Abstract
Background
The Myc oncogene family has been implicated in many human malignancies and is often associated with particularly aggressive disease, suggesting Myc as an attractive prognostic marker and therapeutic target. However, for epithelial ovarian cancer (EOC), there is little consensus on the incidence and clinical relevance of Myc aberrations. Here we comprehensively investigated alterations in gene copy number, expression, and activity for Myc and evaluated their clinical significance in EOC.
Methods
To address inconsistencies in the literature regarding the definition of copy number variations, we developed a novel approach using quantitative polymerase chain reaction (qPCR) coupled with a statistical algorithm to estimate objective thresholds for detecting Myc gain/amplification in large cohorts of serous (n = 150) and endometrioid (n = 80) EOC. MYC, MYCN, and MYCL1 mRNA expression and Myc activity score for each case were examined by qPCR. Kaplan–Meier and Cox-regression analyses were conducted to assess clinical significance of Myc aberrations.
Results
Using a large panel of cancer cell lines (n = 34), we validated the statistical algorithm for determining clear thresholds for Myc gain/amplification. MYC was the most predominantly amplified of the Myc oncogene family members, and high MYC mRNA expression levels were associated with amplification in EOC. However, there was no association between prognosis and increased copy number or gene expression of MYC/MYCN/MYCL1 or with a pan-Myc transcriptional activity score, in EOC, although MYC amplification was associated with late stage and high grade in endometrioid EOC.
Conclusion
A systematic and comprehensive analysis of Myc genes, transcripts, and activity levels using qPCR revealed that although such aberrations commonly occur in EOC, overall they have limited impact on outcome, suggesting that the biological relevance of Myc oncogene family members is limited to certain subsets of this disease.
Epithelial ovarian cancer (EOC) is a major contributor to human cancer mortality and is molecularly and clinically diverse (1–3). Recent integrated genomic analyses of EOC revealed one of the most common focal amplifications to be in the 8q24 region containing MYC (2), which belongs to the Myc proto-oncogene family that encodes basic helix-loop-helix/leucine zipper transcription factors, including also MYCN and MYCL1, and plays a key role in many human malignancies (4,5). However, the reported frequency of MYC deregulation ranges from 15% to 76% of EOC cases (6–9), limiting an appropriate assessment of the prognostic value of these aberrations in EOC. In addition to MYC, amplification and/or overexpression of MYCL1 (10) and MYCN (11) have been reported in this disease, which suggests that more than one Myc family member may play a biological role in this malignancy. Moreover, subsequent gene expression profiling studies identified molecular subtypes of this disease that were characterized by activation of MYC signaling (1,3), and our previous study demonstrated that high pan-Myc transcriptional activity defined by signature score using a quantitative polymerase chain reaction (qPCR)-based 18-gene Myc activity signature had association with clinical outcome only in a discrete “high-MYCN” molecular subtype of serous tumors (12), highlighting the molecular complexity of EOC. Therefore, a systematic analysis of Myc aberrations is urgently needed and would assist in understanding the importance of each Myc family member in tumor progression and clinical outcome in this heterogeneous disease.
The application of different analytical methodologies and criteria for defining Myc aberrations, especially for copy number elevations, is a likely cause of the inconsistent findings on their clinical significance. For gene amplification, the most widely used techniques include fluorescence in situ hybridization (FISH) (8,9), microarray-based comparative genomic hybridization (aCGH) (2,13,14), and, to a lesser extent, qPCR (8,15). Although MYC amplification or gain has largely been studied using FISH, thresholds are often arbitrarily chosen and this will inevitably affect the ability to reliably determine associations with clinical outcome. Recently developed high-resolution microarray techniques coupled with computational analysis tools allow simultaneous analysis of global copy number variations or expression profiling of thousands of genes, but verification of the results is essential (16). Due to its high sensitivity, accuracy, and specificity, qPCR is widely considered the “gold standard” for validation of such data (17). Hence, here we aimed to comprehensively assess and document the incidence and prognostic impact of increased gene copy number, as determined by a novel, robust, qPCR-based methodology, mRNA expression levels, and a subsequent change in transcriptional activity for MYC, MYCN, and MYCL1 in EOC.
Materials and Methods
Patient Cohorts
Representative cohorts of 150 patients with serous and 61 patients with endometrioid EOC were obtained from the Australian Ovarian Cancer Study, a population-based, case-controlled study undertaken between 2002 and 2006. An additional 19 endometrioid samples were separately obtained from the Gynecological Oncology Biobank at Westmead Hospital, Sydney, Australia. Fresh-frozen tumor specimens of 150 serous and 80 endometrioid tumors were obtained at the time of surgical debulking, prior to chemotherapeutic exposure. Samples were verified as containing at least 70% tumor tissue. Both serous and endometrioid EOC cohorts consist of tumors from all FIGO stages with median clinical follow-up of 46.4 months. Individual cases were subject to review by either light microscopy assessment of representative formalin-fixed tissue taken adjacent to arrayed tissue material or diagnostic slides, or by extraction of information from the original pathology reports when slides were not available. Clinicopathological characteristics for the serous and endometrioid EOCs are summarized in Supplementary Tables 1 and 2 (available online), respectively. Matched genomic DNA and RNA samples were extracted as previously described for EOC cohorts (11). This project was approved by the University of New South Wales Human Research Ethics Committee (HC12551), and all methods were performed in accordance with relevant guidelines and regulations. Written informed consent was obtained from all patients enrolled in the study.
Determination of Cutoff Points for Gain or Amplification of the Myc Family Members
To estimate objective cutoff points for gain or amplification of MYC, MYCN, or MYCL1, we created a practical algorithm that can utilize the distribution of qPCR-measured copy number values in individual cohorts. Cell lines or tumors first were ranked by level of qPCR-measured relative Myc gene copy numbers for each dataset. Both ranks and copy numbers were then normalized by dividing by their respective maximum values. The normalized copy numbers were then plotted as continuous values against normalized rank. After plotting a reference line where normalized values of copy numbers and rank are equal, the distance (d) between the reference line and the amplification curve was calculated for each sample (Equation 1). The following two steps were used to delineate amplification and gain of each Myc oncogene family member. First, the point where maximum d was measured was used as the optimal threshold for identifying the presence of amplification within a whole cohort (see Supplementary Figure 1, available online, for an example plot). Second, after filtering out samples with gene amplification, the presence of a threshold for gain was further tested using the same procedure for all remaining samples.
| (1) |
Where X and Y, normalized rank number, and copy number for a sample, respectively; Xmax and Ymax, the maximum values of X and Y, respectively.
Additional procedures are described in the Supplementary Methods (available online).
Statistical Analysis
Statistical analysis of associations between Myc copy number gain/amplification or overexpression and prognosis was performed using SPSS software v. 22 (IBM, Sydney, Australia). Receiver operator characteristic (ROC) analysis was conducted to determine whether there was an alternative cut-point that would better differentiate between good and poor prognosis for each cohort than the statistical algorithm that was developed in this study. The area under the ROC curve (AUC), with 95% confidence intervals (CIs), was calculated for progression-free survival (PFS) or overall survival (OS). To assess for Myc family gene expression, the statistical methodology described in detail in London et al. (18) was used to determine the optimal cutoff points between low and high expression in each cohort in this study. Briefly, tumors were repeatedly divided into either “high” or “low” groups with the cutoff at lower quartile, median, or upper quartile of Myc family gene expression and signature score. For each cutoff, a Cox proportional-hazards regression model was used to produce a P value, hazard ratio (HR), and 95% CIs. The cutoff with the highest HR was selected as the optimal cutoff. Kaplan–Meier survival analysis was conducted at this cutoff and compared by two-sided log-rank tests. Comparisons of gene expression values between tumor groups were made using either Welch’s t test or one-way analysis of variance (ANOVA), as appropriate. Fisher’s exact test was used to study the association between clinicopathological characteristics and Myc family gene aberrations. Data visualization of the subgroup tests and correlations were performed with GraphPad Prism v. 6.01 for Windows (GraphPad software, Sydney, Australia). Statistical tests were two-sided unless noted otherwise.
Results
qPCR-Measured Myc Family Oncogene Copy Number Determination in a Panel of Cancer Cell Lines
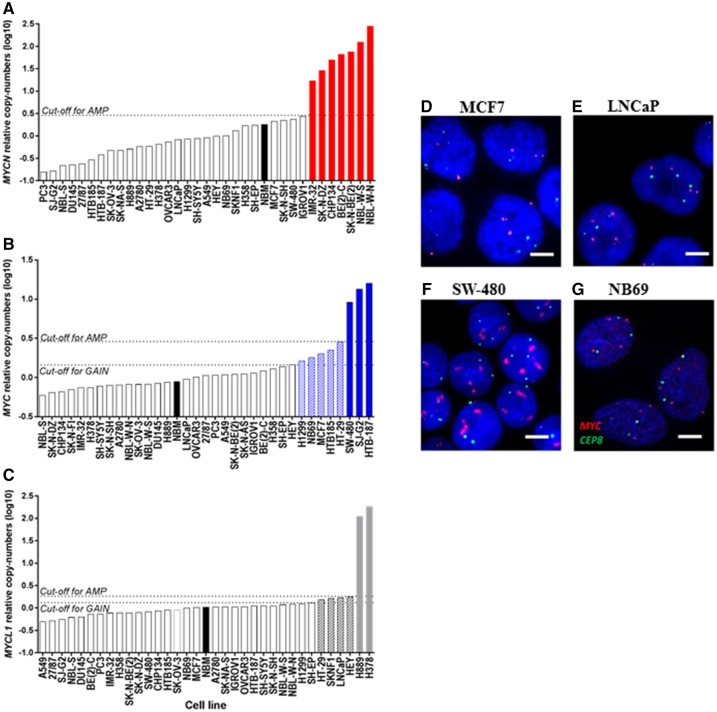
For robust identification of tumor samples with Myc copy-number elevations using qPCR-measured copy numbers, we developed a statistical algorithm to determine an optimal threshold for Myc amplification (Supplementary Figure 1, available online) and validated the algorithm using a large panel of cancer cell lines that included cells for which the status of individual Myc genes had already been established. To do this, we measured MYC, MYCN, and MYCL1 copy numbers by qPCR for each cell line followed by the application of the algorithm to the cell line dataset (Supplementary Table 3, available online). MYCN copy number elevation was identified in cells derived from neuroblastoma, a well-known MYCN-driven cancer, among which all cell lines were independently validated as carrying MYCN amplification, including SK-N-DZ (19), BE(2)-C (20), IMR-32 (20), CHP-134 (21), NBL-W-S (22), and NBL-W-N (22), were classified to a MYCN-amplified group using the algorithm (Figure 1A). The method was also capable of distinguishing cell lines known to carry MYC amplification, including cell lines derived from cancers of brain [SJ-G2 and HTB-187 (23,24)] and colon [SW-480 (15)], from nonamplified cell lines, including EOC cell line, A2780 (11), and prostate cancer cell line, LNCaP (18) (Figure 1B). Two small cell lung cancer cell lines known to harbor MYCL1 amplification, H378 and H889 (15), were also correctly classified with this aberration using our method (Figure 1C), indicating that the algorithm can readily and accurately identify samples with known amplification of Myc family genes.
Figure 1.
Elevation of MYC, MYCN, and MYCL1 copy numbers in a panel of cancer cell lines. A–C)MYCN (A), MYC (B), and MYCL1 (C) copy numbers determined using qPCR and calculated as the geometric means relative to each of three endogenous control genes (Albumin, RNaseP, and NAGK) as described in the Supplementary Methods (available online). The cutoff for amplification was determined using a statistical algorithm. The dotted lines indicate optimal cutoff point for amplification or gain and the black bars indicate DNA of normal bone marrow. The bars with dashed lines or solid fill indicate gain or amplification of Myc gene, respectively, in each graph. D–G) The panels show representative images for FISH performed on cell lines as a tool to validate the qPCR-based method for determining of aberrations for MYC in MCF7 (D), LnCaP (E), SW-480 (F), and NB69 (G) (63x magnification). Vysis probes were used for MYC and CEP 8. The scale bars = 5 µm.
Notably, although several cell lines were determined to have higher MYC, MYCN, or MYCL1 copy numbers than normal bone marrow DNA, which was used as a reference sample carrying a normal copy number of each Myc gene in this analysis, they were not classified to the amplified group (Figure 1A–C), suggesting that of these cells, some may have low-level amplification (gain) of Myc. To examine whether the algorithm was able to distinguish cells with Myc gain from those with normal copy, the algorithm was applied only to these remaining cell lines. Although no line was found to carry MYCN gain (Figure 1A), MYC gain was identified in cell lines derived from tumors of the colon (HT-29), brain (HTB-185), and breast (MCF7) (Figure 1B), each of which had previously been determined to carry MYC gain using FISH and aCGH (24–27), thus corroborating the findings with our method. Additional FISH analysis was performed in some cancer cell lines to validate their MYC alteration status as determined using the qPCR-algorithm method (Supplementary Table 4, available online). In the MCF-7 cell line with MYC gain (qPCR-measured MYC copy number = 2.0), the MYC:CEP8 ratio was 1.7 (Figure 1D), as distinct from a MYC-nonamplified line, LNCaP (qPCR-measured MYC copy number = 1; MYC:CEP8 ratio = 1) (Figure 1E), and from a known MYC-amplified line, SW-480, with multiple red signals for the MYC probe in FISH analysis (Figure 1F). The HT-29 cell line was estimated to have a qPCR measured copy number of 2.9, which was determined by the algorithm to be the upper threshold for gain, and this copy number was in agreement with the MYC:CEP8 ratio of 2.7 determined by FISH (Supplementary Table 4, available online). Additionally, the PCR-algorithm method identified MYC gain in a neuroblastoma cell line, NB69, for which MYC status had not been previously ascertained. FISH analysis of NB69 (qPCR-measured MYC copy number = 1.7) showed that the MYC:CEP8 ratio (1.8) was very similar to that in MCF7, carrying MYC gain (1.7) (Figure 1G), confirming the ability of the PCR-algorithm method to distinguish MYC gain from single copy in samples for which this had not been previously detected. In addition, although it was difficult to quantitate the MYC:CEP8 ratio in MYC amplified cells due to the multiple signals, in those cell lines for which the MYC:CEP8 ratio could be accurately determined by FISH, the numerical values corresponded well to the qPCR-measured copy number values (Supplementary Table 4, available online). Using the method, we also observed MYC gain in H1299, a lung cancer cell line, and MYCL1 gain in cell lines derived from neuroblastoma (SK-N-FI) and colorectal (HT-29), prostate (LNCaP), and ovarian (HEY) tumors (Figure 1C).
Myc Gain/Amplification and Clinical Outcome in EOC
We next applied this method to determine gain and/or amplification of MYC, MYCN, and MYCL1 in cohorts of serous and endometrioid EOC and examined their associations with the clinical outcome. Of 150 serous EOC, 24 (16%) and 11 (7.3%) tumors exhibited MYC amplification and gain, respectively. Ten of 80 endometrioid EOC cases (12.5%) exhibited MYC amplification but none had gain (Table 1). MYCL1 amplification occurred in nine cases of serous and a single case of endometrioid EOC. Neither MYCN amplification nor gain was detected in either histological subtype of EOC.
Table 1.
Univariate analysis of gene copy number and expression levels of MYC, MYCN and MYCL1 in EOC
| Serous EOC (n = 150) |
Endometrioid EOC (n = 80) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PFS |
OS |
PFS |
OS |
|||||||
| Gene | No. of cases | HR (95% CI) | P* | HR (95% CI) | P | No. of cases | HR (95% CI) | P | HR (95% CI) | P |
| MYC | ||||||||||
| Gain | 11 | 0.99 (0.70 to 1.39) | .94 | 0.93 (0.59 to 1.47) | .75 | 0 | — | — | — | — |
| Amplification | 24 | 1.23 (0.78 to 1.96) | .38 | 0.92 (0.49 to 1.70) | .79 | 10 | 1.11 (0.38 to 3.23) | .85 | 0.66 (0.15 to 2.87) | .58 |
| MYCN | ||||||||||
| Gain | 0 | — | — | — | — | 0 | — | — | — | — |
| Amplification | 0 | — | — | — | — | 0 | — | — | — | — |
| MYCL1 | ||||||||||
| Gain | 0 | — | — | — | — | 0 | — | — | — | — |
| Amplification | 9 | 1.64 (0.80 to 3.36) | .18 | 1.56 (0.68 to 3.6) | .29 | 1 | ND | ND | ND | ND |
| Any Myc gene alteration | 44 | 1.28 (0.87 to 1.88) | .20 | 1.04 (0.64 to 1.69) | .88 | 11 | 0.97 (0.33 to 2.82) | .95 | 0.59 (0.14 to 2.57) | .48 |
| Overexpression | ||||||||||
| MYC | MED | 1.04 (0.73 to 1.48) | .84 | 1.12 (0.73 to 1.74) | .60 | MED | 1.37 (0.63 to 2.94) | .43 | 1.69 (0.66 to 4.34) | .27 |
| MYCN | LQ | 0.89 (0.59 to 1.32) | .55 | 1.51 (0.68 to 1.95) | .60 | LQ | 1.92 (0.66 to 5.60) | .22 | 1.02 (0.34 to 3.10) | .97 |
| MYCL1 | LQ | 0.82 (0.55 to 1.23) | .06 | 0.70 (0.42 to 1.12) | .13 | MED | 2.06 (0.92 to 4.64) | .07 | 2.22 (0.87 to 5.63) | .09 |
Log-rank P value. PFS = progression-free survival; OS = overall survival; HR = hazard ratio; CI = confidence interval; ND = not determined; MED = Medium; LQ = Lower Quartile.
When the clinical impact of MYC or MYCL1 gain/amplification was investigated in serous EOC, no association with clinicopathological parameters, including stage, grade, or residual disease, was found (Table 2). Survival analysis indicated that neither MYC amplification nor gain was associated with PFS or OS (Table 1). When tumors with either gain or amplification of MYC were pooled, still no association with clinical outcome was found (P = .48 and P = .7 for PFS and OS, respectively; data not shown). Moreover, MYCL1 amplification was not associated with prognosis in this histological subtype (Table 1). Upon combining cases with amplification or gain of either MYC or MYCL1 into a single group, no relationship between these gene aberrations and prognosis was observed in serous EOC (Table 1). In endometrioid tumors, MYC amplification was found to be associated with late stage and high grade (Table 2), although no impact on patient outcome was observed (Table 1). Neither MYC nor MYCL1 alteration, nor combined MYC/MYCL1 alteration, were predictive of patient outcome according to multivariable analysis that took into account potential confounding effects of debulking status and FIGO stage (Supplementary Table 5, available online).
Table 2.
The association of MYC and MYCL1 aberrations with clinicopathological parameters in 150 serous and 80 endometrioid EOC
| Serous EOC |
Endometrioid EOC |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
MYC |
MYCL1 |
MYC |
||||||||||||
| Gain |
Amplification |
Amplification |
Amplification |
|||||||||||
| Parameters | Total | No | Yes | P* | No | Yes | P | No | Yes | P | Total | No | Yes | P |
| Stage | ||||||||||||||
| I/II | 17 | 16 | 1 | 1.000 | 15 | 2 | 1.000 | 17 | 0 | .598 | 53 | 50 | 3 | .010 |
| III/IV | 133 | 123 | 10 | 111 | 22 | 124 | 9 | 25 | 18 | 7 | ||||
| Grade | ||||||||||||||
| 1 or 2 | 34 | 32 | 2 | 1.000 | 31 | 3 | .288 | 33 | 1 | .685 | 49 | 46 | 3 | .041 |
| 3 | 116 | 107 | 9 | 95 | 21 | 108 | 3 | 31 | 24 | 7 | ||||
| Residual disease | ||||||||||||||
| No | 96 | 89 | 7 | 1.000 | 82 | 14 | .643 | 91 | 5 | .723 | 66 | 57 | 9 | .683 |
| Yes, >1 cm | 54 | 50 | 4 | 44 | 10 | 50 | 4 | 14 | 13 | 1 | ||||
P values are obtained from Fisher’s exact test. EOC = epithelial ovarian cancer.
To further assess the predictive impact of Myc amplification and validate our algorithm, we conducted ROC curve analysis, which can determine whether there was an alternative cutoff point for Myc amplification that would discriminate between patients with good and poor outcome in each EOC cohort. As a measure of accuracy, the AUC value with 95% CIs was calculated for each ROC curve. The ROC analysis for MYC, MYCN, and MYCL1 amplification for serous and endometrioid EOC cohorts showed that the AUC does not differ statistically from 0.5, indicating that Myc amplification is not associated with patient outcome (Supplementary Figure 2, available online). Furthermore, a common approach to identify gene copy number alterations to date is aCGH analysis coupled with computational algorithms, such as Genomic Identification of Significant Targets in Cancer (28). Using this approach, the putative amplification status of MYCN, MYC, or MYCL1 in publicly available TCGA datasets of serous EOC was determined through cBioPortal (http://www.cbioportal.org/faq.jsp). Consistent with our findings, MYC amplification was confirmed to be the most frequent aberration among the Myc family in the serous EOC dataset (n = 557) in which MYC, MYCN, or MYCL1 amplification was identified in 154 (28%), 6 (1.1%), or 50 (9%) cases, respectively. None of these Myc aberrations was associated with poor patient outcome (Supplementary Figure 3. available online).
mRNA Expression Levels and Transcriptional Activity of Myc and Clinical Outcome in EOC
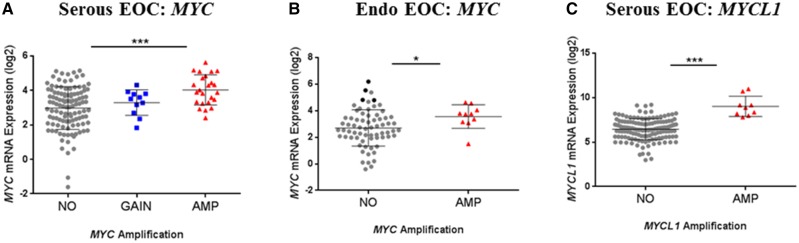
In the absence of any association between Myc gain/amplification and clinical outcome, we examined the relationship of Myc gene expression to copy number and to clinical outcome in EOC. In both histological subtypes, MYC-amplified tumors expressed higher levels of mRNA than tumors without amplification (Figure 2A,B); however, MYC gain, which was observed in some serous tumors, was not associated with an increase of MYC mRNA expression levels (Figure 2A). MYCL1-amplified serous tumors expressed higher levels of MYCL1 as compared with nonamplified tumors (Figure 2C). Despite a trend for high MYCL1 gene expression to be associated with reduced PFS, this failed to achieve statistical significance and no relationship was observed for OS (Table 1). High mRNA expression of MYC, MYCN, or MYCL1 was not predictive of outcome, following adjustment for the debulking status and FIGO stage in multivariable analysis of both serous and endometrioid EOC (data not shown).
Figure 2.
Expression levels of MYC, MYCN, and MYCL1 mRNA according to Myc status in ovarian tumors. In cohorts serous epithelial ovarian cancer (EOC) (A,C), and endometrioid EOC (B), relative gene expression levels of MYC (A,B) and MYCL1 (C) were calculated using the ΔΔCt, method and were compared between tumors with no alteration, gain, and amplification of each Myc gene. P values were calculated using either one-way ANOVA followed by Dunnett’s post-hoc analysis (A) or Welch’s t test (B,C).
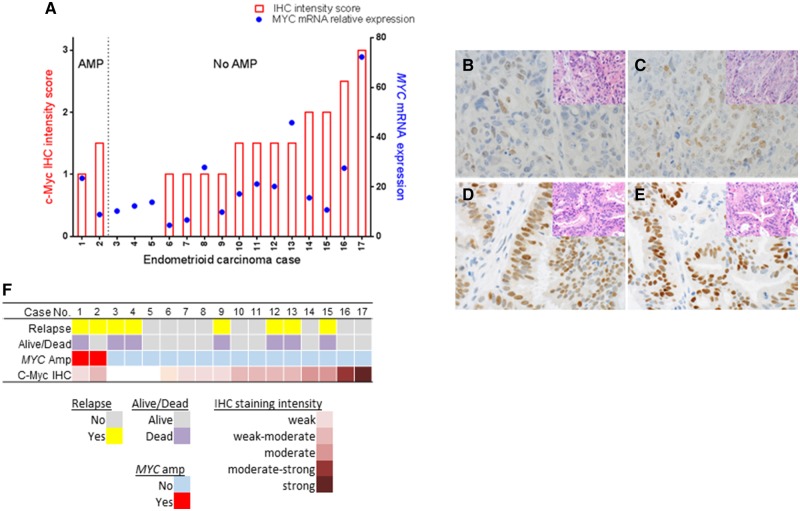
Intriguingly, a proportion of endometrioid tumors expressed very high levels of MYC mRNA in the absence of gene amplification (Figure 2B), indicating that MYC overexpression can occur independently from gene amplification. To further investigate whether these tumors with MYC overexpression but lacking MYC amplification also express high c-Myc protein, 17 of the 80 endometrioid EOC were analyzed for c-Myc protein levels immunohistochemically. The analysis revealed that 14 of the 17 cases were positively stained for nuclear c-Myc (Figure 3A). The discordance between amplification and expression was confirmed, such that four cases without MYC amplification showed high (moderate to strong) staining intensity for c-Myc (highlighted in black in the no amplification group in Figure 2B), whereas the two MYC-amplified tumors exhibited only weak to weak-moderate c-Myc protein staining intensity (Figure 3A). Examples of c-Myc staining characteristics are presented in Figure 3B–E. Despite the lack of correlation between MYC amplification and protein expression levels, a positive and statistically significant correlation between c-Myc protein expression levels (intensity score) and mRNA expression levels was found (Pearson’s r = 0.6067, P = .0098), indicating that overexpression of MYC mRNA can be acquired without the gene amplification and that this generally translates into high protein expression. Nonetheless, high c-Myc protein expression levels appeared to have no relationship with patient outcome (Figure 3F), as found for MYC amplification and mRNA expression.
Figure 3.
Relationship between genomic amplification, mRNA, and protein expression levels of MYC in endometrioid epithelial ovarian cancer (EOC) tumors. Seventeen paraffin-embedded endometrioid EOC tumors were stained with anti c-Myc (Antibody: 9E10). Tumors were scored as having absent, weak, moderate, or strong c-Myc protein staining, as defined in the Supplementary Methods (available online). A) Tumors are aligned according to MYC status and in order of increasing immunohistochemistry score. Also indicated are mRNA expression and clinical information for each sample. B–E) Examples of tumors with different levels of nuclear c-Myc protein expression and MYC amplification status with corresponding hemotoxylin and eosin-stained tissue in inserts are shown (600x magnification). B,C) Cases 1 and 2: MYC amplification but negative protein expression. D) Case 16: No MYC amplification but moderate-strong protein expression. E) Case 17: No MYC amplification but strong protein expression. F) Overview of properties of the 17 endometrioid tumor samples. Different tracks are indicated below.
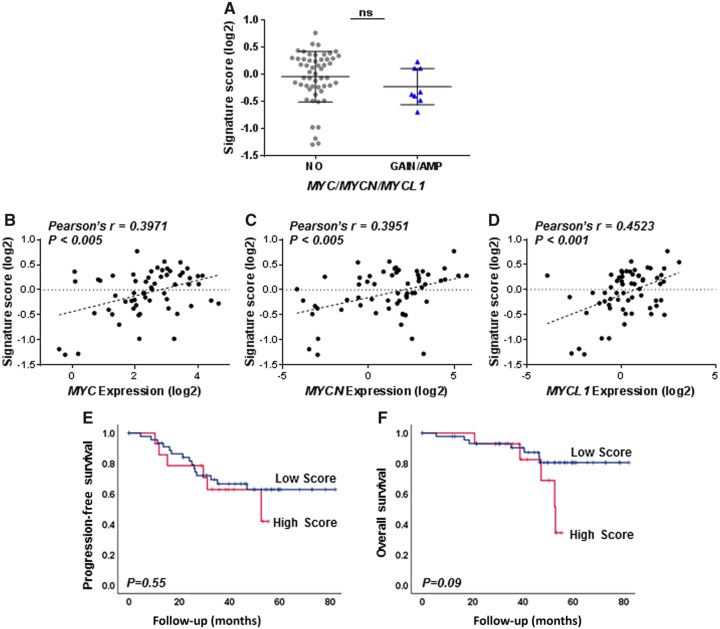
We further investigated the relationship between gain/amplification or expression levels of the Myc family genes and elevated overall MYC/MYCN/MYCL1 transcriptional activity in the endometrioid EOC cohort using a qPCR-based, 18-gene Myc activity signature that we previously developed and showed to have prognostic value in a subset of serous tumors with high MYCN, but not in the overall serous cohort using data available through TCGA (12). Signature score did not differ between tumors based on Myc gain or amplification status (Figure 4A) but was positively correlated with MYC, MYCN, or MYCL1 mRNA expression levels (Figure 4B–D). High signature score was not associated with patient outcome in the endometrioid tumors (Figure 4E,F). Together these analyses indicate that although elevated expression levels of Myc transcripts and/or transcriptional activity can occur without gene amplification, there is no evidence that these factors are determinants of outcome in serous and endometrioid EOC.
Figure 4.
Relationship of Myc activity signature to molecular alterations of Myc family genes and the prognostic significance in endometrioid epithelial ovarian cancer (EOC). A) Myc activity levels were compared between tumors without Myc alteration (NO) and those with either gain or amplification (GAIN/AMP) in endometrioid EOC. Welch’s t test was used to determine different levels of Myc activity. B–D) Expression levels of MYC (B), MYCN (C), or MYCL1 (D) genes were also measured using qPCR and correlated to signature score in EOC. E,F) Kaplan–Meier analysis of signature scores in 61 endometrioid EOC was conducted for PFS (E) and OS (F). Signature scores were dichotomized at upper quartile in the overall cohorts of each tumor. P values were obtained from the log-rank test in each case.
Discussion
Here we undertook to provide complete analyses of aberrations in copy number, gene expression, and oncoprotein activity of MYC/MYCN/MYCL1 in order to determine the relevance of Myc deregulation to clinical outcome in serous and endometrioid tumors, the two most common EOC histological subtypes. Unlike most previous studies either reporting the prognostic significance of aberrations of one of the Myc family members in EOC or using tumor cohorts consisting of all different EOC histological subtypes (6,7,9), our detailed analysis of alterations of each Myc gene showed that MYC amplification was the most common Myc family gene aberration but had no impact on clinical outcome in the overall cohorts of serous and endometrioid tumors. Moreover, elevated MYC mRNA or protein can occur in the absence of gene amplification, highlighting the need to consider other measures of Myc functionality in addition to copy-number aberrations. These findings also suggest a fundamental difference between EOC and clearly Myc-driven malignancies like neuroblastoma, where MYCN amplification, occurring in ∼25% of cases, is associated with the highest levels of MYCN transcription and with the worst clinical outcome (29). Hence, Myc genes may not be a major contributor of EOC tumor progression, but aberrant expression and activity of Myc may represent an epiphenomenon conferred by a complex network of upstream regulators. However, together with several previous studies reporting the effects of direct or indirect targeting of MYC on the malignant phenotypic changes in EOC cells in vitro (30,31), significant associations between MYC amplification and stage or grade in endometrioid EOC, which was demonstrated in this study using a large number of samples compared to a previous study (8), suggest a potential role of this MYC aberration in tumor progression in certain subsets of EOC. In support of this notion, our previous study demonstrated that the association of high Myc activity with poor clinical outcome was limited to a “high-MYCN” molecular subtype of EOC (12). Therefore, given the heterogeneous nature of EOC, being composed of subtypes according to molecular characterization, clinical presentation, and response to therapy, we cannot rule out the possibility that highly activated Myc oncogenes may be confined to tumor subsets. Accordingly, further investigations into Myc oncogenes as prognostic indicators and therapeutic targets for EOC would be best conducted in the context of disease subgroups as well as other less common histological subtypes, including clear cell and mucinous EOC, subject to the availability of adequate sample numbers.
Compared with FISH or aCGH, the statistical algorithm created in this study is a relatively rapid and inexpensive approach to estimate objective cutoffs for gain or amplification using relative copy numbers measured by qPCR. Although qPCR has not been commonly used for determining copy number, a strong correlation between the results of FISH, aCGH, and qPCR in detecting the presence of MYCN amplification supports the utility of qPCR as a suitable rapid, cost-effective, and reliable means of detecting gene aberrations (32). Such methods typically calculate the copy number relative to a normal reference sample, but a robust approach is still needed to distinguish meaningful increases above normal levels and further to discriminate between gain and amplification. Therefore, when used together with the statistical method that can estimate objective thresholds for gain/amplification, qPCR might be a valuable tool in reducing the inconsistencies in this field that exist to date. Although due to the limited availability, the threshold in genomic level using FISH could not be further validated in clinical samples, validation of this method using a large panel of cancer cell lines with existing data derived from different analytical methodologies allowed robust determination of clear thresholds for Myc gain/amplification. Such methodology could be applied to future studies investigating the biological and clinical relevance of Myc gain in numerous other malignancies.
Overall, this comprehensive study could find no relationship between Myc aberrations at the levels of their genes, transcripts, or transcriptional activities and clinical outcome in either EOC histological subtype, suggesting the biological relevance of Myc oncogene family members is limited to certain subsets of this disease. These findings provide new insights into our understanding of the prognostic importance and potential therapeutic relevance of the Myc oncogene family in EOC.
Supplementary Material
Funding
This project was supported by grants from the Cancer Institute New South Wales and the National Health and Medical Research Council (NHMRC) of Australia. Children’s Cancer Institute Australia is affiliated with UNSW Australia and the Sydney Children’s Hospitals Network. We acknowledge the contribution of the Australian Ovarian Cancer Study (AOCS; full list of the institutions that supported that study can be found at http://www.aocstudy.org) and the Gynecological Oncology Biobank at Westmead. The Westmead Gynaecological Oncology Biobank at Westmead was supported by the Cancer Institute NSW (12/RIG/1-17 and 15/RIG/1-16) and NHMRC (ID310670, ID628903). The Australian Ovarian Cancer Study was supported by the U.S. Army Medical Research and Materiel Command under DAMD17-01-1-0729, The Cancer Council Victoria, Queensland Cancer Fund, The Cancer Council New South Wales, The Cancer Council South Australia, The Cancer Foundation of Western Australia, The Cancer Council Tasmania and the NHMRC (ID400413 and ID400281).
Notes
Affiliations of authors: Children’s Cancer Institute Australia, Lowy Cancer Research Centre, UNSW Australia, Kensington, NSW, Australia (MJ, AJR, AJG, SS, MH, MDN, MJH); Department of Gynecological Oncology, Westmead Hospital and Centre for Cancer Research, The Westmead Institute for Medical Research, The University of Sydney, Sydney, NSW, Australia (CK, AD); Department of Anatomical Pathology, Prince of Wales Hospital, Randwick, NSW, Australia (AJG); Peter MacCallum Cancer Centre, Melbourne, VIC, Australia (Australian Ovarian Cancer Study Group); Centre for Big Data Research in Health/School of Women’s and Children’s Health, UNSW Australia, Kensington, NSW, Australia (K-AM); University of New South Wales Centre for Childhood Cancer Research, UNSW Australia, Kensington, NSW, Australia (MDN).
Present affiliation: Garvan Institute of Medical Research, Darlinghurst, NSW, Australia (AJR).
The AOCS gratefully acknowledges additional support from S. Boldeman, the Agar family, Ovarian Cancer Australia, Ovarian Cancer Action (UK), and the Peter MacCallum Foundation. We also acknowledge the contribution of the AOCS study nurses and research assistants and we thank all of the women who participated in the study.
The authors disclose no potential conflicts of interest.
References
- 1. Tan TZ, Miow QH, Huang RY.. Functional genomics identifies five distinct molecular subtypes with clinical relevance and pathways for growth control in epithelial ovarian cancer. EMBO Mol Med. 2013;5(7):983–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. The Cancer Genome Altas Research Network Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Verhaak RG, Tamayo P, Yang JY, Hubbard D, Zhang H, Creighton CJ.. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J Clin Invest. 2013;123(1):517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beltran H. The N-myc oncogene: maximizing its targets, regulation, and therapeutic potential. Mol Cancer Res. 2014;12(6):815–822. [DOI] [PubMed] [Google Scholar]
- 5. Meyer N, Penn LZ.. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8(12):976–990. [DOI] [PubMed] [Google Scholar]
- 6. Darcy KM, Brady WE, Blancato JK, et al. Prognostic relevance of c-MYC gene amplification and polysomy for chromosome 8 in suboptimally-resected, advanced stage epithelial ovarian cancers: a Gynecologic Oncology Group study. Gynecol Oncol. 2009;114(3):472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Diebold J, Suchy B, Baretton GB, et al. DNA ploidy and MYC DNA amplification in ovarian carcinomas—correlation with p53 and bcl-2 expression, proliferative activity and prognosis. Virchows Archiv. 1996;429(4-5):221–227. [DOI] [PubMed] [Google Scholar]
- 8. Dimova I, Raitcheva S, Dimitrov R, Doganov N, Toncheva D.. Correlations between c-myc gene copy-number and clinicopathological parameters of ovarian tumours. Eur J Cancer. 2006;42(5):674–679. [DOI] [PubMed] [Google Scholar]
- 9. Wang ZR, Liu W, Smith ST, Parrish RS, Young SR.. c-myc and chromosome 8 centromere studies of ovarian cancer by interphase FISH. Exp Mol Pathol. 1999;66(2):140–148. [DOI] [PubMed] [Google Scholar]
- 10. Wu R, Lin L, Beer DG, et al. Amplification and overexpression of the L-MYC proto-oncogene in ovarian carcinomas. Am J Pathol. 2003;162(5):1603–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Helland A, Anglesio MS, George J, et al. Deregulation of MYCN, LIN28B and LET7 in a molecular subtype of aggressive high-grade serous ovarian cancers. PLoS One. 2011;6(4):e18064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jung M, Russell AJ, Liu B, et al. A Myc activity signature predicts poor clinical outcomes in Myc-associated cancers. Cancer Res. 2017;77(4):971–981. [DOI] [PubMed] [Google Scholar]
- 13. TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim YH, Girard L, Giacomini CP, et al. Combined microarray analysis of small cell lung cancer reveals altered apoptotic balance and distinct expression signatures of MYC family gene amplification. Oncogene. 2006;25(1):130–138. [DOI] [PubMed] [Google Scholar]
- 15. Hook KE, Garza SJ, Lira ME, et al. An integrated genomic approach to identify predictive biomarkers of response to the aurora kinase inhibitor PF-03814735. Mol Cancer Ther. 2012;11(3):710–719. [DOI] [PubMed] [Google Scholar]
- 16. Wang Y, Barbacioru C, Hyland F, et al. Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays. BMC Genomics. 2006;7(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. D’haene B, Vandesompele J, Hellemans J.. Accurate and objective copy number profiling using real-time quantitative PCR. Methods. 2010;50(4):262–270. [DOI] [PubMed] [Google Scholar]
- 18. Savinainen KJ, Linja MJ, Saramäki OR, et al. Expression and copy number analysis of TRPS1, EIF3S3 and MYC genes in breast and prostate cancer. Br J Cancer. 2004;90(5):1041–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hossain MM, Banik NL, Ray SK.. N-Myc knockdown and apigenin treatment controlled growth of malignant neuroblastoma cells having N-Myc amplification. Gene. 2013;529(1):27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carr J, Bown NP, Case MC, Hall AG, Lunec J, Tweddle DA.. High-resolution analysis of allelic imbalance in neuroblastoma cell lines by single nucleotide polymorphism arrays. Cancer Genet Cytogenet. 2007;172(2):127–138. [DOI] [PubMed] [Google Scholar]
- 21. Mosse YP, Greshock J, Margolin A, et al. High-resolution detection and mapping of genomic DNA alterations in neuroblastoma. Genes Chromosomes Cancer. 2005;43(4):390–403. [DOI] [PubMed] [Google Scholar]
- 22. Manohar CF, Salwen HR, Brodeur GM, Cohn SL.. Co-amplification and concomitant high levels of expression of a DEAD box gene with MYCN in human neuroblastoma. Genes Chromosomes Cancer. 1995;14(3):196–203. [DOI] [PubMed] [Google Scholar]
- 23. Fruhwald M, O’Dorisio M, Rush L, et al. Gene amplification in PNETs/medulloblastomas: mapping of a novel amplified gene within the MYCN amplicon. J Med Genet. 2000;37(7):501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Siu I-M, Lal A, Blankenship JR, Aldosari N, Riggins GJ.. c-Myc promoter activation in medulloblastoma. Cancer Res. 2003;63(16):4773–4776. [PubMed] [Google Scholar]
- 25. Rummukainen J, Kytölä S, Karhu R, Farnebo F, Larsson C, Isola JJ.. Aberrations of chromosome 8 in 16 breast cancer cell lines by comparative genomic hybridization, fluorescence in situ hybridization, and spectral karyotyping. Cancer Genet Cytogenet. 2001;126(1):1–7. [DOI] [PubMed] [Google Scholar]
- 26. Shadeo A, Lam WL.. Comprehensive copy number profiles of breast cancer cell model genomes. Breast Cancer Res. 2006;8(1):R9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Augenlicht LH, Wadler S, Corner G.. Low-level c-myc amplification in human colonic carcinoma cell lines and tumors: a frequent, p53-independent mutation associated with improved outcome in a randomized multi-institutional trial. Cancer Res. 1997;57(9):1769–1775. [PubMed] [Google Scholar]
- 28. Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G.. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maris JM, Hogarty MD, Bagatell R, Cohn SL.. Neuroblastoma. Lancet. 2007;369(9579):2106–2120. [DOI] [PubMed] [Google Scholar]
- 30. Prathapam T, Aleshin A, Guan Y, Gray JW, Martin GS.. p27Kip1 mediates addiction of ovarian cancer cells to MYCC (c-MYC) and their dependence on MYC paralogs. J Biol Chem. 2010;285(42):32529–32538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baratta MG, Schinzel AC, Zwang Y, et al. An in-tumor genetic screen reveals that the BET bromodomain protein, BRD4, is a potential therapeutic target in ovarian carcinoma. Proc Natl Acad Sci USA. 2015;112(1):232–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Malakho SG, Korshunov A, Stroganova AM, Poltaraus AB.. Fast detection of MYCN copy number alterations in brain neuronal tumors by real-time PCR. J Clin Lab Anal. 2008;22(2):123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.