Abstract
The RNA World hypothesis posits that RNA was once responsible for genetic information storage and catalysis. However, a prebiotic mechanism has yet to be reported for the replication of duplex RNA that could have operated before the emergence of polymerase ribozymes. Previously, we showed that a viscous solvent enables information transfer from one strand of long RNA duplex templates, overcoming ‘the strand inhibition problem'. Here, we demonstrate that the same approach allows simultaneous information transfer from both strands of long duplex templates. An additional challenge for the RNA World is that structured RNAs (like those with catalytic activity) function poorly as templates in model prebiotic RNA synthesis reactions, raising the question of how a single sequence could serve as both a catalyst and as a replication template. Here, we show that a viscous solvent also facilitates the transition of a newly synthesized hammerhead ribozyme sequence from its inactive, duplex state to its active, folded state. These results demonstrate how fluctuating environmental conditions can allow a ribozyme sequence to alternate between acting as a template for replication and functioning as a catalyst, and illustrate the potential for temporally changing environments to enable molecular processes necessary for the origin of life.
INTRODUCTION
The RNA World hypothesis posits that an early form of life utilized RNA for both genetic information storage and catalysis, before the advent of coded proteins and DNA (1,2). Support for this hypothesis includes experimental demonstrations of enzyme-free template-directed RNA synthesis and the evolution of ribozymes with complex functions, including oligonucleotide ligation and nucleotide polymerization. However, an experimental system has yet to be demonstrated where a duplex serves as a template for RNA replication without the aid of a highly-evolved ribozyme and where the same sequence in the duplex can alternate between acting as a template for replication and exhibiting functionality (e.g. simple catalysis)—two expected roles in the early stages of an RNA World.
There are at least three major challenges to demonstrating a plausible prebiotic system in which a ribozyme alternates between acting as a template for replication and performing catalysis. First, ribozymes and other functional RNAs adopt folded intramolecular structures that are critical to their catalytic activity, but can severely limit their ability to act as templates in non-enzymatic template-directed synthesis reactions (3,4). Even with the help of an RNA polymerase ribozyme, the rate and efficiency of RNA synthesis from structured templates are too low to compete with substrate and template degradation (4). Oligonucleotides appear to have been an enabling component of prebiotic RNA replication, as oligonucleotides allow the transfer of information from RNA sequences that is otherwise extremely inefficient, or even impossible, by mononucleotide substrates. The more favorable free energy of oligonucleotide binding to complementary sequences facilitates information transfer by both increasing the equilibrium concentration of substrates bound to a template strand (5–7) and by providing the free energy necessary to unfold the intramolecular secondary structures of a template strand that would otherwise block information transfer (8). Despite the advantages provided by oligonucleotide-mediated information transfer, additional kinetic and thermodynamic challenges still limit polymerase-free RNA replication (4).
A second challenge arises when a template-directed RNA synthesis reaction proceeds successfully, resulting in the production of the template-product duplex. This duplex impedes both the original template strand and the new product strand from functioning as templates in subsequent synthesis reactions, a challenge known as the strand inhibition problem. Thermal denaturation of the duplex can release the single strands, but when the temperature decreases, re-annealing of a gene-length duplex is thermodynamically favored over mononucleotide or oligonucleotide substrate binding to the duplex strands (4,8). Consequently, the majority of model prebiotic studies of non-enzymatic, template-directed RNA synthesis have utilized a simplified model system where a single RNA strand directs the synthesis of its complementary strand (5,9,10). In contrast, a general mechanism for RNA replication that allows full (both strand) copying of duplex templates of variable length and sequence, and does not rely upon highly-evolved ribozymes, has not been demonstrated.
A third major challenge arises when considering how, following replication, a ribozyme sequence would transition from the duplex state to its single stranded, catalytically active structure while in the presence of its complementary sequence. In water, this transition is typically thermodynamically disfavored. Though previous studies have shown that an RNA sequence can direct the synthesis of a catalytic RNA sequence (5,11,12), none have demonstrated that the newly synthesized ribozyme can fold into its catalytically active state in the presence of its complementary sequence.
Previously, we demonstrated that the problem of strand inhibition can be overcome by heating and cooling RNA and DNA duplexes in a viscous solvent. As we had hypothesized, solvent viscosity can enable information transfer from a single strand of a long duplex template (545 bp and 3 kb) by providing oligonucleotides with a kinetic advantage for diffusion and binding to the single stranded templates compared to the long template strands (8). However, additional exploration of the ability for viscosity to circumvent the strand inhibition problem is necessary if this approach is to be seriously considered as a plausible prebiotic mechanism for RNA replication. True duplex replication requiressynthesis of both strands of a duplex, which presents challenges that go beyond those associated with the synthesis of one strand of an existing duplex. In particular, copying both strands requires having oligonucleotides substrates present in the reaction that are complementary to each strand of the duplex. Consequently, the set of oligonucleotide substrates that are required for the copying of both strands will have some degree of complementarity between them. Thus, there is the potential that duplex formation between the substrates could also inhibit substrate binding to the individual strands of the duplex template, essentially representing another form of the strand inhibition problem that is also unsolved. Additionally, previous model prebiotic RNA replication studies have shown that an RNA sequence that functions well as a template can have a complementary sequence that is an extremely poor template, a situation that would have limited the prebiotic replication of functional RNA sequences, which typically adopt stable secondary (folded) structures. It is therefore necessary to demonstrate that any proposed prebiotic mechanism for information transfer from an RNA duplex is able to transfer information from both strands.
Here, we demonstrate the replication of RNA duplexes ranging in length from ∼100 to 600 bp by template-directed synthesis reactions in which heating and cooling in a viscous solvent allows oligonucleotides to circumvent the strand inhibition problem for both strands of the template duplexes. Additionally, the RNA duplex templates each contain a catalytically active ribozyme sequence along one strand; the hammerhead (HH) ribozyme sequence. The same heating and cooling process in the viscous solvent is shown to trap the newly synthesized RNA duplex products as single strands with the HH ribozyme sequence folded in its catalytically active form, as demonstrated by its ability to cleave a substrate oligonucleotide. Neither replication nor catalysis from the RNA duplex occurs in water, highlighting the utility of viscous solvents for overcoming some long-standing thermodynamic challenges facing RNA replication in model prebiotic reactions by altering the kinetics of RNA hybridization and folding in a length-dependent manner (8,13,14).
MATERIALS AND METHODS
Assay of Cy3S4 and Cy5S4′ binding to r613 template
Agarose gel electrophoretic mobility was used to monitor the binding of oligonucleotide substrates to the RNA template duplex, following a protocol described previously (8). Briefly, mixtures of oligonucleotides S1–S10 (containing fluorescent Cy3 cyanine dye tagged Cy3S4), complementary oligonucleotides S1′-S10′ (containing fluorescent Cy5 cyanine dye tagged Cy5S4′), and the r613 duplex template were prepared in a 20:1 molar ratio of oligonucleotides to the template strands (with the template duplex being present at a concentration of 2.7 × 10−8 molal). These samples were heated to temperatures sufficiently high for complete duplex denaturation: specifically, 95°C for aqueous buffer and 80°C for glycholine, a 4:1 mixture of glycerol and choline chloride. Melting temperature determination is shown in Supplementary Figure S1. The samples were then cooled to room temperature at a rate of 4°C/min. After this heating and cooling cycle, sample aliquots were loaded into a 2% agarose gel and subjected to electrophoresis. The agarose gel was then imaged on a Typhoon FLA 9500 laser scanner (GE Healthcare) at a resolution of 50 μm. Cy3 fluorescence was measured using an excitation wavelength of 532 nm and the bandpass green (BPG1) emission filter. Cy5 fluorescence was measured using an excitation wavelength of 635 nm and the long pass red (LPR) emission filter. The gel was then stained with ethidium bromide in 1× TAE buffer with gentle shaking for 15 min and imaged using an excitation wavelength of 532 nm and the long pass green (LPG) emission filter.
Ligation of assembled oligonucleotide substrates
Samples were prepared as 2 mg aliquots of glycholine containing the RNA template at a concentration of 4 × 10−7 molal (for all template lengths) and with each of the RNA oligonucleotides present in a 4:1 molar ratio with the template (5′-phosphorylated to enable ligation by T4 RNA ligase 2). Samples were heated to denaturing temperatures (95°C for aqueous buffer; 80°C for glycholine) and cooled to 20°C at a constant rate over 20 min. RNA samples were diluted with water, 10× T4 RNA ligase 2 buffer (to a concentration of 1×), and 10 U of T4 RNA ligase 2 (New England Biolabs) to a reaction volume of 20 μl. After incubating for 1 hour at 37°C, 6 μl of each reaction mixture was combined with 6 μl of 2× loading dye (concentration at 2× is: 95% formamide, 0.01% SDS, 0.5 mM EDTA pH 8.0). The resulting sample was heated to 95°C for 3 min and then placed on ice before loading into a 10% denaturing polyacrylamide gel (8 M urea, 1× TBE buffer). Prior to sample loading, gels were pre-run for >30 min at 14 W and 45 V/cm. After sample loading, gels were run at these same conditions for ∼1 h and imaged for Cy3 and Cy5 fluorescence before staining with SYBR Gold (as described above).
Assay of cleavage by r613 duplex in the presence of oligonucleotide substrates for replication
Samples were prepared as 2 mg aliquots of glycholine containing RNA template at a concentration of 2 × 10−6 molal and FAM-labeled substrate in a 4:1 molar ratio with the template. Samples with 2 × 10−6 molal concentration of the RNA oligonucleotides used for the ligation assay were added to a control sample to verify they had no effect on the cleavage activity of the template containing the HH ribozyme. Samples were then heated to 80°C for 2 min and cooled to 20°C at a constant rate over 20 min. RNA samples were diluted with water, and 10× T4 RNA ligase 2 buffer was added (to a concentration of 1×) for a final reaction volume of 20 μl. After incubating for 1 h at 20°C, the reaction was quenched by adding EDTA to a final concentration of 2.5 mM and adjusting pH to 5 with a sodium acetate buffer. Then, 6 μl of each reaction mixture was combined with 6 μl of 2× loading dye. The resulting sample was heated to 85°C for 2 min and then placed on ice before loading into a 20% denaturing polyacrylamide gel (8 M urea, 1× TBE buffer). Prior to sample loading, gels were pre-run for >30 min at 14 W and 50 V/cm. After sample loading, gels were run at 20 W for around 1 h and imaged for FAM fluorescence before staining with SYBR Gold (as described above in Assay of Cy3S4 and Cy5S4′ binding to r613 template).
Purification of r613 replication products and cleavage assay
One goal of this study was to demonstrate that newly synthesized replication products from r613 template replication can exhibit hammerhead cleavage activity. To generate sufficient r613 replication product to assay hammerhead cleavage activity, ninety r613 template replication reactions were carried out in parallel, each being approximately 5 μl in volume (5 mg of glycholine as solvent), with 750 ng (4 × 10−7 molal) of r613 template, and each of the RNA oligonucleotides (32 nt each) in a 4:1 molar ratio with the template. The same protocol described above for the ligation of assembled oligonucleotide substrates was followed for these replication reactions. After these reactions were performed, all ninety were mixed together and their combined products were concentrated using Zymo Oligo Clean & Concentrator columns. The replication products between 192 and 320 bp (as visualized by Cy3 and Cy5 fluorescence) were purified by PAGE in denaturing conditions (1× TBE, 10% acrylamide). The purified replication products were then assayed for hammerhead cleavage activity using the same protocol described in the previous section.
RESULTS
A model cycle for viscosity-mediated replication and catalysis from a ribozyme
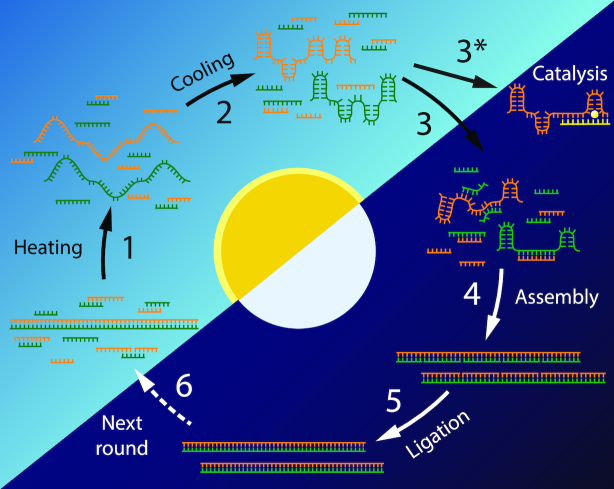
Our model for prebiotic RNA replication is divided into six steps (Figure 1). First, a viscous solution containing a long duplex template and shorter oligonucleotide substrates is heated to completely denature the duplex (Step 1). Upon cooling, the solvent viscosity increases and the template strands form intramolecular structures (Step 2). The oligonucleotides, which diffuse more quickly than the long template strands, bind to the template strands (Step 3), invading and unfolding template intramolecular structure (Step 4). Ligation of the template-assembled oligonucleotide substrates completes replication of the original duplex (Step 5), producing two duplexes ready to undergo further rounds of replication (Step 6).
Figure 1.
Cycle for viscosity-mediated replication of duplex RNA. In a viscous solvent, heating and cooling of an RNA duplex (Steps 1–2) leads to kinetic trapping of the RNA in a folded, single stranded state. Shorter, more mobile oligonucleotides assemble on the template strands (Steps 3 and 4). Ligation of the assembled substrates (Step 5) completes the replication cycle, and another round of replication can begin (Step 6). The dashed arrow at Next round (Step 6) indicates that the experiments described in the current work do not continue around the cycle for a second round of duplex replication. The kinetic trapping of intramolecular structure also allows catalytic RNA sequences to access their active forms (Step 3*), which are inaccessible in the duplex state that follows replication.
The same process of heating and cooling promotes catalysis by a ribozyme sequence located on one of the strands in the RNA duplex. Step 3* represents the potential for heating and cooling (Steps 1–2) to promote catalysis by ‘releasing’ a ribozyme sequence from a duplex after replication and kinetically trapping the ribozyme in its single stranded, folded and active conformation.
To test the feasibility of our model prebiotic RNA replication cycle, we utilized double stranded RNA (dsRNA) templates (∼100–600 bp) with sequences taken from a bacterial plasmid (SI). All templates contain a 43 nt hammerhead (HH) nuclease ribozyme sequence on the sense strand, which cleaves a 21 nt HH substrate (15). The dsRNA template names refer to their lengths (e.g. the r613 template is 613 bp). As in our previous study of viscosity-enabled, RNA template-directed synthesis (8), we used glycholine, a 4:1 mixture of glycerol and choline chloride (13), as a model viscous solvent that is miscible with water and composed of small, low-volatility organic molecules. Solvents of this type are of particular interest as part of our model prebiotic cycle for RNA replication as such solvents could have been periodically generated by the evaporation of water from small bodies of water on the prebiotic Earth that had dissolved organics with a propensity to form low-water, high viscosity solvents. We note that glycerol is generally considered a plausible prebiotic molecule (16,17), and has recently been shown to form in model interstellar ices (18). The synthesis of choline chloride, on the other hand, has not been reported in a model prebiotic reaction. Nevertheless, it is conceivable that viscous solvents similar to glycholine could have regularly appeared on the surface of the prebiotic Earth, as a growing number of biological and related small molecules are being shown to form eutectic liquids that are miscible with water, with some having high viscosity (19,20).
Assembly of oligonucleotide substrates on both strands of an RNA duplex
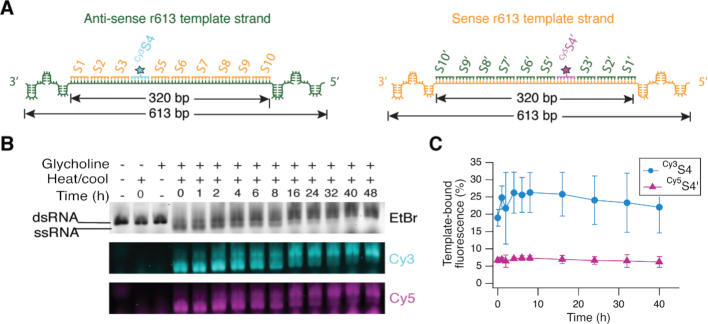
First, we demonstrate that glycholine enables the assembly of oligonucleotide substrates on both strands of a dsRNA template (Figure 1, Steps 1–4). These experiments utilize two sets of 32 nt substrates: S1 through S10 are complementary to a region of the template anti-sense strand, and S1′ through S10′ are complementary to the corresponding region of the template sense strand. A single oligonucleotide in each set was tagged with a cyanine dye, Cy3 or Cy5, as a fluorescent marker (designated Cy3S4 and Cy5S4′) (Figure 2A). Oligonucleotides of 32 nt in length were used because the duplexes they form with the template strands have melting temperatures in glycholine that are well above room temperature (the lowest temperature used in this study). Shorter oligonucleotides could be used if samples are cooled below room temperature.
Figure 2.
Solvent viscosity enables oligonucleotide assembly on both strands of an RNA duplex. (A) RNA oligonucleotides S1–S10 and S1′–S10′ were designed to coat a 320 bp region of the r613 template duplex. Oligonucleotides Cy3S4 and Cy5S4′ are fluorescent markers for template binding. (B) Agarose gel showing the kinetics of oligonucleotide binding to the RNA template. The Cy3 and Cy5 fluorescence images indicate that Cy3S4 and Cy5S4′ bind to both the ss and dsRNA templates after heating and cooling in glycholine, but not in water. Full gel image is shown in Supplementary Figure S2. Note that the upper bands that appear in the EtBr stained after heating and cooling in glycholine (lanes marked 2–48 h) do not necessarily represent only dsRNA. The slower mobility and more diffuse nature of these bands (compared to the band in the third lane) suggest that additional non-duplex structures continue to be present in the RNA that was annealed in glycholine, even after 48 h. (C) Kinetics of Cy3S4 and Cy5S4′ binding to the r613 template. Error bars represent standard deviations based on three experimental replicates. Values plotted are provided in Supplementary Table S1.
Samples containing the r613 template and oligonucleotide substrates were heated above the template-denaturing temperature and then cooled to 20°C over a period of 20 min (details in Materials and Methods). Oligonucleotide binding kinetics at 20°C were followed using agarose gel electrophoresis (Figure 2B, Supplementary Figure S2). Ethidium bromide (EtBr) staining indicates that heating and cooling in glycholine causes r613 to become kinetically trapped as single strands for up to 24 h (Figure 2B, EtBr). During this time period, oligonucleotides Cy3S4 and Cy5S4′ bind to the template, migrating with both the ssRNA and dsRNA template bands (Figure 2B). Cy5S4′ exhibits lower template binding affinity than Cy3S4 (Figure 2C, Supplementary Table S1), consistent with the prediction of more stable intramolecular structure on the template sense strand (Supplementary Figure S3). In aqueous buffer, the r613 duplex reforms within less than 1 min after cooling, preventing Cy3S4 and Cy5S4′ binding, and illustrating the strand inhibition problem in water (Figure 2B, second lane).
Full replication of RNA duplexes with various lengths
Following assembly of oligonucleotide substrates on the RNA template in glycholine, replication of the RNA duplex templates was completed by covalently linking the pre-assembled oligonucleotides. For the model experimental cycle presented here, a ligase enzyme (T4 RNA ligase 2) is used to covalently link oligonucleotide substrates, with all other steps of the cycle (Figure 1, Steps 1–5) being enzyme-free. The ligase is not introduced to the system until after oligonucleotide assembly on both template strands, and was chosen as a robust and efficient means of linking the oligonucleotides so that the degree of substrate assembly can be visualized. Presumably, this final step of the cycle would have originally been promoted by non-enzymatic reactions on the early Earth (e.g. by prebiotic condensing agents).
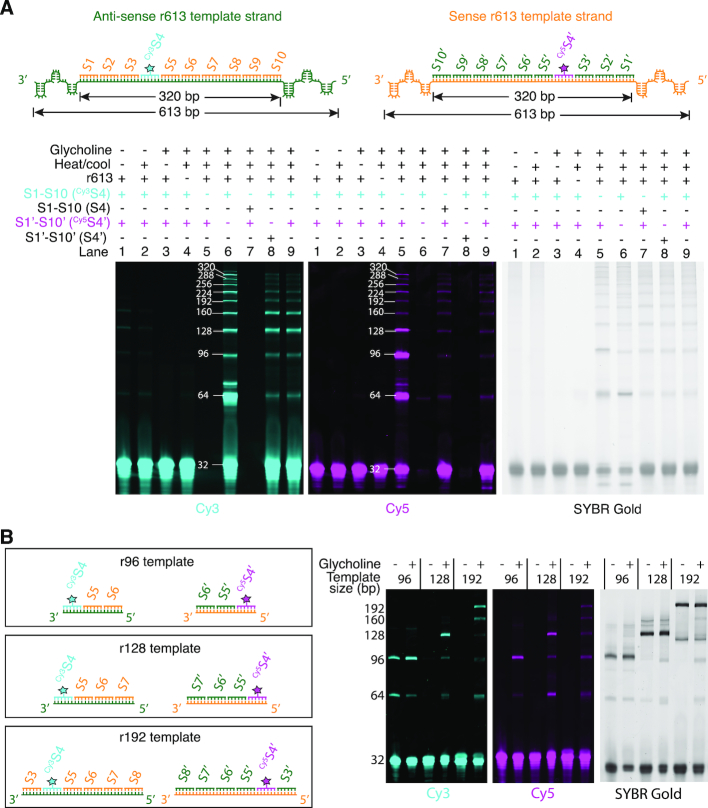
Glycholine samples containing the r613 template duplex and oligonucleotide substrates S1–S10 and S1′–S10′ were heated, cooled, ligated, and visualized on a denaturing polyacrylamide gel (Figure 3A). Cy3S4 is a marker for synthesis of the sense strand, while Cy5S4′ tracks synthesis of the anti-sense strand. A ladder of synthesis products up to 320 nt is observed in both the Cy3 and Cy5 images, demonstrating that complementary strands were simultaneously synthesized from both strands of the original dsRNA template (Figure 3A). No synthesis products are observed in glycholine without heating and cooling, or in aqueous buffer with/without heating and cooling (Figure 3A, lanes 1–3). Replication from the r613 duplex was also observed in two other solvents, glycerol and reline (urea and choline chloride in a 2:1 ratio) (Supplementary Figure S4), demonstrating that viscosity-mediated replication is not particular to the solvent glycholine.
Figure 3.
Replication of RNA duplex templates in glycholine. (A) Samples containing the r613 template duplex and oligonucleotides S1–S10 and S1′–S10′ were heated, cooled and then ligated. The Cy3 and Cy5 images track synthesis of the RNA sense and anti-sense strands, respectively. In glycholine, copying from both strands of the r613 duplex is observed. Band intensities, calculated yields, and standard deviations (based on three experiments) of all replication products in lanes 5, 6 and 9 are provided in Supplementary Table S2. (B) Glycholine enables replication from a range of shorter RNA template duplexes (96–192 bp). Yields determined for full-length replication products from all template sizes tested (based on two experiments) are provided in Supplementary Table S3.
To further explore the prebiotic relevance of our system, we carried out replication using templates of varying lengths. Because the typical length of known aptamers and ribozymes (ca. 20–200 nt) is much shorter than 613 nt, we tested our method on duplexes derived from the same plasmid sequence as the r613 duplex, but with lengths of only 96, 128 and 192 bp (corresponding to replication by the ligation of three, four, and six of the 32 nt oligonucleotide substrates) (Figure 3B). All possible replication products were observed using these templates, illustrating that duplexes over a wide range of lengths can be replicated by this approach. Additionally, we carried out replication of the r613 duplex template with a mixture of oligonucleotides composed of our designed oligonucleotide substrates along with random sequence oligonucleotides, and observed no significant reduction in replication efficiency (Supplementary Figure S5).
Catalytic activity from a newly synthesized ribozyme sequence
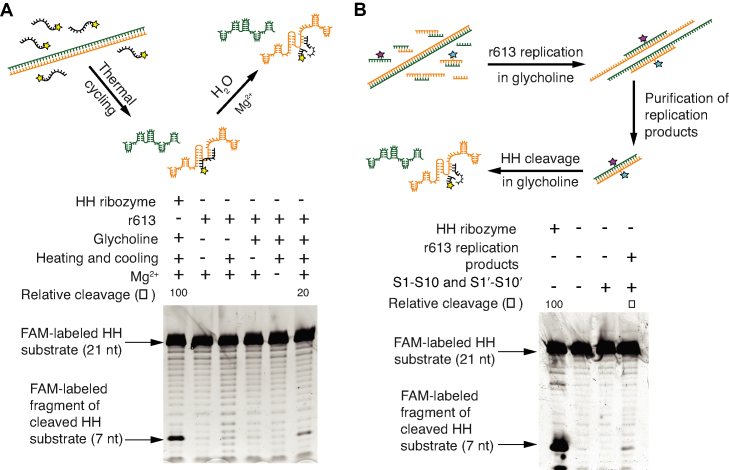
If replication in the RNA World was proceeded by template-directed synthesis, then following replication, ribozyme function would have been hampered by duplex formation with its complementary strand. We hypothesized that RNA replication in a viscous solvent could also facilitate the transition of a ribozyme sequence from its duplex form to its active, single stranded form (Figure 1, Step 3*). To test this hypothesis, we first verified that heating and cooling of dsRNA templates in glycholine can kinetically trap the HH ribozyme sequence in its catalytically active form (Figure 4A). When the r613 dsRNA template is heated and cooled with a FAM-labeled, 21 nt HH ribozyme substrate in glycholine, HH substrate cleavage is observed (Figure 4A). The HH ribozyme present on the r613 sense strand exhibits 20% of the cleavage yield observed by the 43 nt HH ribozyme sequence alone. This reduced but still robust cleavage yield indicates that the HH ribozyme is catalytically active within the longer r613 template, despite the r613 template secondary structure that may interfere with binding of the HH substrate to the HH ribozyme sequence. These results indicate that glycholine enables the transition of a HH ribozyme from a duplex state to a folded, catalytically active state, a transition not seen in aqueous buffer.
Figure 4.
HH substrate cleavage from a kinetically trapped RNA duplex. (A) Schematic illustrating cleavage of the HH substrate (black) by an RNA template strand containing the HH ribozyme (orange) after heating and cooling in glycholine. A denaturing polyacrylamide gel shows that an RNA duplex can cleave the HH substrate. Densitometry profiles are shown in Supplementary Figure S6. (B) Cleavage of the HH substrate is observed from products of viscosity-mediated replication with r613 and oligonucleotides S1–S10 and S1′–S10′. Densitometry profiles are provided in Supplementary Figure S7.
Next, we show that RNA sequences directly synthesized from duplex replication in glycholine can cleave the HH substrate. Beginning with the r613 template duplex, we heated and cooled oligonucleotides S1–S10 and S1′–S10′ in glycholine to generate replication products up to 320 bp in length. These Cy3- and Cy5-tagged replication products were PAGE purified, mixed with the FAM-labeled HH substrate as well as oligonucleotides S1–S10 and S1′–S10′, and then heated and cooled in glycholine (Figure 4B). After dilution with aqueous buffer, a band at the position of the cleaved HH substrate (7 nt) appeared, indicating cleavage of the HH substrate by the replication products (Figure 4B). In the presence of its oligonucleotide substrates, the r613 duplex replication products exhibit a HH cleavage yield that is 9% of that exhibited by the minimal 43 bp HH ribozyme sequence (Figure 4B, final lane). This yield is close to one half of the 20% cleavage yield noted above for the HH sequence that is initially within a duplex. Thus, the HH ribozyme sequences synthesized by duplex replication have an activity that is almost one half of the maximum that can be expected for this sequence within the r613 duplex when released by heating and cooling in glycholine. Additionally, a control experiment in which the sense strand was synthesized from the anti-sense strand confirmed that the cleavage activity exhibited by the sample of synthesized RNA is not a result of contamination from the original r613 template (Supplementary Figure S9). Taken together, these results show how replication and catalysis from an RNA duplex— a key event in the RNA World hypothesis—is promoted by kinetic trapping of RNA in glycholine.
DISCUSSION
We have demonstrated that viscous solvents can enable the replication of gene-length (∼100–600 bp) RNA duplexes by a process that could have been driven on the prebiotic Earth by fluctuations in local environmental conditions that included oscillations in temperature and water activity. All results from our model systems are consistent with increased solvent viscosity being a physical property that facilitates replication by limiting the mobility of long RNA templates relative to shorter oligonucleotide substrates. Heating and cooling in a viscous solvent allows the assembly of these shorter substrates onto the longer, kinetically trapped template strands. Previously we reported that viscous solvents enable information transfer from one strand of a long, heterogeneous duplex RNA template (8). The current study goes further by demonstrating the simultaneous synthesis of both strands of an RNA duplex template, illustrating that two additional challenges to prebiotic nucleic acid replication can be overcome by the inclusion of a viscous solvent. First, replication proceeds even when the pool of oligonucleotide substrates required for the copying of both strands contain perfectly complementary sequences. Second, our demonstration that both strands of an RNA duplex can be copied in the same reaction shows that viscosity-enabled information transfer is not limited to one strand of a duplex (such as the strand with a particular nucleotide composition or less internal structure).
To our knowledge, this is the first study to demonstrate replication of an RNA duplex (i.e. reciprocal synthesis from both strands) without the aid of a polymerase enzyme. Unlike previous studies of non-enzymatic template-directed RNA synthesis that begin with single stranded RNA templates, we have shown that our viscosity-mediated approach can facilitate replication of double stranded RNA templates with a range of lengths and degrees of internal structure. In this study, we carried out a single round of replication (Steps 1 through 5). We did not proceed to a second round because our current method for oligonucleotide ligation (Step 5) requires an irreversible change in solution conditions (i.e., addition of ligase enzyme and MgCl2). It is hypothesized that a prebiotic system that relied on a different chemistry for oligonucleotide ligation, such as chemical condensing agents or phosphodiester exchange, could have had the ability to repeatedly progress through Steps 1–6 for continuous duplex amplification.
It is important to note that substantial advances have been made in the quest to achieve protein-free replication of RNA, including the demonstration that RNA can act as a processive nucleotide polymerase and as an oligonucleotide ligase (9,21). Those results support the hypothesis that RNA may have, at some point in the early stages of life, functioned as both the polymer responsible for information storage and information transfer (i.e., replication). However, the ribozymes used in such studies are the result of substantial sequence evolution and therefore likely to have only emerged after a simpler mechanism was already operating that allowed for the replication of all RNA sequences (or proto-RNA sequences (22)). In this context, the work presented here relates to earlier stages of nucleic acid evolution, when replication was presumably more reliant on geophysical and geochemical processes than on highly evolved molecules.
We have also shown that a viscous solvent enables transition of a ribozyme sequence from a catalytically inactive duplex form to a folded, catalytically active form. For these studies we utilized the hammerhead (HH) ribozyme sequence as an example of the relatively simple ribozymes (as compared to a ligase or a processive polymerase) that would have emerged during the earlier stages of nucleic acid evolution, before replication was taken over by more highly-evolved ribozymes and/or protein enzymes. Just as a viscous solvent can enable replication of an RNA duplex, heating and cooling RNA duplexes in viscous solvents can result in the trapping of the HH ribozyme sequence in its single stranded folded state, enabling activity in the presence of its complementary sequence. These results support our hypothesis that solvent viscosity also provides a solution to the ‘replicator-catalyst’ paradox: the structural properties necessary for RNA catalysis—stable intramolecular folding—also inhibit the ability of a sequence to serve as a template for replication in water (3,9). While stable template intramolecular structure poses an obstacle to replication in water (2), this property may provide an advantage for template-directed replication in a viscous solvent. That is, viscosity could facilitate replication from a template duplex by favoring intramolecular template structures, which slow down re-annealing of the template duplex and permit binding of oligonucleotide substrates. Thus, the emergence of functional RNA sequences in the origins of life may have been aided by viscous solvents that promote replication of templates with intramolecular structure.
Finally, we reiterate that glycholine, composed of glycerol and choline chloride, was chosen for this study because it fulfills the viscosity and water-miscibility requirements of our model system, but may not have been present on the prebiotic Earth. Nevertheless, given the complex mixture of molecules that are generated in model prebiotic reactions, it is easy to envision that other organic and mixed-aqueous organic solvents with high viscosity could have been regularly generated on the surface of early Earth by the evaporation of water from a pond or small lake. Indeed, additional results showing RNA duplex replication in glycerol (Supplementary Figure S4), which has been formed in model interstellar ices (18), indicate that other viscous solvents could have facilitated replication on the early Earth. With the broadest perspective, this study and other studies (23–27) highlight the important role that non-aqueous or microenvironments—generated by cyclic changes in physical/geochemical conditions—may have played in driving prebiotic chemical reactions that are not thermodynamically or kinetically favorable in water.
Supplementary Material
ACKNOWLEDGEMENTS
We thank G. Newnam for technical assistance. We thank the Petit Institute for Bioengineering and Biosciences for use of core facilities.
Notes
Present address: Christine He, Innovative Genomics Institute, University of California, Berkeley, 2151 Berkeley Way, Berkeley, CA 94704, USA.
Present address: Isaac Gállego, MRC Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge Biomedical Campus, Cambridge CB2 0QH, UK.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the NSF and the NASA Astrobiology Program under the NASA/NSF Center for Chemical Evolution [CHE-1504217]; National Science Foundation Graduate Research Fellowship [DGE-1148903 to C.H.]; Consejo Nacional de Ciencia y Tecnologia (CONACYT) scholarship [382818 to A.L.C.]. Funding for open access charge: CCE
Conflict of interest statement. None declared.
REFERENCES
- 1. Gesteland R.F., Cech T.R., Atkins J.F.. The RNA World. 2006; 3rd edn. NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 2. Robertson M.P., Joyce G.F.. The origins of the RNA world. Cold Spring Harbor Perspect. Biol. 2012; 4:doi:10.1101/cshperspect.a003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ivica N.A., Obermayer B., Campbell G.W., Rajamani S., Gerland U., Chen I.A.. The paradox of dual roles in the RNA world: Resolving the conflict between stable folding and templating ability. J. Mol. Evol. 2013; 77:55–63. [DOI] [PubMed] [Google Scholar]
- 4. Szostak J.W. The eightfold path to non-enzymatic RNA replication. J. Syst. Chem. 2012; 3:2. [Google Scholar]
- 5. Prywes N., Blain J.C., Del Frate F., Szostak J.W.. Nonenzymatic copying of RNA templates containing all four letters is catalyzed by activated oligonucleotides. eLife. 2016; 5:e17756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. James K.D., Ellington A.D.. Surprising fidelity of template-directed chemical ligation of oligonucleotides. Chem. Biol. 1997; 4:595–605. [DOI] [PubMed] [Google Scholar]
- 7. Deck C., Jauker M., Richert C.. Efficient enzyme-free copying of all four nucleobases templated by immobilized RNA. Nat. Chem. 2011; 3:603–608. [DOI] [PubMed] [Google Scholar]
- 8. He C., Gallego I., Laughlin B., Grover M.A., Hud N.V.. A viscous solvent enables information transfer from gene-length nucleic acids in a model prebiotic replication cycle. Nat. Chem. 2017; 9:318–324. [DOI] [PubMed] [Google Scholar]
- 9. Attwater J., Wochner A., Holliger P.. In-ice evolution of RNA polymerase ribozyme activity. Nat. Chem. 2013; 5:1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Joyce G.F., Orgel L.E.. Non-enzymatic template-directed synthesis on RNA random copolymers: Poly(C,A) templates. J. Mol. Biol. 1988; 202:677–681. [DOI] [PubMed] [Google Scholar]
- 11. Wochner A., Attwater J., Coulson A., Holliger P.. Ribozyme-catalyzed transcription of an active ribozyme. Science. 2011; 332:209–212. [DOI] [PubMed] [Google Scholar]
- 12. Attwater J., Raguram A., Morgunov A.S., Gianni E., Holliger P.. Ribozyme-catalysed RNA synthesis using triplet building blocks. eLife. 2018; 7:e35255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gállego I., Grover M.A., Hud N.V.. Folding and imaging of DNA nanostructures in anhydrous and hydrated deep-eutectic solvents. Angew. Chem. Int. Ed. 2015; 54:6765–6769. [DOI] [PubMed] [Google Scholar]
- 14. Lannan F.M., Mamajanov I., Hud N.V.. Human telomere sequence DNA in water-free and high-viscosity solvents: G-quadruplex folding governed by Kramers rate theory. J. Am. Chem. Soc. 2012; 134:15324–15330. [DOI] [PubMed] [Google Scholar]
- 15. Martick M., Scott W.G.. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006; 126:309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Apel C.L., Deamer D.W.. The formation of glycerol monodecanoate by a dehydration/condensation reaction: Increasing the chemical complexity of amphiphiles on the early earth. Orig. Life Evol. Biol. 2005; 35:323–332. [DOI] [PubMed] [Google Scholar]
- 17. Joyce G.F., Schwartz A.W., Miller S.L., Orgel L.E.. The case for an ancestral genetic system involving simple analogs of the nucleotides. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:4398–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaiser R.I., Maity S., Jones B.M.. Synthesis of prebiotic glycerol in interstellar ices. Angew. Chem. Int. Ed. 2015; 54:195–200. [DOI] [PubMed] [Google Scholar]
- 19. Choi Y.H., van Spronsen J., Dai Y.T., Verberne M., Hollmann F., Arends I., Witkamp G.J., Verpoorte R.. Are natural deep eutectic solvents the missing link in understanding cellular metabolism and physiology. Plant Physiol. 2011; 156:1701–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smith E.L., Abbott A.P., Ryder K.S.. Deep eutectic solvents (DESs) and their applications. Chem. Rev. 2014; 114:11060–11082. [DOI] [PubMed] [Google Scholar]
- 21. Horning D.P., Joyce G.F.. Amplification of RNA by an RNA polymerase ribozyme. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:9786–9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hud N.V., Cafferty B.J., Krishnamurthy R., Williams L.D.. The origin of RNA and “my grandfather's axe”. Chem. Biol. 2013; 20:466–474. [DOI] [PubMed] [Google Scholar]
- 23. Frankel E.A., Bevilacqua P.C., Keating C.D.. Polyamine/nucleotide coacervates provide strong compartmentalization of Mg2+, nucleotides, and RNA. Langmuir. 2016; 32:2041–2049. [DOI] [PubMed] [Google Scholar]
- 24. Burcar B., Pasek M., Gull M., Cafferty B.J., Velasco F., Hud N.V., Menor-Salván C.. Darwin's warm little pond: A one-pot reaction for prebiotic phosphorylation and the mobilization of phosphate from minerals in a urea-based solvent. Angew. Chem. Int. Ed. Engl. 2016; 55:13249–13253. [DOI] [PubMed] [Google Scholar]
- 25. Kreysing M., Keil L., Lanzmich S., Braun D.. Heat flux across an open pore enables the continuous replication and selection of oligonucleotides towards increasing length. Nat. Chem. 2015; 7:203–208. [DOI] [PubMed] [Google Scholar]
- 26. Forsythe J.G., Yu S.S., Mamajanov I., Grover M.A., Krishnamurthy R., Fernandez F.M., Hud N.V.. Ester-Mediated amide bond formation driven by Wet-Dry cycles: A possible path to polypeptides on the prebiotic earth. Angew. Chem., Int. Ed. Engl. 2015; 54:9871–9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Monnard P.A., Szostak J.W.. Metal-ion catalyzed polymerization in the eutectic phase in water-ice: a possible approach to template-directed RNA polymerization. J. Inorg. Biochem. 2008; 102:1104–1111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.