Abstract
Maintaining manganese (Mn) homeostasis is important for the virulence of numerous bacteria. In the human respiratory pathogen Streptococcus pneumoniae, the Mn-specific importer PsaBCA, exporter MntE, and transcriptional regulator PsaR establish Mn homeostasis. In other bacteria, Mn homeostasis is controlled by yybP-ykoY family riboswitches. Here, we characterize a yybP-ykoY family riboswitch upstream of the mgtA gene encoding a PII-type ATPase in S. pneumoniae, suggested previously to function in Ca2+ efflux. We show that the mgtA riboswitch aptamer domain adopts a canonical yybP-ykoY structure containing a three-way junction that is compacted in the presence of Ca2+ or Mn2+ at a physiological Mg2+ concentration. Although Ca2+ binds to the RNA aptamer with higher affinity than Mn2+, in vitro activation of transcription read-through of mgtA by Mn2+ is much greater than by Ca2+. Consistent with this result, mgtA mRNA and protein levels increase ≈5-fold during cellular Mn stress, but only in genetic backgrounds of S. pneumoniae and Bacillus subtilis that exhibit Mn2+ sensitivity, revealing that this riboswitch functions as a failsafe ‘on’ signal to prevent Mn2+ toxicity in the presence of high cellular Mn2+. In addition, our results suggest that the S. pneumoniae yybP-ykoY riboswitch functions to regulate Ca2+ efflux under these conditions.
INTRODUCTION
Small non-coding regulatory RNAs (sRNAs) are ubiquitous and found in all domains of life and play myriad roles in regulating gene expression (1,2). In bacteria, these sRNAs exist as short transcripts with lengths of 30–500 nucleotides (3,4). Bacterial sRNAs are broadly classified into two categories: cis-acting (riboswitches) or trans-acting (independent transcripts) RNAs. Trans-acting sRNAs modulate the stability or translation of mRNA transcript(s) by imperfect base-pairing interactions (5,6) at or near the ribosome binding site (RBS) on the target transcript. Riboswitches, on the other hand, are highly structured regulatory segments located within the 5′ untranslated regions (UTRs) of mRNAs that interact with small molecules, such as metals and other ligands, resulting in a change in transcription or translation mediated directly by environmental changes. sRNAs may also interact with RNA-binding proteins thereby modifying their activities (7,8).
Bacterial sRNAs function in many cellular processes, including the response to environmental stresses, e.g. those mediated by the immune system, and maintaining homeostasis (5,9). Accumulating evidence suggests that sRNAs are differentially expressed in bacteria when transitioning from colonization to an active infection which suggests that sRNAs are key players in mediating invasive disease (10–13). Streptococcus pneumoniae is a Gram-positive bacterium that commonly colonizes the nasopharynx of healthy individuals, persisting as a harmless bacterial commensal (14). Despite a largely asymptomatic colonization, S. pneumoniae remains a leading causative agent of sinusitis, otitis media (middle ear infection), and the life-threatening invasive diseases pneumonia, bacteremia, and meningitis globally (15,16). Transition from the harmless commensal to pathogen often occurs after a viral respiratory tract infection and is triggered by numerous factors, many of which are unknown (14). Progression into the lungs and intrusion into the circulatory system, exposes the pneumococcus to numerous stress conditions and environmental changes (17), including transition metal fluctuations, an important mediator in the ‘fight or flight’ response during infection (18,19). The capacity for bacteria to quickly adapt to these different environments may derive in part from sRNAs modulating the expression of virulence factors.
Multiple (>100) sRNAs have been identified in S. pneumoniae (20–25), but the functional role for most of these remains unknown. One particular candidate sRNA, SN44, identified in S. pneumoniae TIGR4 using genome-tilling arrays (21) is of interest here. SN44 is predicted on the basis of sequence homology to be a cis-acting yybP-ykoY family response element (riboswitch) that is conserved among different Streptococcus species (21) (Supplementary Figure S1). The yybP-ykoY motif defines a ubiquitous class of riboswitches that are upstream of genes that encode uncharacterized or poorly characterized proteins many of which have been linked to Mn2+ homeostasis. To date, yybP-ykoY-regulated gene products, MntP of Escherichia coli (26), MntX of Neisseria spp. (27), and YaoB (a predicted P-type II ATPase) of Lactococcus lactis (28), have been independently implicated as Mn2+ efflux transporters capable of relieving cellular Mn toxicity. Other studies show that several of these associated yybP-ykoY family riboswitches respond to high Mn2+in vitro and in vivo modulating transcription and translation of downstream gene(s) within the transcript (28–30). Although yybP-ykoY riboswitches have generally been observed to bind Mn2+in vitro with highest affinity among other transition metals examined, none thus far exhibit strict Mn2+ specificity (30).
The yybP-ykoY riboswitch aptamer domain adopts a four-helix junction architecture with P2 stacked on P1 and P4 stacked on P3, with the transition metal sensing pocket formed by nucleotides in the L1 loop (in P1) and L3 loop in P3 (Figure 1A). P2 and P4 are not strictly conserved and can be replaced by a short single-stranded connecting region or a longer helical stem, just above the cross-over point (Figure 1). A recent structural analysis of the L. lactis yybP-ykoY riboswitch aptamer domain suggests that the metal site may well be plastic and capable of adopting a range of coordination structures and nuclearity, with binuclear and trinuclear metal (Cd2+) complexes observed in that study (30). The binuclear site in a previous structure of the L. lactis yaoB riboswitch (28) was modeled as Mg2+ or Mn2+ in the MA site and Mn2+ in the MB site (Figure 2A). The MB site is thought to provide specificity for Mn2+ given the presence of five inner-sphere coordination bonds, including the N7 of A41 in the L3 loop (Figure 2A–C).
Figure 1.
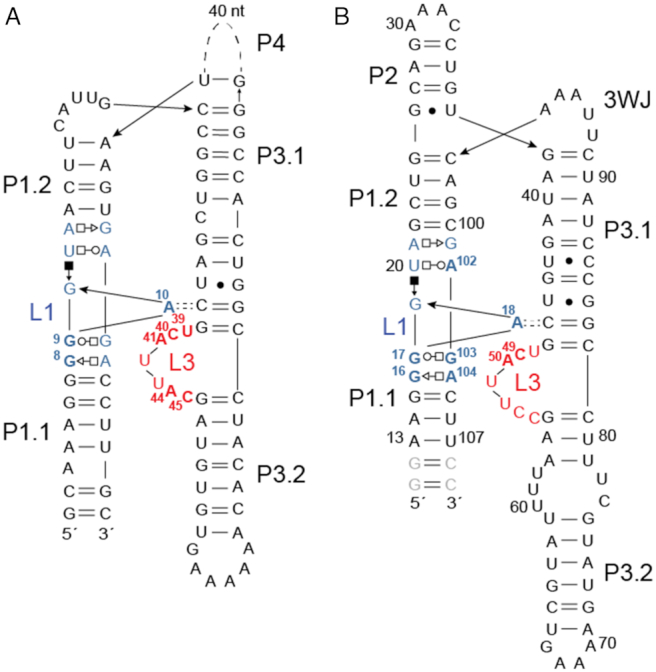
Secondary structure diagrams of the aptamer domains of (A) E. coli alx sequence (30) and (B) Streptococcus pneumoniae D39 mgtA riboswitch domains. In panel A, nucleotides in L1 (blue) and in L3 (red) that correspond to those shown in the structural models (Figure 2) are highlighted in bold, and correspond to the numbering convention of the L. lactis yybP-ykoY riboswitch (see Figure 2A) (28). In panel B, we used the natural nucleotide residue numbers, and those nucleotides targeted for substitution in this work in L1 (blue) and L3 (red) are highlighted in bold. For example, the loop L1 G8-G9-A10 L. lactis system corresponds to the G16-G17-A18 in the S. pneumoniae RNA, while the L. lactis C40-A41 L3 sequence corresponds to the C49-A50 sequence in S. pneumoniae. A41 in the L. lactis RNA (A50 in the mgtA RNA) makes an inner sphere coordination bond with the MnB metal (see Figure 2A). The gray nucleotides in panel B are non-native and were added for in vitro transcription.
Figure 2.
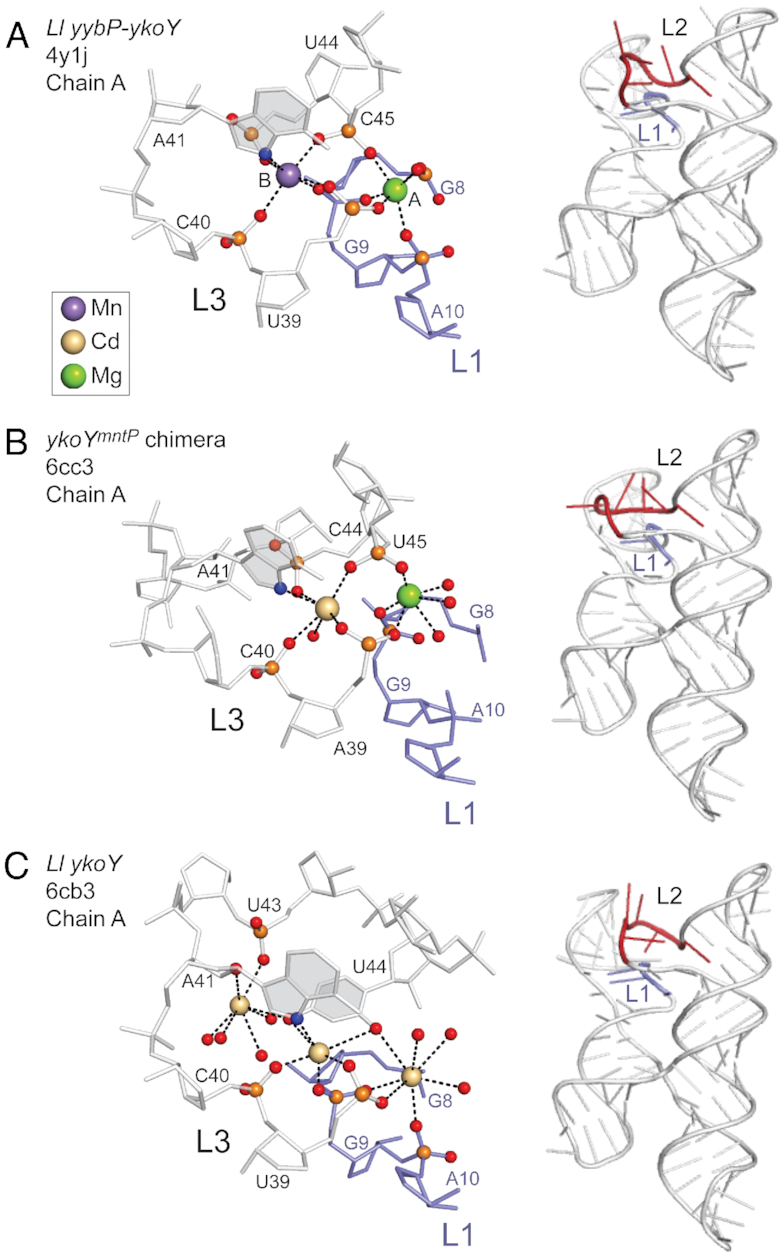
Regulatory metal coordination site region (left) and global ribbon diagrams (right) of the various yybP-ykoY riboswitch aptamer domains. (A) L. lactis yybP-ykoY riboswitch bound to Mg2+ (site A) and the Mn2+ (site B) (28); (B) chimeric E. coli ykoYmntP riboswitch bound to Mg2+ (site A) and Cd2+ (site B) (30) and (C) the three metal (Cd2+)-bound form of the L. lactis yaoB riboswitch (30).The nucleotide numbering convention in panels B and C are as adopted for the L. lactis yybP-ykoY riboswitch (panel A). L1 residues are highlighted in blue, and L3 residues are shown in gray (left) or red (right) of panels A-C. The nucleotide base of L3 nucleotide A41 is shaded gray since N7 donates a coordination bond to the Mn2+ (A) and Cd2+ (B, C) ions in these structures.
Like other annotated yybP-ykoY family riboswitches, the putative SN44 riboswitch overlaps the RBS within the target gene transcript and features a transcription terminator helix just 5′ to the RBS (Supplementary Figure S1). The SN44 RNA in S. pneumoniae D39 serotype 2 is encoded within the 5′ UTR of the mgtA (spd_1383; previously caxP or PacL) transcript, encoding a PII-type ATPase transporter. MgtA was previously suggested to function in Ca2+ efflux (25,31), while in a different report was suggested to be implicated in the response against Zn2+ and Mn2+ toxicity also (32). The idea that MgtA in S. pneumoniae is not restricted to Ca2+ and could also transport Mn2+ is also supported by the fact that the MgtA protein is encoded from the negative-sense DNA strand immediately upstream of the mntE gene, which encodes the constitutively expressed Mn2+-specific cation diffusion facilitator exporter MntE (33,34). Both MgtA and MntE are important for the virulence of S. pneumoniae (31,33). Despite intensive study, the physiological roles of many genes critical for virulence, including those encoding divalent metal transporters like MgtA, remain unclear.
In this study, we examine the metal binding and metal-induced folding of the mgtA aptamer and the regulation of mgtA in S. pneumoniae D39 strain. We find that the S. pneumoniae D39 mgtA aptamer RNA binds stoichiometric Mn2+ and Ca2+ in a background of physiological Mg2+ and each is capable of stabilizing the same compact conformation; however, Mn2+ stimulates in vitro read-through transcription to a far greater extent than Ca2+. We establish for the first time that the mgtA mRNA is upregulated during Mn-stress in this organism, and that this increased transcription in vivo requires the mgtA (yybP-ykoY family) riboswitch. Further investigation reveals that MgtA may export Mn2+ under conditions of extreme Mn-stress thereby protecting cells from Mn2+ toxicity. The mgtA riboswitch may also function to regulate Ca2+ export under these conditions.
MATERIALS AND METHODS
See Supplementary Material detailing methods for bacterial strain and plasmid construction, bacterial growth, disk diffusion assay, β-galactosidase activity assay, RNA isolation and quantitative real-time PCR, western blot, yybP-ykoY RNA synthesis, and inductively coupled plasma-mass spectrometry (ICP-MS) for measurement of total cell-associated divalent cationic metals. Bacterial strains and plasmids used in this study are listed in Supplementary Table S1 and relevant DNA oligonucleotide primers in Supplementary Tables S2 and S3.
Native polyacrylamide gel electrophoresis (PAGE) analysis of yybP-ykoY RNAs (35)
Purified RNA (2 μg) was heated at 90°C for 2 min followed by incubation at room temperature for 3 min, then subsequently allowed to refold in 50 mM HEPES [pH 7.5], 50 mM NaCl at room temperature for 10 min. Various divalent metal ion concentrations ranging from 0.01 to 5 mM were added and RNA mixture was left at room temperature for an additional 30 min prior to mixing with loading dye (10% glycerol and 0.01% xylene cyanol). RNA was separated by native PAGE (8% gel prepared with 19:1 acrylamide/bisacrylamide) at 4°C in running buffer containing 34 mM Tris, 66 mM HEPES [pH 7.5], and 3 mM MgCl2, which was recirculated every hour. RNA was stained with ethidium bromide and observed with a Gbox (Biorad).
SHAPE probing
SHAPE was performed as previously described using 1-methyl-7-nitroisatoic anhydride (1M7) derivatization reagent (36–38). RNA constructs consisted of the wild-type sequence from the S. pneumoniae D39 mgtA 5′ UTR (containing the yybp-ykoY aptamer domain) flanked by SHAPE flanking sequences (37). RNAs were in vitro transcribed from PCR templates and purified similarly to that described above for RNA synthesis. Purified RNAs (10 pmol) was refolded in 50 mM HEPES, pH 7.5, 50 mM NaCl buffer before 1M7 derivatization. Reverse transcription of the derivatized RNA was performed with a FAM 6-labeled reverse primer. Unmodified RNA (2 pmol) used as a sequencing ladder was synthesized using reverse transcriptase, NED-labeled reverser primer, and ddCTP or ddGTP. An equal volume of sequencing ladder and reaction mixture with and without 1M7 was precipitated with 0.3 M sodium acetate, pH 5.2 and 100% ethanol, washed with 70% ethanol, air-dried, and re-dissolved in 15 μl water. Fragment analysis was performed using capillary electrophoresis by GENEWIZ (Plainfield, NJ, USA). Data processing was performed using QuShape following a protocol as previously described (39).
Small angle X-ray scattering (SAXS)
Small angle and wide angle X-ray scattering data were acquired at three different RNA concentrations (0.8, 1.6 and 3.2 mg/ml) of select RNAs folded in buffer containing 50 mM HEPES, pH 7.5, 50 mM NaCl, 3 mM MgCl2 and 0.5 mM of the metal ion indicated. Data were collected at beam line 12-ID of the Advanced Photon Source at the Argonne National Laboratory. The wavelength (λ) of X-ray radiation was set to 1.033 Å. Procedures for SAXS/WAXS measurement were similar to those previously described (40,41). Thirty images were collected for each RNA concentration and its corresponding background buffer.
SAXS data were averaged, and the background was subtracted using the NCI-SAXS program package. The averaged scattering profiles of three RNA concentrations were merged using PRIMUS in the ATSAS program package (http://www.embl-hamburg.de/biosaxs/). A GUINIER plot was generated as ln(I(q)) versus q2 to check sample quality and to obtain I0 and the radius of gyration (Rg) within the range of qmax*Rg < 1.3. The data from each RNA concentration were normalized with I0. Conformation of the RNAs was examined using the Kratky plot for q < 0.3 Å−1. Scattering profiles of RNAs were then Fourier-transformed using GNOM of the ATSAS package to obtain the normalized pair distance distribution graph.
Ab in initio modeling was performed using the program DAMMIN in a slow mode (42). For each RNA, 10 models were obtained, filtered, and averaged using the DAMPUP, DAMFILT and DAMAVER of the ATSAS package (http://www.embl-hamburg.de/bioSAXS). Normalized spatial discrepancy (NSD) between each pair of the models was computed. The model with the lowest NSD value was selected as the reference model for superimposing onto other models. Outliner models (two models) with an NSD above mean +2* variation were removed before averaging.
Isothermal titration calorimetry
ITC experiments were performed following standard protocol previously described (30). Briefly, RNAs were heated at 95°C for 2 min, cooled down at room temperature for 3 min, diluted into ITC buffer (30 mM HEPES [pH 6.8], 150 mM NaCl), and incubated at room temperature for an additional 10 min. MgCl2 was then added to a final concentration of 3 mM and the RNA was subsequently incubated at room temperature for 30 min. RNAs were dialyzed into ITC buffer containing 3 mM MgCl2, filtered using a 3 kDa MWCO column, and degassed prior to ITC measurement. Divalent metal cations were dissolved in ITC buffer, filtered, and degassed. Experiments were performed with 50–100 μM RNA and 0.5 to 2 mM metal cation at 25°C, depending on affinity and magnitude of enthalpy. Divalent metal cation was injected with 0.5 to 1 ul volume every 300 s. Data were corrected for heats of dilution by subtracting the enthalpies of cations titrated into the ITC buffer from raw data and analyzed using an independent binding model by Nanoanlyze (TA instrument).
IVT termination assay
In vitro transcription termination assays were performed using standard protocols (28,43). To construct the in vitro transcription plasmid, the S. pneumoniae mgtA 5′-UTR (containing the aptamer domain, intrinsic terminator, and the first 45-nt of the MgtA protein coding sequence) was fused to a B. subtilis glyQS promoter. An ApC dinucleotide sequence was added to the 5′ end of the mgtA aptamer domain with no cytosines present in the next 12 nucleotides. DNA templates were produced by PCR and spin column purified (Qiagen). To assemble stalled RNA polymerase complexes, 20 mM template was mixed with initiation buffer (20 mM Tris–HCl [pH 7.5], 50 mM NaCl, 250 μM MgCl2, 1 mM DTT and 5% glycerol), 0.15 μCuries/μl 32P ATP, 20 μM each of unlabeled ATP/CTP/GTP, and 0.01 U/μl Escherichia coli RNA polymerase holoenzyme. Reactions were incubated at 37°C for 15 min for elongation, then transferred to ice for 2 min. For synchronized transcription at each condition, 10 μl of stalled RNA polymerase was mixed with 1.5 μl 10× metal solution, and 1.5 μl 10× elongation buffer (4.5 mg/ml, 650 μM each of unlabeled ATP/CTT/GTP/UTP, 100 mM Tris [pH 7.5], 2 mM DTT, 10% glycerol and 2 mM MgCl2. Reactions were incubated at 37°C for an additional 15 min, then terminated by addition of 10 μl RNA loading dye (95% formamide, 20 mM EDTA [pH 8.0], supplemented with xylene cyanol). Transcripts (10 μl) were separated by 6% urea denaturing PAGE and analyzed by a phosphorimager. The sizes of the terminated and full-length RNA products were confirmed by RNA ladder (data not shown). Bands were quantified with ImageJ and each reaction was converted to the fraction of full-length product over total RNA transcribed. The data were fit using Origin 8 data analysis software to the Hill equation (n = 1), where x is the concentration of metal and k is the concentration at which the change in read-through is half maximal. Each assay was performed in triplicate.
Statistical analysis
When appropriate, P-values were determined relative to non-stressed parent or WT type strains using unpaired t-tests or a one-way ANOVA with Dunnett's post-test determined by GraphPad Prism software.
RESULTS
The S. pneumoniae mgtA riboswitch is comprised of a three-way helical junction that is conserved among Streptococcus spp.
As described above, the crystallographic structure of the Mn2+ bound L. lactis yaoB 5′ UTR (yybP-ykoY family) aptamer domain reveals a four-way helical junction (4WJ) comprising of tandem coaxially stacked helices (P2 on P1 and P4 on P3) with the two ‘legs’ of the H-like structure docked at a conserved interface between loop (L) L1 and L3 (Figure 1A) (28). A single Mn2+ ion (MB) is coordinated by six inner-sphere interactions from nucleotides in loops L1 and L3 including an A41 N7-Mn2+ coordination bond; a Mg2+ ion is also found nearby in MA (28) (Figure 2). The binding of a single Mn2+ ion influences stability of the L. lactis yaoB riboswitch like that of a fluoride-sensing riboswitch (44) but in a way that is distinct from the Mg2+-sensing riboswitch (or M-box), in which multiple Mg2+ ions cooperatively impact the stability of the riboswitch (45,46).
The yybP-ykoY family riboswitch located upstream of the mgtA gene in Streptococcus pneumoniae D39 reveals, in contrast to the L. lactis structure, a three-way helical junction (3WJ) with P4 replaced by an invariant UNAAA (N, can be U or G) sequence (Figure 1B; Supplementary Figure S1C) followed by a transcription termination hairpin with a U-rich sequence (Supplementary Figure S1A). This UNAAA sequence is also found close to a 3WJ in the twister ribozyme (AUAAA), where it induces a sharp turn in the backbone at a helical junction (47). The 5′ end of L1 loop is highly conserved, while the 3′ end contains an invariable AGA nucleotide sequence (nucleotides 101–104) preceded by a variable nucleotide at position 101 that is only conserved as a guanosine among Streptococcus spp (Figure 1B; Supplementary Figure S1C). These nucleotides are anticipated to contribute to the metal binding pocket of L1 and L3 in the mgtA riboswitch. The metal coordinating nucleotides C49 and A50 as defined by the previous structural work as C49 and A41 in the L. lactis RNA (Figure 2) (28,30) within the L3 bulge are also invariant. Finally, the C46-G83 base-pair in the P3.1 stem above L3 is anticipated to hydrogen bond to A18 to form a cross-helix A minor interaction as in other yybP-ykoY family riboswitches (Figure 1).
Mn2+ and Ca2+ to the mgtA riboswitch alters an RNA conformational equilibrium
A comparison of the E. coli mntP riboswitch in the divalent metal-bound and metal-free (Na+ only) apo states suggest that the aptamer domain is pre-organized into a conformation that is quite close to the metal-bound form (Supplementary Figure S2), with major differences occurring in L3, which is largely unstructured in the absence of metal (28). The extent to which this is true in solution, however, and for this particular riboswitch specifically, is unclear. To address this, we monitored the folding equilibrium of the S. pneumoniae mgtA riboswitch aptamer domain by native PAGE, evaluating the impact of divalent metal cations Mn2+, Ca2+ and Mg2+ (Figure 3).
Figure 3.
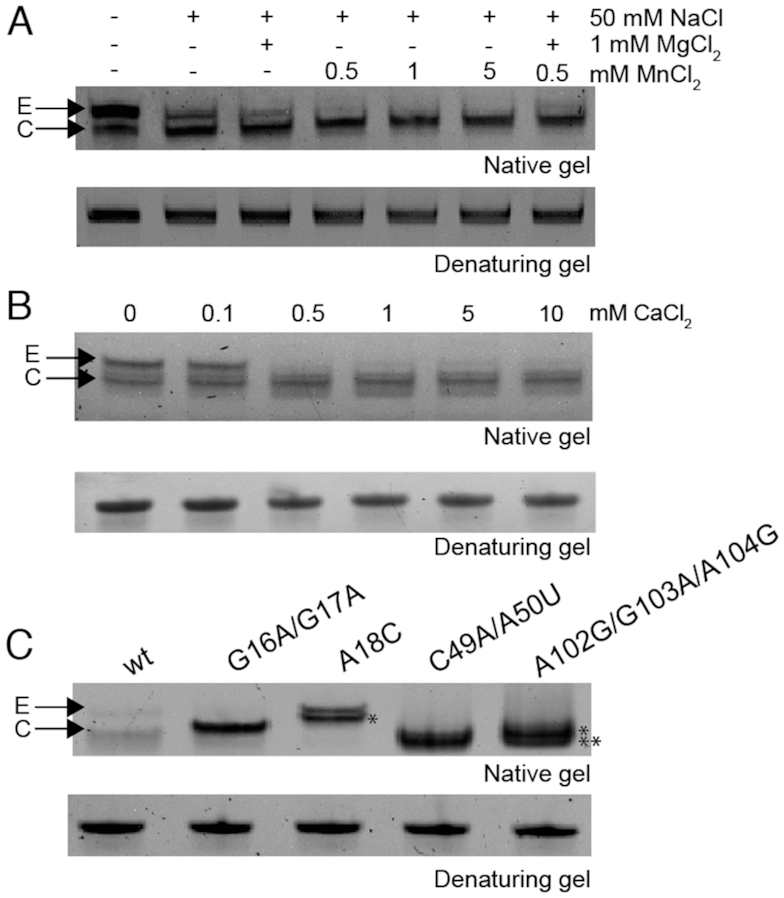
Mn2+ or Ca2+ binding to S. pneumoniae mgtA riboswitch induces conformational change and promotes compact folding. Global hydrodynamic analysis of the RNA aptamer in the presence of metal ions by native-PAGE. (A) Influence of salt solution, Mg2+ and Mn2+ on folding of the wild-type RNA aptamer domain. (B) Influence of Ca2+ in a background of 0.5 mM NaCl. (C) Effects of key metal-binding nucleotides in L1 and L3 that contribute to the overall global structure and compaction of the RNA in a background of 0.5 mM NaCl. Representative gel is shown from at three independent experiments. E, extended form; C, compact form; * and ** denote the two independent folded conformations for the triple mutant RNA, where * is a new conformation.
In these experiments, the RNA aptamer domain was denatured and refolded at room temperature in a step-wise progression by first adding a monovalent salt (50 mM NaCl), followed by divalent metal cation. Two conformations were resolvable by native PAGE run with Mg2+ in the running buffer: an extended or open form (E, upper band) and a more compact form (C, lower band) (Figure 3A). In the absence of added salt, the majority of the RNA (≥80%) is found in the extended form, while inclusion of the 50 mM monovalent salt drives the RNA towards a compacted form. Addition of 1 mM Mg2+ to 50 mM Na+ shifts the equilibrium further to the compacted form but not completely (Figure 3A). The addition of as little as 0.5 mM Mn2+ is efficient at compacting the RNA with ≈90% of the RNA adopting the compact form (Figure 3A and Supplementary Figure S3A). As anticipated from a cation competition model, the aptamer appears to fold less efficiently when 1 mM Mg2+ and 0.5 mM Mn2+ are simultaneously added into the folding buffer relative to folding in the presence of Mn2+ alone (Figure 3A). Indeed, nucleotides in L1 and L3 of the L. lactis yaoB riboswitch can coordinate binding of either Mn2+ or Mg2+ (28). The degree to which increasing concentrations of Mn2+ shifts the conformational equilibrium to a compact conformation can be modelled assuming that the apo form in 3 mM Mg2+ exists as a mixture of extended and compact and Mn2+-bound form is exclusively compact. This model of the relative concentrations of the two different RNA conformers gives an effective equilibrium dissociation constant (Kd) of 0.14 ± 0.02 mM (Supplementary Figure S3A and B) in 3 mM Mg2+, or ≈6-fold weaker than that reported for L. lactis yaoB riboswitch (Kd of 25 μM for Mn2+) using isothermal titration calorimetry (ITC) in a background of 5 mM Mg2+ (30). Quantitative ITC data for the binding of the pneumococcal mgtA riboswitch aptamer to divalent cations is presented below.
Although the likely cognate metal ion for the S. pneumoniae mgtA riboswitch is Mn2+ given its classification as a yybP-ykoY family riboswitch, the mgtA riboswitch incorporates the ribosome binding site of the mgtA gene, which encodes PII-type ATPase transporter (MgtA) that has been implicated in Ca2+ efflux (31); furthermore, biophysical experiments show that the analogous transporter from Listeria monocytogenes (LMCA1) transports Ca2+ (48). Ca2+ binding to this family of RNAs has not yet been investigated. Using native-PAGE, we show that 0.5 mM Ca2+ also affects the global hydrodynamics of the RNA aptamer, driving the RNA to the more compact form that is similar in mobility to that obtained upon incubation with Mn2+ (Figure 3B).
Mutant mgtA riboswitch aptamers have altered structures or altered Mn2+ sensitivities
To assess which nucleotides coordinate Mn2+ in the S. pneumoniae mgtA RNA aptamer, we mutated conserved nucleotides in L1 and L3 that comprise the metal binding site region as detailed in other yybP-ykoY structures (Figure 2) and tested the impact of these substitutions on the RNA aptamer folding equilibrium by native-PAGE (Figure 3C). These include G16, G17 and A18 on the 5′ side of L1, C49 and A50 in L3 and A102, G102 and A104 on the 3′ side of L1 (S. pneumoniae numbering) (see Figure 1B). The G16A/G17A and C49A/A50U RNAs show no change in electrophoretic mobility by native-PAGE over the same range of Mn2+ concentration that stabilizes the compacted (C) form of the wild-type RNA (Figure 3C and Supplementary Figure S3C and D), suggesting a loss of Mn2+ binding ability. We note that the G16A/G17A mutant RNA appears to migrate slightly faster than the extended (E) form of the WT RNA (Figure 3C). Although the C49A/A50U mutant has lost its ability to bind metal (Supplementary Figure S3D), this mutant RNA migrates similarly with the compacted WT RNA. The triple A102G/G103A/A104G (L1) mutant RNA, on the other hand, simultaneously adopts two RNA conformations that migrate closely with that of the compacted WT RNA, but each is formed independent of Mn2+ concentration (Supplementary Figure S3E, Figure 3C). Finally, targeting the predicted cross-strand A-minor interaction (nucleotide A18; Figure 1B) with a cytosine substitution (A18C) shifts the E-to-C equilibrium far toward E, as expected, in a background of 3 mM Mg2+ (Supplementary Figure S3F and G) while also increasing the apparent Kd for Mn ≈6.5-fold to 0.90 ± 0.01 mM.
Thus, all four mutant RNAs perturb the RNA conformational ensemble, at least as observed by native-PAGE, and only one of four RNAs (A18C) is responsive to the presence of Mn2+, albeit at much higher metal concentrations. These data suggest that Mn2+ binding may not be required to globally fold the S. pneumoniae mgtA riboswitch aptamer into an ‘X-like’ or undocked structure in the presence of physiological (low mM) Mg2+, as all mutant RNAs adopt an E- or E-like state, a finding consistent with recent single-molecule FRET experiments (49).
SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) probing of the wild-type aptamer
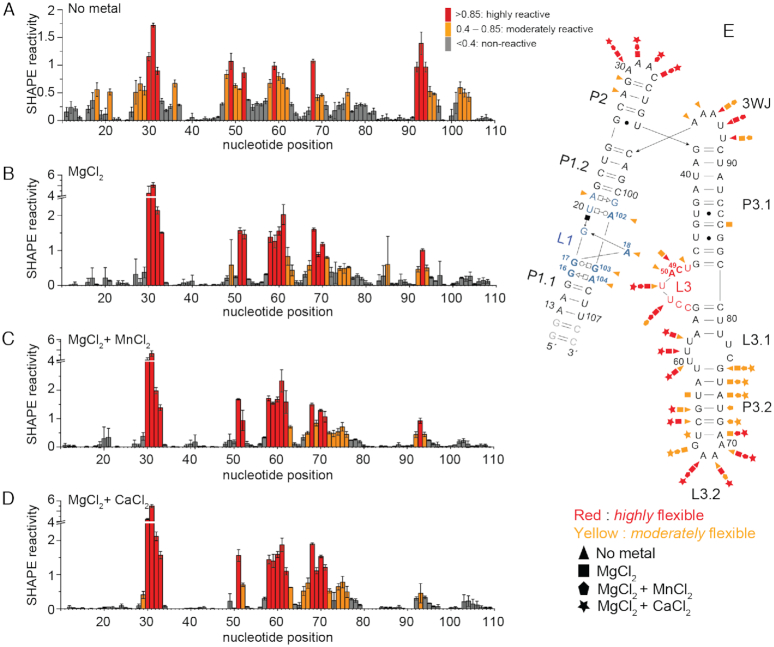
The impact of divalent metal binding on the structure of the S. pneumoniae mgtA RNA aptamer was further investigated by the structural probing method SHAPE. SHAPE experiments measure the relative rates of reactivity of 1-methyl-7-nitroisatoic anhydride (1M7) to 2′-hydroxyls and generally report on local flexibility of an individual nucleotide. In the absence of divalent metal (50 mM Na+ only), nucleotides in L1 (16–21; 102–104) and L3 (residues 48–50) show high to moderate reactivity (Figure 4A), suggesting that no metal-binding pocket is formed. A18 of the A-minor interaction is also moderately flexible in the absence of metal ions. The addition of Mg2+ stabilizes base-pairing in the P2 stem (residues 28–29), destabilizing the base of the P3.2 stem, while quenching flexibility of all nucleotides implicated in metal binding, including both sides of L1 and C49 and A50 in L3 (Figure 4B). The modest reduction in flexibility observed for nucleotides A18 in L1 and C49 and A50 in L3 suggests that the A-minor interaction forms in the presence of Mg2+ alone. Addition of Mn2+ to a background of Mg2+ leads only to a small change in the flexibility of nucleotides C49 and A50, which are predicted to directly interact with the Mn2+ MB ion (vide infra), making them more rigid (Figure 4C). The addition of Ca2+ results in a SHAPE profile (Figure 4D) that is statistically indistinguishable from that obtained upon addition of Mn2+. These data taken together reveal that the RNA conformation obtained in the presence of the 3 mM Mg2+ alone, Mg/Mn and Mg/Ca are very similar and that major differences in reactivity relative to the Na+-only structure are localized to the metal-binding region. These findings are consistent with native-PAGE analysis which reveals roughly two distinct conformations with distinguishable, yet rather similar, electrophoretic mobilities.
Figure 4.
Mn2+ and Ca2+ impact nucleotide flexibility in the mgtA RNA aptamer. (A) SHAPE reactivity of the S. pneumoniae mgtA RNA aptamer domain in the presence of various divalent metals. Regions of low reactivity (<0.4) are colored gray, moderate reactivity (0.4–0.85) are yellow, and high reactivity (>0.85) are red. Each bar represents a single nucleotide in the sequence as numbered on x-axis. (B) Summary of the flexibility of nucleotides in the RNA aptamer.
Small angle X-ray scattering (SAXS) analysis of mgtA riboswitch aptamers
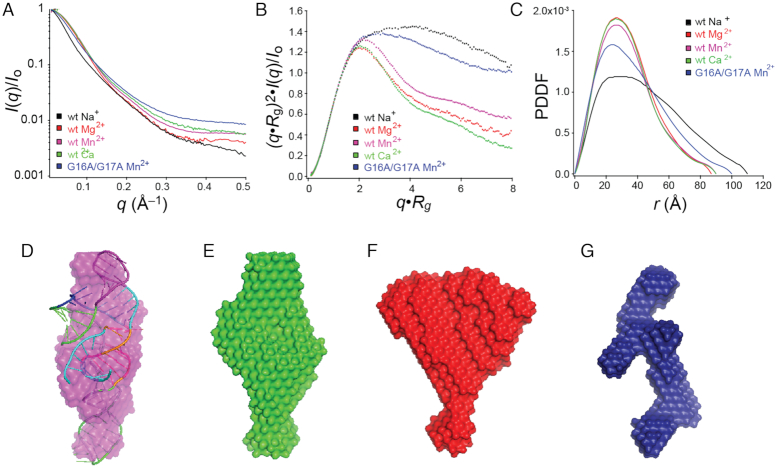
Small angle X-ray scattering (SAXS) was next used to obtain additional insights into the global fold of the wild-type mgtA RNA as a function of divalent metal ion status, compared to the G16A/G17A L1 mutant which fails to adopt the compact or C-state. Refolding of the aptamer in buffer with NaCl only and no divalent metal ions results in the largest radius of gyration (Rg) (Supplementary Figure S4; ≈37 Å) with clear evidence of conformational heterogeneity in this state. Addition of Mg2+ results in a significant increase in RNA compaction, with the Rg some 10 Å smaller, to 26.7 ± 0.5 Å. Addition of Mn2+ to the Mg2+-folded structure results in a similarly compacted structure, that is perhaps slightly more extended (Rg = 27.6 ± 0.4 Å) with no significant change observed with the addition of Ca2+ to the Mg2+ folded RNA. The dimensionless Kratky plots also reveal that the mgtA aptamer folds upon the addition of Mg2+, while inclusion of other transition metals tested only change the overall compaction of the RNA slightly (Figure 5A and B). A significant change in the pair-wise distance distribution is observed with Mg2+ showing a larger number of shorter distances (20–40 Å) within the RNA (Figure 5C), consistent with a folded form of the mgtA aptamer RNA. Only a minor change in the pair-wise distance distribution results when Mn2+ or Ca2+ are present in a background of Mg2+ (Figure 5C). Taken together these data are consistent with the native-PAGE and SHAPE results, demonstrating that different divalent metal ions can lead to conformational change and compaction of the mgtA RNA aptamer.
Figure 5.
Global structure analysis of the mgtA RNA aptamer with divalent metal ions. (A) Scattering profiles collected at separate RNA concentrations 0.8, 1.6 and 3.2 mg/ml with indicated metal, then merged. (B) Dimensionless Kratky plots. (C) Pair-wise distance distribution plots for all RNAs. (D) Simulated 3D structure of the RNA aptamer by SimRNA embedded in the envelope obtained from SAXS in the presence of Mn2+. SAXS simulated envelope models for WT RNA with Ca2+ (E), Mg2+ (F) and the G16A/G17A mutant RNA that is unable to bind metal (G).
We next used these data to calculate a three-dimensional (3D) envelope (or bead model) of the mgtA RNA aptamer, which could only be determined in the presence of divalent metal ions, a finding consistent with the conformational heterogeneity in the metal-free form of the RNA as evidenced by the non-linear Guinier plot (Supplementary Figure S4A). As expected, the resulting averaged envelopes obtained in the presence of Mg2+ plus Mn2+ or Mg2+ plus Ca2+ are similar (Figure 5D and E), but yet distinct from that of Mg2+ alone (Figure 5F). A simulated structural model of the aptamer embedded into the SAXS envelope in the presence of Mg2+/Mn2+ or Mg2+/Ca2+ fits well (Figure 5D), showing that in the presence of Mn2+ or Ca2+, the three hairpins likely organize into two coaxially stacked helices (P2 on P1) connected to a third helix (P3) at the three-way junction (3WJ).
In contrast to the WT RNA aptamer, the non-metal binding G16A/G17A mutant RNA that fails to adopt the compact form at any Mn2+ concentration by native-PAGE analysis (Supplementary Figure S3C) adopts a more extended average conformation (Rg = 29.3 ± 0.4 Å), independent of Mn2+ (Figure 5B and C). The resulting averaged SAXS-derived bead model yields a ‘Y-like’ topology, suggesting that the G16A/G17A mutant RNA is characterized by an open or extended conformation minimally comprising three helices connected by a 3WJ (Figure 5G).
The mgtA riboswitch aptamer binds Ca2+ more tightly than Mn2+
The genomic location of the S. pneumoniae mgtA riboswitch as well as our own hydrodynamic data (Figures 3–5) suggest that Mn2+ and possibly Ca2+ may be regulatory. We next used ITC to monitor the binding of Mn2+ and Ca2+ directly, monitored at what we anticipate is near physiological conditions of Mg2+ (3 mM). The binding of Mn2+ to the mgtA RNA aptamer is exothermic, whereas the binding of Ca2+ is endothermic, where Mn2+ binding is more strongly entropically-driven relative to Ca2+ (Figure 6 and Table 1). Both metals bind to the mgtA riboswitch with an apparent stoichiometry, n, ≈1, likely reporting on the displacement of Mg2+ bound to the MB site by Ca2+ or Mn2+ (Figure 2A) (28). The effective Kd for Ca2+ is low micromolar (1.72 ± 0.03 μM), which is 50-fold higher affinity than for Mn2+ (Kd = 54 ± 26 μM). The Kd for Mn2+ obtained from ITC is somewhat tighter than that derived from native-PAGE folding equilibrium analysis, although both are in the 10−4 M Mn2+ range (Supplementary Figure S3A and B).
Figure 6.
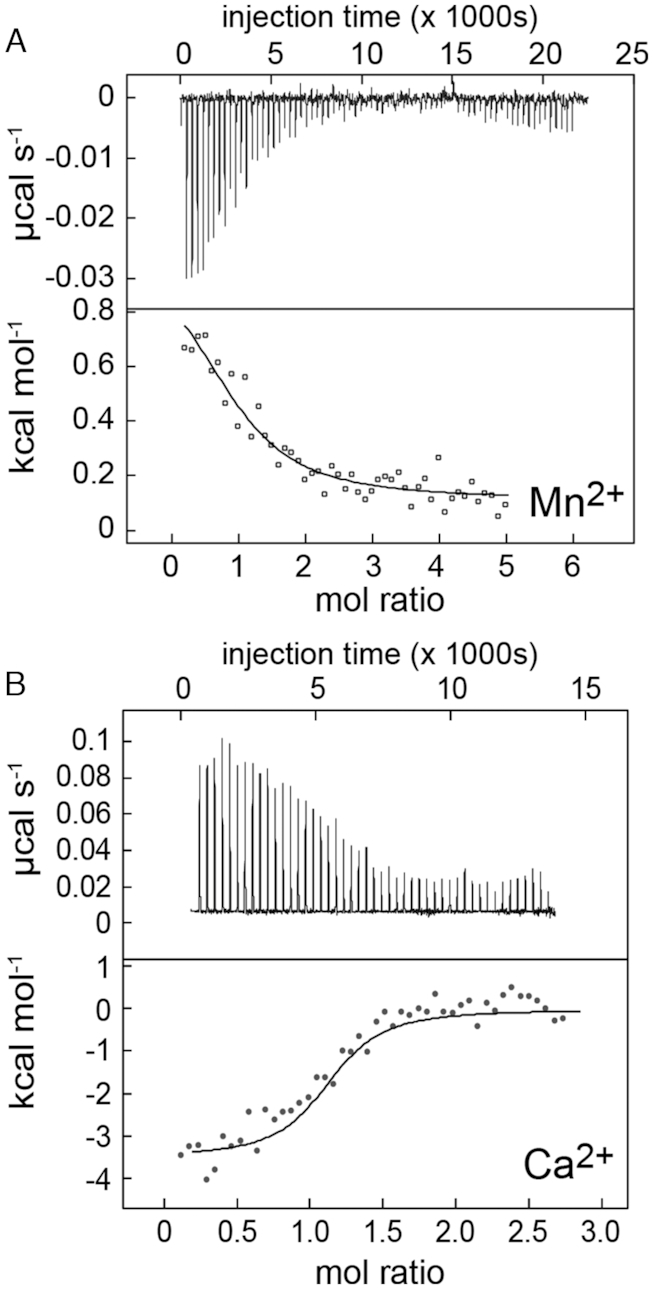
Thermograms of the interaction of the mgtA aptamer with Mn2+ (A) or Ca2+ (B) by isothermal titration calorimetry. For each panel, the upper panel shows the raw ITC data plotted as the change in power (μcal s−1) required to maintain equal temperature between the sample and reference cells as the function of time; lower panel shows intergated heat normalized for mol of injectant added. The thermodynamic parameters for metal binding defined by the continuous lines drawn through the data are summarized in Table 1.
Table 1.
Thermodynamic parameters of cation interactions with the mgtA RNA aptamer
| Parametera | Mn2+ | Ca2+ |
|---|---|---|
| K d (μM) | 54 ± 26 | 1.72 ± 0.03 |
| n | 1.0 ± 0.0 | 1.07 ± 0.0 |
| ΔH (kcal/mol) | 1.1 ± 0.1 | –3.6 ± 0.1 |
| TΔS (kcal/mol) | 7.0 ± 0.2 | 4.3 ± 0.1 |
| ΔG (kcal/mol) | –5.9 ± 0.02 | –7.9 ± 0.2 |
aMean and standard deviations are obtained from duplicated independent experiments. All experiments were performed in a background of 3 mM MgCl2 at 25°C. Data reported here are calculated from a one-site random binding model.
A significant number of the structural models of metal-bound yybP-ykoY riboswitches have been reported and are often used to interpret the sensing mechanism of riboswitches (Figure 2B and C). Moreover the recent reported affinities for Cd2+, Kd in the sub-mM range depending on the construct (30), suggest that Cd2+ could potentially turn on the yybP-ykoY riboswitches. To test the significance of those models in the context of the mgtA riboswitch, we also determined Cd2+ binding to the mgtA riboswitch. Although the mgtA RNA aptamer is compacted in the presence of 500 μM Cd2+ in a background of 3 mM Mg2+ (Supplementary Figure S5A), higher concentrations of Cd2+ drive the RNA aptamer into a more slowly migrating conformation(s) by native-PAGE. This suggests that at low concentrations, Cd2+ may bind in the same pocket (MB) (see Figure 2B) as Mn2+ and Ca2+. Both SHAPE (Supplementary Figure S5C) and SAXS (data not shown) analyses suggest that an extended or more open conformation is formed at high Cd2+ concentrations. It is also worth noting that low Cd2+ concentrations (≤500 μM) fail to activate transcription read-through and that higher Cd2+ concentrations are inhibitory (data not shown). Collectively, these data suggest that Cd2+ can bind and affect global change of the mgtA riboswitch, however the functional outcome is not likely physiologically relevant.
The mgtA riboswitch responds more robustly to Mn2+in vitro
Previously, it was demonstrated by in vitro transcription (IVT) termination experiments that the L. lactis yaoB riboswitch is activated by 0.5 mM Mn2+ but not by other metal ions tested (Fe2+, Co2+, Ni2+ and Ca2+); Mg2+ also induced transcription read-through, but only when provided at high concentrations (10 mM), suggesting that Mg2+ is not a preferred metal (28). Likewise, an L. lactis yaoB riboswitch chimera harboring L3 mutations to match the E. coli mntP and alx riboswitch L3 nucleotides, respond to Mn2+ as do other transition metals tested (Cd2+, Co2+, Cu2+, Fe2+ and Ni2+); however, transcription read-through was not induced by Ca2+, Mg2+ and Zn2+ (30). Other experiments demonstrate that these yybP-ykoY family riboswitches can also mediate Mn2+-dependent regulation in vivo (28,29). Thus, it is not possible to predict which metal induce transcription for the mgtA RNA aptamer based solely on it being part of the yybP-ykoY family riboswitches, nor on the selectivity in terms of binding for different metal ions. This observation motivated a series of in vitro transcription termination assays to determine which metal induces transcription for the mgtA RNA.
The binding of Mn2+ to the S. pneumoniae RNA aptamer is hypothesized to destabilize the terminator hairpin and permit read-through transcription of full-length mgtA mRNA transcript (Supplementary Figure S1C). Indeed, titration of increasing Mn2+ concentrations into the reaction led to the synthesis of full-length transcripts by E. coli RNA polymerase (Figure 7A) in a background of 250 μM Mg2+, revealing that Mn2+ binding to the aptamer activates transcription read-through. Ca2+ is also capable of activating transcription read-through (Figure 7B), but to a much lower extent compared to Mn2+ (Figure 7C). The half-maximal transcriptional activation by Mn2+ (0.23 ± 0.1 mM; Hill number, nH = 1) reveals that 5-fold less Mn2+ is needed to activate transcription read-through when compared to Ca2+, which is half-maximal at 1.1 ± 0.1 mM; Hill nH = 2.5 ± 0.6. Thus, despite the fact that Ca2+ binds more tightly to the RNA aptamer than Mn2+ (Figure 6) and appears to yield a similar conformation in solution, it stimulates read-through transcription to a far lesser degree. We found that Mg2+ alone is also capable of stimulating transcription read-through of mgtA (Supplementary Figure S6) with the addition of 3 mM Mg2+ resulting in ≈50% maximum read-through (Supplementary Figure S6B), yielding an effective Kd of 1.7±0.8 mM (Hill nH = 1.0 ± 0.8). Thus, Mn2+ is the most effective activator of mgtA transcription read-through and is uniquely functional at sub-mM Mn2+ in a concentration range comparable to the KdMn.
Figure 7.
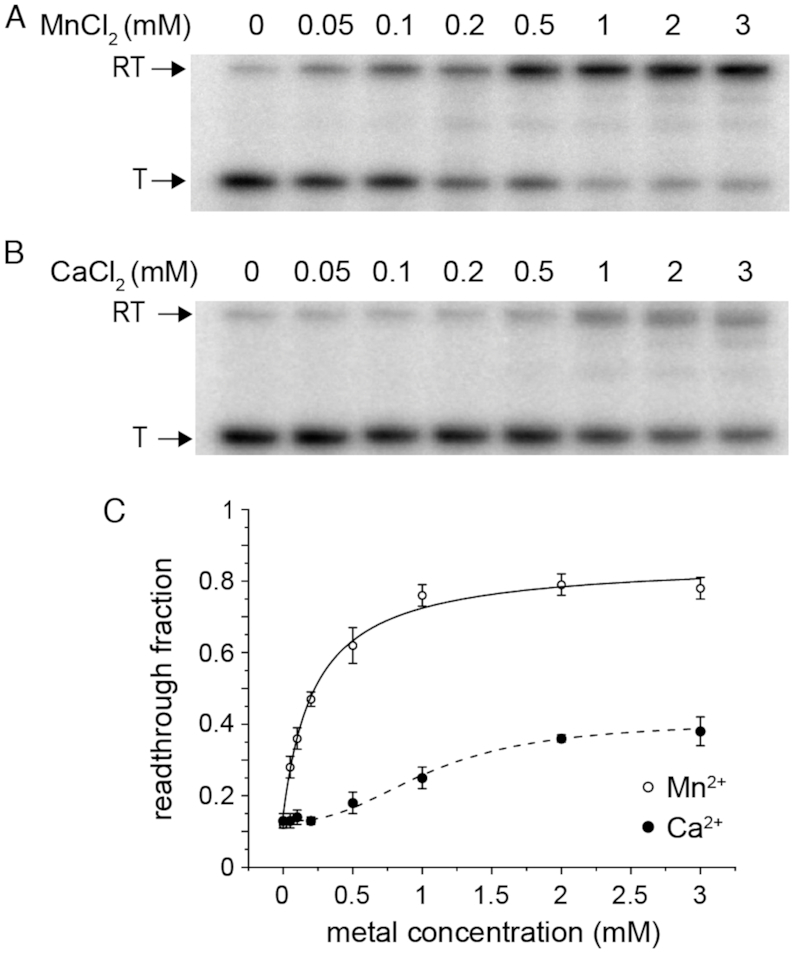
Mn2+ activates in vitro transcription of mgtA better than Ca. 32P labeled products produced in presence of Mn (A) and Ca (B) (in a background of 250 μM Mg2+) were separated by PAGE. Termination (T) products reflect terminator formation. Read-through (RT) products form as a result of metal-binding and stabilizing the aptamer thereby preventing formation of the terminator. (C) Quantification of in vitro transcription termination assays, with the continuous lines fit to a Hill model of activation (see text for details).
The S. pneumoniae mgtA riboswitch responds more robustly to Mn2+in vivo
We next determined if the S. pneumoniae mgtA riboswitch could also mediate Mn2+- and Ca2+-dependent regulation in vivo. S. pneumoniae cells lacking the Mn-specific exporter MntE (encoded by mntE) were used to induce Mn-stress growth conditions; ΔmntE cells accumulate high levels of intracellular Mn compared to wild type (WT) cells (34,50). During cellular Mn stress, mgtA mRNA and MgtA protein levels increase 5- and 4-fold, respectively, for ΔmntE mutants compared to WT cells (Figure 8 and Supplementary Figure S7). Clotrimazole, known to inhibit Ca2+ transport of eukaryotic sarco/endoplasmatic reticulum Ca2+-ATPase (SERCA PII-type ATPase), served as a suitable agent for imposing Ca2+-stress in S. pneumoniae WT cells (Supplementary Figure S8A). We note that clotrimazole treatment did not significantly affect intracellular Mn levels (Supplementary Figure S8B). We found that MgtA protein synthesis remained unchanged during Ca2+-stress, despite a 4-fold increase in cellular Ca2+ levels (Supplementary Figure S8C). No further metal activation studies were performed in S. pneumoniae, since we subsequently discovered that deletion of mgtA and possibly lower cellular MgtA protein levels are detrimental to cell viability (see below).
Figure 8.
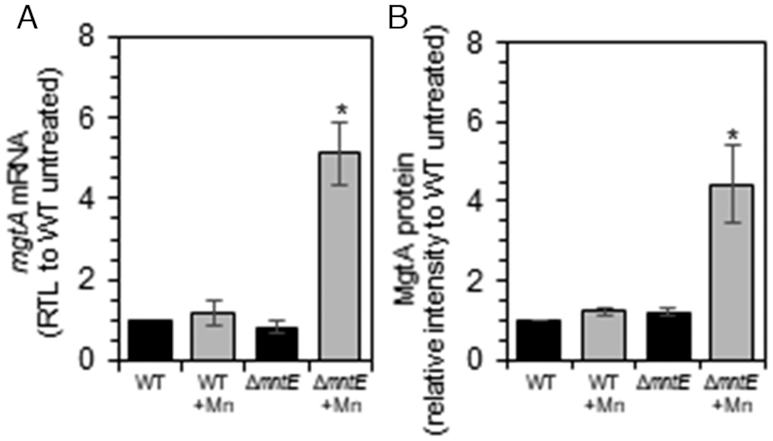
mgtA is induced during Mn-stress. Exponentially growing S. pneumoniae cells were diluted into pre-warmed BHI broth with 0 (black) or 200 μM (gray) Mn. Cells were harvested after 3.5 h growth. (A) Relative transcript levels of mgtA RNA. (B) Relative intensities of FLAG-tagged MgtA protein. The mean of at least three independent cultures ± SEM are shown. *P-value ≤ 0.05.
As an alternative method, we fused the S. pneumoniae mgtA leader region containing the mgtA riboswitch to a lacZ gene and monitored metal activation in Bacillus subtilis WT and ΔmntR strains. The latter strain experiences Mn toxicity, because the Mn-repressor MntR coordinates transcriptional regulation of the Mn uptake transporters (MntH and MntABC) and the Mn efflux pumps (MneP and MneS) (51,52). In the B. subtilis ΔmntR strain, the mgtA-lacZ mRNA level increases several-fold with the addition of Mn2+ to the growth medium (Figure 9A). Likewise, increasing Mn2+ concentrations stimulates expression of β-galactosidase up to 3.5-fold (black solid circles, Figure 9B). Removal of the terminator hairpin led to significantly higher activity (red solid squares), while deletion of the aptamer domain resulted in a minimal change in activity (grey solid triangles, Figure 9B). No response was observed in the absence of the 100 nt region upstream of the predicted mgtA riboswitch (open symbols, Figure 9B).
Figure 9.
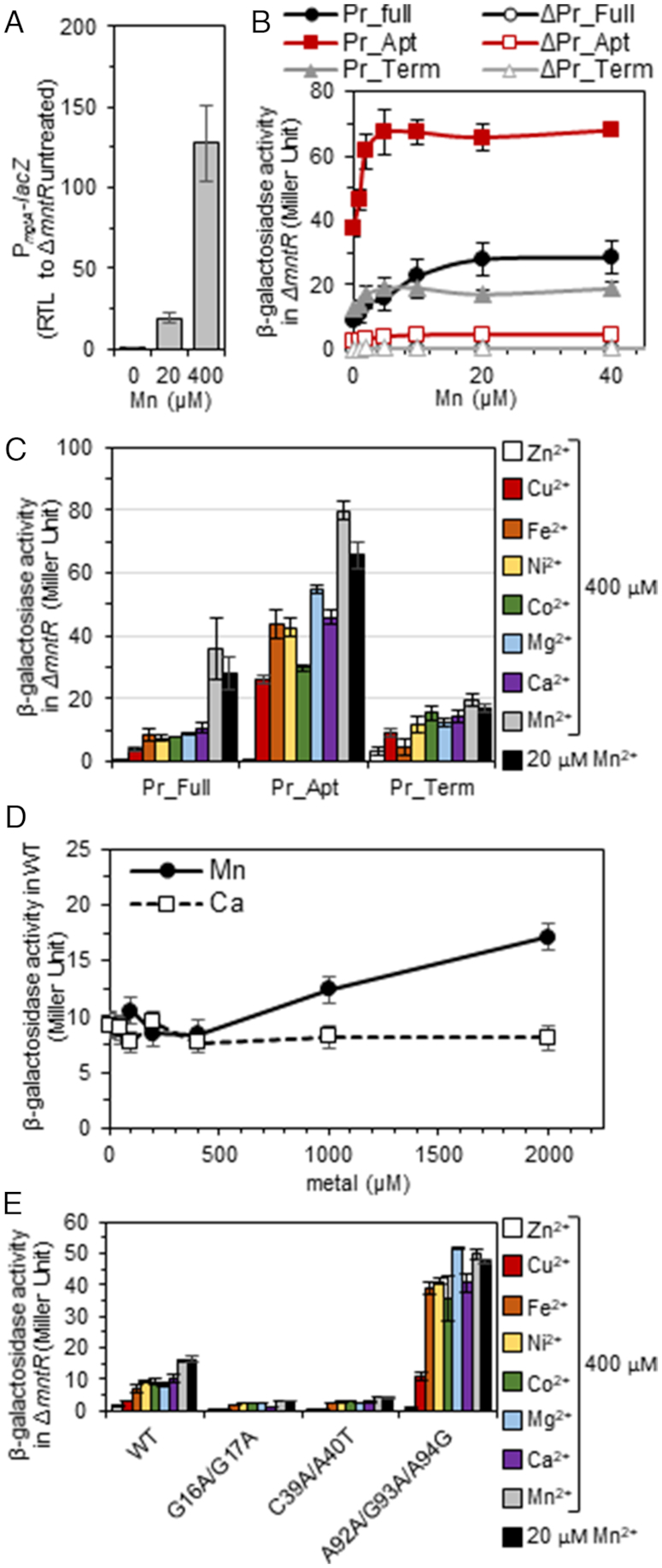
The S. pneumoniae mgtA riboswitch senses and responds specifically to intracellular Mn in B. subtilis. Exponentially B. subtilis cells growing in LB treated with indicated metal for 2 h, then harvested. (A) Relative transcription of PmgtA-lacZ fusion in ΔmntR cells grown with increasing concentrations of Mn. (B) β-Galactosidase activity in ΔmntR carrying various promoter-lacZ fusions in response to increasing concentrations of Mn. (C) β-galactosidase activity in response to various metal cations. (D) β-Galactosidase activity in WT cells containing WT-mgtA leader sequence in response to Mn and Ca. (E) β-galactosidase activity in ΔmntR carrying Pr_full (WT) and mutant forms the mgtA riboswitch in response to various metal cations. The mean of at least three independent cultures ± SEM are shown. Pr_Full, promoter plus intact 5′ UTR (185 nt; see Supplementary Figure S1); Pr_Apt, promoter plus aptamer (nucleotides 1–124); and Pr_Term, promoter plus terminator hairpin (nucleotides 101–185).
In contrast to Mn2+, the addition of other metal ions (Zn2+, Cu2+, Fe2+, Ni2+, Co2+, Mg2+ or Ca2+) to the growth medium had little to no effect on β-galactosidase expression (Figure 9C). Again, no response was observed in the absence of the 100 nt region upstream of the aptamer (Supplementary Figure S9). Deletion of the transcriptional terminator hairpin led to several-fold higher activation, while loss of the aptamer domain showed little if any change (Figure 9C). In B. subtilis WT cells, ≥400 μM Mn2+ was required to observe activation of β-galactosidase expression (Figure 9D). No activation was observed in WT cells with other metal ions tested (Supplementary Figure S9B), including Ca2+ (Figure 9D) even when added up to 10 mM (data not shown). Together, these data demonstrate that the S. pneumoniae mgtA riboswitch functions as a Mn2+-specific ‘on’ switch in vivo, inducing expression of MgtA.
To further connect aptamer folding with function and confirm metal specificity in vivo, key nucleotides within the S. pneumoniae mgtA RNA aptamer shown to disrupt folding of the aptamer were mutated in the context of the 5′ UTR and tested for functionality in vivo in a heterologous B. subtilis expression system. Both L1 (G16A/G17A) and L3 (C49A/A50U) substitutions reduced expression of β-galactosidase for all metal ions tested (Figure 9E). In contrast, the triple mutation of the 3′ side of L1 (A102C/G103A/G104G) results in high-level expression for all metals tested except for Zn2+ and Cu2+, which inhibit growth under these conditions, to a level consistent with unregulated expression, i.e. that obtained with aptamer alone (Figure 9C). This mutant 5′ UTR RNA may mis-fold in the cell and lead to a loss or destabilization of the terminator hairpin (see Supplementary Figure S1B). Taken collectively, these data correlate well with our global conformational analyses (Figures 3–5).
The mgtA riboswitch regulates a PII-type ATPase that protects cells against Mn toxicity
Our results so far indicate that the S. pneumoniae mgtA riboswitch functions as a Mn2+-specific sensor that turns on a predicted PII-type ATPase metal effluxer, MgtA, that we reasoned might play a role in cellular Mn homeostasis. To investigate this, we constructed S. pneumoniae D39 strains harboring clean deletions of mgtA and mntE, singularly or in combination. Transformation efficiency for the ΔmgtA strain appeared typical, despite a two-thirds reduction in colony size, while the ΔmgtA ΔmntE strain had the lowest efficiency, when selected on TSAII+SBA medium containing antibiotic under 5% CO2 atmosphere. Colony appearance was uniform in size and shape (data not shown). Absence of oxygen did not improve colony size of ΔmgtA mutants during the selection process, suggesting that the colony phenotype was not a result of reactive oxygen species. Furthermore, ΔmgtA isolates failed to reach 0.2 OD in 8–12 h when inoculated in brain-heart infusion (BHI) broth. Prolonged growth (>24 h) in BHI yielded fast-growing suppressors in isolated ΔmgtA harboring strains. Supplementation of BHI with exogenous Mg2+, Ca2+, Mn2+ or Fe2+ did not affect the growth pattern observed, nor did treatment with metal chelators EDTA and desferrioxamine (data not shown). These data support the idea that MgtA plays a role in Mn homeostasis in S. pneumoniae, since combined ΔmgtA and ΔmntE mutations showed the most pronounced colony defect during strain construction. We note that a ΔmgtA mutation was reported previously in S. pneumoniae D39 (53); however, we suspect that suppressor mutations were present in this strain, given the discrepancies with other published papers (25,31).
Because of this apparent requirement for pneumococcal MgtA under metal replete conditions, we tested the ability of MgtA to rescue Mn-sensitivity in a B. subtilis ΔmntR mutant. The S. pneumoniae mgtA gene was placed under an IPTG-inducible promoter and integrated into the chromosome of B. subtilis in single copy. Induction of MgtA synthesis has no effect on wild-type cell growth over time in LB with or without Mn (Figure 10A and Supplementary Figure S10). B. subtilis ΔmntR mutants fail to grow in the presence of Mn2+ compared to wild-type cells (Figure 10A and Supplementary Figure S10) due to Mn accumulation (Figure 10B). This Mn sensitivity is largely eliminated by induction of MgtA synthesis (Figure 10A). Total cell-associated Mn levels do not rise in ΔmntR mutants when MgtA is overexpressed, despite treatment with exogenous Mn2+ (Figure 10A). Interestingly, Ca2+ levels are significantly lower only in ΔmntR treated with Mn2+ only when MgtA is induced (Figure 10B). All other metals (Fe2+, Zn 2+, Cu2+) examined remained similar to WT levels (Supplementary Figure S11); Ni2+ and Co2+ were below the level of detection. These data indicate that the MgtA P-type ATPase transporter is capable of transporting both Ca2+ and Mn2+ under conditions of extreme Mn2+ toxicity. Further studies are required to fully understand the physiological importance of Ca2+ transport by MgtA in cells.
Figure 10.
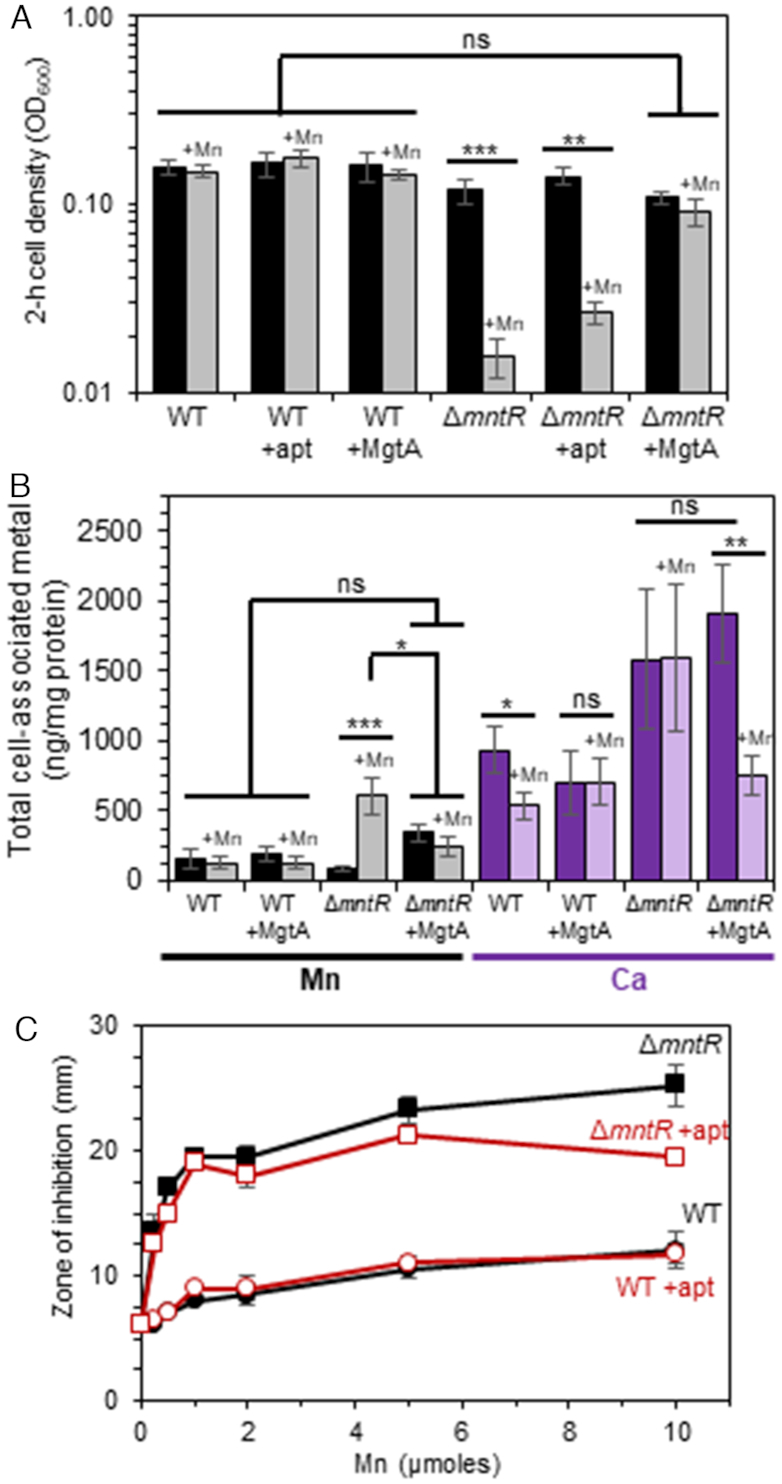
S. pneumoniae MgtA protein that is associated with the yybP-ykoY riboswitch rescues Mn-sensitivity in B. subtilis. Sensitivity of Mn2+ was monitored in B. subtilis WT and ΔmntR cells harboring the mgtA RNA aptamer sequence or MgtA in LB broth containing 0 (darker bars) or 50 μM (lighter bars) Mn overtime. Isopropyl β-D-thiogalactopyranoside (1mM) was added to induce MgtA protein synthesis. (A) 2-h cell density. (B) Total cell associated Mn and Ca determined by IPC-MS after 2.5 h growth. (C) Disk diffusion assay. The mean of at least three independent cultures ± SEM are shown for each experiment. *P-value ≤ 0.05; **P-value ≤ 0.01; ***P-value ≤ 0.001; ns, no significance.
DISCUSSION
Metal cation homeostasis is essential for colonization and pathogenesis of bacteria. As such, the mammalian host has evolved mechanisms to both restrict or intoxicate invading bacteria with specific metals, depending on the microenvironmental niche. In response, bacteria have evolved complex regulatory strategies to adapt in an effort to maintain homeostasis of essential metal ions. Recent work provides support for the idea that individual metal ions in a bacterial cell are buffered to an extent that is largely determined by the absolute sensitivities or set-points of metallosensors that regulate the expression of downstream genes, which often encode metal transporters (54,55). These metallosensors detect metal activities (free metal concentrations) around a relatively narrow concentration window defined by their KdMe (Me, metal), a range in concentration that is often far less than total cell-associated metal (54,55). Of importance here is that Mg2+ levels are thought to be maintained in the low mM range, primarily controlled by the Mg2+-sensing ‘M-box’ riboswitch (in B. subtilis, for example) (45), while Mn2+ concentrations are transcriptionally controlled by Mn2+-sensing protein repressors MntR in B. subtilis (56) and a related repressor PsaR, in S. pneumoniae (57). The M-box riboswitch is tuned to ≈mM affinity (55). The KdMn for the regulatory ‘sensing’ site in PsaR is in the low μM range (1.3 μM), while the Salmonella MntR KdMn is about 10-fold lower affinity (13 μM at pH 8.0) (55). This ‘set-point’ model suggests that Mn2+ may be buffered in the low μM range, which in S. pneumoniae may be further restricted by the constitutive expression of MntE, to which effluxes Mn2+ from the pneumococcal cell (50). In contrast to what is known about Mn2+ homeostasis, intracellular bacterial Ca2+ homeostasis remains largely unexplored, and to our knowledge a bacterial Ca2+-specific metallosensor or two-component response regulatory system has yet to be described.
In this report, we describe the physical and functional characterization of a Mn2+-sensing riboswitch that is found upstream of the mgtA gene. Using a combination of native gel mobility experiments, SHAPE probing, SAXS analysis and in vitro transcription experiments, we show that this riboswitch aptamer domain is capable of binding Mn2+ and Ca2+ in the presence of mM Mg2+, and that all three metals, including Mg2+ alone stabilize a compact state, relative to an extended or ‘open’ conformation. All three metals stimulate transcription read-through in vitro using E. coli RNA polymerase, but Mn2+ is clearly the most potent activating metal in these experiments, both in terms of maximal read-through activity and metal sensitivity (≤100 μM metal). The half-maximal Mn2+ concentration required to shift the folding equilibrium of the RNA to the compact form and stimulate in vitro transcription is within a factor of four of the KdMn measured by calorimetry (50–200 μM); furthermore, the response in all three experiments conducted in 3 mM Mg2+ fits well to a rectangular hyperbolic function, consistent with a single Mn2+-sensing site on the riboswitch, which we speculate is the MnB site (see Figure 2A). As anticipated from previous work, mutations in the metal-site pocket abrogate or greatly weaken (A18C) Mn2+-dependent folding, and when incorporated into the mgtA 5′ UTR in cells, render them functionally inactive.
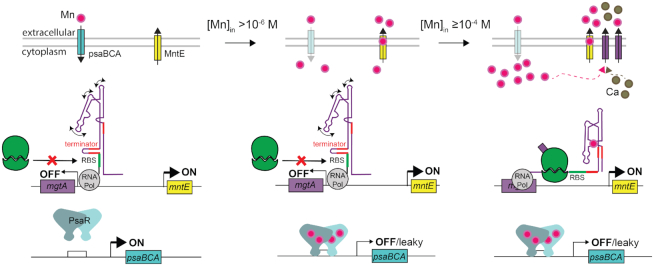
These physical characteristics are largely recapitulated in pneumococcal cells and in a heterologous B. subtilis host, with Mn2+ the only divalent metal ion capable of read-through transcription in either cellular background, which in S. pneumoniae results in a corresponding increase in the cellular amount of MgtA protein. Ca2+ is not a strong mgtA inducer in vivo. However, significant Mn2+-dependent mRNA or protein expression in either bacterial host is only observed in a strain that is extremely sensitive to Mn2+ toxicity, i.e. in ΔmntR B. subtilis and in ΔmntE S. pneumoniae. Under these conditions where mgtA mRNA and MgtA protein levels are higher, MgtA is capable of lowering total cell-associated Mn2+, consistent with a role in Mn2+ efflux by MgtA. These physiological findings are entirely consistent with the observed in vitro metal sensitivity of the riboswitch itself, which is in the 50–200 μM Mn2+ range, or set ≥5–10-fold higher than the metalloregulator proteins MntR and PsaR; unfortunately, the metal binding affinity of the MntE has not yet been determined but might be expected to be in the low μM range. Thus, if these systems are present and functioning, they would prevent intracellular Mn2+ from reaching the ≈100 μM range, and thus this riboswitch and subsequent MgtA protein production would simply not fire. Thus, our data support the hypothesis that the MgtA production functions as a ‘fail-safe’ or back-up system to allow S. pneumoniae to adapt to acute phases of Mn2+ toxicity in the host when Mn2+ uptake regulated by PsaR and Mn2+ efflux by MntE fail to avoid cellular toxicity (Figure 11) (50).
Figure 11.
Threshold model for Mn2+ sensing and detoxification in S. pneumoniae. The Mn2+ binding affinity of the S. pneumoniae mgtA riboswitch is such that there is sufficient free intracellular Mn2+, governed by the relative affinities of the transcriptional Mn uptake repressor PsaR and the constitutively expressed Mn-specific efflux pump MntE (34,57), to ensure MgtA is not induced and that key intracellular Mn-dependent enzymes are active. At concentrations >1 μM Mn2+, PsaR will bind Mn2+, increasing its affinity to DNA and repress psaBCA transcription, thereby reducing Mn import; excess Mn2+ will continue to be effluxed by MntE. Our data suggest that as free Mn2+ rises to ≥100 μM, the mgtA riboswitch functions as a failsafe ‘on’ signal inducing MgtA expression to prevent Mn2+ toxicity. In addition, the riboswitch also functions to regulate Ca2+ efflux under this condition.
The functional significance of Ca2+ binding by this riboswitch and Ca2+ efflux by MgtA under conditions of extreme Mn2+ stress (Figure 10B) are not yet known. MgtA is clearly capable of effluxing Ca2+ from cells and clotrimazole and extracellular Ca2+ interfere with this process (Supplementary Figure S8), but under what conditions this might occur during the course of a bacterial infection are not known. mgtA is the first gene in a co-transcribed two-gene operon with a downstream, non-essential gene (spd_1382) encoding a glutathione S-transferase (25). Consequently, the defective growth of ΔmgtA mutants, irrespective of Mn2+ status, could be due to a polarity on Spd_1382 expression and/or a second role of MgtA in cellular metabolism possibly in maintaining Ca2+ homeostasis. The former is less likely as the replacement of the native mgtA gene with a construct encoding MgtA triple-FLAG-tagged protein linked to an antibiotic resistance cassette did not show defective growth (data not shown). The putative essentiality of mgtA was not studied further here, but an implication of this operon arrangement is that expression of the Spd_1382 glutathione S-transferase is also controlled by the mgtA riboswitch aptamer in response to Mn2+ and Ca2+. One possibility is that under metal-replete conditions, small amounts of MgtA produced via leaky expression function in Ca2+ efflux, perhaps required to efficiently metallate an obligatory Ca2+-requiring enzyme on the outside of the pneumococcal cell; indeed, such a role for P1B-type ATPases in metallating periplasmic, extracellular or membrane-anchored client proteins is not without precedent for Cu+-specific P1B-ATPases in other bacteria (58–60). Alternatively, MgtA might play an important role in maintaining Ca2+ homeostasis, but total levels of Ca2+ simply do not change much under transition metal-replete conditions. This would be consistent with the classification and biochemical characterization of the bacterial MgtA-like transporters as primarily Ca2+ transporters (48). In this model, only under conditions of acute Mn2+ toxicity does MgtA function as a Mn2+ transporter, a condition bolstered by increased accumulation of the MgtA in the membrane to effect efflux of this transition metal.
As a classic PII-type ATPase, MgtA is predicted to transport two metal cations per reaction cycle, which could in theory involve any combination of Mn2+ or Ca2+ (48). This observation is not intuitive in terms of the basis of metal ion recognition by the transporter based on the significant chemical differences between Mn2+ and Ca2+. However, it is not unprecedented. The dual metal specificity of MgtA offers striking functional parallels to the secretory pathway Ca2+-ATPases (SPCAs) previously identified in yeast and vertebrates and known to transport both Mn2+ and Ca2+ into the Golgi from the cytosol. Here, the metal is used to metallate key client enzymes in this compartment or is simply secreted via this route (61–63). MgtA in fact shows significant sequence similarity to SPCAs and may well harbor the same Q747A substitution that enhances Mn2+ transport relative to Ca2+ (Supplementary Figure S12). Biochemical studies of S. pneumoniae MgtA coupled with more extensive physiological characterization of the functional role of MgtA are clearly required to further elucidate the metal-dependence and functional role of the MgtA transport cycle.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J.D. Helmann (Cornell University, NY) for providing the necessary plasmids and B. subtilis strains to make the lacZ reporter fusions and IPTG inducible constructs, H. Niu (Indiana University, IN) for providing all necessary reagents for the IVT assays, and the Y.X. Wang group (National Cancer Institute) for help collecting the SAXS data. We appreciate the support of the A.E. Simon's group (University of Maryland, College Park), and a special thanks to J. May for providing the necessary training for us to perform SHAPE profiling.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Idaho State University start up fund (to J.E.M.); Institutional Development Award from the National Institutes of General Medical Sciences [P20GM103408 to Idaho State University]; National Institutes of General Medical Sciences [R35 GM118157 to D.P.G., R01 GM042569-25S1 to J.E.M, R01 GM127715 to M.E.W.]. Funding for open access charge: Idaho State University start up funds (to J.E.M.).
Conflict of interest statement. The authors certify that they have no affiliations with or involvement in any organization or entity with any financial interests or non-financial interests in the materials discussed in this manuscript.
REFERENCES
- 1. Barrick J.E., Corbino K.A., Winkler W.C., Nahvi A., Mandal M., Collins J., Lee M., Roth A., Sudarsan N., Jona I. et al.. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:6421–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meyer M.M., Hammond M.C., Salinas Y., Roth A., Sudarsan N., Breaker R.R.. Challenges of ligand identification for riboswitch candidates. RNA Biol. 2011; 8:5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 2004; 58:303–328. [DOI] [PubMed] [Google Scholar]
- 4. Storz G., Vogel J., Wassarman K.M.. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell. 2011; 43:880–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Waters L.S., Storz G.. Regulatory RNAs in bacteria. Cell. 2009; 136:615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wagner E.G.H., Romby P.. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet. 2015; 90:133–208. [DOI] [PubMed] [Google Scholar]
- 7. Brantl S. Bacterial chromosome-encoded small regulatory RNAs. Future Microbiol. 2009; 4:85–103. [DOI] [PubMed] [Google Scholar]
- 8. Repoila F., Darfeuille F.. Small regulatory non-coding RNAs in bacteria: physiology and mechanistic aspects. Biol. Cell. 2009; 101:117–131. [DOI] [PubMed] [Google Scholar]
- 9. Majdalani N., Vanderpool C.K., Gottesman S.. Bacterial small RNA regulators. Crit. Rev. Biochem. Mol. Biol. 2005; 40:93–113. [DOI] [PubMed] [Google Scholar]
- 10. Song J., Lays C., Vandenesch F., Benito Y., Bes M., Chu Y., Lina G., Romby P., Geissmann T., Boisset S.. The expression of small regulatory RNAs in clinical samples reflects the different life styles of Staphylococcus aureus in colonization vs. infection. PLoS One. 2012; 7:e37294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xia L., Xia W., Li S., Li W., Liu J., Ding H., Li J., Li H., Chen Y., Su X. et al.. Identification and expression of small non-coding RNA, L10-Leader, in different growth phases of Streptococcus mutans. Nucleic Acid Ther. 2012; 22:177–186. [DOI] [PubMed] [Google Scholar]
- 12. Ahmed W., Zheng K., Liu Z.F.. Small non-coding RNAs: new insights in modulation of host immune response by intracellular bacterial pathogens. Front. Immunol. 2016; 7:431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kröger C., Dillon S.C., Cameron A.D., Papenfort K., Sivasankaran S.K., Hokamp K., Chao Y., Sittka A., Hébrard M., Händler K. et al.. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E1277–E1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weiser J.N., Ferreira D.M., Paton J.C.. Streptococcus pneumoniae: transmission, colonization and invasion. Nat. Rev. Microbiol. 2018; 16:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. O’Brien K.L., Wolfson L.J., Watt J.P., Henkle E., Deloria-Knoll M., McCall N., Lee E., Mulholland K., Levine O.S., Cherian T. et al.. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet. 2009; 374:893–902. [DOI] [PubMed] [Google Scholar]
- 16. Collaborators, G.L.R.I. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 2018; 18:1191–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chao Y., Marks L.R., Pettigrew M.M., Hakansson A.P.. Streptococcus pneumoniae biofilm formation and dispersion during colonization and disease. Front. Cell Infect. Microbiol. 2014; 4:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Honsa E.S., Johnson M.D., Rosch J.W.. The roles of transition metals in the physiology and pathogenesis of Streptococcus pneumoniae. Front. Cell Infect. Microbiol. 2013; 3:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Becker K.W., Skaar E.P.. Metal limitation and toxicity at the interface between host and pathogen. FEMS Microbiol. Rev. 2014; 38:1235–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Livny J., Brencic A., Lory S., Waldor M.K.. Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res. 2006; 34:3484–3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kumar R., Shah P., Swiatlo E., Burgess S.C., Lawrence M.L., Nanduri B.. Identification of novel non-coding small RNAs from Streptococcus pneumoniae TIGR4 using high-resolution genome tiling arrays. BMC Genomics. 2010; 11:350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tsui H.C., Mukherjee D., Ray V.A., Sham L.T., Feig A.L., Winkler M.E.. Identification and characterization of noncoding small RNAs in Streptococcus pneumoniae serotype 2 strain D39. J. Bacteriol. 2010; 192:264–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Acebo P., Martin-Galiano A.J., Navarro S., Zaballos A., Amblar M.. Identification of 88 regulatory small RNAs in the TIGR4 strain of the human pathogen Streptococcus pneumoniae. RNA. 2012; 18:530–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sinha D., Zimmer K., Cameron T.A., Rusch D.B., Winkler M.E., De Lay N.R.. Redefining the sRNA transcriptome inStreptococcus pneumoniae Serotype 2 Strain D39. J. Bacteriol. 2019; doi:10.1128/JB.00764-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Slager J., Aprianto R., Veening J.W.. Deep genome annotation of the opportunistic human pathogen Streptococcus pneumoniae D39. Nucleic Acids Res. 2018; 46:9971–9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Waters L.S., Sandoval M., Storz G.. The Escherichia coli MntR miniregulon includes genes encoding a small protein and an efflux pump required for manganese homeostasis. J. Bacteriol. 2011; 193:5887–5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Veyrier F.J., Boneca I.G., Cellier M.F., Taha M.K.. A novel metal transporter mediating manganese export (MntX) regulates the Mn to Fe intracellular ratio and Neisseria meningitidis virulence. PLoS Pathog. 2011; 7:e1002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Price I.R., Gaballa A., Ding F., Helmann J.D., Ke A.. Mn(2+)-sensing mechanisms of yybP-ykoY orphan riboswitches. Mol. Cell. 2015; 57:1110–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dambach M., Sandoval M., Updegrove T.B., Anantharaman V., Aravind L., Waters L.S., Storz G.. The ubiquitous yybP-ykoY riboswitch is a manganese-responsive regulatory element. Mol. Cell. 2015; 57:1099–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bachas S.T., Ferré-D’Amaré A.R.. Convergent use of heptacoordination for cation selectivity by RNA and protein metalloregulators. Cell Chem. Biol. 2018; 25:962–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rosch J.W., Sublett J., Gao G., Wang Y.D., Tuomanen E.I.. Calcium efflux is essential for bacterial survival in the eukaryotic host. Mol. Microbiol. 2008; 70:435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Neef J., Andisi V.F., Kim K.S., Kuipers O.P., Bijlsma J.J.. Deletion of a cation transporter promotes lysis in Streptococcus pneumoniae. Infect. Immun. 2011; 79:2314–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rosch J.W., Gao G., Ridout G., Wang Y.D., Tuomanen E.I.. Role of the manganese efflux system mntE for signalling and pathogenesis in Streptococcus pneumoniae. Mol. Microbiol. 2009; 72:12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martin J.E., Giedroc D.P.. Functional determinants of metal ion transport and selectivity in paralogous cation diffusion facilitator transporters CzcD and MntE in Streptococcus pneumoniae. J. Bacteriol. 2016; 198:1066–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lilley D.J.M. Analysis of global conformation of branched RNA species using electrophoresis and fluorescence. Methods Enzymol. 2000; 317:368–393. [DOI] [PubMed] [Google Scholar]
- 36. Mortimer S.A., Weeks K.M.. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 2007; 129:4144–4145. [DOI] [PubMed] [Google Scholar]
- 37. Wilkinson K.A., Merino E.J., Weeks K.M.. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006; 1:1610–1616. [DOI] [PubMed] [Google Scholar]
- 38. Rice G.M., Busan S., Karabiber F., Favorov O.V., Weeks K.M.. SHAPE analysis of small RNAs and riboswitches. Methods Enzymol. 2014; 549:165–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Karabiber F., McGinnis J.L., Favorov O.V., Weeks K.M.. QuShape: rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA. 2013; 19:63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen B., Zuo X., Wang Y.X., Dayie T.K.. Multiple conformations of SAM-II riboswitch detected with SAXS and NMR spectroscopy. Nucleic Acids Res. 2012; 40:3117–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Y.X., Zuo X., Wang J., Yu P., Butcher S.E.. Rapid global structure determination of large RNA and RNA complexes using NMR and small-angle X-ray scattering. Methods. 2010; 52:180–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Svergun D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999; 76:2879–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Blouin S., Lafontaine D.A.. A loop loop interaction and a K-turn motif located in the lysine aptamer domain are important for the riboswitch gene regulation control. RNA. 2007; 13:1256–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ren A., Rajashankar K.R., Patel D.J.. Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch. Nature. 2012; 486:85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dann C.E. 3rd, Wakeman C.A., Sieling C.L., Baker S.C., Irnov I., Winkler W.C.. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007; 130:878–892. [DOI] [PubMed] [Google Scholar]
- 46. Wakeman C.A., Ramesh A., Winkler W.C.. Multiple metal-binding cores are required for metalloregulation by M-box riboswitch RNAs. J. Mol. Biol. 2009; 392:723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Panja S., Hua B., Zegarra D., Ha T., Woodson S.A.. Metals induce transient folding and activation of the twister ribozyme. Nat. Chem. Biol. 2017; 13:1109–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dyla M., Terry D.S., Kjaergaard M., Sorensen T.L., Andersen Lauwring, Andersen J., J.P. Rohde Knudsen, Altman C., Nissen R.B., Blanchard S.C.. Dynamics of P-type ATPase transport revealed by single-molecule FRET. Nature. 2017; 551:346–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sung H.L., Nesbitt D.J.. Single molecule FRET kinetics of the Mn(2+) riboswitch: evidence for allosteric Mg(2+) control of “induced fit” vs. “conformal selection” folding pathways. J. Phys. Chem. B. 2019; 123:2005–2015. [DOI] [PubMed] [Google Scholar]
- 50. Martin J.E., Lisher J.P., Winkler M.E., Giedroc D.P.. Perturbation of manganese metabolism disrupts cell division in Streptococcus pneumoniae. Mol. Microbiol. 2017; 104:334–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huang X., Shin J.H., Pinochet-Barros A., Su T.T., Helmann J.D.. Bacillus subtilis MntR coordinates the transcriptional regulation of manganese uptake and efflux systems. Mol. Microbiol. 2017; 103:253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Que Q., Helmann J.D.. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol. Microbiol. 2000; 35:1454–1468. [DOI] [PubMed] [Google Scholar]
- 53. Neef J., Andisi V.F., Kim K.S., Kuipers O.P., Bijlsma J.J.. Deletion of a cation transporter promotes lysis in Streptococcus pneumoniae. Infect. Immunity. 2011; 79:2314–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Osman D., Foster A.W., Chen J., Svedaite K., Steed J.W., Lurie-Luke E., Huggins T.G., Robinson N.J.. Fine control of metal concentrations is necessary for cells to discern zinc from cobalt. Nat. Commun. 2017; 8:1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Osman D., Martini M.A., Foster A.W., Chen J., Scott A.J.P., Morton R.J., Steed J.W., Lurie-Luke E., Huggins T.G., Lawrence A.D. et al.. Bacterial sensors define intracellular free energies for correct enzyme metalation. Nat. Chem. Biol. 2019; 15:241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Glasfeld A., Guedon E., Helmann J.D., Brennan R.G.. Structure of the manganese-bound manganese transport regulator of Bacillus subtilis. Nat. Struct. Biol. 2003; 10:652–657. [DOI] [PubMed] [Google Scholar]
- 57. Lisher J.P., Higgins K.A., Maroney M.J., Giedroc D.P.. Physical characterization of the manganese-sensing pneumococcal surface antigen repressor from Streptococcus pneumoniae. Biochemistry. 2013; 52:7689–7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Padilla-Benavides T., George Thompson A.M., McEvoy M.M., Arguello J.M.. Mechanism of ATPase-mediated Cu+ export and delivery to periplasmic chaperones: the interaction of Escherichia coli CopA and CusF. J. Biol. Chem. 2014; 289:20492–20501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Osman D., Patterson C.J., Bailey K., Fisher K., Robinson N.J., Rigby S.E., Cavet J.S.. The copper supply pathway to a Salmonella Cu,Zn-superoxide dismutase (SodCII) involves P(1B)-type ATPase copper efflux and periplasmic CueP. Mol. Microbiol. 2013; 87:466–477. [DOI] [PubMed] [Google Scholar]
- 60. Utz M., Andrei A., Milanov M., Trasnea P.I., Marckmann D., Daldal F., Koch H.G.. The Cu chaperone CopZ is required for Cu homeostasis in Rhodobacter capsulatus and influences cytochrome cbb3 oxidase assembly. Mol. Microbiol. 2018; 111:764–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Durr G., Strayle J., Plemper R., Elbs S., Klee S.K., Catty P., Wolf D.H., Rudolph H.K.. The medial-Golgi ion pump Pmr1 supplies the yeast secretory pathway with Ca2+ and Mn2+ required for glycosylation, sorting, and endoplasmic reticulum-associated protein degradation. Mol. Biol. Cell. 1998; 9:1149–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vanoevelen J., Dode L., Van Baelen K., Fairclough R.J., Missiaen L., Raeymaekers L., Wuytack F.. The secretory pathway Ca2+/Mn2+-ATPase 2 is a Golgi-localized pump with high affinity for Ca2+ ions. J. Biol. Chem. 2005; 280:22800–22808. [DOI] [PubMed] [Google Scholar]
- 63. Mukhopadhyay S., Linstedt A.D.. Identification of a gain-of-function mutation in a Golgi P-type ATPase that enhances Mn2+ efflux and protects against toxicity. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:858–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.