Abstract
The eIF4E-homologous protein (4EHP) is a translational repressor that competes with eIF4E for binding to the 5′-cap structure of specific mRNAs, to which it is recruited by protein factors such as the GRB10-interacting GYF (glycine-tyrosine-phenylalanine domain) proteins (GIGYF). Several experimental evidences suggest that GIGYF proteins are not merely facilitating 4EHP recruitment to transcripts but are actually required for the repressor activity of the complex. However, the underlying molecular mechanism is unknown. Here, we investigated the role of the uncharacterized Drosophila melanogaster (Dm) GIGYF protein in post-transcriptional mRNA regulation. We show that, when in complex with 4EHP, Dm GIGYF not only elicits translational repression but also promotes target mRNA decay via the recruitment of additional effector proteins. We identified the RNA helicase Me31B/DDX6, the decapping activator HPat and the CCR4–NOT deadenylase complex as binding partners of GIGYF proteins. Recruitment of Me31B and HPat via discrete binding motifs conserved among metazoan GIGYF proteins is required for downregulation of mRNA expression by the 4EHP–GIGYF complex. Our findings are consistent with a model in which GIGYF proteins additionally recruit decapping and deadenylation complexes to 4EHP-containing RNPs to induce translational repression and degradation of mRNA targets.
INTRODUCTION
During translation initiation, the small ribosomal subunit is recruited to mRNA by the eukaryotic initiation factor 4F (eIF4F) complex, a heterotrimeric complex that recognizes the cap structure at the 5′ end of the mRNA. Cap binding is accomplished by the eIF4E subunit of the complex, a direct binding partner of the scaffold subunit eIF4G that in turn associates with the RNA helicase eIF4A. eIF4G also mediates recruitment of the preinitiation complex, consisting of the 40S ribosomal subunit and associated factors, to trigger a series of steps leading to initiation of protein synthesis (1,2).
Several translational control mechanisms regulate assembly of the eIF4F complex at the mRNA cap structure (1,2). The eIF4E-homologous protein (4EHP or eIF4E-2), is a cap-binding protein that, unlike eIF4E, is unable to interact with eIF4G and thus blocks translation initiation (3,4). 4EHP regulates translation in a transcript-specific manner as it is recruited via interactions with specific RNA-binding proteins (RBPs). These interactions are mediated by the conserved motif YXYX4LΦ (where Y, X, L and Φ represent Tyr, any amino acid, Leu, and hydrophobic residue, respectively), termed the canonical (C) 4EHP-binding motif, present on 4EHP-binding proteins (5,6).
In metazoans, 4EHP complexes regulate mRNA expression in a variety of biological processes. Translational control by 4EHP is crucial for the specification of Drosophila melanogaster (Dm) embryonic patterning (5,7), for viability and for completion of development (8). During fly embryogenesis, the RBPs Bicoid and Brain Tumor recruit 4EHP to repress the translation of caudal and hunchback mRNAs, respectively, at precise spatial locations in the embryo (5,7). Murine 4EHP binds to Prep1 and inhibits the translation of Hoxb4 mRNA during oogenesis (9). 4EHP was also shown to associate with the mammalian miRNA-induced silencing complex (miRISC) together with the 4EHP-interaction partner known as eIF4E-transporter (4E-T) and the CCR4–NOT deadenylase complex (10,11).
4EHP also forms a translational repressor complex with the GIGYF1/2 proteins [Grb10-interacting GYF (glycine-tyrosine-phenylalanine) protein 1 and 2]. Metazoan GIGYF proteins were first described as regulators of the insulin signaling pathway (12) and are characterized by the presence of a small globular GYF domain that binds to proteins containing the proline-rich motif PPGФ [where Ф is any hydrophobic amino acid with the exception of Trp (13)]. To bind specifically to 4EHP, GIGYF1/2 proteins use, in addition to the canonical 4EHP-binding motif, noncanonical (NC) and auxiliary (A) motifs that interact with three binding surfaces on 4EHP [dorsal (D), lateral (L) and 4EHP-specific (S), respectively; Supplementary Figure S1A; (14)]. Like 4EHP, GIGYF1/2 are required for proper embryonic development as gene disruption causes perinatal lethality in mice (6,15).
The mammalian 4EHP–GIGYF complex can be recruited to target mRNAs via association with proteins containing PPGФ motifs. Namely, the zinc finger protein ZNF598 directs the complex to mRNAs essential for normal mouse development (6). Moreover, binding of 4EHP–GIGYF2 to tristetraprolin (TTP) modulates the expression of mRNAs with AU-rich elements in the 3′ untranslated region [UTR; (16,17)]. GIGYF2 has also been identified as a miRISC-associated factor that binds to TNRC6 proteins, facilitating the repression of miRNA-targets (18,19).
Recent evidence suggests that GIGYF1/2 proteins do not act solely as 4EHP mRNA recruiting factors but can directly participate in the repressor function of the complex. In fact, regulation of translation initiation by 4EHP is compromised in GIGYF1/2-null cells (14). Human GIGYF2 also binds directly to a subset of mRNAs and recruits the CCR4–NOT complex to control their expression (20). Therefore, a detailed knowledge of the molecular mechanisms mediated by GIGYF proteins would provide valuable insight into the alternative regulation of translation initiation by the cap binding protein 4EHP.
In this work, we investigated the molecular functions of the uncharacterized Dm 4EHP–GIGYF repressor complex and demonstrate that it couples translational repression with the regulation of mRNA stability. In this context, the GIGYF protein is essential for the repressor function of the complex as it acts as a scaffold protein that mediates interactions with multiple effector proteins. In addition to 4EHP, GIGYF recruits the RNA helicase Me31B/DDX6, the decapping activator HPat and the CCR4–NOT deadenylase complex to induce translational repression and decay of a target mRNA. We further report that recruitment of these effector proteins is conserved among metazoan GIGYF proteins and mediated by discrete binding motifs. Our data suggest a conserved and previously unappreciated mechanism for the regulation of translation initiation and mRNA decay by the 4EHP–GIGYF repressor complex.
MATERIALS AND METHODS
DNA constructs
Plasmids for the expression of green fluorescent protein (GFP)-tagged and λN-hemagglutinin (HA)-tagged Dm 4EHP were obtained by inserting the cDNA corresponding to the 4EHP ORF into the EcoRI and XbaI restriction sites of the pAC5.1B-EGFP and pAC5.1B-λN-HA vectors. Plasmids for the expression of GFP-tagged and λN-HA-tagged Dm GIGYF were obtained by inserting the cDNA corresponding to the GIGYF ORF into the EcoRI and NotI restriction sites of the pAC5.1B-EGFP and pAC5.1B-λN-HA vectors. GIGYF N-terminal (residues M1-N640) and C-terminal (residues I641-P1574) fragments were amplified by PCR, using the plasmid containing full length (FL) GIGYF as a template, and cloned into the same vectors and restriction sites as the FL construct. Double-stranded RNA (dsRNA)-resistant GIGYF proteins were obtained by insertion of an optimized (resistant to the GIGYF dsRNA) synthetic cDNA fragment (GeneArt), composed of the first 700 bp of GIGYF sequence, into the pAC5.1B-λN-HA-GIGYF vector by site directed mutagenesis. This mutagenesis reaction also removed the λN sequence from the final DNA construct (HA-GIGYF dsRNA resistant). The plasmid for the expression of GFP-tagged Dm Bicoid was obtained by inserting the cDNA corresponding to the Bicoid ORF into the EcoRV and XhoI restriction sites of the pAC5.1B-EGFP vector. The plasmid for the expression of λN-HA-tagged Dm Brat was obtained by inserting the cDNA of the corresponding ORF into the HindIII and NotI restriction sites of the pAC5.1B-λN-HA vector. The plasmid for the expression of GFP-tagged Dm Tis11 was obtained by inserting the corresponding cDNA into the EcoRI and NotI restriction sites of the pAC5.1B-EGFP vector. The plasmid for the expression of GFP-tagged ZNF598 was obtained by inserting the corresponding ORF cDNA into the EcoRV and NotI restriction sites of the pAC5.1B-EGFP vector. The plasmids used for expression of the luciferase reporters and the GFP-, V5- or HA-tagged subunits of the Dm CCR4–NOT and PAN2-PAN3 deadenylase complexes, the decapping factors, Dm XRN1, MBP, Dm GW182, Dm 4E-T, MS2-HA-Hs GIGYF2 or Hs PATL1 were previously described (14,21–28).
The plasmid for the expression of V5-Streptavidin binding protein (SBP)-tagged Hs GIGYF1 was obtained by inserting the corresponding cDNA into the XhoI and EcoRI restriction sites of the pT7-V5-SBP-C1 vector. The V5-SBP-Hs GIGYF2 construct was obtained by inserting the corresponding cDNA into the XhoI and BamHI sites of the pT7-V5-SBP-C1 vector. The plasmid for the expression of the MS2-HA-Hs GIGYF1 construct was obtained by inserting the corresponding cDNA into the XhoI and NotI sites of the pcDNA3.1-MS2-HA vector.
To generate plasmids for expression in Escherichia coli, DNA fragments coding for the Dm GIGYF GYF domain (residues P553-H621) were inserted into the NdeI and NheI restriction sites of the pnYC-NpG-GB1 vector. This construct expresses the GIGYF GYF domain fused N-terminally to a Glutathione S-Transferase (GST)-tag cleavable by the HRV3C protease (29) and C-terminally, following a ASG (Ala-Ser-Gly) linker, to the DNA encoding the B1 domain of immunoglobulin-domain protein G (GB1). The DNA sequence coding for the P-rich region of Dm HPat (P-reg; residues G57-Y499) was cloned into the NdeI and NheI restriction sites of the pnEA-NpM-GB1 vector which allows the expression of HPat P-reg with an N-terminal MBP tag cleavable by the HRV3C protease (29), and a C-terminal ASG peptide fused to the GB1 domain.
All of the mutants used in this study were generated by site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis kit (Stratagene). All of the constructs and mutations were confirmed by sequencing and are listed in Supplementary Table S1.
Tethering assays and dsRNA interference
For the λN-tethering assay, 2.5 × 106 S2 cells were cotransfected in 6-well plates, using Effectene (Qiagen). The transfection mixtures contained 0.1 μg of reporter plasmid [firefly luciferase (F-Luc)-5BoxB, F-Luc-V5 or F-Luc-5BoxB-A95C7-HhR)], 0.4 μg of the Renilla luciferase (R-Luc)-A90-HhR control reporter and different amounts of the plasmids expressing the λN-HA-fusion proteins. In the experiments with the λN-HA-4EHP fusion proteins the following amounts were used: 0.05 μg of wild-type (WT), 0.08 μg of D*, L* and S* mutants and 0.05 μg of the CAP* mutant. The λN-HA-GIGYF plasmids were transfected together with the F-Luc-5BoxB or the F-Luc-V5 reporters according to the following amounts: 0.01 μg of WT, C-term and C*, GYF*, C*+GYF*, ΔMBM or ΔMBM+GYF* mutants, and 0.005 μg of N-term. In the experiments containing the F-Luc reporter with an internal poly(A) tail, 0.005 μg of all plasmids expressing the λN-HA-GIGYF proteins were used in the transfection reaction. In the experiment shown in Figure 2E and F, the transfection mixtures also contained plasmids expressing λN-HA-GW182 (0.01 μg), GFP-V5 (0.025 μg) or the V5-tagged DCP1 GSSG-mutant (1.0 μg). The R-Luc control reporter (R-Luc-A90-HhR) contains an internal polyadenosine stretch of 90 nucleotides followed by a self-cleaving hammerhead ribozyme in place of the cleavage and polyadenylation site to prevent deadenylation and mRNA decay (25).
Figure 2.
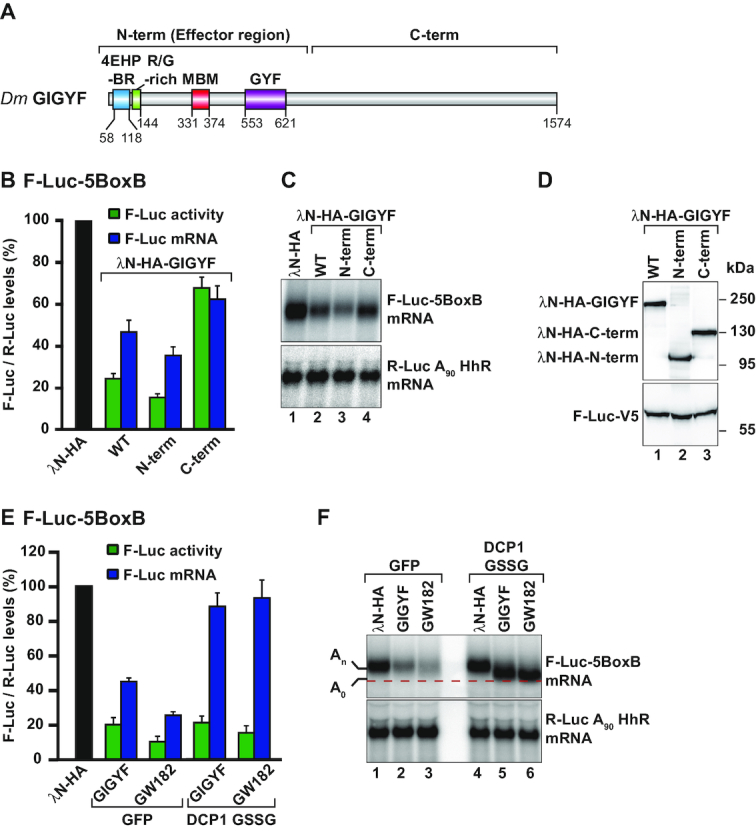
Dm GIGYF represses translation and induces mRNA decay. (A) Schematic representation of the Dm GIGYF protein. The N-term contains a 4EHP-binding region (4EHP-BR), an Arg/Gly-rich sequence adjacent to the 4EHP-BR, a Me31B binding motif (MBM) and a glycine-tyrosine-phenylalanine (GYF) domain of the Smy2-type (13). The C-term does not contain any known domain and is predicted to contain primarily α-helices. The amino acid positions at the domain/motif boundaries are indicated below the protein. (B–D) Tethering assay using the F-Luc-5BoxB reporter and the indicated λN-HA-tagged proteins in S2 cells. Samples were analyzed as described in Figure 1A. Relative luciferase activity (green bars), relative reporter mRNA levels (blue bars) and representative northern and western blot images are depicted in panels B, C and D, respectively. (E, F) A tethering assay with the F-Luc-5BoxB reporter and the indicated λN-HA-tagged proteins performed in control cells (GFP-V5-expressing cells) and in cells expressing a DCP1 GSSG mutant. GW182 was used as a positive control for deadenylation-dependent mRNA decapping. Samples were analyzed as described in Figure 1A. In panel F, the red dashed line marks the position of the deadenylated (A0) F-Luc-5BoxB reporter mRNA after northern blot analysis of representative RNA samples. An indicates the position of the adenylated reporter mRNA.
In the complementation assay presented in Figure 5, 0.03 μg of the plasmid expressing WT or a mutant dsRNA-resistant version of the GIGYF proteins was added to the transfection mixture.
Figure 5.
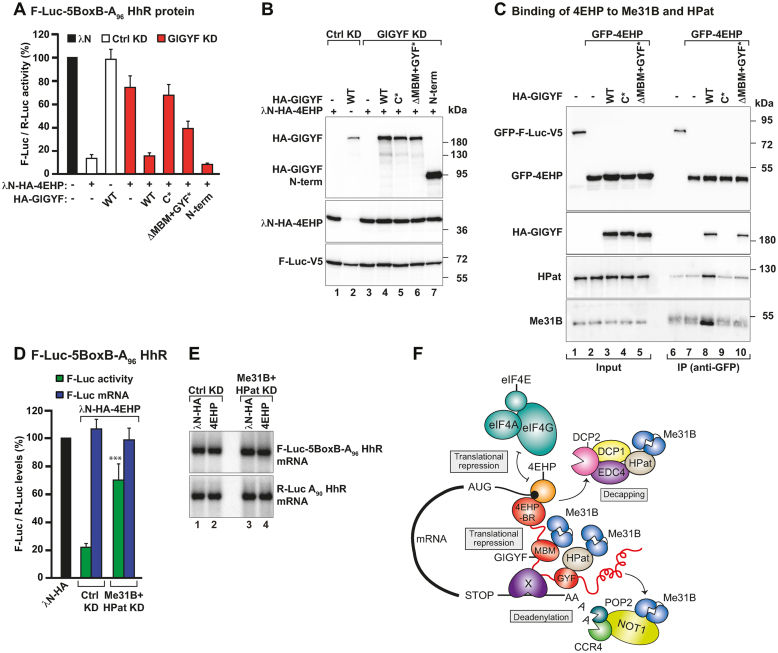
GIGYF recruits Me31B and HPat to 4EHP-repressor complexes. (A) A complementation assay using the F-Luc-5BoxB-A96-HhR reporter and λN-HA-4EHP was performed in Ctrl and GIGYF-depleted cells expressing HA-GIGYF proteins (dsRNA-resistant and either WT, mutants or the N-term). A plasmid expressing R-Luc-A90-HhR served as a transfection control. Samples were analyzed as described in Figure 1A. (B) Western blot analysis showing the expression of the λN-HA-4EHP, HA-GIGYF and F-Luc-V5 proteins used in the complementation assay. (C) Interaction of GFP-4EHP with endogenous Me31B and HPat proteins assayed by anti-GFP immunoprecipitation in the presence or absence of HA-GIGYF (WT or mutants). GFP-F-Luc-V5 was used as a negative control. The inputs (4% for GFP- and HA-tagged proteins and 0.1% for Me31B and HPat) and bound fractions (5% for GFP- and HA-tagged proteins and 20% for Me31B and HPat) were analyzed by western blotting using anti-GFP, anti-HA, anti-Me31B and anti-HPat antibodies. (D, E) S2 cells were treated with dsRNA targeting neomycin (Ctrl KD) or Me31B and HPat mRNAs (Me31B+HPat KD) and transfected with a mixture of three plasmids coding for F-Luc-5BoxB-A96-HhR, R-Luc-A90-HhR and the indicated λN-HA-tagged proteins. In panel D, F-Luc activity and mRNA levels were normalized to that of R-Luc in the presence of the different tethered proteins. Samples were analyzed as described in Figure 1A. Panel E shows a representative northern blot. The P value (***P < 0.0005) was determined using the two-tailed Student's t test. (F) Model for the assembly of a 4EHP-repressor complex. 4EHP (orange) is the cap-binding protein that blocks eIF4F mRNA recruitment. GIGYF (red) provides binding sites for 4EHP (4EHP-BR), Me31B (MBM), HPat (GYF domain) and the CCR4–NOT deadenylase complex. Additionally, GIGYF might also bind an unknown RNA-binding protein (X) or directly interact with the target mRNA. HPat and the CCR4–NOT complex also can interact with Me31B (24,40,41). The formation of this complex ensures that multiple mechanisms are employed to robustly suppress target mRNA expression: translational repression, deadenylation and decapping, which is subsequently followed by degradation of the mRNA.
Knockdowns using dsRNA were performed as previously described (21). Cells were harvested three days after transfection and the F-Luc and R-Luc luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega), and total RNA was isolated using TriFast (Peqlab Biotechnologie). RNA samples were analyzed by northern blot as described previously (21).
To measure the mRNA half-lives, cells were treated with actinomycin D (5 μg/ml final concentration) three days post-transfection and collected at the indicated time points. mRNA reporter levels were normalized to the levels of endogenous rp49 mRNA. These values were set to 100 at time point zero. Data points from three independent experiments were plotted in a graph with logarithmic scale in the y axis.
Coimmunoprecipitation assays and Western blotting
For coimmunoprecipitation (coIP) assays, S2 cells (2.5 × 106 cells per well in six-well plates and two wells per condition) were transfected using Effectene reagent (Qiagen). Cells were harvested three days after transfection and coIP assays were performed in the presence of RNaseA as described previously (22). The majority of our Dm coIP assays were performed with a homemade polyclonal anti-GFP antibody with the exception of Supplementary Figures S1H, S4F and G and S7A. In Supplementary Figure S4F and G, we used an anti-V5 antibody to immunoprecipitate as baits the low abundant Dm DCP2 and XRN1 proteins. These proteins have an increased stability and expression levels in S2 cells when tagged with V5 compared to GFP (our unpublished data). All Western blots were developed with the ECL Western Blotting Detection System (GE Healthcare) according to the manufacturer's recommendations. Antibodies used in this study are listed in Supplementary Table S2. All coIP and pulldown assays in HEK293T cells were performed in the presence of RNaseA as described previously (28).
Pulldown assays
The pulldown assays were performed as previously described (30,31). Briefly, bacterial lysates expressing recombinant GST-Dm GIGYF GYF-GB1 were incubated with glutathione agarose beads (Macherey-Nagel) for 30 min in buffer containing 50 mM sodium phosphate (pH 7.5), 200 mM sodium chloride, 2 mM of β-mercaptoethanol and 40 mM imidazole. The immobilized GST-GIGYF GYF was then incubated for 30 min with bacterial lysates expressing the Dm HPat P-rich region tagged N-terminally with MBP and C-terminally with GB1 or MBP alone as a control. The proteins associated with GIGYF GYF domain were eluted with glutathione and analysed by SDS-PAGE followed by Coomassie Blue staining.
RESULTS
Dm 4EHP represses translation and induces decay of bound mRNAs
To investigate the mechanism by which Dm 4EHP–GIGYF inhibits the expression of target mRNAs, we made use of a reporter assay. We tethered 4EHP fused to the N protein from the bacteriophage λ (λN-HA-4EHP) to a firefly luciferase (F-Luc) reporter mRNA. The reporter contains five BoxB hairpins (5BoxB) in the 3′ UTR to which the λN peptide binds with high affinity (32). Expression of the F-Luc-5BoxB mRNA in Drosophila S2 cells was analyzed in the absence (λN-HA) and the presence of the λN-HA-4EHP fusion protein by monitoring protein levels (F-Luc activity, green bars in the graphs) and steady-state mRNA levels (F-Luc mRNA, blue bars in the graphs). In all experiments a plasmid encoding Renilla luciferase (R-Luc) was included as a transfection and normalization control. We found that in the presence of λN-HA-4EHP F-Luc activity was repressed to 20% relative to that of λN-HA alone (set to 100%; Figure 1A). The repression was specific as the expression of a similar F-Luc reporter lacking the BoxB hairpins (F-Luc-V5), or of the R-Luc reporter, were unaffected by λN-HA-4EHP (Supplementary Figure S1B–E). Thus, under these experimental conditions, 4EHP cannot repress translation without recruitment to mRNA. Interestingly, the reduction in F-Luc activity was accompanied by a 50% decrease in F-Luc mRNA steady-state levels, as determined by northern blotting (Figure 1A and B, lanes 2 versus 1), suggesting that 4EHP also elicits degradation of the tethered mRNA. However, the reduction in F-Luc mRNA levels (50%) was not as strong as the decrease in F-Luc activity (to 20%; Figure 1A) indicating that 4EHP might primarily act as a translational repressor.
Figure 1.
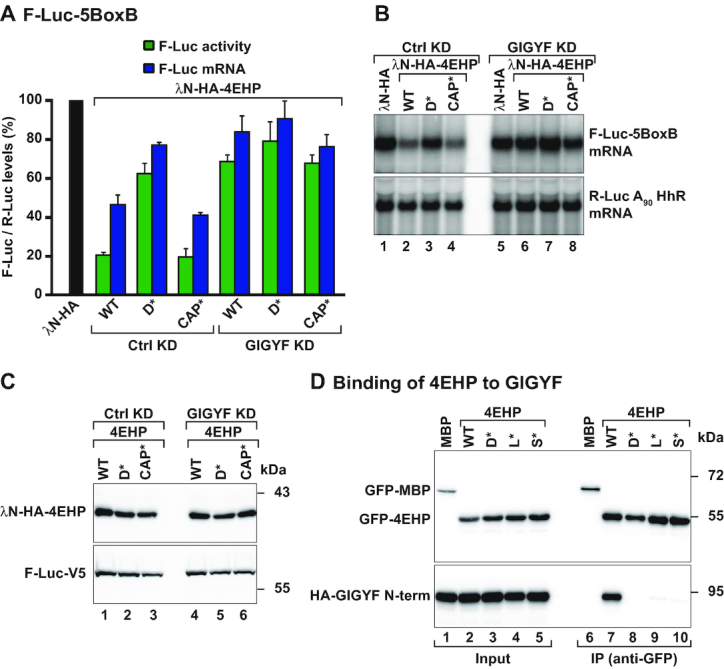
Dm 4EHP requires Dm GIGYF to downregulate reporter mRNA expression. (A) Tethering assay using the firefly luciferase (F-Luc)-5BoxB reporter and the indicated λN-HA-tagged proteins in control [Ctrl knockdown (KD): neomycin dsRNA-treated cells] or GIGYF-depleted Dm S2 cells (GIGYF KD). A plasmid expressing Renilla luciferase (R-Luc)-A90-HhR served as a transfection control. The F-Luc activity (green bars) and mRNA levels determined by northern blotting (blue bars) were normalized to those of the R-Luc transfection control and set to 100% in cells expressing the λN-HA peptide (black bar). Bars represent the mean values and error bars denote the standard deviation from at least three independent experiments. (B) Northern blot analysis of a representative tethering experiment shown in (A). (C) Western blot showing the expression levels of the tethered proteins. (D) Immunoprecipitation assay showing the interaction between GFP-tagged 4EHP WT or the indicated mutants (dorsal: D*; lateral: L*; and 4EHP-specific: S*) and HA-tagged GIGYF N-term. The proteins were immunoprecipitated using an anti-GFP antibody. GFP-MBP (maltose binding protein) served as a negative control. The input (2.8% of the total lysate) and bound fractions (10%) were analyzed by western blotting using anti-GFP and anti-HA antibodies.
Dm 4EHP requires GIGYF to repress translation and induce mRNA decay
We then tested the association of overexpressed and tagged 4EHP (either HA or GFP)- with known binding partners using coIP assays. Specifically, we analyzed whether Dm 4EHP can associate with Dm Bicoid, Dm Brat, or the fly orthologs of human 4E-T and GIGYF1/2 (5–7,33). In Dm S2 cells, 4EHP efficiently interacted with Dm GIGYF, which is 21/23% identical and 32/36% similar to human GIGYF1 and GIGYF2, respectively [CG11148; Supplementary Figures S1F, lane 8 and S1G, lane 6; DRSC Integrative Orthologue Prediction Tool, Harvard Medical School; (34)]. In contrast, Dm 4EHP did not bind to Brat, Bicoid and only weakly with 4E-T (Supplementary Figure S1F–H). The binding of Bicoid and Brat to 4EHP in embryos is likely to be indirect (7,14) and possibly mediated by a protein that is absent in S2 cells. Human 4E-T binds both eIF4E and 4EHP (33,35). In our coIP assay, overexpressed Dm 4E-T preferentially interacted with HA-eIF4E (Supplementary Figure S1H). It is unclear why the Dm and the human 4E-T proteins differ in terms of binding to 4EHP.
In an attempt to disrupt binding of 4EHP to GIGYF, we introduced specific mutations in the dorsal (D*), lateral (L*) and 4EHP-specific (S*) binding surfaces using the crystal structure of the human 4EHP–GIGYF complex as a model [Supplementary Figures S1A and S2A; (14)]. All the substitutions disrupted the interaction of GFP-tagged 4EHP with an N-terminal (N-term) fragment of GIGYF which contains the 4EHP-binding region (Figure 1D, lanes 7 versus 8–10 and Figure 2A). These results indicate that Dm GIGYF interacts with 4EHP via distinct binding motifs termed canonical, noncanonical and auxiliary, as observed for the human homologous proteins [Supplementary Figure S1A; (14)].
To test if the interaction with Dm GIGYF was required for 4EHP to regulate expression of the reporter mRNA, we tethered the 4EHP D* mutant to the F-Luc-5BoxB reporter. We observed that this mutant was strongly impaired in its ability to reduce F-Luc protein and mRNA levels (Figure 1A and B, lanes 3 versus 1 and 2). We also examined if cap binding was necessary for 4EHP to regulate the expression of the reporter mRNA. Therefore, we used a 4EHP protein carrying amino acid changes in the cap-binding pocket [CAP*; Supplementary Figure S2A, (7)]. Tethering the 4EHP CAP* mutant protein, which still interacts with GIGYF (Supplementary Figure S2B, lane 8), reduced F-Luc activity and induced mRNA degradation as efficiently as the WT protein (Figure 1A and B, lanes 4 versus 1). All 4EHP proteins were expressed at comparable levels (Figure 1C, lanes 1–3) and did not affect the expression of the F-Luc reporter missing the BoxB elements (Supplementary Figure S1B–E). Recruitment of 4EHP to the F-Luc reporter mRNA via the BoxB elements probably circumvents its RNA/cap-binding function, a property that might be essential when considering natural mRNA targets. Upon mRNA binding, the interaction of 4EHP with GIGYF then leads to translational repression and mRNA decay. These results therefore suggest that 4EHP requires direct binding to GIGYF to control mRNA expression.
To investigate if GIGYF is the only 4EHP-binding partner mediating translational repression and mRNA decay, we examined the repressive function of 4EHP in cells depleted of endogenous GIGYF [GIGYF knockdown (KD)]. As there are no antibodies against Dm GIGYF, we estimated the efficacy and specificity of the knockdown by analyzing the expression of HA-tagged GIGYF in cells treated with dsRNA targeting either neomycin as the control (Ctrl) or the Dm GIGYF mRNA (Supplementary Figure S2C). In cells depleted of GIGYF, tethered 4EHP was unable to repress expression of the F-Luc-5BoxB reporter (F-Luc activity and mRNA levels) to the same extent as in control cells, even though λN-HA-4EHP levels did not change. In fact, F-Luc activity decreased only to 70% instead of the 20% observed in the presence of GIGYF (Figure 1A–C). Furthermore, the 4EHP CAP* mutant also repressed F-Luc activity and induced mRNA degradation in a GIGYF-dependent manner (Figure 1A–C). Thus, our results suggest that Dm GIGYF is essential for the function of the 4EHP–GIGYF complex as a repressor of mRNA expression, as seen in human cells (14,20).
Dm GIGYF represses translation and triggers mRNA degradation
To investigate the role of GIGYF in 4EHP-mediated regulation of mRNA expression, we tethered λN-HA-GIGYF to the F-Luc 5BoxB reporter. We observed that GIGYF efficiently reduced F-Luc activity and mRNA levels (Figure 2B–D) in a manner that was comparable to the tethering of 4EHP. The binding of GIGYF to the reporter mRNA decreased F-Luc activity to 25% and the abundance of the reporter mRNA to 50% (Figure 2B and C) relative to the λN-HA control protein. Reporter mRNA repression was specific, as it required binding to the target mRNA and could not be observed for an F-Luc reporter lacking the BoxB hairpins (Supplementary Figure S3A and B).
To determine whether the reduction of reporter mRNA levels by GIGYF was a consequence of reduced mRNA stability rather than transcriptional blockage, we treated the transfected cells with a transcriptional inhibitor (actinomycin D) and monitored the levels of F-Luc-5BoxB mRNA over time. The stable rp49 mRNA was used as a normalization control. In the presence of GIGYF the half-life of the F-Luc-5BoxB mRNA was significantly shorter (17 ± 4.0 min) than the half-life of the same RNA in cells expressing the control λN-HA (130 ± 26.5 min; Supplementary Figure S3C and D). Together, these results indicate that Dm GIGYF represses translation and accelerates the degradation of a bound mRNA.
The N-terminal fragment of GIGYF defines the effector region of the protein
GIGYF proteins consist of a mainly unstructured N-terminal half, comprising the 4EHP-binding region (4EHP-BR) and the GYF-domain, and a long α-helical C-terminal half (Figure 2A). We analyzed the potential of separate N- or C-terminal (C-term) protein fragments to repress the expression of F-Luc-5BoxB reporter (Figure 2A and Supplementary Table S1). We observed that the N-term reduced F-Luc activity (to less than 20%) and reporter mRNA levels (to 40%) to a similar extent as full length GIGYF (Figure 2B and C). The C-term GIGYF fragment, expressed at a comparable level, reduced expression of the reporter mRNA only partially (to 70%) and had a similar effect on F-Luc activity and mRNA levels (Figure 2B–D). None of the proteins repressed an F-Luc reporter lacking the BoxB structures (Supplementary Figure S3A and B). Since full length (WT) and the N-term of GIGYF induced a stronger reduction in F-Luc activity compared to mRNA level, which was also observed for 4EHP (Figure 2B, F-Luc activity versus F-Luc mRNA), it appears that the N-term of Dm GIGYF defines the major effector region of the protein.
GIGYF promotes deadenylation-dependent mRNA decapping
One of the mRNA decay mechanisms operating in the cytoplasm of eukaryotic cells involves the shortening of the poly(A) tail at the 3′ end of the mRNA by the CCR4–NOT deadenylase complex and the hydrolysis of the cap structure through the activity of the decapping complex (which includes the decapping enzyme DCP2 and its associated factors). Once decapped, mRNAs are rapidly degraded by the exonuclease XRN1 in a 5′-to-3′ direction (36).
As human GIGYF2 controls the expression of a subset of mRNAs via the recruitment of the CCR4–NOT complex (20), we hypothesized that Dm GIGYF might cause deadenylation-dependent decapping of bound mRNAs. To test this possibility, we performed tethering assays in cells overexpressing a mutant version of the decapping activator DCP1 (DCP1 GSSG, Supplementary Table S1), which blocks DCP2 activity in a dominant negative manner (27). In these cells, degradation of the reporter mRNA by tethering GIGYF or GW182 [the latter acting as a positive control known to trigger deadenylation-dependent decapping (21,37)] was inhibited and resulted in the accumulation of a mRNA decay intermediate lacking the poly(A) tail (A0, Figure 2E and F, lanes 2–3 versus 5–6). The deadenylated reporter mRNA was therefore translationally silenced (Figure 2E and Supplementary Figure S3E).
Dm GIGYF interacts with deadenylation and decapping factors
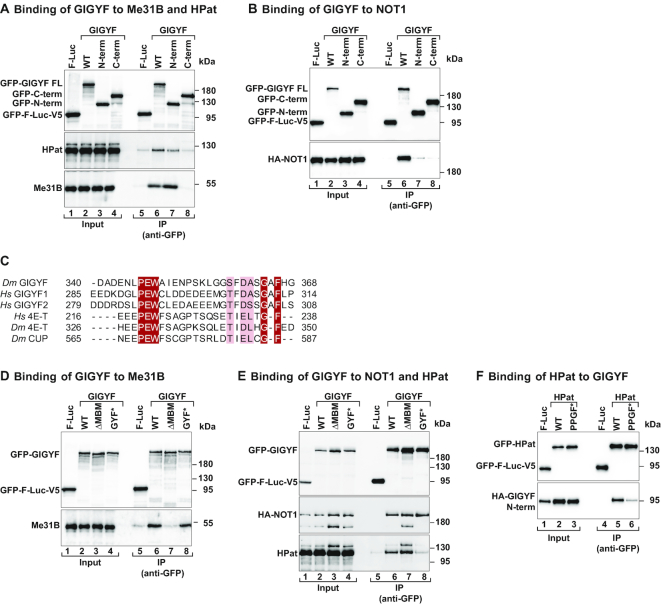
Given that GIGYF triggers decay of a bound reporter mRNA, we performed coIP experiments to analyze its interaction with endogenous or overexpressed proteins involved in cytoplasmic mRNA deadenylation and decapping. We observed that overexpressed Dm GIGYF associated with several decapping activators, namely, Me31B/DDX6, HPat, EDC3 and EDC4 (Figure 3A, Supplementary Figure S4A and D, Supplementary Table S3), and different subunits of the CCR4–NOT deadenylase complex such as NOT1, NOT3 and POP2 (Figure 3B, Supplementary Figure S5A and B and Supplementary Table S3). Notably, these interactions are conserved, as the human GIGYF1 and GIGYF2 proteins also bind to the decapping factors PatL1 (Dm HPat ortholog, Supplementary Table S3 and Supplementary Figure S4B and C) and DDX6 [Dm Me31B ortholog, Supplementary Table S3 and Supplementary Figure S5C and (20)], and to the CCR4–NOT complex subunits CNOT1, CNOT2 and CNOT3 [Supplementary Table S3 and Supplementary Figure S5C and (20)]. In agreement with its role in mRNA decay, Dm GIGYF weakly coimmunoprecipitated other components of the deadenylation complexes, such as CCR4, NOT2, PAN2 and PAN3 (Supplementary Table S3 and Supplementary Figure S5B and D). Dm GIGYF however did not bind to DCP1, DCP2 and XRN1 as well as CAF40 (Supplementary Table S3, Supplementary Figure S4E–G and Supplementary Figure S5E).
Figure 3.
Dm GIGYF interacts with components of the decapping and deadenylation machineries. (A) Immunoprecipitation assay showing the interaction between GFP-GIGYF full length (WT), and the indicated fragments, and endogenous HPat and Me31B. The proteins were immunoprecipitated in the presence of RNaseA using an anti-GFP antibody. GFP-F-Luc-V5 served as a negative control. The input (3% for GFP-tagged proteins, 0.2% for HPat and 0.4% for Me31B) and bound fractions (30% for GFP-tagged proteins and HPat, and 40% for Me31B) were analyzed by western blotting using anti-GFP, anti-HPat and anti-Me31B antibodies. (B) The interaction among full length (WT) and fragments of GFP-GIGYF and HA-NOT1 was determined by anti-GFP immunoprecipitation in S2 cell lysates. GFP-F-Luc-V5 served as a negative control. The input (3% for GFP-tagged proteins and 1% for HA-tagged proteins) and bound fractions (15% for GFP-tagged proteins and 30% for HA-tagged proteins) were analyzed by western blotting using anti-GFP and anti-HA antibodies. (C) Sequence alignment of the MBM present in Drosophila melanogaster (Dm) GIGYF and human (Hs) GIGYF1 and 2 proteins with the CUP-homology region from the Hs and Dm 4E-T proteins or the Dm CUP protein. Identical residues are highlighted in red boxes and printed in white whereas residues with >70% similarity are shown with a light color background. (D) Immunoprecipitation assay showing the interaction between GFP-tagged GIGYF proteins (WT and indicated mutants) and endogenous Me31B. The proteins were immunoprecipitated using anti-GFP antibodies. GFP-F-Luc-V5 served as a negative control. The input (4% for GFP-tagged proteins and 0.18% for endogenous Me31B) and bound fractions (40% for GFP-tagged proteins and 45% for endogenous Me31B) were analyzed by western blotting using anti-GFP, anti-Me31B antibodies. (E) The interaction of HA-NOT1 and endogenous HPat with WT and mutant GIGYF proteins was analyzed by immunoprecipitation using anti-GFP antibodies. GFP-F-Luc-V5 served as a negative control. The input (4% for GFP-tagged proteins and 0.8% for HA-tagged proteins and endogenous HPat) and bound fractions (15% for GFP-tagged proteins and 35% for HA-NOT1 and endogenous HPat) were analyzed by western blotting using anti-GFP, anti-HA and anti-HPat antibodies. (F) Western blot analysis of the interaction of GFP-HPat [WT and PPGF motif mutant (PPGF*)] with HA-GIGYF N-term. The input (2% for GFP-tagged proteins and 0.8% for HA-tagged proteins) and bound fractions (10% for GFP-tagged proteins and 20% for HA-tagged proteins) were analyzed by western blotting using anti-GFP and anti-HA antibodies.
Next, we defined the regions of GIGYF that mediate interactions with the deadenylation and decapping factors. We observed that GIGYF N-term, the effector region of the protein, was capable of binding endogenous Me31B and HPat (Figure 3A), although binding to HPat was reduced compared to full length (WT) GIGYF. We were unable to determine the regions of GIGYF required for the interaction with NOT1 as both the N- and C-term of the protein bound only weakly (Figure 3B), indicating that GIGYF uses multiple NOT1 interacting elements to achieve efficient binding. Thus, the effector N-term region of GIGYF interacts with 4EHP, Me31B, HPat and possibly NOT1. These interactions link GIGYF to translational inhibition and mRNA degradation, providing a plausible mechanism for the repressor function of the protein.
The GIGYF effector region contains a conserved Me31B-binding motif (MBM)
Interestingly, an alignment of GIGYF proteins reveals a conserved motif (residues L331-K374) in the effector region of Dm GIGYF that has sequence similarity with the CUP-homology domain (CHD) present in 4E-T-like proteins (Figures 2A and 3C). The CHD of 4E-T mediates the interaction with Me31B/DDX6 [Supplementary Table S3; (38,39)]. To determine if this conserved motif constitutes a Me31B-binding motif (MBM), we generated a GIGYF deletion mutant (ΔMBM, Supplementary Table S1) and performed coIP assays. Indeed, the interaction of Dm GIGYF with Me31B was abolished when the MBM was removed (Figure 3D, lanes 7 versus 6). Importantly, the same deletion did not affect the interaction of GIGYF with HPat or NOT1 (Figure 3E, lanes 7 versus 6), two known Me31B/DDX6-binding proteins (24,40,41), indicating that these proteins bind to GIGYF independently of Me31B.
HPat is a GYF-domain binding protein
The GYF domain of GIGYF proteins forms a hydrophobic pocket that constitutes the binding site for a proline-rich motif (PPGΦ, where Ф is any hydrophobic amino acid with the exception of Trp) present in GYF-domain interacting proteins (13). Interestingly, Dm HPat contains two PPGF motifs. We introduced point mutations in the GYF domain of GIGYF to prevent binding to GYF-domain interacting proteins [GYF* mutant, Supplementary Table S1; (13)] and tested the interaction with HPat. These amino acid substitutions compromised the interaction of GIGYF with HPat without affecting binding to NOT1 or Me31B (Figure 3D and E). Likewise, an HPat protein without the PPGF motifs (PPGF mutant, PPGF*) showed reduced binding to the GIGYF N-term compared to WT HPat (Figure 3F) and the GYF domain of Dm GIGYF bound directly to the P-rich region (P-reg) of Dm HPat in pulldown assays using recombinant proteins (Supplementary Figure S4H).
We also tested the interaction of Dm GIGYF with other GYF-domain binding proteins known to recruit human GIGYF1/2 to specific mRNAs: TTP [Dm Tis11, Supplementary Table S3; (17)], ZNF598 (6) and GW182 (18). In contrast to human GIGYF1/2, none of the proteins were able to interact with Dm GIGYF (Supplementary Figure S6A–C). Analysis of the protein sequence revealed that Dm TTP/Tis11 differs from the human TTP as it does not contain the proline-rich motifs that mediate the interaction with the GYF domain; thus, it is not surprising that it could not interact with Dm GIGYF. Dm ZNF598 contains two PPGΦ motifs (PPPGF and PPGL), whereas only one PPGΦ motif (PPPGL) is present in GW182. However, they did not bind to Dm GIGYF in our assays. Our findings point to the decapping activator HPat as the only known interactor of the GYF domain of Dm GIGYF.
GIGYF requires both Me31B and HPat to block translation and induce mRNA decay
To identify which protein partners determine the repressor function of the GIGYF effector region, we performed a series of tethering assays with different GIGYF mutant proteins. To test the effect of 4EHP-binding, we also introduced substitutions into the canonical motif of GIGYF (C*, Supplementary Table S1) that disrupt the GIGYF–4EHP interaction [Supplementary Figure S7A, lanes 8 versus 7, (6,14)]. We observed that the GIGYF C*, GIGYF GYF* and the combined C* + GYF* proteins repressed F-Luc activity and induced degradation of the reporter mRNA to the same extent as the WT protein, although the GYF* mutants were marginally less active (Supplementary Figure S7B and C). None of the mutants affected the expression of the reporter mRNA without the BoxB hairpins (Supplementary Figure S7D–F). Therefore, tethered Dm GIGYF can repress translation and induce reporter mRNA decay in the absence of interactions with 4EHP or HPat.
The GIGYF protein that no longer interacts with Me31B (ΔMBM mutant, Supplementary Table S1) was impaired in its repressor function. Importantly, when bound to the reporter mRNA, this mutant protein or a combined Me31B- and HPat-binding mutant (ΔMBM+GYF* mutant, Supplementary Table S1) were less active and reduced F-Luc activity to 40–50% instead of the 20% observed with WT GIGYF (Supplementary Figure S8A–C). F-Luc mRNA was also less degraded (Supplementary Figure S8A and B). This result indicates that Me31B contributes to GIGYF-mediated translational repression and mRNA decay. Since none of the mutations in the binding sites completely blocked the repressor function of GIGYF, it is likely that redundancy in the network of protein-protein interactions or additional factors, e.g. the CCR4–NOT complex (20), or yet unknown proteins binding to the GIGYF C-term also participate in the repressive mechanism.
We also tested the ability of GIGYF to regulate mRNA expression in cells depleted of Me31B and HPat. Single depletions of Me31B or HPat (Me31B KD or HPat KD) had only minor effects on degradation of the GIGYF-bound reporter, which remained translationally silenced (Supplementary Figures S8D-K and S9A-C). Nevertheless, in the absence of both decapping factors (Me31B+HPat KD), GIGYF-mediated translational repression and mRNA decay were significantly compromised; compared to control cells, both a two-fold increase in F-Luc activity and a reduction in reporter mRNA decay were observed (Supplementary Figure S9D–F). Likewise, in the absence of Me31B (Me31B KD), inhibition of F-Luc activity and decay of the reporter mRNA by the GIGYF GYF* protein were also less efficient than WT GIGYF (Supplementary Figure S8D and E). The repressor function of the GIGYF ΔMBM protein was slightly reduced in Me31B KD cells relative to control cells (Supplementary Figure S8H-K). These data indicate that both Me31B and HPat act in concert with GIGYF to regulate reporter mRNA expression.
Both Me31B and HPat participate in GIGYF-mediated translational repression
In the experiment where mRNA degradation was blocked by overexpression of the DCP1 GSSG mutant (Figure 2E and F), the reporter mRNA is deadenylated and translationally silenced in the presence of GIGYF. This experiment suggests that in the absence of mRNA decay GIGYF still acts as a translational repressor. However, this repressor function is altered in cells codepleted of Me31B and HPat (Supplementary Figure S9D). As the data pointed towards a role of Me31B and HPat in GIGYF-mediated translational repression, we examined the translational regulation of an F-Luc-5BoxB reporter mRNA that cannot be deadenylated and degraded by GIGYF. This reporter, F-Luc-5BoxB-A96-HhR, is refractory to deadenylation-dependent decapping because the cleavage and polyadenylation sites were replaced by an internal polyadenosine stretch of 96 nucleotides, followed by seven cytosine bases and a self-cleaving hammerhead ribozyme (26). Tethering of GIGYF efficiently reduced F-Luc protein levels (F-Luc activity) without changes in the reporter mRNA levels (Figure 4A and B). Deletion of the Me31B-binding motif (ΔMBM) or single depletion of Me31B (Me31B KD) impaired GIGYF-mediated regulation of F-Luc activity, which increased to 40% compared to 20% in control conditions (Figure 4A and D). These results indicate that Me31B is an important player in the mechanism of translational repression by GIGYF. Furthermore, interfering with the binding of GIGYF to both Me31B and HPat, such as when tethering a combined GIGYF ΔMBM+GYF* mutant (Figure 4A), tethering the GIGYF GYF* mutant in Me31B KD cells (Figure 4D) or tethering GIGYF in cells codepleted of Me31B and HPat (Figure 4F), the repression of F-Luc activity by GIGYF decreased to 60–80% instead of the 20% observed in control conditions. Once again, mRNA levels did not change (Figure 4A-G). A combined GIGYF C+ΔMBM+GYF* mutant repressed F-Luc activity to the same extent as the GIGYF ΔMBM+GYF* mutant (Supplementary Figure S10A and B). Additionally, we observed that in cells depleted of Me31B, the GIGYF ΔMBM mutant repressed F-Luc translation to ∼50% compared to the 40% observed in control cells (Supplementary Figure S10C–F). This data indicates that Me31B contributes to GIGYF-mediated translational repression via direct binding to the MBM and indirectly via the association to other GIGYF-binding partners, such as HPat or the CCR4–NOT complex.
Figure 4.
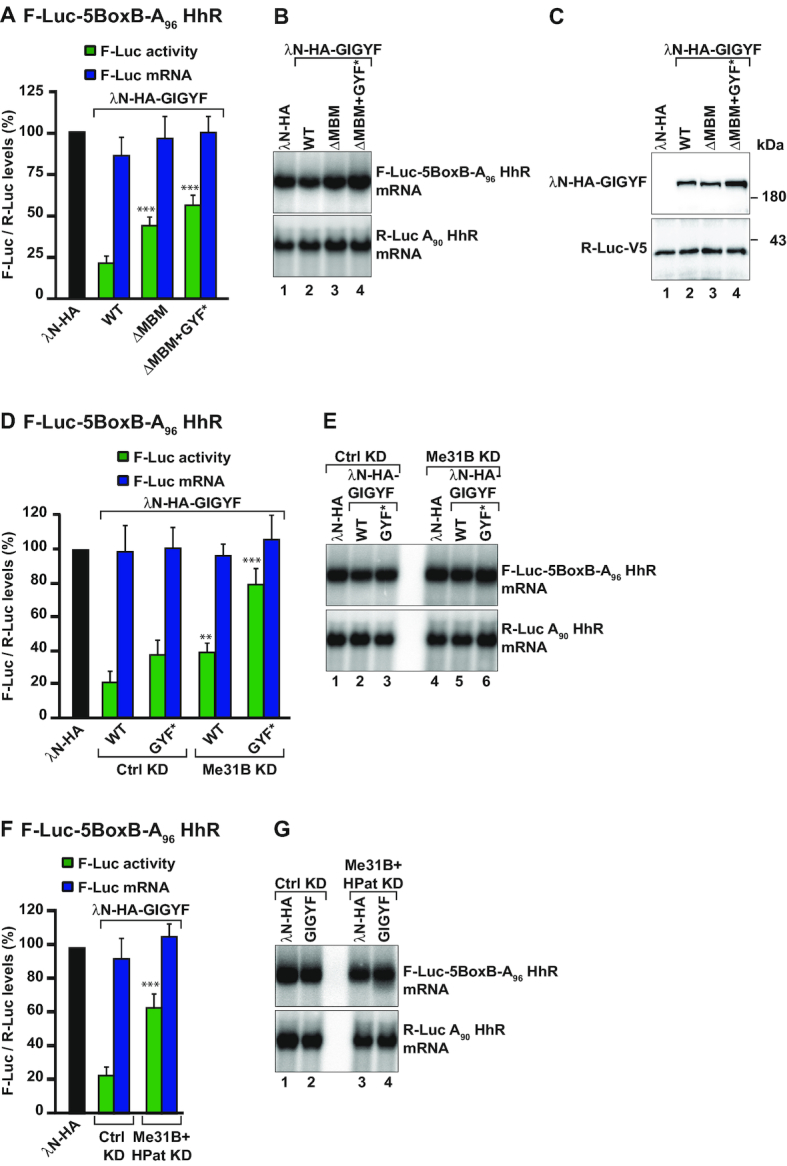
Translational repression by GIGYF requires the decapping factors Me31B and HPat. (A–C) S2 cells were transfected with the F-Luc-5BoxB reporter in which the cleavage and polyadenylation signal were replaced by a polyadenosine stretch of 96 residues followed by seven cytosines and a self-cleaving Hammerhead ribozyme (F-Luc-5BoxB-A96-HhR). λN-HA-GIGYF WT, Me31B-binding mutant (ΔMBM), or double ΔMBM+GYF* mutant and the transfection control R-Luc-A90-HhR plasmids were cotransfected with the F-Luc reporter. Luciferase activity and mRNA levels were analyzed as described in Figure 1A. The P value (***P < 0.0005) was determined using the two-tailed Student's t test between WT GIGYF and the other GIGYF proteins. A representative northern blot is shown next to the corresponding graph (B). Panel C shows expression levels of the tethered proteins as analyzed by western blotting. (D–G) S2 cells were treated with the indicated dsRNAs and transfected with a mixture of three plasmids coding for F-Luc-5BoxB-A96-HhR, R-Luc-A90-HhR and the indicated λN-HA-tagged proteins. In panels D and F, F-Luc activity and mRNA levels were normalized to that of R-Luc in the presence of the different tethered proteins. Panels E and G show representative northern blots. The P value (**P < 0.005, ***P < 0.0005) was determined using the two-tailed Student's t test between WT GIGYF in Ctrl KD and GIGYF proteins in the other experimental conditions.
In contrast, single depletion of HPat (HPat KD) or tethering of GIGYF GYF* mutant had not effect or only weakly interfered with the regulation of mRNA translation by GIGYF, respectively (Figure 4D and Supplementary Figure S10G and H).
Altogether, our data indicate that both Me31B and HPat are recruited by GIGYF to induce translational repression.
GIGYF links 4EHP to Me31B and HPat
Our results show that GIGYF recruits multiple proteins to mediate translational repression, deadenylation and decay of a bound mRNA target. To understand if the multiple interactions established by GIGYF are required for the function of the 4EHP–GIGYF complex as a translational repressor, we tethered λN-HA-4EHP to the F-Luc-5BoxB-A96-HhR reporter in control or GIGYF-depleted cells (GIGYF KD) and assessed the ability of 4EHP to repress translation in cells complemented with WT or mutant GIGYF proteins (C*, ΔMBM+GYF* or N-term). Upon tethering, 4EHP efficiently reduced translation of the F-Luc-5BoxB-A96-HhR reporter (Figure 5A). Once more, in the absence of GIGYF (GIGYF KD), 4EHP-mediated translational repression was strongly impaired (Figure 5A and B). Transient expression of the WT HA-tagged GIGYF or the effector region (N-term), but not of the GIGYF canonical mutant (C*), restored the ability of 4EHP to suppress F-Luc activity (Figure 5A and B). Notably, 4EHP was also less efficient in the inhibition of F-Luc activity in cells expressing the GIGYF ΔMBM+GYF* mutant (Figure 5A and B). Thus, 4EHP requires GIGYF, which in turn recruits Me31B and HPat, to repress mRNA translation.
In agreement with these findings, we observed that overexpressed GFP-4EHP does not efficiently coimmunoprecipitate with endogenous Me31B and HPat unless HA-GIGYF is coexpressed (Figure 5C, lanes 8 versus 7). The assembly of the 4EHP–GIGYF–Me31B–HPat complex is dependent on GIGYF, as overexpression of HA-GIGYF mutants that no longer bind to GFP-4EHP (C*) or to Me31B and HPat (ΔMBM+GYF*) do not allow GFP-4EHP to efficiently coimmunoprecipitate the full complex (Figure 5C, lanes 9, 10 versus 8). Hence, GIGYF bridges 4EHP to the decapping factors Me31B and HPat.
In parallel, we also observed that depletion of Me31B and HPat also strongly impaired the ability of tethered 4EHP to repress translation of the F-Luc-5BoxB-A96-HhR reporter (Figure 5D and E), suggesting that both Me31B and HPat are important players in the control of translation by 4EHP.
Overall, our results indicate that GIGYF adds additional mechanisms to the control of gene expression by the cap-binding protein 4EHP. Translational repression of a 4EHP-bound mRNA occurs not only through competition with eIF4E for cap binding, as reported previously (5,7) but also via the interaction with GIGYF, Me31B and HPat. The repressed mRNA is further deadenylated and degraded to ensure the shutdown of mRNA expression (Figure 5F).
DISCUSSION
Through direct competition with eIF4E for cap binding, 4EHP blocks the interaction of the eIF4F complex with mRNA, thus inhibiting translation (3,4,42). Here, we show that independent of cap binding, the interaction of Dm 4EHP with GIGYF promotes translational repression and induces deadenylation and decay of bound mRNA targets. Moreover, in the absence of interaction with GIGYF, tethered 4EHP loses its repressor function [this study and (14)].
Previous work has shown that GIGYF proteins bind several proteins known to elicit translational repression and mRNA decay such as the miRISC components Ago2 and TNRC6C or the CCR4–NOT deadenylase complex (18–20). Moreover, GIGYF proteins also link 4EHP to specific mRNAs via interaction with RNA-binding proteins such as TTP and the ZNF598 (6,17). In this study, we identified novel Dm GIGYF binding partners: the RNA helicase Me31B, the decapping activator HPat and different subunits of the CCR4–NOT deadenylase complex. These interactions are required for post-transcriptional mRNA regulation and reveal a new mechanism employed by the 4EHP–GIGYF complex to regulate the expression of its mRNA targets at the level of protein synthesis.
Regulation of translation and mRNA decay by GIGYF proteins
Human GIGYF2 was recently described as an RBP, which regulates mRNA expression by two distinct mechanisms. Recruitment of the 4EHP–GIGYF2 complex by RBPs promotes cap-dependent translational repression, whereas 4EHP-independent and direct mRNA binding of GIGYF leads to recruitment of the CCR4–NOT complex and subsequent mRNA degradation (20). Our studies on the uncharacterized Dm 4EHP–GIGYF complex support a revised model in which binding of the 4EHP–GIGYF complex to an mRNA, either directly or indirectly, leads to translational repression followed by mRNA decay (Figure 5F). This mechanism of downregulation of mRNA expression is possible because GIGYF proteins engage in multiple protein interactions that are mediated by distinct binding elements. In fact, we describe for the first time that GIGYF bridges 4EHP to Me31B, HPat and the CCR4–NOT complex to repress mRNA expression. These multiple interactions provide redundancy and link translational repression to mRNA decay, ensuring an efficient inhibition of mRNA expression. Notably, the fact that the binding elements and interactions identified in this study for Dm GIGYF are preserved by human homologous proteins and protein complexes [this work and (20)], indicates a conservation of the mechanism by which the 4EHP–GIGYF complex represses mRNA expression across metazoans.
Repression of translation by the 4EHP–GIGYF complex relies on Me31B and HPat
Our data further highlight that the concurrent interaction of Dm GIGYF with Me31B and HPat via the effector region is crucial for induction of translational repression of a 4EHP–GIGYF-bound mRNA. In fact, the effector region of Dm GIGYF is sufficient to rescue 4EHP-mediated translational repression in S2 cells depleted of GIGYF. Moreover, codepletion of Me31B and HPat significantly impaired the function of 4EHP and GIGYF as translational repressors (Figures 4 and 5). In the absence of interaction with only one of these factors, the repression of the mRNA by 4EHP and GIGYF is only slightly changed, probably reflecting the redundancy of the protein interactions mediating the repression mechanism.
Although the mechanism by which Me31B and HPat repress translation is still unclear, both proteins have been implicated in the control of translation (43–45). For instance, recruitment of Me31B orthologs by different translational repressors and RNA-associated proteins is a common mechanism for the control of translation and mRNA stability in different pathways, including microRNA (miRNA)-mediated gene silencing (40,41,46–49). In particular, binding of human DDX6 to the translational repressor eIF4E-Transporter (4E-T), which also associates with 4EHP (33), is promoted by the miRISC/CCR4–NOT complex to induce translational repression and decay of miRNA targets (10). Therefore, the functional cooperation of 4EHP and Me31B orthologs in different RNPs appears to be a widespread mechanism associated with several post-transcriptional regulatory pathways. On the other hand, Pat1 proteins are also key factors in the regulation of post-transcriptional gene expression. They act as scaffold or remodeling proteins in multiple RNPs and induce translational repression, deadenylation and decay [reviewed in (44,45)]. Although the interaction of each protein with GIGYF is independent of the other, HPat and Me31B bind to each other (Figure 5F). Additional studies are necessary to understand whether Me31B and HPat function as complex to repress translation or if each protein independently blocks translation by different but redundant mechanisms (Figure 5F).
Even though Me31B and HPat are recruited by Dm GIGYF to induce translational repression of a bound mRNA, additional factors also appear to contribute to the repressor function of the 4EHP–GIGYF complex. In the absence of interactions with Me31B and HPat, the repressor function of Dm 4EHP and GIGYF is not completely abolished. Additional factors that might explain the remaining GIGYF repressor function include the CCR4–NOT deadenylase complex [this work and (20)] or other proteins that may bind to its C-term which was partially active in our assays.
GIGYF proteins contain discrete sequence elements to mediate DDX6- and HPat binding
In this study we reveal that Dm GIGYF binds to Me31B via a sequence motif that is only partially similar to the CHD of 4E-T [Figure 3C; (39)]. The presence of the Me31B/DDX6-binding motif (MBM) in GIGYF proteins suggests that GIGYF and 4E-T have mutually exclusive interactions with the RNA helicase and are part of distinct RNPs, as observed for the large majority of Me31B-binding proteins (50). The MBM is conserved in GIGYF proteins from nematodes to humans and agrees with large proteomic interaction studies that have identified the human GIGYF2 or 4EHP as members of Me31B/DDX6 RNPs (10,20,51).
Our data also describe Dm HPat as a novel Dm GIGYF GYF-domain binding protein and identified in Dm HPat the PPGF motifs that mediate the interaction (Figure 3E and F). Although this protein-protein interaction is conserved in human cells (Supplementary Figure S4B and C), human PatL1 does not contain the PPGF motifs to bind to the GYF domain of the GIGYF1/2 proteins. Thus, binding to PatL1 might be mediated by a different sequence motif or another GIGYF1/2-interacting partner. Human GIGYF1/2 proteins instead are recruited to specific mRNAs upon binding to GYF-domain interacting proteins such as TTP, ZNF598 and GW182 (6,17,18). Despite the presence of PPGΦ in Dm ZNF598 and Dm GW182, Dm GIGYF also differs from the human orthologs as it does not associate with these GYF-domain interacting proteins (Supplementary Figure S6). Although the precise mechanism is unknown, post-translational modifications or other protein factors may control binding to the GYF domain of Dm GIGYF. In the absence of interactions with these RNA-associated proteins, it is also unclear how the Dm 4EHP–GIGYF complex is specifically recruited to mRNA targets or how it is regulated. Thus, new studies are required to determine if the GYF domain of Dm GIGYF interacts with other proteins besides HPat, as observed for the human GIGYF1/2. Alternatively, similar to human GIGYF2, Dm GIGYF can also directly bind to mRNA (20).
The biological role of Dm GIGYF
We show that GIGYF proteins are regulators of mRNA translation and stability using reporter assays in Dm cells. However, the consequences and the natural targets of GIGYF-mediated mRNA regulation are still poorly characterized. In Dm cells, GIGYF associates with centrosomes during mitosis (52). GIGYF-null flies have locomotor defects and a short life span due to the dysregulation of autophagy that causes accumulation of ubiquitinated proteins and dysfunctional mitochondria in neuronal and muscle tissues (53). However, these cellular functions have not been shown to be related to the control of mRNA expression described in this or other studies (6,13,17,20). Elucidation of the Dm 4EHP–GIGYF mRNA targets and functions in different tissues or developmental stages would clarify the cellular functions of this repressor complex. Moreover, it is also possible that GIGYF proteins have additional functions unrelated to translational repression and mRNA decay. For example, in this study we have not addressed the role of the C-term of the protein which is almost 1000 amino acids long and predicted to be α-helical. This long protein region might function in coupling RNA metabolism to other regulatory pathways.
Altogether, we show for the first time that the recruitment of two decapping activators (Me31B and HPat) by GIGYF proteins to induce translational repression adds a new layer to the mechanistic understanding of the inhibition of mRNA expression by the 4EHP cap-binding protein.
Supplementary Material
ACKNOWLEDGEMENTS
We dedicate this manuscript to our mentor Elisa Izaurralde who passed away too early. We are grateful to P. Natalin, I. Behm-Asmant, A. Eulalio, E. Huntzinger and L. Gasse for generating the Brat, Bicoid, 4EHP and Tis11 and DCP1 GSSG expression plasmids, respectively. We thank C. Weiler and M. Y. Chung for excellent technical assistance and all the members of the Izaurralde lab for insightful comments on the manuscript.
Author Contributions: E.I. and C.I. conceived the project. V.R., P.B., D.P. and S.H. performed experiments and analyzed data together with E.I. and C.I. C.I. wrote the manuscript with the help of all authors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Max Planck Society. Funding for open access charge: Max Planck Society.
Conflict of interest statement. None declared.
REFERENCES
- 1. Jackson R.J., Hellen C.U., Pestova T.V.. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010; 11:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hershey J.W., Sonenberg N., Mathews M.B.. Principles of translational control: an overview. Cold Spring Harb. Perspect. Biol. 2012; 4:a011528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rom E., Kim H.C., Gingras A.C., Marcotrigiano J., Favre D., Olsen H., Burley S.K., Sonenberg N.. Cloning and characterization of 4EHP, a novel mammalian eIF4E-related cap-binding protein. J. Biol. Chem. 1998; 273:13104–13109. [DOI] [PubMed] [Google Scholar]
- 4. Joshi B., Cameron A., Jagus R.. Characterization of mammalian eIF4E-family members. Eur. J. Biochem. 2004; 271:2189–2203. [DOI] [PubMed] [Google Scholar]
- 5. Cho P.F., Poulin F., Cho-Park Y.A., Cho-Park I.B., Chicoine J.D., Lasko P., Sonenberg N.. A new paradigm for translational control: inhibition via 5′-3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell. 2005; 121:411–423. [DOI] [PubMed] [Google Scholar]
- 6. Morita M., Ler L.W., Fabian M.R., Siddiqui N., Mullin M., Henderson V.C., Alain T., Fonseca B.D., Karashchuk G., Bennett C.F. et al.. A novel 4EHP–GIGYF2 translational repressor complex is essential for mammalian development. Mol. Cell Biol. 2012; 32:3585–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cho P.F., Gamberi C., Cho-Park Y.A., Cho-Park I.B., Lasko P., Sonenberg N.. Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila embryos. Curr. Biol. 2006; 16:2035–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Valzania L., Ono H., Ignesti M., Cavaliere V., Bernardi F., Gamberi C., Lasko P., Gargiulo G.. Drosophila 4EHP is essential for the larval-pupal transition and required in the prothoracic gland for ecdysone biosynthesis. Dev. Biol. 2016; 410:14–23. [DOI] [PubMed] [Google Scholar]
- 9. Villaescusa J.C., Buratti C., Penkov D., Mathiasen L., Planaguma J., Ferretti E., Blasi F.. Cytoplasmic Prep1 interacts with 4EHP inhibiting Hoxb4 translation. PLoS One. 2009; 4:e5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chapat C., Jafarnejad S.M., Matta-Camacho E., Hesketh G.G., Gelbart I.A., Attig J., Gkogkas C.G., Alain T., Stern-Ginossar N., Fabian M.R. et al.. Cap-binding protein 4EHP effects translation silencing by microRNAs. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:5425–5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jafarnejad S.M., Chapat C., Matta-Camacho E., Gelbart I.A., Hesketh G.G., Arguello M., Garzia A., Kim S.H., Attig J., Shapiro M. et al.. Translational control of ERK signaling through miRNA/4EHP-directed silencing. Elife. 2018; 7:e35034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Giovannone B., Lee E., Laviola L., Giorgino F., Cleveland K.A., Smith R.J.. Two novel proteins that are linked to insulin-like growth factor (IGF-I) receptors by the Grb10 adapter and modulate IGF-I signaling. J Biol Chem. 2003; 278:31564–31573. [DOI] [PubMed] [Google Scholar]
- 13. Ash M.R., Faelber K., Kosslick D., Albert G.I., Roske Y., Kofler M., Schuemann M., Krause E., Freund C.. Conserved beta-hairpin recognition by the GYF domains of Smy2 and GIGYF2 in mRNA surveillance and vesicular transport complexes. Structure. 2010; 18:944–954. [DOI] [PubMed] [Google Scholar]
- 14. Peter D., Weber R., Sandmeir F., Wohlbold L., Helms S., Bawankar P., Valkov E., Igreja C., Izaurralde E.. GIGYF1/2 proteins use auxiliary sequences to selectively bind to 4EHP and repress target mRNA expression. Genes Dev. 2017; 31:1147–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Giovannone B., Tsiaras W.G., de la Monte S., Klysik J., Lautier C., Karashchuk G., Goldwurm S., Smith R.J.. GIGYF2 gene disruption in mice results in neurodegeneration and altered insulin-like growth factor signaling. Hum. Mol. Genet. 2009; 18:4629–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tao X., Gao G.. Tristetraprolin recruits eukaryotic initiation factor 4e2 to repress translation of AU-Rich element-containing mRNAs. Mol. Cell Biol. 2015; 35:3921–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fu R., Olsen M.T., Webb K., Bennett E.J., Lykke-Andersen J.. Recruitment of the 4EHP-GYF2 cap-binding complex to tetraproline motifs of tristetraprolin promotes repression and degradation of mRNAs with AU-rich elements. RNA. 2016; 22:373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schopp I.M., Amaya Ramirez C.C., Debeljak J., Kreibich E., Skribbe M., Wild K., Bethune J.. Split-BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes. Nat. Commun. 2017; 8:15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kryszke M.H., Adjeriou B., Liang F., Chen H., Dautry F.. Post-transcriptional gene silencing activity of human GIGYF2. Biochem. Biophys. Res. Commun. 2016; 475:289–294. [DOI] [PubMed] [Google Scholar]
- 20. Amaya Ramirez C.C., Hubbe P., Mandel N., Bethune J.. 4EHP-independent repression of endogenous mRNAs by the RNA-binding protein GIGYF2. Nucleic Acids Res. 2018; 46:5792–5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E.. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006; 20:1885–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tritschler F., Eulalio A., Helms S., Schmidt S., Coles M., Weichenrieder O., Izaurralde E., Truffault V.. Similar modes of interaction enable Trailer Hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes. Mol. Cell Biol. 2008; 28:6695–6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Braun J.E., Tritschler F., Haas G., Igreja C., Truffault V., Weichenrieder O., Izaurralde E.. The C-terminal alpha-alpha superhelix of Pat is required for mRNA decapping in metazoa. EMBO J. 2010; 29:2368–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haas G., Braun J.E., Igreja C., Tritschler F., Nishihara T., Izaurralde E.. HPat provides a link between deadenylation and decapping in metazoa. J. Cell Biol. 2010; 189:289–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Igreja C., Izaurralde E.. CUP promotes deadenylation and inhibits decapping of mRNA targets. Genes Dev. 2011; 25:1955–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zekri L., Kuzuoglu-Ozturk D., Izaurralde E.. GW182 proteins cause PABP dissociation from silenced miRNA targets in the absence of deadenylation. EMBO J. 2013; 32:1052–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chang C.T., Bercovich N., Loh B., Jonas S., Izaurralde E.. The activation of the decapping enzyme DCP2 by DCP1 occurs on the EDC4 scaffold and involves a conserved loop in DCP1. Nucleic Acids Res. 2014; 42:5217–5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peter D., Igreja C., Weber R., Wohlbold L., Weiler C., Ebertsch L., Weichenrieder O., Izaurralde E.. Molecular architecture of 4E-BP translational inhibitors bound to eIF4E. Mol. Cell. 2015; 57:1074–1087. [DOI] [PubMed] [Google Scholar]
- 29. Diebold M.L., Fribourg S., Koch M., Metzger T., Romier C.. Deciphering correct strategies for multiprotein complex assembly by co-expression: application to complexes as large as the histone octamer. J. Struct. Biol. 2011; 175:178–188. [DOI] [PubMed] [Google Scholar]
- 30. Gruner S., Peter D., Weber R., Wohlbold L., Chung M.Y., Weichenrieder O., Valkov E., Igreja C., Izaurralde E.. The Structures of eIF4E-eIF4G Complexes Reveal an Extended Interface to Regulate Translation Initiation. Mol. Cell. 2016; 64:467–479. [DOI] [PubMed] [Google Scholar]
- 31. Igreja C., Peter D., Weiler C., Izaurralde E.. 4E-BPs require non-canonical 4E-binding motifs and a lateral surface of eIF4E to repress translation. Nat. Commun. 2014; 5:4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gehring N.H., Kunz J.B., Neu-Yilik G., Breit S., Viegas M.H., Hentze M.W., Kulozik A.E.. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol. Cell. 2005; 20:65–75. [DOI] [PubMed] [Google Scholar]
- 33. Kubacka D., Kamenska A., Broomhead H., Minshall N., Darzynkiewicz E., Standart N.. Investigating the consequences of eIF4E2 (4EHP) interaction with 4E-transporter on its cellular distribution in HeLa cells. PLoS One. 2013; 8:e72761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B., Perrimon N., Mohr S.E.. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2011; 12:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ferraiuolo M.A., Basak S., Dostie J., Murray E.L., Schoenberg D.R., Sonenberg N.. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J. Cell Biol. 2005; 170:913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Franks T.M., Lykke-Andersen J.. The control of mRNA decapping and P-body formation. Mol. Cell. 2008; 32:605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kuzuoglu-Ozturk D., Bhandari D., Huntzinger E., Fauser M., Helms S., Izaurralde E.. miRISC and the CCR4–NOT complex silence mRNA targets independently of 43S ribosomal scanning. EMBO J. 2016; 35:1186–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kamenska A., Lu W.T., Kubacka D., Broomhead H., Minshall N., Bushell M., Standart N.. Human 4E-T represses translation of bound mRNAs and enhances microRNA-mediated silencing. Nucleic Acids Res. 2014; 42:3298–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ozgur S., Basquin J., Kamenska A., Filipowicz W., Standart N., Conti E.. Structure of a human 4E-T/DDX6/CNOT1 complex reveals the different interplay of DDX6-Binding proteins with the CCR4–NOT complex. Cell Rep. 2015; 13:703–711. [DOI] [PubMed] [Google Scholar]
- 40. Chen Y., Boland A., Kuzuoglu-Ozturk D., Bawankar P., Loh B., Chang C.T., Weichenrieder O., Izaurralde E.. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol. Cell. 2014; 54:737–750. [DOI] [PubMed] [Google Scholar]
- 41. Mathys H., Basquin J., Ozgur S., Czarnocki-Cieciura M., Bonneau F., Aartse A., Dziembowski A., Nowotny M., Conti E., Filipowicz W.. Structural and biochemical insights to the role of the CCR4–NOT complex and DDX6 ATPase in microRNA repression. Mol Cell. 2014; 54:751–765. [DOI] [PubMed] [Google Scholar]
- 42. Hernandez G., Altmann M., Sierra J.M., Urlaub H., Diez del Corral R., Schwartz P., Rivera-Pomar R.. Functional analysis of seven genes encoding eight translation initiation factor 4E (eIF4E) isoforms in Drosophila. Mech Dev. 2005; 122:529–543. [DOI] [PubMed] [Google Scholar]
- 43. Ostareck D.H., Naarmann-de Vries I.S., Ostareck-Lederer A.. DDX6 and its orthologs as modulators of cellular and viral RNA expression. Wiley Interdiscip. Rev. RNA. 2014; 5:659–678. [DOI] [PubMed] [Google Scholar]
- 44. Jonas S., Izaurralde E.. The role of disordered protein regions in the assembly of decapping complexes and RNP granules. Genes Dev. 2013; 27:2628–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marnef A., Standart N.. Pat1 proteins: a life in translation, translation repression and mRNA decay. Biochem. Soc. Trans. 2010; 38:1602–1607. [DOI] [PubMed] [Google Scholar]
- 46. Chu C.Y., Rana T.M.. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006; 4:e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rouya C., Siddiqui N., Morita M., Duchaine T.F., Fabian M.R., Sonenberg N.. Human DDX6 effects miRNA-mediated gene silencing via direct binding to CNOT1. RNA. 2014; 20:1398–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Radhakrishnan A., Chen Y.H., Martin S., Alhusaini N., Green R., Coller J.. The DEAD-Box protein Dhh1p Couples mRNA decay and translation by monitoring codon optimality. Cell. 2016; 167:122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Freimer J.W., Hu T.J., Blelloch R.. Decoupling the impact of microRNAs on translational repression versus RNA degradation in embryonic stem cells. Elife. 2018; 7:e38014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Valkov E., Jonas S., Weichenrieder O.. Mille viae in eukaryotic mRNA decapping. Curr. Opin. Struct. Biol. 2017; 47:40–51. [DOI] [PubMed] [Google Scholar]
- 51. Ayache J., Benard M., Ernoult-Lange M., Minshall N., Standart N., Kress M., Weil D.. P-body assembly requires DDX6 repression complexes rather than decay or Ataxin2/2L complexes. Mol. Biol. Cell. 2015; 26:2579–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Muller H., Schmidt D., Steinbrink S., Mirgorodskaya E., Lehmann V., Habermann K., Dreher F., Gustavsson N., Kessler T., Lehrach H. et al.. Proteomic and functional analysis of the mitotic Drosophila centrosome. EMBO J. 2010; 29:3344–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim M., Semple I., Kim B., Kiers A., Nam S., Park H.W., Park H., Ro S.H., Kim J.S., Juhasz G. et al.. Drosophila Gyf/GRB10 interacting GYF protein is an autophagy regulator that controls neuron and muscle homeostasis. Autophagy. 2015; 11:1358–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.