Abstract
Many cell culture and animal models have been used to study hepatitis B virus (HBV) replication and its effects in the liver; these have facilitated development of strategies to control and clear chronic HBV infection. We discuss the advantages and limitations of systems for studying HBV and developing antiviral agents, along with recent advances. New and improved model systems are needed. Cell culture systems should be convenient, support efficient HBV infection, and reproduce responses of hepatocytes in the human body. We also need animals that are fully permissive to HBV infection, convenient for study, and recapitulate human immune responses to HBV and effects in the liver. High-throughput screening technologies could facilitate drug development based on findings from cell and animal models.
Keywords: Treatment, Mouse, Immune Response, Viral Infection
Hepatitis B virus (HBV) is a hepatotropic virus with strict specificity for human hepatocytes.1,2 This species tropism and the difficulties in culturing human hepatocytes have limited systems for studying HBV. We review the HBV life cycle (Figure 1) and discuss cell culture and animal model systems for studying HBV replication and its mechanisms of pathogenesis. We also discuss strategies for development of anti-HBV agents based on findings from model systems.
Figure 1.

HBV replication. 1. Virus binding and entry into the host cell (large rectangle). 2. Intracellular trafficking and delivery of rcDNA to the nucleus (large circle). 3. Conversion of rcDNA to cccDNA, or integration of the double-stranded linear (DSL) DNA into cell DNA (3a). 4. and 4a. Transcription to synthesize viral RNAs (wavy lines), including the core (C) mRNA for both the core and the reverse transcriptase (RT) proteins; large surface (LS) mRNA for the large (L) surface or envelope protein; small (S) mRNA for the middle (M) and small (S) envelope proteins; X mRNA for the X protein; and PreC mRNA for the PreCore protein. The C mRNA is also the pgRNA: the template for producing progeny viral DNA via reverse transcription. 5. Translation to synthesize viral proteins. 6. Assembly of the pgRNA- (and RT-) containing NC, or alternatively, empty capsids (6a). 7. Reverse transcription of pgRNA to synthesize the (–) strand single-stranded (SS) DNA and then rcDNA. 8. Nuclear recycling of progeny rcDNA to form more cccDNA (intracellular cccDNA amplification). 9. Envelopment of the rcDNA-containing NC and secretion of complete virions, or alternatively, secretion of empty virions (9b) or HBsAg spheres and filaments (9a). Processing of the PreCore protein and secretion of HBeAg are shown in 9c. The secretion of putative RNA virions is not yet resolved (9?). The different viral particles outside the cell are shown with their approximate concentrations in the blood of infected persons indicated: the complete, empty, or RNA virions as large circles (outer envelope) with an inner diamond shell (capsid), with or without rcDNA (gapped, double concentric circle) or RNA (wavy line) inside the capsid, respectively; HBsAg spheres and filament as small circles and cylinders. Approximate concentrations of the different viral particles in the blood of infected patients are indicated but can vary among patients and over time in the same patient. Intracellular capsids, diamonds, with either viral pgRNA, SS ([–] strand) DNA (straight line), rcDNA, or empty. Dashed lines of the diamond in the rcDNA-containing mature NC and the complete virion, destabilization of the mature NC. Boxed letters, viral proteins translated from the mRNAs. Filled small circle inside the capsids, the RT protein binding to the 5′ end of the pgRNA and covalently attached to the 5′ end of the (–) strand DNA. Twisted circle, cccDNA. The thin dashed line and arrow indicate that HBeAg (small orange circles outside the cell) is frequently decreased or lost late in infection. For simplicity, synthesis of the minor DSL form of the genomic DNA in the mature NC, its secretion in virions, and infection of DSL DNA-containing virions are not depicted here, as are the functions of X. Adapted from Hu and Liu,177 with the Creative Commons Public License, https://creativecommons.org/licenses/by/4.0/legalcode.
Early steps in the HBV replication cycle, from cell binding through formation of covalently closed circular DNA (cccDNA), were difficult to study until the virus’s receptor, the solute carrier family 10 member 1 (SLC10A1, also called NTCP),3 was identified on human hepatocytes. This discovery led to cell culture systems to study these processes. Later steps of the replication cycle, from virus transcription to virion secretion, had been extensively studied in established cell culture systems, although many questions remain.
Because of the strict species specificity of HBV infection, animal models for studying the host response to the virus and disease pathogenesis have been suboptimal. Currently approved antiviral agents target only the DNA synthesis activity of the HBV reverse transcriptase protein, so these block viral DNA synthesis from pregenomic RNA (pgRNA) (step 7, Figure 1),4,5 with the exception of the cytokine type I interferon, which appears to inhibit multiple steps of the HBV life cycle and promote the antivirus immune response.5,6 Efforts to develop antiviral agents against viral genome reservoir or cccDNA, or to promote the patient’s antivirus response, have been limited by the lack of optimal model systems.
Cell Culture Systems
A number of cell culture systems have been used to study HBV replication (Table 1). What are the advantages and disadvantages of each of these systems? No system is ideal for all studies, but findings from different systems can be complementary.
Table 1.
Advantages and Limitations of Cell Culture Systems for Studying HBV
| Systems | Advantages | Limitations |
|---|---|---|
| Human hepatoma cell lines (HepG2, Huh7) | • Readily available • Robust in supporting steps in replication after transcription |
• Fail to support infection • Oncogenic transformation: only partially mimic normal hepatocytes |
| HepaRG | • Support infection • Mimic normal hepatocytes better than other hepatoma cell lines |
• Low efficiency of infection • Complicated conditions for differentiation |
| HepG2-NTCP | • Readily available | • Oncogenic transformation: only partially mimic normal hepatocytes |
| Primary human hepatocytes | • Support HBV infection • Mimic normal hepatocytes the best • Support complete HBV life cycle |
• Low efficiency of infection • Limited in supply • High cost • Donor variability |
| iHep cells | • Unlimited in supply • Support complete HBV life cycle • Mimic normal hepatocytes better than hepatoma cell lines |
• Complicated conditions for differentiation • Induced hepatocytes may not fully mimic mature hepatocytes |
Human Liver Cell Lines
The hepatoma cell lines HepG2 (derived from a hepatoblastoma) and Huh7 (derived from a hepatocellular carcinoma [HCC]), have been widely used to study HBV replication, especially the later stages, from transcription and virion production. These cells lack the HBV receptor and are therefore not susceptible to HBV infection. However, transfection of these cells with cloned HBV DNA in over-length constructs, which serve as functional mimics of cccDNA to direct virus transcription, leads to expression of all virus gene products, assembly of intracellular empty capsids and nucleocapsids, and formation and secretion of virion particles, including complete (infectious) and empty virions and HB surface antigen (HBsAg) particles (Figure 1).7–11 Virtually unlimited in supply and relatively stable genetically, the hepatoma cultures are convenient systems for studying these stages of HBV replication. Transient transfection with cloned HBV DNA and stably transfected cell lines derived from these hepatoma cells have been widely used to study HBV replication. Although transient transfection affords the flexibility of testing a large variety of virus mutants, the stable transfection system allows high efficiency of virus replication, including temporally controlled viral gene expression and replication.
Hepatoma cell lines allow not only for constitutive virus expression and replication, but inducible expression and replication of HBV. The HepAD38 and HepDE19 cell lines direct HBV pgRNA expression under control of the tetracycline-repressible promoter instead of the native viral core promoter.12,13 HepG2 cells, and to a lesser-degree Huh7 cells, also support an intracellular recycling pathway that produces cccDNA following production of intracellular mature nucleocapsids (NCs), which contains relaxed circular DNA (rcDNA), the precursor to cccDNA (Figure 1, steps 8 and 3).12–15 The heterologous promoter can be switched off following virus gene expression, DNA replication, and cccDNA production, so that cccDNA-supported viral gene expression and replication can be monitored, including the use of sensitive reporter systems.16,17 However, cccDNA-directed transcription in these system is inefficient,17,18 presumably due to the lack of appropriate transcription factors and/or epigenetic regulation, which promotes high levels of virus transcription in hepatocytes of infected individuals. In addition to not expressing the HBV receptor and providing a suboptimal environment for virus transcription, infected hepatoma cells fail to recapitulate many aspects of the responses in the infected hepatocytes in vivo, including potential innate immune response.19–21
The HepaRG cell line is a bi-potent liver progenitor cell line derived from a hepatoma in a patient infected with hepatitis C virus (HCV). These cells differentiate into hepatocyte-like and bile duct epithelium-like cells in culture. In contrast to all other human hepatoma cell lines tested, HepaRG cells, on differentiation, support HBV infection and the subsequent steps of HBV replication.22,23 Furthermore, HBV-infected cells appear to mount an innate immune response that is able to suppress HBV replication.20 Their response to interferons, including the suppression of HBV, is similar to that of primary human hepatocytes and better than that of HepG2 cells.21
However, there is controversy over the innate immune response to HBV infection. HBV does not induce a strong innate response, such as that mediated by type I interferon in humans or chimpanzees.6,24–27 However, under certain circumstances, such as on exposure of virus DNA to cytosolic DNA-sensing mechanisms, due to enhanced or accelerated uncoating of viral nucleocapsids,19 HBV can induce an innate immune response. A shortcoming of the HepaRG cells is that HBV infection is inefficient (even with high multiplicity of infection and differentiation conditions required for cells to become susceptible to infection), complex, and time-consuming (requires 4 weeks). Also, there is little to no amplification of cccDNA (Figure 1, steps 8 and 9) after HBV infection of HepaRG cells,23 which is also true for HepG2 cells engineered to express NTCP (see later in this article). This could be due to the low levels of HBV replication in these cells, and consequently, the low levels of rcDNA.
Transgenic expression of NTCP in HepG2 cells3 allows them to become infected with HBV, so one block to infection in these cells is therefore virus binding and entry. Transgenic expression of NTCP in Huh7 cells results in inefficient infection by HBV28; Huh7 cells are less efficient than HepG2 cells in supporting cccDNA formation via intracellular recycling (Figure 1, steps 8 and 3).15 The lower infection efficiency of Huh7-expressing NTCP (Huh7-NTCP) cells vs. HepG2-NTCP cells could result from the lower capacity of Huh7 cells to support cccDNA formation (Figure 1, steps 2 and 3).
HepG2-NTCP cells are more convenient to carry in culture than HepaRG cells. HBV infection of HepG2-NTCP is sufficiently robust to allow for studies of the early stages of HBV infection, from cell attachment to cccDNA establishment and maintenance,29–33 as well as for testing antiviral agents.34,35 The infection may spread among cells cultured in the presence of polyethylene glycol (PEG),36 which is routinely added during the infection period (overnight) and then removed. It is not clear how PEG, or dimethyl sulfoxide (DMSO, also used to reduce cell proliferation), increase cell infection by HBV. NTCP-HepG2 cells are transformed, so they lack many features of human hepatocytes, which affects their ability to support virus infection and replication or mount an immune response.37,38
Primary Human Hepatocytes
Primary human hepatocytes are considered to be the most physiologically relevant culture system for HBV infection in vitro, as they most closely represent the human hepatocytes in the liver, the natural target cells that are infected by HBV. Thus, they are best for studying metabolic and innate immune responses to HBV infection. Primary human hepatocytes do support the complete life cycle of HBV.39–41 However, they are expensive, scarce, do not proliferate in culture, have a limited life span (1 month or shorter), vary among batches, and have low infection efficiency. They support only a limited spread of infection due to their rapid dedifferentiation in culture.42,43 Human fetal hepatocytes have also been shown to support HBV infection. These cells have greater viability and proliferative capacity than adult hepatocytes.44,45 In immune-deficient mice, endogenous hepatocytes can be destroyed and replaced with human hepatocytes, which expand following transplantation to generate mice with humanized livers (or chimeric mice).46 The progeny of these human hepatocytes can be isolated from the chimeric mice and cultured for HBV infection. This approach to carrying primary human hepatocytes could increase reproducibility among studies; however, the procedure is time-consuming and technically challenging.
Efforts have been made constantly to improve systems for culture of primary human hepatocytes. These include methods to promote proliferation and maintain or increase differentiation, using an appropriate extracellular matrix, co-culture with nonparenchymal cells, incubation with DMSO, and 3-dimensional organization.43,45,47,48 Primary human hepatocytes cultured in 3-dimensions stably express hepatocyte genes and function for a month or longer, and can be infected with HBV at low multiplicity of infection without the need for PEG or DMSO.27 These improvements, if reproducible and widely adopted, could lead to wider application of primary human hepatocytes for HBV studies.
Induced Human Hepatocyte-like Cells
Developments in cell reprogramming techniques have led to production of induced human hepatocyte-like (iHep) cells. iHep cells or human hepatocyte-like cells are obtained via induced differentiation of human embryonic stem cells at least some aspects of the innate immune response in or induced pluripotent stem cells. They have more physiologic features of hepatocytes in humans than cell lines, and can support the complete life cycle of HBV, including virus spread among cells due to long-term maintenance of hepatic differentiation.47,49,50 iHep cells are virtually unlimited in supply, amenable to genetic manipulation, and recapitulate infected hepatocytes in humans.47 Strategies to optimize or transdifferentiate pluripotent stem cells51–55 might lead to development of a convenient and relevant system for studying HBV infection.
Nonhepatic and Nonhuman Cells
The human kidney embryonic cell line HEK293 and the chicken hepatoma cell line LMH each support later stages of HBV replication, including cccDNA formation via the intra-cellular recycling pathway (Figure 1).14,56 It is therefore clear that no strictly liver- or human-specific factors are required for cccDNA formation. However, nonhepatic cells such as HEK293 are unable to support HBV transcription, due to the lack of appropriate transcription factors in these cells.57–59 Therefore, strong heterologous promoters such as human cytomegalovirus immediate early enhancer/promoter are used to express HBV genes in these cells.60 HEK293 cells and LMH cells can be transfected at high efficiency, and along with human hepatocyte-derived cells, they can be useful for comparative studies of the contributions of tissue- and species-specific factors to HBV replication.
Primary hepatocytes from macaques and pigs, but not mice, rats, or dogs, can support HBV infection following ectopic expression of the human NTCP, indicating the existence of host species-specific factors, beyond the receptor, that are required to support HBV infection.61,62 Interestingly, hepatocytes from tree shrews (Tupaia)63 can be infected with HBV, without the need for transgenic expression of human NTCP. These cells were instrumental in identification of NTCP as the HBV receptor.3
Of particular interest to the issue of species specificity of HBV infection, and the development of a small animal (mouse) model of HBV infection (see below), is the observation that mouse hepatocytes are unable to support HBV cccDNA formation, despite their high levels of viral transcription and subsequent steps through virion production.64 A human-mouse heterokaryon assay attributed this deficiency to the lack of a factor in mouse cells that is needed to support cccDNA formation.65 It was reported almost 2 decades ago that livers of mice with disruption of the hepatocyte nuclear factor 1 alpha gene (Hnf1a) support low levels of HBV cccDNA formation.66 Although the underlying mechanism for this observation has remained unclear, this result did suggest that it might be possible to manipulate mouse cells to allow for HBV cccDNA formation. Indeed, the mouse hepatocyte cell line AML12, derived from transgenic mice that express human tumor growth factor alpha, support cccDNA formation as efficiently as human HepG2 cells, when it expresses the HBV pgRNA under control of a tetracycline-repressible promoter (AML12HBV10 cells).67 Increased formation of cccDNA in AML12HBV10 cells correlates with an increase in uncoating of nucleocapsids, which releases rcDNA into the cell nucleus, a prerequisite for cccDNA formation (Figure 1, steps 2 and 8). Mature nucleocapsids, which contain rcDNA, are normally destabilized in human cells, compared with immature nucleocapsids, which is likely to be a priming event for uncoating68; this destabilizing increases in AML12HBV10 cells. AML12 cells that express human NTCP, in contrast to other mouse hepatocyte cell lines or primary mouse hepatocytes, are susceptible to HBV infection.54,69,70
These findings indicate that a major block in mouse hepatocytes to HBV infection is at the stage of nucleocapsid uncoating. Interestingly, mutations of the virus core protein increase cccDNA formation in human hepatoma cells and are associated with increased nucleocapsid uncoating.15 cccDNA formation might therefore be accomplished in mouse hepatocytes either by manipulation of the mouse cells or modification of the viral capsid protein.
Animal Models
There are many stages to chronic HBV infection.71–74 Features of chronic HBV infection include formation of cccDNA, assembly of infectious viral particles for dissemination, and persistence. The immune response (innate and adaptive) involves seroconversion to HBsAg and HBe antigen (HBeAg) and eventual clearance of or tolerance to the virus. Chronic infection can lead to hepatitis, liver cirrhosis, and hepatocellular carcinoma. Studies of the mechanisms by which the immune response to HBV infection leads to hepatitis and processes of liver injury and repair require animal models.
Humans have a unique immune response to HBV similar to chimpanzees. Both do not have a conventional innate immune response (interferons, tumor necrosis factor, interleukin 1 beta) to HBV; however, HBV clearance in chimpanzees involves T-cell expression of genes that are regulated by interferon gamma and therefore suggest a nonconventional innate immune response.24 Furthermore, clearance of HBV infection in humans varies with their age: human newborns exposed to HBV do not clear the virus, but almost all exposed adults eliminate HBV.75 Animal models are used to systemically study interactions between HBV and the liver microenvironment, the immune response, and interactions between gut and liver.
Some animals are naturally infected with hepadnaviruses that are similar to HBV, such as woodchucks (infected with woodchuck hepatitis virus) and ducks (infected with duck HBV); these animals develop many features of human HBV infection. Chimpanzees are the only nonhuman immune-competent animals that are naturally susceptible to HBV infection. Chimpanzee studies were instrumental in the development of HBV vaccines in the 1970s for evaluation of vaccine safety and efficacy.76 Natural animal models are also useful in verifying the safety and efficacy of antiviral nucleot(s)ides, small interfering RNAs (siRNAs), or other molecules.77–83 Limitations to their study include genetic variation, lack of tools for studying the immune response, longer study periods, challenging laboratory techniques, and ethical considerations.84
Mice have well-characterized immune systems and are easy to handle and manipulate, but cannot be infected with HBV. Therefore, multiple genetically modified mouse models of HBV infection have been developed, using transgenic, transfection, and humanized liver technologies. Mice are useful and reliable for studying progression of HBV infection and the immune response. What animal models are available and what have we learned from them (see Table 2)?
Table 2.
Animal Models of Chronic HBV Infection
| Immune response |
Preclinical studies |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adaptive immunity |
Pathogenesis |
|||||||||||
| HBV infection |
B cell |
T cell |
||||||||||
| Animal | System | cccDNA | Infectability | Persistence | Innate immunity |
Seroconversion to HBeAg and HBsAg seroconversion |
Tolerance and clearance |
Hepatitis | Cirrhosis | HCC | Antiviral | Immune modulator |
| Transgenic mice | Expression of HBV transgenes | − | − | + | − | −/− | Tolerance | − | − | +/− | + | + |
| Expression of human NTCP | − | +/− | − | − | −/− | NA | − | − | − | − | − | |
| Transfected mice | Hydro-dynamic injection | − | − | +/− | + | +/+ or +/− | Tolerance and clearance | − | − | − | + | + |
| AAV injection | − | − | + | + | −/− | Tolerance | − | − | + | + | + | |
| AdV injection | − | − | +/− (dose–dependent) | + | −/− or +/+ | Tolerance or clearance | − | − | − | + | + | |
| Mice with humanized liver | human hepatocytes | + | + | + | +/− | −/− | tolerance | + | − | − | + | − |
| Human hepatocytes and human hematopoietic cells | + | + | + | + | −/− | Tolerance | + | + | − | + | + | |
| Other animals | Woodchuck | + | + | + | − | +/+ | Tolerance and age-dependence | + | + | + | + | + |
| Tupaia | + | + | +/− | +/− | +/− | Tolerance and clearance | +/− | +/− | +/− | ? | ? | |
| Macaque | +/− | +/− | +/− | − | −/− | Tolerance | − | − | − | − | − | |
| Macaque expressing human NTCP transgene | + | + | + | + | +/+ | Tolerance and clearance | ? | ? | − | ? | ? | |
NOTE. +/– signs indicate the presence/absence or occurrence/non-occurrence of the corresponding characteristics, whereas question marks (?) indicate the status of the specific characteristics are currently not sure unclear.
AAV, adeno-associated virus; NA, not applicable.
Transgenic Mice
HBV transgenic mice that express single virus proteins (envelope, core, precore, or X) or all viral genes have been generated. Infectious HBV virions produced by mouse hepatocytes are morphologically indistinguishable from human-derived virions. Because these mice are transgenic for HBV, they do not have the potential of viral clearance.64 However, infected mice can be used to study features of the innate immune response, such as HBV inhibition of interferon and tumor necrosis factor.85,86 Adoptive transfer of immune cells from syngeneic mice immunized against HBV can be used to study the adaptive immune response. These mice do not develop liver fibrosis or cirrhosis, and only a few strains of transgenic mice, such as those that express large HBsAg or overexpress HBx protein, develop HCC.87,88 HBV transgenic mice are used to study the effects of cytokines or drugs that might prevent HBV progression, and for development of strategies for breaking immune tolerance to HBV.89–96
NTCP is the receptor not only for HBV, but also for hepatitis D virus (HDV).3 HDV, but not HBV, can infect transgenic mice that express human NTCP.70 Similarly, rat or dog hepatocytes that express human NTCP are susceptible to HDV but not HBV infection.61 This indicates that additional factors are required for HBV replication and for induction of an antiviral immune response.
Transfected Mice
To overcome the limitations of HBV transgenic mice, vectors are used for the introduction of HBV DNA in livers of mice. Hydrodynamic injection is used to transfer genetic material to mouse liver; it involves rapid injection of a large volume of liquid via the tail vein. Hydrodynamic injection of plasmids carrying an overlength HBV 1.3X results in high levels of HBV replication, high blood levels of HBV DNA, and an adaptive immune response that clears the HBV.97 The system was modified to establish HBV persistence to allow studies of immune tolerance to the virus.98 Chou et al99 reported rapid clearance of HBV from BALB/cJ and NOD/ShiLtJ mice, but HBV persisted for longer times in C57BL/6J and C3H/HeN mice. The authors demonstrated the age-dependence of HBV clearance in C3H/HeN mice, which involved age-related establishment of the gut microbiota and the toll-like receptor 4. The mouse immune response to HBV can be studied to identify genes that regulate the innate and adaptive immune response in the liver and the effects of the intestinal microbiota.
HBV DNA can also be delivered into mouse hepatocytes via adenovirus (AdV)- or adeno-associated virus–based vectors. HBV protein production was observed for up to 3 months after intravenous injection of AdV-HBV, followed by HBV clearance by B-cell– and T-cell–mediated responses.100 Hydrodynamic injections and AdV vectors can induce innate immune responses, so findings from these systems should be interpreted with caution.101 Finally, these systems can duplicate HBeAg or HBsAg seroconversion without manifestation of liver diseases.
Hepatocytes from transfected mice do not contain HBV cccDNA, but further genetic manipulation of mice might allow this to occur (see above). In addition to its role for studying immune responses during acute HBV infection, the hydrodynamic injection and adeno-associated virus–HBV systems have also been used to evaluate antiviral compounds and strategies to induce an anti-HBV immune response.94,102–104 Hydrodynamic injection of an siRNA that knocks down expression of a region of HBV DNA that is conserved among genotypes,105 or an siRNA that knocks down expression of conserved regions of the X gene, reduced levels of HBsAg in mice.106
Humanized Chimera Mice
Mice with humanized livers are used to study HBV infection.107 SCID mice that express urokinase-type plasminogen activator under control of an albumin promoter (alb-uPA) have subacute liver failure as newborns, but their liver can be reconstituted with human hepatocytes. Although these hepatocytes can reconstitute up to 70% of liver, the alb-uPA SCID mice develop kidney and hematologic disorders and have a low breeding efficiency and narrow time period for liver manipulation. Human hepatocytes can also be transplanted into livers of Rag2–/– (immune-deficient) mice with disruption of the fumarylacetoacetate hydrolase gene (Fah–/–) and interleukin 2 receptor gamma chain (Il2rg–/–) (FRG chimeras). FAH deficiency leads to hepatocellular injury, which can be prevented by injections of 2-(2-nitro-4-trifluoro-methyl-benzoyl) 1,3-cyclohexedione (NTBC); loss of RAG2 and IL2RG deficiencies provides the immune-deficient back-ground required for engraftment of human hepatocytes.
FRG mice can receive transplants of human hepatocytes at any age, and the degree of liver damage can be controlled by adjusting NTBC administration; serial transplantations gradually improve the efficiency of engraftment of human hepatocytes. The chimeras can be infected with HBV virions, and its infected grafted human hepatocytes generate functional HBV cccDNA and viral spreading. Because these mice do not have an immune system, they are not used to study the antivirus immune response. However, human T cells can be transferred to the mice to study their response to infection of human hepatocytes with HBV.108,109 Some drugs cannot be studied in these mice because of the administration of NTBC or other treatments during the process. Chimeric mice therefore provide unique platforms to study the mechanisms of HBV cccDNA formation and direct antiviral agent studies.110–118 For example, myrcludex B, a synthetic lipopeptide derived from HBV envelope preS1 domain, was found to block de novo infection of human hepatocytes with HBV and virus spread in FRG chimeras.118,119
Mice with humanized immune systems were developed by transplantation of human hematopoietic cells or fetal liver cells.120–123 AFC8-hu HSC/Hep and A2/NSG-hu-HSC/Hep mice have humanized immune systems and livers following co-transplantation of human CD34+ hematopoietic stem cells and human hepatocyte progenitors (or fetal liver cells). A2/NSG-hu-HSC/Hep mice can be infected with HBV, which persists for at least 4 months with limited antiviral innate and adaptive immune responses. Some mice even develop hepatitis and liver fibrosis following transplantation of fetal liver cells.122 These mice can be used to study the antiviral immune response and its contribution to liver disease. A2/NSG-hu-HSC/Hep mice have T-cell– and B-cell–mediated immune responses to HBV infection, but these differ from those of humans due to the cellular configuration of the liver microenvironment. Chimeric mice contain fewer hepatocytes and have lower levels of virus replication than humans,107 but are still useful for studying the mechanisms by which HBV causes liver disease.
Nonmouse Models
It has been a challenge to develop other models of HBV infection, because most animals are not infected by HBV or do not develop disease. Although chimpanzees are susceptible to HBV infection and develop an antiviral immune response, studies have been restricted mainly due to ethical concerns. The hepadnavirus capuchin monkey HBV was recently reported to infect Brazilian capuchin monkeys, without causing inflammation, and infect human hepatocytes via NTCP.124 The capuchin monkey system requires further study. WHV is similar to HBV in terms of morphology, genome structure, replication, epidemiology, course of infection, and development of HCC with integrated WHV DNA. Most WHV integrations observed in HCC samples occurred in proximity to N-myc genes; similar HBV insertions are also found near TERT, MLL4, and CCNE1 in HCC samples from patients with HBV infection. Studies based on HCC tissues have observed both WHV and HBV to integrate in the genome at specific yet distinct sites,125–127 as a consequence of simulating clone expansion. However, the viruses cause liver disease via distinct mechanisms and several treatment strategies, such as core protein allosteric modulators, are specific to HBV. Capuchin monkeys are difficult to handle, have limited availability, are of outbred origin, and have insufficiently characterized immune systems.128–131
Tupaia is the only nonprimate animal found susceptible to HBV infection. HBV infection of Tupaia is transient with low levels of HBV replication. Liver fibrosis and HCC have been observed in some studies, whereas rapid clearance of HBV was reported in others. Tupaia hepatocytes were used to discover the first HBV receptor, NTCP.3,63,132
Mauritian cynomolgus macaques were reported to have persistent HBV infection.133 Although rhesus macaques are not naturally susceptible to HBV infection, transgenic expression of human NTCP by their hepatocytes allows for infection by HBV. HBV infection persisted in 2 transgenic macaques, leading to induction of HBV-specific T-cell responses, whereas HBV was rapidly cleared from 2 other macaques. These macaques could be a useful, immune-competent primate for studies of HBV infection.61,62
Application for Drug Development
Systems to transfect cells with HBV DNA and study HBV replication have been available for more than 3 decades, and to transfect mice for studies of HBV replication have been available for 2 decades, so many agents have been identified that target the later stages of the HBV life cycle.8,10,64 These include inhibitors of HBV DNA transcription, RNA stability, capsid assembly, RNaseH digestion, virion secretion, HBsAg secretion, and reverse transcription (reference: article in this issue about Drug Development). Drugs approved by the Food and Drug Administration for treatment of chronic HBV infection inhibit HBV reverse transcriptase. Agents such as GLS4, JNJ-379, and NVR3–778, have been developed to target core proteins, capsid assembly, and other steps of virus replication.134–136 Immune-modulating drugs have been studied in animal models, including an agonist of the toll-like receptor 7 (GS-9620 and RO6864018), therapeutic vaccines (GS4774 and NASVAC), and the activator of retinoic acid-inducible gene-I and nucleotide binding oligo-merization domain containing 2 (NOD2) (SB9200).137–140
Cell models of HBV infection can be screened to identify agents that disrupt the early stages of the HBV life cycle, especially the entry process. Myrcludex B, which was characterized in HepaRG cells and uPA SCID mice with humanized livers, is being tested in a phase 2 trial.132,141,142 Myrcludex B is a lipopeptide that mimics the HBV preS1 2–48 aa region and disrupts interaction between NTCP and HBV.3,28,132,142,143 Studies of HepG2-NTCP cells and other cells that express an NTCP transgene have led to identification of other inhibitors of HBV entry (see Table 3). Bile acids, the endogenous substrates of NTCP, block entry of HBV into cells. Tauroursodeoxycholic acid blocks binding of HBV to NTCP at a half maximal inhibitory concentration of approximately 1 µM.28,144–146
Table 3.
Identified Anti-HBV Entry Inhibitors by Application of New Models
| Name | Structure | Model system for identification |
Reference |
|---|---|---|---|
| bile acids tauroursodeoxycholic acid (TUDCA) etc. |  |
HepaRG Huh7-NTCP HepG2-NTCP PHHs PTHs |
28, 144, 146, 153 |
| Cyclosporin A | 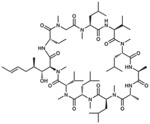 |
HepaRG Huh7-NTCP HepG2-NTCP PHHs |
150, 153 |
| Ezetimibe | 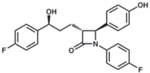 |
HepaRG HepG2-NTCP PHHs PTHs |
144, 149 |
| Irbesartan | 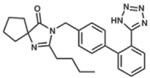 |
HepG2-NTCP | 148, 152 |
| Proscillaridin A | 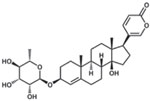 |
HepG2-NTCP | 151 |
| Zafirlukast | HepaRG | 147 | |
| Vanitaracin A |  |
HepG2-NTCP PHHs |
157, 158 |
| NPD8716 | HepG2-NTCP PHHs |
156 | |
| SCY995 | 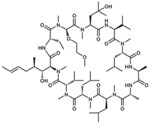 |
HepG2-NTCP PHHs |
35 |
| WD1 |  |
HepG2-NTCP PHHs |
159 |
| Ro41–5253 | 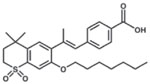 |
HepaRG PHHs |
162 |
| Epigallocatechin gallate | 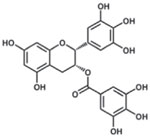 |
HuS-E/2 Huh7-NTCP |
161 |
| Proanthocyanidin | 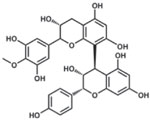 |
HepG2-NTCP PHHs |
164 |
| Oolonghomobisflavan | 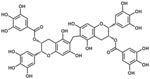 |
HepG2-NTCP PHHs |
164 |
| Genistein | 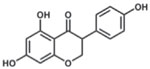 |
iHeps PHHs |
49 |
| PA452 |  |
iHeps PHHs HepG2-NTCP |
49 |
| Bexarotene | 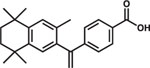 |
HepG2-NTCP HepaRG PTHs |
163 |
| Fasiglifam | HepG2-NTCP | 155 | |
| Rapamycin |  |
HepG2-NTCP | 154 |
| Myrcludex-B (lipopeptide) | Myristoyl-GTNLSVPNPLGFFPDHQLDPAFGANSNNPDWDF NPNKDHWPEANQVG | HepaRG PTHs Humanized mouse |
118, 142 |
Some drugs approved by the Food and Drug Administration impair NTCP transporter function and inhibit HBV entry into cells. This class of drugs includes cyclosporin A, ezetimibe, irbesartan, proscillaridin A, rapamycin, fasiglifam, and zafirlukast.144,147–155 Small molecules that interact with NTCP have been identified from natural products or other sources using NTCP-reconstituted cell lines. This class includes vanitaracin A (a tricyclic polyketide), NTI-007 (an imidazopyridine derivative), NPD8716 (a coumarin derivative), SCY995 (a cyclosporin derivative), and WD1 (a macrocyclic peptide).35,156–160 Some small molecules identified in cell culture experiments have been shown to reduce expression of NTCP, including Ro41–5253 (a synthetic retinoid), epigallocatechin gallate (a flavonoid), and glabridin (a licorice-derived polyphenol).161,162 Bexarotene, an anti-neoplastic agent that activates retinoid X receptor, inhibits HBV infection of cells.163 Expression of the phospholipase A2 group IIA gene (PLA2G2A), which is regulated by retinoid X receptor and activates arachidonic acid and eicosanoid biosynthesis pathways, reduces HBV infection by unclear mechanisms. Some compounds target HBV particles themselves, such as proanthocyanidin and oolong homobisflavan (derived from extracts of grape seed and Chinese tea). These inhibit virus entry without modifying NTCP-mediated bile acid metabolism.164 Anti-HBV compounds identified from screens of iHep cells include genistein and PA452, which have unclear mechanisms.165 Cells that are susceptible to HBV infection have facilitated the identification of anti-HBV drugs. In addition to entry inhibitors, these cells might be used to identify agents that block cccDNA formation or maintenance or HBV integration into the genome.
Reporter Systems for Evaluating the HBV Infection Process
Anti-HBV agents have been primarily identified in screens (not necessarily high-throughput) to identify agents that reduce HBV DNA or antigens in infected cells, quantified by real-time polymerase chain reaction (PCR) or enzyme-linked immunosorbent assay. However, expression of reporter genes (such as those encoding luciferase or green fluorescent protein) in infected cells has been used in high-throughput screens for identifying inhibitors of HIV and HCV infection.166–169 What Are the Examples of Using Reporters With HBV Infection?
The HCV entry process has been extensively analyzed using HCV pseudoparticles: retroviral- or lentiviral-based virions that display HCV envelope proteins on the surface and carry a luciferase gene as part of the incorporated viral genome. Meredith et al38 prepared lentiviral-based HBV pseudoparticles that carried HBs antigens on the viral envelope. These pseudoparticles entered cells in an NTCP-dependent manner, and this system is expected for using a high-throughput screen to identify HBV inhibitors.
HBV-carrying NanoLuc particles were prepared by transfecting HepG2 cells with a plasmid encoding HBV in which the core region was replaced by a gene encoding NLuc and a helper plasmid that carried an HBV genome that was defective in packaging.170 The resulting HBV/NLuc particles entered target HepG2-NTCP cells and primary human hepatocytes in an NTCP-dependent manner and produced NLuc for at least 12 days after infection.171 NLuc quantification theoretically evaluates the virus life cycle from the entry and cccDNA formation through core promoter-mediated transcription. In this system KX2–391 was identified as an inhibitor of the transcription mediated by a HBV precore promoter.172 These HBV infection–based reporter systems are expected to be useful for large-scale screening to identify anti-HBV agents that target early processes of the HBV life cycle.
Systems for Identifying cccDNA-targeting Agents
Agents that target cccDNA are of the highest priority to be desired aiming for curing HBV infection. The most reliable method for detection of cccDNA is Southern blot analysis, which requires larger amounts of samples than those obtained from 96- or 384-well plates. Southern blotting is therefore not useful in high-throughput screens. Real-time PCR with a cccDNA-specific primer probe set is a practical alternative for cccDNA detection; however, it is not clear whether real-time PCR detects cccDNA with a high level of specificity in the presence of excess amounts of rcDNA. In addition to the demand of a standard protocol to specifically detect cccDNA, techniques that mimic or could be used to monitor cccDNA at high specificity, sensitivity, and throughput, along with decreased cost and time, would facilitate drug development. Several studies have described plasmid-based systems that mimic cccDNA and can be used to evaluate its dynamics.
Several groups have developed recombinant cccDNA produced from a precursor plasmid converted by either Cre/loxP-mediated or ϕC31 integrase-mediated recombination technology.29,165,173,174 Following transfection of cells or hydrodynamic injection of mice, these recombinant cccDNAs supported sustained viral replication for extended periods. One system provided viral replication beyond 70 days in mice.165 Li et al175 used a recombination minicircle system to transfer into cells a luciferase-encoding reporter that generates a precursor plasmid containing the entire HBV sequence, in which the core region was fused with a split Gaussia luciferase (GLuc)-encoding gene. This precursor plasmid could be converted (through DNA recombination in Escherichia coli) to eliminate the backbone vector and yield a minicircle HBV-reporter carrying the entire HBV genome fused with the full GLuc gene and a splicing site.175 This minicircle HBV cccDNA-reporter, which generated a GLuc-containing pgRNA transcript after splicing and eventually yielded nucleocapsids, was maintained for a minimum of 23 days in HepG2 cells. In this system, luciferase activity serves as a surrogate reporter for evaluating cccDNA stability. Constructs of this type are expected to be of use in high-throughput screening.
HepDE19 cells, which can induce HBV replication and subsequent recycling and cccDNA formation upon tetracycline depletion, permit monitoring of cccDNA levels based on measurement of HBeAg; HBeAg production in this cell line depends on cccDNA level.176 However, the use of this cell line is limited by the cross-reactivity of HBeAg with virus core protein in enzyme-linked immunosorbent assay analysis. To address this limitation, Cai et al16 produced a second-generation version of this cell line, HepBHAe82, which encodes an HA-tagged HBe. An assay using this improved cell line detected cccDNA with high sensitivity and low background, so it could be of great value for identifying cccDNA-targeting inhibitors.
Future Directions
No system is optimal for HBV research or drug development; efforts are needed to develop better models. Cell culture systems that most closely resemble normal hepatocytes in the human body but are more convenient, less expensive, unlimited in supply or more readily available, and allow more efficient amplification of infection and spread would be highly desirable. Further improvement of the primary human hepatocyte culture conditions and optimization of iHep cells are of particular interest.
Animal models of HBV infection have increased our understanding of the antiviral immune response and liver disease progression. However, we urgently need mice that are fully permissive to HBV infection, including the establishment and amplification of cccDNA, perhaps starting with mice that express the NTCP transgene in liver. Further discoveries of factors required for virus entry or replication and spread could lead to such a model. This knowledge can be applied to macaque or Tupaia models, to generate alternative outbred nonprimate or primate models for drug development.
These systems can be used not only to study the process of chronic HBV infection, but also in drug discovery. Reporter systems that quickly identify changes in specific features of the HBV life cycle would facilitate discovery of new antiviral agents. Systems for discovery of agents that target cccDNA could facilitate efforts to find a cure for chronic HBV infection.
Acknowledgments
Funding
J.H. was supported by grants AI043453, AI074982, AI77532, and AI27670 from the National Institutes of Health. P.J.C. was supported by grants MOST-106-2321-B-002-011-, MOST-106-2811-B-002-074-, and MOST-107-3017-F-002-002 from the Ministry of Science and Technology, Executive Yuan, Taiwan; NTU-107L9014-1 from the Center of Precision Medicine, National Taiwan University; and the Liver Disease Prevention & Treatment Research Foundation. P.J.C. acknowledges financial support from the “Center of Precision Medicine” from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan. K.W. was supported by the Japan Society for the Promotion of Science KAKENHI (JP17H04085, JP66KT0111), JST CREST program, and the Japan Agency for Medical Research and Development (AMED) (18fk0310114j0002, 18fk0310103j0202, 18fk0310101j1002, 18fk0210036j0001, 18fm0208019j0002). T.W. was supported by the Japan Agency for Medical Research and Development (AMED) (18fk0310103j0002, 18fk0210009j0003, 18fk0210015j0203, 18fk0210020j1002, 18jk0210003j0001) and by the Japan Society for the Promotion of Science KAKENHI (JP17K09447).
Abbreviations used in this paper:
- AdV
adenovirus
- alb-uPA
urokinase-type plasminogen activator under control of an albumin promoter
- cccDNA
covalently closed circular DNA
- DMSO
dimethyl sulfoxide
- GLuc
Gaussia luciferase
- HBeAg
HB e surface antigen
- HBsAg
HB surface antigen
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HDV
hepatitis D virus
- iHep
induced human hepatocyte-like
- NC
nucleocapsid
- NTCP
solute carrier family 10 member 1
- PCR
polymerase chain reaction
- PEG
polyethylene glycol
- pgRNA
pregenomic RNA
- rcDNA
relaxed circular DNA
- siRNA
small interfering RNA.
Footnotes
Conflicts of interest
The authors disclose no conflicts.
References
- 1.Seeger C, Mason WS. Molecular biology of hepatitis B virus infection. Virology 2015;479–480:672–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu J Hepatitis B virus virology and replication. In: Liaw Y-F, Zoulim F, eds. Hepatitis B virus in human diseases London: Humana Press, 2016:1–34. [Google Scholar]
- 3.Yan H, Zhong G, Xu G, et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 2012;1:e00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark DN, Hu J. Hepatitis B virus reverse transcriptase: target of current antiviral therapy and future drug development. Antiviral Res 2015;123:132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zoulim F, Durantel D. Antiviral therapies and prospects for a cure of chronic hepatitis B. Cold Spring Harb Perspect Med 2015;5:a021501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maini MK, Gehring AJ. The role of innate immunity in the immunopathologyand treatment of HBV infection. J Hepatol 2016;64:S60–S70. [DOI] [PubMed] [Google Scholar]
- 7.Ning X, Nguyen D, Mentzer L, et al. Secretion of genome-free hepatitis B virus—single strand blocking model for virion morphogenesis of para-retrovirus. PLoS Pathogens 2011;7:e1002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sureau C, Romet-Lemonne JL, Mullins JI, et al. Production of hepatitis B virus by a differentiated human hepatoma cell line after transfection with cloned circular HBV DNA. Cell 1986;47:37–47. [DOI] [PubMed] [Google Scholar]
- 9.Acs G, Sells MA, Purcell RH, et al. Hepatitis B virus produced by transfected Hep G2 cells causes hepatitis in chimpanzees. Proc Natl Acad Sci U S A 1987; 84:4641–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci U S A 1987;84:1005–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsurimoto T, Fujiyama A, Matsubara K. Stable expression and replication of hepatitis B virus genome in an integrated state in a human hepatoma cell line transfected with the cloned viral DNA. Proc Natl Acad Sci U S A 1987;84:444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ladner SK, Otto MJ, Barker CS, et al. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother 1997;41:1715–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo H, Jiang D, Zhou T, et al. Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation. J Virol 2007;81:12472–12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao W, Hu J. Formation of hepatitis B virus covalently closed circular DNA: removal of genome-linked protein. J Virol 2007;81:6164–6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui X, Luckenbaugh L, Bruss V, et al. Alteration of mature nucleocapsid and enhancement of covalently closed circular DNA formation by hepatitis B virus core mutants defective in complete-virion formation. J Virol 2015; 89:10064–10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai D, Wang X, Yan R, et al. Establishment of an inducible HBV stable cell line that expresses cccDNA-dependent epitope-tagged HBeAg for screening of cccDNA modulators. Antiviral Res 2016;132:26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou T, Guo H, Guo JT, et al. Hepatitis B virus e antigen production is dependent upon covalently closed circular (ccc) DNA in HepAD38 cell cultures and may serve as a cccDNA surrogate in antiviral screening assays. Antiviral Res 2006;72:116–124. [DOI] [PubMed] [Google Scholar]
- 18.Chou YC, Jeng KS, Chen ML, et al. Evaluation of transcriptional efficiency of hepatitis B virus covalently closed circular DNA by reverse transcription-PCR combined with the restriction enzyme digestion method. J Virol 2005;79:1813–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui X, Clark DN, Liu K, et al. Viral DNA-dependent induction of innate immune response to hepatitis B virus in immortalized mouse hepatocytes. J Virol 2015;90:486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lucifora J, Durantel D, Testoni B, et al. Control of hepatitis B virus replication by innate response of HepaRG cells. Hepatology 2010;51:63–72. [DOI] [PubMed] [Google Scholar]
- 21.Shen F, Li Y, Wang Y, et al. Hepatitis B virus sensitivity to interferon-alpha in hepatocytes is more associated with cellular interferon response than with viral genotype. Hepatology 2018;67:1237–1252. [DOI] [PubMed] [Google Scholar]
- 22.Gripon P, Rumin S, Urban S, et al. Infection of a human hepatoma cell line by hepatitis B virus. Proc Natl Acad Sci U S A 2002;99:15655–15660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hantz O, Parent R, Durantel D, et al. Persistence of the hepatitis B virus covalently closed circular DNA in Hep-aRG human hepatocyte-like cells. J Gen Virol 2009; 90:127–135. [DOI] [PubMed] [Google Scholar]
- 24.Wieland S, Thimme R, Purcell RH, et al. Genomic analysis of the host response to hepatitis B virus infection. Proc Natl Acad Sci U S A 2004;101:6669–6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng X, Xia Y, Serti E, et al. Hepatitis B virus evades innate immunity of hepatocytes but activates cytokine production by macrophages. Hepatology 2017;66: 1779–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guidotti LG, Isogawa M, Chisari FV. Host-virus interactions in hepatitis B virus infection. Curr Opin Immunol 2015;36:61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ortega-Prieto AM, Skelton JK, Wai SN, et al. 3D microfluidic liver cultures as a physiological preclinical tool for hepatitis B virus infection. Nat Commun 2018; 9:682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ni Y, Lempp FA, Mehrle S, et al. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 2014;146:1070–1083.e6. [DOI] [PubMed] [Google Scholar]
- 29.Qi Y, Gao Z, Xu G, et al. DNA polymerase kappa is a key cellular factor for the formation of covalently closed circular DNA of hepatitis B virus. PLoS Pathog 2016; 12:e1005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long Q, Yan R, Hu J, et al. The role of host DNA ligases in hepadnavirus covalently closed circular DNA formation. PLoS Pathog 2017;13:e1006784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo J, Cui X, Gao L, et al. Identification of Intermediate in hepatitis B virus CCC DNA formation and sensitive and selective CCC DNA detection. J Virol 2017;91: e00539–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song M, Sun Y, Tian J, et al. Silencing retinoid X receptor alpha expression enhances early-stage hepatitis B virus infection in cell cultures. J Virol 2018;92: e01771–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li M, Sohn JA, Seeger C. Distribution of hepatitis B virus nuclear DNA. J Virol 2018;92:e01391–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seeger C, Sohn JA. Targeting hepatitis B virus with CRISPR/Cas9. Mol Ther Nucleic Acids 2014;3:e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimura S, Watashi K, Fukano K, et al. Cyclosporin derivatives inhibit hepatitis B virus entry without interfering with NTCP transporter activity. J Hepatol 2017;66: 685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michailidis E, Pabon J, Xiang K, et al. A robust cell culture system supporting the complete life cycle of hepatitis B virus. Sci Rep 2017;7:16616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verrier ER, Colpitts CC, Bach C, et al. A targeted functional RNA interference screen uncovers glypican 5 as an entry factor for hepatitis B and D viruses. Hepatology 2016;63:35–48. [DOI] [PubMed] [Google Scholar]
- 38.Meredith LW, Hu K, Cheng X, et al. Lentiviral hepatitis B pseudotype entry requires sodium taurocholate co-transporting polypeptide and additional hepatocyte-specific factors. J Gen Virol 2016;97:121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gripon P, Diot C, Theze N, et al. Hepatitis B virus infection of adult human hepatocytes cultured in the presence of dimethyl sulfoxide. J Virol 1988;62:4136–4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rijntjes PJ, Moshage HJ, Yap SH. In vitro infection of primary cultures of cryopreserved adult human hepatocytes with hepatitis B virus. Virus Res 1988;10:95–109. [DOI] [PubMed] [Google Scholar]
- 41.Rumin S, Gripon P, Le Seyec J, et al. Long-term productive episomal hepatitis B virus replication in primary cultures of adult human hepatocytes infected in vitro. J Viral Hepat 1996;3:227–238. [DOI] [PubMed] [Google Scholar]
- 42.Ni Y, Urban S. Stem cell-derived hepatocytes: a promising novel tool to study hepatitis B virus infection. J Hepatol 2017;66:473–475. [DOI] [PubMed] [Google Scholar]
- 43.Thomas E, Liang TJ. Experimental models of hepatitis B and C—new insights and progress. Nat Rev Gastro-enterol Hepatol 2016;13:362–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ochiya T, Tsurimoto T, Ueda K, et al. An in vitro system for infection with hepatitis B virus that uses primary human fetal hepatocytes. Proc Natl Acad Sci U S A 1989; 86:1875–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou M, Zhao F, Li J, et al. Long-term maintenance of human fetal hepatocytes and prolonged susceptibility to HBV infection by co-culture with non-parenchymal cells. J Virol Methods 2014;195:185–193. [DOI] [PubMed] [Google Scholar]
- 46.Ishida Y, Yamasaki C, Yanagi A, et al. Novel robust in vitro hepatitis B virus infection model using fresh human hepatocytes isolated from humanized mice. Am J Pathol 2015;185:1275–1285. [DOI] [PubMed] [Google Scholar]
- 47.Shlomai A, Schwartz RE, Ramanan V, et al. Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell-derived hepatocellular systems. Proc Natl Acad Sci U S A 2014;111:12193–12198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winer BY, Huang TS, Pludwinski E, et al. Long-term hepatitis B infection in a scalable hepatic co-culture system. Nat Commun 2017;8:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xia Y, Carpentier A, Cheng X, et al. Human stem cell-derived hepatocytes as a model for hepatitis B virus infection, spreading and virus-host interactions. J Hepatol 2017;66:494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaneko S, Kakinuma S, Asahina Y, et al. Human induced pluripotent stem cell-derived hepatic cell lines as a new model for host interaction with hepatitis B virus. Sci Rep 2016;6:29358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takebe T, Sekine K, Enomura M, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 2013;499:481–484. [DOI] [PubMed] [Google Scholar]
- 52.Levy G, Bomze D, Heinz S, et al. Long-term culture and expansion of primary human hepatocytes. Nat Bio-technol 2015;33:1264–1271. [DOI] [PubMed] [Google Scholar]
- 53.Shan J, Schwartz RE, Ross NT, et al. Identification of small molecules for human hepatocyte expansion and iPS differentiation. Nat Chem Biol 2013;9:514–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, Zhuang Q, Wang Y, et al. HBV life cycle is restricted in mouse hepatocytes expressing human NTCP. Cell Mol Immunol 2014;11:175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Becker JS, McCarthy RL, Sidoli S, et al. Genomic and proteomic resolution of heterochromatin and its restriction of alternate fate genes. Mol Cell 2017;68:1023–1037. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kock J, Rosler C, Zhang JJ, et al. Generation of covalently closed circular DNA of hepatitis B viruses via intracellular recycling is regulated in a virus specific manner. PLoS Pathog 2010;6:e1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quasdorff M, Protzer U. Control of hepatitis B virus at the level of transcription. J Viral Hepat 2010;17:527–536. [DOI] [PubMed] [Google Scholar]
- 58.Seeger C, Zoulim F, Mason WS. Hepadnaviruses. In: Knipe DM, Howley PM, eds. Fields virology Philadelphia: Lippincott, Williams & Wilkins, 2013:2185–2221. [Google Scholar]
- 59.Tang H, McLachlan A. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc Natl Acad Sci U S A 2001;98:1841–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seeger C, Baldwin B, Tennant BC. Expression of infectious woodchuck hepatitis virus in murine and avian fibroblasts. J Virol 1989;63:4665–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lempp FA, Wiedtke E, Qu B, et al. Sodium taurocholate cotransporting polypeptide is the limiting host factor of hepatitis B virus infection in macaque and pig hepatocytes. Hepatology 2017;66:703–716. [DOI] [PubMed] [Google Scholar]
- 62.Burwitz BJ, Wettengel JM, Muck-Hausl MA, et al. Hepatocytic expression of human sodium-taurocholate cotransporting polypeptide enables hepatitis B virus infection of macaques. Nat Commun 2017;8:2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walter E, Keist R, Niederost B, et al. Hepatitis B virus infection of tupaia hepatocytes in vitro and in vivo. Hepatology 1996;24:1–5. [DOI] [PubMed] [Google Scholar]
- 64.Guidotti LG, Matzke B, Schaller H, et al. High-level hepatitis B virus replication in transgenic mice. J Virol 1995;69:6158–6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lempp FA, Mutz P, Lipps C, et al. Evidence that hepatitis B virus replication in mouse cells is limited by the lack of a host cell dependency factor. J Hepatol 2016;64: 556–564. [DOI] [PubMed] [Google Scholar]
- 66.Raney AK, Eggers CM, Kline EF, et al. Nuclear covalently closed circular viral genomic DNA in the liver of hepatocyte nuclear factor 1 alpha-null hepatitis B virus transgenic mice. J Virol 2001;75:2900–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cui X, Guo JT, Hu J. Hepatitis B virus covalently closed circular DNA formation in immortalized mouse hepato-cytes associated with nucleocapsid destabilization. J Virol 2015;89:9021–9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cui X, Ludgate L, Ning X, et al. Maturation-associated destabilization of hepatitis B virus nucleocapsid. J Virol 2013;87:11494–11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lempp FA, Qu B, Wang YX, et al. Hepatitis B virus infection of a mouse hepatic cell line reconstituted with human sodium taurocholate cotransporting polypeptide. J Virol 2016;90:4827–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.He W, Ren B, Mao F, et al. Hepatitis D virus infection of mice expressing human sodium taurocholate co-transporting polypeptide. PLoS Pathog 2015; 11:e1004840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Croagh CM, Lubel JS. Natural history of chronic hepatitis B: phases in a complex relationship. World J Gastro-enterol 2014;20:10395–10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fattovich G, Bortolotti F, Donato F. Natural history of chronic hepatitis B: special emphasis on disease progression and prognostic factors. J Hepatol 2008;48: 335–352. [DOI] [PubMed] [Google Scholar]
- 73.Liaw YF. HBeAg seroconversion as an important end point in the treatment of chronic hepatitis B. Hepatol Int 2009;3:425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ganem D, Prince AM. Hepatitis B virus infection—natural history and clinical consequences. N Engl J Med 2004; 350:1118–1129. [DOI] [PubMed] [Google Scholar]
- 75.Chang MH, Sung JL, Lee CY, et al. Factors affecting clearance of hepatitis B e antigen in hepatitis B surface antigen carrier children. J Pediatr 1989;115:385–390. [DOI] [PubMed] [Google Scholar]
- 76.Wieland SF. The chimpanzee model for hepatitis B virus infection. Cold Spring Harb Perspect Med 2015;5: a201469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Menne S, Cote PJ. The woodchuck as an animal model for pathogenesis and therapy of chronic hepatitis B virus infection. World J Gastroenterol 2007;13:104–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ohshita H, Tateno C. Propagation of human hepatocytes in uPA/SCID mice: producing chimeric mice with humanized liver. Methods Mol Biol 2017;1506:91–100. [DOI] [PubMed] [Google Scholar]
- 79.Agmon-Levin N, Arango MT, Kivity S, et al. Immunization with hepatitis B vaccine accelerates SLE-like disease in a murine model. J Autoimmun 2014;54:21–32. [DOI] [PubMed] [Google Scholar]
- 80.Uchida T, Hiraga N, Imamura M, et al. Human cytotoxic T lymphocyte-mediated acute liver failure and rescue by immunoglobulin in human hepatocyte transplant TK-NOG mice. J Virol 2015;89:10087–10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Menne S, Tumas DB, Liu KH, et al. Sustained efficacy and seroconversion with the Toll-like receptor 7 agonist GS-9620 in the Woodchuck model of chronic hepatitis B. J Hepatol 2015;62:1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lutgehetmann M, Mancke LV, Volz T, et al. Humanized chimeric uPA mouse model for the study of hepatitis B and D virus interactions and preclinical drug evaluation. Hepatology 2012;55:685–694. [DOI] [PubMed] [Google Scholar]
- 83.Vanwolleghem T, Libbrecht L, Hansen BE, et al. Factors determining successful engraftment of hepatocytes and susceptibility to hepatitis B and C virus infection in uPA-SCID mice. J Hepatol 2010;53:468–476. [DOI] [PubMed] [Google Scholar]
- 84.Allweiss L, Dandri M. Experimental in vitro and in vivo models for the study of human hepatitis B virus infection. J Hepatol 2016;64:S17–S31. [DOI] [PubMed] [Google Scholar]
- 85.Gao LF, Sun WS, Ma CH, et al. Establishment of mice model with human viral hepatitis B. World J Gastro-enterol 2004;10:841–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Uprichard SL, Wieland SF, Althage A, et al. Transcriptional and posttranscriptional control of hepatitis B virus gene expression. Proc Natl Acad Sci U S A 2003; 100:1310–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chisari FV, Filippi P, McLachlan A, et al. Expression of hepatitis B virus large envelope polypeptide inhibits hepatitis B surface antigen secretion in transgenic mice. J Virol 1986;60:880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim CM, Koike K, Saito I, et al. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature 1991;351:317–320. [DOI] [PubMed] [Google Scholar]
- 89.Julander JG, Sidwell RW, Morrey JD. Characterizing antiviral activity of adefovir dipivoxil in transgenic mice expressing hepatitis B virus. Antiviral Res 2002; 55:27–40. [DOI] [PubMed] [Google Scholar]
- 90.Weber O, Schlemmer KH, Hartmann E, et al. Inhibition of human hepatitis B virus (HBV) by a novel non-nucleosidic compound in a transgenic mouse model. Antiviral Res 2002;54:69–78. [DOI] [PubMed] [Google Scholar]
- 91.Julander JG, Colonno RJ, Sidwell RW, et al. Characterization of antiviral activity of entecavir in transgenic mice expressing hepatitis B virus. Antiviral Res 2003;59: 155–161. [DOI] [PubMed] [Google Scholar]
- 92.Wieland SF, Guidotti LG, Chisari FV. Intrahepatic induction of alpha/beta interferon eliminates viral RNA-containing capsids in hepatitis B virus transgenic mice. J Virol 2000;74:4165–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McClary H, Koch R, Chisari FV, et al. Relative sensitivity of hepatitis B virus and other hepatotropic viruses to the antiviral effects of cytokines. J Virol 2000;74:2255–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McCaffrey AP, Nakai H, Pandey K, et al. Inhibition of hepatitis B virus in mice by RNA interference. Nat Bio-technol 2003;21:639–644. [DOI] [PubMed] [Google Scholar]
- 95.Ebert G, Poeck H, Lucifora J, et al. 5’ Triphosphorylated small interfering RNAs control replication of hepatitis B virus and induce an interferon response in human liver cells and mice. Gastroenterology 2011;141: 696–706.e1–3. [DOI] [PubMed] [Google Scholar]
- 96.Buchmann P, Dembek C, Kuklick L, et al. A novel therapeutic hepatitis B vaccine induces cellular and humoral immune responses and breaks tolerance in hepatitis B virus (HBV) transgenic mice. Vaccine 2013;31: 1197–1203. [DOI] [PubMed] [Google Scholar]
- 97.Yang PL, Althage A, Chung J, et al. Hydrodynamic injection of viral DNA: a mouse model of acute hepatitis B virus infection. Proc Natl Acad Sci U S A 2002; 99:13825–13830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huang LR, Wu HL, Chen PJ, et al. An immunocompetent mouse model for the tolerance of human chronic hepatitis B virus infection. Proc Natl Acad Sci U S A 2006; 103:17862–17867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chou HH, Chien WH, Wu LL, et al. Age-related immune clearance of hepatitis B virus infection requires the establishment of gut microbiota. Proc Natl Acad Sci U S A 2015;112:2175–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sprinzl MF, Oberwinkler H, Schaller H, et al. Transfer of hepatitis B virus genome by adenovirus vectors into cultured cells and mice: crossing the species barrier. J Virol 2001;75:5108–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hartman ZC, Kiang A, Everett RS, et al. Adenovirus infection triggers a rapid, MyD88-regulated transcriptome response critical to acute-phase and adaptive immune responses in vivo. J Virol 2007;81:1796–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lin SR, Yang HC, Kuo YT, et al. The CRISPR/Cas9 system facilitates clearance of the intrahepatic HBV templates in vivo. Mol Ther Nucleic Acids 2014;3:e186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dion S, Bourgine M, Godon O, et al. Adeno-associated virus-mediated gene transfer leads to persistent hepatitis B virus replication in mice expressing HLA-A2 and HLA-DR1 molecules. J Virol 2013;87:5554–5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang D, Liu L, Zhu D, et al. A mouse model for HBV immunotolerance and immunotherapy. Cell Mol Immunol 2014;11:71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wu HL, Huang LR, Huang CC, et al. RNA interference-mediated control of hepatitis B virus and emergence of resistant mutant. Gastroenterology 2005;128:708–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huang Y. Preclinical and clinical advances of GalNAc-decorated nucleic acid therapeutics. Mol Ther Nucleic Acids 2017;6:116–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sun S, Li J. Humanized chimeric mouse models of hepatitis B virus infection. Int J Infect Dis 2017;59: 131–136. [DOI] [PubMed] [Google Scholar]
- 108.Giersch K, Allweiss L, Volz T, et al. Hepatitis Delta co-infection in humanized mice leads to pronounced induction of innate immune responses in comparison to HBV mono-infection. J Hepatol 2015;63:346–353. [DOI] [PubMed] [Google Scholar]
- 109.Kah J, Koh S, Volz T, et al. Lymphocytes transiently expressing virus-specific T cell receptors reduce hepatitis B virus infection. J Clin Invest 2017;127:3177–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dandri M, Lutgehetmann M. Mouse models of hepatitis B and delta virus infection. J Immunol Methods 2014; 410:39–49. [DOI] [PubMed] [Google Scholar]
- 111.Rhim JA, Sandgren EP, Palmiter RD, et al. Complete reconstitution of mouse liver with xenogeneic hepatocytes. Proc Natl Acad Sci U S A 1995;92:4942–4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Petersen J, Dandri M, Gupta S, et al. Liver repopulation with xenogenic hepatocytes in B and T cell-deficient mice leads to chronic hepadnavirus infection and clonal growth of hepatocellular carcinoma. Proc Natl Acad Sci U S A 1998;95:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tateno C, Yoshizane Y, Saito N, et al. Near completely humanized liver in mice shows human-type metabolic responses to drugs. Am J Pathol 2004;165:901–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lutgehetmann M, Bornscheuer T, Volz T, et al. Hepatitis B virus limits response of human hepatocytes to interferon-alpha in chimeric mice. Gastroenterology 2011;140:2074–2083.e1–2. [DOI] [PubMed] [Google Scholar]
- 115.Dandri M, Petersen J. Chimeric mouse model of hepatitis B virus infection. J Hepatol 2012;56:493–495. [DOI] [PubMed] [Google Scholar]
- 116.Azuma H, Paulk N, Ranade A, et al. Robust expansion of human hepatocytes in Fah–/–/Rag2–/–/Il2rg–/– mice. Nat Biotechnol 2007;25:903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bissig KD, Le TT, Woods NB, et al. Repopulation of adult and neonatal mice with human hepatocytes: a chimeric animal model. Proc Natl Acad Sci U S A 2007; 104:20507–20511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Petersen J, Dandri M, Mier W, et al. Prevention of hepatitis B virus infection in vivo by entry inhibitors derived from the large envelope protein. Nat Biotechnol 2008; 26:335–341. [DOI] [PubMed] [Google Scholar]
- 119.Volz T, Allweiss L, Ben MM, et al. The entry inhibitor Myrcludex-B efficiently blocks intrahepatic virus spreading in humanized mice previously infected with hepatitis B virus. J Hepatol 2013;58:861–867. [DOI] [PubMed] [Google Scholar]
- 120.Bility MT, Cheng L, Zhang Z, et al. Hepatitis B virus infection and immunopathogenesis in a humanized mouse model: induction of human-specific liver fibrosis and M2-like macrophages. PLoS Pathog 2014; 10:e1004032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bility MT, Zhang L, Washburn ML, et al. Generation of a humanized mouse model with both human immune system and liver cells to model hepatitis C virus infection and liver immunopathogenesis. Nat Protoc 2012;7:1608–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Irudayaswamy A, Muthiah M, Zhou L, et al. Long-term fate of human fetal liver progenitor cells transplanted in injured mouse livers. Stem Cells 2018;36:103–113. [DOI] [PubMed] [Google Scholar]
- 123.Washburn ML, Bility MT, Zhang L, et al. A humanized mouse model to study hepatitis C virus infection, immune response, and liver disease. Gastroenterology 2011;140:1334–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.de Carvalho Dominguez Souza BF, Konig A, Rasche A, et al. A novel hepatitis B virus species discovered in capuchin monkeys sheds new light on the evolution of primate hepadnaviruses. J Hepatol 2018;68:1114–1122. [DOI] [PubMed] [Google Scholar]
- 125.Arbuthnot P, Kew M. Hepatitis B virus and hepatocellular carcinoma. Int J Exp Pathol 2001;82:77–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fourel G, Trepo C, Bougueleret L, et al. Frequent activation of N-myc genes by hepadnavirus insertion in woodchuck liver tumours. Nature 1990;347:294–298. [DOI] [PubMed] [Google Scholar]
- 127.Sung WK, Zheng H, Li S, et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet 2012;44:765–769. [DOI] [PubMed] [Google Scholar]
- 128.Tennant BC, Gerin JL. The woodchuck model of hepatitis B virus infection. ILAR J 2001;42:89–102. [DOI] [PubMed] [Google Scholar]
- 129.Rogler CE, Hino O, Su CY. Molecular aspects of persistent woodchuck hepatitis virus and hepatitis B virus infection and hepatocellular carcinoma. Hepatology 1987;7:74S–78S. [DOI] [PubMed] [Google Scholar]
- 130.Cummings IW, Browne JK, Salser WA, et al. Isolation, characterization, and comparison of recombinant DNAs derived from genomes of human hepatitis B virus and woodchuck hepatitis virus. Proc Natl Acad Sci U S A 1980;77:1842–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Millman I, Southam L, Halbherr T, et al. Woodchuck hepatitis virus: experimental infection and natural occurrence. Hepatology 1984;4:817–823. [DOI] [PubMed] [Google Scholar]
- 132.Glebe D, Urban S, Knoop EV, et al. Mapping of the hepatitis B virus attachment site by use of infection-inhibiting preS1 lipopeptides and tupaia hepatocytes. Gastroenterology 2005;129:234–245. [DOI] [PubMed] [Google Scholar]
- 133.Dupinay T, Gheit T, Roques P, et al. Discovery of naturally occurring transmissible chronic hepatitis B virus infection among Macaca fascicularis from Mauritius Island. Hepatology 2013;58:1610–1620. [DOI] [PubMed] [Google Scholar]
- 134.Arends JE, Lieveld FI, Ahmad S, et al. New viral and immunological targets for hepatitis B treatment and cure: a review. Infect Dis Ther 2017;6:461–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Klumpp K, Shimada T, Allweiss L, et al. Efficacy of NVR 3–778, alone and in combination with pegylated interferon, vs entecavir in uPA/SCID mice with humanized livers and HBV infection. Gastroenterology 2018; 154:652–662.e8. [DOI] [PubMed] [Google Scholar]
- 136.Wu G, Liu B, Zhang Y, et al. Preclinical characterization of GLS4, an inhibitor of hepatitis B virus core particle assembly. Antimicrob Agents Chemother 2013;57: 5344–5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Korolowicz KE, Iyer RP, Czerwinski S, et al. Antiviral efficacy and host innate immunity associated with SB 9200 treatment in the woodchuck model of chronic hepatitis B. PLoS One 2016;11:e0161313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lanford RE, Guerra B, Chavez D, et al. GS-9620, an oral agonist of Toll-like receptor-7, induces prolonged suppression of hepatitis B virus in chronically infected chimpanzees. Gastroenterology 2013;144:1508–1517. e1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lobaina Y, Hardtke S, Wedemeyer H, et al. In vitro stimulation with HBV therapeutic vaccine candidate Nasvac activates B and T cells from chronic hepatitis B patients and healthy donors. Mol Immunol 2015;63:320–327. [DOI] [PubMed] [Google Scholar]
- 140.Lok AS, Pan CQ, Han SH, et al. Randomized phase II study of GS-4774 as a therapeutic vaccine in virally suppressed patients with chronic hepatitis B. J Hepatol 2016;65:509–516. [DOI] [PubMed] [Google Scholar]
- 141.Bogomolov P, Alexandrov A, Voronkova N, et al. Treatment of chronic hepatitis D with the entry inhibitor myrcludex B: first results of a phase Ib/IIa study. J Hepatol 2016;65:490–498. [DOI] [PubMed] [Google Scholar]
- 142.Gripon P, Cannie I, Urban S. Efficient inhibition of hepatitis B virus infection by acylated peptides derived from the large viral surface protein. J Virol 2005;79: 1613–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Barrera A, Guerra B, Notvall L, et al. Mapping of the hepatitis B virus pre-S1 domain involved in receptor recognition. J Virol 2005;79:9786–9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Konig A, Doring B, Mohr C, et al. Kinetics of the bile acid transporter and hepatitis B virus receptor Na+/taurocholate cotransporting polypeptide (NTCP) in hepatocytes. J Hepatol 2014;61:867–875. [DOI] [PubMed] [Google Scholar]
- 145.Oehler N, Volz T, Bhadra OD, et al. Binding of hepatitis B virus to its cellular receptor alters the expression profile of genes of bile acid metabolism. Hepatology 2014; 60:1483–1493. [DOI] [PubMed] [Google Scholar]
- 146.Yan H, Peng B, Liu Y, et al. Viral entry of hepatitis B and D viruses and bile salts transportation share common molecular determinants on sodium taurocholate cotransporting polypeptide. J Virol 2014;88:3273–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Donkers JM, Zehnder B, van Westen GJP, et al. Reduced hepatitis B and D viral entry using clinically applied drugs as novel inhibitors of the bile acid transporter NTCP. Sci Rep 2017;7:15307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ko C, Park WJ, Park S, et al. The FDA-approved drug irbesartan inhibits HBV-infection in HepG2 cells stably expressing sodium taurocholate co-transporting polypeptide. Antivir Ther 2015;20:835–842. [DOI] [PubMed] [Google Scholar]
- 149.Lucifora J, Esser K, Protzer U. Ezetimibe blocks hepatitis B virus infection after virus uptake into hepatocytes. Antiviral Res 2013;97:195–197. [DOI] [PubMed] [Google Scholar]
- 150.Nkongolo S, Ni Y, Lempp FA, et al. Cyclosporin A inhibits hepatitis B and hepatitis D virus entry by cyclophilin-independent interference with the NTCP receptor. J Hepatol 2014;60:723–731. [DOI] [PubMed] [Google Scholar]
- 151.Okuyama-Dobashi K, Kasai H, Tanaka T, et al. Hepatitis B virus efficiently infects non-adherent hepatoma cells via human sodium taurocholate cotransporting poly-peptide. Sci Rep 2015;5:17047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang XJ, Hu W, Zhang TY, et al. Irbesartan, an FDA approved drug for hypertension and diabetic nephropathy, is a potent inhibitor for hepatitis B virus entry by disturbing Na(+)-dependent taurocholate cotransporting polypeptide activity. Antiviral Res 2015;120:140–146. [DOI] [PubMed] [Google Scholar]
- 153.Watashi K, Sluder A, Daito T, et al. Cyclosporin A and its analogs inhibit hepatitis B virus entry into cultured hepatocytes through targeting a membrane transporter, sodium taurocholate cotransporting polypeptide (NTCP). Hepatology 2014;59:1726–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Saso W, Tsukuda S, Ohashi H, et al. A new strategy to identify hepatitis B virus entry inhibitors by AlphaScreen technology targeting the envelope-receptor interaction. Biochem Biophys Res Commun 2018;501:374–379. [DOI] [PubMed] [Google Scholar]
- 155.Nio Y, Akahori Y, Okamura H, et al. Inhibitory effect of fasiglifam on hepatitis B virus infections through suppression of the sodium taurocholate cotransporting polypeptide. Biochem Biophys Res Commun 2018; 501:820–825. [DOI] [PubMed] [Google Scholar]
- 156.Kaneko M, Futamura Y, Tsukuda S, et al. Chemical array system, a platform to identify novel hepatitis B virus entry inhibitors targeting sodium taurocholate cotransporting polypeptide. Sci Rep 2018;8:2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kaneko M, Watashi K, Kamisuki S, et al. A novel tricyclic polyketide, vanitaracin A, specifically inhibits the entry of hepatitis B and D viruses by targeting sodium taurocholate cotransporting polypeptide. J Virol 2015; 89:11945–11953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Matsunaga H, Kamisuki S, Kaneko M, et al. Isolation and structure of vanitaracin A, a novel anti-hepatitis B virus compound from Talaromyces sp. Bioorg Med Chem Lett 2015;25:4325–4328. [DOI] [PubMed] [Google Scholar]
- 159.Passioura T, Watashi K, Fukano K, et al. De novo macrocyclic peptide inhibitors of hepatitis B virus cellular entry. Cell Chem Biol 2018;25:906–915.e5. [DOI] [PubMed] [Google Scholar]
- 160.Zhang J, Fu LL, Tian M, et al. Design and synthesis of a novel candidate compound NTI-007 targeting sodium taurocholate cotransporting polypeptide [NTCP]-APOA1-HBx-Beclin1-mediated autophagic pathway in HBV therapy. Bioorg Med Chem 2015;23:976–984. [DOI] [PubMed] [Google Scholar]
- 161.Huang HC, Tao MH, Hung TM, et al. (–)-Epigallocatechin-3-gallate inhibits entry of hepatitis B virus into hepatocytes. Antiviral Res 2014;111:100–111. [DOI] [PubMed] [Google Scholar]
- 162.Tsukuda S, Watashi K, Iwamoto M, et al. Dysregulation of retinoic acid receptor diminishes hepatocyte permissiveness to hepatitis B virus infection through modulation of sodium taurocholate cotransporting polypeptide (NTCP) expression. J Biol Chem 2015;290:5673–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Song M, Sun Y, Tian J, et al. Silencing retinoid X receptor alpha expression enhances early-stage hepatitis B virus infection in cell cultures. J Virol 2018;92:e01771–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tsukuda S, Watashi K, Hojima T, et al. A new class of hepatitis B and D virus entry inhibitors, proanthocyanidin and its analogs, that directly act on the viral large surface proteins. Hepatology 2017;65:1104–1116. [DOI] [PubMed] [Google Scholar]
- 165.Yan Z, Zeng J, Yu Y, et al. HBVcircle: a novel tool to investigate hepatitis B virus covalently closed circular DNA. J Hepatol 2017;66:1149–1157. [DOI] [PubMed] [Google Scholar]
- 166.Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med 2003;197:633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gervaix A, West D, Leoni LM, et al. A new reporter cell line to monitor HIV infection and drug susceptibility in vitro. Proc Natl Acad Sci U S A 1997;94:4653–4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hsu M, Zhang J, Flint M, et al. Hepatitis C virus glyco-proteins mediate pH-dependent cell entry of pseudo-typed retroviral particles. Proc Natl Acad Sci U S A 2003; 100:7271–7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J Virol 1992; 66:2232–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Nishitsuji H, Ujino S, Shimizu Y, et al. Novel reporter system to monitor early stages of the hepatitis B virus life cycle. Cancer Sci 2015;106:1616–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nishitsuji H, Harada K, Ujino S, et al. Investigating the hepatitis B virus life cycle using engineered reporter hepatitis B viruses. Cancer Sci 2018;109:241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Harada K, Nishitsuji H, Ujino S, et al. Identification of KX2–391 as an inhibitor of HBV transcription by a recombinant HBV-based screening assay. Antiviral Res 2017;144:138–146. [DOI] [PubMed] [Google Scholar]
- 173.Guo X, Chen P, Hou X, et al. The recombined cccDNA produced using minicircle technology mimicked HBV genome in structure and function closely. Sci Rep 2016; 6:25552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wu M, Li J, Yue L, et al. Establishment of Cre-mediated HBV recombinant cccDNA (rcccDNA) cell line for cccDNA biology and antiviral screening assays. Antiviral Res 2018;152:45–52. [DOI] [PubMed] [Google Scholar]
- 175.Li F, Cheng L, Murphy CM, et al. Minicircle HBV cccDNA with a Gaussia luciferase reporter for investigating HBV cccDNA biology and developing cccDNA-targeting drugs. Sci Rep 2016;6:36483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Cai D, Mills C, Yu W, et al. Identification of disubstituted sulfonamide compounds as specific inhibitors of hepatitis B virus covalently closed circular DNA formation. Anti-microb Agents Chemother 2012;56:4277–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hu J, Liu K. Complete and incomplete hepatitis B virus particles: formation, function, and application. Viruses 2017;9(3). [DOI] [PMC free article] [PubMed] [Google Scholar]


