Figure 4.
Multiple meiotic and post-meiotic defects of mlks2 mutants.
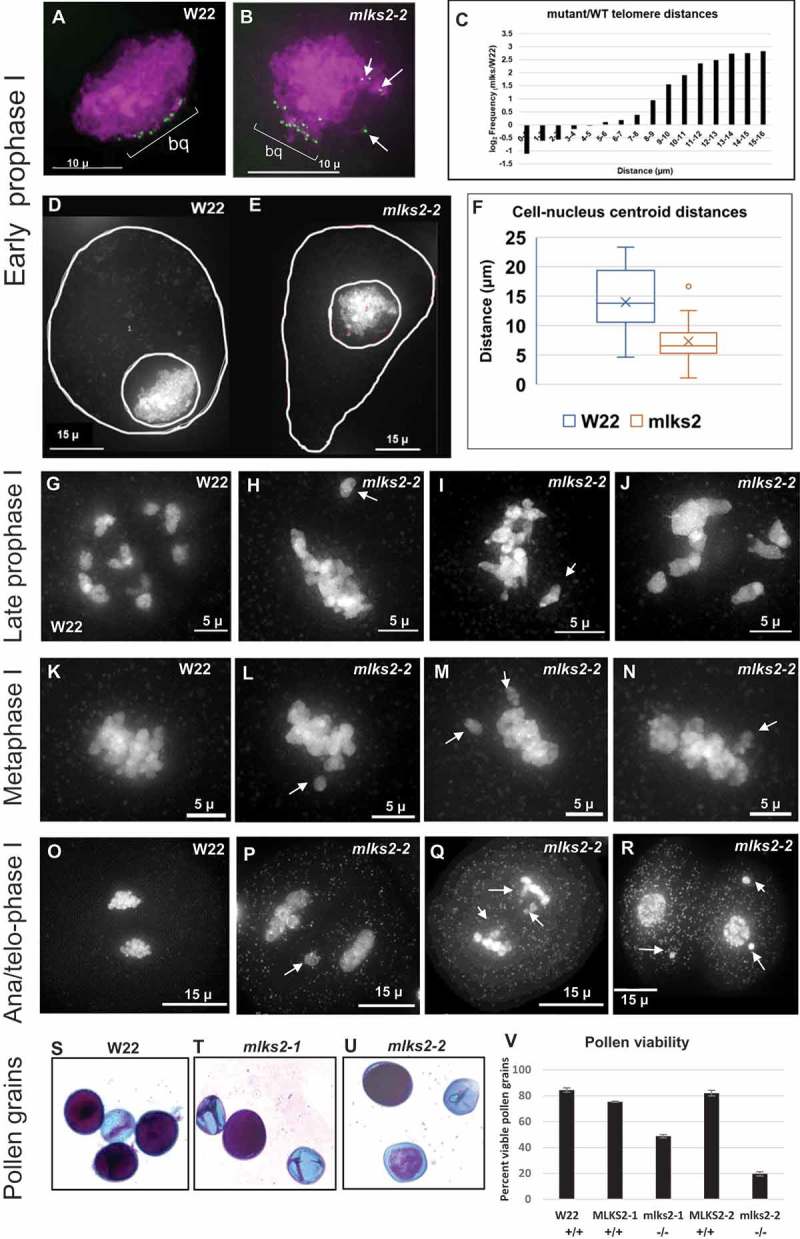
Cytological defects of mlks2-2 during and after male meiosis. a) Early prophase I stage W22 meiocyte nucleus showing a typical bouquet (green dots, ‘bq’) of NE-associated telomeres visualized in the nucleus (DAPI, shown in magenta) using the 3D acrylamide oligo FISH method [78]. b) Partial telomere bouquet in mlks2-2 mutant at meiotic prophase, showing unusually distant telomeres (arrows) relative to the main bouquet (bq) telomere cluster region. c) Histogram showing bouquet-stage telomere pairwise distance distributions plotted as log2 fold change of mutant/wild-type per 1 micron distance bins (n = 6 W22, n = 11 mlks2-2). The mutants show a pronounced increase in the longer telomere-to-telomere distance bins. Nuclear position phenotypes for normal d) or mutant e) cells shown as projections from the middle-most 1/5 of the optical sections through the nuclei stained with DAPI, including traces around the cell and nuclear peripheries to ascertain 2D centroid locations. f) Eccentricity plots showing the distribution of distances of between the pairwise centroids of nuclei and cells for normal (W22) and mutant (mlks2) meiocytes at at the bouquet stage, using the same nuclei as those analyzed in a-c. g) Late prophase I stage W22 meiocyte showing bivalents spread throughout the volume of the nucleus. h-j) Late prophase I stage mlks2-2 meiocytes showing clumping of bivalents. k) W22 meiocyte showing bivalents on a normal meiosis I metaphase plate. l-n) Mutant mlks2-2 meiocytes showing one or more chromosomes (arrows) not located in the meiosis I metaphase plate. O) W22 meiocyte at late anaphase I or early telophase I. p-q) Mutant mlks2-2 late anaphase I or early telophase I showing irregularly positioned ‘laggard’ chromosomes (arrows). r) Mutant mlks2-2 at telophase after meiosis I, before meiosis II, and showing micronuclei (arrows) that are associated with failure of chromosomes or chromosomal fragments to reach the spindle poles. s-u) Pollen viability stains for wild-type s) or mutant t, u) pollen. Dark purple indicates viable pollen, light blue indicates inviable pollen. v) Quantification of pollen viability with n = 1,000 or more for each genotype.
