Abstract
SOX8, SOX9 and SOX10 compose the SOXE transcription factor group. They govern cell fate and differentiation in many lineages, and mutations impairing their activity cause severe diseases, including campomelic dysplasia (SOX9), sex determination disorders (SOX8 and SOX9) and Waardenburg-Shah syndrome (SOX10). However, incomplete knowledge of their modes of action limits disease understanding. We here uncover that the proteins share a bipartite transactivation mechanism, whereby a transactivation domain in the middle of the proteins (TAM) synergizes with a C-terminal one (TAC). TAM comprises amphipathic α-helices predicted to form a protein-binding pocket and overlapping with minimal transactivation motifs (9-aa-TAD) described in many transcription factors. One 9-aa-TAD sequence includes an evolutionarily conserved and functionally required EΦ[D/E]QYΦ motif. SOXF proteins (SOX7, SOX17 and SOX18) contain an identical motif, suggesting evolution from a common ancestor already harboring this motif, whereas TAC and other transactivating SOX proteins feature only remotely related motifs. Missense variants in this SOXE/SOXF-specific motif are rare in control individuals, but have been detected in cancers, supporting its importance in development and physiology. By deepening understanding of mechanisms underlying the central transactivation function of SOXE proteins, these findings should help further decipher molecular networks essential for development and health and dysregulated in diseases.
INTRODUCTION
The diversification and sophistication of cell types that has occurred during evolution has been possible thanks to the multiplication and specialization of many types of genes and regulatory factors. In particular, the SOX family has evolved to exert master roles in cell fate determination and differentiation in progenitor and stem cells as well as differentiated cells (1–3). SOX proteins are defined as harboring a high-mobility-group (HMG)-type domain that is at least 50% identical to that of the family founder, the sex-determining region on the Y chromosome (SRY). This domain binds and bends DNA at sequences matching or resembling the C[A/T]TTG[T/A][T/A] motif. It also features nuclear import and export signals and interacts with various proteins. Based on sequence identity, SOX proteins are distributed into eight groups, A to H (4). Members of the same group share close to 100% identity in the HMG domain and also share a high degree of identity in other functional domains, including dimerization, transactivation and transrepression motifs, whereas proteins belonging to distinct groups share no or minimal identity outside the HMG domain (1). Whereas the HMG domain has been characterized in great detail, current knowledge of the structure/function properties of the other cardinal attributes of SOX proteins remains generally meager. We here set out to increase knowledge of the transactivation domains of the SOXE proteins.
Humans and most vertebrates possess three SOXE proteins: SOX8, SOX9 and SOX10. Their genes overlap in expression and are either uniquely, additively, or redundantly needed in such key processes as chondrogenesis (SOX9), sex determination and differentiation (SOX8 and SOX9), melanogenesis (SOX9 and SOX10) (5), neural crest development (SOX8, SOX9 and SOX10), and neuronal and glial differentiation (SOX8, SOX9 and SOX10) (6–8). In humans, SOX8 mutations cause a spectrum of female and male reproductive anomalies (9), while SOX9 mutations cause Campomelic Dysplasia, a severe skeletal malformation syndrome, as well as XY sex reversal (10–12), and SOX10 mutations cause Waardenburg-Shah syndrome (13). Furthermore, SOX9 and SOX10 overexpression are poor or favorable prognosis markers in many cancers, such as glioma, melanoma and breast, colorectal, pancreas and prostate cancer (5,14). These findings point to critical roles for SOXE proteins in various developmental, physiological and pathological processes. Reaching deep understanding of the structural organization and modes of actions of these proteins is thus fundamental to uncover how they function normally and can be dysregulated in diseases.
Transactivation is a focal activity of SOXE proteins. For instance, SOX8 and SOX9 transactivate Sertoli cell-specific genes (15); SOX9 also transactivates chondrocyte-specific genes (16,17); and SOX10 transactivates oligodendrocyte- and melanocyte-specific genes (18,19). A current conundrum is that compelling evidence of redundant and additive activities in multiple processes contrasts with data suggesting that SOXE proteins utilize different transactivation domains. SOX8 was indeed proposed to transactivate through a centrally located sequence (20,21), which we will refer to as TAM (transactivation domain in the middle of the protein). In contrast, SOX9 has a key transactivation domain at its C-terminus (22), which we will refer to as TAC, and might enhance its transactivation activity through a PQA-rich domain, which does not exist in SOX8 and SOX10 (23). SOX10, like SOX9, possesses a potent TAC domain (24,25), and also possesses a so-called K2 domain, matching SOX8 TAM and contributing to transactivation in an apparently cell type-specific manner (26). These data raise questions on whether SOX8 possesses a functional TAC and SOX9 a functional TAM, and whether the SOX9 PQA and SOX10 TAM have autonomous transactivation activity or only potentiate the activity of TAC.
We show here that TAM and TAC are autonomous and synergistic transactivation domains in each SOXE protein and that PQA may help mediate SOX9 transactivation in specific contexts. Focusing on TAM, we identify a unique EΦ[D/E]QYΦ sequence that is required for transactivation, is remarkably conserved in SOXE and SOXF proteins, and is predicted to participate in a binding pocket that likely interacts with transcriptional co-activators or basal transcriptional machinery components.
MATERIALS AND METHODS
SOX protein sequence analyses
SOX protein sequences were downloaded from NCBI (Supplementary Table S1) and aligned with the ClustalW tool embedded in MacVector16 software (MacVector, Apex, NC, USA). Hydropathy plots were generated using the Kyte-Doolittle scale (27). The presence of 9-aa-TAD motifs was determined using the Piskacek tool (28). Secondary and tertiary structures were predicted for the SOX9 TAM-CD region using SWISS-MODEL (29), I-TASSER (30) and PEP-FOLD3 (31). The best scoring models were exported in PDB format and processed using UCSF Chimera v1.11.2 (32) to generate high-quality images. Synonymous and missense variants in SOXE and SOXF sequences in control human individuals were downloaded from the gnomAD database (33) and somatic missense variants detected in cancers from the COSMIC database (34).
Generation of wild-type and mutant SOX protein expression plasmids
Human SOXE and SOX17 expression plasmids were generated by cloning full-length coding sequences in frame with an N-terminal 3FLAG epitope (35) in the pcDNA3.1(+) vector (Thermo Fisher Scientific, Waltham, MA, USA). These sequences were amplified by PCR using PfuUltra High-Fidelity DNA Polymerase (Agilent Technologies, Santa Clara, CA, USA) and human cDNA using forward and reverse primers containing BamHI and EcoRI sites, respectively (Supplementary Table S2). Plasmids encoding GAL4DBD/SOXE fusion proteins were generated by cloning SOXE cDNA segments into the pBIND plasmid (Promega, Madison, WI, USA). These segments were generated by PCR using custom-made primers (Supplementary Table S3). Missense mutations were introduced in SOX sequences by QuikChange Site-Directed Mutagenesis (Stratagene, San Diego, CA, USA) using tailored primers (Supplementary Table S4). The integrity of all plasmid inserts was verified by Sanger sequencing.
Reporter assays
HEK-293 (CRL-1573; ATCC, Manassas, VA, USA) and SW-1353 (HTB-94; ATCC) cells were cultured in monolayer in 2 ml DMEM supplemented with 10% FBS (Life Technologies, Carlsbad, CA, USA). Cells (0.3 million) were plated in each well of six-well plates and transfected 4–6 h later with a mixture made of 100 μl DMEM, 3 μl FuGENE6 (Promega) and 1 μg plasmids. The latter included 500 ng of reporter plasmid (Col2a1 [5x48]-p89Luc (36), Acan [4xA1]-p89Luc (37), pG5Luc (Promega), 6FXO-p89Luc (38) or TOP-Flash (39)), 100 ng of pSVβGal plasmid (reporter used to measure transfection efficiency) (40), and 400 ng of expression plasmids (various combinations of empty pCDNA 3.1, pCDNA 3.1-SOXE, pCDNA 3.1-SOX17, pCDNA 3.1-SOX5 and pCDNA 3.1-SOX6, pBind-GAL4DBD/SOXE, or constitutively stabilized β-catenin/CS2 plasmid (37,41)). Cell extracts were prepared in Tropix lysis buffer (0.2% Triton X-100, 100 mM potassium phosphate, pH 7.8, 1 mM DTT) 20–24 h after the start of transfection and assayed for luciferase and β-galactosidase activities using the Dual-Light Luciferase & β-Galactosidase Reporter Gene Assay System (Applied Biosystems, Foster City, CA, USA) and a GloMax Explorer Multimode Microplate Reader (Promega). Reporter activities were normalized for transfection efficiency by calculating the ratios of luciferase versus β-galactosidase activities.
Western blot
The levels of SOX and GAL4DBD/SOX proteins produced from expression plasmids were determined by subjecting cell extracts to 10% SDS-PAGE and transferring proteins to PVDF membranes using iBLOT 2 Gel Transfer Device (Thermo Fisher Scientific). Membranes were blocked in Tris-Buffered Saline with 0.1% (v/v) Tween 20 (TBST) and 5% (w/v) nonfat dry milk for 1 h and then incubated overnight at 4°C in blocking solution containing anti-FLAG M2-peroxidase-conjugated antibody (A8592, Sigma-Aldrich, St. Louis, MO, USA) at a 1:12000 dilution or peroxidase-conjugated GAL4 antibody (sc-510, Santa Cruz Biotechnology, Dallas, TX, USA) at a 1:500 dilution. Peroxidase-generated signals were detected using ECL Prime Western Blotting Detection Reagent (GE Healthcare, Chicago, IL, USA) or SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) on a ChemiDoc Imaging System (Bio-Rad Laboratories, Hercules, CA, USA).
RNA isolation and qRT-PCR assay
Lipofectamine 3000 (Thermo Fisher Scientific) was used to transfect mouse chondrogenic ATDC5 cells (42) with various combinations of expression plasmids for mouse SOX5, mouse SOX6, and human full-length or mutant SOX9. Total RNA was prepared 24 h later using TRIzol (Life Technologies) and following manufacturer's instructions. cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). qPCR was performed using the StepOne Plus Real Time PCR system (Thermo Fisher Scientific), SYBR Green PCR Master Mix (Thermo Fisher Scientific) and custom-designed primers (Integrated DNA Technologies) (Supplementary Table S5). Col2a1 and Acan mRNA levels were calculated relative to those of Hprt according to the ΔΔCt method.
Statistical analyses
Differences between datasets were evaluated using the Student's t-test. Differences that reached P values lower than 0.05 were considered significant.
RESULTS
Sequence conservation suggests key roles for the SOXE TAM and TAC regions
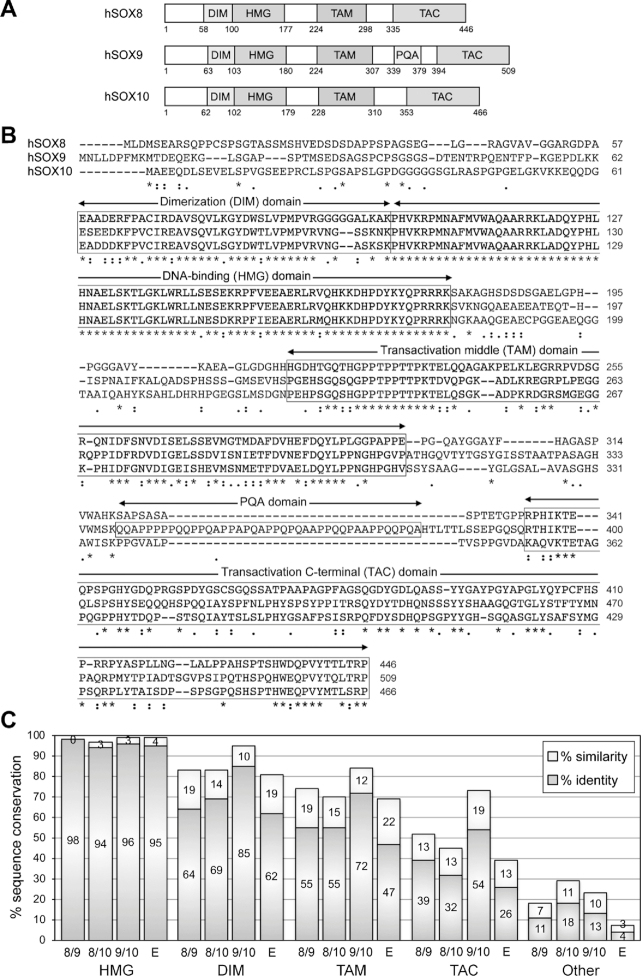
Alignment of the human SOXE protein sequences showed, as expected, that the HMG domain is the most conserved region among the three proteins, with 97–99% identity/similarity (Figure 1A–C). The homodimerization domain (DIM) comes next, with 81–95% identity/similarity among the three proteins. TAM, that is, the reported transactivation domain of SOX8 and K2 domain of SOX10, is third, with 70–84% similarity between the proteins. TAC, that is, the C-terminal region that includes the main transactivation domain reported for SOX9 and SOX10, is fourth, with only 45–73% similarity among the proteins. Other regions are only 7–29% identical/similar. Sequence comparisons for various vertebrate species revealed that SOX8 orthologs are under tighter evolutionary constraint to conserve TAM than TAC, whereas SOX9 and SOX10 orthologs are under similar constraints for TAM and TAC (Supplementary Figure S1A and B). Overall, SOXE protein orthologs are conserved at 34% in TAM, but only 17% in TAC. Together, these data suggest that both TAM and TAC may have key functions in all three SOXE proteins.
Figure 1.
Sequence conservation among the human SOXE proteins. (A) Schematic showing the domain organization of the three human SOXE proteins. Conserved domains are shown with boxes and the amino acids at the boundaries of the protein and domains are indicated with numbers. DIM, homodimerization domain; HMG, DNA-binding domain; TAM, transactivation domain located in the middle of the proteins; PQA, P-, Q- and A-rich domain in SOX9; TAC, carboxy-terminal transactivation domain. (B) ClustalW alignment of the amino acids of the human SOXE proteins. The dimerization (DIM), DNA-binding (HMG), middle transactivation (TAM) and C-terminal transactivation (TAC) domains are boxed. Stars indicate identical residues and dots indicate similar residues. Numbers indicate residue positions within the proteins. (C) Graph showing the degrees of protein conservation. The percentages of sequence identity and similarity were calculated by ClustalW alignment. They are shown for each conserved domain and for the rest of the protein sequences (other). SOX8 is compared to SOX9 (8/9), SOX8 to SOX10 (8/10), SOX9 to SOX10 (9/10) and the three proteins together (E).
PQA may contribute to SOX9 stability and transactivation in specific contexts
In the first investigation of SOX9 domains involved in transactivation, Südbeck and colleagues showed that a fusion protein made with the GAL4 DNA-binding domain (GAL4DBD) and a SOX9 segment containing only PQA and flanking sequences was unable to activate transcription, and that a fusion protein containing only TAC was 4 times as active as a protein featuring both PQA and TAC (22). These data suggested that PQA is not a transactivation domain and that it interferes with TAC activity. In contrast, McDowall and colleagues reported that deletion of PQA weakened the ability of SOX9 to transactivate a reporter containing tandemly repeated SOX binding sites, thus suggesting that PQA could be a weak transactivation domain potentiating the activity of TAC (23,43). In view of these results, we attempted to clarify the role of PQA.
Comparing SOX9 orthologs, we found that PQA is made of 35–45 residues only consisting of prolines (42%), glutamines (39%), and alanines (18%) (Supplementary Figure S2A and B). In contrast, it has only 4–15 residues in lower vertebrates, with only a few glutamines in ancient fish. Only one SOXE protein exists in most invertebrates, and this protein contains a region poorly enriched in P, Q and A residues (Supplementary Figure S2C). The PQA domain was thus gradually acquired following SOX9 emergence in vertebrates from a SOXE ancestor.
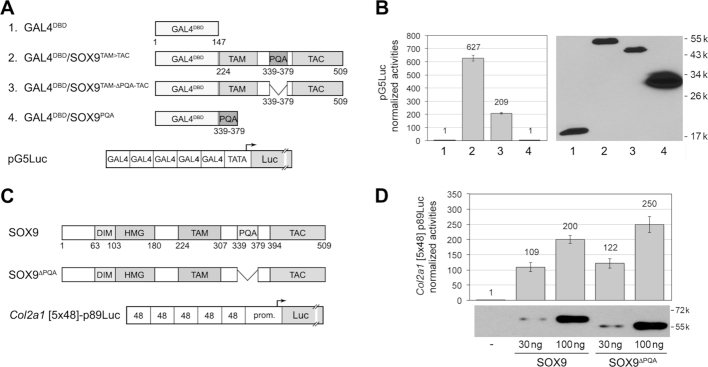
We investigated the function of PQA using two distinct assays. The first one tested whether the domain is sufficient for transactivation, i.e. capable on its own to interact with transcriptional co-activators or basal transcription machinery. We constructed plasmids encoding GAL4DBD/SOX9 fusion proteins and transfected them in HEK-293 cells along with pG5Luc, a reporter plasmid containing a tandem of five GAL4DBD-binding sites (Figure 2A). A fusion protein containing GAL4DBD and the SOX9 TAM-to-TAC segment appeared to be about three times more active than the same protein lacking PQA, but this difference was due in part to differences in protein amount (Figure 2B). A protein made of GAL4DBD and only PQA was inactive, advocating that PQA lacks autonomous transactivation capability. Our second assay tested whether PQA is necessary for SOX9 transactivation. We used expression plasmids encoding the full-length human SOX9 protein fused to an N-terminal 3FLAG epitope and another plasmid encoding SOX9 without PQA, and we tested them with Col2a1 [5x48]-p89Luc, a reporter featuring five tandem copies of a 48-bp Col2a1 enhancer (Figure 2C). This enhancer is a bona fide SOX9 target: it contains a SOX9 consensus binding site, i.e. a pair of inverted SOX-like recognition sites separated by four nucleotides, and it is directly bound by SOX9 in chondrocytes in vivo (16,17,44). SOX9 and SOX9ΔPQA were produced at even levels in HEK-293 cells and transactivated the reporter equally potently (Figure 2D). Similar results were obtained in SW-1353 chondrosarcoma cells with Acan [4xA1]-p89Luc, a reporter containing a 359-bp Acan enhancer (Supplementary Figure S2D). This enhancer features SOX9 and SOX5/6 consensus binding sites and is directly bound by the SOX trio in chondrocytes in vivo (16,17,37)
Figure 2.
Transactivation capability of the SOX9 PQA domain. (A) Top, schematic of fusion proteins made using the GAL4 DNA-binding domain and SOX9 domains. These proteins are numbered 1 to 4, as in panel D. Bottom, schematic of the pG5Luc reporter used to functionally test the fusion proteins. The reporter has 5 tandem copies of the GAL4 DNA-binding site upstream of a TATA box and the firefly luciferase gene. (B) Left, plot showing the ability of GAL4DBD/SOX9 fusion proteins to transactivate pG5Luc upon transient transfection in HEK-293 cells. Reporter activities are presented for one representative experiment as the mean ± standard deviation obtained for triplicate cultures per condition. Data were normalized for transfection efficiency and are reported as fold increase relative to the activity of the reporter in the presence of an empty expression plasmid. Right, western blot showing the levels of the respective proteins present in cell lysates at the end of the experiment. Note that the lower amount of protein in lane 3 compared to lane 2 may explain in part why deletion of PQA reduced the ability of the SOX9 TAM-to-TAC region to activate the pG5Luc reporter. These results were reproduced in multiple experiments. (C) Top, schematic of the SOX9 full-length protein and a mutant protein lacking the PQA domain. Bottom, schematic of the reporter used to functionally test these proteins. The reporter contains five tandem copies of a 48-bp mouse Col2a1 enhancer, which features a SOX9 consensus binding site, and the –89/+6 Col2a1 promoter upstream of the firefly luciferase gene. (D) Top, plot comparing the ability of SOX9 and SOX9ΔPQA to transactivate the Col2a1 reporter. HEK-293 cells were transfected with 30 or 100 ng of SOX9 expression plasmids. Reporter activities are presented as described for panel B. Bottom, western blot of cell lysates prepared at the end of the experiment show that deletion of PQA had no obvious effect on SOX9 protein production and stability.
In conclusion, these data suggested that PQA has no major role in SOX9 transactivation, but did not rule out that SOX9 acquired this unique domain during evolution to enhance its stability or activity in specific contexts.
TAM and TAC are synergistic transactivation domains in all three SOXE proteins
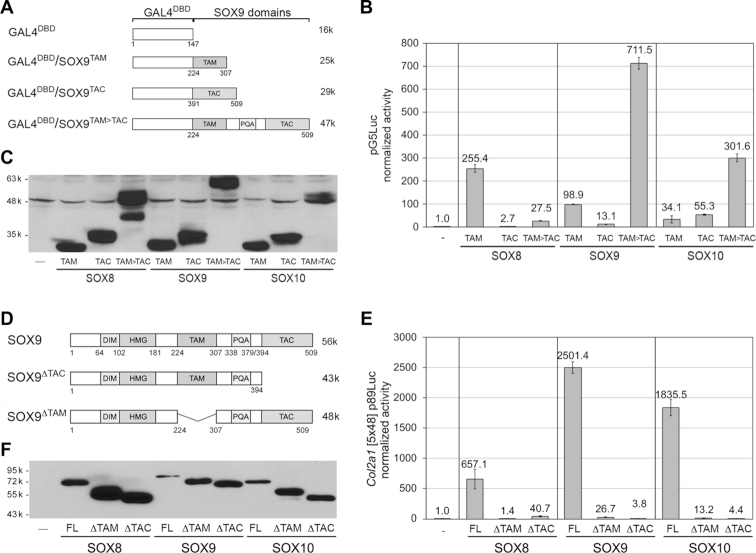
To determine whether and how TAM and TAC contribute to SOXE transactivation, we first constructed plasmids encoding GAL4DBD fused to the TAM, TAC or TAM-to-TAC domains of the human proteins (Figure 3A and Supplementary Figure S3A) and we transfected them in HEK-293 cells along with pG5Luc (Figure 3B and C). All TAM and TAC domains were able to activate transcription, although with different performance levels. SOX8TAM was more potent than SOX9TAM (2.7×) and SOX10TAM (7.4×). In contrast, SOX8TAC was less potent than SOX9TAC (4.5×) and SOX10TAC (12×). While SOX8TAM>TAC was less active than SOX8TAM (19.3×), SOX9TAM>TAC and SOX10TAM>TAC were several times more active than their respective TAM and TAC domains alone. We next tested the requirement of TAM and TAC for transactivation in the natural context of SOXE proteins. We constructed plasmids encoding the full-length human proteins or proteins lacking TAM or TAC (Figure 3D and Supplementary Figure S3B) and transfected them in HEK-293 cells (Figure 3E and F). All full-length proteins powerfully activated the Col2a1 reporter, but their activities were drastically reduced in the absence of TAM or TAC (16- to 658-fold). Similar results were obtained with SW-1353 cells and using the Acan reporter (Supplementary Figure S3C–F). In conclusion, the first assay indicated that TAM and TAC are able to work as independent transactivation domains, and the second assay indicated that the two domains work synergistically in the context of each SOXE protein.
Figure 3.
Transactivation capabilities of the SOXE TAM and TAC domains. (A) Schematics of fusion proteins containing GAL4DBD and SOX9 domains. See Supplementary Figure S3A for fusion proteins of GAL4DBD with SOX8 and SOX10 domains. (B) Reporter assay comparing the abilities of GAL4DBD/SOXE fusion proteins to activate pG5Luc. HEK-293 cells were transfected with pG5Luc and expression plasmids for the fusion proteins shown in panel A. Reporter activities are presented for one experiment as the mean ± standard deviation obtained for triplicate cultures per condition. Data were normalized for transfection efficiency and are reported as fold increase relative to the activity of the reporter in the presence of an empty expression plasmid. These results were reproduced multiple times. (C) Western blot of cell lysates prepared at the end of the experiment showing that all protein forms were made in similar amounts. The blot was made with lysate amounts normalized for transfection efficiency. The lower band seen in the GAL4DBD/SOX8TAM>TAC lane likely reflects partial degradation of the protein. (D) Schematics of the SOX9 full-length protein and mutant proteins lacking either TAC or TAM. See Supplementary Figure S3B for equivalent SOX8 and SOX10 schematics. (E) Reporter assay comparing the abilities of the three SOXE proteins to activate the Col2a1 [5x48]-p89Luc reporter in HEK-293 cells, and effects of deleting their TAM or TAC domain. Reporter activities are presented as described in panel B. (F) Western blot of cell lysates prepared at the end of the experiment showing that all protein forms were made. Major differences in reporter activities (panel E) are not due to variations in relative amounts of the proteins and must thus genuinely reflect differences in functional capabilities. The blots were made with lysate amounts normalized for transfection efficiency.
Since the relative activities of TAM and TAC greatly differed in the GAL4DBD/SOXE assay depending on their SOXE origin, we asked whether swapping SOX9TAM and SOX9TAC with the corresponding SOX8 and SOX10 domains would affect SOX9 activity. We constructed expression plasmids accordingly and tested them in HEK-293 cells using the Col2a1 reporter (Supplementary Figure S4A). Although differential activities were observed that were consistent with the low activities of SOX10TAM and SOX8TAC and high activity of SOX10TAC in the GAL4DBD/SOXE assay, all chimeric proteins efficiently activated the reporter, indicating that the two domains, regardless of SOXE origin, were able to synergize in the context of the full-length SOX9 protein (Supplementary Figure S4B and C).
The C-terminal half of TAM (TAM-CD) is a potent transactivation domain
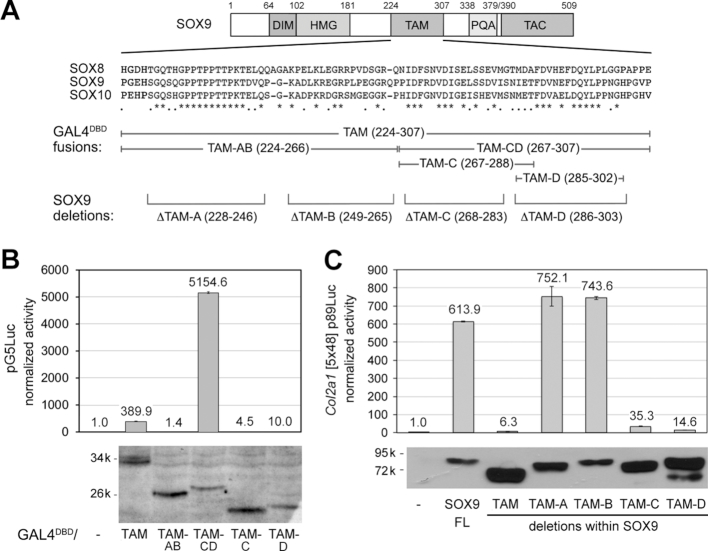
Henceforth, we focused on the TAM domain. To reveal which segment of the domain is involved in transactivation, we generated plasmids encoding fusions of GAL4DBD with halves or quarters of TAM, and plasmids encoding SOX9 proteins lacking most of each TAM quarter (Figure 4A). These quarters were named TAM-A, TAM-B, TAM-C and TAM-D. In transfection of HEK-293 cells with pG5Luc, TAM-AB, TAM-C and TAM-D failed to transactivate, whereas TAM-CD was very potent (Figure 4B). In transfection of HEK-293 and SW-1353 cells with the Col2a1 or Acan reporter, TAM-A or TAM-B deletion was inconsequential, whereas TAM-C or TAM-D deletion virtually abrogated SOX9 activity (Figure 4C and Supplementary Figure S5A). Taken together, these data suggested that TAM-AB is dispensable and that residues within TAM-C and TAM-D are necessary and sufficient for transactivation.
Figure 4.
Identification of subdomains of SOX9 TAM mediating transactivation. (A) From top to bottom, schematic of the SOX9 protein; alignment of the TAM sequences of the three human SOXE proteins; segments of TAM fused to GAL4DBD; and TAM-A to TAM-D sequences deleted in the SOX9 protein. (B) Reporter assay comparing the abilities of proteins made by fusing GAL4DBD with subdomains of SOX9 TAM to transactivate pG5Luc. Reporter activities are presented for one representative experiment as the mean ± standard deviation obtained for triplicate cultures per condition. Data were normalized for transfection efficiency and are reported as fold increase relative to the activity of the reporter in the presence of an empty expression plasmid. The western blot of cell lysates shows that all protein forms were efficiently made in the cells and thus that major differences in reporter activities among proteins genuinely reflect intrinsic differences in transactivation capabilities. These results were reproduced in multiple experiments. (C) Reporter assay comparing the abilities of wild-type SOX9 and SOX9 proteins lacking the whole TAM or TAM segments to transactivate the Col2a1 reporter. Reporter activities are presented as described in panel B. The western blot of cell lysates shows that all protein forms were efficiently made in the cells and thus that major differences in reporter activities among proteins genuinely reflect intrinsic differences in transactivation capabilities. These results were reproduced in multiple experiments.
Since all data were obtained so far using reporter assays, we next asked whether SOX9 also requires TAM and TAC to activate the endogenous Col2a1 and Acan genes. In transfection of ATDC5 cells, a chondrogenic cell line derived from a mouse teratoma and frequently used to study chondrocyte differentiation in vitro (45), full-length SOX9 successfully cooperated with SOX5 and SOX6 to enhance Col2a1 and Acan expression (3.7× and 2.5×, respectively) (Supplementary Figure S5B). In contrast, SOX9 lacking either TAC or TAM-D was unable to do so, lending further evidence that both TAM and TAC are critical for SOX9 functions.
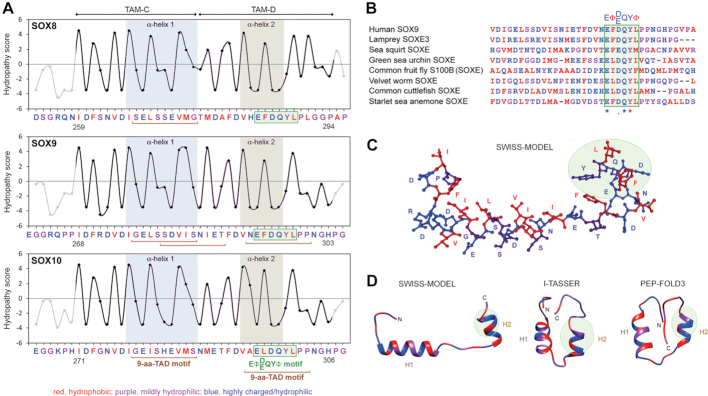
TAM-CD exhibits characteristic features of acidic transactivation domains and a unique, highly conserved EΦ[D/E]QYΦ motif
Transactivation domains are categorized based on amino acid composition (46). For instance, the SOXE TAC is a non-acidic PQS-rich transactivation domain (22). TAM-CD examination revealed numerous acidic (Asp and Glu) and other hydrophilic amino acids alternating with hydrophobic residues (Ile, Leu, Met, Phe and Val) (Figure 5A). This pattern is reminiscent of the minimal nine-amino-acid-transactivation-domain motif (9-aa-TAD) described for acidic transactivation domains in many transcription factors, including GAL4 (yeast), P53, NFAT and NF-kB proteins (mammals) and VP16 (human herpes virus) (28). Accordingly, the Piskacek algorithm identified one such motif in all SOXE TAM-C regions, one overlapping TAM-C and TAM-D in SOX9, and one in SOX9 and SOX10 TAM-D (Figure 5A). In many cases, 9-aa-TAD sequences contain a ΦXXΦΦ core motif (Φ, hydrophobic residue; X, any residue) that interacts with basal transcription machinery components, such as hTAFII31 (47). We found a partially related, but distinct motif in TAM-D in the three human SOXE proteins (Figure 5A). This motif responds to an EΦ[D/E]QYΦ consensus and is remarkably conserved not only in all SOX8, SOX9, and SOX10 vertebrate sequences (Supplementary Figure S1), but also in the lamprey SOXE3 protein and in the sole SOXE protein existing in invertebrates (Figure 5B). Since the P53 ΦXXΦΦ motif was shown to transit from a random coil to an α-helix upon binding to hTAFII31 (47), we used SWISS-MODEL and I-TASSER, which are template-based structure prediction software, and PEP-FOLD3, a de novo program predicting protein structures directly from amino acid sequences, to predict the secondary and tertiary structures of TAM-CD. The SWISS-MODEL model that reached the highest quality score (QMEAN, –1.36; sequence identity with the template, 23.53%) was built according to a region of the CdiI Immunity protein from Yersinia kristensenii (PDB ID: 4ZQV). The best I-TASSER model (C-score, –1.95; sequence identity with the template, 19%) was based on a glycosylated calcitonin growth factor from Anguilla japonica (PDB ID: 1BYV). Of 10 models proposed by PEP-FOLD3, we retained the best one (sOPEP score: –45.011). All models concurred that TAM-CD could form two α-helices, one using the TAM-C 9-aa-TAD motif and the other one using most of the TAM-D 9-aa-TAD sequence and EΦ[D/E]QYΦ motif (Figure 5A, C and D). These helices would fold into a protein-binding pocket coated externally with polar residues and internally with hydrophobic and aromatic residues.
Figure 5.
In silico analysis of transactivation domain features of the SOXE TAM-CD region. (A) Plots of the hydropathy scores of TAM-CD and flanking residues in human SOX8, SOX9 and SOX10. Amino acids are typed in colors according to the nature of their side chains, as indicated underneath the plots. Two predicted α-helices are shaded in the plots; 9-aa-TAD motifs are delineated with brown brackets underneath the sequences; and the conserved EΦ[D/E]QYΦ motif is highlighted with a green box. (B) ClustalW sequence alignment showing a high degree of conservation of the EΦ[D/E]QYΦ motif in SOXE proteins from various vertebrate and invertebrate species. (C and D) Binding-pocket structure of the SOX9 TAM-CD domain predicted by SWISS-MODEL, I-TASSER and PEP-FOLD3. Top, ball-and-stick representation showing amino acid cores and side chains. Bottom, cartoon representations. The N- and C-termini of the domain are marked. The α-helices are indicated as H1 and H2. The EΦ[D/E]QYΦ motif is highlighted with a green bubble. The color code is otherwise the same as for the sequences in the panel A.
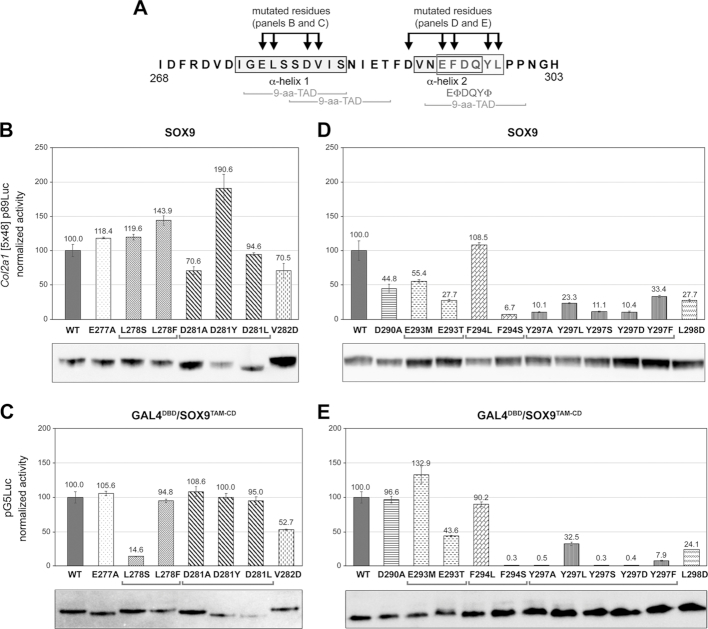
Specific residues in the EΦ[D/E]QYΦ motif are critical for transactivation
We introduced a series of missense mutations in TAM-CD to test the importance of highly conserved residues in transactivation (Figure 6A). These mutations were selected to significantly alter the hydropathicity, polarity or size of amino acid side chains. Overall, mutations in residues participating in the α-helix 1 had no drastic effect on SOX9 activity (Figure 6B). In the GAL4 assay, where transactivation is only driven by TAM-CD and is thus more sensitive, mutations of residues with hydrophobic side chains protruding inside the binding pocket (Leu278 and Val282) were deleterious, whereas mutations in residues with acidic side chains projecting outwards (Glu277 and Asp281) were inconsequential (Figure 6C).
Figure 6.
Test of the effects of amino acid substitutions in TAM-CD on transactivation. (A) Schematic showing the SOX9 TAM-CD residues substituted in GAL4DBD/SOX9TAM-CD and SOX9 expression plasmids. (B–E) Reporter assays comparing the abilities of wild-type and variant SOX9 and GAL4DBD/SOX9TAM-CD proteins to transactivate their respective Col2a1 and pG5Luc target reporters upon transfection in HEK-293 cells. Normalized reporter activities are presented for one representative experiment as the mean ± standard deviation obtained for triplicate cultures and in percentage of the activities obtained with wild-type proteins. Western blots of cell lysates prove that major differences in reporter activities are not due to differences in relative amounts of the various proteins. These results were reproduced in multiple experiments.
Replacing the first residue of the EΦ[D/E]QYΦ motif with the residues present in the ΦXXΦΦ motif of VP16 or P53 (Glu293Met or Glu293Thr, respectively) impaired the activity of full-length SOX9 and one of the mutations also affected TAM-CD activity, explaining that this acidic residue (or Asp) is conserved in all SOXE proteins (Figure 6D and E). All mutations introduced in other residues of the EΦ[D/E]QYΦ motif dramatically reduced SOX9 and TAM-CD activities, except F294L, which resulted in an aromatic to aliphatic side chain change, but did not significantly change the hydropathy index. These findings further supported the conclusion that hydrophobic residues projecting inside the binding pocket are critical for transactivation and that even non-hydrophobic residues composing the EΦ[D/E]QYΦ motif are critical too.
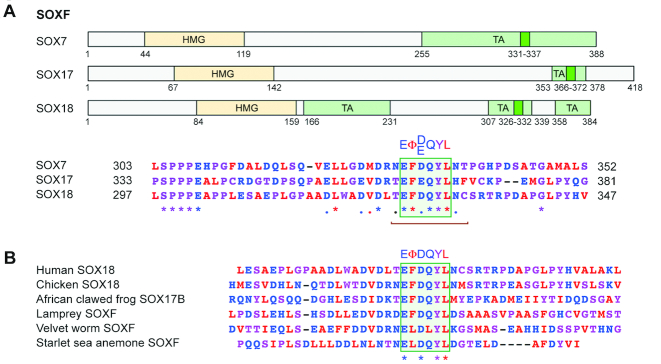
The EΦ[D/E]QYΦ motif is conserved in both SOXE and SOXF proteins
The discovery that the EΦ[D/E]QYΦ motif of TAM-CD is fully evolved in invertebrates prompted us to determine whether an identical or similar motif and its associated features are also present in the SOXE TAC domain and in other transactivating SOX proteins. We used three criteria: (i) sequence conservation between group members; (ii) presence of 9-aa-TAD domains and (iii) presence of ΦXXΦΦ or EΦ[D/E]QYΦ-like motifs. The SOXE TAC domains contained 9-aa-TAD motifs and a VYXXL sequence resembling a ΦXXΦΦ, but no EΦ[D/E]QYΦ-like sequence (Supplementary Figure S6A). SOXB1 (SOX1, SOX2 and SOX3) and SOXC (SOX4, SOX11 and SOX12) proteins featured 9-aa-TAD, and ΦXXΦΦ/EΦ[D/E]QYΦ-like sequences in their transactivation domains, but these sequences were very different from those of the SOXE TAM-CD domains (Supplementary Figure S6B). Interestingly, SOXF proteins (SOX7, SOX17 and SOX18) featured a 9-aa-TAD sequence with an EΦ[D/E]QYL motif fully matching the SOXE EΦ[D/E]QYΦ consensus in their transactivation domains (Figure 7A). This motif is also remarkably conserved from invertebrates to humans (Figure 7B). This finding pairs with the fact that the HMG domains of the SOXE and SOXF proteins are more closely related to one another than to those of other SOX proteins (48). Altogether, the data suggest that the SOXE and SOXF groups emerged from a common ancestor that was already featuring an EΦ[D/E]QYΦ motif. We looked for the presence of an EΦ[D/E]QYΦ motif in all other SOX proteins, but did not find any (data not shown).
Figure 7.
Identification of an EΦ[D/E]QYΦ motif in SOXF proteins. (A) Schematics showing the locations of the HMG and transactivation regions (TA, pale green boxes) of SOXF proteins and alignment of TA regions that show a high degree of conservation among group members, a 9-aa-TAD motif (brown bracket) and an EΦ[D/E]QYL motif. The TA regions were previously delineated for SOX7 (63), SOX17 (64) and SOX18 (59,65). (B) ClustalW sequence alignment showing the high degree of conservation of the EΦ[D/E]QYL motif in SOXF proteins from various vertebrate and invertebrate species.
The EΦ[D/E]QYL motif was previously recognized in SOXF proteins, and SOX17 was shown to require it to activate endodermal genes on its own and in synergy with β-catenin (49) as well as to reprogram somatic cells into induced pluripotent stem cells (50). Further, this domain was shown to help SOX17 bind to β-catenin and prevent constitutively stabilized β-catenin from activating TOP-Flash, a reporter gene classically used as a readout of canonical WNT signaling (51). SOX9 was also shown to inhibit β-catenin transcriptional activity, but to use its TAC domain to bind to β-catenin (52). We therefore decided to directly compare the contributions of the SOX9 and SOX17 EΦ[D/E]QYL motifs to the protein activities. As expected, deletion of the motif significantly reduced the abilities of SOX9 and SOX17 to activate reporter genes (Supplementary Figure S7A and B). When tested with TOP-Flash, wild-type SOX9 and SOX17 inhibited the activity of constitutively stabilized β-catenin in a dose-dependent manner (Supplementary Figure S7C and D). Deletion of the whole TAM or only its EFDQYL motif slightly reversed SOX9 inhibition of β-catenin, whereas deletion of TAC totally reversed this inhibition, and whereas deletion of the EFEQYL motif from SOX17 effectively reversed the inhibition. These data suggest that the EΦ[D/E]QYΦ motif might contribute to inhibition of canonical WNT signaling by both SOXE and SOXF proteins, but that TAC has a dominant role in SOXE proteins and that SOXF proteins might feature specific sequences around EFEQYL that potentiate its inhibitory activity. The latter proposition is supported by evidence that deletion of its entire C-terminus prevented SOX17 from binding to β-catenin, whereas the sole deletion of the EFEQYL motif (located in the C-terminus) only had a partial effect (49).
The SOXE/SOXF EΦ[D/E]QYΦ motif is highly conserved in the human population
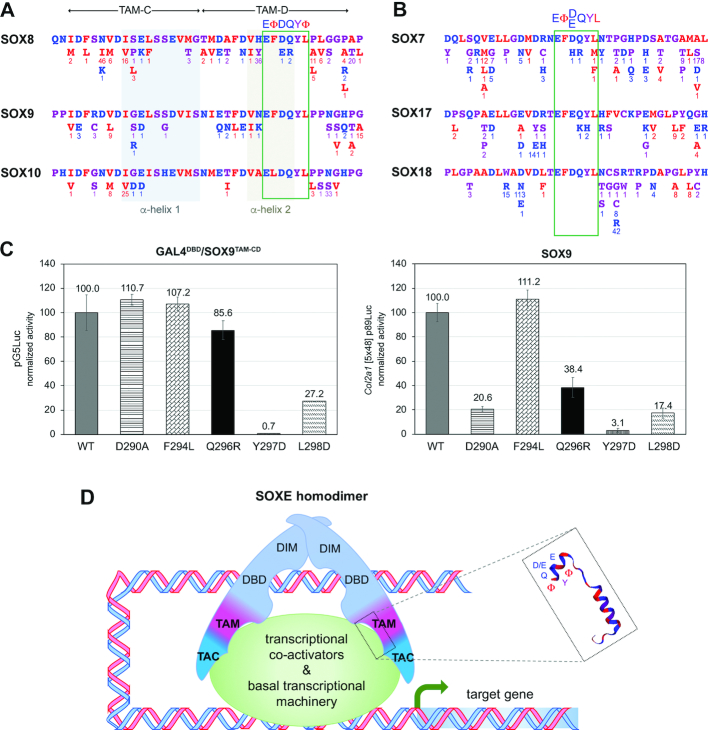
The outstanding degree of conservation of the SOXE/SOXF EΦ[D/E]QYΦ motif suggests that mutations in this motif would be incompatible with healthy development and adult life. To test this hypothesis, we searched for literature reports of missense mutations or other in-frame micro-alterations in this domain in SOXE and SOXF genes that were linked to a human disease, but did not find any. We then searched GnomAD, a database of genomic sequences from >140 000 unrelated control individuals, and COSMIC, a catalog of somatic mutations in cancer. Detailed analysis of SOX9 revealed that synonymous and missense variants affected similar proportions of residues throughout all domains of the protein, except the HMG domain, where significantly fewer missense variants were detected in gnomAD individuals compared to synonymous variants in the same cohort and compared to missense variants in cancers (Supplementary Figure S8A and B). This finding suggests that a particularly tight sequence conservation constraint exists for this domain in healthy individuals and that this constraint is lifted in cancer and participates in tumorigenesis. The occurrence of missense variants was lower in the TAM than in the HMG domain in COSMIC samples, but the difference did not reach statistical significance. We therefore closely examined the types of missense variants present in the SOXE and SOXF EΦ[D/E]QYΦ motifs in gnomAD and COSMIC samples.
Interestingly, gnomAD missense variants affected many residues around the EΦ[D/E]QYΦ motifs of SOXE and SOXF proteins, but none occurred within the SOX9, SOX10 and SOX18 EΦ[D/E]QYΦ motifs and only a few occurred within the SOX8, SOX7 and SOX17 EΦ[D/E]QYΦ motifs (Figure 8A and B). The SOX8 variants detected in this motif represented conservative changes (D286E and Q287R) in the residues that occupy the X positions in the related ΦXXΦΦ sequence of other transcription factors. The D286E change is unlikely to be consequential since both D and E are highly acidic and occupy the third position of the SOXE/SOXF EΦ[D/E]QYΦ motif. The other variant, Q287R, might be consequential since Q is highly conserved in SOXE and SOXF proteins and since it is uncharged whereas R is positively charged. To test whether this variant could affect protein activity, we introduced an equivalent mutation (Q296R) in the SOX9 and GAL4DBD/SOX9TAM-CD proteins. We observed that both proteins still retained significant activity in their respective assays (Figure 8C), suggesting that the SOX8 Q287R variant is not detrimental enough to cause a disease.
Figure 8.
Analysis of SOXE and SOXF missense variants in the human control population and model for the SOXE bipartite transactivation mechanism. (A) Missense variants listed in gnomAD in SOXE TAM-CD are presented underneath the domain sequences. The numbers of alleles detected in over 140 000 unrelated individuals are indicated for each variant. (B) Missense variants listed in gnomAD in the SOXF EΦ[D/E]QYL motif and flanking residues. (C) Test of the effect of a Q296R variant detected in SOX8 in healthy human individuals on transactivation. The variant was introduced in the GALDBD/SOX9TAM-CD and SOX9 proteins. The proteins were then tested in HEK-293 cells upon co-transfection with the pG5Luc or Col2a1 reporter. Other missense variants (as described in Figure 6) were tested in parallel for comparison. Data were calculated and are presented as in similar assays in previous figures. They were reproduced in multiple experiments. (D) Model of the current view for the mechanism used by SOXE proteins to activate target genes. Previous studies demonstrated that SOXE proteins homodimerize upon binding to inverted recognition sites present in the enhancers or promoter of their target genes. These events involve their HMG and dimerization domains. The present study uncovered that they transactivate these genes through a bi-partite mechanism involving a centrally located domain (TAM) synergizing with the protein C-terminal region (TAC). The functional core of TAM is predicted to fold into binding pocket-like structure upon interaction with specific transcriptional co-activators or components of the basal transcription machinery. It contains a highly conserved EΦ[D/Q]YΦ motif that is critical for transactivation and is thus likely involved in recognizing and firmly binding functional partners. Not shown here is a PQA-rich domain present solely in SOX9 that may facilitate transactivation in specific contexts.
Unlike the gnomAD database, the COSMIC database did contain missense variants in the SOX9 EFDQYL motif (Supplementary Figure S8C). We tested them along with two variants in the α-helix 1. The latter almost doubled the activity of SOX9, whereas the two variants affecting the E residue of the EFDQYL motif greatly decreased SOX9 activity and the variant affecting the D residue of the motif modestly decreased SOX9 activity (Supplementary Figure S8D). Beside confirming the importance of EFDQYL for SOX9 activity, these data add support to the notion that the SOXE/SOXF EΦ[D/E]QYΦ motif is critical for the function of SOXE proteins and suggest that mutations in this motif could underlie congenital or acquired human diseases.
DISCUSSION
This study has brought unity and new information on the mechanisms whereby the SOXE proteins achieve transactivation. It has provided evidence that each protein carries two synergistic transactivation domains, TAM and TAC (Figure 8D). By deploying a bipartite transactivation mechanism, SOXE proteins may engage exclusive sets of transcriptional partners to effectively activate target genes. The C-terminal half of TAM is predicted to form a protein-binding pocket. Its EΦ[D/E]QYΦ core motif is functionally crucial and is highly conserved evolutionarily and in human healthy individuals not only in SOXE proteins but also in SOXF proteins.
The unique PQA domain of SOX9 gained in length and P-, Q- and A-enrichment upon mammalian evolution, but its role remains unclear. Glutamine-rich sequences exist in many types of proteins, especially in transactivation domains, and there is evidence that they help stabilize proteins and strengthen protein-protein interactions (53). The SOX9 PQA domain was not found in our study nor in a previous study by Südbeck et al. to have autonomous transactivation function (22). McDowall et al. reported that PQA enhanced the ability of SOX9 to transactivate an artificial pS10E1bCat reporter, which likely bound SOX9 monomers (23). We failed to replicate this finding using bona fide Col2a1 and Acan reporters that bind SOX9 homodimers, but observed that GAL4DBD/SOX9TAM>TAC was less abundant and less active in transactivating pG5Luc when it lacked PQA. This reporter is also likely to bind GAL4DBD/SOX9 monomers. One can thus envision that PQA enhances SOX9 stability and transactivation efficiency when the protein binds target genes as a monomer, as in Sertoli cells, but may not be necessary when the protein binds targets as a homodimer, as in other cell types.
Different transactivation mechanisms were previously suggested for the three SOXE proteins, mostly because most studies only used the GAL4DBD assay to map transactivation domains and did not test the consequences of deletions or point mutations in the protein sequences. The use of both assays led us to find two functional domains in each protein. This finding was not unexpected considering the high degree of sequence conservation and redundancy of SOXE proteins in many processes. What was unexpected was to find that the two domains synergize with one another. Schreiner et al. indeed showed that deletion of SOX10 TAM in the mouse led to a milder disease than total inactivation of Sox10 (26). Melanogenesis and enteric nervous system development were significantly impaired, but neural crest and oligodendrocyte development were only slightly affected. This led the authors to suggest that SOX10 TAM contributes to SOX10 activity in a cell type-specific context rather than being mandatory in all processes to synergize with TAC. It remains possible, however, that TAM contributes to SOX10 activity in all cell types, but that its deletion is pathogenic only in cells critically dependent on its dosage. Supporting this model is the fact that humans with heterozygous mutations inactivating one SOX10 allele have deficiencies primarily in melanogenesis and enteric nervous system development. Synergy implies that TAC and TAM domains may cooperatively contact the same protein or may contact different components of transcriptional complexes. CBP/p300, a major transcriptional co-activator, was shown to interact with SOX9 via TAC, but apparently not through TAM (54). TRAP230, a member of the transcriptional mediator, was shown to interact with the SOX9 TAC domain, and was mentioned to interact also with SOX8 TAM and SOX10 TAC (55,56). P53 and other proteins contact CBP/p300 through 9-aa-TAD domains (57), but other transcriptional partners have also been identified for 9-aa-TAD and ΦXXΦΦ domains, such hTAFII31 (47). All this suggests that SOXE proteins may use their two transactivation domains to contact various partners and to do so cooperatively or independently of one another.
Dissection of functional segments revealed that the C-terminal half of TAM (TAM-CD) is both needed and sufficient for transactivation. TAM-CD has characteristic features of acidic transactivation domains: enrichment for acidic residues that may promote an unstructured conformation prone to encounter transcriptional partners, and enrichment for aromatic and bulky hydrophobic residues that could favor the formation of a specific structure upon interaction with these partners (58). Protein modeling tools supported this view by proposing that TAM-CD would indeed fold into a binding pocket. Functional assays added further support to this view by showing that non-conservative substitutions were most deleterious when occurring in hydrophobic residues predicted to line the inner surface of the binding pocket.
We found an EΦ[D/E]QYΦ sequence to be the most conserved segment of TAM-CD among all SOXE proteins from invertebrates to humans and also to be the most critical segment for transactivation. Its conservation in SOXF proteins unsettles a longstanding concept that proteins from different SOX groups share no significant identity outside the HMG domain and strongly suggests that the two SOX groups recently evolved from a common ancestor. It is intriguing, however, that the two groups diverged in their expression patterns and gene targetome to control different cell types, but nevertheless conserved a core motif for transactivation. Missense mutations introduced in the EΦ[D/E]QYΦ sequence of SOX18 were previously shown to impair transactivation, showing that this motif is critical in SOX18 too (59). Previous studies also showed that this sequence facilitates interaction of SOX17 with β-catenin and may thereby allow SOX17 to cooperate with β-catenin in the activation of endodermal genes in Xenopus embryos (49) and to reprogram fibroblasts into pluripotent stem cells with higher efficiency than SOX2 (50). These data, however, contrast with evidence that SOX17 can also repress β-catenin/TCF activity and interact with β-catenin to promote its degradation (60). Similarly, SOX9 and β-catenin were shown to physically interact with one another and to have reciprocal antagonistic activities, including induction of mutual degradation (52). TAC was required for interaction of SOX9 with β-catenin. In direct comparisons, we found that the EF[D/E]QYL motif is more critical for transactivation by SOX9 than by SOX17, but is more potent in SOX17 than SOX9 to inhibit β-catenin. Altogether, conservation, structural and functional data point to the EΦ[D/E]QYΦ motif as being a pivotal sequence that combines with other sequences, likely distinct in SOXE and SOXF proteins, to engage in protein interactions with either transcriptional or non-transcriptional consequences.
SOXE and SOXF mutations cause severe developmental diseases, namely Campomelic Dysplasia and XY sex reversal for SOX9, Waardenburg-Shah syndrome for SOX10, and hypotrichosis-lymphedema-telangiectasia-renal defect syndrome for SOX18 (61). The mutations vary from entire gene deletions to point mutations. Among the latter, nonsense mutations within the SOXE TAC domain have demonstrated the critical importance of this domain. Missense mutations are almost always located in the HMG domain or SOXE dimerization domain. The only two missense mutations reported in the SOX9 TAC domain (R394G and R437C) caused testicular dysgenesis, but no sex reversal and no campomelic dysplasia, thus a less severe disease than that caused by allele deletions (62). To our knowledge, no missense mutations or in-frame microdeletions causing a developmental disease have been reported for the SOXE TAM domain and for the SOXE/SOXF EΦ[D/E]QYΦ motif. Interestingly, this motif is essentially devoid of missense variants in SOXE and SOXF genes in control individuals in the gnomAD database, unlike surrounding residues, whereas several missense variants abrogating the activity of this motif were found in SOX9 in cancers in the COSMIC database. This suggests, along with the fact that mice lacking SOX10 TAM have a milder disease than Sox10-null mice, that germline mutations in the EΦ[D/E]QYΦ motif or surrounding residues may cause only benign developmental diseases and that somatic mutations may impact the progression of such adult-onset diseases as cancers. By providing novel insights on SOXE structure/function relationships, this study should contribute to a better understanding of the mechanisms whereby the proteins achieve pivotal functions in development, physiology and pathology, and eventually to the design of precision-medicine disease treatments.
DATA AVAILABILITY
The Genome Aggregation Database (gnomAD) is a collection of exome and genome sequencing data from >140k unrelated individuals used as control subjects in various studies. It is available online (https://gnomad.broadinstitute.org). The 9-aa-TAD prediction tool developed by Piskacek is a freely available online computational resource to identify 9-aa-TAD motifs in protein sequences: (http://www.med.muni.cz/9-aa-TAD/). SWISS-MODEL is an automated protein structure homology-modeling server available worldwide (https://:swissmodel.expasy.org). I-TASSER (Iterative Threading ASSEmbly Refinement) is a freely accessible hierarchical approach to protein structure and function prediction (https://zhanglab.ccmb.med.umich.edu/I-TASSER/). PEP-FOLD3 is a freely accessible de novo approach aimed at predicting peptide structures from amino acid sequences (http://bioserv.rpbs.univ-paris-diderot.fr/services.html).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ash Zawerton and J. Paige Yeager for expert technical assistance and other members of the Lefebvre laboratory for helpful advice throughout this study and manuscript preparation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) [AR046249, AR072649 to V.L.]. Funding for open access charge: NIAMS [AR072649 to V.L.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Kamachi Y., Kondoh H.. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013; 140:4129–4144. [DOI] [PubMed] [Google Scholar]
- 2. Julian L.M., McDonald A.C., Stanford W.L.. Direct reprogramming with SOX factors: masters of cell fate. Curr. Opin. Genet. Dev. 2017; 46:24–36. [DOI] [PubMed] [Google Scholar]
- 3. Guth S.I., Wegner M.. Having it both ways: Sox protein function between conservation and innovation. Cell Mol. Life Sci. 2008; 65:3000–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schepers G.E., Teasdale R.D., Koopman P.. Twenty pairs of sox: extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev. Cell. 2002; 3:167–170. [DOI] [PubMed] [Google Scholar]
- 5. Harris M.L., Baxter L.L., Loftus S.K., Pavan W.J.. Sox proteins in melanocyte development and melanoma. Pigment Cell Melanoma Res. 2010; 23:496–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lefebvre V., Dvir-Ginzberg M.. SOX9 and the many facets of its regulation in the chondrocyte lineage. Connect. Tissue Res. 2017; 58:2–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. She Z.Y., Yang W.X.. Sry and SoxE genes: how they participate in mammalian sex determination and gonadal development. Semin. Cell Dev. Biol. 2017; 63:13–22. [DOI] [PubMed] [Google Scholar]
- 8. Weider M., Wegner M.. SoxE factors: transcriptional regulators of neural differentiation and nervous system development. Semin. Cell Dev. Biol. 2017; 63:35–42. [DOI] [PubMed] [Google Scholar]
- 9. Portnoi M.F., Dumargne M.C., Rojo S., Witchel S.F., Duncan A.J., Eozenou C., Bignon-Topalovic J., Yatsenko S.A., Rajkovic A., Reyes-Mugica M. et al.. Mutations involving the SRY-related gene SOX8 are associated with a spectrum of human reproductive anomalies. Hum. Mol. Genet. 2018; 27:1228–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Foster J.W., Dominguez-Steglich M.A., Guioli S., Kwok C., Weller P.A., Stevanovic M., Weissenbach J., Mansour S., Young I.D., Goodfellow P.N. et al.. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature. 1994; 372:525–530. [DOI] [PubMed] [Google Scholar]
- 11. Wagner T., Wirth J., Meyer J., Zabel B., Held M., Zimmer J., Pasantes J., Bricarelli F.D., Keutel J., Hustert E. et al.. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell. 1994; 79:1111–1120. [DOI] [PubMed] [Google Scholar]
- 12. Unger S., Scherer G., Superti-Furga A.. Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A. 1993; Seattle (WA)GeneReviews((R)). [Google Scholar]
- 13. Pingault V., Ente D., Dastot-Le Moal F., Goossens M., Marlin S., Bondurand N.. Review and update of mutations causing Waardenburg syndrome. Hum. Mutat. 2010; 31:391–406. [DOI] [PubMed] [Google Scholar]
- 14. Jo A., Denduluri S., Zhang B., Wang Z., Yin L., Yan Z., Kang R., Shi L.L., Mok J., Lee M.J. et al.. The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis. 2014; 1:149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rahmoun M., Lavery R., Laurent-Chaballier S., Bellora N., Philip G.K., Rossitto M., Symon A., Pailhoux E., Cammas F., Chung J. et al.. In mammalian foetal testes, SOX9 regulates expression of its target genes by binding to genomic regions with conserved signatures. Nucleic Acids Res. 2017; 45:7191–7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu C.F., Lefebvre V.. The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis. Nucleic Acids Res. 2015; 43:8183–8203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ohba S., He X., Hojo H., McMahon A.P.. Distinct transcriptional programs underlie Sox9 regulation of the mammalian chondrocyte. Cell Rep. 2015; 12:229–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Klum S., Zaouter C., Alekseenko Z., Bjorklund A.K., Hagey D.W., Ericson J., Muhr J., Bergsland M.. Sequentially acting SOX proteins orchestrate astrocyte- and oligodendrocyte-specific gene expression. EMBO Rep. 2018; 19:e46635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seberg H.E., Van Otterloo E., Cornell R.A.. Beyond MITF: multiple transcription factors directly regulate the cellular phenotype in melanocytes and melanoma. Pigment Cell Melanoma Res. 2017; 30:454–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schepers G.E., Bullejos M., Hosking B.M., Koopman P.. Cloning and characterisation of the Sry-related transcription factor gene Sox8. Nucleic Acids Res. 2000; 28:1473–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pfeifer D., Poulat F., Holinski-Feder E., Kooy F., Scherer G.. The SOX8 gene is located within 700 kb of the tip of chromosome 16p and is deleted in a patient with ATR-16 syndrome. Genomics. 2000; 63:108–116. [DOI] [PubMed] [Google Scholar]
- 22. Sudbeck P., Schmitz M.L., Baeuerle P.A., Scherer G.. Sex reversal by loss of the C-terminal transactivation domain of human SOX9. Nat. Genet. 1996; 13:230–232. [DOI] [PubMed] [Google Scholar]
- 23. McDowall S., Argentaro A., Ranganathan S., Weller P., Mertin S., Mansour S., Tolmie J., Harley V.. Functional and structural studies of wild type SOX9 and mutations causing campomelic dysplasia. J. Biol. Chem. 1999; 274:24023–24030. [DOI] [PubMed] [Google Scholar]
- 24. Kuhlbrodt K., Schmidt C., Sock E., Pingault V., Bondurand N., Goossens M., Wegner M.. Functional analysis of Sox10 mutations found in human Waardenburg-Hirschsprung patients. J. Biol. Chem. 1998; 273:23033–23038. [DOI] [PubMed] [Google Scholar]
- 25. Pusch C., Hustert E., Pfeifer D., Sudbeck P., Kist R., Roe B., Wang Z., Balling R., Blin N., Scherer G.. The SOX10/Sox10 gene from human and mouse: sequence, expression, and transactivation by the encoded HMG domain transcription factor. Hum. Genet. 1998; 103:115–123. [DOI] [PubMed] [Google Scholar]
- 26. Schreiner S., Cossais F., Fischer K., Scholz S., Bosl M.R., Holtmann B., Sendtner M., Wegner M.. Hypomorphic Sox10 alleles reveal novel protein functions and unravel developmental differences in glial lineages. Development. 2007; 134:3271–3281. [DOI] [PubMed] [Google Scholar]
- 27. Kyte J., Doolittle R.F.. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982; 157:105–132. [DOI] [PubMed] [Google Scholar]
- 28. Piskacek M., Havelka M., Rezacova M., Knight A.. The 9aaTAD transactivation domains: From Gal4 to p53. PLoS One. 2016; 11:e0162842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bienert S., Waterhouse A., de Beer T.A., Tauriello G., Studer G., Bordoli L., Schwede T.. The SWISS-MODEL repository-new features and functionality. Nucleic Acids Res. 2017; 45:D313–D319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y.. The I-TASSER suite: protein structure and function prediction. Nat. Methods. 2015; 12:7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lamiable A., Thevenet P., Rey J., Vavrusa M., Derreumaux P., Tuffery P.. PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016; 44:W449–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 33. Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B. et al.. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016; 536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Forbes S.A., Beare D., Boutselakis H., Bamford S., Bindal N., Tate J., Cole C.G., Ward S., Dawson E., Ponting L. et al.. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017; 45:D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lobbestael E., Reumers V., Ibrahimi A., Paesen K., Thiry I., Gijsbers R., Van den Haute C., Debyser Z., Baekelandt V., Taymans J.M.. Immunohistochemical detection of transgene expression in the brain using small epitope tags. BMC Biotechnol. 2010; 10:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lefebvre V., Mukhopadhyay K., Zhou G., Garofalo S., Smith C., Eberspaecher H., Kimura J.H., de Crombrugghe B.. A 47-bp sequence of the first intron of the mouse pro alpha 1(II) collagen gene is sufficient to direct chondrocyte Expression. Ann. N. Y. Acad. Sci. 1996; 785:284–287. [DOI] [PubMed] [Google Scholar]
- 37. Han Y., Lefebvre V.. L-Sox5 and Sox6 drive expression of the aggrecan gene in cartilage by securing binding of Sox9 to a far-upstream enhancer. Mol. Cell Biol. 2008; 28:4999–5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dy P., Penzo-Mendez A., Wang H., Pedraza C.E., Macklin W.B., Lefebvre V.. The three SoxC proteins–Sox4, Sox11 and Sox12–exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 2008; 36:3101–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Veeman M.T., Slusarski D.C., Kaykas A., Louie S.H., Moon R.T.. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr. Biol. 2003; 13:680–685. [DOI] [PubMed] [Google Scholar]
- 40. MacGregor G.R., Caskey C.T.. Construction of plasmids that express E. coli beta-galactosidase in mammalian cells. Nucleic Acids Res. 1989; 17:2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee E., Salic A., Kirschner M.W.. Physiological regulation of [beta]-catenin stability by Tcf3 and CK1epsilon. J. Cell Biol. 2001; 154:983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Atsumi T., Miwa Y., Kimata K., Ikawa Y.. A chondrogenic cell line derived from a differentiating culture of AT805 teratocarcinoma cells. Cell Differ. Dev. 1990; 30:109–116. [DOI] [PubMed] [Google Scholar]
- 43. Barrionuevo F., Scherer G.. SOX E genes: SOX9 and SOX8 in mammalian testis development. Int. J. Biochem. Cell Biol. 2010; 42:433–436. [DOI] [PubMed] [Google Scholar]
- 44. Lefebvre V., Huang W., Harley V.R., Goodfellow P.N., de Crombrugghe B.. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol. Cell Biol. 1997; 17:2336–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yao Y., Wang Y.. ATDC5: an excellent in vitro model cell line for skeletal development. J. Cell Biochem. 2013; 114:1223–1229. [DOI] [PubMed] [Google Scholar]
- 46. Frietze S., Farnham P.J.. Transcription factor effector domains. Sub cell Biochem. 2011; 52:261–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Uesugi M., Verdine G.L.. The alpha-helical FXXPhiPhi motif in p53: TAF interaction and discrimination by MDM2. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:14801–14806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reiprich S., Wegner M.. From CNS stem cells to neurons and glia: Sox for everyone. Cell Tissue Res. 2015; 359:111–124. [DOI] [PubMed] [Google Scholar]
- 49. Sinner D., Rankin S., Lee M., Zorn A.M.. Sox17 and beta-catenin cooperate to regulate the transcription of endodermal genes. Development. 2004; 131:3069–3080. [DOI] [PubMed] [Google Scholar]
- 50. Aksoy I., Jauch R., Eras V., Chng W.B., Chen J., Divakar U., Ng C.K., Kolatkar P.R., Stanton L.W.. Sox transcription factors require selective interactions with Oct4 and specific transactivation functions to mediate reprogramming. Stem Cells. 2013; 31:2632–2646. [DOI] [PubMed] [Google Scholar]
- 51. Zorn A.M., Barish G.D., Williams B.O., Lavender P., Klymkowsky M.W., Varmus H.E.. Regulation of Wnt signaling by Sox proteins: XSox17 alpha/beta and XSox3 physically interact with beta-catenin. Mol. Cell. 1999; 4:487–498. [DOI] [PubMed] [Google Scholar]
- 52. Akiyama H., Lyons J.P., Mori-Akiyama Y., Yang X., Zhang R., Zhang Z., Deng J.M., Taketo M.M., Nakamura T., Behringer R.R. et al.. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev. 2004; 18:1072–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schaefer M.H., Wanker E.E., Andrade-Navarro M.A.. Evolution and function of CAG/polyglutamine repeats in protein-protein interaction networks. Nucleic Acids Res. 2012; 40:4273–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tsuda M., Takahashi S., Takahashi Y., Asahara H.. Transcriptional co-activators CREB-binding protein and p300 regulate chondrocyte-specific gene expression via association with Sox9. J. Biol. Chem. 2003; 278:27224–27229. [DOI] [PubMed] [Google Scholar]
- 55. Zhou R., Bonneaud N., Yuan C.X., de Santa Barbara P., Boizet B., Schomber T., Scherer G., Roeder R.G., Poulat F., Berta P.. SOX9 interacts with a component of the human thyroid hormone receptor-associated protein complex. Nucleic Acids Res. 2002; 30:3245–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rau M.J., Fischer S., Neumann C.J.. Zebrafish Trap230/Med12 is required as a coactivator for Sox9-dependent neural crest, cartilage and ear development. Dev. Biol. 2006; 296:83–93. [DOI] [PubMed] [Google Scholar]
- 57. Piskacek M., Vasku A., Hajek R., Knight A.. Shared structural features of the 9aaTAD family in complex with CBP. Mol. Biosyst. 2015; 11:844–851. [DOI] [PubMed] [Google Scholar]
- 58. Ravarani C.N., Erkina T.Y., De Baets G., Dudman D.C., Erkine A.M., Babu M.M.. High-throughput discovery of functional disordered regions: investigation of transactivation domains. Mol. Syst. Biol. 2018; 14:e8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sandholzer J., Hoeth M., Piskacek M., Mayer H., de Martin R.. A novel 9-amino-acid transactivation domain in the C-terminal part of Sox18. Biochem. Biophys. Res. Commun. 2007; 360:370–374. [DOI] [PubMed] [Google Scholar]
- 60. Sinner D., Kordich J.J., Spence J.R., Opoka R., Rankin S., Lin S.C., Jonatan D., Zorn A.M., Wells J.M.. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol. Cell Biol. 2007; 27:7802–7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Valenzuela I., Fernandez-Alvarez P., Plaja A., Ariceta G., Sabate-Rotes A., Garcia-Arumi E., Vendrell T., Tizzano E.. Further delineation of the SOX18-related Hypotrichosis, Lymphedema, Telangiectasia syndrome (HTLS). Eur. J. Med Genet. 2018; 61:269–272. [DOI] [PubMed] [Google Scholar]
- 62. Katoh-Fukui Y., Igarashi M., Nagasaki K., Horikawa R., Nagai T., Tsuchiya T., Suzuki E., Miyado M., Hata K., Nakabayashi K. et al.. Testicular dysgenesis/regression without campomelic dysplasia in patients carrying missense mutations and upstream deletion of SOX9. Mol. Genet. Genomic Med. 2015; 3:550–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Takash W., Canizares J., Bonneaud N., Poulat F., Mattei M.G., Jay P., Berta P.. SOX7 transcription factor: sequence, chromosomal localisation, expression, transactivation and interference with Wnt signalling. Nucleic Acids Res. 2001; 29:4274–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Francois M., Koopman P., Beltrame M.. SoxF genes: key players in the development of the cardio-vascular system. Int. J. Biochem. Cell Biol. 2010; 42:445–448. [DOI] [PubMed] [Google Scholar]
- 65. Hosking B.M., Muscat G.E., Koopman P.A., Dowhan D.H., Dunn T.L.. Trans-activation and DNA-binding properties of the transcription factor, Sox-18. Nucleic Acids Res. 1995; 23:2626–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Genome Aggregation Database (gnomAD) is a collection of exome and genome sequencing data from >140k unrelated individuals used as control subjects in various studies. It is available online (https://gnomad.broadinstitute.org). The 9-aa-TAD prediction tool developed by Piskacek is a freely available online computational resource to identify 9-aa-TAD motifs in protein sequences: (http://www.med.muni.cz/9-aa-TAD/). SWISS-MODEL is an automated protein structure homology-modeling server available worldwide (https://:swissmodel.expasy.org). I-TASSER (Iterative Threading ASSEmbly Refinement) is a freely accessible hierarchical approach to protein structure and function prediction (https://zhanglab.ccmb.med.umich.edu/I-TASSER/). PEP-FOLD3 is a freely accessible de novo approach aimed at predicting peptide structures from amino acid sequences (http://bioserv.rpbs.univ-paris-diderot.fr/services.html).