Figure 2.
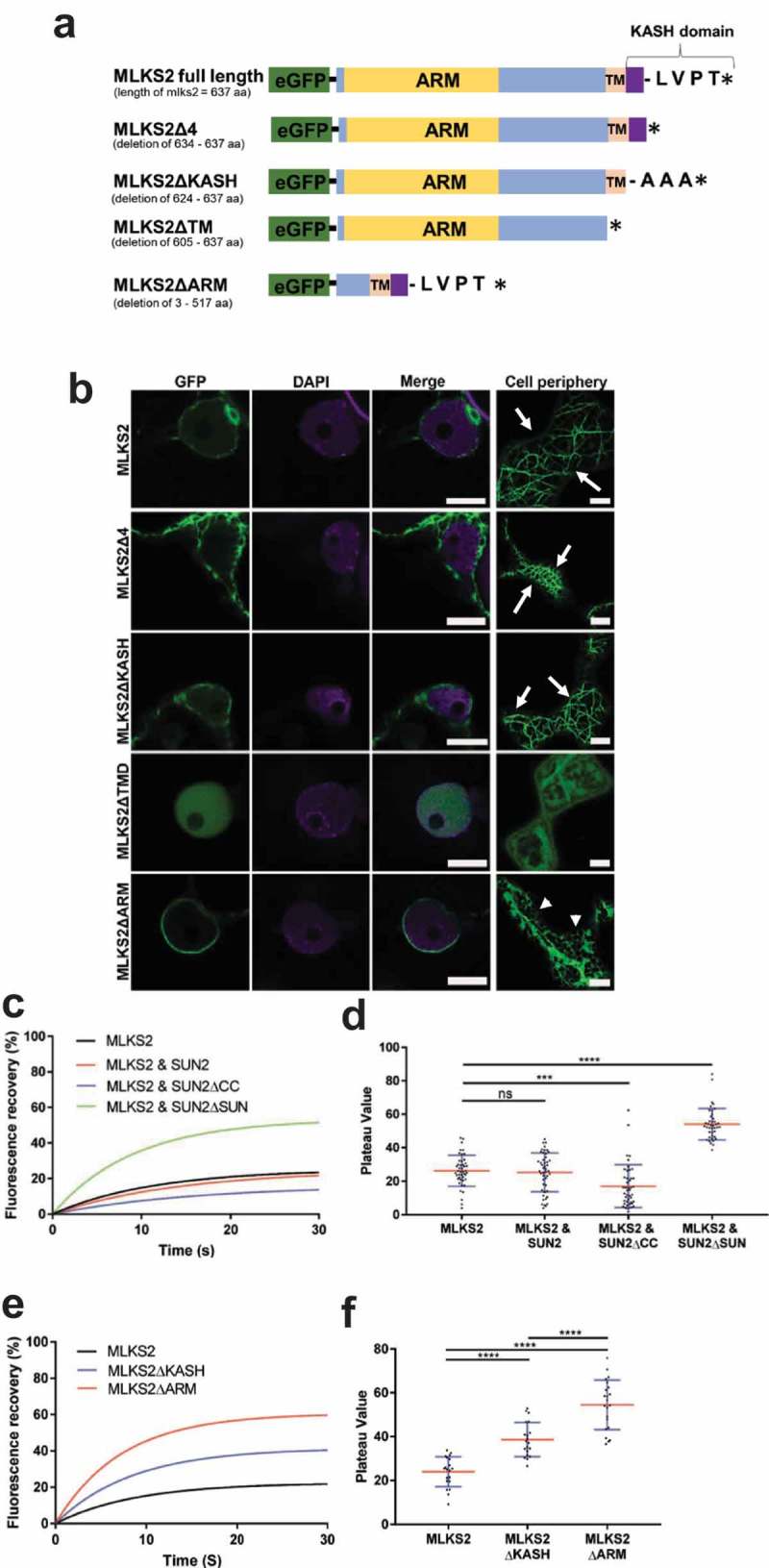
MLKS2 cellular localization and ZmSUN2-interaction.
Subcellular localization of full length and deletion mutants of GFP-MLKS2 in transiently expressing N. benthamiana leaf cells as previously described [35]. a) Domain deletion diagram of all constructs used showing the ARM domain (yellow), disordered regions (blue), transmembrane domain (TM), KASH domain (purple), terminal AA residues, and stop codon (*). b) Full length GFP-MLKS2 (green) and all but one deletion mutant localize to the NE around DAPI-stained chromatin (magenta). GFP-MLKS2ΔTM appears soluble and distributed throughout the nucleoplasm. A cell periphery network-like pattern was observed for all constructs, with GFP-MLKS2ΔTM appearing as soluble in the cytoplasm and GFP-MLKS2ΔARM appearing as associated with the ER. c) Fluorescence recovery curve from MLKS2 FRAP alone or when co-expressed with mCherry-ZmSUN2, mCherry-ZmSUN2ΔCC (without coiled coil domain), or mCherry-ZmSUN2ΔSUN (without SUN domain). d) FRAP recovery plateau values for fluorescence recovery curves shown in c). e) Fluorescence recovery curves showing increased mobile fraction in the NE for KASH or ARM domain deletion variants of GFP-MLKS2. f) FRAP recovery plateau values of MLKS2, MLKS2ΔKASH, and MLKS2ΔARM shown in d). Scale bar denotes 10 µm. For whisker plots, blue lines denote SD error bars, red lines denote mean. Nuclei (N = ≥30) imaged across three experimental repeats per treatment. ANOVA statistical test used where Ns = P ≥ 0.05, *** = P ≤ 0.001 and **** = P ≤ 0.0001.
