ABSTRACT
The recent developments in Cathepsin protease research have unveiled a number of key observations which are fundamental to further our understanding of normal cellular homeostasis and disease. By far, the most interesting and promising area of Cathepsin biology stems from how these proteins are linked to the fate of living cells through the phenomenon of Lysosomal Leakage and Lysosomal Membrane Permeabilisation. While extracellular Cathepsins are generally believed to be of central importance in tumour progression, through their ability to modulate the architecture of the Extracellular Matrix, intracellular Cathepsins have been established as being of extreme significance in mediating cell death through Apoptosis. With these two juxtaposed key research areas in mind, the focus of this review highlights recent advancements in how this fast-paced area of Cathepsin research has recently evolved in the context of their mechanistic regulation in cancer research.
Abbreviations : ECM, Extracellular Matrix; MMP, Matrix Metalloproteases; LL, Lysosomal Leakage; LMP, Lysosomal Membrane Permeabilisation; LMA, Lysosomorphic Agents; BC, Breast Cancer; ASM, Acid Sphingomyelinase; TNF-α, Tumor Necrosis Factor-alpha; LAMP, Lysosomal Associated membrane Protein; PCD, Programmed Cell Death; PDAC, Pancreatic Ductal Adenocarcinoma; ROS, Reactive Oxygen Species; aa, amino acids.
KEYWORDS: Cathepsin, Cystatins, cancer, metastasis, lysosome, apoptosis
Introduction
Tumor metastasis has been long known to be of central importance in malignancy and represents itself as a key step in tumor progression that may be amenable to therapeutic intervention in the fight against cancer. In line with neoplastic transformation of cells, the emergence of the Extracellular Matrix (ECM) has been given great significance over the last 20 years with a view to targeting it for therapeutic benefits based upon it promoting tumor cell proliferation, chemotherapeutic resistance, angiogenesis and providing the microenvironment essential for tumor survival and metastasis[1]. The basis of these efforts stemmed from observations that tumor cells possess enhanced proteolytic activity which permits them to breakdown ECM components and thus take on greater motility through the basal lamina during tumor metastasis [2].
Generally speaking, the extracellular proteins responsible for enhanced proteolytic activity of tumor cells include the Matrix Metallo- (MMPs)-[3] and additional serine-, threonine-, aspartic- and cysteine-proteases [4]. In the absence of any effective MMP-directed therapeutics having emerged [5], mainly due to their broad activity and toxic side effects [6], it is therefore not surprising that as an alternative approach, the Cathepsin proteases have recently developed into an intense area of study with a view to dissecting their regulatory mechanisms in cancer cell biology with therapeutic development as a foresight. While the superfamily of Cathepsin proteins has developed over a long time span, each member has been shown to possess distinct (and sometimes over-lapping) roles in their contribution to tumor progression based on their level and distribution of expression either at the subcellular level or at the extracellular level within tissues. In large, such studies have also given arise to a number of interesting research avenues that have focused on defining with greater clarity the substrate-specific protein sequences cleaved by well-characterized Cathepsins and Cathepsin-directed regulatory and effector signaling pathways. Consequently, areas of research that have flourished more recently include harnessing the potential of the Cathepsins in the design of novel cancer therapeutics based on Cathepsin-specific cleavage sites, with a view to harnessing their activity against tumor cells in conventional chemotherapy and more so against cells that exhibit chemotherapeutic resistance or metastasis.
As the role of intracellular Cathepsins in recent efforts have provided some very novel and exciting insights into firmly establishing their role in cell demise, the emergence of Lysosomal Leakage (LL) and Lysosomal Membrane Permeabilisation (LMP) has simultaneously gathered importance in the sense that this presents a key (and distinct) step in the initiation and execution of Cathepsin-dependent cell death. Initially, most of the limited evidence to support these beliefs were derived from the use of Lysosomorphic Agents (LMA), which destabilize the lysosomal membraine, resulting in the release of Cathepsin proteases into the cytoplasm and thus triggering cell death through caspase-dependent apoptosis (or Lysosomal Cell Death, LCD) [7]. However, such mechanisms have been verified and extended to offer greater biological significance in systems utilizing naturally-occurring apoptosis-inducing agents such as TNF, FAS, TRAIL and products of oxidative stress-all of which have offered a clearer insight into the biological, physiological and regulatory relationship between LL, LCD and the Cathepsins [8–12]. Moreover, such results have been arrived at with some interesting (and sometimes surprising) observations that have helped to drive this area of research into looking at what other key signaling cascades may also be connected to the Cathepsins before, during and after the initiation of LL in disease.
Based upon these collective findings as a backdrop, in which intracellular Cathepsins share a central importance in LCD while extracellular Cathepsins potentiate ECM remodeling and tumor metastasis, the regulation of Cathepsins may offer a more focused picture for improving the therapeutic development of these proteases further and which forms the core of this review article.
The Cathepsins and cancer
Over the years the central importance of the Cathepsins in disease have been intensely explored, particularly in the context of tumor proliferation, metastasis, invasion, ECM degradation, angiogenesis [13] and inflammation [14]. Consequently, it is with these perspectives in mind that the driving force behind Cathepsin research has broadly diversified with significant progress being made in certain areas of applied research, such as antibody-drug conjugates, diagnostic imaging and their use in targeted drug delivery [14,15]. More importantly, the focus of Cathepsin protease regulation has synergistically evolved to address their role in modulating the immune response and inflammatory cells (such as macrophages), both of which are central to maintaining the tumor microenvironment and the ECM during cancer and some inflammatory diseases (for an excellent review, see Kramer et al. [14]).
To date, the Cathepsin super-family of proteases constitutes a family of 15 lysosomal proteases which can be broadly classified into aspartic (D, E)-, serine (A, G)- and cysteine (B, C, F, H, K, L, O, S, V, Z/X, W)-proteases subtypes. Furthermore, such proteases can be subdivided based on their proteolytic activity into endo-peptidases (S, K, V, F, L) and both endo- and exo-peptidase (B, H, Z/X, C), of which Cathepsins C, B,H, K, O, L, Z/X are expressed broadly and which have been the most extensively studied to unveil general mechanism in substrate cleavage, lysosome research and cell death in disease [16] (see Table 1).
Table 1.
Amino acid (aa) length and tissue-specific distribution of expression for the Cathepsins (Liver, Kidney, Thyroid Gland, Spleen, Placenta, Brain, Heart, Skeletal Muscle, Testis, Ovary, Macrophages, Cytotoxic T Lymphocyte, Osteoclasts, Epithelial cells of the GastroIntestine, Embryonic Respiratory and Urinary Tract and Foetuses, Lung, Lymph Nodes, Antigen Presenting Cells, Cornea, Thymus, Skin, Monocytes, Neutrophils, Melanoma, Plasma, Platelets, Intestine, Stomach, Erythrocytes, Prostate).
| Cathepsin | (aa) | Expression | Reference |
|---|---|---|---|
| B | 339 | L,K,TG,Sp | Chan et al.[17] |
| C | 463 | Lu,Sp,K,P,CTL | Paris et al.[18] |
| F | 484 | B,H,SM,T,O,M | Wang et al.[19] |
| L | 333 | L,TG,K | Joseph et al.[20] |
| H | 335 | L,K,Sp | Fuchs et al.[21] |
| K | 329 | Os,M,EGI,ERUTF,Lu | Inaoka et al.[22] |
| O | 321 | L,K,P,O | Santamaria et al.[23] |
| S | 331 | Sp,LN,APC,H | Shi et al.[24] |
| V | 334 | C,T,Th | Bromme et al.[25] |
| W | 376 | Sp,LN,CTL | Linnever et al.[26] |
| Z/X | 303 | L,K,P,Lu | Nagler et al.[27] |
| A | 480 | B,S,P,PL,L | Galjart et al.[28] |
| G | 225 | S,Mo,N | Salvesen et al.[29] |
| D | 412 | Sp,K,L,Me,Pla,Plat | Faust et al.[30] |
| E | 401 | B,I,S,Er,LN,S,Sp,Lu | Azuma et al. [31] |
As seen from expression profiling studies, a number of Cathepsins have been shown to be highly expressed during tumor invasion and metastasis [32] (Table 2). For example, Cathepsin A was observed to be highly expressed in metastatic lesions of Malignant Melanoma [33], increased Cathepsin B protein expression was seen at the invasive edges of B16 Melanomas cancer cells [34], in patients with enhanced inter-pulmonary metastases [35] and Cathepsin D was seen to be over-expressed in metastatic Breast Cancer (BC) cells [36]. A number of studies have also supported the central role of Cathepsins in tumor invasion and metastasis using a variety of approaches. For example, overexpression of Cathepsin D in fibroblast cells resulted in increased fibroblast motility and invasion [37], Cathepsin Z/X upregulation was correlated with higher invasiveness in vitro [38], the importance of Cathepsin H in glioblastoma cell line invasion was demonstrated using Matrigel assays [39], Cathepsin K had enhancing effects on breast tumor epithelial cell development in Cathepsin K-positive fibroblast co-cultures [40] and Cathepsin S knockout and mutant Cathepsin S expression resulted in reduced invasiveness of pancreatic cancer [2].
Table 2.
Expression levels of the Cathepsin proteins have been documented to cause Tumor Invasion (TI), Metastasis (M), Tumor Growth (TG), Extracellular Matrix Degradation (ECMD) or Angiogenesis (ANG) inMalignant Melanoma, Lung, Breast, Colon, GlioBlastoma, Hepatocarcinoma, Glioma, Ovarian, Squamous Cell Carcinoma, Basal Cell Carcinoma, Gastric, Prostate Cancer and Hepatocarcinoma .
Of importance has been Cathepsin B, the expression of which is elevated at the genomic, proteomic [50] and serum levels while often being linked to tumor progression in advanced stage cancer patients [51] such as in BC malignancy [52]. Herein, the underlying involvement of Cathepsin B has been verified by a number of approaches using overexpression, antisense [53], siRNA and shRNA studies [54] that have characterized and established the importance of Cathepsin B at various stages of cancer development. Moreover, as to be expected from a large family of proteases, some Cathepsins share some redundancy with regards to their modes of activity as seen with Cathepsins B and Z/X in cancer invasiveness [55]. As seen in overexpression studies, MCF7 cells underwent Epithelial-Mesenchymal Transition (EMT) while knockdown expression resulted in Mesenchymal-Epithelial Transition. Additionally, using such approaches, Cathepsin B has been further characterized in its role as promoting or reducing angiogenesis. Here, Cathepsin B knockout leukocytes showed reduced CD18 shedding and an accumulation in angiogenic vessel with reduced extravasal activity [56]. Collectively, such studies do indeed highlight the potential pleiotropic input of the Cathepsins in a variety of important processes in disease that extend from cellular transformation of cells to angiogenesis of tumor metastases and further work is indeed warranted in this area to define more clearly the key regulatory steps that give rise to Cathepsin-mediated activity.
Likewise, Cathepsin L research is also emerging as an interesting area of cancer research as Cathepsin L knockdown studies demonstrated abrogated tumor proliferation, growth and invasiveness in BC cells [57,58] while also rendering Glioma cells sensitive to radiotherapy treatment [58]. Interestingly, such silencing studies have also revealed the contribution that the Cathepsins can potentially make to chemoresistance, as seen in the case of Cathepsin L, in which silencing sensitized BC cells [59,60] and A549 lung cancer cells [61] to Paclitaxel and Cisplatin treatments. Such studies therefore suggest an important aspect of Cathepsin expression in connection with their contribution to conventional chemotherapeutic resistance while highlighting the importance of maintaining efforts on novel Cathepsin-specific therapeutic design.
In light of chemoresistance, the recent significance of Cathepsins expression in cancer stem cells have also gained momentum. As a developing area of interest, aberrantly expressed Cathepsins B and D transcripts and proteins were identified in Tongue Squamous Stem Cells using the techniques of Immunohistochemistry (IHC) staining and Nanostring/CISH mRNA analysis [62] and enhanced expression in liver metastases from adenocarcinoma OCT4-positive cells [63]. In further developments, Cathepsin B activity was also found to be enriched in Glioblastoma stem cells from hypoxic niches using IHC and fluorogenic substrates in CD133-positive cells [64] indicating the possible importance of Cathepsin B as a potential marker in stem cell biology and cancer prognosis. Collectively, the potential involvement of the Cathepsins in cancer stem cell regulation appears to have significant and more research is therefore warranted in determining more precisely what mechanistic role they play in tumor signaling, progression and chemotherapeutic resistance in such systems.
Signalling networks and Cathepsin expression
Cathepsin transcriptional regulation
Based upon the observations that Cathepsin overexpression correlates strongly with tumor growth and progression, more recent efforts have therefore been directed at defining with greater clarity Cathepsin gene regulation and the contribution it may make to disease progression.
Consequently, the regulation of Cathepsin B was found to be modulated at the transcriptional level under INF-γ [65] or IL-6 cytokine stimulatory conditions [66] and transcription factors such as SP1 (-Cathepsin B) [67], SP3 (-Cathepsin L) [68] and Upstream Stimulatory Factors, USF (-Cathepsins B and L) [69,70] observed to be key regulators of Cathepsin gene expression. Similarly, using expression and knockdown studies of transcription factor FOXO3a in a gastric cancer model, Yu et al. (2016) demonstrated its ability to positively regulate the Cathepsin L promoter and protein expression, which had the effect of promoting EMT of cells by it reducing E-Cadherin protein levels [71]. More recently, developing efforts have also identified a number of other key transcription factors and the involvement of micro-RNAs in regulating a number of Cathepsin transcripts. For example, transcription factor FOXM1 was found to bind and activate the Cathepsin B promoter in gastric cancer cells [72] and as reported by Luan et al. (2018) [73], Cathepsins B and L were over-expressed in an MZF1 transcription factor-dependent manner, in BC cells [73]. Finally, Cathepsins B and L protein secretion was also dependent on active transcription factors Ets1, SP1, NF-κB during EMT in primary melanoma invasiveness [74]. While such findings clearly demonstrate a link between signal transduction pathways and the regulation of Cathepsin genes at the transcriptional level, greater clarification is needed on whether such signaling cascades (and intermediates) may affect the activity of the Cathepsins at the protein level.
More recently, Cathepsin L transcriptional repression has been demonstrated to be regulated by mir-152 in gastrointestinal tumors and the expression of which induced apoptosis and inhibited proliferation, migration, and invasion of gastrointestinal stromal cells [75]. Similarly, Cathepsin A transcription was targeted by expressed mir106b-5p causing suppression of Colorectal Carcinoma (CRC) cell migration and invasion [76]. Interestingly, mir200c over-expression reduced Cathepsin L expression in lung A549 cells, resulting in enhanced susceptibility to Paclitaxel-mediated cell death and EMT suppression [77]. While, such recent studies link Cathepsin transcriptional expression to an alternative mode of regulation through the miRNAs, more importantly, such studies also highlight the relative importance of miRNA as a potential therapeutic in overcoming Cathepsin-mediated chemoresistance and Cathepsin-mediated EMT regulation [77]. As seen previously, mir152 has been linked to regulating Wnt-mediated EMT inhibition [78] and mir106b-5p with the regulation of key signaling intermediates such as GSK3B, VEGFA and PTK2 in colon and cervical cancers [79] and in both of which, Cathepsins A and L are seen to play a vital regulatory role. Thus, it is conceivable that these miRNAs could be regulating these Cathepsins and other key signaling pathway intermediates simultaneously, or exclusively in a time-dependent and coordinated manner. In similar studies, mir200C was found to inhibit EMT (or lung cancer cell invasion via HMGB1 signaling) [80] while also being able to reverse EMT [81]-both of which may have possible regulatory inputs from Cathepsin L expression via this microRNA.
As an emerging area of interest, further exploration of Cathepsin gene regulation by microRNAs that also modulate other key oncogenes or tumour suppressors and which may act in concert (or crosstalk) with the Cathepsin (at the protein or transcriptional level) is therefore much needed.
As an additional feature of Cathepsin gene transcription, alternative exon splicing [82] and exon skipping [83] can give rise to protein forms translated from additional downstream ATG [84] start codons that do not enter the secretory pathway (due to them lacking the required secretory signal) and can therefore enter the nucleus.
Post translational activation of Cathepsins
As the Cathepsins superfamily members vary in their polypeptide lengths, general tissue expression or distribution and their intracellular (or extracellular) localization in normal or disease states (Tables 1 and 2), for simplicity we will aim to focus on the most characterized of these to highlight the general pathways responsible for protein maturation after translation. For example, Cathepsin B is synthesized in the Rough Endoplasmic Reticulum (RER) as a 339 amino acid (aa) protein containing a 17 aa signal peptide [85]. Following insertion into the RER lumen, the signal peptide is removed and the 43/46 KDa inactive pro-Cathepsin B precursor is glycosylated and transported to the Golgi where it is further glycosylated with phosphor-mannose residues. Following this, it binds the Mannose-6-Phosphate Receptor (M6PR) allowing transportation from the trans-Golgi to the late endosome (where the acidic environment permits the intermolecular pro-domain removal [86] and then on to the lysosomal compartment. It is through this key endosomal cleavage (and regulatory) step, that Cathepsin D (an aspartic protease) can also activate Cathepsin B [87], as can Cathepsin G, urokinase-type Plasminogen Activator (uPAR) and tissue-type plasminogen activator and elastasase proteins [88,89]. Further cleavage of Cathepsin B generates a double chain form consisting of a 25 KDa heavy and 5 KDa light chain [90]. Interestingly, Cathepsins can also undergo transport from the Golgi to the lysosome independently of the M6PR. For example, Sortilin was found to transport Cathepsin D and Cathepsin H in this manner [91] and in a recent study by Boonen et al. (2016), the protein SEZ6L2 was discovered to be a novel substrate for Cathepsin D and which could transport Cathepsin D to the endosomes (from the Golgi and cell surface) in Hela and neuroblastoma cells [92] (see Figure 1). Once activated (in the late endosomes), Cathepsin activity can also be negatively (and tightly) regulated by naturally-occurring endogenous inhibitors, of which there are four main groups named the Stefins, Cystatins, Kininogens and non-inhibitory homologues [16]. Structurally, Cathepsin B is bilobal [93] and can act as an endo-peptidase at neutral pH or an exo-peptidase (with carboxypeptidase activity) at acidic pH [94]. Like Cathepsin B, Cathepsin H can also be trans-activated (by Cathepsin L) and for both of which strong supporting data has been derived from structural studies [95,96].
Figure 1.
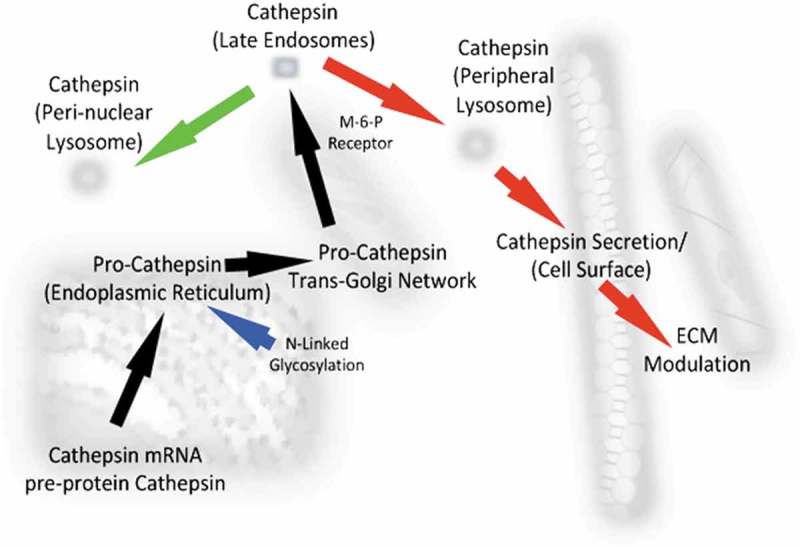
Schematic showing the synthesis, trafficking and maturation of Cathepsin proteins in normal cells (green arrows) and cancer cells (red arrows). Pro Cathepsin mRNAs are translated, inserted into the Endoplasmic Reticulum where they are glycosylated and transported to the Golgi network. Upon further glycosylation, they are transported in a Mannose-6-Phosphate (M-6-P)-dependent manner to the late Endomes after which they can enter the Perinuclear Lysosome or the Peripheral Lysosome and be secreted to modulate the Extracellular Matrix (ECM, red arrows).
Secretion of Cathepsins
In contrast to normal cells, during malignancy the Cathepsins have been observed to localize at the cell periphery and in exocytic vessicles after the traversing the trans-Golgi [97]. More recently, the respective importance of cell surface V-ATPase and extracellular acidification have been reported for the secretion of active or non-active Cathepsin B [98] and Cathepsin D [99] in metastatic BC cells [98]. Such studies offer greater clarity in defining broad regulatory steps involved in active Cathepsin secretion during cancer progression and identify what other protein intermediates regulate this key step in Cathepsin localization. Collectively, these observations offer support to the belief that in cancer cells, lysosomes behave differently by relocating from a perinuclear location to the cell periphery allowing their contents to be readily localized to the ECM [100]. However, more studies are required to ascertain mechanistically which proteins are directly involved in anterograde and retrograde trafficking of the active Cathepsins, particularly in cells that contain dysfunctional lysosomes or cells derived from different tumor compartments or stages of tumor development.
Lysosomal leakage as a key regulatory mechanism for cell death
While lysosomes were discovered over 50 years ago as subcellular organelles responsible for the degradation of extracellular materials (taken up by cells through phagocytosis) or the intracellular degradation of complex molecules (such as DNA, proteins, lipids and carbohydrates), they were also described as “suicide bags” that can cause tissue autolysis upon rupture [101,102]. From these initial observations, compelling evidence has accumulated to support their distinct and unique role in regulating apoptosis through the activation of the Caspases, thus giving them center stage in the areas of auto-inflammatory disease and cancer research [103]. However, in the field of Cathepsin research, we are presented with a paradox in the sense that while secreted extracellular Cathepsins can give rise to metastatic disease [32,104], deregulated intracellular Cathepsin expression (derived from LL) is of equal importance in determining the apoptotic fate of tumor cells [10,105]. Consequently, the lysosomal localized portion of Cathepsins may offer an axis of regulation that can be exploited for therapeutic purposes and has therefore generated great interest over the years [104,106].
Defining the involvement of the lysosome at the molecular level and its involvement in cell fate with greater clarity is an area that has indeed developed more recently through the understanding of LL which occurs as a consequence of LMP. In comparison to normal cells, cancer cells are believed to have larger, more fragile lysosomes [107] and which are more sensitive to disruption by agents that result in LMP. Agents which can trigger this change have given rise to a wealth of knowledge in how the lysosome is linked to apoptosis in cancer cells (See Figure 2).
Figure 2.
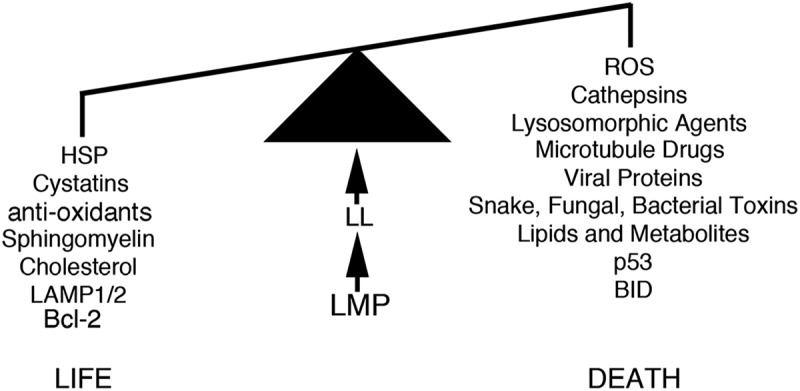
Lysosomal Membrane Permeability (LMP) and Lysosomal Leakage (LL) determine cell survival based on the cellular expression levels of Heat Shock Proteins (HSP), Cystatins, anti-oxidants, Sphingomyelin, Cholesterol, LAMP1/2 and Bcl-2. Cell death can also be induced by the lysosome, based upon cellular levels of Reactive Oxidative Species (ROS), Cathepsin, Lysosomorphic Agents, Lysosomal Leakage, Microtubule Disrupting drugs, Viral Proteins, Toxins, Lipids and Metabolites, protein p53 and protein BID (see text for more details).
Firstly, lysosomes are sensitized to Reactive Oxygen Species (ROS), which are produced at higher levels in cancer cells [108] due to a range of stimuli such as drugs, heavy metals [109] and conditions such as ischaemia and inflammation [109].
Secondly, the study of LMAs have yielded some interesting insights into how LMP can be selectively induced in cancer cells [110]. Good examples of these include amines with hydrophilic side chains (for example, imidazole [102], Ciprofloxacin [105], Sphingosine [111] and Siramesine [112,113]). More recently, a Riccardin D-derivative was also seen to trigger LMP and Cathepsin B release in Prostate Cancer cell apoptosis. Interestingly, it also caused Cathepsin B to relocate from the lysosomes to the nucleus where it was observed to potentiate DNA damage by suppressing BRCA1 activity, thus highlighting a novel role for nuclear Cathepsin B activity [114].
Thirdly, Sphingosine accumulation due to the activation of lysosomal Acid Sphingomyelinase (ASM) and acid Ceramide production can also activate LMP. While other signaling intermediates which trigger LMP (including phospholipase-A2) have been long known about [115], more recently the treatment of rat hepatocytes with Tumor Necrosis Factor-alpha (TNF-α) was also seen to induce LMP [116]. In support of this, inhibition of Sphingokinase 1 (SPK1, which converts sphingosine to sphingosine-1-phosphate, SP1) also induced LMP, with the loss of S1P giving rise to abnormal lysosomes [117]. The finding that Sphingokinase 1 is also a Cathepsin B substrate offers additional support to the alternative manner in which Cathepsin B can exert its death-inducing activity [118,119].
Fourthly, p53 is believed to induce LMP in Myeloid leukemia cells [120] and TNF-α treated Fibrosarcoma cells [121]. Here, the dependency of LMP activation was influenced by the presence of phospho-ser15-p53 at the lysosomal membraine [121] and the involvement of any other isoforms of p53 in LMP activation are yet to be defined [122]. As an unexaustive list, other agents are also capable of inducing LMP such as microtubule targeting drugs [123,124], viral entry proteins [125] and cobra snake venom toxins [126].
Conversely, we must not disregard the role of proteins and intermediates which can also stabilize and safeguard the lysosome against the above damaging agents [127], such as Heat Shock Proteins (HSP, HSP70 and HSP90), Mcl-1/BclXL, anti-oxidants (such as Vitamins C and E, Super Oxide Dismutase, Glutathione Peroxidases and Catalases), Cholesterols, Sphingomyelin and Lysosomal Associated Membraine Proteins (LAMP) 1 and 2 [127]. Generally speaking, deregulated expression of these products may give rise to resistance in Cathepsin-mediated death under circumstances where Cathepsin expression levels are detected as being relatively high enough to trigger the onset of LMP (under normal conditions). Good evidence to support the effect of Cathepsin expression and LMP onset also comes from treating rat hepatocytes with TNF-α (or Sphingosine) in cells lacking Cathepsin B and in which LMP was observed to be ablated [116]. Similarly, Cathepsin D was also shown to induce LMP upon TNF-R1 internalization in a Caspase-8 and −7 dependent manner and which activated ASM [128]. Mechanistically, Cathepsin-induced LMP is believed to occur upon the intra-lysosomal degradation of highly-glycosylated LAMPs, which form a glycocalyx shield on the inner side of the lysosomal membraine [129,130].
Lysosomal Membrane Permeabilisation regulation by the Cathepsins
Lysosomal-mediated cell death can take on a number of forms depending on the types of agents that adversely affect the integrity of the lysosome [110,131].
Upon robust LMP induction, the morphological features of cell death are not witnessed and uncontrolled necrosis occurs instead. Conversely, in the presence of limited LMP, apoptosis [111,132] or caspase-independent death can be activated [133] and readily visualized. Under the former of these circumstances, the Cystatins fail to attenuate cell death unless they are over-expressed suggesting the central importance of Cathepsin expression levels (in relation to endogenous Cystatins) in death pathway induction. Nevertheless, LMP-induced apoptosis is activated through Mitochondrial Outer Membrane Permeabilization (or MOMP) and Cytochrome C release [134], which can be brought about by Cathepsin-mediated BID cleavage (and its activation to pro-apoptotic tBID) or by inhibiting the cleavage of anti-apoptotic Bcl2, BCl-xl and Mcl-1 proteins [135–137]. Moreover, the caspases can be activated directly upon their cleavage by Cathepsins or by cleavage of their inhibitors such as the XIAP proteins [137] (Figure 3).
Figure 3.
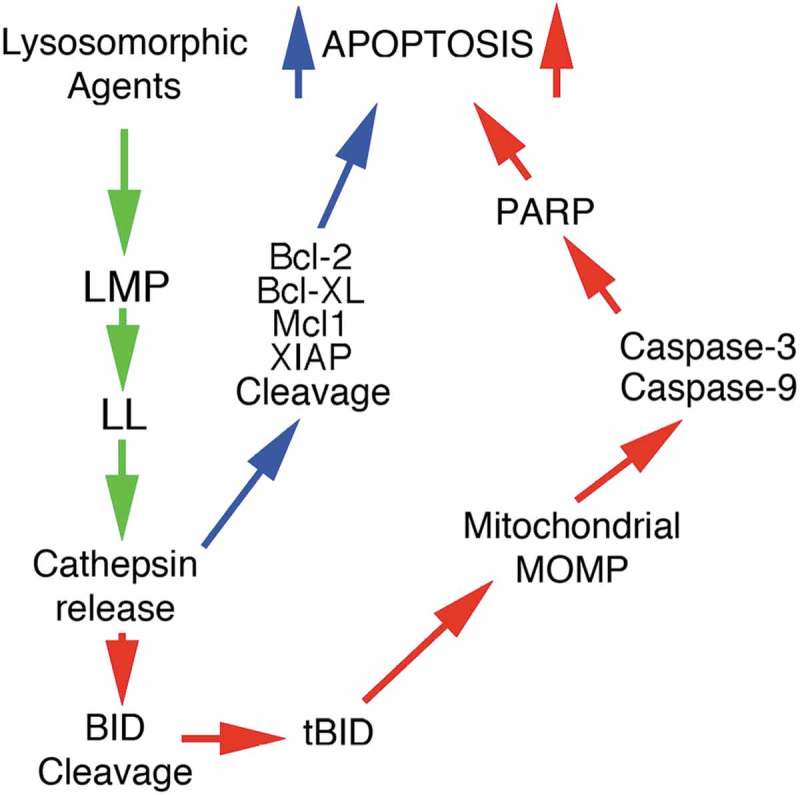
Schematic showing the events leading to Cathepsin protein release (green arrows) following the induction of Lysosomal Membrane Permeabilisation (LMP) and Lysosomal Leakage (LL) which result in apoptosis, through Mitochondrial Outer Membrane Permeabilization (MOMP) and Poly (ADP-ribose) polymerase (PARP) activation (red arrows) via Caspases-3 and −9. The blue arrows highlight enhanced apoptosis through the activity of Cathepsin-mediated cleavage of anti-apoptosis proteins (see text for more details).
LMP can also cause cell death with no caspase or little caspase activation (even in the instance of HSP70 depletion [138]), upon hypochlorous acid [139] and Siramesine treatment of cells [112,113]. Here, inhibition of activated caspases did not seem to reduce cell death suggesting that the Cathepsins can take on a role as alternative executors of cell death [16] and that other uncharacterized signaling mechanisms may be involved at the molecular level of Cathepsin regulation and activity.
Additionally, it is also worth noting the unique relationship that appears to be developing based on the recent observations that Cathepsin expression contributes to cell death [118,119] and how silencing of Cathepsins can sensitise cells to conventional chemotherapy and DNA-damaging agents [59–61]. While these present potentially conflicting roles for the Cathepsins, it clearly highlights the importance of precisely delineating the contribution of the Cathepsins to other key life-death signaling pathways (or vice versa) in future studies.
Signal transduction intermediates and Cathepsin regulation
As the field of Cathepsin research has developed at a phenomenal pace over the last 5 years, it therefore comes as little surprise that the regulation of these proteins at the transcriptional and the post-translational level are being viewed in a broader context with regards to how they may crosstalk or interact with other signaling cascades (or intermediates). For example, Cathepsin D was found to cleave Bcl-2 causing the sensitization of BC cells to TRAIL-mediated death, thus linking Cathepsin D activity with cleavage of negative (or pro-survival) regulators of PCD [140]. Moreover, Fritsch et al. (2016) confirmed BID cleavage by Cathepsin D (resulting in Caspase-9 activation) and identified a novel substrate for Cathepsin D (using proteomics) as HSP90 [141]. Analysis of HSP90-Cathepsin D cleavage-null mutants, partially prevented apoptosis of U937 and Jurkat cells highlighting a central role for Cathepsin D in HSP90 level regulation in LL, LMP and apoptosis. Meanwhile, Burton et al. (2017) linked nuclear Cathepsin L with CUX1 proteolytic processing and thus linked Cathepsin L with EMT (and MET) progression in BC and PC cells [142]. Interestingly, in this study subcellular Cathepsin L was also observed to translocate from the nucleus to the cytoplasm, highlighting a novel protein trafficking-function for Cathepsin L. Kim et al. (2018) showed that Cathepsin S could bind BRCA1 and facilitate it’s ubiquitination and breakdown, thus suppressing DNA repair in BC development, while knocked-down expression of Cathepsin S stabilized BRCA1 levels, thus collectively linking Cathepsin S to BRCA1 signaling transduction [143]. As a novel substrate, Bian et al. (2015) found Cathepsin B to colocalise and target p27kip for degradation in CRC cells, thus linking it to cell cycle modulation [144].
Bannoud et al. (2018) found that Cathepsin D can bind the M6PR and are co-transported in an estradiol stimulatory manner through MCF-7 cells and which linked Cathepsin D trafficking with estradiol-dependent EMT progression (and possible chemoresistence) [145]. Additionally, chemokines CXCL-9 and −10 were seen to upregulate Cathepsin B expression and secretion in BC cells via CXCR3 signaling [146], while the tumour suppressor Protilin was identified as a novel substrate for Cathepsin Z/X, the cleavage of which reduced its binding to Clathrin, thus linking Cathepsin Z/X activity and regulation of Clathrin-dependent endocytosis [147]. Recently, Shao et al. (2018) showed that NEDD4 was needed for EGFR-dependent lung cancer cell migration and EGF-induced Cathepsin B secretion, as seen from a lack of Cathepsin B secretion in analysing a ligase-dead mutant of NEDD4 and in NEDD4 knockdown experiments [148]. Finally, BAG2 was observed to process pro-Cathepsin B auto-activation and facilitated the secretion of pro-Cathepsin B to the cell surface, thereby increasing its metastatic potential in triple-negative breast cancer cells and thus linking BAG2 oncogenic signaling with Cathepsin B activation and tumor invasiveness [149].
Collectively, all these findings suggest the importance of Cathepsin regulation during cancer progression and which may be more complex than once thought. However, novel mechanisms are being unveiled through which the Cathepsins may impose their proteolytic activity on other signaling intermediates and thus modulate (or be modulated by) other key signaling and protein trafficking cascades that have already been firmly implicated in disease or cell survival. When taken together, all of the above findings contribute greatly to our clinical understanding of the Cathepsins and allows researchers to view them in a broader context either in model systems or in disease.
Reflective and future directions
The last 3 years in Cathepsin research and cancer biology have presented themselves as being incredibly diverse and hugely productive. In reflection, and based on the literature published, there are many new and exciting areas of basic research that are emerging or coming into fruition and which appear to be moving basic research towards a translational phase at a phenomenal pace (Figure 4). In reflection, what started off as a cytoplasmic organelle responsible for the general disposal and degradation of subcellular matter is slowly emerging as an organelle that has many other key regulatory functions in normal cellular homeostasis. As reviewed herein, the peripheral lysosome can indeed contribute to apoptosis while simultaneously being the source of secreted Cathepsin proteins and thus offers a clearer understanding of how the Cathepsins can be more realistically targeted during therapeutic design.
Figure 4.
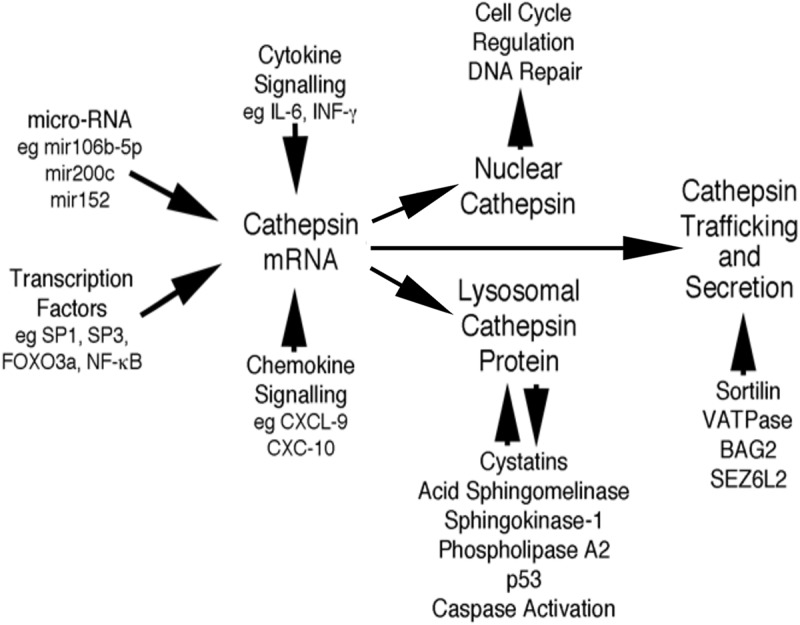
Schematic highlighting the regulatory inter-relationships between transcription factors, micro-RNAs (mir), Cytokine/Chemokine signalling, Cathepsin inhibitors and signalling intermediates, DNA repair and cell cycle regulators and trafficking proteins with Cathepsin gene regulation, protein trafficking and secretion (see text for details).
Taken with the identification of novel substrates (or binding partners) for the Cathepsins and crosstalk between key signaling, transcriptional or protein trafficking pathways, the future of research in this area holds great promise in unravelling with even greater clarity the regulation of the Cathepsins while identifying more potential therapeutic targets.
Funding Statement
This work was supported by the Russian Science Foundation [16-15-10410].
Acknowledgments
I would like to dedicate this article to Aaditya Raj Lazar Soond. I must also say ‘Thank You’ to Ms Joginder Kaur, Mr Malkiat Soond, Raj Kumar Soond and Alena Savikova for their ‘greatness’ during the hours I spent writing.
Disclosure statement
No potential conflict of interest was reported by the authors.
Ethical standards
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008. The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional guides on the care and use of laboratory animals.
References
- [1].Holle AW, Young JL, Spatz JP.. In vitro cancer cell-ECM interactions inform in vivo cancer treatment. Adv Drug Deliv Rev. 2016;97:270–279. [DOI] [PubMed] [Google Scholar]
- [2].Gocheva V, Zeng W, Ke D, et al. Distinct roles for cysteine cathepsin genes in multistage tumorigenesis. Genes Dev. 2006;20:543–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yadav L, Puri N, Rastogi V, et al. Matrix metalloproteinases and cancer - roles in threat and therapy. Asian Pac J Cancer Prev. 2014;15:1085–1091. [DOI] [PubMed] [Google Scholar]
- [4].Hamalisto S, Jaattela M. Lysosomes in cancer-living on the edge (of the cell). Curr Opin Cell Biol. 2016;39:69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fingleton B. MMPs as therapeutic targets–still a viable option? Semin Cell Dev Biol. 2008;19:61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kruger A, Kates RE, Edwards DR. Avoiding spam in the proteolytic internet: future strategies for anti-metastatic MMP inhibition. Biochim Biophys Acta. 2010;1803:95–102. [DOI] [PubMed] [Google Scholar]
- [7].Wang F, Gomez-Sintes R, Boya P. Lysosomal membrane permeabilization and cell death. Traffic. 2018;19:918–931. [DOI] [PubMed] [Google Scholar]
- [8].Woo SM, Min KJ, Seo BR, et al. YM155 sensitizes TRAIL-induced apoptosis through cathepsin S-dependent down-regulation of Mcl-1 and NF-kappaB-mediated down-regulation of c-FLIP expression in human renal carcinoma Caki cells. Oncotarget. 2016;7:61520–61532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Seo BR, Min K-J, Woo SM, et al. Inhibition of Cathepsin S induces Mitochondrial ROS that sensitizes TRAIL-mediated apoptosis through p53-mediated downregulation of Bcl-2 and c-FLIP. Antioxid Redox Signal. 2017;27:215–233. [DOI] [PubMed] [Google Scholar]
- [10].Foghsgaard L, Wissing D, Mauch D, et al. Cathepsin B acts as a dominant execution protease in tumor cell apoptosis induced by tumor necrosis factor. J Cell Biol. 2001;153:999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Brunk UT, Dalen H, Roberg K, et al. Photo-oxidative disruption of lysosomal membranes causes apoptosis of cultured human fibroblasts. Free Radic Biol Med. 1997;23:616–626. [DOI] [PubMed] [Google Scholar]
- [12].Brunk UT, Svensson I. Oxidative stress, growth factor starvation and Fas activation may all cause apoptosis through lysosomal leak. Redox Rep. 1999;4:3–11. [DOI] [PubMed] [Google Scholar]
- [13].Pogorzelska A, Zolnowska B, Bartoszewski R. Cysteine cathepsins as a prospective target for anticancer therapies-current progress and prospects. Biochimie. 2018;151:85–106. [DOI] [PubMed] [Google Scholar]
- [14].Kramer L, Turk D, Turk B. The future of cysteine Cathepsins in disease management. Trends Pharmacol Sci. 2017;38:873–898. [DOI] [PubMed] [Google Scholar]
- [15].Olson OC, Joyce JA. Cysteine cathepsin proteases: regulators of cancer progression and therapeutic response. Nat Rev Cancer. 2015;15:712–729. [DOI] [PubMed] [Google Scholar]
- [16].Turk V, Stoka V, Vasiljeva O, et al. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim Biophys Acta. 2012;1824:68–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chan SJ, San Segundo B, McCormick MB, et al. Nucleotide and predicted amino acid sequences of cloned human and mouse preprocathepsin B cDNAs. Proc Natl Acad Sci U S A. 1986;83:7721–7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Paris A, Strukelj B, Pungercar J, et al. Molecular cloning and sequence analysis of human preprocathepsin C. FEBS Lett. 1995;369:326–330. [DOI] [PubMed] [Google Scholar]
- [19].Wang B, Shi GP, Yao PM, et al. Human cathepsin F. Molecular cloning, functional expression, tissue localization, and enzymatic characterization. J Biol Chem. 1998;273:32000–32008. [DOI] [PubMed] [Google Scholar]
- [20].Joseph LJ, Chang LC, Stamenkovich D, et al. Complete nucleotide and deduced amino acid sequences of human and murine preprocathepsin L. An abundant transcript induced by transformation of fibroblasts. J Clin Invest. 1988;81:1621–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Fuchs R, Machleidt W, Gassen HG. Molecular cloning and sequencing of a cDNA coding for mature human kidney cathepsin H. Biol Chem Hoppe-Seyler. 1988;369:469–475. [DOI] [PubMed] [Google Scholar]
- [22].Inaoka T, Bilbe G, Ishibashi O, et al. Molecular cloning of human cDNA for cathepsin K: novel cysteine proteinase predominantly expressed in bone. Biochem Biophys Res Commun. 1995;206:89–96. [DOI] [PubMed] [Google Scholar]
- [23].Santamaria I, Pendas AM, Velasco G, et al. Genomic structure and chromosomal localization of the human cathepsin O gene (CTSO). Genomics. 1998;53:231–234. [DOI] [PubMed] [Google Scholar]
- [24].Shi GP, Munger JS, Meara JP, et al. Molecular cloning and expression of human alveolar macrophage cathepsin S, an elastinolytic cysteine protease. J Biol Chem. 1992;267:7258–7262. [PubMed] [Google Scholar]
- [25].Bromme D, Li Z, Barnes M, et al. Human cathepsin V functional expression, tissue distribution, electrostatic surface potential, enzymatic characterization, and chromosomal localization. Biochemistry. 1999;38:2377–2385. [DOI] [PubMed] [Google Scholar]
- [26].Linnevers C, Smeekens SP, Bromme D. Human cathepsin W, a putative cysteine protease predominantly expressed in CD8+ T-lymphocytes. FEBS Lett. 1997;405:253–259. [DOI] [PubMed] [Google Scholar]
- [27].Nagler DK, Zhang R, Tam W, et al. Human cathepsin X: A cysteine protease with unique carboxypeptidase activity. Biochemistry. 1999;38:12648–12654. [DOI] [PubMed] [Google Scholar]
- [28].Galjart NJ, Gillemans N, Harris A, et al. Expression of cDNA encoding the human “protective protein” associated with lysosomal beta-galactosidase and neuraminidase: homology to yeast proteases. Cell. 1988;54:755–764. [DOI] [PubMed] [Google Scholar]
- [29].Salvesen G, Farley D, Shuman J, et al. Molecular cloning of human cathepsin G: structural similarity to mast cell and cytotoxic T lymphocyte proteinases. Biochemistry. 1987;26:2289–2293. [DOI] [PubMed] [Google Scholar]
- [30].Faust PL, Kornfeld S, Chirgwin JM. Cloning and sequence analysis of cDNA for human cathepsin D. Proc Natl Acad Sci U S A. 1985;82:4910–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Azuma T, Pals G, Mohandas TK, et al. Human gastric cathepsin E. Predicted sequence, localization to chromosome 1, and sequence homology with other aspartic proteinases. J Biol Chem. 1989;264:16748–16753. [PubMed] [Google Scholar]
- [32].Mohamed MM, Sloane BF. Cysteine cathepsins: multifunctional enzymes in cancer. Nat Rev Cancer. 2006;6:764–775. [DOI] [PubMed] [Google Scholar]
- [33].Kozlowski L, Rucinska M, Wojtukiewicz MZ. [Metastases of malignant melanoma of unknown primary site: an important diagnostic and therapeutic problem]. Pol Merkur Lekarski. 2000;8:486–488. [PubMed] [Google Scholar]
- [34].Sloane BF, Dunn JR, Honn KV. Lysosomal cathepsin B: correlation with metastatic potential. Science. 1981;212:1151–1153. [DOI] [PubMed] [Google Scholar]
- [35].Fujise N., Nanashima A., Taniguchi Y., et al. Prognostic impact of cathepsin B and matrix metalloproteinase-9 in pulmonary adenocarcinomas by immunohistochemical study. Lung Cancer. 2000;27:19–26. [DOI] [PubMed] [Google Scholar]
- [36].Dian D, Vrekoussis T, Shabani N, et al. Expression of cathepsin-D in primary breast cancer and corresponding local recurrence or metastasis: an immunohistochemical study. Anticancer Res. 2012;32:901–905. [PubMed] [Google Scholar]
- [37].Pruitt F. L., He Y., Franco O.E., et al. Cathepsin D acts as an essential mediator to promote malignancy of benign prostatic epithelium. Prostate. 2013;73:476–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Krueger S, Kalinski T, Wolf H, et al. Interactions between human colon carcinoma cells, fibroblasts and monocytic cells in coculture–regulation of cathepsin B expression and invasiveness. Cancer Lett. 2005;223:313–322. [DOI] [PubMed] [Google Scholar]
- [39].Sivaparvathi M, Sawaya R, Gokaslan ZL, et al. Expression and the role of cathepsin H in human glioma progression and invasion. Cancer Lett. 1996;104:121–126. [DOI] [PubMed] [Google Scholar]
- [40].Kleer CG, Bloushtain-Qimron N, Chen Y-H, et al. Epithelial and stromal cathepsin K and CXCL14 expression in breast tumor progression. Clin Cancer Res. 2008;14:5357–5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Aggarwal N, Sloane BF, Cathepsin B. multiple roles in cancer. Proteomics Clin Appl. 2014;8:427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sudhan DR, Pampo C, Rice L, et al. Cathepsin L inactivation leads to multimodal inhibition of prostate cancer cell dissemination in a preclinical bone metastasis model. Int J Cancer. 2016;138:2665–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sudhan DR, Rabaglino MB, Wood CE, et al. Cathepsin L in tumor angiogenesis and its therapeutic intervention by the small molecule inhibitor KGP94. Clin Exp Metastasis. 2016;33:461–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Verbovsek U, Van Noorden CJ, Lah TT. Complexity of cancer protease biology: cathepsin K expression and function in cancer progression. Semin Cancer Biol. 2015;35:71–84. [DOI] [PubMed] [Google Scholar]
- [45].Wilkinson RD, Williams R, Scott CJ, et al. therapeutic, diagnostic, and prognostic potential. Biol Chem. 2015;396:867–882. [DOI] [PubMed] [Google Scholar]
- [46].Nagler DK, Krüger S, Kellner A, et al. Up-regulation of cathepsin X in prostate cancer and prostatic intraepithelial neoplasia. Prostate. 2004;60:109–119. [DOI] [PubMed] [Google Scholar]
- [47].Wang J, Chen L, Li Y, et al. Overexpression of cathepsin Z contributes to tumor metastasis by inducing epithelial-mesenchymal transition in hepatocellular carcinoma. PLoS One. 2011;6:e24967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kozlowski L, Wojtukiewicz MZ, Ostrowska H. Cathepsin A activity in primary and metastatic human melanocytic tumors. Arch Dermatol Res. 2000;292:68–71. [DOI] [PubMed] [Google Scholar]
- [49].Pranjol MZI, Gutowski NJ, Hannemann M, et al. Cathepsin D non-proteolytically induces proliferation and migration in human omental microvascular endothelial cells via activation of the ERK1/2 and PI3K/AKT pathways. Biochim Biophys Acta Mol Cell Res. 2018;1865:25–33. [DOI] [PubMed] [Google Scholar]
- [50].Jedeszko C, Sloane BF. Cysteine cathepsins in human cancer. Biol Chem. 2004;385:1017–1027. [DOI] [PubMed] [Google Scholar]
- [51].Ebert MP, Krüger S, Fogeron M-L, et al. Overexpression of cathepsin B in gastric cancer identified by proteome analysis. Proteomics. 2005;5:1693–1704. [DOI] [PubMed] [Google Scholar]
- [52].Bengsch F, Buck A, Günther SC, et al. Cell type-dependent pathogenic functions of overexpressed human cathepsin B in murine breast cancer progression. Oncogene. 2014;33:4474–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Krueger S, Haeckel C, Buehling F, et al. Inhibitory effects of antisense cathepsin B cDNA transfection on invasion and motility in a human osteosarcoma cell line. Cancer Res. 1999;59:6010–6014. [PubMed] [Google Scholar]
- [54].Withana NP, Blum G, Sameni M, et al. Cathepsin B inhibition limits bone metastasis in breast cancer. Cancer Res. 2012;72:1199–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Mitrovic A, Mirkovic B, Sosic I, et al. Inhibition of endopeptidase and exopeptidase activity of cathepsin B impairs extracellular matrix degradation and tumour invasion. Biol Chem. 2016;397:165–174. [DOI] [PubMed] [Google Scholar]
- [56].Nakao S, Zandi S, Sun D, et al. Cathepsin B-mediated CD18 shedding regulates leukocyte recruitment from angiogenic vessels. Faseb J. 2018;32:143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Qin G, Cai Y, Long J, et al. Cathepsin L is involved in proliferation and invasion of breast cancer cells. Neoplasma. 2016;63:30–36. [DOI] [PubMed] [Google Scholar]
- [58].Wang W, Long L, Wang L, et al. Knockdown of Cathepsin L promotes radiosensitivity of glioma stem cells both in vivo and in vitro. Cancer Lett. 2016;371:274–284. [DOI] [PubMed] [Google Scholar]
- [59].Zhang H, Zhang L, Wei L, et al. Knockdown of cathepsin L sensitizes ovarian cancer cells to chemotherapy. Oncol Lett. 2016;11:4235–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Han ML, Zhao Y-F, Tan C-H, et al. Cathepsin L upregulation-induced EMT phenotype is associated with the acquisition of cisplatin or paclitaxel resistance in A549 cells. Acta Pharmacol Sin. 2016;37:1606–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Sui H, Shi C, Yan Z, et al. Overexpression of Cathepsin L is associated with chemoresistance and invasion of epithelial ovarian cancer. Oncotarget. 2016;7:45995–46001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Featherston T., Marsh R.W., van Schaijik B., et al. Expression and localization of cathepsins b, d, and g in two cancer stem cell subpopulations in moderately differentiated oral tongue squamous cell carcinoma. Front Med (Lausanne). 2017;4:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Mehrotra S, Wickremesekera SK, Brasch HD, et al. Expression and localization of Cathepsins B, D and G in cancer stem cells in liver metastasis from colon adenocarcinoma. Front Surg. 2018;5:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Breznik B, Limbaeck Stokin C, Kos J, et al. Cysteine cathepsins B, X and K expression in peri-arteriolar glioblastoma stem cell niches. J Mol Histol. 2018;49:481–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Storm Van’s Gravesande K, Layne MD, Ye Q, et al. IFN regulatory factor-1 regulates IFN-gamma-dependent cathepsin S expression. J Immunol. 2002;168:4488–4494. [DOI] [PubMed] [Google Scholar]
- [66].Ibrahim SA, El-Ghonaimy EA, Hassan H, et al. Hormonal-receptor positive breast cancer: IL-6 augments invasion and lymph node metastasis via stimulating cathepsin B expression. J Adv Res. 2016;7:661–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Yan S, Berquin IM, Troen BR, et al. Transcription of human cathepsin B is mediated by Sp1 and Ets family factors in glioma. DNA Cell Biol. 2000;19:79–91. [DOI] [PubMed] [Google Scholar]
- [68].Jean D, Guillaume N, Frade R. Characterization of human cathepsin L promoter and identification of binding sites for NF-Y, Sp1 and Sp3 that are essential for its activity. Biochem J. 2002;361:173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Yan S, Jane DT, Dufresne MJ, et al. Transcription of cathepsin B in glioma cells: regulation by an E-box adjacent to the transcription initiation site. Biol Chem. 2003;384:1421–1427. [DOI] [PubMed] [Google Scholar]
- [70].Yan S, Sloane BF. Isolation of a novel USF2 isoform: repressor of cathepsin B expression. Gene. 2004;337:199–206. [DOI] [PubMed] [Google Scholar]
- [71].Yu S, Yu Y, Zhang W, et al. FOXO3a promotes gastric cancer cell migration and invasion through the induction of cathepsin L. Oncotarget. 2016;7:34773–34784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Yang L, Cui M, Zhang L, et al. FOXM1 facilitates gastric cancer cell migration and invasion by inducing Cathepsin D. Oncotarget. 2017;8:68180–68190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Luan H, Mohapatra B, Bielecki TA, et al. Loss of the nuclear pool of ubiquitin ligase CHIP/STUB1 in breast cancer unleashes the MZF1-cathepsin pro-oncogenic program. Cancer Res. 2018;78:2524–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Tripathi R, Fiore LS, Richards DL, et al. Abl and Arg mediate cysteine cathepsin secretion to facilitate melanoma invasion and metastasis. Sci Signal. 2018;11 DOI: 10.1126/scisignal.aao0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Lu HJ, Yan J, Jin P-Y, et al. MicroRNA-152 inhibits tumor cell growth while inducing apoptosis via the transcriptional repression of cathepsin L in gastrointestinal stromal tumor. Cancer Biomark. 2018;21:711–722. [DOI] [PubMed] [Google Scholar]
- [76].Ni S, Weng W, Xu M, et al. miR-106b-5p inhibits the invasion and metastasis of colorectal cancer by targeting CTSA. Onco Targets Ther. 2018;11:3835–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Zhao YF, Han M-L, Xiong Y-J, et al. A miRNA-200c/cathepsin L feedback loop determines paclitaxel resistance in human lung cancer A549 cells in vitro through regulating epithelial-mesenchymal transition. Acta Pharmacol Sin. 2018;39:1034–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Friedrich M, Pracht K, Mashreghi M-F, et al. The role of the miR-148/-152 family in physiology and disease. Eur J Immunol. 2017;47:2026–2038. [DOI] [PubMed] [Google Scholar]
- [79].Yi Y, Liu Y, Wu W, et al. The role of miR-106p-5p in cervical cancer: from expression to molecular mechanism. Cell Death Discov. 2018;4:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Liu PL, Liu W-L, Chang J-M, et al. MicroRNA-200c inhibits epithelial-mesenchymal transition, invasion, and migration of lung cancer by targeting HMGB1. PLoS One. 2017;12:e0180844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Gao HX, Yan L, Li C, et al. miR-200c regulates crizotinib-resistant ALK-positive lung cancer cells by reversing epithelial-mesenchymal transition via targeting ZEB1. Mol Med Rep. 2016;14:4135–4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Abudula A, Rommerskirch W, Weber E, et al. Splice variants of human cathepsin L mRNA show different expression rates. Biol Chem. 2001;382:1583–1591. [DOI] [PubMed] [Google Scholar]
- [83].Muntener K, Zwicky R, Csucs G, et al. Exon skipping of cathepsin B: mitochondrial targeting of a lysosomal peptidase provokes cell death. J Biol Chem. 2004;279:41012–41017. [DOI] [PubMed] [Google Scholar]
- [84].Goulet B, Baruch A, Moon N-S, et al. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol Cell. 2004;14:207–219. [DOI] [PubMed] [Google Scholar]
- [85].Mort JS, Buttle DJ. Cathepsin B. Int J Biochem Cell Biol. 1997;29:715–720. [DOI] [PubMed] [Google Scholar]
- [86].Mach L, Mort JS, Glossl J. Noncovalent complexes between the lysosomal proteinase cathepsin B and its propeptide account for stable, extracellular, high molecular mass forms of the enzyme. J Biol Chem. 1994;269:13036–13040. [PubMed] [Google Scholar]
- [87].van der Stappen JW, Williams AC, Maciewicz RA, et al. Activation of cathepsin B, secreted by a colorectal cancer cell line requires low pH and is mediated by cathepsin D. Int J Cancer. 1996;67:547–554. [DOI] [PubMed] [Google Scholar]
- [88].Dalet-Fumeron V, Boudjennah L, Pagano M. Competition between plasminogen and procathepsin B as a probe to demonstrate the in vitro activation of procathepsin B by the tissue plasminogen activator. Arch Biochem Biophys. 1996;335:351–357. [DOI] [PubMed] [Google Scholar]
- [89].Dalet-Fumeron V, Guinec N, Pagano M. In vitro activation of pro-cathepsin B by three serine proteinases: leucocyte elastase, cathepsin G, and the urokinase-type plasminogen activator. FEBS Lett. 1993;332:251–254. [DOI] [PubMed] [Google Scholar]
- [90].Frosch BA, Berquin I, Emmert-Buck MR, et al. Molecular regulation, membrane association and secretion of tumor cathepsin B. APMIS. 1999;107:28–37. [DOI] [PubMed] [Google Scholar]
- [91].Canuel M, Korkidakis A, Konnyu K, et al. Sortilin mediates the lysosomal targeting of cathepsins D and H. Biochem Biophys Res Commun. 2008;373:292–297. [DOI] [PubMed] [Google Scholar]
- [92].Boonen M, Staudt C, Gilis F, et al. Cathepsin D and its newly identified transport receptor SEZ6L2 can modulate neurite outgrowth. J Cell Sci. 2016;129:557–568. [DOI] [PubMed] [Google Scholar]
- [93].Musil D, Zucic D, Turk D, et al. The refined 2.15 A X-ray crystal structure of human liver cathepsin B: the structural basis for its specificity. Embo J. 1991;10:2321–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Quraishi O, Nägler DK, Fox T, et al. The occluding loop in cathepsin B defines the pH dependence of inhibition by its propeptide. Biochemistry. 1999;38:5017–5023. [DOI] [PubMed] [Google Scholar]
- [95].Hao Y, Purtha W, Cortesio C, et al. Crystal structures of human procathepsin H. PLoS One. 2018;13:e0200374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Sosnowski P, Turk D. Caught in the act: the crystal structure of cleaved cathepsin L bound to the active site of Cathepsin L. FEBS Lett. 2016;590:1253–1261. [DOI] [PubMed] [Google Scholar]
- [97].Gocheva V, Joyce JA. Cysteine cathepsins and the cutting edge of cancer invasion. Cell Cycle. 2007;6:60–64. [DOI] [PubMed] [Google Scholar]
- [98].Uhlman A, Folkers K, Liston J, et al. Effects of vacuolar H(+)-ATPase inhibition on activation of Cathepsin B and Cathepsin L secreted from MDA-MB231 breast cancer cells. Cancer Microenviron. 2017;10:49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Achour O, Ashraf Y, Bridiau N, et al. Alteration of cathepsin D trafficking induced by hypoxia and extracellular acidification in MCF-7 breast cancer cells. Biochimie. 2016;121:123–130. [DOI] [PubMed] [Google Scholar]
- [100].Koblinski JE, Ahram M, Sloane BF. Unraveling the role of proteases in cancer. Clin Chim Acta. 2000;291:113–135. [DOI] [PubMed] [Google Scholar]
- [101].de Duve C. The lysosome turns fifty. Nat Cell Biol. 2005;7:847–849. [DOI] [PubMed] [Google Scholar]
- [102].Firestone RA, Pisano JM, Bonney RJ. Lysosomotropic agents. 1. Synthesis and cytotoxic action of lysosomotropic detergents. J Med Chem. 1979;22:1130–1133. [DOI] [PubMed] [Google Scholar]
- [103].Aits S, Jaattela M. Lysosomal cell death at a glance. J Cell Sci. 2013;126:1905–1912. [DOI] [PubMed] [Google Scholar]
- [104].Palermo C, Joyce JA. Cysteine cathepsin proteases as pharmacological targets in cancer. Trends Pharmacol Sci. 2008;29:22–28. [DOI] [PubMed] [Google Scholar]
- [105].Boya P, Andreau K, Poncet D, et al. Lysosomal membrane permeabilization induces cell death in a mitochondrion-dependent fashion. J Exp Med. 2003;197:1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Fehrenbacher N, Jaattela M. Lysosomes as targets for cancer therapy. Cancer Res. 2005;65:2993–2995. [DOI] [PubMed] [Google Scholar]
- [107].Vasiljeva O, Reinheckel T, Peters C, et al. Emerging roles of cysteine cathepsins in disease and their potential as drug targets. Curr Pharm Des. 2007;13:387–403. [DOI] [PubMed] [Google Scholar]
- [108].Eaton JW, Qian M. Molecular bases of cellular iron toxicity. Free Radic Biol Med. 2002;32:833–840. [DOI] [PubMed] [Google Scholar]
- [109].Kurz T, Terman A, Gustafsson B, et al. Lysosomes and oxidative stress in aging and apoptosis. Biochim Biophys Acta. 2008;1780:1291–1303. [DOI] [PubMed] [Google Scholar]
- [110].Dielschneider RF, Henson ES, Gibson SB. Lysosomes as oxidative targets for cancer therapy. Oxid Med Cell Longev. 2017;2017:3749157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Kagedal K, Zhao M, Svensson I, et al. Sphingosine-induced apoptosis is dependent on lysosomal proteases. Biochem J. 2001;359:335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Ostenfeld MS, Fehrenbacher N, Høyer-Hansen M, et al. Effective tumor cell death by sigma-2 receptor ligand siramesine involves lysosomal leakage and oxidative stress. Cancer Res. 2005;65:8975–8983. [DOI] [PubMed] [Google Scholar]
- [113].Ostenfeld MS, Høyer-Hansen M, Bastholm L, et al. Anti-cancer agent siramesine is a lysosomotropic detergent that induces cytoprotective autophagosome accumulation. Autophagy. 2008;4:487–499. [DOI] [PubMed] [Google Scholar]
- [114].Wang Y., Niu H., Hu Z., et al. Targeting the lysosome by an aminomethylated Riccardin D triggers DNA damage through cathepsin B-mediated degradation of BRCA1. J Cell Mol Med. 2018. DOI: 10.1111/jcmm.14077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Zhao M, Antunes F, Eaton JW, et al. Lysosomal enzymes promote mitochondrial oxidant production, cytochrome c release and apoptosis. Eur J Biochem. 2003;270:3778–3786. [DOI] [PubMed] [Google Scholar]
- [116].Ullio C, Casas J, Brunk UT, et al. Sphingosine mediates TNFalpha-induced lysosomal membrane permeabilization and ensuing programmed cell death in hepatoma cells. J Lipid Res. 2012;53:1134–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Mora R, Dokic I, Kees T, et al. Sphingolipid rheostat alterations related to transformation can be exploited for specific induction of lysosomal cell death in murine and human glioma. Glia. 2010;58:1364–1383. [DOI] [PubMed] [Google Scholar]
- [118].Taha TA, Kitatani K, Bielawski J, et al. Tumor necrosis factor induces the loss of sphingosine kinase-1 by a cathepsin B-dependent mechanism. J Biol Chem. 2005;280:17196–17202. [DOI] [PubMed] [Google Scholar]
- [119].Yuan A, Yu C-J, Luh K-T, et al. Aberrant p53 expression correlates with expression of vascular endothelial growth factor mRNA and interleukin-8 mRNA and neoangiogenesis in non-small-cell lung cancer. J Clin Oncol. 2002;20:900–910. [DOI] [PubMed] [Google Scholar]
- [120].Yuan XM, Li W, Dalen H, et al. Lysosomal destabilization in p53-induced apoptosis. Proc Natl Acad Sci U S A. 2002;99:6286–6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Li N, Zheng Y, Chen W, et al. Adaptor protein LAPF recruits phosphorylated p53 to lysosomes and triggers lysosomal destabilization in apoptosis. Cancer Res. 2007;67:11176–11185. [DOI] [PubMed] [Google Scholar]
- [122].Vieler M, Sanyal S. p53 Isoforms and Their Implications in Cancer. Cancers (Basel). 2018;10 DOI: 10.3390/cancers10090288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Broker LE, Huisman C, Span SW, et al. Cathepsin B mediates caspase-independent cell death induced by microtubule stabilizing agents in non-small cell lung cancer cells. Cancer Res. 2004;64:27–30. [DOI] [PubMed] [Google Scholar]
- [124].Groth-Pedersen L, Aits S, Corcelle-Termeau E, et al. Identification of cytoskeleton-associated proteins essential for lysosomal stability and survival of human cancer cells. PLoS One. 2012;7:e45381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Maier O, Galan DL, Wodrich H, et al. An N-terminal domain of adenovirus protein VI fragments membranes by inducing positive membrane curvature. Virology. 2010;402:11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Feofanov AV, Sharonov GV, Astapova MV, et al. Cancer cell injury by cytotoxins from cobra venom is mediated through lysosomal damage. Biochem J. 2005;390:11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Johansson AC, Appelqvist H, Nilsson C, et al. Regulation of apoptosis-associated lysosomal membrane permeabilization. Apoptosis. 2010;15:527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Edelmann B, Bertsch U, Tchikov V, et al. Caspase-8 and caspase-7 sequentially mediate proteolytic activation of acid sphingomyelinase in TNF-R1 receptosomes. Embo J. 2011;30:379–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Eskelinen EL, Tanaka Y, Saftig P. At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol. 2003;13:137–145. [DOI] [PubMed] [Google Scholar]
- [130].Fehrenbacher N, Bastholm L, Kirkegaard-Sørensen T, et al. Sensitization to the lysosomal cell death pathway by oncogene-induced down-regulation of lysosome-associated membrane proteins 1 and 2. Cancer Res. 2008;68:6623–6633. [DOI] [PubMed] [Google Scholar]
- [131].Mrschtik M, Ryan KM. Lysosomal proteins in cell death and autophagy. Febs J. 2015;282:1858–1870. [DOI] [PubMed] [Google Scholar]
- [132].Kagedal K, Johansson U, Ollinger K. The lysosomal protease cathepsin D mediates apoptosis induced by oxidative stress. Faseb J. 2001;15:1592–1594. [DOI] [PubMed] [Google Scholar]
- [133].Kirkegaard T, Jaattela M. Lysosomal involvement in cell death and cancer. Biochim Biophys Acta. 2009;1793:746–754. [DOI] [PubMed] [Google Scholar]
- [134].Green DR, Llambi F. Cell Death Signaling. Cold Spring Harb Perspect Biol. 2015;7 DOI: 10.1101/cshperspect.a006080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Appelqvist H, Johansson A-C, Linderoth E, et al. Lysosome-mediated apoptosis is associated with cathepsin D-specific processing of bid at Phe24, Trp48, and Phe183. Ann Clin Lab Sci. 2012;42:231–242. [PubMed] [Google Scholar]
- [136].Cirman T, Oresić K, Mazovec GD, et al. Selective disruption of lysosomes in HeLa cells triggers apoptosis mediated by cleavage of Bid by multiple papain-like lysosomal cathepsins. J Biol Chem. 2004;279:3578–3587. [DOI] [PubMed] [Google Scholar]
- [137].Droga-Mazovec G, Bojic L, Petelin A, et al. Cysteine cathepsins trigger caspase-dependent cell death through cleavage of bid and antiapoptotic Bcl-2 homologues. J Biol Chem. 2008;283:19140–19150. [DOI] [PubMed] [Google Scholar]
- [138].Nylandsted J, Brand K, Jaattela M. Heat shock protein 70 is required for the survival of cancer cells. Ann N Y Acad Sci. 2000;926:122–125. [DOI] [PubMed] [Google Scholar]
- [139].Yap YW, Whiteman M, Bay BH, et al. Hypochlorous acid induces apoptosis of cultured cortical neurons through activation of calpains and rupture of lysosomes. J Neurochem. 2006;98:1597–1609. [DOI] [PubMed] [Google Scholar]
- [140].Jancekova B, Ondrouskova E, Knopfova L, et al. Enzymatically active cathepsin D sensitizes breast carcinoma cells to TRAIL. Tumour Biol. 2016;37:10685–10696. [DOI] [PubMed] [Google Scholar]
- [141].Fritsch J, Fickers R, Klawitter J, et al. TNF induced cleavage of HSP90 by cathepsin D potentiates apoptotic cell death. Oncotarget. 2016;7:75774–75789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Burton LJ, Henderson V, Liburd L, et al. Snail transcription factor NLS and importin beta1 regulate the subcellular localization of Cathepsin L and Cux1. Biochem Biophys Res Commun. 2017;491:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Kim S, Jin H, Seo HR, et al. Regulating BRCA1 protein stability by cathepsin S-mediated ubiquitin degradation. Cell Death Differ. 2018. DOI: 10.1038/s41418-018-0153-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Bian B, Mongrain S, Cagnol S, et al. Cathepsin B promotes colorectal tumorigenesis, cell invasion, and metastasis. Mol Carcinog. 2016;55:671–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Bannoud N, Carvelli FL, Troncoso M, et al. Cation-dependent mannose-6-phosphate receptor expression and distribution are influenced by estradiol in MCF-7 breast cancer cells. PLoS One. 2018;13:e0201844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Bronger H, Karge A, Dreyer T, et al. Induction of cathepsin B by the CXCR3 chemokines CXCL9 and CXCL10 in human breast cancer cells. Oncol Lett. 2017;13:4224–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Pecar Fonovic U, Kos J. Cathepsin X Cleaves Profilin 1 C-Terminal Tyr139 and Influences Clathrin-Mediated Endocytosis. PLoS One. 2015;10:e0137217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Shao G, Wang R, Sun A, et al. The E3 ubiquitin ligase NEDD4 mediates cell migration signaling of EGFR in lung cancer cells. Mol Cancer. 2018;17:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Yang KM, Bae E, Ahn SG, et al. Co-chaperone BAG2 Determines the Pro-oncogenic Role of Cathepsin B in Triple-Negative Breast Cancer Cells. Cell Rep. 2017;21:2952–2964. [DOI] [PubMed] [Google Scholar]


