Abstract
MicroRNAs (miRNAs) have emerged as the promising molecular biomarkers for early diagnosis and enhanced understanding of the molecular pathogenesis of cancers as well as certain diseases. Here, a facile, label-free, and amplification-free electrochemical biosensor was developed to detect miRNA by using DNA origami nanostructure-supported DNA probes, with methylene blue (MB) serving as the hybridization redox indicator, for the first time. Specifically, the use of cross-shaped DNA origami nanostructures containing multiple single-stranded DNA probes at preselected locations on each DNA nanostructure could increase the accessibility and the recognition efficiency of the probes (due to the rational controlled density of DNA probes). The successful immobilization of DNA origami probes and their hybridization with targeted miRNA-21 molecules was confirmed by electrochemical impedance spectroscopy and cyclic voltammetry methods. A differential pulse voltammetry technique was employed to record the oxidation peak current of MB before and after target hybridization. The linear detection range of this biosensor was from 0.1 pM to 10.0 nM, with a lower detection limit of 79.8 fM. The selectivity of the miRNA biosensor was also studied by observing the discrimination ability of single-base mismatched sequences. Because of the larger surface area and unprecedented customizability of DNA origami nanostructures, this strategy demonstrated great potential for sensitive, selective, and label-free determination of miRNA for translational biomedical research and clinical applications.
1. Introduction
MicroRNAs (miRNAs), a group of short noncoding RNAs (∼19–23 nucleotide long), play a vital role in cell growth, gene expression, and apoptosis.1 Recently, an increasing number of studies have been reported in which improper miRNA expression levels are clearly associated with the development of many diseases, including cancers,2,3 thereby triggering research interest in utilizing miRNAs as potential biomarkers for detecting and monitoring various diseases. To date, a variety of methodologies have been proposed for miRNA detection and quantification, such as quantitative polymerase chain reaction, northern blotting, next-generation sequencing, microarrays, and so forth.4−9 However, lack of accuracy, low efficiency, and/or excessive time have limited the implementation of these approaches. Electrochemical biosensors have recently emerged as a promising technique for the rapid and easy operational detection of miRNAs.10−18 To improve the sensitivity and measured detection limits of electrochemical miRNA biosensing, various electrode modification materials, including carbon nanotubes,10,11 graphene,13,14 metal nanoparticles,16−19 and MoS2,20 have been incorporated into a modification layer that enhances the electron-transfer rate and amplifies the sensor signal.
Recently, because of the biocompatibility and complex technologies of electrode surface modification (via those inorganic materials), DNA nanostructures21,22 have been considered as a biocompatible alternative for miRNA detection processes. For example, Pei et al. demonstrated the use of a DNA tetrahedral probe, combined with varied amplification strategies (such as redox enzyme incorporation and hybridization chain reaction), to detect miRNAs with high sensitivity and specificity.23−33 In this design, DNA tetrahedral nanostructures were immobilized on the electrode surface, via the strong Au–S interaction between three thiol-modified vertices and the gold surface. A single-stranded DNA (ssDNA) probe, protruding out from the fourth vertex, was used to realize target recognition.23 This strategy, which resulted in enhanced probe accessibility and reduced the effects of surface crowding, represents a new paradigm for designing biocompatible biosensors for clinical research and disease diagnosis. Nevertheless, the signal amplification process required enzyme biomolecules, specialized reagents, and multiple chemical reactions, which greatly increased the complexity of experiments and exerted environmental limitations on the advanced application of such biosensors. Therefore, a facile, cost-effective, and amplification-free biosensing platform would be highly desirable for miRNA analysis.
Over the past decade, DNA origami has attracted extensive interest because of its diverse structural engineering capability, large surface area, and unprecedented customizability for precisely arranging target-binding sites at the nanometer scale.34−36 This makes DNA origami an excellent candidate for fabrication of biosensing systems. Herein, we have designed a novel, label-free, and amplification-free miRNA biosensor, based on a cross-shaped DNA origami and using methylene blue (MB) as a hybridization redox indicator. In our design, a DNA origami was utilized, for the first time, as the miRNA sensing platform to provide a large number of free-standing DNA probes to capture target nucleic acids with enhanced probe accessibility and high sensitivity. In addition, the immobilization of DNA origami probes on a working electrode (WE) surface was realized, via electrostatic adsorption between a cationic and biodegradable chitosan film37,38 and negatively charged DNA origami nanostructures. This method offers the advantage of not only providing a simple yet stable attachment of DNA origami on an electrode surface because of the large contact surface area of the DNA origami, but also avoiding the use of modified thiol-DNA or biotin-DNA. With the facile fabrication method that does not require laborious labeling, probe immobilization, and signal amplification, our strategy promises the aforementioned advantages in determining the concentration of miRNA.
2. Experimental Section
2.1. Materials and Chemicals
All chemically synthesized DNA strands and miRNAs were purchased from Integrated DNA Technologies, Inc. (www.idtdna.com), and their detailed sequences are included in the Supporting Information. The extended staple strands, serving as a probe for miRNA recognition, were purified with denaturing PAGE, while other staple strands were used without purification. The human serum and all other chemicals used were purchased from Sigma-Aldrich and were used, as received, without further purification. 1× TAE/Mg2+ buffer (40 mM Tris-HCl; 20 mM acetic acid; 2 mM EDTA; 11.5 mM magnesium acetate; pH 8.0) was used for DNA origami annealing. 10 mM of Tris-HCl, containing 1 mM EDTA buffer (pH 8.0), was used for DNA origami immobilization. 2× saline sodium citrate (SSC) buffer (300 mM NaCl, 30 mM sodium citrate, pH 8.0) was used as a hybridization buffer, and 0.2× SSC was used as a washing buffer. To protect miRNAs from RNase degradation, all solutions were treated with diethyl pyrocarbonate (0.1%) before use.
2.2. Assembly of ssDNA Probe-Decorated DNA Origami
The cross-shaped DNA origami, containing protruding ssDNA probes, was constructed by annealing a mixture of M13mp18 viral DNA and staple strands that included the ssDNA probe strands (1:5 molar ratio) in 1× TAE/Mg2+ buffer, from 90 to 15 °C over 12 h in a thermocycler. The DNA origami was then purified, using 100 kDa MWCO centrifugal filters purchased from Pall, Inc., to remove excess staple strands.
2.3. Fabrication of the Modified Electrode for miRNA Biosensors
The bare gold electrode (3 mm in diameter) was carefully polished to a mirror-like finish with a 0.3 and 0.05 μm alumina suspension and polishing cloth and was sonicated for 10 min in Milli-Q water. The polished gold electrode was then cleaned in 0.5 M sulfuric acid by an electrochemical method, with an applied potential ranging from 0.0 to 1.5 V, until the standard cyclic peak was observed. Finally, the mirror-finish gold electrode was dried with nitrogen and ready for further modification.
Subsequently, the cleaned electrode was dipped into a chitosan solution (1% chitosan solution in 1% acetic acid) to achieve a uniform surface coating, which was then stabilized by immersing the electrode into 0.1 M NaOH, followed by air-drying. Afterward, the DNA origami (1 nM) immobilization process was carried out in the immobilization buffer at room temperature for 4 h. Upon immobilization, the modified electrode was gently rinsed with the 0.1 M phosphate buffer to remove unbound origami structures and kept in the immobilization buffer for further use.
2.4. miRNA-21 Detection
The modified electrode containing ssDNA probes was incubated with miRNA-21 in the 2.0× SSC hybridization buffer at room temperature for 2 h. The electrode was then extensively washed with 0.2× SSC solution to remove nonhybridized miRNA from the electrode surface. For the MB-binding process, the electrode modified with miRNA/ssDNA hybrids was immersed in MB solution [4.0 μM MB; 0.2 M NaCl; 0.1 M phosphate-buffered saline (PBS)] for 5 min. The excess MB was washed thoroughly with washing buffer to make sure that the obtained current signal was only caused by MB bound in the sequences.
2.5. Electrochemical Experiments
Electrochemical measurements were performed with IVIUM CompactStat (Netherlands). The conventional three-electrode system, composed of a modified gold WE, a Pt mesh counter electrode, and an Ag/AgCl (3.0 M KCl) reference electrode, was employed. The biosensor fabrication process and the miRNA detection were characterized by cyclic voltammetry (CV) and electrochemical impedance spectroscopy (EIS). CV curves were performed in 5.0 mM [Fe(CN)6]3–/4– containing 0.1 M KCl with a scan range from −0.2 to +0.6 V. EIS experiments were performed with a frequency range from 0.1 Hz to 10 kHz. All experimental results were analyzed in terms of Randle’s equivalent circuit model. Linear sweep voltammetry (LSV) and differential pulse voltammetry (DPV) experiments were carried out at room temperature.
3. Results and Discussion
3.1. Mechanism of the Electrochemical Biosensor
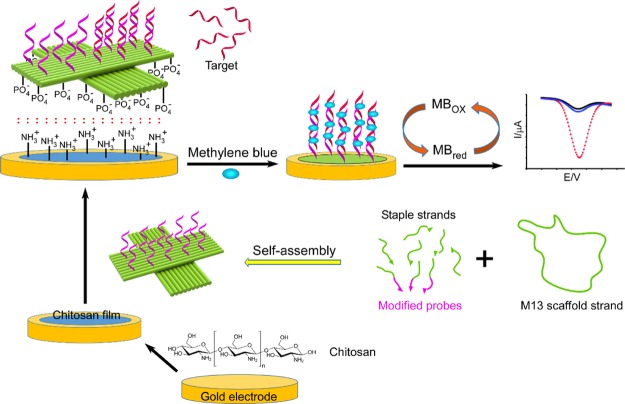
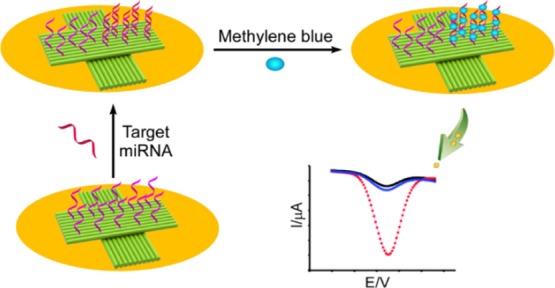
The working principle and electron transfer process of the DNA origami modified electrode are shown in Scheme 1. The precoated thin film of chitosan on the gold electrode was used to electrostatically absorb the DNA origami platforms, containing 10 preselected ssDNA probes by selectively extending the staple strands (see Supporting Information for detail). The successful formation of the cross-shaped DNA origami nanostructure was studied with an atomic force microscope (AFM), as shown in Figure S1. When the hybrids of the DNA probe–miRNA were formed, the MB molecules were bound to the hybridization chain, and the corresponding oxidation peak current of MB was directly proportional to the miRNA concentration. This relationship can be employed in the sensitive detection of miRNA.
Scheme 1. Illustration of a DNA Origami-Assisted Electrochemical Biosensor for miRNA Detection.
The cross-shaped DNA origami, containing preselected ssDNA probes, was immobilized on a gold electrode surface, via chitosan adsorption. MB molecules were used as redox indicators to show the differences before and after miRNA hybridization.
3.2. Electrochemical Characterization of Modified Electrodes
EIS and CV, as effective electrochemical characterization techniques, were employed to verify the stepwise electrode modification process. Figure 1 shows the EIS Nyquist plots that were recorded for each step of electrode modification, where the semicircle portion at higher frequencies corresponds to the electron transfer-limited process, and whose diameter is associated to electron-transfer resistance (Ret), and the linear portion at lower frequencies is related to the diffusion-limited process.39,40 It can be seen that the bare gold electrode demonstrated a very small semicircle (Ret = 1050 Ω), indicating a rapid electron transfer process of the redox probe ([Fe(CN)6]3–/4–) on the gold electrode.
Figure 1.
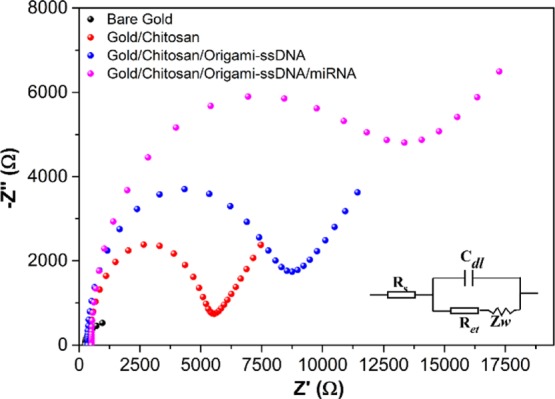
EIS curves of the bare gold electrode, the modification of gold electrode with chitosan (gold/chitosan), gold/chitosan/origami-ssDNA, and gold/chitosan/origami-ssDNA/miRNA. EIS was performed in 5.0 mM [Fe(CN)6]3–/4– containing 0.1 M KCl, with a frequency range from 0.1 Hz to 10 kHz.
Upon coating chitosan on the gold electrode surface (gold/chitosan), the Ret value was drastically increased to 5500 Ω, indicating that the chitosan was successfully coated on the electrode surface, which effectively hindered the negatively charged [Fe(CN)6]3–/4– to reach to the electrode surface. When the cross-shaped DNA origami nanostructures (1 nM) were further added (see Figure S2 for optimization of the concentration of DNA origami platforms) and immobilized onto gold/chitosan (defined as gold/chitosan/origami-ssDNA), the Ret value increased to 9020 Ω, which reflected the successful immobilization of the DNA origami on the gold/chitosan electrode surface. This increase in the Ret value could be attributed to the fact that the negatively charged DNA origami layer on the chitosan-modified electrode surface produced electrostatic repulsion and, thus, hindered the electron transfer between the redox probe ([Fe(CN)6]3–/4–) and the modified electrode surface. After hybridization of the miRNA-21 target and the gold/chitosan/origami-ssDNA probe, an even larger semicircle was observed, suggesting a further increase in Ret (14 700 Ω) and, thereby, a slower electron-transfer rate. This observation was made after the formation of miRNA/ssDNA probe hybrids on the DNA origami, which led to increased negative charges on the electrode surface and, therefore, amplified electrostatic repulsion [Fe(CN)6]3–/4–. All EIS results demonstrated that the miRNA biosensor had been successfully fabricated.
The EIS results were further validated by CV measurements carried out in 5.0 mM ([Fe(CN)6]3–/4–) solution containing 0.1 M KCl. The CV curves provided information regarding the electrochemical processes that happened at the electrode/solution interface. Figure 2 illustrates the CV behavior of the redox probe [Fe(CN)6]3–/4– on the gold electrode surface before and after each electrode modification step and the hybridization with the miRNA-21 target. When the chitosan was coated on the electrode, a decrease in peak current, as compared with the bare gold electrode, was observed, indicating that the chitosan film had partially obstructed the electron transfer. Upon immobilization of the cross-shaped DNA origami on the gold/chitosan electrode, the current decreased more, as a result of the negatively charged DNA backbone, as well as the spatial blockage of origami nanostructures between [Fe(CN)6]3–/4– and the electrode surface. After incubating with the solution of target miRNAs, the redox peak current decreased significantly, suggesting that the recognition of the target miRNAs with the ssDNA probes on the origami surface had reduced the conductivity and the efficiency of electron transfer between the electrode and redox solution. These results were in good accordance with those observed from the EIS measurements and, therefore, confirmed the successful fabrication of the biosensor.
Figure 2.
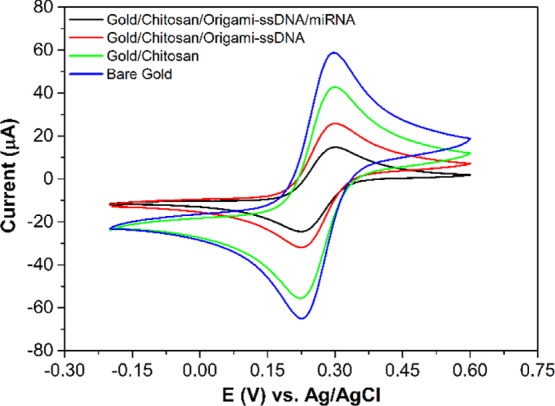
CV curves of the bare gold electrode, modifications of the gold electrode: gold/chitosan, gold/chitosan/origami-ssDNA, and gold/chitosan/origami-ssDNA/miRNA. CV curves were performed in 5.0 mM [Fe(CN)6]3–/4– containing 0.1 M KCl with a scan range from −0.2 to +0.6 V.
3.3. miRNA Detection Using MB as the Redox Indicator
To investigate the feasibility of the proposed miRNA detection, MB was utilized as an electrochemical indicator to monitor the hybridization interaction with the target miRNA. It has been demonstrated that the MB, a phenothiazine dye, can exhibit different binding affinities for ssDNA and double-stranded DNA (dsDNA) because MB can bind with ssDNA through the electrostatic interaction, while the effect of intercalations and electrostatic interactions coexist between MB and dsDNA.41 These differences, therefore, lead to different electrochemical responses upon MB interacting with ssDNA and dsDNA.42−44Figure 3A presents the LSV curves of gold/chitosan/origami-ssDNA and gold/chitosan/origami-ssDNA/miRNA after incubation with MB in PBS solution containing 0.1 M KCl. When the target miRNA-21 was absent, a peak current was observed at 3.99 μA (red). However, the peak current greatly increased to 6.72 μA, after hybridization of the gold/chitosan/origami-ssDNA probe with the miRNA-21 (green). This increase in electrochemical response indicated that more MB molecules were absorbed on the probe surface, which strongly supported the existence of ssDNA/miRNA hybrids on the electrode surface. This result was consistent with the previous studies.45,46 In addition, the effect of the scan rate on the CV responses of the MB on the fabricated electrode before and after miRNA target hybridization was investigated. As presented in Figure 3B, in both cases, the plots of the anodic and cathodic peak currents (Ipa and Ipc) against the scan rate (v) exhibit a linear relationship in the range of 0.05–1.0 V s–1, suggesting a surface-confined electrode process. Based on the slope of the curve of Ip versus v, the surface coverage of electroactive MB (ΓMB) on the electrode surface can be estimated using eq 1(47)
| 1 |
where F is Faraday’s constant (96 485 C mol–1), n is the number of electrons transferred (n = 2 for MB), A is the area of the electrode (cm2), Γ is the surface coverage, v is the scan rate (V s–1), R is the gas constant (8.314 J K–1 mol–1), and T is the temperature. From the slope of Ipc versus v shown in Figure 3B, the calculated ΓMB for the gold/chitosan/origami-ssDNA/miRNA was 918.9 pmol/cm2, which is, as expected, much higher than that for the gold/chitosan/origami-ssDNA (213.1 pmol/cm2). This indicates that more MB could bind to miRNA/ssDNA probe duplexes, facilitating the electron transfer from the MB molecules to the electrode. Therefore, these results confirm that this miRNA biosensor is suitable for detection of miRNA, according to the reduction response of MB.
Figure 3.
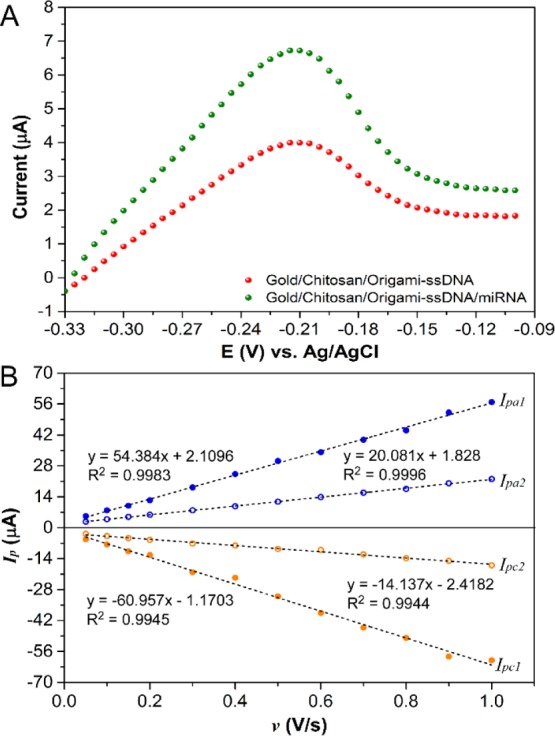
Representative LSV curves, recorded in 1× PBS buffer containing 0.1 M KCl for ssDNA probes (red curve) and miRNA/ssDNA probes (green curve), after incubation with 4 μM MB solution for 5 min (A), dependence of the MB redox peak current on v for ssDNA and miRNA/ssDNA (B). Ipa1 and Ipc1 are for gold/chitosan/origami-ssDNA/miRNA and Ipa2 and Ipc2 are for gold/chitosan/origami-ssDNA.
3.4. Sensitivity and Selectivity of the miRNA Analysis
To investigate the detectable efficiency of the proposed gold/chitosan/origami-ssDNA biosensor, a sensitivity study was conducted via DPV measurements at various concentrations of the target miRNA-21. As illustrated in Figure 4A, the oxidation currents of MB noticeably increased with an increase in the miRNA-21 concentrations. The resulting logarithmic plot of miRNA-21 concentrations versus MB oxidation currents, as shown in Figure 4B, exhibits a good linear relationship over a range of 100 fM to 10 nM, with a regression equation of ΔI (μA) = 1.3702 log C + 18.084 (n = 3), where ΔI represents the oxidation current difference between the presence and absence of the miRNA-21 target, and C denotes the concentration of miRNA-21. The detection limit of the proposed biosensor was estimated to be as low as 79.8 fM, based on the equation 3σ/a, where “σ” is the standard deviation, and “a” is the slope of the linear regression. This relatively wide measuring range and lower detection limit indicated that the fabricated miRNA-21 biosensor, based on a cross-shaped DNA nanostructure and coupled with MB as a hybridization redox indicator, can be prepared for miRNA-21 assays.
Figure 4.
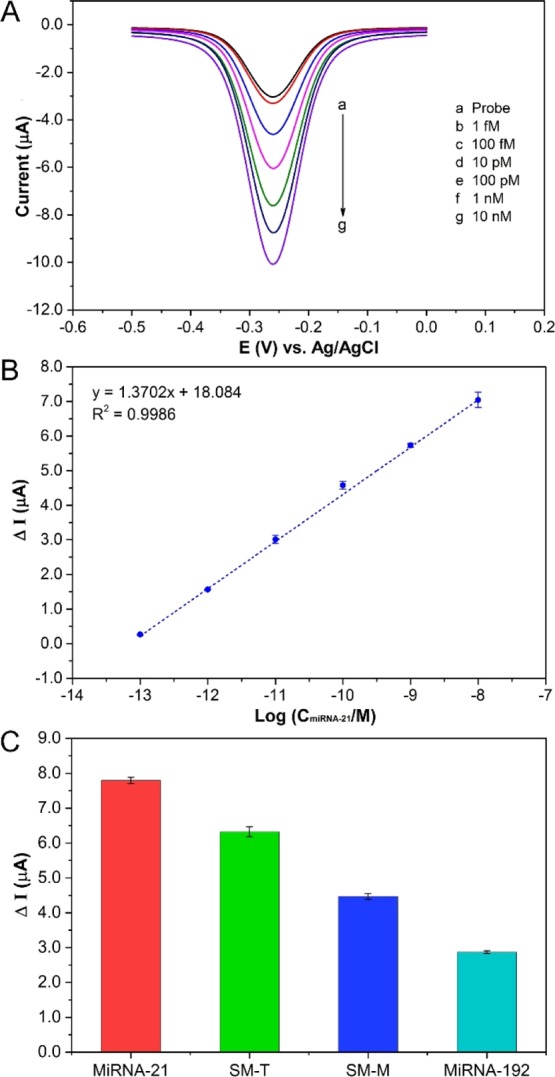
DPV responses for oxidation of MB at different concentrations of miRNA (A), the relationship between ΔI and logarithm of miRNA-21 concentration (B), and selectivity of the proposed biosensor for the detection of target and mismatch miRNA (C).
Furthermore, the DNA origami-based electrochemical biosensor presents a facile and cost-effective detection strategy as compared to using complex multistep fabrication and expensive enzymes (see Table 1). In addition, it was noted that the background noise of the ssDNA probe was relatively high due to the binding interaction between MB and DNA origami platforms (Figure S3). In order to decrease the background noise and improve the detection limitation of the miRNA biosensor, optimization of the structural design of the DNA origami platform and the density of ssDNA probes are needed.
Table 1. Comparison with the Reported DNA Nanostructure-Based miRNA Detection.
| strategy | analytical technique | linear range (pM) | LOD (fM) | ref |
|---|---|---|---|---|
| avidin-HRP/DNA-thiols/gold | CV | 1 | (26) | |
| Klenow/AgNP/DNA-thiols/gold | LSV | 0.001–1000 | 0.4 | (24) |
| poly-HRP/DNA-thiols/gold | amperometric | 0.01–1000 | 10 | (48) |
| AgNP/AuNP/DNA-thiols/gold | LSV | 0.0001–100 | 0.002 | (49) |
| hemin/DNA-thiols/gold | amperometric | 0.5–10 000 | 176 | (50) |
| avidin-HRP/DNA-thiols/gold | amperometric | 0.01–10 000 | 0.01 | (27) |
| avidin-HRP/DNA-thiols/gold | chronoamperometric | 0.001–1 | 0.00093 | (28) |
| ferrocene/DNA-thiols/AuNPs/gold | DPV | 100–1 000 000 | 10 | (51) |
| MB/DNA-thiols/gold | DPV | 1–100 | 1000 | (52) |
| MB/DNA origami/chitosan/gold | DPV | 0.1–10 000 | 79.8 | this study |
In addition, the selectivity of the developed miRNA-21 biosensor was examined by comparing the DPV’s current response of the target miRNA-21 with that of a noncomplementary miRNA (miRNA-192), and single-base mismatched sequences including at terminal position (SM-T) and middle position (SM-M) under the same conditions. As can be seen in Figure 4C, the peak current variation (ΔI) of the complementary miRNA-21 is much higher than that of the noncomplementary miRNA-192 sequence, SM-T, and SM-M. These results confirmed that this constructed biosensor had good specificity and could distinguish the target miRNA-21 from single-mismatched sequences and other analogous interferences.
3.5. miRNA-21 Analysis in Human Serum
Human biological fluids contain many species interfering with sensitive miRNA analysis. In this study, human serum was utilized as real biological samples for target miRNA-21 detection. Three concentrations of miRNA-21 (1, 10, and 1000 pM) spiked into 1% human serum were tested by the developed biosensor. The resulted relative standard deviation (RSD) and percentage of recovery are calculated and listed in Table 2. For every concentration of miRNA-21 in human serum, the recovery percentage was in nearly quantitative yield with low RSD, indicating that the DNA origami-based biosensor is a good analytical tool in miRNA assay, which possesses excellent practical application in cancer clinics and diagnosis.
Table 2. Recovery Detection of miRNA-21 in Human Serum with the Proposed Biosensor (n = 3).
| sample | added (pM) | detected (pM) | RSD (%) | recovery (%) |
|---|---|---|---|---|
| 1 | 1 | 1.02, 1.06, 0.93 | 6.64 | 100.33 |
| 2 | 10 | 9.85, 10.1, 9.92 | 1.30 | 99.57 |
| 3 | 1000 | 1003.03, 976.42, 986.32 | 1.36 | 98.86 |
4. Conclusions
In summary, we have fabricated a label-free and amplification-free electrochemical biosensor, based on cross-shaped DNA origami nanostructures, for miRNA-21 detection. This was coupled with MB as a hybridization redox indicator. Instead of using covalent immobilization of DNA nanostructures on gold electrodes, via thiol–Au interactions, the physical absorption of DNA origami by the chitosan film was utilized for the first time. The stable and effective immobilization of the DNA origami was confirmed by EIS and CV experiments. The electrochemical signal of the DNA biosensor originated from the different responses of the MB molecules that were bound with ssDNA and the hybridization of miRNA/DNA. The fabrication method of this biosensor was simple and highly sensitive, with a 79.8 fM detection limit of miRNA by using the DPV method. In addition, the free-standing ssDNA probes that were supported by the DNA origami not only enhanced probe accessibility but also provided a possibility of detecting multiple targets and larger molecules (proteins, virus, and aptamers) because of the rationally controlled number and density of DNA probes on the DNA origami surface. Furthermore, the biosensor is effective to distinguish single-base mismatched sequences from target miRNA-21, as well as exhibits satisfactory performance in human serum, indicating its great potential for application in disease diagnosis and assessment.
Acknowledgments
This work was supported by the National Science Foundation under grants CCF-1814797. We thank Dr. Manashi Nath, and Dr. Jahangir Masud, at Chemistry of Missouri University S&T for providing valuable discussion, access, and training to IVIUM CompactStat (Netherlands).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.9b01166.
AFM images, optimization of the concentration of DNA probes, the background noise caused by MB binding, and DNA sequences (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Treiber T.; Treiber N.; Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. 10.1038/s41580-018-0059-1. [DOI] [PubMed] [Google Scholar]
- Yokoi A.; Matsuzaki J.; Yamamoto Y.; Yoneoka Y.; Takahashi K.; Shimizu H.; Uehara T.; Ishikawa M.; Ikeda S. I.; Sonoda T.; Kawauchi J.; Takizawa S.; Aoki Y.; Niida S.; Sakamoto H.; Kato K.; Kato T.; Ochiya T. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat. Commun. 2018, 9, 4319. 10.1038/s41467-018-06434-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardsen E.; Andersen S.; Melbo-Jorgensen C.; Rakaee M.; Ness N.; Al-Saad S.; Nordby Y.; Pedersen M. I.; Donnem T.; Bremnes R. M.; Busund L. T. MicroRNA 141 is associated to outcome and aggressive tumor characteristics in prostate cancer. Sci. Rep. 2019, 9, 386. 10.1038/s41598-018-36854-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D.-C.; Li Q.-G.; Ding X.-W.; Ding Y.-T. Circulating microRNAs: potential biomarkers for cancer. Int. J. Mol. Sci. 2011, 12, 2055–2063. 10.3390/ijms12032055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farran B.; Dyson G.; Craig D.; Dombkowski A.; Beebe-Dimmer J. L.; Powell I. J.; Podgorski I.; Heilbrun L.; Bolton S.; Bock C. H. A study of circulating microRNAs identifies a new potential biomarker panel to distinguish aggressive prostate cancer. Carcinogenesis 2018, 39, 556–561. 10.1093/carcin/bgy025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. W.; Li Z.; Moore P. S.; Monaghan A. P.; Chang Y.; Nichols M.; John B. A sensitive non-radioactive northern blot method to detect small RNAs. Nucleic Acids Res. 2010, 38, e98 10.1093/nar/gkp1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.; Zhang S.; Ouyang C.; Wang Z.; Wu D.; Liu Y.; Jiang Y.; Wu Z.-S. DNA nanostructures from palindromic rolling circle amplification for the fluorescent detection of cancer-related microRNAs. Talanta 2019, 192, 175–181. 10.1016/j.talanta.2018.07.090. [DOI] [PubMed] [Google Scholar]
- Zeng G.; Xun W.; Wei K.; Yang Y.; Shen H. MicroRNA-27a-3p regulates epithelial to mesenchymal transition via targeting YAP1 in oral squamous cell carcinoma cells. Oncol. Rep. 2016, 36, 1475–1482. 10.3892/or.2016.4916. [DOI] [PubMed] [Google Scholar]
- de Ronde M. W. J.; Ruijter J. M.; Lanfear D.; Bayes-Genis A.; Kok M. G. M.; Creemers E. E.; Pinto Y. M.; Pinto-Sietsma S.-J. Practical data handling pipeline improves performance of qPCR-based circulating miRNA measurements. RNA 2017, 23, 811–821. 10.1261/rna.059063.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.; Peng J.; Wang J.; Tang H.; Tan L.; Xie Q.; Yao S. Carbon nanotube-based label-free electrochemical biosensor for sensitive detection of miRNA-24. Biosens. Bioelectron. 2014, 54, 158–164. 10.1016/j.bios.2013.10.061. [DOI] [PubMed] [Google Scholar]
- Tian Q.; Wang Y.; Deng R.; Lin L.; Liu Y.; Li J. Carbon nanotube enhanced label-free detection of microRNAs based on hairpin probe triggered solid-phase rolling-circle amplification. Nanoscale 2015, 7, 987–993. 10.1039/c4nr05243a. [DOI] [PubMed] [Google Scholar]
- Liu S.; Yang Z.; Chang Y.; Chai Y.; Yuan R. An enzyme-free electrochemical biosensor combining target recycling with Fe3O4/CeO2@Au nanocatalysts for microRNA-21 detection. Biosens. Bioelectron. 2018, 119, 170–175. 10.1016/j.bios.2018.08.006. [DOI] [PubMed] [Google Scholar]
- Song Y.; Xu T.; Xu L.-P.; Zhang X. Nanodendritic gold/graphene-based biosensor for tri-mode miRNA sensing. Chem. Commun. 2019, 55, 1742–1745. 10.1039/c8cc09586h. [DOI] [PubMed] [Google Scholar]
- Wang J.; Zhang L.; Lu L.; Kang T. Molecular beacon immobilized on graphene oxide for enzyme-free signal amplification in electrochemiluminescent determination of microRNA. Mikrochim. Acta 2019, 186, 142. 10.1007/s00604-019-3252-9. [DOI] [PubMed] [Google Scholar]
- Cui L.; Wang M.; Sun B.; Ai S.; Wang S.; Zhang C.-y. Substrate-free and label-free electrocatalysis-assisted biosensor for sensitive detection of microRNA in lung cancer cells. Chem. Commun. 2019, 55, 1172–1175. 10.1039/c8cc09688k. [DOI] [PubMed] [Google Scholar]
- Qiu X.; Hildebrandt N. Rapid and Multiplexed MicroRNA Diagnostic Assay Using Quantum Dot-Based Forster Resonance Energy Transfer. ACS Nano 2015, 9, 8449–8457. 10.1021/acsnano.5b03364. [DOI] [PubMed] [Google Scholar]
- Bharti A.; Agnihotri N.; Prabhakar N. A voltammetric hybridization assay for microRNA-21 using carboxylated graphene oxide decorated with gold-platinum bimetallic nanoparticles. Microchim. Acta 2019, 186, 185. 10.1007/s00604-019-3302-3. [DOI] [PubMed] [Google Scholar]
- Zhang T.; Chai H.; Meng F.; Guo Z.; Jiang Y.; Miao P. DNA-Functionalized Porous Fe3O4 Nanoparticles for the Construction of Self-Powered miRNA Biosensor with Target Recycling Amplification. ACS Appl. Mater. Interfaces 2018, 10, 36796–36804. 10.1021/acsami.8b15419. [DOI] [PubMed] [Google Scholar]
- Qi L.; Xiao M.; Wang X.; Wang C.; Wang L.; Song S.; Qu X.; Li L.; Shi J.; Pei H. DNA-encoded Raman-active anisotropic nanoparticles for microRNA detection. Anal. Chem. 2017, 89, 9850–9856. 10.1021/acs.analchem.7b01861. [DOI] [PubMed] [Google Scholar]
- Xiao M.; Man T.; Zhu C.; Pei H.; Shi J.; Li L.; Qu X.; Shen X.; Li J. MoS2 nanoprobe for microRNA quantification based on duplex-specific nuclease signal amplification. ACS Appl. Mater. Interfaces 2018, 10, 7852–7858. 10.1021/acsami.7b18984. [DOI] [PubMed] [Google Scholar]
- Seeman N. The design and engineering of nucleic acid nanoscale assemblies. Curr. Opin. Struct. Biol. 1996, 6, 519–526. 10.1016/s0959-440x(96)80118-7. [DOI] [PubMed] [Google Scholar]
- Kuzuya A.; Wang R.; Sha R.; Seeman N. C. Six-helix and eight-helix DNA nanotubes assembled from half-tubes. Nano Lett. 2007, 7, 1757–1763. 10.1021/nl070828k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H.; Lu N.; Wen Y.; Song S.; Liu Y.; Yan H.; Fan C. A DNA nanostructure-based biomolecular probe carrier platform for electrochemical biosensing. Adv. Mater. 2010, 22, 4754–4758. 10.1002/adma.201002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao P.; Wang B.; Chen X.; Li X.; Tang Y. Tetrahedral DNA nanostructure-based microRNA biosensor coupled with catalytic recycling of the analyte. ACS Appl. Mater. Interfaces 2015, 7, 6238–6243. 10.1021/acsami.5b01508. [DOI] [PubMed] [Google Scholar]
- Ge Z.; Lin M.; Wang P.; Pei H.; Yan J.; Shi J.; Huang Q.; He D.; Fan C.; Zuo X. Hybridization chain reaction amplification of microRNA detection with a tetrahedral DNA nanostructure-based electrochemical biosensor. Anal. Chem. 2014, 86, 2124–2130. 10.1021/ac4037262. [DOI] [PubMed] [Google Scholar]
- Lin M.; Wen Y.; Li L.; Pei H.; Liu G.; Song H.; Zuo X.; Fan C.; Huang Q. Target-responsive, DNA nanostructure-based E-DNA sensor for microRNA analysis. Anal. Chem. 2014, 86, 2285–2288. 10.1021/ac500251t. [DOI] [PubMed] [Google Scholar]
- Wen Y.; Pei H.; Shen Y.; Xi J.; Lin M.; Lu N.; Shen X.; Li J.; Fan C. DNA Nanostructure-based Interfacial engineering for PCR-free ultrasensitive electrochemical analysis of microRNA. Sci. Rep. 2012, 2, 867. 10.1038/srep00867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; Huang J.; Zhang S.; Mo F.; Su S.; Li Y.; Fang L.; Deng J.; Huang H.; Luo Z.; Zheng J. Electrochemical Biosensor for DNA Methylation Detection through Hybridization Chain-Amplified Reaction Coupled with a Tetrahedral DNA Nanostructure. ACS Appl. Mater. Interfaces 2019, 11, 3745–3752. 10.1021/acsami.8b20144. [DOI] [PubMed] [Google Scholar]
- Dong S.; Zhao R.; Zhu J.; Lu X.; Li Y.; Qiu S.; Jia L.; Jiao X.; Song S.; Fan C.; Hao R.; Song H. Electrochemical DNA Biosensor Based on a Tetrahedral Nanostructure Probe for the Detection of Avian Influenza A (H7N9) Virus. ACS Appl. Mater. Interfaces 2015, 7, 8834–8842. 10.1021/acsami.5b01438. [DOI] [PubMed] [Google Scholar]
- Wen Y.; Pei H.; Wan Y.; Su Y.; Huang Q.; Song S.; Fan C. DNA nanostructure-decorated surfaces for enhanced aptamer-target binding and electrochemical cocaine sensors. Anal. Chem. 2011, 83, 7418–7423. 10.1021/ac201491p. [DOI] [PubMed] [Google Scholar]
- Abi A.; Lin M.; Pei H.; Fan C.; Ferapontova E. E.; Zuo X. Electrochemical switching with 3D DNA tetrahedral nanostructures self-assembled at gold electrodes. ACS Appl. Mater. Interfaces 2014, 6, 8928–8931. 10.1021/am501823q. [DOI] [PubMed] [Google Scholar]
- Xu F.; Dong H.; Cao Y.; Lu H.; Meng X.; Dai W.; Zhang X.; Al-Ghanim K. A.; Mahboob S. Ultrasensitive and Multiple Disease-Related MicroRNA Detection Based on Tetrahedral DNA Nanostructures and Duplex-Specific Nuclease-Assisted Signal Amplification. ACS Appl. Mater. Interfaces 2016, 8, 33499–33505. 10.1021/acsami.6b12214. [DOI] [PubMed] [Google Scholar]
- Qu X.; Xiao M.; Li F.; Lai W.; Li L.; Zhou Y.; Lin C.; Li Q.; Ge Z.; Wen Y.; Pei H.; Liu G. Framework nucleic acid-mediated pull-down microRNA detection with hybridization chain reaction amplification. ACS Appl. Bio Mater. 2018, 1, 859–864. 10.1021/acsabm.8b00278. [DOI] [PubMed] [Google Scholar]
- Rothemund P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- Liu W.; Zhong H.; Wang R.; Seeman N. C. Crystalline two-dimensional DNA-origami arrays. Angew. Chem., Int. Ed. 2011, 50, 264–267. 10.1002/anie.201005911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz H.; Douglas S. M.; Shih W. M. Folding DNA into Twisted and Curved Nanoscale Shapes. Science 2009, 325, 725–730. 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranwal A.; Kumar A.; Priyadharshini A.; Oggu G. S.; Bhatnagar I.; Srivastava A.; Chandra P. Chitosan: An undisputed bio-fabrication material for tissue engineering and bio-sensing applications. Int. J. Biol. Macromol. 2018, 110, 110–123. 10.1016/j.ijbiomac.2018.01.006. [DOI] [PubMed] [Google Scholar]
- Mishra S. K.; Kannan S. Development, mechanical evaluation and surface characteristics of chitosan/polyvinyl alcohol-based polymer composite coatings on titanium metal. J. Mech. Behav. Biomed. Mater. 2014, 40, 314–324. 10.1016/j.jmbbm.2014.08.014. [DOI] [PubMed] [Google Scholar]
- Li X.; Shen L.; Zhang D.; Qi H.; Gao Q.; Ma F.; Zhang C. Electrochemical impedance spectroscopy for study of aptamer-thrombin interfacial interactions. Biosens. Bioelectron. 2008, 23, 1624–1630. 10.1016/j.bios.2008.01.029. [DOI] [PubMed] [Google Scholar]
- Tlili C.; Korriyoussoufi H.; Ponsonnet L.; Martelet C.; Jaffrezicrenault N. Electrochemical impedance probing of DNA hybridisation on oligonucleotide-functionalised polypyrrole. Talanta 2005, 68, 131–137. 10.1016/j.talanta.2005.04.069. [DOI] [PubMed] [Google Scholar]
- Zhu L.; Zhao R.; Wang K.; Xiang H.; Shang Z.; Sun W. Electrochemical behaviors of methylene blue on DNA modified electrode and its application to the detection of PCR product from NOS sequence. Sensors 2008, 8, 5649–5660. 10.3390/s8095649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y.; Yao X.; Liu Q.; Li J. Hairpin DNA probe based electrochemical biosensor using methylene blue as hybridization indicator. Biosens. Bioelectron. 2007, 22, 1126–1130. 10.1016/j.bios.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Silvestrini M.; Fruk L.; Moretto L. M.; Ugo P. Detection of DNA Hybridization by Methylene Blue Electrochemistry at Activated Nanoelectrode Ensembles. J. Nanosci. Nanotechnol. 2015, 15, 3437–3442. 10.1166/jnn.2015.10214. [DOI] [PubMed] [Google Scholar]
- Li Y.; Chang Y.; Yuan R.; Chai Y. Highly Efficient Target Recycling-Based Netlike Y-DNA for Regulation of Electrocatalysis toward Methylene Blue for Sensitive DNA Detection. ACS Appl. Mater. Interfaces 2018, 10, 25213–25218. 10.1021/acsami.8b08545. [DOI] [PubMed] [Google Scholar]
- Yan F.; Erdem A.; Meric B.; Kerman K.; Ozsoz M.; Sadik O. A. Electrochemical DNA Biosensor for the Detection of Specific Gene Related to Microcystis Species. Electrochem. Commun. 2001, 3, 224–228. 10.1016/s1388-2481(01)00149-7. [DOI] [Google Scholar]
- Gu J.; Lu X.; Ju H. DNA Sensor for Recognition of Native Yeast DNA Sequence with Methylene Blue as an Electrochemical Hybridization Indicator. Electroanalysis 2002, 14, 949–954. . [DOI] [Google Scholar]
- Bard A. J.; Faulkner L. R.. Electrochemical Methods: Fundamentals and Applications, 2nd ed.; John Wiley & Sons, Inc.: New York, 2001. [Google Scholar]
- Zeng D.; Wang Z.; Meng Z.; Wang P.; San L.; Wang W.; Aldalbahi A.; Li L.; Shen J.; Mi X. DNA tetrahedral nanostructure-based electrochemical miRNA biosensor for simultaneous detection of multiple miRNAs in pancreatic carcinoma. ACS Appl. Mater. Interfaces 2017, 9, 24118–24125. 10.1021/acsami.7b05981. [DOI] [PubMed] [Google Scholar]
- Miao P.; Tang Y.; Yin J. MicroRNA detection based on analyte triggered nanoparticle localization on a tetrahedral DNA modified electrode followed by hybridization chain reaction dual amplification. Chem. Commun. 2015, 51, 15629–15632. 10.1039/c5cc05499k. [DOI] [PubMed] [Google Scholar]
- Huang Y. L.; Mo S.; Gao Z. F.; Chen J. R.; Lei J. L.; Luo H. Q.; Li N. B. Amperometric biosensor for microRNA based on the use of tetrahedral DNA nanostructure probes and guanine nanowire amplification. Microchim. Acta 2017, 184, 2597–2604. 10.1007/s00604-017-2246-8. [DOI] [Google Scholar]
- Liu S.; Su W.; Li Z.; Ding X. Electrochemical detection of lung cancer specific microRNAs using 3D DNA origami nanostructures. Biosens. Bioelectron. 2015, 71, 57–61. 10.1016/j.bios.2015.04.006. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Huang Y.; Li C. A reusable electrochemical sensor for one-step biosensing in complex media using triplex-forming oligonucleotide coupled DNA nanostructure. Anal. Chim. Acta 2019, 1055, 90–97. 10.1016/j.aca.2018.12.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.