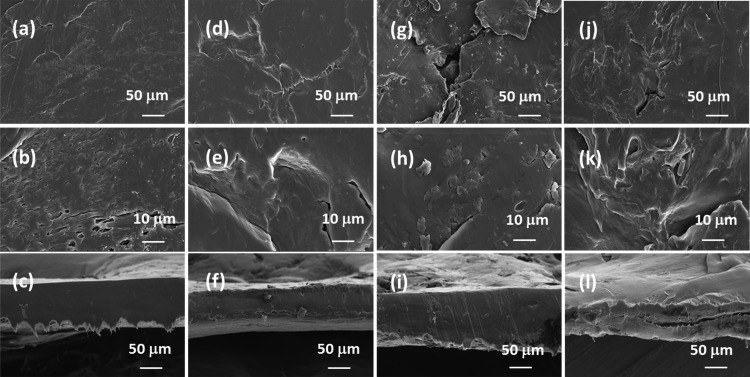
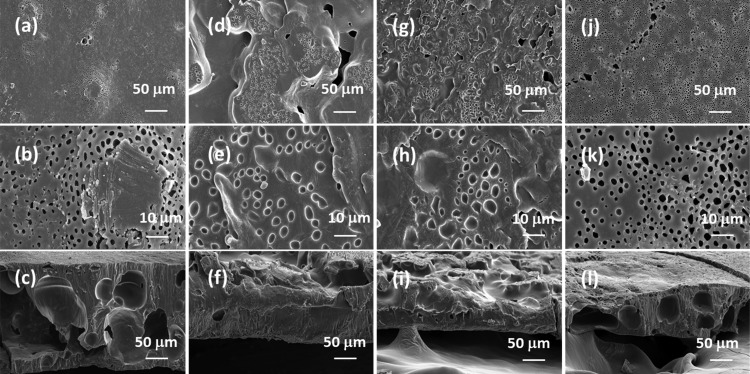
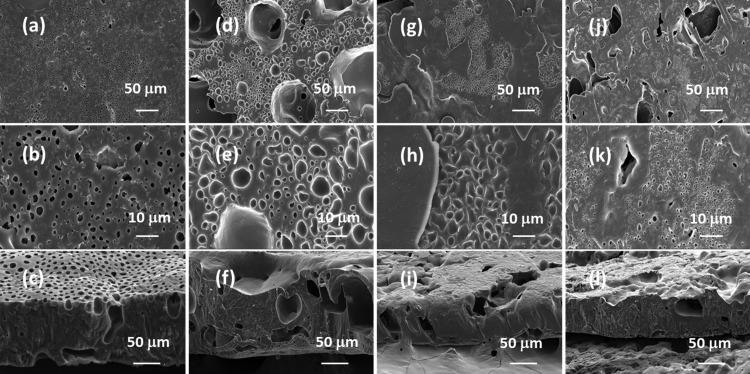
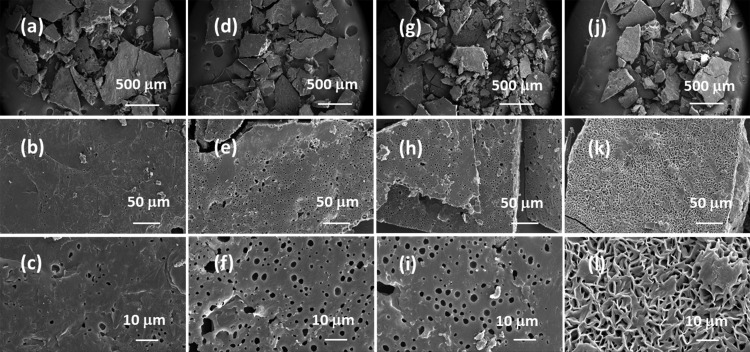
FE-SEM images on the lipase-embedded PCL films were obtained with different quantities (2 to 8 wt %) of lipase-loaded samples against degradation time to monitor the surface and cross-sectional morphological changes. Figure 5 presents the FE-SEM images of lipase-embedded control samples before subjecting to enzymatic degradation. It can be seen from the images that the nondegraded samples possessed a relatively smooth surface morphology as well as cross sections. The cross-sectional views also revealed the average thickness of the films in the range of ∼100 to 120 μm. After 2 days of polymer degradation, the FE-SEM imaging on 2 and 4 wt % lipase loaded PCL films revealed a little degradation on the surface and cross-sectional morphology (Figure 6). However, the 6 and 8 wt % lipase-loaded PCL films possessed relatively a greater amount of degraded polymer that was evidenced by the cracks on the surface and cross-sectional views. After 4 days, the polymer degradation was more evident with all the lipase loadings (Figure 7). In this case, the surface of the polymer films was found to have macropores that were in the range of ∼2 to 3 μm. The cross-sectional imaging revealed a greater degree of polymer degradation in 4 days. Macropores, whose dimensions are several tens of micrometers, were also observed. Similar surface and cross-sectional morphologies were also observed in polymer samples that were subjected to 6 days of degradation (Figure 8). In our earlier work, we employed a pouring strategy of enzymes on the polymer surface that did not result in formation of any pores in the bulk of the PCL film.31 It is obvious that such a strategy results in slower polymer degradation, as the enzymes have to penetrate from the surface in a two-dimensional fashion. Such surface degradation was employed for enzymatic lithography to fabricate micron-to-submicron-scale patterns in PCL. Using microcontact printing and polymer pen lithography techniques, enzymes have been applied over selective regions on the PCL film surface, which resulted in enzymatic hydrolysis to create patterns through surface degradation.38,39 On the other hand, in the embedded approach, the enzymes present in the bulk of the polymer film start to degrade the matrix in a three-dimensional fashion, resulting in micron-scale pores. This was evidenced by the enzyme leaching studies, which indicate that the lipase can freely approach the surface and the bulk of the polymer, thereby leading to faster degradation. It is known that water is essential for enzymatic hydrolysis of polyesters. Ganesh and Gross reported that as the humidity was increased from 20 to 95 %, the enzymatic polymer degradation was found to be accelerated.30 They have also reported that at 75 and 95% relative humidity, the percentage water content in the enzyme-embedded PCL film were found to be 0.30 and 0.82, respectively. Under these conditions, the enzymatic polymer degradation was found to be highly efficient. Thus, it has been shown that the water molecules could diffuse through the enzyme-embedded PCL matrix and assist in hydrolysis. Because we also employed a relative humidity of 75 % in our study, the mechanism of water diffusion to the bulk of the polymer film and the subsequent hydrolysis can be correlated to that reported in the literature. The PCL samples subjected to 8 days of degradation were found to be highly brittle and therefore could not be obtained as free-standing films for the FE-SEM observation. Hence, the obtained crumbled pieces were used for morphological analyses (Figure 9). In this case also, the surface of the remaining crystalline powders possessed micron-scale pores. In all the cases, the 6 and 8 wt % lipase loaded samples exhibited relatively higher degree of degradation than the lower enzyme loadings. All these results corroborated the TGA and DTA analyses. The degradation of the PCL films into crumbled pieces by 8 days of incubation clearly indicated the high efficacy of the lipase-embedded degradation approach.
Figure 5.
FE-SEM images of the (a–c) 2%, (d–f) 4%, (g–i) 6%, and (j–l) 8% lipase-embedded PCL films before subjecting to enzymatic degradation. Top row ((a), (d), (g), and (j)), middle row ((b), (e), (h), and (k)), and bottom row ((c), (f), (i), and (l)) images correspond to the respective low and high magnifications of the surface and cross-section of the PCL film sections.
Figure 6.
FE-SEM images of the (a–c) 2%, (d–f) 4%, (g–i) 6%, and (j–l) 8% lipase-embedded PCL films after subjecting to 2 days of enzymatic degradation. Top row ((a), (d), (g), and (j)), middle row ((b), (e), (h), and (k)), and bottom row ((c), (f), (i), and (l)) images correspond to the respective low and high magnifications of the surface and cross-section of the PCL film sections.
Figure 7.
FE-SEM images of the (a–c) 2%, (d–f) 4%, (g–i) 6%, and (j–l) 8% lipase-embedded PCL films after subjecting to 4 days of enzymatic degradation. Top row ((a), (d), (g), and (j)), middle row ((b), (e), (h), and (k)), and bottom row ((c), (f), (i), and (l)) images correspond to the respective low and high magnifications of the surface and cross-section of the PCL film sections.
Figure 8.
FE-SEM images of the (a–c) 2%, (d–f) 4%, (g–i) 6%, and (j–l) 8% lipase-embedded PCL films after subjecting to 6 days of enzymatic degradation. Top row ((a), (d), (g), and (j)), middle row ((b), (e), (h), and (k)), and bottom row ((c), (f), (i), and (l)) images correspond to the respective low and high magnifications of the surface and cross-section of the PCL film sections.
Figure 9.
FE-SEM images of the (a–c) 2%, (d–f) 4%, (g–i) 6%, and (j–l) 8% lipase-embedded PCL films after subjecting to 8 days of enzymatic degradation. Top row ((a), (d), (g), and (j)), middle row ((b), (e), (h), and (k)), and bottom row ((c), (f), (i), and (l)) images correspond to the respective ultra-low, low, and high magnifications of the PCL film surfaces.
Gel permeation chromatography (GPC) analyses were carried out with 8% lipase-embedded PCL samples to understand the change in molecular weight as a function of polymer degradation. Table 2 lists the number-average molecular weight (Mn), weight-average molecular weight (Mw), and polydispersity index (PDI) of the lipase-embedded PCL films before and after subjecting to varying times of incubation. It can be seen from the table that the control sample exhibited Mn and Mw values of 36500 and 53030, respectively. With the leftover PCL films after enzymatic degradation, the Mn and Mw values were found to be significantly lower than the control, indicating efficient degradation of the polymer films.
Table 2. GPC analyses of 8% lipase-embedded PCL films before (control) and after 2 to 8 days of enzymatic degradation.
| sample | number-average molecular weight (Mn) | weight-average molecular weight (Mw) | polydispersity Index (PDI) |
|---|---|---|---|
| control | 36500 | 53030 | 1.5 |
| 2 days | 21200 | 35060 | 1.7 |
| 4 days | 25210 | 36890 | 1.5 |
| 6 days | 10250 | 28030 | 2.7 |
| 8 days | 17260 | 27600 | 1.6 |
Conclusions
PCL films embedded with 2 to 8 wt % lipase derived from a probiotic source L. plantarum were studied for their enzymatic degradation. TGA analyses showed a substantial decrease (∼100 °C) in the onset thermal decomposition temperature, when compared between the lipase-embedded PCL film before and after subjecting to 8 days of incubation. With the increase in lipase content from 2 to 8%, the thermal decomposition temperature for complete mass loss was found to decrease from 460 to 395 °C. It was found from the DTA analyses that the lipase-embedded control PCL films exhibited percentage crystallinity in the range of 30 to 39%. Among the different loading of lipase, 8% exhibited the highest enzymatic activity on the amorphous regions of the PCL films that resulted in increasing the crystallinity from 39% to 75% by 2 days and to 95% by 8 days. XRD analyses on the 8% lipase-embedded PCL film confirmed that the crystallite sizes of the respective (110) and (200) peaks calculated using Scherrer’s formula were increased by ∼1.5 to 2.1 times after enzymatic polymer degradation. The enzyme release kinetics revealed leaching of ∼50 to 65% of the embedded lipase into the buffer solution by 2 days. Such leaching was found to be beneficial in generating micron-scale pores in the bulk of the polymer film, which provided greater accessibility to the enzymes to freely approach the surface as well as the bulk of the PCL film. The gravimetric analyses revealed that the lowest mass loss of 11% was exhibited by the 2% lipase-embedded polymer film after 2 days and the highest mass loss of 73% was observed with 8% lipase loading after 8 days of incubation. FE-SEM studies revealed a three-dimensional fashion of polymer degradation through surface and cross-sectional morphological imaging. The micron-scale pores were clearly visible in the case of 4 and 6 days of incubated samples, whereas the PCL films incubated for 8 days were found to be crumbled that indicated efficient polymer degradation through this enzyme embedded approach. GPC analyses further corroborated the efficient enzymatic polymer degradation through the substantial decrease in the number-average molecular weight of the 8% lipase-embedded control PCL film from 36500 to 17260 after degradation. It is noteworthy that the present work reports the enzymatic polymer degradation under static conditions, wherein no further lipase was added apart from the initial loading. Thus, the polymer degradation rates reported in this work can substantially be improved by employing shaking conditions and replenishing with additional enzymes periodically. Furthermore, the enzyme-embedded polymer degradation approach shows potential to be extended for polymers containing ester functionalities using lipases from various microbial sources.
Acknowledgments
The authors would like to thank BITS, Pilani Hyderabad campus for their financial support. XRD and FE-SEM facilities of Central Analytical Laboratory of BITS Pilani Hyderabad campus are greatly acknowledged. The authors thank Dr. N. Anbananthan from ion-exchange India Ltd for GPC analyses.
Author Contributions
‡ Both authors contributed equally.
The authors declare no competing financial interest.
References
- Nair L. S.; Laurencin C. T. Biodegradable Polymers as Biomaterials. Prog. Polym. Sci. 2007, 32, 762–798. 10.1016/j.progpolymsci.2007.05.017. [DOI] [Google Scholar]
- Ulery B. D.; Nair L. S.; Laurencin C. T. Biomedical Applications of Biodegradable Polymers. J. Polym. Sci., Part B: Polym. Phys. 2011, 49, 832–864. 10.1002/polb.22259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechty W. B.; Kryscio D. R.; Slaughter B. V.; Peppas N. A. Polymers for Drug Delivery Systems. Annu. Rev. Chem. Biomol. Eng. 2010, 1, 149–173. 10.1146/annurev-chembioeng-073009-100847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y.; Yanful E. K.; Bassi A. S. A Review of Plastic Waste Biodegradation. Crit. Rev. Biotechnol. 2005, 25, 243–250. 10.1080/07388550500346359. [DOI] [PubMed] [Google Scholar]
- Mülhaupt R. Green Polymer Chemistry and Bio-Based Plastics: Dreams and Reality. Macromol. Chem. Phys. 2013, 214, 159–174. 10.1002/macp.201200439. [DOI] [Google Scholar]
- Zaikov G. E.; Lomakin S. M. Ecological Issue of Polymer Flame Retardancy. J. Appl. Polym. Sci. 2002, 86, 2449–2462. 10.1002/app.10946. [DOI] [Google Scholar]
- Royer S.-J.; Ferrón S.; Wilson S. T.; Karl D. M. Production of Methane and Ethylene from Plastic in the Environment. PLoS One 2018, 13, e0200574 10.1371/journal.pone.0200574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimao M. Biodegradation of Plastics. Curr. Opin. Biotechnol. 2001, 12, 242–247. 10.1016/S0958-1669(00)00206-8. [DOI] [PubMed] [Google Scholar]
- Cerniglia C. E. Biodegradation of Polycyclic Aromatic Hydrocarbons. Curr. Opin. Biotechnol. 1993, 4, 331–338. 10.1016/0958-1669(93)90104-5. [DOI] [Google Scholar]
- Bhardwaj H.; Gupta R.; Tiwari A. Communities of Microbial Enzymes Associated with Biodegradation of Plastics. J. Polym. Environ. 2013, 21, 575–579. 10.1007/s10924-012-0456-z. [DOI] [Google Scholar]
- Loredo-Treviño A.; Gutiérrez-Sánchez G.; Rodríguez-Herrera R.; Aguilar C. N. Microbial Enzymes Involved in Polyurethane Biodegradation: A Review. J. Polym. Environ. 2012, 20, 258–265. 10.1007/s10924-011-0390-5. [DOI] [Google Scholar]
- Borja J.; Taleon D. M.; Auresenia J.; Gallardo S. Polychlorinated Biphenyls and Their Biodegradation. Process Biochem. 2005, 40, 1999–2013. 10.1016/J.PROCBIO.2004.08.006. [DOI] [Google Scholar]
- Suzuki M.; Tachibana Y.; Oba K.; Takizawa R.; Kasuya K. Microbial degradation of poly(ε-caprolactone) in a coastal environment. Polym. Degrad. Stab. 2018, 149, 1–8. 10.1016/j.polymdegradstab.2018.01.017. [DOI] [Google Scholar]
- Vroman I.; Tighzert L. Biodegradable Polymers. Materials 2009, 2, 307–344. 10.3390/ma2020307. [DOI] [Google Scholar]
- Khan I.; Dutta J. R.; Ganesan R. Enzymes’ Action on Materials: Recent Trends. J. Cell. Biotechnol. 2016, 1, 131–144. 10.3233/JCB-15013. [DOI] [Google Scholar]
- Miao Z.-M.; Cheng S.-X.; Zhang X.-Z.; Wang Q.-R.; Zhuo R.-X. Degradation and Drug Release Property of Star Poly(ε-Caprolactone)s with Dendritic Cores. J. Biomed. Mater. Res., Part B 2007, 81, 40–49. 10.1002/jbm.b.30634. [DOI] [PubMed] [Google Scholar]
- Labet M.; Thielemans W. Synthesis of Polycaprolactone: A Review. Chem. Soc. Rev. 2009, 38, 3484–3504. 10.1039/b820162p. [DOI] [PubMed] [Google Scholar]
- Shah A. A.; Hasan F.; Hameed A.; Ahmed S. Biological Degradation of Plastics: A Comprehensive Review. Biotechnol. Adv. 2008, 26, 246–265. 10.1016/j.biotechadv.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Banerjee A.; Chatterjee K.; Madras G. Enzymatic Degradation of Polymers: A Brief Review. Mater. Sci. Technol. 2014, 30, 567–573. 10.1179/1743284713Y.0000000503. [DOI] [Google Scholar]
- Yeniad B.; Naik H.; Heise A. Lipases in Polymer Chemistry. Adv. Biochem. Eng. Biotechnol. 2011, 125, 69–95. 10.1007/10_2010_90. [DOI] [PubMed] [Google Scholar]
- Jaeger K.-E.; Ransac S.; Dijkstra B. W.; Colson C.; Heuvel M.; Misset O. Bacterial Lipases. FEMS Microbiol. Rev. 1994, 15, 29–63. 10.1111/j.1574-6976.1994.tb00121.x. [DOI] [PubMed] [Google Scholar]
- Uppada S. R.; Akula M.; Bhattacharya A.; Dutta J. R. Immobilized Lipase from Lactobacillus Plantarum in Meat Degradation and Synthesis of Flavor Esters. J. Genet. Eng. Biotechnol. 2017, 15, 331–334. 10.1016/J.JGEB.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I.; Nagarjuna R.; Ray Dutta J.; Ganesan R. Towards Single Crystalline, Highly Monodisperse and Catalytically Active Gold Nanoparticles Capped with Probiotic Lactobacillus Plantarum Derived Lipase. Appl. Nanosci. 2018, 10.1007/s13204-018-0735-7. [DOI] [Google Scholar]
- Kobayashi S.; Uyama H.; Takamoto T. Lipase-Catalyzed Degradation of Polyesters in Organic Solvents. A New Methodology of Polymer Recycling Using Enzyme as Catalyst. Biomacromolecules 2000, 1, 3–5. 10.1021/bm990007c. [DOI] [PubMed] [Google Scholar]
- a Gunatillake P. A.; Adhikari R. Biodegradable synthetic polymers for tissue engineering. Eur Cell Mater. 2003, 5, 1–16. 10.22203/eCM.v005a01. [DOI] [PubMed] [Google Scholar]; b Pathak V. M.; Navneet Review on the Current Status of Polymer Degradation: A Microbial Approach. Bioresour. Bioprocess. 2017, 4, 15. 10.1186/s40643-017-0145-9. [DOI] [Google Scholar]
- Dash T. K.; Konkimalla V. B. Poly-ϵ-Caprolactone Based Formulations for Drug Delivery and Tissue Engineering: A Review. J. Control. Release 2012, 158, 15–33. 10.1016/J.JCONREL.2011.09.064. [DOI] [PubMed] [Google Scholar]
- Malikmammadov E.; Tanir T. E.; Kiziltay A.; Hasirci V.; Hasirci N. PCL and PCL-Based Materials in Biomedical Applications. J. Biomater. Sci. Polym. Ed. 2018, 29, 863–893. 10.1080/09205063.2017.1394711. [DOI] [PubMed] [Google Scholar]
- Gan Z.; Yu D.; Zhong Z.; Liang Q.; Jing X. Enzymatic Degradation of Poly(ε-Caprolactone)/Poly(Dl-Lactide) Blends in Phosphate Buffer Solution. Polymer 1999, 40, 2859–2862. 10.1016/S0032-3861(98)00549-7. [DOI] [Google Scholar]
- Ganesh M.; Dave R. N.; L’Amoreaux W.; Gross R. A. Embedded Enzymatic Biomaterial Degradation. Macromolecules 2009, 42, 6836–6839. 10.1021/ma901481h. [DOI] [Google Scholar]
- Ganesh M.; Gross R. A. Embedded Enzymatic Biomaterial Degradation: Flow Conditions & Relative Humidity. Polymer 2012, 53, 3454–3461. 10.1016/j.polymer.2012.06.017. [DOI] [Google Scholar]
- Khan I.; Ray Dutta J.; Ganesan R. Lactobacillus Sps. Lipase Mediated Poly (ε-Caprolactone) Degradation. Int. J. Biol. Macromol. 2017, 95, 126–131. 10.1016/j.ijbiomac.2016.11.040. [DOI] [PubMed] [Google Scholar]
- Schäler K.; Achilles A.; Bärenwald R.; Hackel C.; Saalwächter K. Dynamics in Crystallites of Poly(ε-Caprolactone) As Investigated by Solid-State NMR. Macromolecules 2013, 46, 7818. 10.1021/ma401532v. [DOI] [Google Scholar]
- Obregon N.; Agubra V.; Pokhrel M.; Campos H.; Flores D.; de la Garza D.; Mao Y.; Macossay J.; Alcoutlabi M. Effect of Polymer Concentration, Rotational Speed, and Solvent Mixture on Fiber Formation Using Forcespinning®. Fibers 2016, 4, 20. 10.3390/fib4020020. [DOI] [Google Scholar]
- Eldsäter C.; Erlandsson B.; Renstad R.; Albertsson A.-C.; Karlsson S. The Biodegradation of Amorphous and Crystalline Regions in Film-Blown Poly(ϵ-Caprolactone). Polymer 2000, 41, 1297–1304. 10.1016/S0032-3861(99)00278-5. [DOI] [Google Scholar]
- Yoshie N.; Oike Y; Kasuya K.-i.; Doi Y.; Inoue Y. Change of surface structure of poly (3-hydroxybutyrate) film upon enzymatic hydrolysis by PHB depolymerase. Biomacromolecules 2002, 3, 1320–1326. 10.1021/bm020077a. [DOI] [PubMed] [Google Scholar]
- Gupta K. K.; Kundan A.; Mishra P. K.; Srivastava P.; Mohanty S.; Singh N. K.; Mishra A.; Maiti P. Polycaprolactone Composites with TiO2 for Potential Nanobiomaterials: Tunable Properties Using Different Phases. Phys. Chem. Chem. Phys. 2012, 14, 12844. 10.1039/C2CP41789H. [DOI] [PubMed] [Google Scholar]
- Pang X.; Jiang Y.; Xiao Q.; Leung A. W.; Hua H.; Xu C. pH-Responsive Polymer–drug Conjugates: Design and Progress. J. Control. Release 2016, 222, 116–129. 10.1016/j.jconrel.2015.12.024. [DOI] [PubMed] [Google Scholar]
- Ganesh M.; Nachman J.; Mao Z.; Lyons A.; Rafailovich M.; Gross R. Patterned enzymatic degradation of poly (ε-caprolactone) by high-affinity microcontact printing and polymer pen lithography. Biomacromolecules 2013, 14, 2470–2476. 10.1021/bm400552u. [DOI] [PubMed] [Google Scholar]
- Mao Z.; Ganesh M.; Bucaro M.; Smolianski I.; Gross R. A.; Lyons A. M. High throughput, high resolution enzymatic lithography process: Effect of crystallite size, moisture, and enzyme concentration. Biomacromolecules 2014, 15, 4627–4636. 10.1021/bm501475n. [DOI] [PubMed] [Google Scholar]