Abstract
Although transposable elements are an important source of regulatory variation, their genome-wide contribution to the transcriptional regulation of stress-response genes has not been studied yet. Stress is a major aspect of natural selection in the wild, leading to changes in the transcriptional regulation of a variety of genes that are often triggered by one or a few transcription factors. In this work, we take advantage of the wealth of information available for Drosophila melanogaster and humans to analyze the role of transposable elements in six stress regulatory networks: immune, hypoxia, oxidative, xenobiotic, heat shock, and heavy metal. We found that transposable elements were enriched for caudal, dorsal, HSF, and tango binding sites in D. melanogaster and for NFE2L2 binding sites in humans. Taking into account the D. melanogaster population frequencies of transposable elements with predicted binding motifs and/or binding sites, we showed that those containing three or more binding motifs/sites are more likely to be functional. For a representative subset of these TEs, we performed in vivo transgenic reporter assays in different stress conditions. Overall, our results showed that TEs are relevant contributors to the transcriptional regulation of stress-response genes.
INTRODUCTION
Transposable elements (TEs) represent a large portion of eukaryotic genomes. They are repetitive sequences that have the ability to move around in the genome making new copies of themselves in the process. Some TEs contain regulatory sequences such as promoters, transcription start sites (TSSs) and transcription factor binding sites (TFBSs) that can affect the expression of nearby genes (1,2). Multiple examples of individual TE copies affecting gene expression have been described in a wide-range of organisms (3). More recently, genome-wide approaches have been used to explore the overall contribution of TEs to gene regulation (4,5). In particular, several studies have found that TEs contain binding sites for a variety of transcription factors (TFs) involved in very relevant cellular processes such as cell pluripotency, placenta development or immune response (6–9). These studies also found that it is one or a few TE families the ones that contribute more to the TFBS repertoire.
Most of the genome-wide approaches aimed at identifying TFBSs in TEs are based on the analysis of chromatin immunoprecipitation sequencing (ChIP-seq) that provide experimental evidence for the binding of a particular TF to a discrete genomic region. However, ChIP-seq only provides information for the binding sites occurring in the particular conditions in which the experiment is performed. Because it is impossible to assay all tissue types and developmental stages under all conditions, combining binding site predictions using ChIP-seq with transcription factor binding motif (TFBM) predictions using bioinformatic tools should help identify a more complete dataset of binding sites (10). The binding profiles for an increasing number of TFs are available in dedicated databases such as JASPAR, including the newer ones based on hidden Markov models named transcription factor flexible models (TFFM) (11,12). Several genomic features, such as chromatin accessibility or epigenetic marks, are often used to evaluate the regulatory potential of the genomic sequences containing TFBS (13,14). In any case, functional validation of the identified TFBSs is needed to conclude that the predicted binding sites are functional.
Most stress-related TFs are conserved across organisms (15). Stress is a major aspect of natural selection in the wild that leads to changes in the transcriptional regulation of a variety of genes. Both in humans and in the fruitfly Drosophila melanogaster, adaptation to high altitude, toxic environments, high temperature environments, and pathogen exposure has already been described (16,17). These adaptations are related with hypoxia, xenobiotic, heavy-metal, oxidative, heat, and immune stress. However, the contribution of TEs to the binding sites of stress-related TFs remains largely unexplored.
In this work, we used bioinformatic tools to predict the presence of binding motifs in TEs, and available ChIP-seq data to identify binding sites for several TFs involved in six stress responses (Table 1) (18–25). Enhancer and/or promoter features such as open chromatin regions, active histone marks and co-binding of stress-related proteins, and other genomic features such as location regarding nearby genes and function of nearby genes were also investigated. Besides genomic information, population-level information was also used to identify the subset of TEs more likely to contain functional TFBSs. Finally, in vivo enhancer assays were performed for a diverse set of TEs. Our results showed that TEs are likely to contribute a significant fraction of stress-related transcription factor binding sites in humans and in D. melanogaster.
Table 1.
Transcription factors analyzed in this study. Description of the stress-related transcription factors, including the identifier (ID) of the position weight matrix (PWM) or transcription factor flexible model (TFFM) used. The stresses analyzed were: HSE: Heat Shock element; ARE: Antioxidant response element; HRE: Hypoxia response element; IRE: Immunity response element; MRE: Metal response element; and XRE: Xenobiotic response element. In D. melanogaster, only TFFM IDs are provided for models built based on a D. melanogaster PWMs. *For HSF, HIF1, MTF-1, and XBP1 a vertebrate PWM was used.
| Drosophila melanogaster | Human | |||||||
|---|---|---|---|---|---|---|---|---|
| Stress | Transcription Factors | PWM ID | Score threshold (PWM) | TFFM ID | Transcription factors | PWM ID | Score threshold (PWM) | TFFM ID |
| HSE/ARE/HRE/ IRE/MRE/XRE | HSF (18) | MA0486.2* | 10.04 | NA | HSF1 (18) | MA0486.2 | 10.09 | TFFM0048.1 |
| ARE/IRE | DL (19) | MA0022.1 | 8.48 | TFFM0158 | NFKB1 (20) | MA0105.4 | 10.35 | |
| HRE | HIF1 (HIF1A, tango-HIF1B) (21) | MA0259.1* | 9.43 | NA | EGR1 (22) | MA0162.2 | 9.79 | TFFM0020.1 |
| SP1 (20) | MA0079.3 | 10.48 | TFFM0097.1 | |||||
| MRE | MTF-1 (23) | PB0044.1* | 9.27 | NA | – | – | – | – |
| IRE | CAD (19) | MA0216.2 | 10.12 | TFFM0159 | – | – | – | – |
| DEAF1 (24) | MA0185.1 | 8.33 | NA | – | – | – | – | |
| NUB (78) | MA0197.2 | 8.99 | NA | – | – | – | – | |
| XBP1 (25) | MA0844.1* | 9.69 | NA | – | – | – | – | |
| ARE/XRE | CNC (20) | MA0530.1 | 9.97 | NA | – | – | – | – |
| ARE | – | – | – | – | NFE2L2 (20) | MA0150.2 | 9.56 | TFFM0071.1 |
| ARE/HRE | – | – | – | – | NRF1 (20) | MA0506.1 | 10.04 | TFFM0082.1 |
| – | – | – | – | CREB1 (20) | MA0018.2 | 7.96 | TFFM0012.1 | |
| HRE/XRE | – | – | – | – | AP1 (FOS) (20) | MA0476.1 | 10.46 | TFFM0032.1 |
Description of the stress-related transcription factors, including the identifier (ID) of the position weight matrix (PWM) or transcription factor flexible model (TFFM) used. The stresses analyzed were: HSE: Heat Shock element; ARE: Antioxidant response element; HRE: Hypoxia response element; IRE: Immunity response element; MRE: Metal response element; and XRE: Xenobiotic response element. In D. melanogaster, only TFFM IDs are provided for models built based on a D. melanogaster PWMs. *For HSF, HIF1, MTF-1 and XBP1 a vertebrate PWM was used.
MATERIALS AND METHODS
Trancription factor binding motifs (TFBMs) predictions based on PWMs
To determine the relative contribution of TEs to the six stress regulatory networks analyzed, we first quantified the presence of motifs for several stress-related transcription factors (TFs) in D. melanogaster and in humans, at a genome-wide level (Table 2). We then checked how many of those TFBMs were located in the TEs annotated in the reference genomes of the two species. To test whether there was enrichment of TFBMs in TEs, we used the binomial test and Bonferroni correction. We compared the number of motifs/sites predicted in TEs with the number expected if motifs/sites were distributed randomly in the genome, taking into account that 5.45% of the D. melanogaster genome and 45.5% of the human genome are TEs. Besides the P-value, we also considered the fold enrichment, as the number of motifs/sites predicted is very high.
Table 2.
Prediction of binding motifs (TFBMs) and binding sites (TFBSs) in D. melanogaster and humans
| TFBMs | TFBSs | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| PWMs | TFFMs | Chip-seq | |||||||
| Transcription factors | Number (TEs/Genome) | % | P-value | TEs | Ratio TE / background | Number (TEs/genome) | % | P-value | Merged TFBMs/TFBSs |
| (A) D. melanogaster | |||||||||
| CNC | 1832/ 34 558 | 5.3 | 1 | – | – | – | – | 1573 | |
| DEAF1 | 10 735/ 219 557 | 4.89 | 5.72e−31 | – | – | – | – | 9042 | |
| MTF-1* | 2223/ 29 964 | 7.42 | 2.62e−45 | – | – | – | – | 1839 | |
| NUB | 8666/ 181 721 | 4.77 | 5.70e−38 | – | – | – | – | 7335 | |
| XBP1* | 528/ 10 402 | 5.08 | 0.86 | – | – | – | – | 458 | |
| caudal | 7068/ 123 046 | 5.74 | 5.87e−05 | 1519 | 0.64 | 5907 / 35 630 | 16.58 | >1e−323 | 8567 |
| dorsal | 5427/ 116 125 | 4.67 | 7.46e−32 | 4579 | 1.16 | 985 / 2883 | 34.17 | >1e−323 | 7555 |
| HSF* | 480/ 7354 | 6.52 | 6.78e−4 | 734 | 1.86 | 1643 / 4493 | 36.57 | >1e−323 | 2191 |
| tango (HIF1B)* | 2754/ 62 228 | 4.43 | 3.32e−30 | 1119 | 1.97 | 4349 / 15 238 | 28.54 | >1e−323 | 4382 |
| Total | 39 713/ 784 955 | 5.06 | 2.2e−16 | 7995 | – | 12 884 / 58 244 | 22.33 | 2.2e-16 | 42 942 |
| (B) Humans | |||||||||
| CREB1 | 1 462 850/ 2 434 226 | 60.10 | 3.95e−322 | 308 156 | 0.89 | 2317/ 15 908 | 14.56 | 3.95e−323 | 1 627 554 |
| EGR1 | 434 593/ 1 169 693 | 37.15 | 2.77e−322 | 196 187 | 1.15 | 9972/ 36 982 | 26.96 | 3.95e−323 | 509 377 |
| FOS | 324 072/ 747 204 | 43.37 | 1.36e−309 | 630 618 | 0.89 | 45 748/ 92 352 | 49.54 | 4.4e−130 | 370 407 |
| HSF1 | 83 286/ 211 771 | 39.33 | 1.18e−322 | 338 290 | 0.69 | 343/ 1432 | 23.95 | 3.82e−63 | 325 915 |
| NFE2L2 | 298 168/ 571 695 | 52.16 | 1.98e−322 | 377 740 | 0.95 | 639/ 744 | 85.89 | 1.42e−115 | 505 947 |
| NFKB1 | 30 447/ 49 199 | 61.89 | 3.95e−323 | 180 383 | 1.47 | 12 638/ 28 678 | 44.07 | 4.62e−6 | 161 213 |
| NRF1 | 26 327/ 127 953 | 20.58 | 7.9e−323 | 28 857 | 0.88 | 259/ 4511 | 5.74 | 3.95e−323 | 37 708 |
| SP1 | 903 287/ 1 929 185 | 46.82 | 1.53e−279 | 138 185 | 1.94 | 4463/ 15 104 | 29.55 | 3.95e−323 | 847 478 |
| Total | 3 563 030/ 7 240 926 | 45.54 | 2.2e−16 | 2 198 416 | – | 76 379 / 195 711 | 39.02 | 2.2e−16 | 4 385 599 |
*TFs for which a vertebrate PWM was used.
Number of PWMs and ChIP-seq peaks (TFBSs) predicted in TEs/number predicted in the genome. For TFFMs, the number of predictions in TEs, and the ratio of predictions in TE versus background sequences is given. The merged TFBMs/TFBSs column shows the number of unique motifs/sites after considering the overlapping of coordinates between PWM, TFFM and ChIP-seq peaks predictions.
We made motif predictions using TFBSTools (26) against version 6.04 of D. melanogaster genome, including all 5416 annotated TEs, and against version hg38 of the human genome. We downloaded the repetitive elements track from UCSC for hg38. After filtering out low complexity regions, simple repeats and other non-TE sequences such as snRNA and tRNA, we ended up with a dataset of 4 510 651 annotated TEs. These TEs belong to 1084 different families, and 36 superfamilies.
Each TF motif has a different length and different information content, thus we will obtain more predictions just by chance for shorter motifs, or motifs with several positions with low information content such as DEAF1 or caudal (Supplementary Figure S1). Thus, a single threshold score for all TFs will not suffice. We calculated an adjusted score threshold for each TF, which takes into account the background nucleotide frequencies of the genome and the relation between false positives and false negatives (Table 1). This threshold has a relation between the false-positive rate and the false-negative rate of 1000 (fnr/fpr = 1000). The threshold calculation was done using the ‘motifs’ library included in BioPython. Motif plots (PWM, TFFM) were done using the ggseqlogo R package (27).
In D. melanogaster, we also extracted the coordinates of genes in the areas surrounding TEs from the Flybase annotation (28). We obtained the gene structure (promoters, UTR’s, exons, and introns) along with the parental relations between genes and their transcripts parsing the Flybase annotation with an in-house script (28).
Construction of TFFMs for D. melanogaster and predictions based on TFFMs
We built a TFFM for each of the four datasets with available ChIP-seq data in D. melanogaster (Supplementary Table S1) and downloaded the TFFMs built for stress-related TFs in humans (11). TFFMs were constructed using a seed motif that is trained with ChIP-seq peaks enriched for motifs of the desired TF (see ChIP-seq data processing section below). For each peak detected by MACS2, we extracted 500-bp, 250-bp region at each side of the summit, using an in-house python script and we used RepeatMasker to mask the repetitive regions. With those sequences, we run meme-chip (29) to obtain the enriched motifs. Meme-chip uses the central 100 bp of input sequences to look for motifs and the rest of the sequence as background. We also enabled the -centrimo-local option and used the JASPAR_CORE_2016 as a target database for CentriMo and TOMTOM (30). In addition, we limited the number of sequences to pass to MEME suite to 2000 (-nmeme). Taking the best motif found by MEME suite, and the sequences from the ChIP-seq peaks we generated the TFFMs.
We used the different TFFMs to run predictions in each of the TEs annotated in D. melanogaster and human genomes and kept predicted TFBMs with a score better than 0.90. If two predictions were overlapping, we kept the one with the best score. We also predicted TFBMs using TFFMs in a set of background sequences matching the GC content of the TEs. We generated the background sequences using BEDtools (bedtools random -l 1000 -n 2000000) and the BiasAway script to adjust for GC content (31). The ratio TE / background was obtained dividing the number of TFBMs every 10 kb in both datasets. If we found the same number of TFBMs in background sequences and in TEs, the ratio is 1. A higher ratio means we found more TFBMs in TEs and a ratio lower than one means that we found more TFBMs in background sequences.
ChIP-seq data processing
For D. melanogaster, we processed the raw data instead of just using the TFBSs regions reported by the authors to ensure fair comparisons across datasets, and to overcome one of the main limitations of ChIP-seq traditional pipelines: the use of uniquely mapping reads that make it very difficult to detect binding regions in TEs. Multi-mapping read allocation allows the detection of binding regions in repetitive regions and improves detection of peaks in mappable regions (32). This approach is based on allocating multi-reads or reads that map to multiple location as fractional counts weighting every alignment.
We found high quality ChIP-seq datasets for four D. melanogaster TFs. We classify a ChIP-seq as high quality if (i) it has good quality reads, (ii) it has no major red flags (FastQC inspection), (iii) it includes an ‘input’ in the experimental setup and (iv) the cross-correlation profile (SPP package) yielded a clear fragment length to continue with the analysis. ChIP-seq experiments for each TF and its corresponding control were downloaded from NCBI (Supplementary Table S2). We mapped the reads to version 6.04 of D. melanogaster genome using Bowtie (-v = 2, m = 99) (33). We used CSEM to assign multi-mapping reads (32). For each sample, we run a cross-correlation analysis using SPP R package for ChIP-seq experiment quality assessment and choose an appropriate fragment length for running MACS2 peak calling software (34). The peak calling with MACS2 was done using the BAM files processed with CSEM for the ChIP-seq experiment (-t) and the input as control. We enabled the –no-model and -extsize 200 parameters. For one experiment (caudal), we used the calculated fragment length (123) instead of 200, because it yielded a higher number of peaks with identifiable motifs. For each TF, we merged peaks retrieved from replicas or different developmental stages into one single file using BEDtools.
For humans, we downloaded eight TFs ChIP-seq datasets from the ENCODE project along with the TFFMs that were constructed based on them (Table 1). All the narrowPeak files coordinates were converted from hg19 to hg38. We calculated the overlap with our set of human TEs with an in-house python script and BEDTools.
TE family enrichment
To calculate the enrichment score we use, the following formula as in Sundaram et al. (2014) (5):
lor = log2((Number of TFBS in all TE copies / Total length of TE family (Kb)) / (Number of TFBS in the genome / genome size (Kb)))
In D. melanogaster, we removed nested TEs to avoid counting twice the same TFBS, ending up with a dataset of 3768 TEs. We focused on the 55 families with high copy number: at least 20 genomic copies. We also required a total length for the TE family of 1 Kb. For the ChIP-seq family enrichment, we also required a minimum of five ChIP-seq peaks in a family. We only consider peaks to belong to a TE if the peak overlaps at least 75% with the TE. In humans, we followed the same strategy used in D. melanogaster, but we required a family to have at least 50 copies. In total, we analyzed 1084 families. In both species, we used a threshold of 1.5 in lor score, which equals 2.83 more TFBSs in TEs than in the rest of the genome.
Overlap of TFBMs and TFBSs
We used BEDTools (35) and an in-house python script to merge the TFBMs/TFBSs coordinates from the three different sources, PWM, TFFM and ChIP-seq peaks, into single regions.
Open chromatin and CBP binding experimental data
We collected up to 12 ATAC-seq and FAIRE-seq experiments and one ChIP-seq CBP experiment (36,37). We converted the coordinates to the v6 D. melanogaster genome and checked which TEs overlap with known open-chromatin regions or contain a CBP peak. Overlapping with open chromatin regions and permutation tests were done using regioneR (38).
Epigenetic marks experimental data
The histone modification regions come from ChIP-seq data with very high coverage (39). The peaks were called by Jung et al. (2014) (39) using 100 million uniquely mapping reads for H3K4me3, H3K36me3 and the input. We converted the ChIP-seq peak coordinates to v6.04 of the D. melanogaster genome. These experiments were done in the Oregon strain. However, in this work we focused on the TEs annotated in the reference strain (y1;cn1, bw1, sp1). To obtain a list of TEs present in the Oregon strain, we run the presence module from T-lex2 (40) with DNA-seq data from modENCODE for the three experiments done with the Oregon strain (SRP045325). We consider a TE to overlap an epigenetic mark if it shares nucleotides with the TE and also with the nucleotides located left or right of that TE (±1000 bp). Currently, we can only estimate with confidence the presence of 3894 out of the 5416 TEs annotated in the reference genome. We found that 2798 of these TEs were present in the Oregon strain; for those TEs, we analyzed the presence of epigenetic marks.
Evidence of selection
We used the list of TEs with evidence of selection reported in Rech et al. (2019) (41). In addition, we also considered TEs with evidence of positive selection based on iHS, H12, nSL and/or FST and located in low recombination regions that were not included in Rech et al. (2019) and were identified using exactly the same procedure (Supplementary Table S3) (41).
TFBS ratio
The TFBS ratio was calculated dividing the expected TFBS in a TE given its length, using the glm from Supplementary Figure S2, by the number of TFBS found in a TE. For example, a TFBS ratio of 1.2 means that we find 20% more TFBS than expected in a given TE.
In vivo enhancer assays
Fly husbandry
Flies were kept at 25°C, with 12-h light and dark cycles, and 60% humidity. DGRP (Drosophila Genetic Reference Panel) strains were used for generating the transgenic constructs (42).
Construct design
For three TEs, FBti0019012, FBti0061428 and FBti0019309, we amplified only the TFBS containing part of the TE. For FBti0019197 and FBti0019985, we amplified all the TE sequence. For FBti0019978, FBti0019082, FBti0061578, FBti0019386 and FBti0019453, we amplified the intergenic region containing the TE and the intergenic region without the TE. In both cases, the intergenic region was the 500 bp region on both sides of the insertion. Finally, for FBti0018880 we cloned three regions: only the TFBS containing part of the TE, the intergenic region with the TE and the intergenic region without the TE. We checked the polymorphism in several DGRP lines with and without these insertions using the online database POPDROWSER, and chose the two most similar strains for the amplification (43). For the fixed TEs, we amplified the two sides of the insertion separately and joined them with a PCR step.
We also generated transgenic flies to be used as positive controls for immunity (44), heat-shock (45) and for oxidative stress (46) (Supplementary Table S4). As negative controls, we generated transgenic flies with the empty vectors. The primers used to amplify all the regions under study are reported in Supplementary Table S4.
Genomic DNA was extracted with the Puregene Cell and Tissue Kit (QIAGEN) and expand high fidelity Taq DNA polymerase was used for DNA amplification (Sigma).
Embryo microinjections
We purified the vector with the GeneEluteTM Plasmid Miniprep kit (Sigma) and prepared the injection mix at 6 μg vector concentration diluted with injection buffer (5 mM KCl, 0.1 mM sodium phosphate, pH 6.8) We microinjected the constructs with the Eppendord Femtojet 4i microinjector into a D. melanogaster strain with a stable docking site (Bloomington Stock number: 24749). Flies were crossed until homozygous flies for the insertion were obtained. The insertion of the construct was verified by PCR and sequencing. We generated three independent stocks that were used as biological replicates for the qPCR experiments.
Stress experiments
All experiments were performed with three biological replicates of thirty 5 to 8 day-old females.
Oxidative stress
Flies were placed on 1.5% agar and 5% sucrose with (stress) and without (non-stress) 10 mM Paraquat (Fisher Scientific) and kept at 25°C for 12 h. After that, guts were dissected, flash frozen in liquid nitrogen and stored at −80°C until RNA extraction.
Xenobiotic stress response
Scintillation vials (Labbox) were coated with a solution containing 200 μl of acetone and 50 μg dichlorodiphenyltrichloroethane (DDT) mixture. Each vial was rolled until the acetone evaporated. The vials were sealed with cotton balls soaked with 1 ml of 5% sucrose solution as a source of food and water (47). Flies were then kept at 21°C for 1,5 h because the efficiency of the DDT is higher at lower temperature (48). RNA was extracted from the whole fly.
Immune stress
Flies were infected with Pseudomonas entomophila, a gram-negative bacteria that infects D. melanogaster in the wild (49). Prior to the infection, flies were starved for 2 h. Then, they were placed in vials containing food and a piece of filter paper soaked with 1.25% of sucrose and bacterial pellet. The bacterial preparation was adjusted to a final OD600 = 50–100 (50). Flies were placed to the optimal infection conditions of the bacteria (29°C and 65% humidity) for 10–12 h. The non-infected flies were exposed to LB medium and 1.25% sucrose on the filter paper. After 10–12 h depending on the strain, guts were dissected, flash frozen in liquid nitrogen and stored at −80°C until RNA extraction.
Heat-shock stress
Flies were placed in empty vials in a water bath at 36°C followed by a 1 h recovery time at room temperature (25°C) (51). After the treatment, flies were flash frozen with liquid nitrogen. The non-treated flies were kept at room temperature for the same period of time and were flash frozen with liquid nitrogen. Samples were stored at −80°C until the RNA extraction. RNA was extracted from the whole fly.
RNA extraction and cDNA synthesis
RNA was extracted using GenElute™ Mammalian Total RNA Miniprep Kit (Sigma Aldrich). We treated the RNA with DNAse I (Thermo Fisher Scientific) after the extraction. cDNA was synthesized from 500 to 1000 ng of RNA using the NZY First-Strand cDNA Synthesis Kit (NZYTECH).
qRT-PCR
Expression was measured using SYBR Green master mix (BioRad) on an iQ5 Thermal cycler. Results were analyzed using the ddCT method and the two-tailed Student’s t-test (52).
RESULTS
We focused on six evolutionary conserved stress responses that are relevant for D. melanogaster and human adaptation: heat-shock, oxidative, hypoxia, immune, xenobiotics and heavy-metal stress (Table 1). Through literature searches, we identified the transcription factors (TFs) involved in these six stress responses. In Drosophila, we analyzed nine TFs available in JASPAR (12). When the D. melanogaster motif was not available, we used the vertebrate motif, as stress-related TFs are thought to be highly conserved (15). For example, there is functional data showing that human MTF-1 can restore to a large extent metal tolerance to flies lacking their own MTF-1 gene (53). Indeed, we found that genes that have previously been reported as heavy-metal responsive in D. melanogaster contained binding motifs for MTF-1 predicted with the human motif (Supplementary Table S2). In humans, we analyzed the eight TFs that were available in the ENCODE project, and PWMs were downloaded from HomerMotifDB (54) (Table 1).
Besides predicting transcription factor binding motifs (TFBMs), when available we used ChIP-seq data to identify transcription factor binding sites (TFBSs). All the TFBS predictions generated in this work are available at http://dx.doi.org/10.20350/digitalCSIC/8590
TEs contain stress-related TFBMs in D. melanogaster and in humans
We used two different approaches to identify TFBMs: Position Weight Matrices (PWMs) and Transcription Factor Flexible Models (TFFMs) (Table 1, see ‘Materials and Methods’ section). While PWMs consider that the nucleotides within the TFBMs are independent, TFFMs take into account nucleotide interdependencies and allow for gaps, which improve the identification of some TFBMs (11).
PWMs predictions
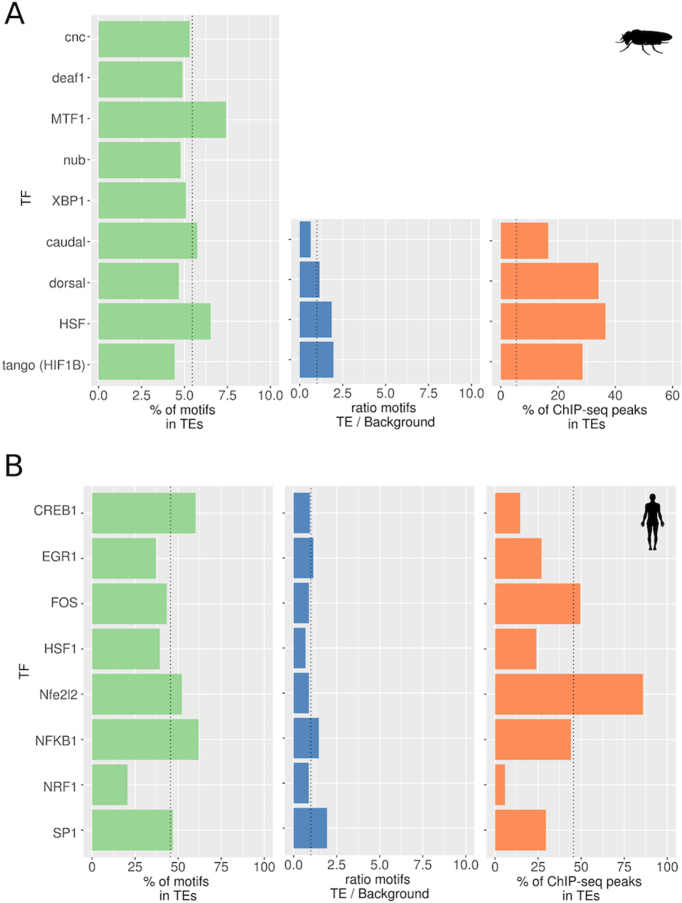
For all TFs, we predicted motifs using PWMs with the software TFBSTools (26), and we adjusted the score threshold for each TF (Table 1 and Supplementary Figure S1A; see ‘Materials and Methods’ section). Overall, the percentage of TFBMs in TEs appears to be small in D. melanogaster (4.43–7.42%, Table 2A). The 5416 TEs annotated in the reference genome represent 5.45% of the euchromatic fraction of the D. melanogaster genome, and on average 5.06% of TFBMs are located in TEs suggesting that overall TEs contained a similar number of TFBMs than expected if motifs were randomly distributed in the genome (Figure 1A and Table 2A). Only MTF-1 motifs were slightly enriched in TEs (1.4-fold enrichment, P-value = 2.62e−45, Table 2A). We tested whether removing the INE-1 family from the analyses affected these results. While the majority of D. melanogaster TE families are active or have been recently active, and contain from a few to 100 copies, the INE-1 family contains ∼2000 copies and has been inactive for the past ∼3- 4.6 million years (55–57). We found that overall, non-INE-1 TEs were not enriched for TFBMs either (Supplementary Figure S3).
Figure 1.
Percentage of transcription factor binding motifs (TFBMs) and ChIP-seq peaks (TFBSs) located in TEs in (A) Drosophila melanogaster and in (B) Humans. In green, motif predictions using position weight matrix (PWMs). The vertical dotted line depicts the expected percentage of motifs in TEs in D. melanogaster (5.45%) and human (45.54%). In blue, ratio of number of motifs predicted in TEs and number of motifs predicted in background sequences with the same properties than TEs. The expected ratio is 1 (vertical dotted line). In orange, percentage of ChIP-seq peaks located in TEs. The expected percentages of TFBSs falling in TEs are represented as vertical dotted lines as in the PWM predictions.
In humans, we also focused on the TEs annotated in the reference genome: 4 510 651 TEs. We found that the percentage of predicted TFBMs inside TEs was quite variable, from 21% to 62% (Figure 1B and Table 2B). Some TFs such as CREB1 or NFKB1 have slightly more TFBMs within TEs than expected considering that TEs constitute 45.5% of the human genome (1.3-fold enrichment, P-value = 3.95e−322 and 1.4-fold enrichment, 3.95e−323, respectively, Figure 1B and Table 2B).
TFFMs predictions
For D. melanogaster, we constructed TFFMs for the four TFs for which ChIP-seq data are available (Supplementary Table S1 and see ‘Materials and Methods’ section). The number of predicted binding motifs in D. melanogaster TEs for all TFs was smaller compared to the PWM predictions except for HSF (Table 2A). This can be partially explained because this motif has one gap at positions 9–10 (Supplementary Figure S1), and as mentioned before, TFFMs are able to handle small gaps. In addition, PWM predictions for HSF were made using the human PWM, while for the TFFM we used D. melanogaster ChIP-seq data. We also predicted motifs in a set of background sequences and estimated the ratio of predictions in TEs versus the background sequences (see ‘Materials and Methods’ section). The ratio was between 0.64 and 1.97 depending on the TF analyzed (see ‘Materials and Methods’ section; Table 2A and Figure 1A).
In humans, TFFMs were available for all eight TFs analyzed (Table 1 and Supplementary Figure S1B). The number of TFBMs predicted using TFFMs was quite variable (Table 2B). Similar to the results obtained with D. melanogaster, the ratio also varied depending on the TF analyzed (see ‘Materials and Methods’ section; Table 2B and Figure 1).
Overall, our results suggest that TEs contain a variable number of bindings sites for the TFs studied in D. melanogaster and in humans. Only for some TFs, we did find a slight enrichment of binding sites in TEs (Figure 1 and Table 2).
TEs are enriched for some stress-related TFBSs in D. melanogaster and humans
Not all the predicted TFBMs will be actively bound by their corresponding TFs (58,59). Thus, besides TFBMs we searched for TFBSs using available ChIP-seq datasets (Supplementary Table S1).
In D. melanogaster, there is ChIP-seq data available in non-stress conditions for four of the nine TFs studied (Table 2A). Based on these data, we retrieved a total of 58 244 TFBSs, of which 12 884 were located within TEs (Table 2A and Figure 1A). This is one order of magnitude less than the total number of predicted motifs with PWMs: 784 955. This suggests that most of the TFBMs predicted would not be bound by the TF, at least in the conditions and developmental stages in which the ChIP-seq experiments were performed. The number of TFBSs varies among TFs, which could be partly explained by the different number of experiments analyzed (Supplementary Table S1). While the number of TFBMs in TEs was overall not higher than expected if motifs were randomly distributed in the genome, when we looked at the ChIP-seq peaks, up to 37% of them occur in TEs (6.7-fold enrichment, P-value < 1 e-323), with an average of 22% (4.1-fold enrichment, P-value = 2.2e−16, Table 2A and Figure 1A).
In humans, there are ChIP-seq data available for all eight TFs studied (see ‘Materials and Methods’ section). Overall, the proportion of TFBSs occurring within TEs is smaller than expected for all TFs, except for NFE2L2 (1.9-fold enrichment, P-value = 1.42e−115, Table 2B and Figure 1B). For HSF1, we also analyzed a ChIP-seq dataset obtained in stress conditions (60). In non-stress conditions, 22.94% (120 out of 523) of the peaks were located inside TEs, while in stress conditions 32.06% (680 out of 2121) of the peaks were inside TEs, suggesting that we might be underestimating the number of peaks in TEs by analyzing non-stress conditions. While more TFBS were identified in stress conditions compared to non-stress conditions, 71% (369 out of 523) of the peaks found in non-stress conditions are present in stress conditions, suggesting that most of the peaks in non-stress conditions were also present in stress conditions.
Overall, we found that TEs are enriched for caudal, dorsal, HSF and tango binding sites in D. melanogaster and for NFE2L2 binding sites in humans (Figure 1 and Table 2). Our results also suggest that we might be underestimating the number of binding sites in TEs since we analyzed ChIP-seq experiments performed in non-stress conditions.
TE families, superfamilies and classes are enriched for different TFBMs/TFBSs in D. melanogaster and humans
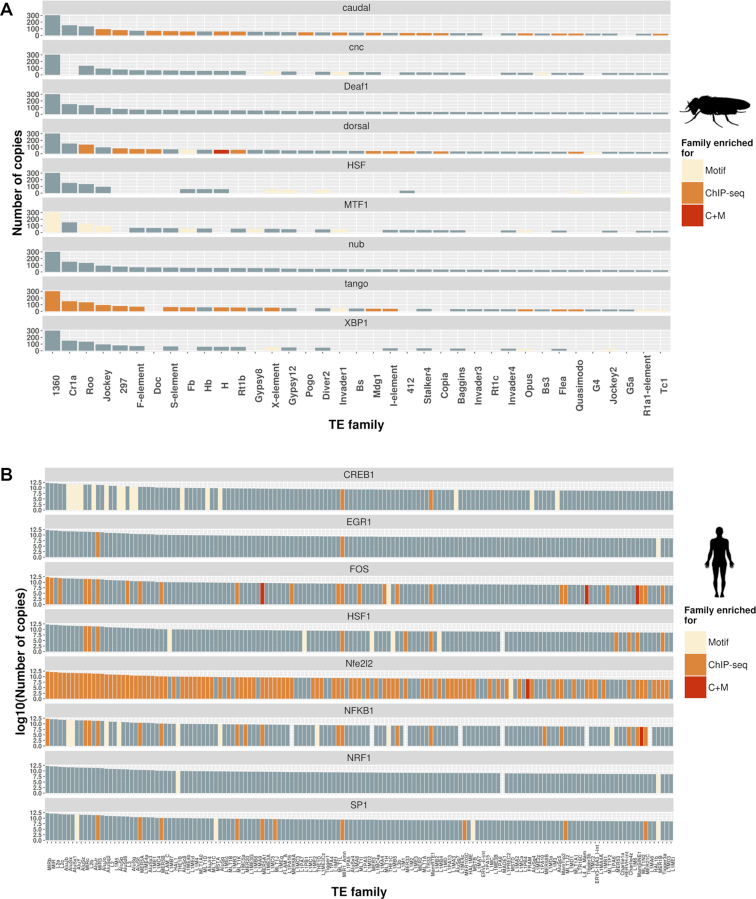
It has been described that particular TE families and TE classes are enriched for TFBSs (5,8). Thus, we measured the enrichment of TFBMs/TFBSs in TEs at the family, superfamily and class levels. In D. melanogaster, we found 14 families enriched both for TFBMs and TFBSs (Supplementary Table S5A and B). If we take into account the copy number of the families enriched for a certain TF, the three largest TE families (1360, Cr1a and roo) were enriched for tango (HIF1B) TFBSs, suggesting that these families could significantly contribute to the spreading of hypoxia response elements (HRE) in the D. melanogaster genome (Figure 2A). At the superfamily level, only TEs that belong to the P and BEL superfamilies were enriched both for TFBMs and TFBSs (Supplementary Table S5C and D). Finally, at the class level, LTRs, nonLTRs, and DNA transposons were enriched for TFBSs for at least one TF (Supplementary Table S5E). Note that LTRs are known to be enriched for TFBSs in human and mouse (5,8).
Figure 2.
Several TE families are enriched for stress-related transcription factor motifs and binding sites. (A) The number of genomic copies for D. melanogaster TE families with at least 25 copies is represented. Families are painted depending on whether they are enriched for motifs, ChIP-seq peaks, or both (C+M). Absent columns for a particular TF indicate that the score could not be calculated due to lack of sufficient motifs or peaks. (B) Equivalent figure for humans. The number of copies is given in log scale due to the high number of copies of some families. Only families with more than 5000 copies are plotted.
In humans, 214 families were enriched both for TFBMs and TFBSs (Supplementary Table S6A and B). The five families with the highest copy numbers were enriched for FOS, NFE2L2 and/or NFKB1 binding sites suggesting that these families could significantly contribute to the spreading of these three response elements (Figure 2B). At the superfamily level, six superfamilies were enriched both for TFBMs and TFBSs (Supplementary Table S6C and D). Finally, SINE were enriched both for TFBMs and TFBSs, while DNA transposons, LTRs, and LINEs were enriched for TFBSs (Supplementary Table S6E and F).
Overall, both in D. melanogaster and in humans, we found enrichment for different TFBMs/TFBSs at the family (Figure 2), superfamily and class levels (Supplementary Tables S5 and S6) suggesting that they could significantly contribute to the TFBMs/TFBSs repertoire in D. melanogaster and in humans.
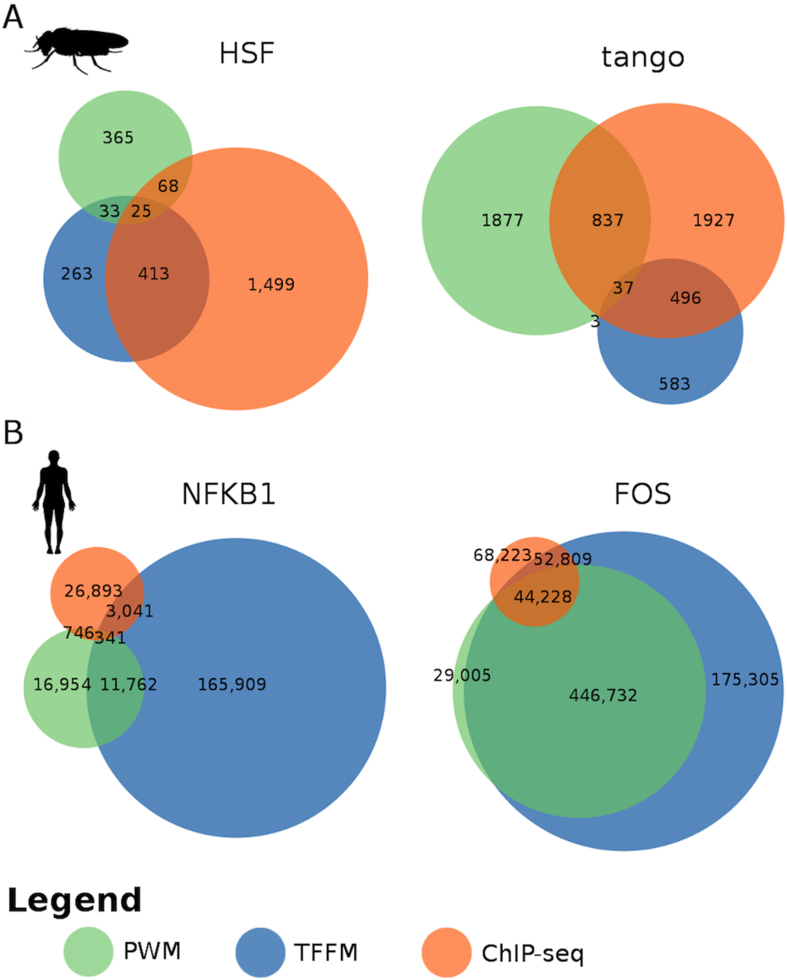
The overlap between TFBMs and TFBSs predictions varies among stress-related TFs in D. melanogaster and humans
To identify the unique TFBMs/TFBSs in TEs, we checked the overlap among the predictions of the three methodologies used. The overlap between PWMs and TFFMs was in general low for all TFs in both species (Figure 3 and Supplementary Figure S4). If we consider the ChIP-seq peaks as true binding events (not necessarily functional), we observed that neither PWM nor TFFM predictions alone are able to predict all binding sites for a given TF (Figure 3 and Supplementary Figure S4). As mentioned above, only for some TFs, such as HSF in D. melanogaster, TFFMs outperformed PWMs at predicting motifs (Figure 3A). In humans, only FOS showed a high overlap between motif predictions and ChIP-seq peaks, while for other TFs the overlap was quite small (Figure 3B and Supplementary Figure S4B).
Figure 3.
Overlap of TFBMs and TFBS predictions. Venn diagrams showing the overlap in the predictions across methods (PWM, TFMM, and ChIP-seq) within TEs for representative transcription factors in panel (A) D. melanogaster and panel (B) humans. A motif/peak is considered as shared if there is overlap in their coordinates. Note that a ChIP-seq peak can overlap with several motifs.
The fraction of ChIP-seq peaks for which we could not predict a motif with either PWMs or TFFMs might be explained by indirect binding through another TF, undiscovered minor motifs, or unspecific binding (5,61). Similarly, the fraction of motifs predicted by PWMs or TFFMs that did not overlap with a ChIP-seq peak could be explained because ChIP-seq data were obtained in non-stress conditions, and for a few developmental stages. Thus, because all three methods have limitations, to obtain the unique number of TFBMs/TFBSs identified, we merged the predictions for those TFs where we had multiple sources of motif and/or binding predictions (Table 2).
For the rest of this work, we focused on D. melanogaster TFBMs/TFBSs predictions as our ultimate goal was to test whether a subset of TFBMs/TFBSs were functional by using in vivo reporter gene assays.
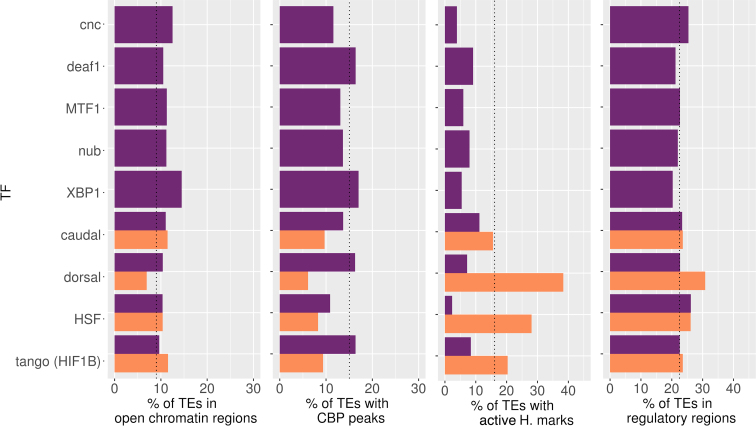
TEs containing TFBMs/TFBSs are not globally enriched for enhancer and/or promoter distinctive features
To further investigate the potential role of TEs with predicted TFBMs/TFBSs as enhancers or promoters, we checked whether these TEs were enriched for several distinctive features associated with these regulatory regions: location in open chromatin, co-binding of CREB-binding protein (CBP), and presence of active histone marks. We also checked the genomic location of the identified TEs. We considered all the TEs with at least one predicted TFBMs/TFBSs (3593 TEs) and the TEs with three or more TFBMs/TFBSs (2183 TEs) as it has been shown that functional regulatory regions tend to be bound by multiple related TFs, usually three or more (58,62,63). Indeed, we found that the number of unique predicted TFBMs/TFBSs correlates very well with TE length: most TEs have at least one motif prediction if they have a minimum length of 220 bp (Supplementary Figure S2).
Active transcription has been linked to changes in nucleosome organization in regulatory elements due to TF binding (36). Thus, identifying motifs located within open chromatin regions should be an effective strategy to identify functional binding sites (64). By combining, all ATAC-seq and FAIRE-seq experiments performed in Davie et al. (2015) and Koenecke et al. (2016), we obtained 36 507 distinct open chromatin regions (36,37). Only 637 open chromatin regions were detected inside TEs, corresponding to 489 unique TEs. This overlap is much smaller than expected by chance (permutation test, P-value = 0.0002, Supplementary Figure S5), suggesting that TEs in D. melanogaster do not tend to be located in open chromatin regions, as has been previously reported in humans (65). Overall, TEs containing one or more TFBMs/TFBSs were not preferentially located in open chromatin regions if we consider each one of the TFs independently (Figure 4 and Supplementary Table S7) or altogether (Supplementary Table S8). The same result was obtained for TEs containing three or more TFBMs/TFBSs (Supplementary Table S8). The only exceptions were TEs containing TFBMs/TFBSs for XBP1, which were slightly enriched in open chromatin regions (14.49% versus 9.04%, P-value = 0.04, Supplementary Table S7).
Figure 4.
Enhancer/promoter genomic characteristics in TEs with predicted TFBMs/TFBSs in D. melanogaster. Percentage of TEs with at least one TFBMs/TFBSs for each one of the nine transcription factors studied overlapping with (A) open chromatin regions, (B) containing a CBP peak, (C) enriched for active histone marks or (D) located in a regulatory region. In purple, merged dataset of TFBMs/TFBSs and in orange dataset with evidence from ChIP-seq. The vertical dotted line showed the expected percentage for each feature.
We also looked for evidence of co-binding of CBP, which has a role as an activator of several TFs, some of them related to different stress responses, such as CNC (66), HSF (67), HIF1A (68), MTF-1 (69), or immune response (70). We identified 815 TEs that contain a CBP-binding region. We did not find significantly more CBP interactions in TEs that have one or more TFBMs/TFBSs (Figure 4 and Supplementary Table S7), while we see a depletion of CBP peaks in TEs that contain one or more ChIP-seq peaks for a stress TF (Figure 4 and Supplementary Table S7). Overall, TEs with one or more TFBMs/TFBSs and TEs with three or more TFBMs/TFBSs are not enriched for CBP binding sites (Supplementary Table S8).
Binding of TFs to TEs has been found to be strongly associated with the epigenetic status of a TE (5,71). Indeed, TEs have been postulated as tissue-specific gene regulators through epigenetic modifications (72). We thus looked for the presence of two key histone modifications in TEs: H3K4me3 and H3K36me3, associated with promoters and transcriptional elongation, respectively (39, see ‘Materials and Methods’ section). We found that 286 TEs contained the H3K4me3 histone mark, and 584 TEs contained the H3K36me3 histone mark (Supplementary Table S9). We found that TEs containing TFBSs for HSF and dorsal were enriched for H3K4me3 and/or H3K36me3 (P-value = 1.11e−16 and 1.57e−18, respectively, Figure 4 and Supplementary Table S7). Note that histone marks are highly variable across cell types and strains; thus, the fraction of TEs with active epigenetic marks is an underestimation, and many more might be identified in other cell types or conditions.
Finally, we also tested whether TEs with TFBMs/TFBSs were located in proximal regulatory regions. We defined the proximal regulatory region of a gene as the 1000 bp upstream the TSS, the 5′UTR, and the first intron. Only TEs containing TFBSs for dorsal were slightly enriched in regulatory regions (P-value = 3.19e−4, Figure 4 and Supplementary Table S7).
Overall, TEs containing one or more, or three or more, TFBMs/TFBSs were not globally enriched for enhancer or promoter features (Figure 4 and Supplementary Table S8). Only TEs containing TFBMs/TFBSs for XBP1, HSF and dorsal were enriched in open chromatin regions, active histone marks and/or regulatory regions (Figure 4 and Supplementary Table S7).
TEs with three or more TFBMs/TFBSs were present at higher population frequencies
We expect TEs with functional TFBMs/TFBSs to be present at high frequencies or fixed in populations due to an increase in fitness of the individuals that carry them. We found that the proportion of TEs with one or more TFBMs/TFBSs present at high frequencies (≥10% to <95%) in populations is significantly higher than the proportion of all TEs present at high frequencies in the genome (16.2% versus 11%, P-value < 2.2e−16, Table 3). This percentage increased when we only considered TEs with three or more TFBMs/TFBSs (25.9%), and it was even higher in the subset of TEs that have ChIP-seq evidence for three or more TFBSs (42.1%, Table 3).
Table 3.
Number of TEs containing one or more, or three of more TFBMs/TFBSs present at high population frequencies or fixed
| High freq TEs | Fixed TEs (non-INE-1) | Fixed TEs (INE-1) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dataset | TE # | TE # | % | P-value | TE # | % | P-value | TE # | % | P-value |
| All TEs with frequency estimations | 3894 | 424 | 11 | NA | 855 | 22 | NA | 2234 | 58 | NA |
| ≥1 TFBSs | 2438 | 396 | 16.2 | <2.2e−16 | 621 | 25.5 | <2.2e−16 | 1086 | 44.5 | <2.2e−16 |
| ≥3 TFBSs | 1314 | 340 | 25.9 | <2.2e−16 | 386 | 29.4 | <2.2e−16 | 275 | 20.9 | <2.2e−16 |
| ≥3 Chip-seq TFBSs | 311 | 131 | 42.1 | <2.2e−16 | 12 | 3.9 | <2.2e−16 | 0 | 0 | <2.2e−16 |
For the fixed non-INE-1 TEs, we observed a small increase in the proportion of TEs with one or more TFBS present at high frequencies in populations (22% versus 25.5%, P-value < 2.2e−16, Table 3). However, we found a significant decrease when we only considered ChIP-seq peaks (3.9%, P-value < 2.2e−16, Table 3). This can be explained by the shorter length of fixed TEs that makes it more difficult to detect three relatively large non-overlapping ChIP-seq peaks in this dataset. Finally, the proportion of TEs from the INE-1 family decreased in the datasets of TEs with TFBMs/TFBSs consistent with these TEs having reached fixation in populations through neutral processes rather than positive selection (Table 3).
Overall, these results suggest that the subset of TEs containing three or more TFBMs/TFBSs, and especially those TEs with evidence coming from ChIP-seq experiments, could be enriched for functional TFBMs/TFBSs, as the proportion of these TEs present at high frequencies in populations is higher compared to all TEs in the genome. On the other hand, INE-1 elements were depleted for TEs with three or more TFBMs/TFBSs.
TEs containing three or more TFBMs/TFBSs and present at high population frequencies were enriched nearby stress-associated genes
We tested whether TEs containing three or more TFBMs/TFBSs were enriched nearby stress-associated genes. Briefly, we considered as stress-associated genes those identified in GWAS, QTL, transcriptomics and/or protein–protein interaction analysis as described in Rech et al. (2019) (41). We first confirmed that the promoters of genes that have been reported as stress-associated are enriched for the corresponding stress-associated TFBMs/TFBSs compared with the promoters of nonstress-associated genes (Supplementary Figure S6 and Supplementary Table S10).
We observed that high frequency TEs were more often located nearby stress-response genes (28.81% versus 18.93, P-value = 1e−6, Supplementary Table S11A). This association was also significant for TEs present at high frequencies and containing three or more TFBMs/TFBSs (30.31% versus 18.83, P-value = 9.16e−7, Supplementary Table S11B). Thus, TEs containing three or more TFBMs/TFBSs and present at high population frequencies are enriched nearby stress-associated genes.
TEs containing TFBMs/TFBSs affect the expression of genes that were already part of a stress regulatory network
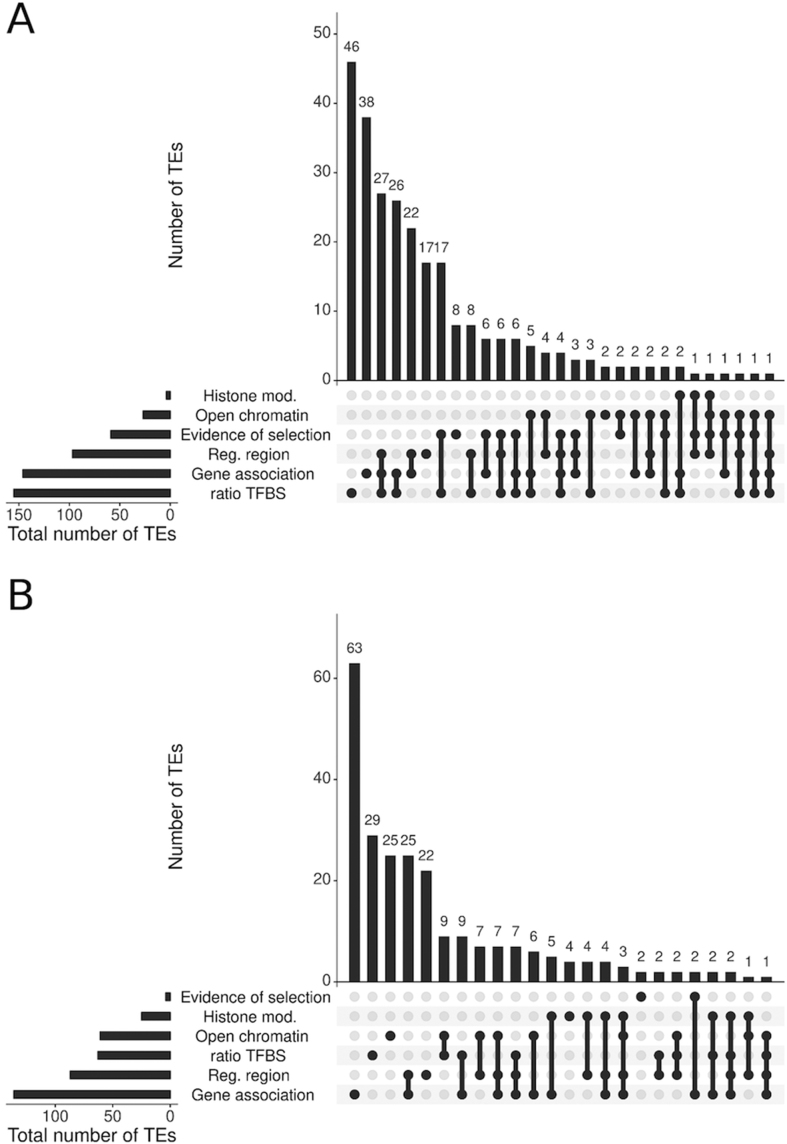
We summarized all the information suggesting that TEs could be adding functional TFBMs/TFBSs (Figure 5 and Supplementary Table S12). Based on our results, we focused on those TEs containing at least three TFBMs/TFBSs, and present at high population frequencies or fixed (non-INE-1). We considered TEs that were (i) enriched for active histone marks, (ii) located in open chromatin regions, (iii) located in regulatory regions, (iv) located nearby stress-associated genes and/or (v) have more TFBSs than expected given their total length (ratio TFBS, see ‘Materials and Methods’ section). In addition, we also considered whether there is evidence suggesting that the region flanking the TE insertion is under positive selection (41, see ‘Materials and Methods’ section). We found that 73 TEs containing at least three TFBMs/TFBSs, and present at high population frequencies or fixed (non-INE-1) showed signatures of selection in their flanking regions (Supplementary Tables S3 and S12).
Figure 5.
Characteristics of TEs containing three or more TFBMs/TFBS present at (A) high frequency or (B) fixed (non-INE-1). Histone mod: TE bears H3K4me3 or H3K36me3 marks associated with active chromatin. Open chromatin: TE is located in an open chromatin region. Evidence of selection: TEs with evidence of selection (41). Reg. region: TE is located in the proximal regulatory region of a gene (promoter, 5′UTR or first intron). Gene association: TEs located nearby stress-associated genes. Ratio TFBS: TE contains 20% more TFBS than expected given their length.
We found that 82.5% (264 out of 320) of the TEs with three or more TFBMs/TFBSs and present at high population frequencies have at least one additional line of evidence suggesting that they might be functional. This percentage is significantly smaller for fixed non-INE-1 TEs suggesting that fixed non-INE TEs are less likely to contain functional TFBMs/TFBSs (63%, 243 out of 386, chi-square P-value < 0.0001).
We chose 11 TEs with at least one additional line of evidence to perform in vivo enhancer reporter assays (Table 4). We also included in Table 4 three TEs, tdn8 (transpac), FBti0020057 (BS) andFBti0018868 (297), which were previously tested in our laboratory (44). All these TEs contained three or more TFBMs/TFBSs, except FBti0019453 (jockey) that contained two and FBti0020057 (BS) that contained one (Table 4). The majority of them were present at high population frequencies, except FBti0019197 (Tc1) and FBti0061578 (baggins) that were fixed. Based on the TFBMs/TFBSs added by the TE and on the functional information available for the nearest gene, we tested the role of these TEs in four stress responses: heat-shock, oxidative, xenobiotic and immune (Table 4). Seven of the 14 TEs were tested in two stress conditions.
Table 4.
Results summary for the in vivo enhancer assays performed
| TFBS/TFBM | q-PCR result (t-test P-value) | |||||||
|---|---|---|---|---|---|---|---|---|
| TE Family Class | TE | Reg. region | Additional evidence | Experimental design | Stress tested | Control | Treated | Reference |
| FBti0019386 Invader4 LTR | DEAF1: 1 CAD: 1 tango:1 | CAD: 2 NUB: 1 DEAF1: 1 dorsal: 1 | Regulatory region CBP TFBS ratio Histone marks Selection evidence | Intergenic | IRE | No | Up-regulation (8.91E-05) | This work |
| FBti0019082 Rt1b non-LTR | CAD: 1 DEAF1: 3 MTF-1: 3 CNC: 3 dorsal: 2 | NUB: 2 XBP1: 1 | Regulatory region Open chromatin CBP Histone marks Selection evidence | Intergenic | IRE | No | Down-regulation (0.033) | This work |
| FBti0019985 roo LTR | DEAF1: 1 NUB: 1 MTF-1: 1 dorsal: 1 | DEAF1: 1 | Regulatory region Selection evidence TFBS ratio | TE/antisense | IRE | No | Up-regulation (0.0126) | This work |
| TE/sense | No | Up-regulation | (44) | |||||
| tdn8 transpac LTR | NUB: 2 DEAF1: 4 | NA | Regulatory region | Intergenic | IRE | No | Up-regulation (0.046) | (44) |
| FBti0020057 BS non-LTR | NUB: 1 | NUB: 3 CAD: 1 DEAF1:1 | Regulatory region Open chromatin Gene: Acbp6 Selection evidence | Intergenic | IRE | Down-regulation (0.0193) | Down-regulation (0.0161) | (44) |
| FBti0019453 jockey non-LTR | NUB: 1 CAD: 1 | NUB: 2 DEAF1:1 | Regulatory region Open chromatin Selection evidence | Intergenic | XRE | No | No | This work |
| IRE | No | Down-regulation (0.007) | This work | |||||
| FBti0019012 Pogo TIR | NUB: 4 HSF:1 tango: 2 CAD: 2 dorsal: 1 | NUB: 3 DEAF1: 1 XBP1: 1 | Regulatory region Gene: mir-31a TFBS ratio | TFBS/sense | IRE | No | No | This work |
| HSE | No | No | This work | |||||
| TFBS/antisense | HSE | No | No | This work | ||||
| FBti0019309 1360 TIR | DEAF1: 2 NUB: 3 MTF-1: 2 tango: 1 | NUB: 3 CAD: 1 DEAF1:2 dorsal:1 | Regulatory region TFBS ratio | TFBS/sense | IRE | No | No | This work |
| TFBS/antisense | HSE | No | No | This work | ||||
| FBti0018880 Bari1 TIR | CNC: 1 DEAF1: 3 NUB: 2 MTF-1: 1 HSF: 2 tango: 1 CAD: 2 dorsal: 3 | MTF-1: 1 DEAF1: 2 | Regulatory region TFBS ratio Gene: Jheh2 Selection evidence | TFBS | ARE | No | No | This work |
| Intergenic | ARE | No | No | This work | ||||
| IRE | No | No | This work | |||||
| FBti0061428 Hobo TIR | dorsal: 3 DEAF1: 3 tango: 2 CAD: 2 CNC: 1 | NA | Open chromatin Histone marks Gene: CG31809 TFBS ratio | TFBS | IRE | No | No | This work |
| HSE | No | No | This work | |||||
| FBti0019197*Tc1 TIR | tango: 1 dorsal: 1 CAD: 1 | MTF-1: 1 NUB: 1 | Regulatory region Histone marks | TE | IRE | No | No | This work |
| ARE | No | No | This work | |||||
| FBti0019978 1360 TIR | MTF-1: 2 tango: 1 CAD: 1 HSF:1 DEAF1: 1 | MTF-1: 1 CAD: 1 DEAF1: 1 | Regulatory region Open chromatin Histone marks | Intergenic | XRE | No | No | This work |
| FBti0061578*baggins non-LTR | DEAF1: 2 tango: 1 | CAD: 1 DEAF1: 1 dorsal: 1 | Regulatory region Histone Marks TFBS ratio Gene:CG2217 | Intergenic | ARE | No | No | This work |
| IRE | No | No | This work | |||||
| FBti0018868 297 LTR | DEAF1: 1 NUB: 1 CAD: 1 | NA | Regulatory region TFBS ratio Gene: TM4SF | TE | IRE | No | No | (44) |
*Fixed TEs; In bold, TFs for which the evidence for the presence of TFBSs in that particular TE comes from ChIP-seq data. Experimental design indicates the region that was cloned in front of the reporter gene. We also included the data for three reporter assays performed previously in the lab (44).
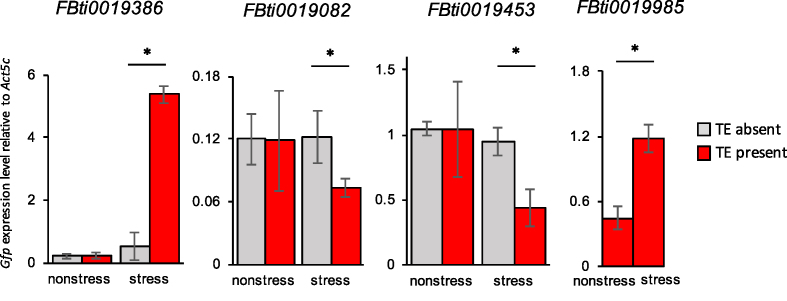
Six of the 14 tested TEs affected the expression of the reporter gene under stress conditions: three TEs up-regulated and three down-regulated the reporter gene (Table 4 and Figure 6; Supplementary Table S13). Four of the six validated TEs added TFBSs that were already present in the promoter region of the nearby gene (Table 4). For five of the six validated TEs, the intergenic region containing the TE was compared with the intergenic region cloned from a strain without the TE insertion, while in the other case only the TE was cloned and significance was determined by comparing with the empty vector (see ‘Materials and Methods’ section). On the other hand, only the TE or only the TE fragment containing the TFBMs/TFBSs was cloned for five of the eight TEs that were not validated (Supplementary Table S13). These results suggest that in most cases the TE sequence is not enough to drive the expression of nearby genes but rather modulates their level of expression.
Figure 6.
Four TE insertions analyzed in this work affect the expression of a reporter gene. qRT-PCR experiments comparing the expression of the gfp reporter gene in transgenic flies containing the genomic region under study without the TE insertion (gray) and with the TE insertions (red), in stress and non-stress conditions. The error bars represent the standard deviation of three biological replicates. Significant results are indicated with *.
Finally, for three insertions, we cloned the TE in sense and in antisense orientation. We did not find differences between the two constructs: FBti0019985 (roo) affected the expression of the reporter gene regardless of the orientation while FBti0019012 (pogo) and FBti0019309 (1360) did not affect the expression in any of the two orientations (Table 4 and Supplementary Table S13).
DISCUSSION
In this work, we showed that transposable elements (TEs) contribute to stress-related transcription factor binding motifs/sites (TFBMs/TFBSs) in D. melanogaster and in humans. This contribution is transcription factor (TF) specific, ranging from 17% to 37% in D. melanogaster and from 6% to 86% in humans (Figure 1 and Table 2). This is consistent with previous reports in humans in which the contribution of TEs was also highly TF specific (5,6). Some of the families with the highest copy number, such as 1360 and Cr1a in D. melanogaster and MIRb and L2c in humans, were enriched for TFBSs suggesting that these families could be significantly contributing to the spreading of particular stress response elements (Figure 2). Indeed, MIRs have previously been shown to contribute to functional enhancers genome-wide in mammals (73).
We showed that while D. melanogaster TEs are not enriched in open chromatin regions, TEs containing binding sites for HSF and dorsal were enriched for active histone marks (Figure 4). Histone marks are often used to identify active regulatory regions at a genome-wide level (72,74,75). Interestingly, SINEs involved in neural gene activation were enriched for active histone marks in control conditions suggesting that these insertions were epigenetically primed prior to neural activation (75). Thus, histone mark enrichment in control conditions, as we have studied in this work, could be informative about the enhancer role of TEs in specific conditions.
We also found that TEs containing three or more TFBSs had a higher proportion of TEs present at high population frequencies (Table 3), and were enriched in the promoter regions of stress-related genes, suggesting that this subset of TEs is likely to be enriched for functional TFBSs. Our results are consistent with previous studies showing that TEs containing three or more TFBSs are more likely to be functional (58,62,63). Indeed, based on the integration of ChIP-seq data for enhancer histone marks and TFs, ERVs have been shown to disproportionally overlap with genomic regions showing combinatorial binding of several TFs (76).
While we could not confirm the functional role of the two TEs that were fixed in all the populations analyzed, six of the 12 TEs present at high population frequencies were validated (Table 4). Five of these six TEs affected the expression of the nearby gene only under stress conditions suggesting that their effect is stress-response specific (Table 4). Most of these TEs, four out of six, add TFBSs that were already present in the promoter region of the nearby gene. This result suggests that rather than recruiting new genes to stress-regulatory networks, these TEs affect the level of expression of genes that were already part of the cellular stress response (Table 4).
Interestingly, all six validated TEs were either LTR or LINE elements, while most of the non-validated TEs, six out of eight, were TIR elements (Table 4). Each validated TE belong to a different family: Invader4, Rt1b, roo, transpac, BS and jockey. These results suggest a different dynamics in D. melanogaster compared with humans or mouse in which often is a particular TE family or subfamily that contributes most of the TFBSs for a given TF (72–73,75,77). It is also noteworthy that five of the six TEs that were functionally validated showed signatures of selection in their flanking regions, suggesting that the changes in expression they induced could have an adaptive effect (Supplementary Table S3). Three of these TEs are associated with down-regulation of the reporter gene (Table 4). These three insertions contain TFBMs related with immune-response TFs, with some of them involved in the negative regulation of genes in response to an immune challenge (78). Finally, we cannot discard that mechanisms other than adding TFBMs/TFBSs could affect the changes in expression of the reporter gene described in this work as TEs have been shown to affect gene expression through a variety of mechanisms (1,2).
While it is possible that the non-validated TEs are false positives, that is, TEs containing non-functional TFBSs, it could also be that these TEs are false negatives. First, in order for some TFs to be able to bind the DNA, they could require genomic context that is missing in the genomic region where the transgene is inserted. For example, it has been reported that binding of HSF to the corresponding motif sequences required the presence of active chromatin marks (79). Moreover, instead of affecting expression of nearby genes, it has been argued that TEs containing TFBSs could provide a buffer of extra binding sites to trap TFs or could serve as a landing pad to allow TFs to scan the DNA (5). Thus, although we cannot discard that the non-validated TEs are indeed non-functional, there are other possible explanations for the lack of effect of these TEs on the expression of the reporter gene. If we extrapolate our validation rate to the subset of TEs with similar characteristics, we can speculate that at least 132 reference TE insertions in the D. melanogaster genome could be adding functional TFBSs to their nearby genes. This is likely an underestimation as we only analyzed binding peaks in non-stress conditions. Thus, our results suggest that TEs are likely to be important contributors to the regulation of stress-response genes in the D. melanogaster genome. Experimental data on binding sites and chromatin features, obtained both under control and stress conditions, should help further quantify their contribution.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Gabriel E. Rech for the analysis of the signatures of selection for reference TEs located in low recombination regions, and for the analysis of the population frequencies of the TEs. We thank the Drosophila Genomics Resource center, supported by NIH grant 2P40OD010949, for sharing flies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministerio de Economia y Competitividad [BFU2014-57779-P]; Ministerio de Ciencia, Innovación y Universidades/AEI [BFU2017-82937-P]; European Commission [H2020-ERC-2014-CoG-647900]. Funding for open access charge: European Commission [H2020-ERC-2014-CoG-647900].
Conflict of interest statement. None declared.
REFERENCES
- 1. Elbarbary R.A., Lucas B.A., Maquat L.E.. Retrotransposons as regulators of gene expression. Science. 2016; 351:aac7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chuong E.B., Elde N.C., Feschotte C.. Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet. 2017; 18:71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Casacuberta E., González J.. The impact of transposable elements in environmental adaptation. Mol. Ecol. 2013; 22:1503–1517. [DOI] [PubMed] [Google Scholar]
- 4. Batut P., Dobin A., Plessy C., Carninci P., Gingeras T.R.. High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res. 2013; 23:169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sundaram V., Cheng Y., Ma Z., Li D., Xing X., Edge P., Snyder M.P., Wang T.. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 2014; 24:1963–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kunarso G., Chia N.Y., Jeyakani J., Hwang C., Lu X., Chan Y.S., Ng H.H., Bourque G.. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 2010; 42:631–634. [DOI] [PubMed] [Google Scholar]
- 7. Lynch V.J., Leclerc R.D., May G., Wagner G.P.. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat. Genet. 2011; 43:1154–1159. [DOI] [PubMed] [Google Scholar]
- 8. Chuong E.B., Elde N.C., Feschotte C.. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 2016; 351:1083–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dunn-Fletcher C.E., Muglia L.M., Pavlicev M., Wolf G., Sun M.A., Hu Y.C., Huffman E., Tumukuntala S., Thiele K., Mukherjee A. et al.. Anthropoid primate-specific retroviral element THE1B controls expression. of CRH in placenta and alters gestation length. PLoS Biol. 2018; 16:e2006337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kantorovitz M.R., Kazemian M., Kinston S., Miranda-Saavedra D., Zhu Q., Robinson G.E., Göttgens B., Halfon M.S., Sinha S.. Motif-blind, genome-wide discovery of cis-regulatory modules in Drosophila and mouse. Dev. Cell. 2009; 17:568–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mathelier A., Wasserman W.W.. The next generation of transcription factor binding site prediction. PLoS Comput. Biol. 2013; 9:e1003214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mathelier A., Fornes O., Arenillas D.J., Chen C.Y., Denay G., Lee J., Shi W., Shyr C., Tan G., Worsley-Hunt R. et al.. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016; 44:D110–D115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shlyueva D., Stampfel G., Stark A.. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 2014; 15:272–286. [DOI] [PubMed] [Google Scholar]
- 14. Venuto D., Bourque G.. Identifying co-opted transposable elements using comparative epigenomics. Dev. Growth Differ. 2018; 60:53–62. [DOI] [PubMed] [Google Scholar]
- 15. Straalen N.MV., Roelofs D.. An Introduction to Ecological Genomics. 2012; UK: Oxford University Press. [Google Scholar]
- 16. Fan S., Hansen M.E., Lo Y., Tishkoff S.A.. Going global by adapting local: A review of recent human adaptation. Science. 2016; 354:54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mackay T.FC., Huang W.. Charting the genotype-phenotype map: lessons from the Drosophila melanogaster Genetic Reference Panel. Wiley Interdiscip Rev. Dev. Biol. 2018; 7:e289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Clos J., Rabindran S., Wisniewski J., Wu C.. Induction temperature of human heat shock factor is reprogrammed in a Drosophila cell environment. Nature. 1993; 364:252–255. [DOI] [PubMed] [Google Scholar]
- 19. Lemaitre B., Hoffmann J.. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007; 25:697–743. [DOI] [PubMed] [Google Scholar]
- 20. Espinosa-Diez C., Miguel V., Mennerich D., Kietzmann T., Sánchez-Pérez P., Cadenas S., Lamas S.. Antioxidant responses and cellular adjustments to oxidative stress. Redox Biol. 2015; 6:183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou D., Haddad G.G.. Genetic analysis of hypoxia tolerance and susceptibility in Drosophila and humans. Annu. Rev. Genomics Hum. Genet. 2013; 14:25–43. [DOI] [PubMed] [Google Scholar]
- 22. Thiel G., Cibelli G.. Regulation of life and death by the zinc finger transcription factor Egr-1. J. Cell Physiol. 2002; 193:287–292. [DOI] [PubMed] [Google Scholar]
- 23. Sims H.I., Chirn G.W., Marr M.T.. Single nucleotide in the MTF-1 binding site can determine metal-specific transcription activation. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:16516–16521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reed D.E., Huang X.M., Wohlschlegel J.A., Levine M.S., Senger K.. DEAF-1 regulates immunity gene expression in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:8351–8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu F., Yu X., Wang H., Zuo D., Guo C., Yi H., Tirosh B., Subjeck J.R., Qiu X., Wang X.Y.. ER stress and its regulator X-box-binding protein-1 enhance polyIC-induced innate immune response in dendritic cells. E. J. Immunol. 2011; 41:1086–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tan G., Lenhard B.. TFBSTools: an R/bioconductor package for transcription factor binding site analysis. Bioinformatics. 2016; 32:1555–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wagih O. ggseqlogo: a versatile R package for drawing sequence logos. Bioinformatics. 2017; 33:3645–3647. [DOI] [PubMed] [Google Scholar]
- 28. Thurmond J., Goodman J.L., Strelets V.B., Attrill H., Gramates L.S., Marygold S.J., Matthews B.B., Millburn G., Antonazzo G., Trovisco V. et al.. FlyBase 2.0: the next generation. Nucleic Acids Res. 2019; 47:D759–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma W., Noble W.S., Bailey T.L.. Motif-based analysis of large nucleotide data sets using MEME-ChIP. Nat. Protoc. 2014; 9:1428–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Worsley H.R., Mathelier A., Del Peso L., Wasserman W.W.. Improving analysis of transcription factor binding sites within ChIP-Seq data based on topological motif enrichment. BMC Genomics. 2014; 15:472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chung D., Kuan P.F., Li B., Sanalkumar R., Liang K., Bresnick E.H., Dewey C., Keleş S.. Discovering transcription factor binding sites in highly repetitive regions of genomes with multi-read analysis of ChIP-Seq data. PLoS Comput. Biol. 2011; 7:e1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Feng J., Liu T., Qin B., Zhang Y., Liu X.S.. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012; 7:1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Davie K., Jacobs J., Atkins M., Potier D., Christiaens V., Halder G., Aerts S.. Discovery of transcription factors and regulatory regions driving in vivo tumor development by ATAC-seq and FAIRE-seq open chromatin profiling. PLos Genet. 2015; 11:e1004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koenecke N., Johnston J., Gaertner B., Natarajan M., Zeitlinger J.. Genome-wide identification of Drosophila dorso-ventral enhancers by differential histone acetylation analysis. Genome Biol. 2016; 17:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gel B., Diez-Villanueva A., Serra E., Buschbeck M., Peinado M.A., Malinverni R.. regioneR: an R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics. 2016; 32:289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jung Y.L., Luquette L.J., Ho J.W., Ferrari F., Tolstorukov M., Minoda A., Issner R., Epstein C.B., Karpen G.H., Kuroda M.I. et al.. Impact of sequencing depth in ChIP-seq experiments. Nucleic Acids Res. 2014; 42:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fiston-Lavier A.S., Barron M.G., Petrov D.A., Gonzalez J.. T-lex2: genotyping, frequency estimation and re-annotation of transposable elements using single or pooled next-generation sequencing data. Nucleic Acids Res. 2015; 43:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rech G.E., Bogaerts-Márquez M., Barrón M.G., Merenciano M., Villanueva-Cañas J.L., Horváth V., Fiston-Lavier A.S., Luyten I., Venkataram S., Quesneville H. et al.. Stress response, behavior, and development are shaped by transposable element-induced mutations in Drosophila. PLos Genet. 2019; 15:e1007900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang W., Massouras A., Inoue Y., Peiffer J., Ràmia M., Tarone A.M., Turlapati L., Zichner T., Zhu D., Lyman R.F. et al.. Natural variation in genome architecture among 205 drosophila melanogaster genetic reference panel lines. Genome Res. 2014; 24:1193–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ramia M., Librado P., Casillas S., Rozas J., Barbadilla A.. PopDrowser: the population drosophila browser. Bioinformatics. 2012; 28:595–596. [DOI] [PubMed] [Google Scholar]
- 44. Ullastres A., Merenciano M., González J.. Natural transposable element insertions drive expression changes in genes underlying Drosophila melanogaster immune response. 2019; bioRxiv doi: 10.1101/655225, 31 May 2019, preprint: not peer reviewed. [DOI]
- 45. Tian S., Haney R.A., Feder M.E.. Phylogeny disambiguates the evolution of heat-shock cis-regulatory elements in Drosophila. PLoS One. 2010; 5:e10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sykiotis G.P., Bohmann D.. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev. Cell. 2008; 14:76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brandt A., Scharf M., Pedra J.H., Holmes G., Dean A., Kreitman M., Pittendrigh B.R.. Differential expression and induction of two Drosophila cytochrome P450 genes near the Rst(2)DDT locus. Insect Mol. Biol. 2002; 11:337–341. [DOI] [PubMed] [Google Scholar]
- 48. Davies T.G., Field L.M., Usherwood P.N., Williamson M.S.. DDT., pyrethrins, pyrethroids and insect sodium channels. IUBMB Life. 2007; 59:151–162. [DOI] [PubMed] [Google Scholar]
- 49. Vodovar N., Vinals M., Liehl P., Basset A., Degrouard J., Spellman P., Boccard F., Lemaitre B.. Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:11414–11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Neyen C., Bretscher A.J., Binggeli O., Lemaitre B.. Methods to study Drosophila immunity. Methods. 2014; 68:116–128. [DOI] [PubMed] [Google Scholar]
- 51. Kristensen T.N., Sorensen P., Pedersen K.S., Kruhoffer M., Loeschcke V.. Inbreeding by environmental interactions affect gene expression in Drosophila melanogaster. Genetics. 2006; 173:1329–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 53. Balamurugan K., Egli D., Selvaraj A., Zhang B., Georgiev O., Schaffner W.. Metal-responsive transcription factor (MTF-1) and heavy metal stress response in Drosophila and mammalian cells: a functional comparison. Biol. Chem. 2004; 385:597–603. [DOI] [PubMed] [Google Scholar]
- 54. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kapitonov V.V., Jurka J.. Molecular paleontology of transposable elements in the Drosophila melanogaster genome. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:6569–6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Singh N.D., Petrov D.A.. Rapid sequence turnover at an intergenic locus in Drosophila. Mol. Biol. Evol. 2004; 21:670–680. [DOI] [PubMed] [Google Scholar]
- 57. Yang H.P., Barbash D.A.. Abundant and species-specific DINE-1 transposable elements in 12 Drosophila genomes. Genome Biol. 2008; 9:R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Spitz F., Furlong E.E.. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012; 13:613–626. [DOI] [PubMed] [Google Scholar]
- 59. Spivakov M. Spurious transcription factor binding: non-functional or genetically redundant. Bioessays. 2014; 36:798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vihervaara A., Sergelius C., Vasara J., Blom M.A., Elsing A.N., Roos-Mattjus P., Sistonen L.. Transcriptional response to stress in the dynamic chromatin environment of cycling and mitotic cells. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E3388–E3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. White M.A., Myers C.A., Corbo J.C., Cohen B.A.. Massively parallel in vivo enhancer assay reveals that highly local features determine the cis-regulatory function of ChIP-seq peaks. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:11952–11957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xie D., Chen C.C., Ptaszek L.M., Xiao S., Cao X., Fang F., Ng H.H., Lewin H.A., Cowan C., Zhong S.. Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome Res. 2010; 20:804–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Paris M., Kaplan T., Li X.Y., Villalta J.E., Lott S.E., Eisen M.B.. Extensive divergence of transcription factor binding in Drosophila embryos with highly conserved gene expression. PLos Genet. 2013; 9:e1003748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Neph S., Vierstra J., Stergachis A.B., Reynolds A.P., Haugen E., Vernot B., Thurman R.E., John S., Sandstrom R., Johnson A.K. et al.. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012; 489:83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vrljicak P., Lucas E.S., Lansdowne L., Lucciola R., Muter J., Dyer N.P., Brosens J.J., Ott S.. Analysis of chromatin accessibility in decidualizing human endometrial stromal cells. FASEB J. 2018; 32:2467–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Katoh Y., Itoh K., Yoshida E., Miyagishi M., Fukamizu A., Yamamoto M.. Two domains of Nrf2 cooperatively bind CBP., a CREB binding protein, and synergistically activate transcription. Genes Cells. 2001; 6:857–868. [DOI] [PubMed] [Google Scholar]
- 67. Takii R., Fujimoto M., Tan K., Takaki E., Hayashida N., Nakato R., Shirahige K., Nakai A.. ATF1 modulates the heat shock response by regulating the stress-inducible heat shock factor 1 transcription complex. Mol. Cell Biol. 2015; 35:11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bhattacharya S., Michels C.L., Leung M.K., Arany Z.P., Kung A.L., Livingston D.M.. Functional role of p35srj, a novel p300/CBP binding protein, during transactivation by HIF-1. Genes Dev. 1999; 13:64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li Y., Kimura T., Huyck R.W., Laity J.H., Andrews G.K.. Zinc-induced formation of a coactivator complex containing the zinc-sensing transcription factor MTF-1, p300/CBP., and Sp1. Mol. Cell Biol. 2008; 28:4275–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Revilla Y., Granja A.. Viral mechanisms involved in the transcriptional CBP/p300 regulation of inflammatory and immune responses. Crit. Rev. Immunol. 2009; 29:131–154. [DOI] [PubMed] [Google Scholar]
- 71. Du J., Leung A., Trac C., Lee M., Parks B.W., Lusis A.J., Natarajan R., Schones D.E.. Chromatin variation associated with liver metabolism is mediated by transposable elements. Epigenetics Chromatin. 2016; 9:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Xie M., Hong C., Zhang B., Lowdon R.F., Xing X., Li D., Zhou X., Lee H.J., Maire C.L., Ligon K.L. et al.. DNA hypomethylation within specific transposable element families associates with tissue-specific enhancer landscape. Nat. Genet. 2013; 45:836–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jjingo D., Conley A.B., Wang J., Marino-Ramirez L., Lunyak V.V., Jordan I.K.. Mammalian-wide interspersed repeat (MIR)-derived enhancers and the regulation of human gene expression. Mob. DNA. 2014; 5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Calo E., Wysocka J.. Modification of enhancer chromatin: what, how, and why. Mol. Cell. 2013; 49:825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Policarpi C., Crepaldi L., Brookes E., Nitarska J., French S.M., Coatti A., Riccio A.. Enhancer SINEs link pol III to pol II transcription in neurons. Cell Rep. 2017; 21:2879–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Teng L., He B., Gao P., Gao L., Tan K.. Discover context-specific combinatorial transcription factor interactions by integrating diverse ChIP-Seq data sets. Nucleic Acids Res. 2014; 42:e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wang J., Xie G., Singh M., Ghanbarian A.T., Rasko T., Szvetnik A., Cai H., Besser D., Prigione A., Fuchs N.V. et al.. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature. 2014; 516:405–409. [DOI] [PubMed] [Google Scholar]
- 78. Dantoft W., Davis M.M., Lindvall J.M., Tang X., Uvell H., Junell A., Beskow A., Engström Y.. The Oct1 homolog Nubbin is a repressor of NF-κB-dependent immune gene expression that increases the tolerance to gut microbiota. BMC Biol. 2013; 11:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lelli K.M., Slattery M., Mann R.S.. Disentangling the many layers of eukaryotic transcriptional regulation. Annu. Rev. Genet. 2012; 46:43–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.