Abstract
The halophyte Suaeda salsa displayed strong resistance to salinity. Up to date, molecular mechanisms underlying tolerance of S. salsa to salinity have not been well understood. In the present study, S. salsa seedlings were treated with 30‰ salinity and then leaves and roots were subjected to Illumina sequencing. Compared with the control, 68,599 and 77,250 unigenes were significantly differentially expressed in leaves and roots in saline treatment, respectively. KEGG enrichment analyses indicated that photosynthesis process, carbohydrate, lipid and amino acid metabolisms were all downregulated in saline treatment, which should inhibit growth of S. salsa. Expression levels of Na+/H+ exchanger, V-H+ ATPase, choline monooxygenase, potassium and chloride channels were upregulated in saline treatment, which could relieve reduce over-accumulation of Na+ and Cl-. Fe-SOD, glutathione, L-ascorbate and flavonoids function as antioxidants in plants. Genes in relation to them were all upregulated, suggesting that S. salsa initiated various antioxidant mechanisms to tolerate high salinity. Besides, plant hormones, especially auxin, ethylene and jasmonic acid signaling transduction pathways were all upregulated in response to saline treatment, which were important to gene regulations of ion transportation and antioxidation. These changes might comprehensively contribute to tolerance of S. salsa to salinity. Overall, the present study provided new insights to understand the mechanisms underlying tolerance to salinity in halophytes.
Introduction
The halophyte Suaeda salsa (L.) Pall is a leaf succulent annual plant. It is widely distributed in Eurasia and has been cultivated as a seawater vegetable in desert and coastal areas in P. R. China [1]. S. salsa shows strong tolerance to salinity. In comparison to the control, treatments with moderate salinities (varied from 50 mM to 200 mM NaCl) promoted CO2 assimilation rate, O2 production, photosynthesis rate and thus plant growth of S. salsa [2–6]. However, high salinities (> 200 mM NaCl) inhibited its growth significantly [6], displaying much higher tolerance to saline stress than other plants.
S. salsa should have special mechanisms underlying tolerance to high salinity. Physiologically, differential regulation of abscisic acid (ABA), gibberellins (GA) homeostasis [7] and Na+ storage [6] might provide strategies for S. salsa plants to survive adverse environmental conditions. Accumulation of proline and decreased content of soluble sugars in leaves promoted osmotic adaptation to saline stress [6]. Molecularly, functions of several genes were characterized. For example, glycerol-3-phosphate acyltransferase (GPAT) [8] and vacuolar H+/Ca2+ transporter [9] enhanced saline tolerance. Activity of plasma membrane aquaporins (AQPs) is involved in leaf succulence in S. salsa and also regulates its tolerance to salinity [10]. Besides, gene expression levels of myo-inositol-1-phosphate synthase (INPS), choline monooxygenase (CMO), betaine aldehyde dehydrogenase (BADH), catalase (CAT), and activities of superoxide dismutase (SOD), peroxidase (POD), CAT, and glutathione peroxidase (GPx) were elevated when S. salsa was treated with salinity, suggesting that these genes/enzymes might also contribute to saline tolerance in S. salsa [11]. Moreover, metabolic profiling revealed that saline treatments depleted contents amino acids, malate, fumarate, choline, phosphocholine, and elevated betaine and allantoin in S. salsa seedlings as well as reduced contents of glucose and fructose but increased contents of proline, citrate, and sucrose in roots [11]. These results revealed molecular mechanisms underlying saline tolerance in S. salsa. However, comprehensive profiles were still lacking.
In other halophytes or model plant species, studies have reported some molecular mechanisms underlying saline tolerance. In response to saline treatment, expansins, phosphatase, ethylene-related pathways were upregulated in Suaeda glauca and Suaeda maritima [12,13]. WRKY transcription factors involved in drought and saline tolerance were upregulated in saline treatments in S. glauca [12], Nicotiana benthamiana [14], Populus tomentosa [15], Jatropha curcas [16] and Arabidopsis thaliana [17]. Choline monooxygenase (CMO) and betaine aldehyde dehydrogenase (BADH) contributed to saline resistance in S. maritima [13,18] and rice [19]. Fatty acid desaturase was required for A. thaliana [20], S. glauca [12] and tomato [21] to resist saline stress. Expression levels of cytochrome P450s increased in salt-treated S. glauca [12], Gossypium hirsutum and Phaseolus vulgaris [22]. MYB genes were elevated in sweet cherry [23], Salicornia brachiate [24] and S. maritima in response to saline treatments [13]. There results indicated that different plants may adopt different molecular strategies to resist saline stress.
Transcriptome sequencing is a powerful tool to explore molecular mechanisms underlying biological process. To the best of our knowledge, no reports have been reported to investigate changes of transcripts of S. salsa in response to saline stress. In the present study, S. salsa seedlings were treated with 30‰ salinity and then leaves and roots were subjected to transcriptome sequencing. Real-time quantitative PCR (qPCR) was performed to validate the results. Bioinformatics analyses were adopted to compare changes at mRNA and pathway levels. These data would be helpful to clarify the molecular mechanisms underlying tolerance of S. salsa to salinity.
Materials and methods
Ethics statement
No specific permit is required for studies on S. salsa in P. R. China. During the experiments, no local regulations or laws were overlooked.
Germination of S. salsa seeds in response to saline treatments
S. salsa seeds were kindly donated by Jiangsu Coastal Area Institute of Agricultural Sciences (Yancheng, P. R. China). Six saline concentrations were prepared, including 0.94‰, 1.88‰, 3.75‰, 7.5‰, 15‰ and 30‰ (geometric sequence by two times), by dissolving commercial sea salts (Tangfeng Slats Company Limited, Tangshan, P. R. China) in deionized water. Deionized water was used as the control. Germination assays were conducted in 9 cm Petri dishes. In each dish, three layers of filter paper were placed at the bottom, 10 ml of saline solution was added and then 100 seeds were placed on filter paper. Seeds were germinated in a light humidified incubator at humidity of 80% RH and dark. The temperature was 25°C at day time (12 h) and 15°C at night (12 h). Germinated seeds were counted daily for 10 days to calculate germination rate. To avoid disturbance of evaporation on salinity, germinated seedlings were discarded and ungerminated seeds were transferred to new dishes with fresh solution every two days. Each assay was repeated four times.
Saline treatments of S. salsa seedlings
S. salsa seeds were germinated in deionized water as described above. When seedlings reached 10 cm approximately, seedlings were transplanted to plastic containers with sands as cultural substrate and Hoagland’s solution as media, and then cultured in a greenhouse at 25°C. The light cycle was 16 hours: 8 hours (light: dark) and light intensity was approximately 14,400 lux. After two weeks, healthy plants were treated with salinity of 30‰ by dissolving commercial sea salts (Tangfeng Slats Company Limited, Tangshan, China) in Hoagland’s solution. Hoagland’s solution was used as the control. Each treatment included 20–30 individuals and repeated three times independently. To supplement evaporated water, distilled water was added every day to keep total volume of culture media consistent. After 10 days, plants were harvested. Roots and top four leaves were collected and immediately frozen in liquid nitrogen for RNA extraction. For each treatment, samples from 5 individuals were pooled.
Transcriptome sequencing
Total RNA was extracted using Biozol reagent (Bioer, Hangzhou, China) according to the manufacture’s protocol. Quality of total RNA was check by 1% agarose gel electrophoresis, NanoPhotometer spectrophotometer (IMPLEN, CA, USA) and Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). RNA samples with RNA integrity number (RIN) higher than 8.0 were considered qualified. RNA concentration was measured using Qubit RNA assay kit on Qubit 2.0 Flurometer (Life Technologies, CA, USA).
To construct sequencing libraries, mRNA was enriched using NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB, USA). Sequencing libraries were prepared using NEBNext mRNA Library Prep Master Mix Set for Illumina (NEB, USA) and NEBNext Multiplex Oligos for Illumina (NEB, USA). Sequencing library was viewed on 1.8% agarose gel to check insert size and quantified using Library Quantification Kit-Illumina GA Universal Kit (Kapa, USA).
Qualified libraries were clustered on a cBot cluster generation system using HiSeq 4000 PE cluster kit (Illumina) and sequenced on an Illumina Hiseq 2500 platform. Three independent samples were sequenced as three biological replicates.
Bioinformatics analyses
Clean reads were achieved by removing adaptors, reads with N ratio higher than 1% and low quality reads (with > 50% bases having Phred quality score ≤ 15). Clean reads were subjected to assembly of unigene library using Trinity v2.0.6 [25]. HTSwq v0.6.0 was applied to calculate FPKM values (expected number of fragments per kilobase of transcript sequence per millions base pairs sequenced) of each unigene. Relative expression levels of each gene were compared among different groups using DESeq2 R package. Fold change ≥ 2 and q value (adjusted P value) < 0.001 were considered significantly changed.
DEGs were mapped to Gene Ontology (GO) database and Kyoto Encyclopedia of Genes and Genomes database (KEGG) [26] for enrichment of GO categories and KEGG pathways using BLAST software [27].
Real-time quantitative PCR
Results of Illumina sequencing were validated using real-time qPCR. cDNA was prepared using BioRT cDNA first strand synthesis kit (Bioer, Hangzhou, China) with oligo(dT) primer. qPCR was carried out using BioEasy master mix (Bioer, Hangzhou, China) on a Line Gene9600 Plus qPCR machine (Bioer, Hangzhou, China). DEGs and primers used for qPCR are listed in S1 Table. Transcriptional elongation factor-1 alpha was used as the internal control. Relative expression levels of each gene between saline treatment and the control were calculated using the typical 2-ΔΔCt method [28]. Three biological replicates were included for each treatment and three technical repeats were performed for each sample.
Results and discussion
Effects of salinity on seed germination
Previously, most experiments employed NaCl to represent saline stress, which might misestimate the effects of other components in sea salts on seed germination and plant growth [29–30]. In the present study, we used sea salts to prepare saline solutions, which should be more similar to real environments.
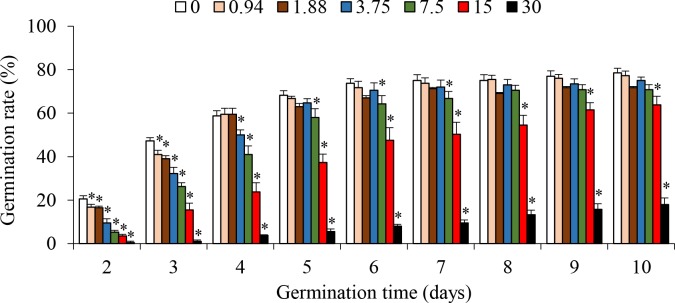
Seeds started to germinate in all treatments after two days. Germination rate significantly decreased in response to saline treatments but increased with germination time. After eight days, in comparison to the control, germination rates were significantly lower in treatments with 30‰ and 15‰, but did not significantly change in other saline treatments (Fig 1). These results suggested that germination of S. salsa seeds could tolerate salinity as high as 7.5‰, which was consistent with previous findings on S. salsa and the general consensus that germination of halophyte seeds is optimum under freshwater but decreases with elevating salinity [31, 32].
Fig 1. Effects of salinity on seed germination rate of Suaeda salsa (mean ± SE).
* significantly different from the control (P < 0.05).
Transcriptome sequencing
The sequencing data were deposited in NCBI with the reference number of PRJNA512222. Illumina sequencing resulted in 65.21 M to 72.28 M of total clean reads and 6.52 G to 7.23 G of total clean bases for each sample (Table 1). The sequencing depth was roughly calculated by the ratio of sequencing data to the size of assembled transcriptome, approximately ranging from 22.37 × to 24.81 ×. The total clean reads were higher than those in previous studies on S. salsa [33]. Besides, all Q20 and Q30 values were higher than 98.09% and 91.78%, respectively (Table 1). These indices suggested that the sequencing was deep and the as-obtained data should be enough for further analyses.
Table 1. Statistics of Illumina sequencing quality for each sample.
| Salinity-Sample | TCR (M) | TCB (G) | Q20 (%) | Q30 (%) | TM (%) | UM (%) |
|---|---|---|---|---|---|---|
| Leaf | ||||||
| Control-1 | 67.88 | 6.79 | 98.49 | 92.78 | 89.02 | 10.51 |
| Control-2 | 67.52 | 6.75 | 98.39 | 92.42 | 89.51 | 10.62 |
| Control-3 | 67.57 | 6.76 | 98.37 | 92.32 | 89.64 | 9.83 |
| 30‰-1 | 67.76 | 6.78 | 98.46 | 92.82 | 90.75 | 10.31 |
| 30‰-2 | 69.85 | 6.99 | 98.45 | 92.80 | 89.89 | 10.99 |
| 30‰-3 | 67.44 | 6.74 | 98.45 | 92.63 | 90.73 | 10.20 |
| Root | ||||||
| Control-1 | 70.26 | 7.03 | 98.52 | 92.91 | 87.74 | 12.26 |
| Control-2 | 65.21 | 6.52 | 98.48 | 92.76 | 86.40 | 12.54 |
| Control-3 | 66.91 | 6.69 | 98.42 | 92.58 | 86.83 | 12.83 |
| 30‰-1 | 70.19 | 7.02 | 98.09 | 91.78 | 87.04 | 12.11 |
| 30‰-2 | 72.28 | 7.23 | 98.15 | 91.88 | 87.00 | 12.53 |
| 30‰-3 | 67.59 | 6.76 | 98.15 | 91.91 | 88.23 | 12.36 |
TCR: total clean reads; TCB: total clean base; TM: total mapping; UM: uniquely mapping.
De novo assembly
Total clean reads of each sample were assembled independently to get the unigene library of each sample. The total number of unigenes ranged from 40,038 to 133,430, with the mean length of unigenes from 798 bp to 1,180 bp (S2 Table). Benchmarking Universal Single-Copy Orthologs (BUSCO) analyses revealed that at least 2.97% BUSCOs were fragmented or missing (S1 Fig). Alternatively, clean reads of all samples were pooled, equal to 1,234.49 M of clean reads, and then subjected to Trinity assembly. Finally, 196,199 unigenes were obtained, with the mean length of 1,673 bp and the N50 length of 2,780 bp (Table 2). BUSCO analyses revealed that only 1.98% BUSCOs were missing and none BUSCOs were fragmented (S1 Fig). Obviously, the as-obtained unigenes showed longer mean length and N50 length as well as less fragmented and missing BUSCOs than those assembled from each sample (S1 Table), suggesting that unigene library generated from all samples had higher quality. The as-obtained unigene library was used for further analyses.
Table 2. Assembly statistics of unigenes and coding sequences (CDS).
| Parameter | Unigene | CDS |
|---|---|---|
| Total clean reads used for assembly (M) | 1,234.49 | 1,234.49 |
| Total number | 196,199 | 122,855 |
| Total length (M bp) | 328.25 | 137.21 |
| Minimum unigene length (bp) | 200 | 297 |
| Maximum unigene length (bp) | 35,045 | 15,345 |
| Average unigene length (bp) | 1,673 | 1,117 |
| N50 length (bp) | 2,780 | 1,431 |
| N90 length (bp) | 836 | 552 |
| GC content (%) | 39.23 | 42.78 |
Functional annotation and analysis
Among the unigene library, more than 65.45% and 45.91%unigenes were longer than 500 bp and 1,000 bp, respectively (S2 Fig). Coding sequence (CDS) prediction revealed 122,855 CDSs, with the total length of 137.21 M bp, the average length of 1,117 bp and the N50 length of 1,431 bp (Table 2). Approximately 72.67% and 36.84% CDSs were longer than 500 bp and 1,000 bp, respectively (S2 Fig).
After blasted against the relevant databases, 61.97%, 51.39%, 43.88%, 47.03%, 48.15%, 44.30% and 14.91% unigenes could hit genes in the RefSeq non-redundant proteins (NR), nucleotide (NT), Swissprot, KEGG, Eukaryotic Orthologous Groups (KOG), Pfam and GO database respectively. Overall, 68.10% unigenes could be annotated to one or more databases.
Differentially expressed genes and qPCR validation
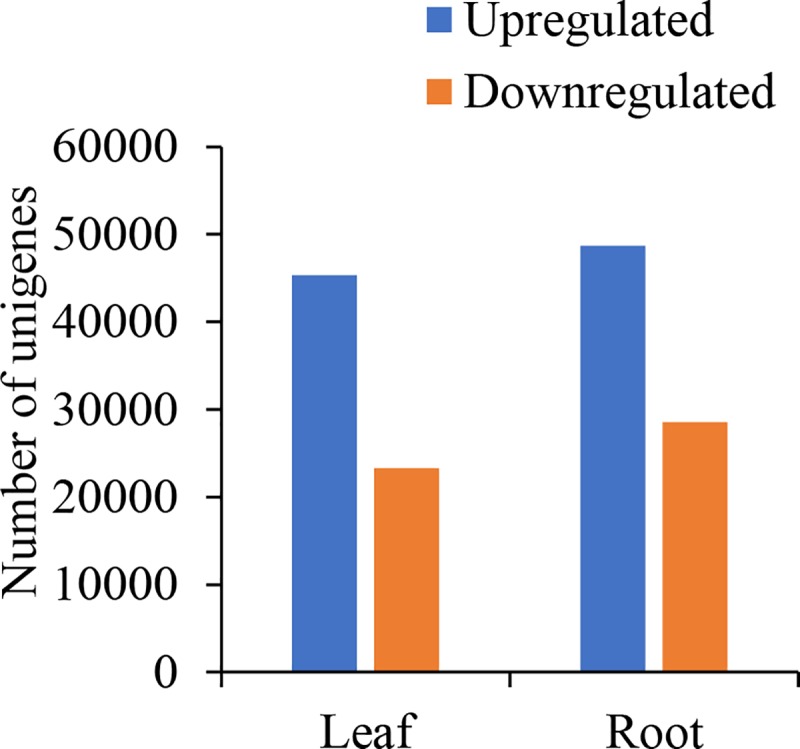
Compared with the control, 68,599 (45,321 upregulated and 23,278 downregulated) and 77,250 unigenes (48,682 upregulated and 28,568 downregulated) were significantly differentially expressed in treatment with 30‰ in leaves and roots, respectively (Fig 2).
Fig 2. Number of differentially expressed genes in Suaeda salsa leaves and roots between treatment with 30‰ and the control.
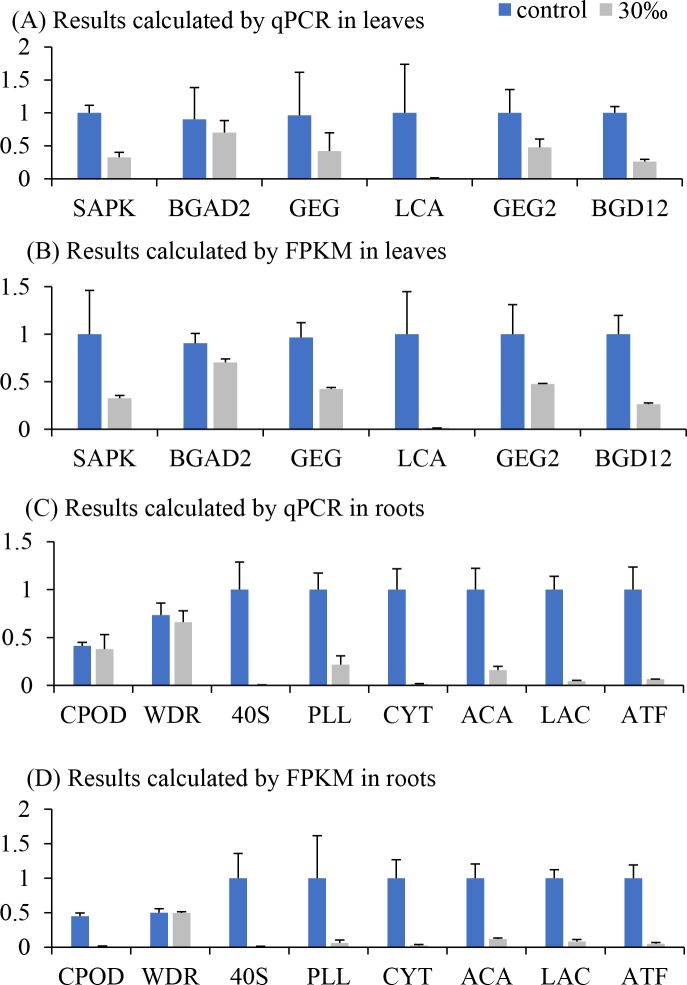
To validate the expression levels predicted by transcriptome sequencing, qPCR was performed for 6 unigenes in leaves and 8 unigenes in roots. The results showed similar tendency to the expression levels calculated by FPKM value (Fig 3), suggesting that DEGs prediction was reliable.
Fig 3. Real-time qPCR validation results (mean ± SE).
Data were normalized by considering the highest value of each gene among three treatments as one. CPOD: Cationic peroxidase 1; WDR: WD repeat-containing protein 87; 40S: 40S ribosomal protein S3a; PLL: Probable linoleate 9S-lipoxygenase 5; CYT: Cytochrome P450 71A6; ACA: Acetyl-CoA acetyltransferase, cytosolic; LAC: Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial; ATF: Branched-chain-amino-acid aminotransferase 5, chloroplastic; SAPK: Serine/threonine-protein kinase; BGAD2: Beta-galactosidase 2; GEG: Glucan endo-1,3-beta-glucosidase; LCA: Lichenase; GEG2: Glucan endo-1,3-beta-glucosidase; BGD12: Beta-glucosidase 12.
Enrichment of GO categories for DEGs
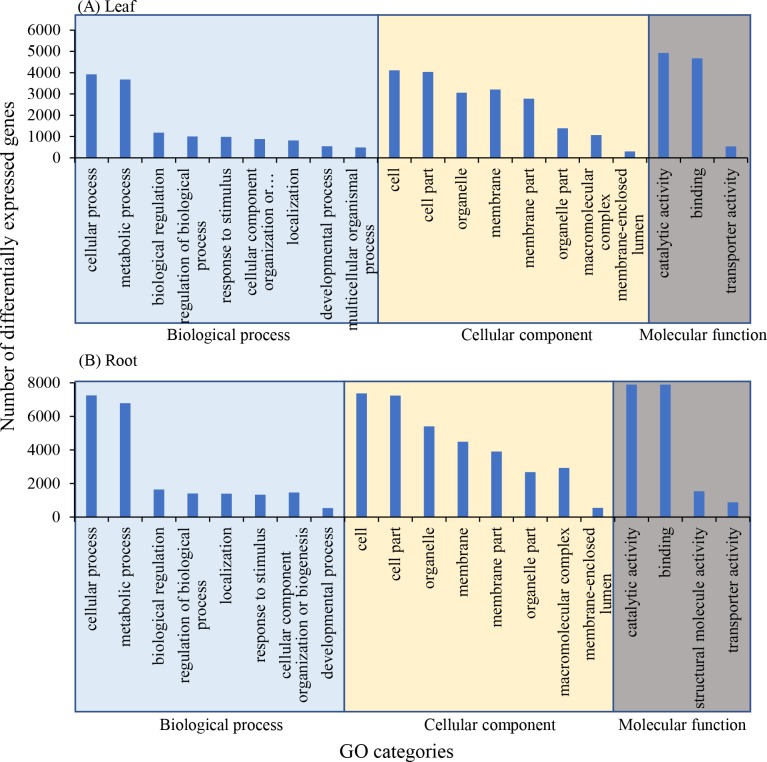
Top 20 GO categories revealed that in leaves and roots, DEGs in comparison between 30‰ and control were mainly assigned to cellular process, metabolic process, cell, cell part, catalytic activity and binding (Fig 4).
Fig 4. Top 20 GO categories of differentially expressed genes in treatment with 30‰ compared with the control.
Enrichment of KEGG pathways for DEGs
KEGG enrichment of significantly downregulated genes showed 24 and 16 pathways in leaves and roots, respectively, which shared nine KEGG pathways, including valine, leucine and isoleucine degradation (ko00280), linoleic acid metabolism (ko00591), glycerophospholipid metabolism (ko00564), circadian rhythm-plant (ko04712), plant hormone signal transduction (ko04075), pentose phosphate pathway (ko00030), riboflavin metabolism (ko00740), nicotinate and nicotinamide metabolism (ko00760) and ascorbate and aldarate metabolism (ko00053). These enriched pathways represented harmful effects of salinity on S. salsa. KEGG enrichment of significantly upregulated genes revealed 17 and 18 pathways in leaves and roots, respectively. Between them, only one pathway (ko00261 monobactam biosynthesis) was shared. These results might provide new insights for investigation of molecular mechanisms underlying adaptation of S. salsa to salinity stress (Tables 3 and 4).
Table 3. Enrichment of KEGG pathway between treatment with 30‰ and the control in Suaeda salsa leaves.
Enriched/total gene: number of DEGs/total genes in the corresponding pathway.
| ID and name of KEGG pathway | Enriched/ total gene |
P value | Q value |
|---|---|---|---|
| Upregulated genes in treatment with 30‰ | |||
| ko00480, Glutathione metabolism | 361/1308 | 0.000 | 0.000 |
| ko00901, Indole alkaloid biosynthesis | 83/242 | 0.000 | 0.000 |
| ko00940, Phenylpropanoid biosynthesis | 550/2219 | 0.000 | 0.000 |
| ko00130, Ubiquinone and other terpenoid-quinone biosynthesis | 161/577 | 0.000 | 0.001 |
| ko00941, Flavonoid biosynthesis | 152/554 | 0.000 | 0.003 |
| ko03018, RNA degradation | 444/1866 | 0.001 | 0.017 |
| ko04144, Endocytosis | 621/2666 | 0.001 | 0.017 |
| ko00300, Lysine biosynthesis | 62/207 | 0.001 | 0.019 |
| ko00460, Cyanoamino acid metabolism | 228/914 | 0.002 | 0.021 |
| ko00073, Cutin, suberine and wax biosynthesis | 71/247 | 0.002 | 0.022 |
| ko00261, Monobactam biosynthesis | 58/195 | 0.002 | 0.022 |
| ko00945, Stilbenoid, diarylheptanoid and gingerol biosynthesis | 80/284 | 0.002 | 0.022 |
| ko04075, Plant hormone signal transduction | 529/2271 | 0.002 | 0.022 |
| ko04120, Ubiquitin mediated proteolysis | 423/1797 | 0.003 | 0.026 |
| ko00966, Glucosinolate biosynthesis | 29/85 | 0.003 | 0.027 |
| ko00400, Phenylalanine, tyrosine and tryptophan biosynthesis | 155/613 | 0.005 | 0.037 |
| Downregulated genes in treatment with 30‰ | |||
| ko00500, Starch and sucrose metabolism | 480/2481 | 0.000 | 0.000 |
| ko00051, Fructose and mannose metabolism | 153/687 | 0.000 | 0.000 |
| ko00940, Phenylpropanoid biosynthesis | 424/2219 | 0.000 | 0.000 |
| ko04075, Plant hormone signal transduction | 434/2271 | 0.000 | 0.000 |
| ko00604, Glycosphingolipid biosynthesis-ganglio series | 51/185 | 0.000 | 0.001 |
| ko00941, Flavonoid biosynthesis | 123/554 | 0.000 | 0.001 |
| ko00740, Riboflavin metabolism | 75/307 | 0.000 | 0.001 |
| ko04016, MAPK signaling pathway—plant | 424/2271 | 0.000 | 0.001 |
| ko00945, Stilbenoid, diarylheptanoid and gingerol biosynthesis | 69/284 | 0.000 | 0.002 |
| ko00010, Glycolysis / Gluconeogenesis | 261/1352 | 0.000 | 0.003 |
| ko00760, Nicotinate and nicotinamide metabolism | 73/311 | 0.000 | 0.003 |
| ko04712, Circadian rhythm—plant | 138/672 | 0.001 | 0.006 |
| ko00564, Glycerophospholipid metabolism | 204/1046 | 0.001 | 0.006 |
| ko00591, Linoleic acid metabolism | 67/299 | 0.002 | 0.012 |
| ko04626, Plant-pathogen interaction | 458/2562 | 0.002 | 0.012 |
| ko00280, Valine, leucine and isoleucine degradation | 190/990 | 0.002 | 0.014 |
| ko00565, Ether lipid metabolism | 73/340 | 0.003 | 0.022 |
| ko00052, Galactose metabolism | 218/1166 | 0.003 | 0.023 |
| ko00531, Glycosaminoglycan degradation | 61/277 | 0.004 | 0.023 |
| ko00520, Amino sugar and nucleotide sugar metabolism | 318/1760 | 0.004 | 0.025 |
| ko00030, Pentose phosphate pathway | 137/707 | 0.005 | 0.031 |
| ko00053, Ascorbate and aldarate metabolism | 193/1033 | 0.006 | 0.032 |
| ko00710, Carbon fixation in photosynthetic organisms | 152/800 | 0.007 | 0.039 |
Table 4. Enrichment of KEGG pathway between treatment with 30‰ and the control in Suaeda salsa roots.
Enriched/total gene: number of DEGs/total genes in the corresponding pathway.
| ID and name of KEGG pathway | Enriched/ total gene |
P value | Q value |
|---|---|---|---|
| Upregulated genes in treatment with 30‰ | |||
| ko03010, Ribosome | 1423/3320 | 0.000 | 0.000 |
| ko00196, Photosynthesis—antenna proteins | 28/43 | 0.000 | 0.000 |
| ko00630, Glyoxylate and dicarboxylate metabolism | 290/856 | 0.000 | 0.000 |
| ko00260, Glycine, serine and threonine metabolism | 261/789 | 0.000 | 0.000 |
| ko01230, Biosynthesis of amino acids | 913/3127 | 0.000 | 0.001 |
| ko01200, Carbon metabolism | 901/3111 | 0.000 | 0.002 |
| ko00072, Synthesis and degradation of ketone bodies | 42/101 | 0.000 | 0.009 |
| ko00220, Arginine biosynthesis | 174/539 | 0.001 | 0.011 |
| ko00195, Photosynthesis | 82/230 | 0.001 | 0.012 |
| ko03008, Ribosome biogenesis in eukaryotes | 385/1294 | 0.001 | 0.016 |
| ko00710, Carbon fixation in photosynthetic organisms | 246/800 | 0.001 | 0.017 |
| ko00190, Oxidative phosphorylation | 373/1263 | 0.002 | 0.023 |
| ko03013, RNA transport | 871/3088 | 0.002 | 0.023 |
| ko04145, Phagosome | 304/1015 | 0.002 | 0.023 |
| ko00920, Sulfur metabolism | 111/342 | 0.004 | 0.041 |
| ko03020, RNA polymerase | 331/1126 | 0.005 | 0.045 |
| ko00261, Monobactam biosynthesis | 67/195 | 0.006 | 0.047 |
| ko00750, Vitamin B6 metabolism | 55/156 | 0.007 | 0.050 |
| Downregulated genes in treatment with 30‰ | |||
| ko00280, Valine, leucine and isoleucine degradation | 308/990 | 0.000 | 0.000 |
| ko00591, Linoleic acid metabolism | 91/299 | 0.000 | 0.000 |
| ko00071, Fatty acid degradation | 169/643 | 0.000 | 0.000 |
| ko00640, Propanoate metabolism | 141/525 | 0.000 | 0.000 |
| ko00592, alpha-Linolenic acid metabolism | 145/547 | 0.000 | 0.000 |
| ko00250, Alanine, aspartate and glutamate metabolism | 192/775 | 0.000 | 0.001 |
| ko00350, Tyrosine metabolism | 152/597 | 0.000 | 0.001 |
| ko00564, Glycerophospholipid metabolism | 247/1046 | 0.000 | 0.002 |
| ko04712, Circadian rhythm—plant | 164/672 | 0.000 | 0.005 |
| ko04075, Plant hormone signal transduction | 493/2271 | 0.001 | 0.009 |
| ko02010, ABC transporters | 185/789 | 0.001 | 0.014 |
| ko00030, Pentose phosphate pathway | 167/707 | 0.001 | 0.015 |
| ko00740, Riboflavin metabolism | 79/307 | 0.002 | 0.023 |
| ko00760, Nicotinate and nicotinamide metabolism | 80/311 | 0.002 | 0.023 |
| ko00053, Ascorbate and aldarate metabolism | 231/1033 | 0.004 | 0.036 |
| ko00511, Other glycan degradation | 124/525 | 0.005 | 0.044 |
Changes of energy metabolism in S. salsa upon saline treatment
Photosynthesis process is quite sensitive to saline stress. In response to saline treatments, photosynthesis process was inhibited in maize [34]and mulberry [35], which should negatively affect accumulation of organic matters and plant growth. Similarly, salinity also displayed negative effects on shoot length of S. salsa [36]. In the present study, carbon fixation in photosynthetic organisms (ko00710) was suppressed in saline-stressed leaves, suggesting that photosynthesis process might be inhibited by salinity, which then should reduce metabolism of organic carbons, displaying downregulation of carbohydrate metabolism (ko00500 starch and sucrose metabolism, ko00051 fructose and mannose metabolism, ko00010, glycolysis/gluconeogenesis, ko00052 galactose metabolism, ko00531 glycosaminoglycan degradation, ko00520 amino sugar and nucleotide sugar metabolism, ko00030 pentose phosphate pathway, ko00640 propanoate metabolism and ko00511 other glycan degradation). Furthermore, downregulation of carbohydrate metabolism would depress lipid metabolism (ko00564, glycerophospholipid metabolism, ko00591 linoleic acid metabolism, ko00071 fatty acid degradation, and ko00592 alpha-linolenic acid metabolism), amino acid metabolism (ko00250 alanine, aspartate and glutamate metabolism and ko00350 tyrosine metabolism) and biosynthesis of complicated polymer compounds (Tables 3 and 4). For example, phenylpropanoid biosynthesis (ko00940), which contributes monolignols, the starting compounds for biosynthesis of lignin [37], was downregulated in the present study. Overall, these changes would comprehensively depress S. salsa growth. These results were partially similar to transcriptomic analyses of S. maritima in treatment with 2% NaCl, which decreased the expression levels of genes related to photosynthesis (particularly the light reaction) and Calvin cycle, but increased most of the genes in the anabolic pathways (such as sucrose and starch synthesis) and catabolic pathways (such as the tricarboxylic acid cycle, glycolysis and the oxidative pentose phosphate pathway). The different changes of sucrose and starch synthesis and glycolysis between these two studies might be attributed to the tested salinities. In the present study, 30‰ salinity was used, which was much higher than that prepared by 2% NaCl.
The KEGG pathway riboflavin metabolism (ko00740) and nicotinate and nicotinamide metabolism (ko00760) is important for biosynthesis of vitamin B2 and B3, respectively. Vitamin B2 is an essential coenzyme to oxidordeuctases (such as succinodehydrogenase, xanthine oxidase and NADH dehydrogenase), participating in degradation of sugars. Vitamin B3 is a major component of coenzyme I (NAD) and coenzyme II (NADPH), which participate in the photosynthesis process, degradation of sugars and lipids. Thus, downregulation of these two pathways could be considered as normal regulation in response to saline-inhibited photosynthesis and accumulation of organic matters.
Changes of ion transportation
To resist saline stress, the first strategy is to relieve harmful accumulation of Na+ and Cl-. In NaCl-treated S. maritima, plasma membrane H+ ATPase (PM-H+ ATPase), Na+/H+ exchanger, Na+/H+ antiporter, vacuolar-type H+ ATPase (V-H+ ATPase), betaine aldehyde dehydrogenase (BADH) and choline monooxygenase (CMO) were upregulated, which were involved in ionic adjustment [13]. Similarly, Na+ influx transporter and the tonoplast Na+/H+ antiporter were upregulated in Suaeda fruticose under saline condition [38]. In the present study, partially similar results were revealed (Table 5). Na+/H+ exchanger, V-H+ ATPase and CMO were significantly upregulated in leaves and roots. These results suggested that S. salsa might share similar mechanisms with other Suaeda species underlying ionic balance (Na+ and H+) in response to saline stress. The H+-ATPase generates an electrochemical membrane potential in the plasma membrane and Na+/H+ exchanger may transport Na+ out of cells via this H+ gradient [39,40]. However, PM-H+ ATPase and Na+/H+ antiporter did not change in response to saline treatment in the present study and also in S. glauca [12], indicating that different Suaeda species might differ in regulation of proteins responsible for ionic balance.
Table 5. FPKM values of selected genes in saline treatment and control in Suaeda salsa.
| Gene name | Leaves | Roots | ||
|---|---|---|---|---|
| Control | 30‰ | Control | 30‰ | |
| Antioxidant enzymes | ||||
| Choline monooxygenase | 56.1 ± 22.2 | 272.3 ± 87.6* | 85.0 ± 12.4 | 119.4 ± 10.4* |
| Copper chaperone for superoxide dismutase | 20.6 ± 1.3 | 47.2 ± 3.9* | 22.0 ± 5.3 | 35.6 ± 9.0 |
| Superoxide dismutase [Cu-Zn] | 87.5 ± 17.6 | 81.2 ± 4.3 | 113.9 ± 19.5 | 172.1 ± 22.0 |
| Superoxide dismutase [Fe] | 27.1 ± 9.5 | 91.8 ± 9.3* | 12.1 ± 2.9 | 57.8 ± 3.8* |
| Ion transportation | ||||
| Cation/H(+) antiporter | 11.7 ± 3.3 | 17.2 ± 2.6 | 32.1 ± 6.3 | 18.5 ± 4.8* |
| K(+) efflux antiporter | 24.6 ± 0.8 | 23.2 ± 1.4 | 25.8 ± 6.6 | 24.6 ± 5.1 |
| Sodium/hydrogen exchanger | 28.9 ± 10.4 | 42.8 ± 4.9* | 15.5 ± 5.9 | 36.8 ± 13.7* |
| Cadmium/zinc-transporting ATPase | 2463.2 ± 173.7 | 3376.2 ± 438.5 | 3263.6 ± 294.8 | 4306.5 ± 1458.9 |
| Calcium-transporting ATPase | 59.2 ± 3.0 | 36.4 ± 2.1* | 186.4 ± 25.2 | 107.9 ± 8.2* |
| Copper-transporting ATPase | 47.3 ± 5.6 | 41.8 ± 4.1 | 54.9 ± 6.9 | 38.5 ± 17.1 |
| Phospholipid-transporting ATPase | 15.4 ± 5.2 | 13.4 ± 3.1 | 16.7 ± 9.3 | 11.8 ± 5.4 |
| Plasma membrane ATPase | 94.1 ± 13.9 | 65.5 ± 5.0 | 48.1 ± 13.8 | 17.1 ± 13.7 |
| Calcium permeable stress-gated cation channel | 156.8 ± 16.7 | 178.8 ± 85.7 | 120.8 ± 10.7 | 119.7 ± 15.3 |
| Chloride channel protein | 171.5 ± 11.1 | 223.7 ± 10.7* | 70.4 ± 3.4 | 75.0 ± 10.5 |
| Cyclic nucleotide-gated ion channel | 34.1 ± 5.8 | 44.0 ± 7.0 | 23.3 ± 5.2 | 18.9 ± 8.0 |
| S-type anion channel | 15.6 ± 4.4 | 45.9 ± 7.5* | 15.9 ± 2.2 | 21.8 ± 8.8 |
| Mechanosensitive ion channel protein | 51.5 ± 7.5 | 62.9 ± 8.0 | 32.1 ± 10.6 | 35.5 ± 6.5 |
| Potassium channel | 9709.0 ± 438.9 | 13596.4 ± 1125.4* | 10829.4 ± 711.0 | 13792.0 ± 3692.7 |
| Probable cyclic nucleotide-gated ion channel | 41.2 ± 3.1 | 28.5 ± 1.8* | 32.1 ± 4.7 | 24.3 ± 2.1 |
| Two pore calcium channel protein | 15.6 ± 5.1 | 26.3 ± 4.9 | 23.2 ± 1.1 | 27.0 ± 7.1 |
| Two-pore potassium channel | 38.8 ± 6.9 | 57.6 ± 5.1* | 63.3 ± 20.9 | 61.1 ± 6.4 |
| Choline monooxygenase | 56.1 ± 22.2 | 272.3 ± 87.6* | 85.0 ± 12.4 | 119.4 ± 10.4* |
| V-type proton ATPase | 348.9 ± 38.8 | 704.5 ± 33.4* | 201.4 ± 14.1 | 398.2 ± 28.0* |
| Indole alkaloid biosynthesis | ||||
| methylesterase | 288.1±77.8 | 778.7±127.6* | 395.8±172.1 | 599.5±236.8 |
| GDSL esterase/lipase | 179.9 ±49.2 | 223.0±22.7 | 371.3±162.2 | 417.0±187.4 |
| Cell wall-associated genes | ||||
| O-acyltransferase WSD1 | 21.8 ± 9.6 | 20.3 ± 6.1 | 10.5 ± 0.6 | 10.6 ± 1.9 |
| Laccase | 13.4 ± 3.5 | 52.7 ± 10.6* | 61.2 ± 19.2 | 121.5 ± 42.7 |
| leucine-rich repeat extensin | 126.8 ± 28.2 | 133.2 ± 8.8 | 132.8 ± 24.5 | 190.1 ± 18.3* |
| Expansin | 427.7 ± 228.4 | 369.5 ± 39.1 | 349.6 ± 91.1 | 531.6 ± 207.4 |
| Cellulose synthase | 157.4 ± 4.3 | 348.3 ± 36.9* | 119.5 ± 5.1 | 358.5 ± 179.1 |
| Cellulose synthase interactive | 54.8 ± 21.1 | 117.8 ± 9.2* | 35.6 ± 9.1 | 91.7 ± 21.6* |
Data represent mean ± standard deviation of FPKM values (n = 3).
* significantly different from the control for the same organ.
Besides, potassium channel, two-pore potassium channel, chloride channel protein, S-type anion channel were significantly upregulated in saline-treated leaves in the present study (Table 5). Similar results have been reported in Zostera marina in which various K+ channels and transporters showed higher uptake capacity of K+ in response to saline treatment than those in the control [41]. Maintenance of K+ supply is a crucial feature of salt tolerance, since transportation of K+ could effectively adjust osmotic pressure among subcellular compartments and cytosol caused by increased vacuolar Na+ concentration [42]. Activity of anion channel is corresponded to chloride channel protein [43]. Upregulation of chloride channel proteins could effectively reduce Cl- accumulation and increase tolerance to salinity in plants [44].
Activation of antioxidant mechanisms in response to saline treatment
Generally, saline treatments induce oxidative stress to plants [45,46]. The halophyte S. salsa should have some mechanisms to avoid detrimental effects of oxidative stress. In the present study, we examined the expression level of superoxide dismutase (SOD), which is the first antioxidant enzyme in response to environmental stress. The results revealed different patterns between two types of SOD. In comparison to the control, expression level of Fe-SOD increased significantly for 3.4 and 4.8 times in saline-treated leaves and roots, respectively, but expression level of Cu/Zn-SOD did not change significantly (Table 5). Consistently, saline treatment drove greater increase of expression level of Fe-SOD than that of Cu-Zn SOD in rice varieties [47]. Although Fe-SOD and Cu/Zn-SOD have similar catalyzing functions, but their amino acid sequences appear to be unrelated [48]. Thus, they may respond to oxidative stress triggered by different stress factors. More investigations are required to distinguish their biological functions.
Comparison between saline treatment and the control suggested that the KEGG pathway glutathione (GSH) metabolism (ko00480) was upregulated in S. salsa leaves. Although GSH functions in nutrient metabolism and regulation of cellular events (including gene expression, DNA and protein synthesis, cell proliferation and apoptosis, signal transduction, cytokine production and immune response, and protein glutathionylation), the major role of GSH occurs in the antioxidant defense [49]. In response to saline treatments, GSH metabolism was activated in reed [50] and Arthrospira platensis [51]. Thus, activation of GSH metabolism might also contribute antioxidant capacity to protect S. salsa from saline-induced oxidation.
GSH is biosynthesized from glutamate, cysteine, and glycine [49]. In roots, saline treatment upregulated the KEGG pathway glycine, serine and threonine metabolism (ko00260) but downregulated alanine, aspartate and glutamate metabolism (ko00250). How the changes of these two pathways affected GSH metabolism still required more investigations.
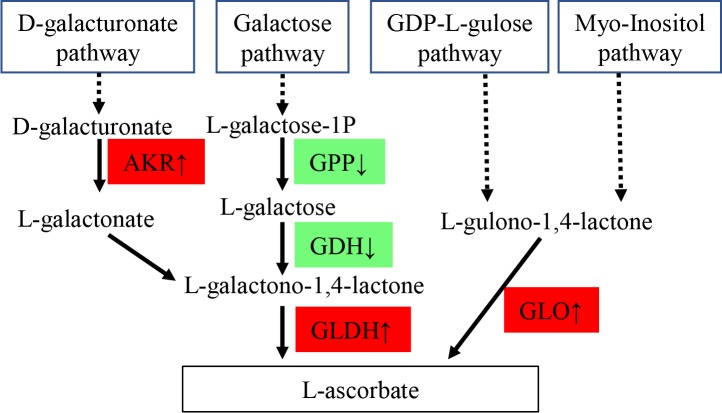
Besides GSH, L-ascorbate (AsA, also known as vitamin C) is another most abundant antioxidant in multicellular organisms [52] and is linked to glutathione metabolism [53]. Biologically, AsA is synthesized from D-galacturonate pathway through aldo-keto reductase (AKR) and L-galactono-1,4-lactone dehydrogenase (GLDH), from galactose pathway through L-galactose-1-phosphate phosphatase (GPP), L-galactose-1-dehydrogenase (GDH) and L-galactono-1,4-lactone dehydrogenase (GLDH), and from GDP-L-gulose pathway and myo-inositol pathway through L-gulono-1,4-lactone oxidase (GLO, Fig 5) [54]. In the present study, compared with the control, saline treatment downregulated expression levels of GPP and GDH, but upregulated expression levels of AKR, GLDH and GLO (S3 Table and Fig 5), suggesting that AsA might be accumulated in saline-stressed S. salsa through D-galacturonate, GDP-L-gulose and/or Myo-Inositol pathways, which might protect plants from harms of oxidation.
Fig 5. Changes of key genes in relation to L-ascorbate biosynthesis.
AKR: aldo-keto reductase; GLDH: L-galactono-1,4-lactone dehydrogenase; GPP: L-galactose-1-phosphate phosphatase; GDH: L-galactose-1-dehydrogenase; GLDH: L-galactono-1,4-lactone dehydrogenase; GLO: L-gulono-1,4-lactone oxidase. Red box: upregulated in treatment with 30‰. Green box: downregulated in treatment with 30‰.
Potential roles of flavonoids in adaptation to salinity
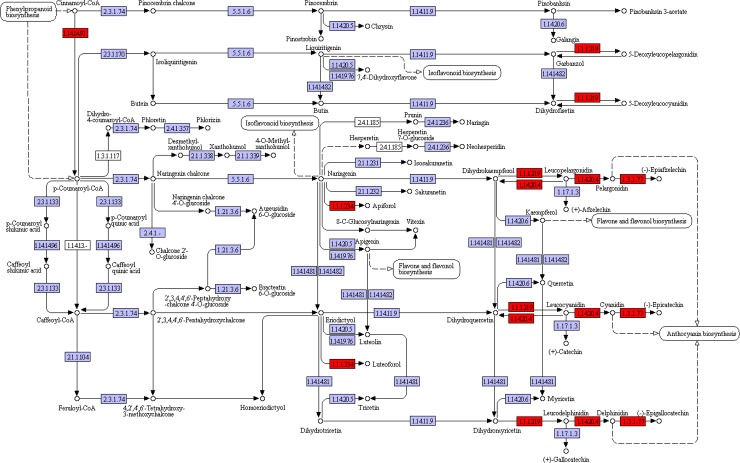
Flavonoids may serve antioxidant functions in response to heavy metals, drought, and salt stresses [55–57] and modifications of flavonoid structure (i.e., glycosylation, prenylation and methylation) could inhibit lipid peroxidation in stressed plants [58,59]. In the present study, the KEGG pathway flavonoid biosynthesis (ko00941) was significantly enriched in leaves no matter upregulated or downregulated genes were subjected to KEGG enrichment analysis. Since enzymes in this pathway had lots of unigenes in the present transcriptome data, to obtain consistent results, unigenes mapped to the same enzyme were pooled and their FPKM values were summed. Finally, the results discovered six differentially expressed enzymes (Fig 6 and S4 Table), including flavonol synthase (EC 1.14.20.6), bifunctional dihydroflavonol 4-reductase/flavanone 4-reductase (EC 1.1.1.219), anthocyanidin synthase (EC 1.14.11.19), anthocyanidin reductase (EC 1.3.1.77), flavanone 4-reductase (EC 1.1.1.234) and trans-cinnamate 4-monooxygenase (EC 1.14.14.91). These results were all upregulated in response to saline treatment, indicating that flavonoid biosynthesis pathway was activated by saline treatment in S. salsa leaves.
Fig 6. Changes of key genes in KEGG pathway of flavonoid biosynthesis.
The basic flow chart was obtained from the website (https://www.kegg.jp/kegg-bin/show_pathway?map00941). Red box: significantly upregulated in treatment with 30‰ compared with the control. Purple box: no significantly change between treatment with 30‰ and the control.
The phenylpropanoid pathway was also upregulated in saline treatment (Table 3), which is the upstream pathway to flavonoid biosynthesis [60]. Thus, phenylpropanoids and flavonoids might function together to protect S. salsa from abiotic stresses [37, 61]. Similar results were observed in various plants, such as rice [62], wheat [63] and S. glauca [12].
Regulation of plant hormones
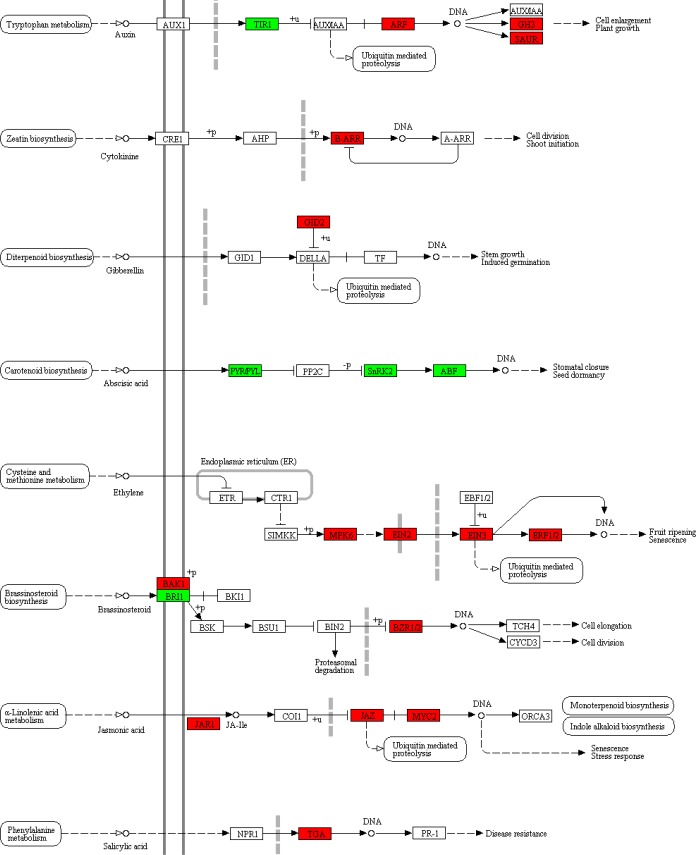
The KEGG pathway plant hormone signal transduction (ko04075) is responsible to transduce signals of hormones (including auxin, cytokinine, gibberellin, abscisic acid, ethylene, brassinosteroid, jasmonic acid and salicylic acid) to downstream performance (including cell enlargement, elongation and division, germination, shoot imitation, plant growth and stress response). KEGG enrichment analyses of upregulated or downregulated genes independently both revealed significantly involvement of plant hormone signal transduction pathway in saline-treated leaves, suggesting that this pathway might play important and complicated roles in saline resistance. The FPKM values of different unigenes mapped to the same protein were summed and statistically re-analyzed by Students’ T-tests (Fig 7 and S5 Table). Only one gene in the signaling transduction of cytokinine, gibberellin and salicylic acid was upregulated. Based on the changing tendency of only one gene, it was unprecise to conclude that these three hormone signals contributed to saline resistance in S. salsa. In the brassinosteroid signaling transduction, three genes significantly differentially expressed, but their changing tendency were contradictory. Compared with the control, BAK1 was upregulated for 1.7 times, BZR1/2 was upregulated for 1.5 times, but BRI was downregulated for 1.4 times in saline treatment. These changes were quite weak and could be ignored.
Fig 7. Changes of key genes in KEGG pathway of plant hormone signal transduction.
The basic flow chart was obtained from the website (https://www.kegg.jp/dbget-bin/www_bget?map04075). Red box: significantly upregulated in treatment with 30‰ compared with the control. Green box: significantly downregulated in treatment with 30‰compared with the control.
Importantly, the signaling transduction of auxin, abscisic acid, ethylene and jasmonic acid involved several genes which mostly showed similar tendency in the present study, suggesting that these signaling pathways might respond to saline treatment in S. salsa and will be discussed in details.
Potential roles of auxin signaling pathway in saline tolerance
The chemical nature of auxin is indole-3-acetic acid. In the present study, the KEGG pathway indole alkaloid biosynthesis (ko00901) was activated in saline-treated leaves. Within this pathway, the unigenes encoding methylesterase (EC 3.1.1.78) and GDSL esterase/lipase (EC 3.1.1.80) were both upregulated in saline treatment in comparison to the control (Table 5), suggesting that concentrations of indole alkaloids might be elevated. Similar results were reported previously, that indole alkaloids were induced by abiotic stresses, including UV-B irradiation [64], PEG-induced drought [65] and salinity [66]. In the auxin pathway which transduces signals of indole-3-acetic acid, auxin response factor (ARF), auxin responsive GH3 (GH3) and small auxin upregulated RNA family protein (SAUR) were also upregulated for1.7, 2.8 and 2.2 times, respectively, in saline-treated leaves (Fig 7 and S5 Table). Taken together, these results suggested that auxin might promote tolerance of S. salsa to saline stress through auxin signaling pathway.
Changes of abscisic acid signaling in response to salinity
In the abscisic acid (ABA) signaling pathways, abscisic acid receptor (PYR/PYL), serine/threonine-protein kinase (SNRK2) and ABA responsive element binding factor (ABF) were downregulated in saline-treated leaves, compared with the control (Fig 7 and S5 Table), suggesting that ABA signaling pathway was inhibited by saline treatment. These results were different from those in non-halophytes but consistent with previous findings in a related halophyte species S. maritima. As previously reported, saline treatment increased ABA concentrations in non-halophytes tobacco [67], tomato [68], grapevine [69]. Exogenous ABA induced saline tolerant in wheat by elevating expression of peroxidase, which might reduce the active oxygen triggered by salinity [70]. Besides, ABA induced stomatal closure to minimize water loss [71]. However, in the halophyte S. maritima, saline treatments significantly decreased ABA concentration in comparison to the control [72]. Slight decrease of ABA concentration was also observed in NaCl-treated Prosopis strombulifera roots and leaves [73]. These results suggested that inhibition of ABA signaling might be a common strategy for halophytes to resist salinity. Jin et al. [12] also reported changes of expression levels of ABA related genes in saline treatment in S. glauca, but the authors did not explain the detailed mechanisms. The underlying mechanisms required further investigations, but one thing was clear. Decreased level of ABA signaling should ensure stomatal opening and water absorption, which are important for S. salsa growth.
Changes of ethylene signaling in response to salinity
In S. salsa leaves, four genes responsible for ethylene signaling were significantly unregulated in saline treatment compared with the control, including mitogen-activated protein kinase (MPK6), ethylene-insensitive protein 2 (EIN2), ethylene-insensitive protein 3 (EIN3) and ethylene responsive factor 1/2 (ERF1/2) (Fig 7 and S5 Table). These results were consistent with the upregulation of ERFs in saline-treated S. glauca [12] and also in accordance with the general understanding that ethylene signaling is indispensable for tolerance to saline stress in Arabidopsis and many other terrestrial plants [74].
Downstream effectors of ethylene signaling could include reactive oxygen species (ROS) scavengers (such as SOD) and ion transporters [74]. In the present study, expression levels of Fe-SOD and ion transporters (including Na+/H+ exchanger, V-H+ ATPase, CMO, potassium channel, two-pore potassium channel and chloride channel protein) were all upregulated in response to saline treatment, which might be mediated by the activation of ethylene signaling pathway.
Moreover, ubiquitin mediated proteolysis (ko04120) is a downstream effector pathway of EIN3. It has been reported that this pathway is involved in flooding, heat, ultravioletradiation, oxidative stresses and diseases in plants [75]. In the present study, ubiquitin mediated proteolysis was upregulated in saline-stressed leaves by KEGG enrichment analysis (Table 3), suggesting that ubiquitin mediated proteolysis process might be activated to degrade salinity-denatured or oxidation-denatured proteins [76]and/or inactivate the repressors in plant hormone regulation pathway [77]. This process might be initiated by ethylene accumulation in S. salsa.
Changes of jasmonic acid signaling in response to salinity
Jasmonic acid (JA) could enhance the tolerance of peanut [78], tomato [79]and barley [80]to saline stress by increasing antioxidant enzyme activities [78], lowering Na+ concentration and improving stomatal conductance [80]. In the present study, three key genes in this pathway, including jasmonic acid-amido synthetase (JAR1), jasmonate ZIM domain-containing protein (JAZ) and transcription factor MYC2 (MYC2), were significantly upregulated in saline treatment compared with the control, suggesting that activation of JA signaling might improve saline tolerance in S. salsa.
Changes of cell wall-associated genes
In response to environmental stresses, modification of cell wall is a common defense mechanism. In both S glauca and S. maritima, genes related to cell wall dynamics, including laccase, expansins, leucine-rich repeat extensins (LRX), wall-associated receptor kinase proteins and/or O-acyltransferase WSD1 were upregulated in saline treatment compared with the control. In the present study, expression levels of expansins, LRX and O-acyltransferase WSD1 did not change significantly between saline treatment and the control, but laccase, and another two wall associated proteins cellulose synthase and cellulose synthase interactive [81] were significantly upregulated in the saline treatment (Table 5), suggesting that cell wall remodeling also took place in S. salsa, which might contribute to saline tolerance in S. salsa as reported in S. glauca and S. maritima [12,13].
Conclusions
In response to treatment with 30‰ salinity, S. salsa displayed suppressed photosynthesis process, carbohydrate, lipid and amino acid metabolisms, which might inhibit growth of S. salsa. To resist saline stress, Na+/H+ exchanger, V-H+ ATPase, choline monooxygenase, potassium and chloride channels were upregulated to ensure ionic balance. Expression levels of Fe-SOD and genes in glutathione metabolism, biosynthesis of L-ascorbate, flavonoids and phenylpropanoids were all upregulated to produce more antioxidants, thus relieving harmful effects of saline-induced oxidation. Plant hormones played essential roles in saline resistance in S. salsa. Importantly, auxin, ethylene and jasmonic acid signaling transduction pathways were upregulated and abscisic acid signaling transduction was inhibited by saline treatment, which might activate antioxidant mechanisms and ionic adjustment.
Supporting information
(TIF)
(TIF)
(DOCX)
(DOCX)
Data represent mean ± standard deviation (n = 3). * significantly different from the control (P < 0.05).
(DOCX)
Data represent mean ± standard deviation (n = 3). * significantly different from the control (P < 0.05).
(DOCX)
Data represent mean ± standard deviation (n = 3). * significantly different from the control (P < 0.05).
(DOCX)
Acknowledgments
We thank Dr. Gen Zhang from the Shenzhen GenProMetab Biotechnology Company Limited for assistants in qPCR experiments and comments on writing.
Data Availability
The sequencing data were deposited in NCBI with the reference number of PRJNA512222.
Funding Statement
This work was supported by the National Natural Science Foundation of China (Su-Ming Guo, No. 51408315), the General Program of National Natural Science Foundation of China (Ying Tan, No. 51478105), a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions and Jiangsu Modern Agricultural Technology System (Jin-Cheng Xing, JATS[2018]139). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Mori S, Kobayashi N, Arao T, Higuchi K, Maeda Y, Yoshiba M, et al. Enhancement of nitrate reduction by chlorine application in Suaeda salsa (L.) Pall. Soil Sci Plant Nutr. 2008;54(6):903–909. [Google Scholar]
- 2.Wang B, Lüttge U, Ratajczak R. Effects of salt treatment and osmotic stress on V‐ATPase and V‐PPase in leaves of the halophyte Suaeda salsa. J Exp Bot. 2001;52(365):2355–2365. 10.1093/jexbot/52.365.2355 [DOI] [PubMed] [Google Scholar]
- 3.Lu C, Qiu N, Lu Q, Wang B, Kuang T. Does salt stress lead to increased susceptibility of photosystem II to photoinhibition and changes in photosynthetic pigment composition in halophyte Suaeda salsa grown outdoors? Plant Sci. 2002;163(5):1063–1068. [Google Scholar]
- 4.Lu C, Qiu N, Wang B, Zhang J. Salinity treatment shows no effects on photosystem II photochemistry, but increases the resistance of photosystem II to heat stress in halophyte Suaeda salsa. J Exp Bot. 2003;54(383):851–860. 10.1093/jxb/erg080 [DOI] [PubMed] [Google Scholar]
- 5.Mori S, Yoshiba M, Tadano T. Growth response of Suaeda salsa (L.) Pall to graded NaCl concentrations and the role of chlorine in growth stimulation. Soil Sci Plant Nutr. 2006;52(5):610–617. [Google Scholar]
- 6.Duan DY, Li WQ, Liu XJ, Ouyang H, An P, Duan DY, et al. Seed germination and seedling growth of Suaeda salsa under salt stress. Ann Bot Fennici. 2007;44(3):161–169. [Google Scholar]
- 7.Li W, Yamaguchi S, Khan MA, An P, Liu X, Tran LSP. Roles of gibberellins and abscisic acid in regulating germination of Suaeda salsa dimorphic seeds under salt stress. Front Plant Sci. 2015;6(1):1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sui N, Tian S, Wang W, Wang M, Fan H. Overexpression of glycerol-3-phosphate acyltransferase from Suaeda salsa improves salt tolerance in Arabidopsis. Front Plant Sci. 2017;8:1337 10.3389/fpls.2017.01337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han N, Lan W, He X, Shao Q, Wang B, Zhao X. Expression of a Suaeda salsa vacuolar H+/Ca2+ transporter gene in Arabidopsis contributes to physiological changes in salinity. Plant Mol Biol Rep. 2011;30(2):470–477. [Google Scholar]
- 10.Qi CH, Chen M, Song J, Wang BS. Increase in aquaporin activity is involved in leaf succulence of the euhalophyte Suaeda salsa, under salinity. Plant Sci. 2009;176(2):200–205. [Google Scholar]
- 11.Wu H, You L, Zhang L, Zhou D, Feng J, Zhao J, et al. Effects of salinity on metabolic profiles, gene expressions, and antioxidant enzymes in halophyte Suaeda salsa. J Plant Growth Regul. 2012;31(3):332–341. [Google Scholar]
- 12.Jin H, Dong D, Yang Q, Zhu D. Salt-Responsive Transcriptome Profiling of Suaeda glauca via RNA Sequencing. Plos One. 2016;11(3): e0150504 10.1371/journal.pone.0150504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gharat SA, Parmar S, Tambat S, Vasudevan M, Shaw BP. Transcriptome analysis of the response to NaCl in Suaeda maritima provides an insight into salt tolerance mechanisms in halophytes. Plos One. 11(9): e0163485 10.1371/journal.pone.0163485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan H, Jia H, Chen X, Hao L, An H, Guo X. The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol. 2014;55(12):2060–2076. 10.1093/pcp/pcu133 [DOI] [PubMed] [Google Scholar]
- 15.Ye S, Jiang Y, Duan Y, Karim A, Fan D, Yang L, et al. Constitutive expression of the poplar WRKY transcription factor PtoWRKY60 enhances resistance to Dothiorella gregaria Sacc. in transgenic plants. Tree Physiol. 2014;34(10):1118–1129. 10.1093/treephys/tpu079 [DOI] [PubMed] [Google Scholar]
- 16.Agarwal P, Dabi M, Agarwal PK. Molecular cloning and characterization of a group II WRKY transcription factor from Jatropha curcas, an important biofuel crop. DNA Cell Biol. 2014;33(8):503–513. 10.1089/dna.2014.2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scarpeci TE, Zanor MI, Mueller-Roeber B, Valle EM. Overexpression of AtWRKY30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana. Plant Mol Biol. 2013;83(3):265–277. 10.1007/s11103-013-0090-8 [DOI] [PubMed] [Google Scholar]
- 18.Sahu BB, Shaw BP. Isolation, identification and expression analysis of salt-induced genes in Suaeda maritima, a natural halophyte, using PCR-based suppression subtractive hybridization. BMC Plant Biol 2009;9:69 10.1186/1471-2229-9-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gharat SA, Shaw BP. NaCl induced changes in the ionic and osmotic components in rice cultivars vis-a-vis that in a natural halophyte. Oryza. 2015;52:46–53. [Google Scholar]
- 20.Zhang JT, Zhu JQ, Zhu Q, Liu H, Gao XS, Zhang HX. Fatty acid desaturase-6 (FAD6) is required for salt tolerance in Arabidopsis thaliana. Biochem Biophys Res Commun. 2009;390(3):469–474. 10.1016/j.bbrc.2009.09.095 [DOI] [PubMed] [Google Scholar]
- 21.Wang HS, Yu C, Tang XF, Zhu ZJ, Ma NN, Meng QW. A tomato endoplasmic reticulum (ER)-type omega-3 fatty acid desaturase (LeFAD3) functions in early seedling tolerance to salinity stress. Plant Cell Rep. 2014;33(1):131–142. 10.1007/s00299-013-1517-z [DOI] [PubMed] [Google Scholar]
- 22.Brankova L., Ivanov S, Alexieva V. The induction of microsomal NADPH:cytochrome p450 and NADH:cytochrome b(5) reductases by long-term salt treatment of cotton (Gossypium hirsutum L.) and bean (Phaseolus vulgaris L.) plants. Plant Physiol Biochem. 2007;45(9):691–695. 10.1016/j.plaphy.2007.07.005 [DOI] [PubMed] [Google Scholar]
- 23.Shen X, Guo X, Guo X, Zhao D, Zhao W, Chen J, et al. PacMYBA, a sweet cherry R2R3-MYB transcription factor, is a positive regulator of salt stress tolerance and pathogen resistance. Plant Physiol Biochem. 2017;112:302–311. 10.1016/j.plaphy.2017.01.015 [DOI] [PubMed] [Google Scholar]
- 24.Shukla PS, Agarwal P, Gupta K, Agarwal PK. Molecular characterization of an MYB transcription factor from a succulent halophyte involved in stress tolerance. AoB Plants. 2015;7:plv054 10.1093/aobpla/plv054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol. 2011;29(7):644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32(Database issue):D277–D280. 10.1093/nar/gkh063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.KJ L, TD S. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 29.Fernandez G, Johnston M, Olivares P. Rol del pericarpio de Atriplex repanda en la germinacion. III. Estudio histological y quimico del pericarpio. Phyton. 1985;45:165–169. [Google Scholar]
- 30.Joshi A, Mali B, Hingalajia H. Halophytes–A good source of forage production under salt stress conditions l. Salvadora persica In: Khan M, Unger I, editors. Biology of Salt Tolerant Plants: Department of Botany, University of Karachi; 1995. pp. 353–360. [Google Scholar]
- 31.Liu X, Qiao H, Li W, Tadano T, Khan MA. Comparative effect of NaCl and seawater on seed germination of Suaeda salsa and Atriplex centralasiatica In: Öztürk M, Waisel Y, Khan MA, Görk G, editors. Biosaline Agriculture and Salinity Tolerance in Plants. Switzerland: Birkhäuser Basel; 2006. p. 45–53. [Google Scholar]
- 32.Khan M, Ungar I. The effect of salinity and temperature on the germination of polymorphic seeds and growth of Atriplex triangularis willd. Am J Bot. 1984;71(4):481–489. [Google Scholar]
- 33.Xu Y, Zhao Y, Duan H, Sui N, Yuan F, Song J. Transcriptomic profiling of genes in matured dimorphic seeds of euhalophyte Suaeda salsa. BMC Genomics. 2017;18(1):727 10.1186/s12864-017-4104-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kan X, Ren J, Chen T, Cui M, Li C, Zhou R, et al. Effects of salinity on photosynthesis in maize probed by prompt fluorescence, delayed fluorescence and P700 signals. Environ Exp Bot. 2017;140:56–64. [Google Scholar]
- 35.Yu C, Huang S, Hu X, Deng W, Xiong C, Ye C, et al. Changes in photosynthesis, chlorophyll fluorescence, and antioxidant enzymes of mulberry (morusssp.) in response to salinity and high-temperature stress. Biologia. 2013;68(3):404–413. [Google Scholar]
- 36.Song J, Fan H, Zhao Y, Jia Y, Du X, Wang B. Effect of salinity on germination, seedling emergence, seedling growth and ion accumulation of a euhalophyte Suaeda salsa in an intertidal zone and on saline inland. Aquat Bot. 2008;88(4):331–337. [Google Scholar]
- 37.Vogt T. Phenylpropanoid biosynthesis, the phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Mol Biol. 2010;27:327–338. [DOI] [PubMed] [Google Scholar]
- 38.Diray-Arce J, Clement M, Gul B, Khan MA, Nielsen BL. Transcriptome assembly, profiling and differential gene expression analysis of the halophyte Suaeda fruticosa provides insights into salt tolerance. BMC genomics. 2015;16:353 10.1186/s12864-015-1553-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaxiola RA, Rao R, Sherman A, Grisafi P, Alper SL, et al. The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc Natl Acad Sci USA. 1999;96(4):1480–1485. 10.1073/pnas.96.4.1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muramatsu Y, Harada A, Ohwaki Y, Kasahara Y, Takagi S, et al. Salt-tolerant ATPase activity in the plasma membrane of the marine angiosperm Zostera marina L. Plant Cell Physiol. 2002;43:1137–1145. 10.1093/pcp/pcf139 [DOI] [PubMed] [Google Scholar]
- 41.Kong F, Li H, Sun P, Zhou Y, Mao Y. De Novo Assembly and Characterization of the Transcriptome of Seagrass Zostera marina Using Illumina Paired-End Sequencing. Plos One. 2014; 9(11):e112245 10.1371/journal.pone.0112245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. 10.1146/annurev.arplant.59.032607.092911 [DOI] [PubMed] [Google Scholar]
- 43.Lurin C, Güclü J, Cheniclet C, Carde JP, Barbier-Brygoo H, Maurel C. CLC-Nt1, a putative chloride channel protein of tobacco, co-localizes with mitochondrial membrane markers. Biochem J. 2000;348:291–295. [PMC free article] [PubMed] [Google Scholar]
- 44.Wei Q, Liu Y, Zhou G, Li Q, Yang C, Peng S. Overexpression of CsCLCc, a chloride channel gene from Poncirus trifoliata, enhances salt tolerance in Arabidopsis. Plant Mol Biol Rep. 2013;31(6):1548–1557. [Google Scholar]
- 45.Sairam RK, Rao KV, Srivastava GC. Differential response of wheat genotypes to long term salinity stress in relation to oxidative stress, antioxidant activity and osmolyte concentration. Plant Sci. 2002;163(5):1037–1046. [Google Scholar]
- 46.Burdon RH, O'Kane D, Fadzillah N, Gill V, Boyd PA, Finch RR. Oxidative stress and responses in Arabidopsis thaliana and Oryza sativa subjected to chilling and salinity stress. Biochem Soc Trans. 1996;24(2):469–472. 10.1042/bst0240469 [DOI] [PubMed] [Google Scholar]
- 47.Singh MP, Singh DK, Rai M. Assessment of growth, physiological and biochemical parameters and activities of antioxidative enzymes in salinity tolerant and sensitive basmati rice varieties. J Agron Crop Sci. 2010;193(6):398–412. [Google Scholar]
- 48.Niketić V, Stojanović S, Nikolić A, Spasić M, Michelson AM. Exposure of Mn and FeSODS, but not Cu/ZnSOD, to NO leads to nitrosonium and nitroxyl ions generation which cause enzyme modification and inactivation: an in vitro study. Free Radic Biol Med. 1999;27(9–10):992–996. [DOI] [PubMed] [Google Scholar]
- 49.Wu G, Fang YZ, Yang S, Lupton JR, Turner ND. Glutathione metabolism and its implications for health. J Nutr. 2004; 134(3):489–492. 10.1093/jn/134.3.489 [DOI] [PubMed] [Google Scholar]
- 50.Chen KM, Gong HJ, Chen GC, Wang SM, Zhang CL. Up-regulation of glutathione metabolism and changes in redox status involved in adaptation of reed (Phragmites communis) ecotypes to drought-prone and saline habitats. J Plant Physiol. 2003;160(3):293–301. 10.1078/0176-1617-00927 [DOI] [PubMed] [Google Scholar]
- 51.Wang H, Yang Y, Chen W, Ding L, Li P, Zhao X, et al. Identification of differentially expressed proteins of Arthrospira (Spirulina) platensis-YZ under salt-stress conditions by proteomics and qRT-PCR analysis. Proteome Sci. 2013;11(1):6 10.1186/1477-5956-11-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Foyer CH, Noctor G. Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell. 2005;17:1866–1875 10.1105/tpc.105.033589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Noctor G, Foyer CH. Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol. 1998;49:249–279. [DOI] [PubMed] [Google Scholar]
- 54.Imai T, Ban Y, Terakami S, Yamamoto T, Moriguchi T. L-Ascorbate biosynthesis in peach: cloning of six L-galactose pathway-related genes and their expression during peach fruit development. Physiol Plant. 2009;136(2):139–149. 10.1111/j.1399-3054.2009.01213.x [DOI] [PubMed] [Google Scholar]
- 55.Izbiańska K, Arasimowicz-Jelonek M, Deckert J. Phenylpropanoid pathway metabolites promote tolerance response of lupine roots to lead stress. Ecotoxicol Environ Saf. 2014;110:61–67. 10.1016/j.ecoenv.2014.08.014 [DOI] [PubMed] [Google Scholar]
- 56.Tattini M, Galardi CP, Massai R, Remorini D, Agati G. Differential accumulation of flavonoids and hydroxycinnamates in leaves of Ligustrum vulgare under excess light and drought stress. New Phytol. 2004;163(3):547–561. [DOI] [PubMed] [Google Scholar]
- 57.Justin MW, Paul JH, Gloria KM. Ethylene-induced flavonol accumulation in guard cells suppresses reactive oxygen species and moderates stomatal aperture. Plant Physiol. 2014;164(4):1707–1717. 10.1104/pp.113.233528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caturla N, Vera-Samper E, Villalain J, Mateo CR, Micol V. The relationship between the antioxidant and antibacterial properties of galloylated catechins and the structure of phospholipid model membranes. Free Rad Biol Med. 2003;34:648–662. [DOI] [PubMed] [Google Scholar]
- 59.Potapovich AI, Kostyuk VA. Comparative study of antioxidant properties and cytoprotective activity of flavonoids. Biochemistry. 2003;68:514–519. [DOI] [PubMed] [Google Scholar]
- 60.Springob K, Nakajima J, Yamazaki M, Saito K. Recent advances in the biosynthesis and accumulation of anthocyanins. Nat Prod Rep. 2003;20(3):288–303. [DOI] [PubMed] [Google Scholar]
- 61.Kim DS, Hwang BK. An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defence response to microbial pathogens. J Exp Bot. 2014;65(9):2295–2306. 10.1093/jxb/eru109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chutipaijit S, Cha-Um S, Sompornpailin K. Differential accumulations of proline and flavonoids in indica rice varieties against salinity. Pak J Bot. 2008;41(5):2497–2506. [Google Scholar]
- 63.Liu C, Li S, Wang M, Xia G. A transcriptomic analysis reveals the nature of salinity tolerance of a wheat introgression line. Plant Mol Biol. 2012;78(1–2):159–169. 10.1007/s11103-011-9854-1 [DOI] [PubMed] [Google Scholar]
- 64.Zhu W, Yang B, Komatsu S, Lu X, Li X, Tian J. Binary stress induces an increase in indole alkaloid biosynthesis in Catharanthus roseus. Front Plant Sci. 2015;6:582 10.3389/fpls.2015.00582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Y, Meng Q, Duan X, Zhang Z, Li D. Effects of PEG-induced drought stress on regulation of indole alkaloid biosynthesis in Catharanthus roseus. J Plant Interact. 2017;12(1):87–91. [Google Scholar]
- 66.Misra N, Gupta AK. Effect of salinity and different nitrogen sources on the activity of antioxidant enzymes and indole alkaloid content in Catharanthus roseus seedlings. J Plant Physiol. 2006;163(1):11–8. 10.1016/j.jplph.2005.02.011 [DOI] [PubMed] [Google Scholar]
- 67.Mizrahi Y, Blumenfeld A, Richmond AE. The role of abscisic acid and salination in the adaptive response of plants to reduced root aeration. Plant Cell Physiol. 1972;13(1):15–21. [Google Scholar]
- 68.Walker MA, Dumbroff EB. Effects of salt stress on abscisic acid and cytokinin levels in tomato. Zeitschrift Für Pflanzenphysiologie. 1981;101(5):461–470. [Google Scholar]
- 69.Downton WJS, Loveys BR. Abscisic acid content and osmotic relations of salt-stressed grapevine leaves. Aust J Plant Physiol 1981;8(5):443–452. [Google Scholar]
- 70.El-Enany AE. Abscisic acid-responsive proteins induce salinity tolerance in wheat seedlings. Acta Physiol Plant. 2000;22(1):53–59. [Google Scholar]
- 71.Cutler S. Rodriguez PL, Finkelstein RR, Abrams SR.Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. 10.1146/annurev-arplant-042809-112122 [DOI] [PubMed] [Google Scholar]
- 72.Clipson NJW, Lachno DR, Flowers TJ. Salt tolerance in the halophyte, Suaeda maritima L. Dum.: abscisic acid concentrations in response to constant and altered salinity. J Exp Bot. 1988;39(10):1381–1388. [Google Scholar]
- 73.Llanes A, Masciarelli O, Ordóñez R, Isla MI, Luna V. Differential growth responses to sodium salts involve different abscisic acid metabolism and transport in Prosopis strombulifera. Biol Plant. 2014;58(1):80–88. [Google Scholar]
- 74.Tao JJ, Chen HW, Ma B, Zhang WK, Chen SY, Zhang JS.) The role of ethylene in plants under salinity stress. Front Plant Sci. 2015;6:1059 10.3389/fpls.2015.01059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yanagawa Y, Komatsu S.Ubiquitin/proteasome-mediated proteolysis is involved in the response to flooding stress in soybean roots, independent of oxygen limitation. Plant Sci. 2012;185-186(4):250–258. [DOI] [PubMed] [Google Scholar]
- 76.Wang W, Teng F, Lin Y, Ji D, Xu Y, Chen C, et al. Transcriptomic study to understand thermal adaptation in a high temperature-tolerant strain of Pyropia haitanensis. Plos One. 2018;13(4):e0195842 10.1371/journal.pone.0195842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Frugis G, Chua NH. Ubiquitin-mediated proteolysis in plant hormone signal transduction. Trends Cell Biol. 2002;12(7):308–311. [DOI] [PubMed] [Google Scholar]
- 78.Kumari GJ, Reddy AM, Naik ST, Kumar SG, Prasanthi J, Sriranganayakulu G, et al. Jasmonic acid induced changes in protein pattern, antioxidative enzyme activities and peroxidase isozymes in peanut seedlings. Biol Plant. 2006;50(2):219–226. [Google Scholar]
- 79.Pedranzani H, Racagni G, Alemano S, Miersch O, Ramírez I, Peña-Cortés H, et al. Salt tolerant tomato plants show increased levels of jasmonic acid. Plant Growth Regul. 2003;41(2):149–158. [Google Scholar]
- 80.Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Close TJ. Large-scale expression profiling and physiological characterization of jasmonic acid-mediated adaptation of barley to salinity stress. Plant Cell Environ. 2007;30(4):410–421. 10.1111/j.1365-3040.2006.01628.x [DOI] [PubMed] [Google Scholar]
- 81.Li S, Bashline L, Zheng Y, Xin X, Huang S, Kong Z, et al. Cellulose synthase complexes act in a concerted fashion to synthesize highly aggregated cellulose in secondary cell walls of plants. Proc Natl Acad Sci. 2016;113(40):11348–11353. 10.1073/pnas.1613273113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(DOCX)
(DOCX)
Data represent mean ± standard deviation (n = 3). * significantly different from the control (P < 0.05).
(DOCX)
Data represent mean ± standard deviation (n = 3). * significantly different from the control (P < 0.05).
(DOCX)
Data represent mean ± standard deviation (n = 3). * significantly different from the control (P < 0.05).
(DOCX)
Data Availability Statement
The sequencing data were deposited in NCBI with the reference number of PRJNA512222.