Abstract
Objective
Diffusion tensor imaging (DTI) of the white matter is a biomarker for neurological disease burden in tuberous sclerosis complex (TSC). To clarify the basis of abnormal diffusion in TSC, we correlated ex vivo high‐resolution diffusion imaging with histopathology in four tissue types: cortex, tuber, perituber, and white matter.
Methods
Surgical specimens of three children with TSC were scanned in a 3T or 7T MRI with a structural image isotropic resolution of 137–300 micron, and diffusion image isotropic resolution of 270‐1,000 micron. We stained for myelin (luxol fast blue, LFB), gliosis (glial fibrillary acidic protein, GFAP), and neurons (NeuN) and registered the digitized histopathology slides (0.686 micron resolution) to MRI for visual comparison. We then performed colocalization analysis in four tissue types in each specimen. Finally, we applied a linear mixed model (LMM) for pooled analysis across the three specimens.
Results
In white matter and perituber regions, LFB optical density measures correlated with fractional anisotropy (FA) and inversely with mean diffusivity (MD). In white matter only, GFAP correlated with MD, and inversely with FA. In tubers and in the cortex, there was little variation in mean LFB and GFAP signal intensity, and no correlation with MRI metrics. Neuronal density correlated with MD. In the analysis of the combined specimens, the most robust correlation was between white matter MD and LFB metrics.
Interpretation
In TSC, diffusion imaging abnormalities in microscopic tissue types correspond to specific histopathological markers. Across all specimens, white matter diffusivity correlates with myelination.
Introduction
Tuberous sclerosis complex (TSC) is a genetic neurocutaneous disorder with prominent neurological involvement. Inactivation of TSC1 or TSC2 genes leads to pathologically enhanced activity of the mechanistic target of rapamycin (mTOR) pathway, with subsequent disinhibition of protein synthesis and cell growth. In the brain, aberrant cellular proliferation, differentiation, and migration lead to various malformations including cortical tubers which contain cells with an ambiguous phenotype of both astrocytic and neuronal lineage.1
Diffusion imaging in TSC reveals abnormalities in white matter appearing normal on structural MRI.2, 3 Fractional anisotropy (FA) and mean diffusivity (MD) measures of major white matter tracts correlate with the neurological phenotype and can be used as a surrogate marker of disease burden.4, 5 These same DTI metrics change in response to treatment with mTOR inhibitors.6, 7 Aberrant myelination inhibition has been reported in both neuronal and oligodendrocyte knockout mouse models of TSC, and can be (partially) prevented with early use of mTOR inhibitors.8, 9 Increased heterotopic and dysplastic neurons has been reported in one of these models, however, and extensive astrocyte pathology is found in tubers and perituber tissue.10, 11 Thus, while we suspect dysmyelination of the white matter may drive abnormal white matter diffusion in human TSC, it is not known whether astrocytosis or neuronal ectopy are responsible too.
In addition, in patients with TSC undergoing epilepsy surgery, epileptogenic tubers and the perituber rim have an increased MD compared to nonepileptogenic regions.12, 13, 14 Pathology‐imaging correlations are needed, as they could be translated into better detection and delineation of subtle lesions, advance the prediction of histopathology from in vivo MRI, and ultimately improve surgical outcomes.15, 16, 17
To elucidate the histopathological basis of diffusion abnormality in TSC, we directly compared quantitative neuropathological data from epilepsy resection specimens to ex vivo high‐resolution structural and diffusion imaging data. Here, we focused on myelination, astrocytosis, and neuronal density in the lesional and perilesional tissues of patients with TSC.
Methods
Subjects
Three children with TSC followed in the Multidisciplinary TSC Program at Boston Children's Hospital underwent epilepsy surgery for refractory seizures in 2015 and 2016. Inclusion criteria included (1) resection performed “en bloc” suitable for imaging and analysis; (2) preoperative diffusion MRI as part of the surgical workup; and (3) diagnosis of TSC by clinical and/or genetic criteria.18 The study was approved by the institution's IRB, and informed consent was obtained in all participants.
Preoperative MR acquisition
Preoperative MRI consisted of 3T MRI and included both structural imaging and diffusion‐weighted imaging, and has been previously described (Data S1).19
Specimen preparation
Surgical resections were performed “en bloc,” while adhering to current standards of pediatric neurosurgical care. Tissue was then immediately fixed in formalin for 24 h prior to imaging, to prevent tissue deformation and decay (Fig. 1). Next, the tissue was transferred to proton‐free Galden® perfluoropolyether (Solvay Solexis, Inc., NJ) to avoid susceptibility artifact15 and ensure no signal in the background during image acquisition.
Figure 1.
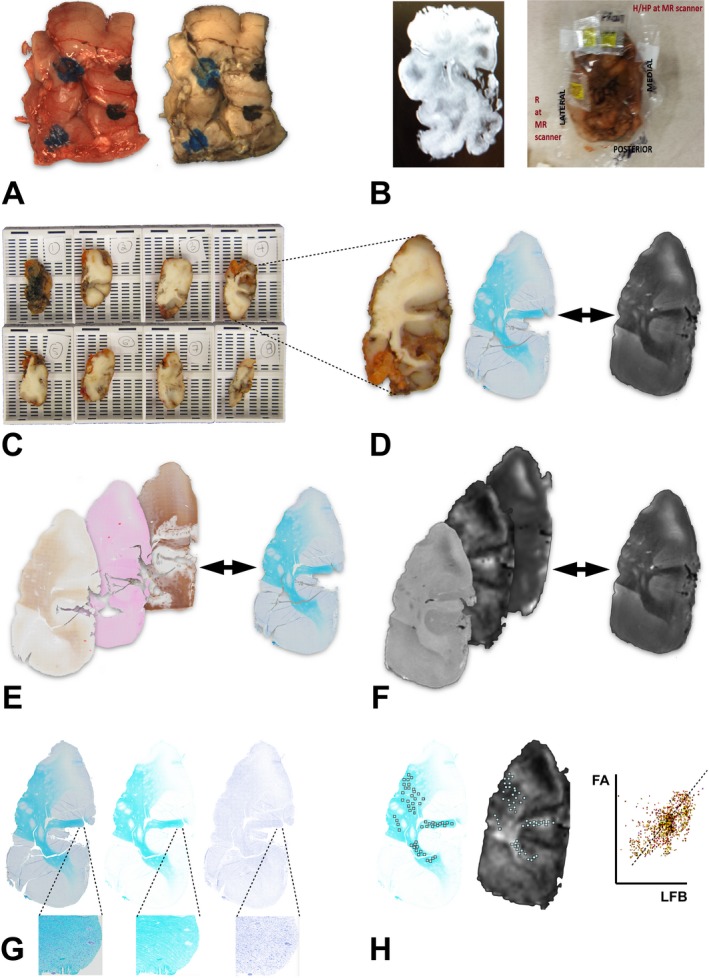
Procedure for comparison of ex vivo histopathology and imaging. (A) Gross resection specimen fresh and after fixation. The blue ink marks the cortical surface. (B) During imaging, potassium pills are placed on the specimen for orientation. A screenshot was made from the scanner display for reference. (C) Block sectioning is done at an orientation that approaches one of the imaging planes. (D) A high‐resolution slide, digitized at 0.686 micron, is registered to a high‐contrast ex vivo MR T2 image at 137 micron resolution. (E) Pathology slides are registered to the reference pathology slide. (F) All imaging sequences are also registered to the reference image. (G) With color deconvolution, the primary color of interest is extracted from the digitized pathology slide (left). In this case, the luxol fast blue (LFB) is isolated from cresyl violet in an 8‐bit monochrome image at the same resolution (middle). The cellular nuclei form a separate image (right), better visible at the higher magnification image inserts. (H) Multiple ROIs are now colocated in corresponding areas of interest in the histopathology and diffusion images. A sample colocalization of intensity values is shown.
While fixation of tissue alters tissue diffusion profiles, the principal eigenvector of the tensor is clearly defined in ex vivo tissue, with good directional correspondence to in vivo data.20, 21 The Computational Radiology Laboratory has previously optimized acquisition methods for ex vivo brain diffusion‐weighted imaging.22
Ex vivo MR acquisition
3 Tesla MRI acquisitions were performed on Siemens Skyra scanner with 64‐channel head coil and a 7 mm loop coil. 7 Tesla MRI acquisitions were performed on Bruker BioSpec 70/30 system with a surface coil, as detailed in the Data S1.
Isotropic resolution for structural imaging ranged from 137 to 300 micron and for diffusion imaging was 1000 micron, 270 micron, and 750 micron for specimens 1, 2, and 3, respectively. T1w, T2w, FA, and MD images were processed on the in‐house developed Computational Radiology imaging toolkit.
Figure 2 illustrates the location of the resected regions in the preoperative MRI, and compares the in vivo images to ex vivo imaging, and to gross pathology, for orientation purposes. Due to the relatively low 2.5 × 2.5 x 2.5 mm resolution of clinically acquired in vivo diffusion images, only visual comparison with high‐resolution ex vivo images was done. Similarly, given complex and variable orientation of the specimen in different scanners, we did not attempt to reorient the gradient directions, and thus limited the analysis to the magnitude of tensor metrics. More detail can be found in the Data S1.
Figure 2.
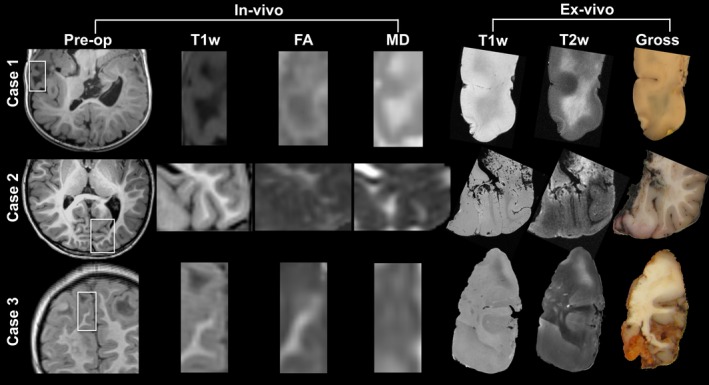
Preoperative imaging and orientation of resection specimens. Resection regions are highlighted in the white rectangle in the preoperative images. Magnified frames of T1‐weighted (T1w), fractional anisotropy (FA), and mean diffusivity (MD) are shown, and compared to the ex vivo images on the right. Note that the imaging plane may be inverted and manipulated to match the ex vivo cutting plane and histopathology slide; for example, case 1 is a pseudo‐axial plane, and images may be inverted along any axis. Pre‐op preoperative MRI; T1w T1‐weighted MRI; T2w T2‐weighted MRI;FA fractional anisotropy; MD mean diffusivity.
Blocking specimen, conventional and immunostaining, digitization
Once imaged, the neuropathologist (HL) blocked the specimen with a thickness of 3–7 mm, accounting for the orientation of acquired MR images (Fig. 1). Tissue was embedded in paraffin using a Leica Peloris tissue processor. Sections were cut at 6 microns (except LFB, at 13 microns) using the Leica RM2255 microtome. Conventional stains were done on a Roche Ventana automated stainer, and immunostaining was done using a Roche BenchMark XT platform with manufacturer protocols optimized for CNS tissue. The following stains were used: Hematoxylin and eosin (H&E; Ventana, Tucson, AZ), Luxol fast blue (LFB; Histo‐Chem, Inc., Jefferson, AR), and immunostaining with glial fibrillary acidic protein (GFAP; Dako, Cambridge, MA, polyclonal, 1:1000) and NeuN neuronal stain (NeuN; Chemicon, Temecula, CA, monoclonal, 1:1000). Whole slide digitization took place on an Olympus VS‐120 Whole Slide Scanner, at 10× magnification at 0.686 micron resolution.
Postprocessing, alignment
MRI slices were compared to digitized histological slides by visual comparison at first, based on anatomical landmarks, tissue edges, and artifacts.23, 24 Neuropathology registration to the 3D MRI volume was performed with the Histolozee tool from the Penn Image Computing and Science Lab (picsl.upenn.edu). The remaining histology slides were nonrigidly aligned to the reference slide with ImageJ (Fig. 1), using anatomical landmarks on the edge of the specimen.25 Note that 3D reconstruction of histology is outside of the scope of this paper.
All ex vivo MR images including diffusion data were registered to the reference isotropic T1w or T2w structural data series, with rigid and nonrigid registration using a block matching paradigm, to compensate for residual distortion.26
Histopathology and MRI registration
In ImageJ, histology images were color deconvoluted to extract the stain of interest, and obtain optical density measures of LFB and GFAP.24, 27 Neuronal density and size within each ROI was calculated with an automated cell‐count in NeuN‐stained histological sections as previously described.28 On the basis of previous work, and if present in the sample, we selected regions of interest (ROIs) in white matter not adjacent to the tuber (“white matter”), transitional tissue between white matter and tuber (“perituber white matter”), and tuber.19, 29
A minimum of 100 ROIs were selected, with variable surface area but each encompassing multiple pixels, to assure accuracy and stability of findings.30 The ROIs together formed tissue‐specific masks at the resolution of the digitized histopathology slide. Further details can be found in the Data S1.
Statistical analysis
Three statistical analyses were done to address three different questions:
In each specimen, regional differences between the histological (optical density of GFAP and LFB, density of neurons) and DTI scalars in the four tissue types were evaluated using the Student t‐test with estimationstats.com software.
Also in each specimen, colocalization was done to study correlations between imaging and histopathology metrics. Details on ImageJ's coloc2 function (imagej.net/coloc_2) are found in the Data S1.
Across all three specimens, linear mixed models (LMM) were used to assess consistent associations between tissue type and histopathology. LMM are an extension of simple linear models which allow for both fixed and random effects. A fixed effect is a parameter that does not vary (an unknown constant) which affects the prediction of the dependent variable. A random effect is nonconstant parameter that introduces systemic variation in the model. Finally, data in the LMM has a hierarchical structure which means that there are clusters of data which are not independent from each other. For more details, please see the Data S1 for a brief layman's explanation of the model.
In the current work, the dependent variable is the tissue type or diffusion metric, the fixed effects (covariates) are the quantitative histopathology measures and the age of subject, and the random effect is the subject. The hierarchy represents the organization of the tissue types, that is, we assumed that the tissue types appear in order from outside (cortex) to inside (tuber, perituber, and then white matter). We opted for this structure as some of the tissue types are hybrid, for example, the perituber tissue is a transitional tissue type between tuber and white matter, and carries histopathological and diffusion imaging features of both.
The parameter estimates of the fixed effects represent the average effect on the dependent variable (e.g., FA) of a one‐unit change in a covariate (e.g., LFB), with other covariates held constant, that is, representing the overall effect of LFB on FA across the specimens. Furthermore, the categorical predictors for cortex, tuber, and perituber are calculated with the white matter as a reference group. For example, perituber tissue has a coefficient of ‐17.15 for FA, confidence interval ‐26.56 to ‐7.74, which means that the mean FA in perituber tissue is 17.15 units lower compared to the mean FA of the white matter.
First, linear mixed models were used to assess associations between tissue type and histopathology:
Here, tissue type (tuber, perituberal white matter, cortex, and white matter) was the dependent variable; age, GFAP, LFB, and neuronal density were introduced as covariates, and subject as a random factor.
Second, the associations between each DTI metric with histopathology values in each tissue type, adjusted for age and subject, were studied in a similar model:
Here, the tissue type (tuber, perituberal white matter, cortex, and white matter) was inserted as a factor; age, GFAP, LFB, and neuronal density as covariates, and subject as a random factor. P‐values < 0.05 were considered significant, after correction for multiple comparisons.
Results
Patient data
Table 1 shows the main clinical characteristics of the patients.
Table 1.
Patient data
| Case | Sex | Age at onset/surgery | Presurgical workup | Resection site and type | Outcome classa |
|---|---|---|---|---|---|
| 1 | Female | 0.1/1.0 year | L temporal | partial lateral lobectomy | Engel IIIb |
| 2 | Female | 1.2/6.5 year | L occipital | occipital lobectomy | Engel III |
| 3 | Male | 1.8/6.2 year | L frontal to right temporal | frontopolar lesionectomy | Engel IVb |
Engel Ia, seizure free since surgery; Engel IIIa, worthwhile seizure reduction; Engel IVb, no appreciable change. See text for case details.
Case 1 became seizure free with a second surgery, extending the prior resection site, and Case 3 became seizure free with vigabatrin.
Visual inspection of neuropathology and MRI
In all modalities, we identified a gradual tissue transition from white matter to perituber tissue, and then to tuber lesion, as previously described.19 Microscopically small satellite lesions, referred to as microtubers, 31 are evident both on the registered microscopy slides and on corresponding high‐resolution ex vivo images (Figs. S1 and S2).
Specimen 1 revealed an extensive subcortical tuber burden in the temporal lobe, and no white matter myelination at age 1, resulting in only faint background LFB uptake in the cortex and homogenous uptake of GFAP across cortex and tuber tissue.
Quantification of neuropathology and MRI metrics
For diffusion MRI measures, tubers had significantly lower FA compared to the perituber tissue, cortex, and white matter – and conversely the MD was typically highest in tubers, followed by cortex, perituber tissue and the white matter. Case 2, however, had a lower FA (and higher MD) of the cortex and perituber white matter than tuber tissue (Fig. 3).
Figure 3.

Cumming plots of DTI and neuropathology metrics. Raw ROI data of mean DTI scalars (FA, MD), of optical density (GFAP and LFB stains), and neuronal density (NeuN stain) are shown for tuber (green) and cortex (blue), perituber (orange), and white matter (red) tissue types. For each tissue type, summary measurements (mean ± standard deviation) are shown as gapped lines, and the number of ROIs is indicated on the x‐axis. Below each scatterplot the mean differences with the shared control (tuber) are plotted as bootstrap sampling distributions. The mean difference is depicted as a dot; the 95% confidence interval is indicated by the ends of the vertical error bar, and the filled gray curve represents the distribution of the effect size. FA fractional anisotropy; MD mean diffusivity; LFB luxol fast blue; GFAP glial fibrillary acidic protein; NeuN neuronal nuclei stain; OD optical density.
For the histopathological metrics, LFB optical density was lowest in tubers, higher in perituber white matter, and maximum in more remote white matter (Cases 2 and 3, not Case 1). LFB uptake in the cortex was variable. GFAP optical density was highest in the tubers, then in perituber white matter, followed by white matter. Neuronal count (NeuN) was highest in the cortex compared to tuber and perituber tissues, but no consistent pattern was found between other tissue types (Fig. 3). Neuronal size did not differ in the tissue types (data not shown).
Composite LMM analysis across all specimens revealed that with increasing distance from the cortex, via tuber and then perituber tissue to the white matter, the OD of the GFAP and LFB increased, whereas the density of neurons decreased (Table 2). Likewise, increasing distance from the cortex was associated with an increase in FA and a decrease in MD (Table 3).
Table 2.
Parameter estimates for linear mixed model for tissue type (cortex, tuber, perituber, white matter)
| Parameter | Estimate | 95% Confidence Interval | P‐value1 | |
|---|---|---|---|---|
| Lower Bound | Upper Bound | |||
| Intercept | 1.2227 | −8.1118 | 10.5571 | 0.3460 |
| Age | −0.1242 | −1.9027 | 1.6544 | 0.5420 |
| Mean LFB | 0.0139 | 0.0133 | 0.0146 | 0.0000* |
| Mean GFAP | 0.0119 | 0.0110 | 0.0128 | 0.0000* |
| Neuronal Density | −190.6483 | −274.1398 | −107.1568 | 0.0000* |
Linear mixed model with tissue type as a factor, age, density of neurons, OD of GFAP and LFB as covariates, and subject as a random factor.
Statistically significant P‐values of ≤ 0.05 are marked with*.
Table 3.
Parameter estimates for linear mixed models of MD and FA
| Parameter | FA | MD | ||||||
|---|---|---|---|---|---|---|---|---|
| Estimate | 95% Confidence Interval | P‐value1 | Estimate | 95% Confidence Interval | P‐value1 | |||
| Lower Bound | Upper Bound | Lower Bound | Upper Bound | |||||
| Intercept | 112.9008 | 103.1775 | 122.6241 | 0.0000 | 86.8769 | −801.7890 | 975.5428 | 0.4360 |
| Age | −7.4141 | −8.4031 | −6.4252 | 0.0000* | ‐0.2783 | −173.2993 | 172.7428 | 0.9870 |
| Mean LFB | 0.2664 | 0.2024 | 0.3304 | 0.0000* | ‐0.0384 | −0.1052 | 0.0284 | 0.2600 |
| Mean GFAP | 0.2077 | 0.1425 | 0.2730 | 0.0000* | ‐0.1519 | −0.2076 | ‐0.0962 | 0.0000* |
| Neuronal Density | −3332.2716 | −7662.2571 | 997.7140 | 0.1310 | 14563.1374 | 10872.6685 | 18253.6064 | 0.0000* |
| Cortex | −16.2705 | −25.5324 | −7.0086 | 0.0010* | 14.3930 | 5.2292 | 23.5568 | 0.0020* |
| Tuber | −39.2181 | −48.9224 | −29.5139 | 0.0000* | 30.2334 | 21.5909 | 38.8760 | 0.0000* |
| Perituber | −17.1500 | −26.5585 | −7.7415 | 0.0000* | 59.6041 | 49.9442 | 69.2641 | 0.0000* |
| White Matter | 02 | 02 | ||||||
Linear mixed model with tissue type as a factor, age, density of neurons, OD of GFAP and LFB as covariates, and subject as a random factor.
This parameter is set to zero because it is redundant. Statistically significant P‐value of ≤ 0.05 are marked with *.
Colocalization and linear mixed modeling of neuropathology and MRI metrics
LFB and diffusion metrics
In the colocalization analysis, the LFB optical density measures in the white matter and perituber white matter correlated negatively with T2 and MD images, as illustrated in Figure 4. By merging the ROIs for the white matter and perituber white matter, the distribution of both LFB and MD values was wider, and we found a consistent negative correlation in each specimen. LFB and FA had a positive but less consistent correlation in these tissue types.
Figure 4.

Example Case 2: Colocalization analysis of histopathology (LFB stain, after color deconvolution) and ex vivo MRI. In the left column, T1‐weighted and T2‐weighted ex vivo structural images and diffusion imaging. On the top row, the regions of interest (ROIs) are placed in the four tissue types: Cortex, tuber, perituber, and white matter. In the last column, two tissue types (perituber and white matter) are merged, yielding a wider distribution of optical density values. In the graphs, the colored dots indicate intensity histograms, and the hotter colors indicate more overlap between MRI metrics (y‐axis) and histopathology optical density values (x‐axis). Spearman's Rho correlations are listed, and regression lines are shown for higher values for illustration purposes. Note that perituber and white matter tissues at times form two different local maxima in the intensity histograms, for example, in the merged white matter mask. This means these tissue types have different mean signal intensities. LFB luxol fast blue; T1 T1‐weighted; T2 T2‐weighted; FA fractional anisotropy; MD mean diffusivity.
The LMM analysis demonstrated a positive correlation between FA and optical densities LFB, and the inverse was found for MD (Table 3). AD and RD did not correlate with LFB, so we could not assess what was driving the correlation with FA (Table S1).
GFAP and diffusion metrics
In the colocalization analysis, the GFAP optical density measures in the white matter correlated positively with T2 and MD images, as illustrated in Figure 5. GFAP and FA had negative but weaker and less consistent correlation in the white matter. No consistent correlation pattern was present in other tissue types.
Figure 5.
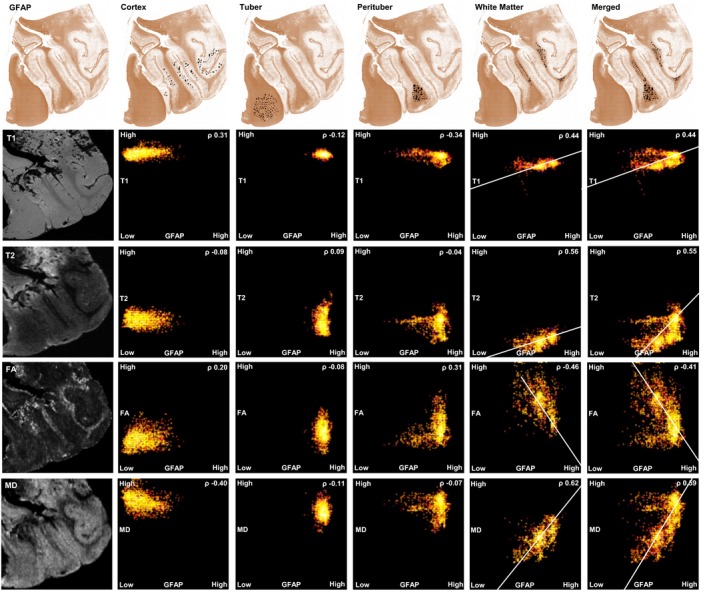
Example Case 2: Colocalization analysis of histopathology (GFAP stain, after color deconvolution) and ex vivo MRI. In the left column, T1‐weighted and T2‐weighted ex vivo structural images and diffusion imaging. On the top row, the regions of interest (ROIs) are placed in the four tissue types: Cortex, tuber, perituber, and white matter. In the last column, two tissue types (perituber and white matter) are merged, yielding a wider distribution of optical density values. In the graphs, the colored dots indicate intensity histograms, and the hotter colors indicate more overlap between MRI metrics (y‐axis) and histopathology optical density values (x‐axis). Spearman's Rho correlations are listed, and regression lines are shown for higher values for illustration purposes. Note that perituber and white matter tissues at times form two different local maxima in the intensity histograms, for example, in the merged white matter mask. This means these tissue types have different mean signal intensities. Legend: GFAP glial fibrillary acidic protein; T1 T1‐weighted; T2 T2‐weighted; FA fractional anisotropy; MD mean diffusivity.
LMM of DTI metrics showed, in contrast, a positive correlation between FA and optical density of GFAP, and the inverse was found for MD (Table 3) – driven by both AD and RD (Table S1).
NeuN and diffusion metrics
For NeuN, the use of neuronal count rather than an optical density measure precluded the colocalization analysis. LMM analysis of DTI metrics showed no correlation between FA and neuronal density, but MD positively correlated with neuronal density (Table 3).
Discussion
In this study, we have correlated 3T and 7T high‐resolution ex vivo structural and diffusion MRI of surgical resection specimens with quantitative neuropathology in TSC. Our protocol included an initial matching of pathology and MRI based on visual review of anatomy,23, 24 followed by more objective rigid and nonrigid registration of the specimens and MRI, which is computationally complex but reproducible.16
Myelination
On visual inspection and quantitatively, LFB optical density measures were highest in white matter, lower in perituber tissue, and lowest in tubers. This gradual change in myelination is in line with ill‐defined rather than discrete borders of tuber pathology,19 and with reduced myelin in dysplastic tissue.24, 32, 33
In both the colocalization analysis and in the LMM, in both the white matter and perituber white matter, quantitative LFB measures correlated with FA, and inversely with MD metrics, suggesting decreased myelination is present in areas with increased diffusion.
In clinical imaging data in children with TSC, we have previously shown DTI measures of the white matter reflect the neurological disease burden. Presence of neurological comorbidities like refractory epilepsy, neurodevelopment and autism spectrum disorder are cumulatively associated with a decreased FA and an increased MD of the corpus callosum.5 Moreover, DTI changes are seen in response to treatment with mTOR inhibitors in patients with TSC, but the underlying mechanism is not known.6, 7 The current work suggests this could represent alterations in myelination, as also seen in rodent models.8, 9, 34, 35
Mechanistically, possible explanations for increased diffusion with reduced myelination are numerous, and include aberrant oligodendrocyte maturation and dysmyelination, reduced numbers of myelinated axons, increased heterotopic cells, enlarged extracellular space from neuronal loss or edema, and demyelination.8, 9, 24, 32, 33, 35, 36, 37
In FCD type IIb, which histopathologically and genetically overlaps with TSC, many such features were seen in an ex vivo MRI‐histopathology study. T2w hyperintensity in the white matter was associated with myelinated fiber loss, high balloon cell density, and dysmyelination. Reduced and dysmorphic oligodendrocytes were also found in these specimens.24 In a mouse model with an inactivated Tsc2 gene in oligodendrocyte precursors, similar decreases in myelinated axon density, myelin thickness, and oligodendrocyte numbers have been reported.8 Although a direct histopathology‐to‐MRI comparison was not made, ex vivo MRI of the callosal white matter in this model showed a reduced FA,8 similar to our work in human patients.5
Clinically, our patients had highly refractory epilepsy which warranted epilepsy surgery. Such uncontrolled and recurrent seizures could constitute an additional “hit” to the already abnormal myelinogenic process in TSC. Longitudinal imaging in children with TSC has revealed diffusion changes and cyst‐like degeneration of tuber tissue which could also represent superimposed injury from seizures over time.6, 19, 38 In FCD type II, seizure effects have not been seen consistently; one study reported a correlation between epilepsy severity or duration and white matter pathology intensity,33 whereas others did not find this correlation.24 A more recent study of 20 epilepsy resection specimens from patients with FCD type II did not report changes in tissue injury markers (including gliosis) in perilesional tissue and white matter with recurrent seizures.39 Our sample was too small to assess a contribution of seizure severity.
Astrocytosis
On visual inspection and quantitatively, reactive astrocytosis, represented by GFAP optical density measures, was typically higher in the tubers than in perituber tissue and white matter, and was lowest in the cortex – consistent with the predominance of glial cells in tuber and perituber tissue,40 and with reports of increased GFAP in FCD‐related white matter.15
Next, in the colocalization analysis of the white matter, GFAP optical density correlated with T2w intensity and with MD. This is different than reported in a study of five resection specimens in FCD, where GFAP correlated with T2w intensity in the cortex, but not in the white matter.15 In a different study of temporal lobe epilepsy, GFAP also did not correlate with white matter T2w intensity, or with FA and MD values.16, 41 The differences with our work could be due to a primary role of astrocyte pathology in TSC.11, 40 In our specimens, obtained from the correlation of GFAP with MD likely represents extension of tuber pathology into the white matter,19, 29, 31 or effects from recurrent seizures (see above). Clinically, the implications of excessive gliosis in TSC could be in the rational design of antiepileptogenic trials, and diffusion imaging could demonstrate target engagement of such targeted therapies.
For tuber and perituber tissue, in the colocalization analysis the GFAP did not consistently correlate with MD or FA. The round shape of tuber data in the intensity histograms (Fig. 5), shows tubers are relatively homogeneous even at this high resolution, with limited variability in both diffusion metrics and GFAP density; as a result, correlations are low.
In the LMM, however, we found a negative correlation between MD and GFAP, and positive correlation between FA and GFAP. We suspect this is a spurious effect from statistical modeling. In all cases, the cortex had the lowest level of gliosis, and the perituber tissue had more gliosis than the adjacent white matter. Case 1 did not have any white matter and in Case 3 the tuber was largely acellular (low gliosis). Thus, in the composite LMM model there appeared to be a low‐to‐high gliosis gradient from cortex to tuber to perituber and then to white matter.
Neurons
The positive association between MD and neuronal density in the LMM likely reflects the presence of abundant cortical neurons, a limited quantity of neurons in the white matter, and a variable and intermediate neuronal count in the tuber and perituber tissues. In a neuropathology study of resection specimen from children with TSC to examine glutamate receptor expression, variations in neuronal size were reported.42 In our sample, we did not study specific neuronal subpopulations in such detail, and there were no consistent differences in neuronal size between the tissue types.
Limitations and future directions
Several limitations decrease the scope of this work. First, only limited conclusions can be drawn from the small number of resection specimens. The wide age range of patients at different maturational stages of myelination rendered findings instable across specimen. The RD and AD did not correlate consistently with neuropathology across specimens, preventing any conclusions about which diffusion direction was driving MD abnormality. Overall, MD was a more robust metric than FA, as previously reported in our longitudinal diffusion imaging study of TSC, too.6
Second, in epilepsy surgery, a good surgical outcome (Engel Class I and II) is considered strong evidence for successful resection of the epileptogenic zone.43 Our poor outcomes prevent us from making inferences about epileptogenic versus nonepileptogenic tubers. Case 2 underwent invasive monitoring, and reactive pathology from surgically implanted hardware could not be differentiated from seizure‐related astrocytosis.15, 16, 41
Finally, comparison with healthy (postmortem) control white was lacking. For intraspecimen consistency, however, we sampled each specimen with multiple ROIs in each tissue type, and analyzed tissue types both separately and combined. In the future, ultrastructural assessment of fiber and oligodendrocyte density and cellular quantification of glial subtypes would complement this work.15, 44, 45
Conflict of Interests
None. The authors report no disclosures or conflicts of interest relevant to this work.
Author Contributions
JP, RS, AP, LV, AS, MT, JB, JS, BS, SP, and MS designed and coordinated the study, executed the imaging and slide digitization, registration, and performed the analysis, and contributed to editing and writing the manuscript. HL is responsible for the histopathology aspects, not including quantification. JM provided the specimen and assisted with marking and orientation of the specimen. OA, BS, and SW directed MR image acquisition and processing, assisted in data interpretation, and edited the manuscript.
Supporting information
Figure S1. Qualitative comparison of histopathology and ex vivo MRI of Case 2
Figure S2. Qualitative comparison of histopathology and ex vivo MRI of Case 3
Table S1. Parameter estimates for linear mixed model of RD and AD
Data S1. Methods
Acknowledgments
We are indebted to our patients for their participation, and to Boston Children's Hospital MRI technical staff members, for their diligent assistance with data acquisition. The authors thank Michelle Ocaña of the HMS Neurobiology Imaging Facility for microscope training and advice on imaging and analysis, and Meera Modi for critical reading of the manuscript.
Funding Information
J. Peters, B. Scherrer, S. Prabhu, M. Sahin, and S. Warfield are supported by NIH R01 NS079788 and U01 NS082320 grants. A. Prohl is supported by Harvard Catalyst | The Harvard Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences, NIH award UL1 TR001102). S. Prabhu is also supported by the Department of Defense W81XWH‐11‐1‐0365. M. Sahin is additionally supported by NIH U54 HD090255 and U54 NS092090 grants. U54 NS092090 is part of the NCATS Rare Diseases Clinical Research Network (RDCRN). RDCRN is an initiative of the Office of Rare Diseases Research (ORDR), NCATS, funded through collaboration between NCATS, NIMH, NINDS, and NICHD. We thank the Harvard Medical School Neurobiology Department and the Neurobiology Imaging Facility for consultation and instrument availability that supported this work. This facility is supported in part by NINDS P30 grant #NS072030.
Funding Statement
This work was funded by U.S. Department of Defense grant W81XWH‐11‐1‐0365; National Institute of Neurological Disorders and Stroke grants P30 NS072030, R01 NS079788, U01 NS082320, U54 HD090255, U54 NS092090, and UL1 TR001102; NCATS grant ; NIMH grant ; NICHD grant .
References
- 1. Ess KC. The neurobiology of tuberous sclerosis complex. Semin Pediatr Neurol 2006;13:37–42. [DOI] [PubMed] [Google Scholar]
- 2. Arulrajah S, Ertan G, Jordan L, et al. Magnetic resonance imaging and diffusion‐weighted imaging of normal‐appearing white matter in children and young adults with tuberous sclerosis complex. Neuroradiology 2009;51:781–786. [DOI] [PubMed] [Google Scholar]
- 3. Makki MI, Chugani DC, Janisse J, Chugani HT. Characteristics of abnormal diffusivity in normal‐appearing white matter investigated with diffusion tensor MR imaging in tuberous sclerosis complex. AJNR Am J Neuroradiol 2007;28:1662–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peters JM, Sahin M, Vogel‐Farley VK, et al. Loss of white matter microstructural integrity is associated with adverse neurological outcome in tuberous sclerosis complex. Acad Radiol 2012;19:17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baumer FM, Peters JM, Clancy S, et al. Corpus callosum white matter diffusivity reflects cumulative neurological comorbidity in Tuberous Sclerosis Complex. Cereb Cortex 2018;28:3665–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peters JM, Prohl A, Kapur K, et al. Longitudinal effects of everolimus on white matter diffusion in tuberous sclerosis complex. Pediatr Neurol 2019;90:24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tillema JM, Leach JL, Krueger DA, Franz DN. Everolimus alters white matter diffusion in tuberous sclerosis complex. Neurology 2012;78:526–531. [DOI] [PubMed] [Google Scholar]
- 8. Carson RP, Kelm ND, West KL, et al. Hypomyelination following deletion of Tsc2 in oligodendrocyte precursors. Ann Clin Transl Neurol 2015;2:1041–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meikle L, Talos DM, Onda H, et al. A mouse model of tuberous sclerosis: neuronal loss of Tsc1 causes dysplastic and ectopic neurons, reduced myelination, seizure activity, and limited survival. J Neurosci 2007;27:5546–5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Meikle L, Pollizzi K, Egnor A, et al. Response of a neuronal model of tuberous sclerosis to mammalian target of rapamycin (mTOR) inhibitors: effects on mTORC1 and Akt signaling lead to improved survival and function. J Neurosci 2008;28:5422–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wong M, Crino PB. Tuberous sclerosis and epilepsy: role of astrocytes. Glia 2012;60:1244–1250. [DOI] [PubMed] [Google Scholar]
- 12. Chandra PS, Salamon N, Huang J, et al. FDG‐PET/MRI coregistration and diffusion‐tensor imaging distinguish epileptogenic tubers and cortex in patients with tuberous sclerosis complex: a preliminary report. Epilepsia 2006;47:1543–1549. [DOI] [PubMed] [Google Scholar]
- 13. Jansen FE, Braun KP, van Nieuwenhuizen O, et al. Diffusion‐weighted magnetic resonance imaging and identification of the epileptogenic tuber in patients with tuberous sclerosis. Arch Neurol 2003;60:1580–1584. [DOI] [PubMed] [Google Scholar]
- 14. Yogi A, Hirata Y, Karavaeva E, et al. DTI of tuber and perituberal tissue can predict epileptogenicity in tuberous sclerosis complex. Neurology 2015;85:2011–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reeves C, Tachrount M, Thomas D, et al. Combined Ex Vivo 9.4T MRI and quantitative histopathological study in normal and pathological neocortical resections in focal epilepsy. Brain Pathol 2016;26:319–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eriksson SH, Free SL, Thom M, et al. Correlation of quantitative MRI and neuropathology in epilepsy surgical resection specimens–T2 correlates with neuronal tissue in gray matter. NeuroImage 2007;37:48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Madan N, Grant PE. New directions in clinical imaging of cortical dysplasias. Epilepsia 2009;50(Suppl 9):9–18. [DOI] [PubMed] [Google Scholar]
- 18. Northrup H, Krueger DA. International tuberous sclerosis complex consensus g. tuberous sclerosis complex diagnostic criteria update: recommendations of the International tuberous sclerosis complex consensus conference. Pediatr Neurol 2012;2013:243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peters JM, Prohl AK, Tomas‐Fernandez XK, et al. Tubers are neither static nor discrete: Evidence from serial diffusion tensor imaging. Neurology 2015;85:1536–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miller KL, McNab JA, Jbabdi S, Douaud G. Diffusion tractography of post‐mortem human brains: optimization and comparison of spin echo and steady‐state free precession techniques. NeuroImage 2012;59:2284–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miller KL, Stagg CJ, Douaud G, et al. Diffusion imaging of whole, post‐mortem human brains on a clinical MRI scanner. NeuroImage 2011;57:167–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Scherrer B, Afacan O, Stamm A, et al. Optimized magnetic resonance diffusion protocol for ex-vivo whole human brain imaging with a clinical scanner, Proc. SPIE 9412, Medical Imaging 2015: Physics of Medical Imaging, 94122U, 2015.
- 23. Eriksson SH, Free SL, Thom M, et al. Reliable registration of preoperative MRI with histopathology after temporal lobe resections. Epilepsia 2005;46:1646–1653. [DOI] [PubMed] [Google Scholar]
- 24. Zucca I, Milesi G, Medici V, et al. Type II focal cortical dysplasia: Ex vivo 7T magnetic resonance imaging abnormalities and histopathological comparisons. Ann Neurol 2016;79:42–58. [DOI] [PubMed] [Google Scholar]
- 25. Schindelin J, Arganda‐Carreras I, Frise E, et al. Fiji: an open‐source platform for biological‐image analysis. Nat Methods 2012;9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Taquet M, Scherrer B, Commowick O, et al. A mathematical framework for the registration and analysis of multi‐fascicle models for population studies of the brain microstructure. IEEE Trans Med Imaging 2014;33:504–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol 2001;23:291–299. [PubMed] [Google Scholar]
- 28. Štajduhar A, Džaja D, Judaš M, Lončarič S. Automatic detection of neurons in NeuN‐stained histological images of human brain. Physica A: Statistical Mechanics and its Applications, 2019. [Google Scholar]
- 29. Ruppe V, Dilsiz P, Reiss CS, et al. Developmental brain abnormalities in tuberous sclerosis complex: a comparative tissue analysis of cortical tubers and perituberal cortex. Epilepsia 2014;55:539–550. [DOI] [PubMed] [Google Scholar]
- 30. Eriksson SH, Free SL, Thom M, et al. Methodological aspects of 3D and automated 2D analyses of white matter neuronal density in temporal lobe epilepsy. Neuropathol Appl Neurobiol 2006;32:260–270. [DOI] [PubMed] [Google Scholar]
- 31. Marcotte L, Aronica E, Baybis M, Crino PB. Cytoarchitectural alterations are widespread in cerebral cortex in tuberous sclerosis complex. Acta Neuropathol 2012;123:685–693. [DOI] [PubMed] [Google Scholar]
- 32. Muhlebner A, Coras R, Kobow K, et al. Neuropathologic measurements in focal cortical dysplasias: validation of the ILAE 2011 classification system and diagnostic implications for MRI. Acta Neuropathol 2012;123:259–272. [DOI] [PubMed] [Google Scholar]
- 33. Shepherd C, Liu J, Goc J, et al. A quantitative study of white matter hypomyelination and oligodendroglial maturation in focal cortical dysplasia type II. Epilepsia 2013;54:898–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. West KL, Kelm ND, Carson RP, et al. Myelin volume fraction imaging with MRI. NeuroImage 2018;182:511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ercan E, Han JM, Di Nardo A, et al. Neuronal CTGF/CCN2 negatively regulates myelination in a mouse model of tuberous sclerosis complex. J Exp Med 2017;214:681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thom M, Sisodiya S, Harkness W. Scaravilli F. Microdysgenesis in temporal lobe epilepsy. A quantitative and immunohistochemical study of white matter neurones. Brain 2001;124:2299–2309. [DOI] [PubMed] [Google Scholar]
- 37. Carson RP, Van Nielen DL, Winzenburger PA, Ess KC. Neuronal and glia abnormalities in Tsc1‐deficient forebrain and partial rescue by rapamycin. Neurobiol Dis 2012;45:369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chu‐Shore CJ, Frosch MP, Grant PE, Thiele EA. Progressive multifocal cystlike cortical tubers in tuberous sclerosis complex: Clinical and neuropathologic findings. Epilepsia 2009. [DOI] [PubMed] [Google Scholar]
- 39. Rossini L, Garbelli R, Gnatkovsky V, et al. Seizure activity per se does not induce tissue damage markers in human neocortical focal epilepsy. Ann Neurol 2017;82:331–341. [DOI] [PubMed] [Google Scholar]
- 40. Sosunov AA, Wu X, Weiner HL, et al. Tuberous sclerosis: a primary pathology of astrocytes? Epilepsia 2008;49(Suppl 2):53–62. [DOI] [PubMed] [Google Scholar]
- 41. Lockwood‐Estrin G, Thom M, Focke NK, et al. Correlating 3T MRI and histopathology in patients undergoing epilepsy surgery. J Neurosci Methods 2012;205:182–189. [DOI] [PubMed] [Google Scholar]
- 42. Talos DM, Kwiatkowski DJ, Cordero K, et al. Cell‐specific alterations of glutamate receptor expression in tuberous sclerosis complex cortical tubers. Ann Neurol 2008;63:454–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brodbeck V, Spinelli L, Lascano AM, et al. Electroencephalographic source imaging: a prospective study of 152 operated epileptic patients. Brain 2011;134:2887–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Muhlebner A, van Scheppingen J, Hulshof HM, et al. Novel Histopathological Patterns in Cortical Tubers of Epilepsy Surgery Patients with Tuberous Sclerosis Complex. PLoS ONE 2016;11:e0157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sosunov AA, McGovern RA, Mikell CB, et al. Epileptogenic but MRI‐normal perituberal tissue in Tuberous Sclerosis Complex contains tuber‐specific abnormalities. Acta Neuropathol Commun 2015;3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Qualitative comparison of histopathology and ex vivo MRI of Case 2
Figure S2. Qualitative comparison of histopathology and ex vivo MRI of Case 3
Table S1. Parameter estimates for linear mixed model of RD and AD
Data S1. Methods


