Abstract
The production and productivity of rice (Oryza sativa L.) are primarily influenced by the application of the critical nutrients nitrogen (N), phosphorus (P), and potassium (K). However, excessive application of these fertilizers is detrimental to the environment and increases the cost of production. Hence, there is a need to develop varieties that simultaneously increase yields under both optimal and suboptimal rates of fertilizer application by maximizing nutrient use efficiency (NuUE). To unravel the hidden genetic variation and understand the molecular and physiological mechanisms of NuUE, three different mapping populations (MPs; BC1F5) derived from three donors (Haoannong, Cheng-Hui 448, and Zhong 413) and recipient Weed Tolerant Rice 1 were developed. A total of three favorable agronomic traits (FATs) were considered as the measure of NuUE. Analysis of variance and descriptive statistics indicated the existence of genetic variation for NuUE and quantitative inheritance of FATs. The genotypic data from single-nucleotide polymorphism (SNP) markers from Tunable Genotyping-By-Sequencing (tGBS) and phenotypic values were used for locating the genomic regions conferring NuUE. A total of 19 quantitative trait loci (QTLs) were detected, out of which 11 QTLs were putative on eight chromosomes, which individually explained 17.02% to 34.85% of the phenotypic variation. Notably, qLC-II_1 and qLC-II_11 detected at zero fertilizer application showed higher performance for LC under zero percentage of NPK fertilizer. The remarkable findings of the present study are that the detected QTLs were associated in building tolerance to low/no nutrient application and six candidate genes on chromosomes 2 and 5 within these putative QTLs were found associated with low nutrient tolerance and related to several physiological and metabolic pathways involved in abiotic stress tolerance. The identified superior introgressed lines (ILs) and trait-associated genetic regions can be effectively used in marker-assisted selection (MAS) for NuUE breeding programs.
Introduction
Ensuring global food security is a significant concern for the rapidly growing world population that is expected to reach 9.8 billion by 2050 [1]. This situation concerning food security is even gloomier in developing countries. To meet the global food demand mainly in Asia, there is a tremendous need to increase rice production by 60–70% by 2050 [2]. At present, the core point of attention for scientists and policymakers is to develop different strategies for improving average yield productivity for sustainability. Plant breeders and biotechnologists follow various strategies to develop varieties with biotic stress resistance, abiotic stress tolerance, and multi-nutrients against malnutrition and cultivars with higher nutrient use efficiency, with the ultimate goal of developing climate-smart rice varieties with higher grain production [3–6]. Several biotic and abiotic stresses, decreasing arable land, labor unavailability, high cost of input fertilizers, water scarcity, and deficiency of nutrient elements in the soil are the major limiting factors for the development of multiple-stress-tolerant, input-use-efficient, and climate-smart rice cultivars [7–11]. Since the beginning of the green revolution, the nutrient elements nitrogen (N), phosphorus (P), and potassium (K) are the key components that maintain nutritional status through an oxidation process and they are significantly associated with increasing global crop productivity [12,13]. These major nutrients play a vital role in cellular mechanisms, enzyme synthesis, and osmotic regulation, and represent several structural components for numerous metabolic pathways to maintain proper plant growth and development [14,15].
Judicious application of fertilizers is the key for sustainable crop production. The improper and unscientific application of fertilizers containing N, P, and K is not only a burden as an extra cost for farmers but is also associated with environmental and human hazards [9]. To date, extensive research in this area has confirmed that the application of higher input fertilizers can cause imbalances in nutrient status in the soil and hence damage soil fertility in a longer period [16,17]. Chinese farmers are applying a higher dose of N fertilizer, particularly in Jiangsu Province. A higher dose of fertilizer of about 305 kg ha−1 is being applied to obtain higher yield vis-à-vis the world’s average dose of N fertilizer at 180 kg ha−1 [18–20]. Recently, Feng et al. [21] mentioned that released rice varieties in China had a greater yield potential, up to 12 t ha-1 or even higher, under high input doses of fertilizer and water. However, a higher dose of fertilizer applications leads to the risk of increased pests and diseases, consequently increasing the cost of pesticide application [22]. To reduce the cost of production and address the environmental safety issues associated with excessive fertilizer application, an appropriate dosage is required to ensure higher yield. There are mainly two different management systems, agricultural and integrated nutrient management systems that determine nutrient uptake, and influences increase in the grain yield and minimize the environmental hazards [23–26]. Green Super Rice (GSR) at the International Rice Research Institute (IRRI) has greatly contributed to minimizing the excessive application of chemical fertilizers by developing rice varieties with high NuUE and that perform better with integrated nutrient management. To date, 42 GSR varieties have been released worldwide and are available for 11 countries in South Asia, Southeast Asia, and Southern and Eastern Africa. Furthermore, these GSR varieties not only showed stable higher yield under low input environments but also showed tolerance of multiple biotic and abiotic stresses [27–31]. More than 130 GSR breeding cultivars are currently undergoing national varietal testing (www.isaaa.org), out of which 60 are being tested in different All India Coordinated Rice Improvement Project (AICRIP) trials in India alone.
Systematic nutrient management studies, site-specific nutrient management, farmers’ fertilizer practices, real-time nitrogen management, fixed-time adjustable-dose nitrogen management, integrated soil-crop system management, and optimal nitrogen management have been tested to increase rice yield under low input conditions [19,20,32–35]. Importantly, a significant yield advantage of 1.51 t ha-1 was achieved by reducing the N fertilizer application by 22.41% [20]. Hence, an organized and convenient procedure is required to establish a stage-specific fertilizer dose application to improve the nutrient balance for increasing agronomic advantages with the least environmental hazards. Three key favorable agronomic traits (FATs), plant height (PH), tiller number (TN), and leaf chlorophyll content (LC), and other yield-attributing traits contribute significantly to better plant architecture [36–39]. These traits were highly influenced by the application of a combination of complete N, P, and K fertilizers. Interestingly, these traits are controlled by the combined effects of several genes (polygenes) with a large influence of environment. Hence, the dissection of these traits that follow non-Mendelian segregation had become difficult and complex, especially when applying traditional breeding tools and methodologies. Recent advances in molecular marker and genomics technology paved the way for dissecting the genetic basis of the complex inheritance of key agronomic traits. Genetic analysis of quantitative trait loci (QTLs) can estimate and identify the influences of different genes responsible for quantitative traits with high statistical power. Further, the identified genes conferring these quantitative traits can be introgressed through precise MAS either singly or in combination with other useful traits/genes through marker-assisted pyramiding approaches [21,40,41].
DNA-based molecular markers such as restriction fragment length polymorphism, amplified fragment length polymorphism, cleaved amplified polymorphic sequence, and simple sequence repeats were used to generate genotypic information to determine the genetic characteristics of the lines. However, the generation of information is time-consuming and labor-intensive [42–44] as these markers are slower in work to provide accurate genotypic information for predicting candidate gene loci due to the low resolution of QTL mapping, environment-dependent expression of QTLs, and gene epistasis [44,45]. Recently developed high-throughput sequencing technologies are robust, accurate, and more informative for generating millions of single nucleotide polymorphism (SNP) markers in a shorter period [43,46,47]. These SNPs are most promising to develop genetic linkage maps for QTL dissection of many traits with higher chromosomal coverage [48–51]. To date, several QTLs have been reported for N, P, and K deficiency tolerance traits by using different QTL mapping methods in different genetic backgrounds of MPs such as recombinant inbred lines (RILs), backcross inbred lines (BILs), introgression lines (ILs), doubled haploids (DHs), chromosome segment substitution lines (CSSLs), and BC2F3 families in rice [10,41,52]. According to a comprehensive literature survey and exploring information available in the Gramene database (http://archive.gramene.org), more than 150 QTLs for N, 130 QTLs for P, and 15 QTLs for K have been associated with deficiency tolerance traits in rice. These significant QTLs are located on eight chromosomes (1, 3, 4, 5 7, 8, 9, and 12) for N [21,53–59]. Similarly, P deficiency tolerance QTLs have been reported on five chromosomes (1, 2, 6, 11, and 12) [10,41,53,60–67]. K deficiency tolerance QTLs are located on three chromosomes (3, 5, and 8) [68,69]. Notably, rather than a deficiency of the individual nutrient, either N or P or K, the combination of the complete N, P, and K deficiency has a significant impact on agronomic traits such as tillering ability, PH, and LC, which are key contributors for determining total grain yield [70,71].
To the best of our knowledge, the identification of QTLs conferring individual N, P, and K deficiency tolerance has been less explored. Interestingly, there was no comprehensive information on QTL data regarding the effect of fertilizer application in different growth interval (DGI) stages and during complete deficiency of N, P, and K fertilizers in rice. Hence, understanding the genetic information of N, P, and K deficiency tolerance of FATs under DGI stages of fertilizer doses is crucial to finding suitable rice cultivars for N, P, and K use efficiency (NuUE). In the current study, we used a set of early backcross MPs derived from a cross of three donors, Haoannong (HAN), Cheng-Hui 448 (CH448), and Zhong 413 (Z413), with a recipient parent, Weed Tolerant Rice 1 (WTR-1), to study the genetic variation for deficiency tolerance for N, P, and K at different stages. In addition, we detected genomic regions conferring N, P, and K defficiency tolerance by using phenotypic evaluation under three different doses of N, P, and K fertilizer, 338.90 kg ha-1 (100%), 271.10 kg ha-1 (80%), and 0 kg ha-1 (0%), and genotyping by tGBS technology to detect the QTLs associated with the trait of low input tolerance. The major objectives of the study were to (i) identify the FAT responses under three different doses of fertilizer application, 100%, 80%, and 0% for N, P, and K conditions; (ii) understand the genetic basis of FAT responses at DGI stages under three different doses of fertilizer application through correlation and heritability estimation; (iii) identification of promising tolerant ILs; and (iv) detect the main-effect QTLs (M-QTLs) at DGI stages under different doses of fertilizer by also estimating the additive effects of M-QTLs (v) enumeration of the candidate genes within the major effect QTLs based on the in silico candidate genes analysis using online databases.
Materials and methods
Plant materials and developing mapping populations
The parental line WTR-1 developed by using a novel Green Super Rice breeding strategy, was used as the recurrent parent while HAN, CH448, and Z413 were used as donor parents [72–74] for the development of MPs for the identification of the genomic regions controlling N, P, and K use efficiency. A set of 120, 67, and 56 BC1F5 plants were generated from the HAN × WTR-1 (DP-1), CH448 × WTR-1 (DP-4), and Z413 × WTR-1 (DP-7) cross combinations to make a total of 243 BC1F5 plants. The parental lines and their MPs along with four checks (Rc 222, SL8, Rc 192, and PSB Rc 82) for each trait underwent phenotypic evaluation to assess the genetic variation and dissect the locus conferring the FATs at DGI stages of fertilizer application under three different N, P, and K fertilizer doses.
Experimental layout
The introgression lines and their parental lines were seeded on the experimental farm at IRRI, Los Baños, Philippines (14°11N, 121°15E), during the wet season of 2017. A total of six individual plots separated by ridges were modified in a grid to maintain two replications. An alpha lattice experimental design was followed to analyze the experimental significance of the laid-out experiment. Each plot size was 245.76 m2 and each entry was planted in 0.4-m rows, with 0.2-m distance between rows and space of 0.2 m between adjacent plants. The 400-μm-thick transparent polyethylene sheets were inserted into the soil to cover the individual plots. At 21 days after seeding, seedlings were transplanted in an area of 491.52 m2 in two replications. The whole experiment was laid out with three different rates of N, P, and K fertilizer. The first rate of fertilizer consisted of complete N, P, and K that is also a recommended dose of fertilizer designated as 100% (N-P2O5-K2O = 100:30:30 kg ha-1); the second rate is a suboptimal dose, designated as 80% (N-P2O5-K2O = 80:30:30 kg ha-1); and the third rate is zero fertilizer dose, designated as 0% (N-P2O5-K2O = 0:0:0 kg ha-1). Furthermore, during crop growth, the fertilizers were applied in four DGI at 5, 20, 35, and 50 days after transplanting (DAT). Table 1 presents the detailed DGI at different fertilizer doses.
Table 1. Time and doses of fertilizer applications during the experiment.
| Time of fertilizer application | Type of fertilizer | Fertilizer | ||
|---|---|---|---|---|
| NH4NO3 | NaH2PO4.2H2O | K2SO4 | (kg ha‒1) | |
| Recommended dose (100%) | ||||
| 5 DATa (basal) | 30 | 30 | 30 | 183.34 |
| 20 DAT | 30 | - | - | 66.67 |
| 35 DAT | 30 | - | - | 66.67 |
| 50 DAT | 10 | - | - | 22.22 |
| Total | 100 | 30 | 30 | 338.90 |
| Suboptimal dose (80%) | ||||
| 5 DAT (basal) | 24 | 24 | 24 | 160.00 |
| 20 DAT | 23 | - | - | 44.44 |
| 35 DAT | 23 | - | - | 44.44 |
| 50 DAT | 10 | - | - | 22.22 |
| Total | 80 | 24 | 24 | 271.10 |
aDays after transplanting.
Phenotypic characterization of the MPs for FATs
The parental lines and their MPs along with checks were evaluated for the key traits PH, TN, and LC. The trait PH was measured from the soil surface to the longest tip of the leaf within a hill. TN was recorded as shoots with at least one noticeable leaf calculated as a tiller. The main stem tiller was counted as one tiller and included in the total tiller number, whereas LC was measured in the middle leaf portion by using the portable, non-destructive chlorophyll meter SPAD-502Plus (Konica Minolta, Japan). The mean LC value indicates the relative chlorophyll content. The phenotypic data were collected from three plants selected from the middle of each entry at four DGI stages every 15 days after fertilizer application. The first fertilizer application was done at 5 DAT and was designated as stage-I. Similarly, stage-II, stage-III, and stage-IV DGIs were designated for fertilizers applied at 21 DAT, 36 DAT, and 51 DAT. The phenotypic data were expressed as mean values for each of the observations recorded. The mean values were further used for the localization of genomic regions conferring these traits.
Genotyping
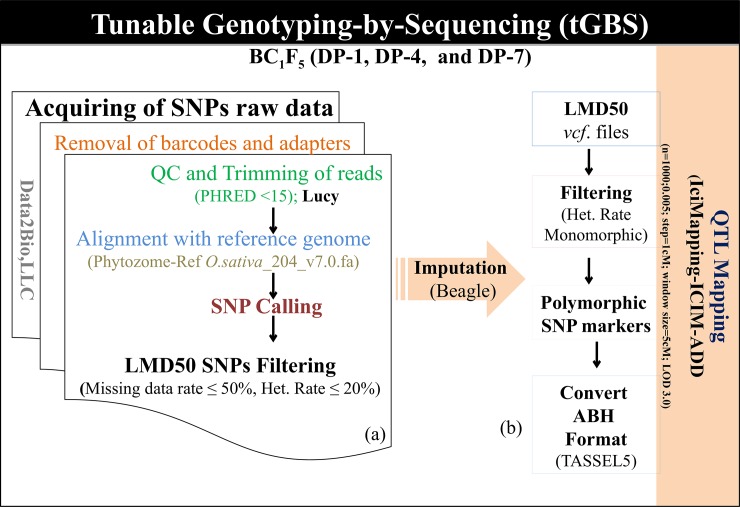
Genomic DNA was extracted from individual plants of the MPs along with the parental lines using the DNeasy Plant Mini Kit (Quiagen, USA). The quality and quantity of the isolated DNA were analyzed using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA) and sent to Data2Bio for tGBS analysis, in which all samples were sequenced through 10 Ion Proton runs. Further, SNP calling and filtration of the generated sequencing information were processed by adopting the methodology developed by Data2Bio, LLC (https://www.data2bio.com), as described by Ali et al. [73]. For the SNP filtration, the threshold of 50% missing rate (LMD50 = minimum calling rate) was set to filter out SNPs having more than 50% missing rate across the MPs. The missing SNPs that were showing ≤ threshold missing rate were imputed using Beagle software ver. 3.3.2 [75]. Additionally, another level of SNP filtering was performed to remove mono and hetero SNPs for the individuals of the MPs using TASSEL ver. 5 and finally exported as HapMap format. The obtained HapMap format SNP marker file was again processed in MS-Excel ver. 2010 to prepare an input file for the QTL analysis [76]. The complete work-flow of tunable GBS data analysis and its use for QTL mapping studies are represented in Fig 1.
Fig 1. A diagramatic representation of the work-flow of tGBS data analysis.
(a) Trimming of nucleotide raw sequence reads, SNP discovery, SNP calling, and removal of low-quality SNPs were performed by Ali et al. [73]. (b) The LMD50 SNP files (low missing SNP data rate of ≤50%) of three genetic backgrounds (DP-1, DP-4, and DP-7 MPs) were filtered SNPs and converted to ABH format for QTL mapping studies.
Construction of linkage map and QTL analysis
The inclusive composite interval mapping (ICIM-ADD) method is available in QTL IciMapping ver. 4.1 software [77] for scanning the FAT data at DGI stages from three cross combinations with different nutrient fertilizer doses (0%, 80%, and 100% N, P, and K fertilizer). Before construction of the linkage map, SNPs were analyzed for segregation distortion using the Chi-square test at a significance of P = 0.05. The SNPs that were showing significant segregation distortion and were co-located in the same physical position were filtered out using the redundant marker removal feature of the software. The processed high-quality SNP markers were used for constructing the genetic map to assign markers on each linkage group. Kosambi mapping function was used to compute the genetic distance using recombination fraction (cM) [78]. The precisely estimated phenotypic data of FATs at different DGIs for each nutrient dose (0%, 80%, and 100% NPK) for each cross combination and high-resolution SNP marker data were used for the QTL analysis. In order to identify the QTLs with high precision and to declare significant QTLs, a 1000 permutation test at the 0.01 level of significance was considered. The linkage map and QTL mapping were constructed for each MP derived from each cross combination (DP-1, DP-4, and DP-7) separately. The detected QTLs which showed >10.00% R2 value were considered as major QTLs.
Candidate gene analysis
To identify the candidate genes within the QTLs, the interval region of both flanking marker positions was considered. The physical position of the SNP flanking markers was used to determine the size of each QTL in kb. Publically accessible MSU Rice Genome Annotation Project (Osa1) release 7 (http://rice.plantbiology.msu.edu/) and the Rice Annotation Project (https://rapdb.dna.affrc.go.jp/) were explored to select the candidate genes residing in each detected QTL. Further, in silico gene expression analysis carried out using the genome wide expression-profiling database, RiceXPro (http://ricexpro.dna.affrc.go.jp). This is the repository of the microarray profiling data of Nipponbare cultivar isolated from different growth and developmental stages.
Statistical analysis
The phenotypic data of each trait at four DGI stages for the three fertilizer doses were analyzed using analysis of variance (ANOVA) for assessing the significance of the experiments at the level of significance P = 0.05. DMRT and Fisher’s t-test were used to ascertain the significant difference between the genotypes and to compare with the check values. The correlation between the traits at different fertilizer doses (0%, 80%, and 100% N, P, and K) was computed by Pearson’s correlation analysis and the correlation matrix was visualized using corrplot package in R (https://github.com/taiyun/corrplot) [79]. The phenotypic mean values of all the traits were prepared using MS-Excel ver. 2010 and the analysis was carried out using PBTools (Version 1.4, http://bbi.irri.org/products). Chi-square goodness of fit was used to analyze the segregation pattern of SNP markers before using these for the QTL analysis. The multiple regression model of maximum likelihood was employed for the composite interval mapping.
Results
The genetic variation of phenotypic traits among different MPs
The early backcross MPs derived from three different genetic backgrounds were evaluated for three FATs under 100% NPK (338.90 kg ha‒1), 80% NPK (271.90 kg ha‒1), and 0% NPK (0 kg ha‒1) treatments at four DGI stages (I, II, III, and IV). The segregation pattern of FATs was determined based on the skewness and kurtosis values. For the MP DP-1, normal distribution was observed for the trait PH at stage II under 100% fertilizer rate; at stages I, II, and III at 80% fertilizer rate; and at stages I and II at 0% fertilizer rate. For the MPs DP-4 and DP-7, most of the traits at different stages and different fertilizer rates showed normal distribution (Table 2 and S1 Fig). ANOVA showed significant variation existing in the three concentrations of N, P, and K fertilizers at specific growth stages for each trait. The F-test statistic values showed the existence of significant variation at the level of significance P = 0.05, indicating a large amount of genetic variation among the MPs (Table 3). Among the MPs, DP-7 exhibited the highest PH at two stages of PH-I and IV (40.51, 92.20 cm), TN at stage IV (43.22 cm), and for LC at two stages, LC-II and LC-IV (44.55 and 45.71 cm, respectively), compared with the other donor and recurrent parents under low input (0% NPK) conditions. The coefficient of variation (CV) revealed the extent of phenotypic variation existing among the different traits for all of the genetic backgrounds of ILs evaluated at DGI and different doses of fertilizer. The highest CVs were observed for TN-III (37.58%), TN-IV (21.82%), and TN-IV (22.90%) in DP-1, DP-4, and DP-7, respectively, under 100% NPK conditions. In contrast, in 80% NPK conditions, TN-IV (32.47% and 38.36%) in DP-1 and DP-4, respectively, and TN-I (22.60%) in DP-7 showed the highest CVs. Under 0% NPK conditions, the highest CV was observed for trait TN at stage III (39.17% and 40.94%) in MPs DP4 and DP7, respectively, followed by CV of 32.84% at stage II in MP DP-1. As expected, the lowest CV values were recorded in the second stage of LC in DP-1, DP-4, and DP-7 in all the different doses of fertilizer. The broad-sense heritability (H2b) was estimated for each trait under different doses of NPK fertilizers at DGI stages. Under 100%, H2b ranged from 15% to 51%. The highest H2b was noted in TN-IV (51%), followed by LC-II and PH-IV, being the same estimated value of 46%. Similarly, in 80% and 0% NPK conditions, H2b ranged from 10% to 65% and 6% to 65%, respectively.
Table 2. Descriptive statistics of three FATs at DGI stages in three different MPs.
| Trait and stage of application of fertilizer | FDa | HAN × WTR-1 | CH448 × WTR-1 | Z413 ×WTR-1 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean±SDb | Range | CVcac | SKd | KTe | Mean±SDb | Range | CVc | SKd | KTe | Mean±SDb | Range | CVc | SKd | KTe | ||
| PH-I (5 DAT) | 100% | 40.49±4.04 | 32.00–55.70 | 9.98 | 0.45 | 0.25 | 40.09±2.87 | 34.25–48.17 | 7.15 | 0.48 | -0.18 | 40.14±3.26 | 27.01–47.17 | 8.12 | -0.88 | 2.61 |
| PH-II (20 DAT) | 64.81±6.80 | 49.53–80.83 | 10.49 | 0.06 | -0.81 | 62.64±5.23 | 52.73–76.23 | 8.35 | 0.13 | -0.29 | 62.21±5.07 | 47.41–72.67 | 8.15 | -0.68 | 0.35 | |
| PH-III (35 DAT) | 83.05±7.66 | 60.67–105.00 | 9.22 | 0.14 | 0.09 | 78.09±7.39 | 54.83–97.93 | 9.46 | -0.07 | 0.21 | 77.92±6.49 | 62.63–93.47 | 8.32 | -0.39 | -0.18 | |
| PH-IV (50 DAT) | 93.89±9.49 | 67.33–122.33 | 10.11 | 0.41 | 0.56 | 86.54±8.16 | 68.67–107.01 | 9.43 | 0.25 | -0.21 | 87.12±6.12 | 76.50–106.25 | 7.02 | 0.63 | 0.56 | |
| TN-I (5 DAT) | 5.58±1.48 | 1.33–11.00 | 26.52 | 0.50 | 0.49 | 5.05±1.02 | 2.67–7.67 | 20.30 | 0.24 | -0.04 | 5.27±1.12 | 2.01–8.11 | 21.49 | 0.62 | 0.74 | |
| TN-II (20 DAT) | 11.46±2.12 | 6.33–17.67 | 18.50 | 0.32 | -0.02 | 15.61±26.38 | 7.45–287.83 | 9.01 | 6.82 | 81.02 | 12.53±1.96 | 7.50–18.01 | 15.64 | 0.44 | 0.01 | |
| TN-III (35 DAT) | 15.91±5.98 | 7.33–48.33 | 37.58 | 2.72 | 9.76 | 14.15±2.61 | 10.87–24.33 | 18.46 | 1.19 | 2.21 | 14.31±2.74 | 8.67–23.50 | 19.15 | 0.65 | 0.88 | |
| TN-IV (50 DAT) | 16.11±4.28 | 10.00–34.33 | 26.58 | 1.47 | 2.95 | 15.14±3.30 | 8.67–25.56 | 21.82 | 0.48 | 0.07 | 15.23±3.50 | 8.67–26.33 | 22.98 | 0.84 | 1.32 | |
| LC-I (5 DAT) | 41.16±2.80 | 33.03–48.00 | 6.80 | -0.45 | 0.06 | 40.68±2.95 | 31.03–47.53 | 7.26 | -0.36 | 0.48 | 43.27±2.75 | 34.70–48.43 | 6.36 | -0.69 | 0.48 | |
| LC-II (20 DAT) | 42.92±2.14 | 37.17–48.90 | 4.99 | -0.31 | -0.19 | 42.97±2.51 | 36.68–48.63 | 5.83 | -0.03 | -0.13 | 44.75±2.27 | 38.37–53.57 | 5.07 | 0.11 | 2.31 | |
| LC-III (35 DAT) | 43.33±2.82 | 33.67–51.33 | 6.51 | -0.35 | 0.29 | 39.76±3.60 | 30.57–49.27 | 9.05 | 0.10 | 0.01 | 41.12±3.22 | 32.37–48.27 | 7.83 | -0.22 | -0.26 | |
| LC-IV (50 DAT) | 44.21±3.40 | 30.17–54.80 | 7.69 | -0.55 | 1.81 | 40.78±4.00 | 26.58–54.97 | 9.81 | 0.01 | 1.72 | 42.81±3.63 | 33.15–50.47 | 8.48 | -0.44 | -0.38 | |
| PH-I (5 DAT) | 80% | 39.55±3.64 | 29.00–52.10 | 9.20 | 0.07 | 0.24 | 38.92±3.79 | 29.01–51.01 | 9.73 | 0.02 | 0.45 | 39.94±3.58 | 31.73–48.01 | 8.97 | -0.27 | -0.53 |
| PH-II (20 DAT) | 64.98±6.41 | 48.30–84.00 | 9.86 | -0.04 | 0.21 | 65.03±5.70 | 55.33–81.30 | 8.77 | 0.83 | 0.24 | 65.85±5.21 | 51.71–76.70 | 7.92 | -0.4 | 0.01 | |
| PH-III (35 DAT) | 76.59±7.93 | 57.00–105.00 | 10.35 | 0.10 | 0.30 | 75.95±7.80 | 60.24–97.78 | 10.27 | 0.34 | 0.12 | 77.60±6.86 | 62.71–92.70 | 8.84 | -0.15 | -0.56 | |
| PH-IV (50 DAT) | 87.81±9.30 | 65.70–120.70 | 10.59 | 0.39 | 0.61 | 85.61±8.08 | 67.23–105.49 | 9.44 | 0.11 | -0.32 | 87.48±7.85 | 57.72–105.01 | 8.98 | -0.58 | 1.53 | |
| TN-I (5 DAT) | 5.13±1.04 | 2.30–8.70 | 20.29 | 0.40 | 0.51 | 4.83±1.04 | 2.87–8.45 | 21.47 | 0.07 | 1.28 | 5.23±1.19 | 3.34–10.11 | 22.66 | 0.88 | 2.02 | |
| TN-II (20 DAT) | 11.54±2.21 | 6.30–19.70 | 19.18 | 0.27 | 0.23 | 11.07±2.19 | 6.70–17.00 | 19.83 | 0.15 | -0.48 | 12.56±2.55 | 7.00–18.30 | 21.26 | 0.43 | -0.01 | |
| TN-III (35 DAT) | 15.82±3.70 | 9.00–30.00 | 23.35 | 1.05 | 1.79 | 15.03±3.15 | 6.07–24.05 | 20.94 | 0.22 | 1.00 | 15.78±3.25 | 9.70–26.70 | 20.62 | 0.93 | 1.06 | |
| TN-IV (50 DAT) | 16.06±5.21 | 8.30–50.00 | 32.47 | 2.80 | 13.93 | 15.58±5.98 | 9.30–65.00 | 38.36 | 5.38 | 40.97 | 15.79±3.45 | 9.11–28.01 | 21.86 | 0.81 | 1.37 | |
| LC-I (5 DAT) | 41.35±3.53 | 27.60–53.10 | 8.53 | -0.68 | 1.74 | 40.60±3.27 | 31.50–47.40 | 8.06 | -0.49 | 0.17 | 42.68±3.77 | 30.5–49.30 | 8.84 | -1.09 | 1.68 | |
| LC-II (20 DAT) | 42.50±3.45 | 29.90–77.10 | 8.13 | 4.44 | 46.70 | 41.91±2.76 | 32.70–47.90 | 6.59 | -0.84 | 1.27 | 44.11±2.19 | 37.4–48.40 | 4.97 | -0.29 | -0.04 | |
| LC-III (35 DAT) | 40.77±3.33 | 21.50–48.30 | 8.17 | -1.20 | 5.33 | 39.54±4.02 | 29.30–49.90 | 10.16 | -0.24 | -0.19 | 41.64±3.63 | 30.3–54.10 | 8.71 | -0.1 | 1.11 | |
| LC-IV (50 DAT) | 41.76±3.80 | 28.70–61.90 | 9.09 | 0.30 | 3.50 | 40.92±3.44 | 33.10–48.60 | 8.41 | -0.18 | -0.75 | 42.87±2.99 | 33.6–47.70 | 6.97 | -0.77 | 0.41 | |
| PH-I (5 DAT) | 0% | 39.85±3.06 | 25.83–49.23 | 7.69 | 0.01 | 1.67 | 40.46±3.23 | 32.70–50.59 | 7.99 | 0.32 | 0.62 | 40.51±3.61 | 32.17–49.83 | 8.92 | -0.15 | -0.05 |
| PH-II (20 DAT) | 61.41±5.74 | 46.90–74.93 | 9.35 | 0.01 | -0.22 | 67.43±14.14 | 50.20–205.30 | 20.97 | 8.09 | 78.98 | 64.97±6.58 | 48.57–83.37 | 10.13 | -0.25 | 0.27 | |
| PH-III (35 DAT) | 77.06±7.32 | 51.83–95.60 | 9.50 | -0.25 | 0.08 | 82.28±7.63 | 62.33–100.67 | 9.27 | 0.12 | -0.03 | 81.66±7.63 | 61.17–98.67 | 9.34 | -0.14 | -0.28 | |
| PH-IV (50 DAT) | 86.78±8.10 | 56.12–112.00 | 9.33 | -0.14 | 0.79 | 92.09±8.70 | 67.33–117.33 | 9.45 | 0.21 | 0.32 | 92.20±8.17 | 77.67–117.67 | 8.86 | 0.74 | 0.73 | |
| TN-I (5 DAT) | 4.99±1.21 | 1.33–12.00 | 24.22 | 0.94 | 5.41 | 5.60±1.31 | 2.67–9.67 | 23.39 | 0.57 | 0.54 | 5.44±1.32 | 2.67–8.33 | 24.25 | 0.19 | -0.35 | |
| TN-II (20 DAT) | 12.61±4.14 | 6.50–65.00 | 32.84 | 9.62 | 120.16 | 11.52±2.16 | 6.67–17.33 | 18.73 | 0.11 | -0.09 | 11.06±2.04 | 7.11–16.33 | 18.43 | 0.25 | -0.18 | |
| TN-III (35 DAT) | 14.48±4.68 | 9.33–55.67 | 32.30 | 5.51 | 42.99 | 16.46±6.45 | 8.00–46.33 | 39.17 | 2.74 | 8.79 | 15.97±6.54 | 8.67–47.14 | 40.94 | 2.84 | 9.78 | |
| TN-IV (50 DAT) | 14.97±3.69 | 8.33–40.00 | 24.66 | 1.85 | 8.84 | 16.26±3.91 | 7.67–31.33 | 24.07 | 0.94 | 1.37 | 16.03±5.56 | 9.33–43.67 | 34.69 | 2.16 | 7.08 | |
| LC-I (5 DAT) | 41.28±2.40 | 34.13–46.50 | 5.80 | -0.21 | -0.14 | 40.99±2.96 | 30.07–47.97 | 7.22 | -0.58 | 1.42 | 43.22±2.95 | 35.86–49.13 | 6.83 | -0.42 | -0.24 | |
| LC-II (20 DAT) | 43.62±2.24 | 37.10–48.80 | 5.15 | -0.36 | 0.18 | 43.05±2.33 | 37.83–49.23 | 5.41 | 0.11 | -0.35 | 44.55±2.35 | 34.53–50.43 | 5.28 | -0.77 | 3.11 | |
| LC-III (35 DAT) | 40.11±3.83 | 28.93–48.10 | 9.55 | -0.46 | -0.06 | 42.59±3.19 | 33.40–48.80 | 7.52 | -0.51 | -0.04 | 44.44±2.74 | 37.93–50.87 | 6.17 | -0.28 | -0.39 | |
| LC-IV (50 DAT) | 41.92±3.23 | 34.20–50.05 | 7.70 | -0.19 | -0.17 | 43.58±3.36 | 33.93–54.50 | 7.71 | -0.02 | 0.41 | 45.71±3.75 | 37.37–59.60 | 8.21 | 1.11 | 2.85 | |
aFertilizer dose
bStandard deviation
cCoefficient of variation
dSkewness
eKurtosis
Table 3. Analysis of variance (ANOVA) and its components for all the FATs in DGI stages under different doses of fertilizer.
| Fertilizer dose | Components | PH-I | TN-I | LC-I | PH-II | TN-II | LC-II | PH-III | TN-III | LC-III | PH-IV | TN-IV | LC-IV |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 100% | DF | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 |
| SS | 4302.66 | 370.93 | 2537.87 | 10817.20 | 1060.06 | 1501.87 | 16348.90 | 7816.03 | 2099.75 | 23329.00 | 5355.86 | 2960.64 | |
| MS | 20.49 | 1.77 | 12.09 | 51.5 | 5.05 | 7.15 | 77.85 | 37.22 | 10.00 | 111.09 | 25.50 | 14.10 | |
| F value | 1.49 | 1.17 | 1.87 | 1.70 | 1.41 | 1.86 | 1.64 | 1.28 | 1.23 | 1.85 | 2.06 | 1.37 | |
| Pr (>F) | 0.00 | 0.13 | 0.00 | 0.00 | 0.01 | 0.00 | 0.00 | 0.04 | 0.07 | 0.00 | 0.00 | 0.01 | |
| H2b (%) | 0.33 | 0.15 | 0.46 | 0.41 | 0.29 | 0.46 | 0.39 | 0.23 | 0.20 | 0.46 | 0.51 | 0.27 | |
| 80% | DF | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 |
| SS | 3490.61 | 255.74 | 2924.77 | 10295.80 | 1218.81 | 2383.19 | 16584.60 | 3332.20 | 2807.13 | 21903.40 | 4973.05 | 3191.20 | |
| MS | 16.70 | 1.22 | 13.99 | 49.26 | 5.83 | 11.40 | 79.35 | 15.94 | 13.43 | 104.80 | 23.79 | 15.27 | |
| F value | 1.75 | 1.43 | 1.33 | 2.16 | 1.51 | 1.31 | 2.31 | 1.41 | 0.97 | 2.83 | 1.32 | 1.47 | |
| Pr (>F) | 0.00 | 0.01 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 | 0.01 | 0.60 | 0.00 | 0.02 | 0.00 | |
| H2b (%) | 0.43 | 0.30 | 0.26 | 0.54 | 0.34 | 0.24 | 0.57 | 0.31 | 0.10 | 0.65 | 0.24 | 0.33 | |
| 0% | DF | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 | 243.00 |
| SS | 2619.52 | 899.32 | 1922.82 | 7728.10 | 862.94 | 1656.49 | 16616.30 | 2971.73 | 3131.77 | 19960.60 | 3029.76 | 3168.06 | |
| MS | 12.53 | 4.30 | 9.20 | 36.93 | 4.13 | 7.93 | 79.50 | 14.22 | 14.98 | 95.51 | 14.50 | 15.16 | |
| F value | 2.13 | 1.07 | 1.61 | 2.05 | 1.27 | 1.91 | 2.37 | 1.13 | 1.21 | 2.89 | 1.95 | 1.37 | |
| Pr (>F) | 0.00 | 0.30 | 0.00 | 0.00 | 0.04 | 0.00 | 0.00 | 0.19 | 0.08 | 0.00 | 0.00 | 0.01 | |
| H2b (%) | 0.53 | 0.06 | 0.38 | 0.51 | 0.21 | 0.48 | 0.58 | 0.11 | 0.20 | 0.65 | 0.49 | 0.27 |
SV = Source of variation; DF = Degrees of freedom; SS = Sum of squares; MS = Mean sum of square; Pr = Probability value-; F value- Significance level; H2b (%) = Broad-sense heritability.
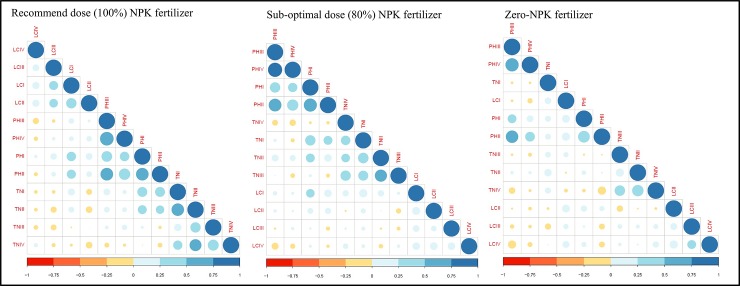
Pearson correlation matrix (PCM) analysis
The PCM was performed in DGI stages in three genetic backgrounds of MPs to assess the correlation among the FATs (PH, LC, and TN). Most of the FATs were found to be significantly positively correlated with each other. The highest significant positive correlation was observed between PH-III and PH-IV (r = 0.68) in 100% N, P, and K. Interestingly, in 80% and 0% NPK conditions, there was no significant positive correlation between the traits. The trait LC-II showed the highest negative correlation with TN-II (r = ‒0.15) and TN-IV (r = ‒0.15) in 100% NPK conditions, followed by LC-IV with PH-III (r = ‒0.17) at 80% NPK, and PH-II with TN-IV (r = ‒0.23) at 0% NPK, as presented in Fig 2.
Fig 2. Heat map showing the PCM of FATs measured in response to different doses of N, P, and K fertilizer at DGI stages.
PH = Plant height; TN = Tiller number; LC = Leaf chlorophyll content.
SNP discovery and construction of saturated linkage map
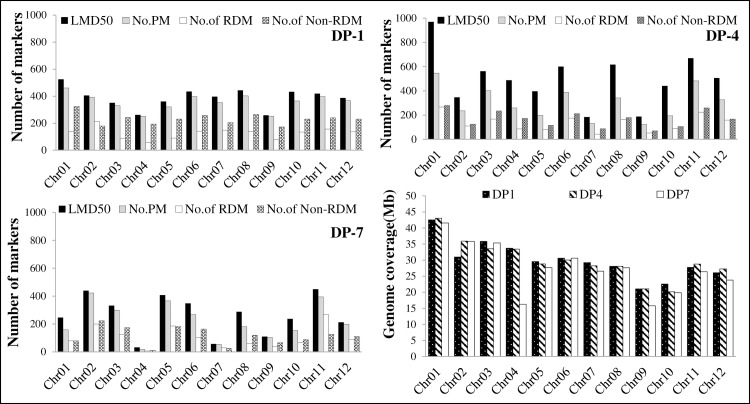
All the genotypes, including parental lines, underwent genotyping by using the advanced and cost-effective genotyping platform tGBS. It was used for the extraction of SNP markers for the localization of the genetic factors influencing FATs under different fertilizer doses. The genotyping platform tGBS yielded extensive sequence information from sequencing three MPs. A total of 10,432, 14,117, and 7,865 raw reads were obtained from the MPs as DP-1, DP-4, and DP-7, respectively (Table 4). A novel SNP calling and filtering method was followed to eliminate missing values (≥50 missing data, LMD 50) from the tunable GBS pipeline (Data2Bio, LLC) (https://www.data2bio.com), as reported in Ali et al. [73]. In the initial SNP filtering, as many as 4,666, 5,957, and 3,147 SNPs were isolated from MPs DP-1, DP-4, and DP-7, respectively, whereas, in the next level of filtering, 4,286 SNPs from DP-1, 3,622 SNPs from DP-4, and 2,610 SNPs from DP-7 were isolated (Table 4). Notably, out of these SNP markers, 35.10% in DP-1, 44.64% in DP-4, and 47.67% in DP-7 were redundant and excluded before further analysis. Finally, the SNPs were filtered out at the final level of filtering based on the segregation distortion regions in the genetic linkage map. A total of 57.80% of the SNPs demonstrated genetic distortion (P = 0.05) in the DP-1 population, followed by 44.63% in DP-4 and 38.72% in DP-7. The SNP markers obtained from all the filtration levels were used for linkage map construction and linkage analysis. In total, a 953.71 cM chromosomal spanning region was covered by 2,782 SNPs in DP-1, 1,398.42 cM by 2,005 SNPs in DP-4, and 834.11 cM by 1,361 SNPs in DP-7. In each population, an average of 230, 167, and 113 SNP markers were distributed across all chromosomes. The highest number of polymorphic markers was present on chromosome 1 (324 SNPs) while the lowest was on chromosome 9 (172) in DP-1, whereas, in DP-4 and DP-7, the highest number of markers was located on chromosome 1 (279) and chromosome 2 (223). However, chromosome 9 (70) and chromosome 4 (11) had the lowest number of polymorphic markers available in DP-4 and DP-7, respectively (Table 5 and Fig 3). The average distance of each SNP marker varied from 0.13 to 0.41 Mb, whereas the longest distance recorded was 1.47 Mb on chromosome 4 in the population DP-7.
Table 4. Processing SNP markers for QTL analysis.
| S. no. | SNP calling | MPs | ||
|---|---|---|---|---|
| HAN × WTR-1 | CH448 × WTR-1 | Z413 × WTR-1 | ||
| 1 | Raw SNPs | 10,432 | 14,117 | 7,865 |
| 2 | Quality base filtering at LMD50 | 4,666 | 5,957 | 3,147 |
| 3 | Removed heterozygous alleles in both parents | 345 | 483 | 454 |
| 4 | Removed mono-morphic alleles in both parents | 35 | 1,852 | 83 |
| 5 | SNP markers for QTL analysis | 4,286 | 3,622 | 2,610 |
| 6 | Exclusion of redundant markers | 1,504 | 1,617 | 1,249 |
| 7 | Number of markers used for linkage map construction | 2,782 | 2,005 | 1,361 |
| 8 | Removed markers with χ2 >6.0 of segregation distortion | 1,608 | 895 | 527 |
| 9 | Number of SNP markers used for ICIM-ADD | 1,174 | 1,110 | 834 |
Table 5. Distribution of SNP markers, coverage, and position of each chromosome.
| Chr | HAN × WTR-1 | CH448 × WTR-1 | Z413 × WTR-1 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| NSM | CL (Mb) |
ADSM (Mb) |
NSM | CL (Mb) |
ADSM (Mb) |
NSM | CL (Mb) |
ADSM (Mb) |
|
| 1 | 324 | 42.53 | 0.13 | 279 | 43.03 | 0.15 | 79 | 41.56 | 0.52 |
| 2 | 179 | 30.97 | 0.17 | 125 | 35.90 | 0.29 | 223 | 35.85 | 0.16 |
| 3 | 242 | 35.83 | 0.15 | 234 | 33.55 | 0.14 | 174 | 35.35 | 0.20 |
| 4 | 194 | 33.72 | 0.17 | 173 | 33.41 | 0.19 | 11 | 16.27 | 1.47 |
| 5 | 231 | 29.54 | 0.13 | 115 | 28.81 | 0.25 | 181 | 27.66 | 0.15 |
| 6 | 257 | 30.61 | 0.12 | 212 | 30.04 | 0.14 | 163 | 30.61 | 0.18 |
| 7 | 216 | 29.21 | 0.14 | 87 | 28.25 | 0.32 | 24 | 26.57 | 1.10 |
| 8 | 265 | 28.07 | 0.11 | 179 | 28.05 | 0.16 | 119 | 27.61 | 0.23 |
| 9 | 172 | 21.04 | 0.12 | 70 | 21.03 | 0.30 | 65 | 15.77 | 0.24 |
| 10 | 231 | 22.57 | 0.10 | 105 | 20.12 | 0.19 | 88 | 19.87 | 0.22 |
| 11 | 240 | 27.75 | 0.12 | 259 | 28.76 | 0.11 | 124 | 26.38 | 0.20 |
| 12 | 231 | 26.08 | 0.11 | 167 | 27.26 | 0.16 | 110 | 23.77 | 0.21 |
| Avg. | 230.83 | 29.82 | 0.13 | 167.08 | 29.85 | 0.20 | 113.41 | 27.27 | 0.41 |
| Total | 2782.00 | 357.92 | 1.57 | 2005.00 | 388.06 | 2.41 | 1361.00 | 327.27 | 4.94 |
Chr = Chromosome; NSM = Number of SNP markers; CL = Chromosome length; ADSM = Average distance of SNP markers
Fig 3. Chromosome-wise distribution of SNP markers in three populations (DP-1, DP-4, and DP-7) extracted from novel tGBS genotyping platform.
Molecular mapping of genomic regions conferring FATs
In order to locate the QTLs influencing the traits PH, TN, and LC at DGI stages under different fertilizer doses, QTL analysis was performed using the ICIM-ADD methodology. A total of 19 main-effect QTLs (M-QTLs) were identified using the composite interval mapping model of QTL IciMapping under LOD threshold of 3.00. The significant QTLs were identified on chromosomes 1, 9, 10, and 12 in 100% NPK conditions; on chromosomes 1, 2, 3, 5, and 12 in 80% NPK conditions; and on chromosomes 2, 3, 4, 5, 6, 8, and 11 in 0% NPK conditions. The detected M-QTLs explained phenotypic variation ranges from 1.89% to 34.85% with LOD score ranges from 3.02 to 28.70. The number of QTLs per trait ranged from one to four, and the highest numbers of QTLs (four QTLs) were associated with LC-II in populations DP-1, DP-4, and DP-7 (S1 Table). A total of four QTLs were discovered for LC-II, TN-I and II, and PH-III in 100% NPK; six QTLs for PH-I, III, and IV, TN-III, and LC-II in 80% NPK; and nine QTLs for PH-I, LC-I and II, and TN-II, III, and IV in 0% of NPK conditions at DGI stages. The highest number of QTLs (nine QTLs) was detected in 0% of NPK conditions, followed by 80% NPK (six QTLs) and 100% NPK (four QTLs). The majority of the negative additive effect of the QTLs was contributed by the donor parental allele source. Among the total QTLs, 57.89% (11 QTLs) were contributed by donor alleles from HAN, CH448, and Z143, whereas 42.10% came from recurrent parent WTR-1. Two M-QTLs (qLC-II_1 and qLC-II_11) contributed highest to the PVE which ranged from 30.81% to 38.22% in DP-7. In all three NPK conditions, five QTLs (four QTLs for LC-II and one QTL for LC-I) were significantly associated with a single trait of LC on chromosomes 1, 2, 8, and 11. The negative additive effect of qLC-II_80%_1 was contributed by Z143, whereas four other QTLs (qLC-II_100%_1, qLC-I_0%_2, qLC-II_0%_8, and qLC-II_0%_11) were contributed by a positive additive effect from WTR-1.
Putative QTLs with high statistical power
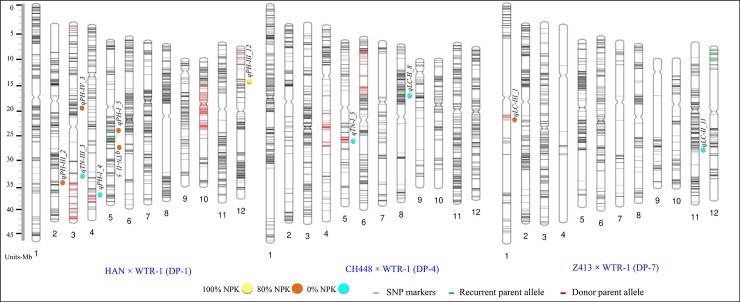
To detect QTLs with higher statistical power, an extreme threshold parameter, 1000 permutation test at P = 0.01, was considered. A total of 11 putative QTLs (≥15% PVE) were detected for the three FATs at DGI stages under three different fertilizer doses (100%, 80%, and 0% NPK). The QTL qPH-III_12 was the only one detected at 100% NPK for the FAT, PH at stage III on chromosome 12 between the markers S12_852867 and S12_2965504. Under suboptimal fertilizer dose, three QTLs for PH, qPH-III_2, qPH-IV_3 and qPH-I_5,were detected on chromosome 2, 3 and 5, while another single QTL for LC (qLC-II_1) and TN (qTN-III_5), were detected on chromosome 1 and 5. Simialrly in zero fertilizer conditions, five QTLs (qTN-III_3, qPH-I_4, qTN-I_5, qLC-II_8 and qLC-II_11) for PH, TN and LC were detected on five chromosomes 3, 4, 5, 8, and 11. However, the significant M-QTLs were identified only from MP DP-1, except four QTLs detected in the cross combination CH448 × WTR-1 (DP-4) and Z143 × WTR-1. The distribution of detected significant main-effect QTLs under three different NPK conditions is depicted in Fig 4. Additionally, minor effects QTLs were also detected for all the traits. The list of total minor effect QTLs including the information of flanking markers along with their LOD score, PVE and the additive effect of parental source of the allele are presented in S1 Table.
Fig 4. The genetic map representing the distribution of polymorphic SNP markers.
Colored circles yellow, orange, and blue indicate 100%, 80%, and 0% nutrient fertilizer doses, respectively. The QTLs highlighted on the genetic map are the major QTLs detected in the study possessing ≥15% phenotypic variation explained. QTLs from different FATs with fertilizer doses are shown in colored circles.
Traits associated with putative QTLs detected at 100%, 80%, and 0% NPK fertilizer dosage
Across the three MPs, a single QTL (qPH-III_12) was identified at 100% NPK fertilizer dose for the trait PH at stage III of fertilizer application. This QTL is located on chromosome 12 and it explained 23.60% of PV. The negative additive effect is contributed by donor parent HAN. Under suboptimal conditions (80% recommended dose of fertilizer), five QTLs (qLC-II_1, qPH-III_2, qPH-IV_3, qPH-1_5, and qTN-III_5) were detected on three chromosomes as 1, 2 and 3. Those QTLs were associated with PH, TN, and LC at DGI stages I, II, III, and IV. Among the 11 main-effect QTLs, qLC-II_1 showed the highest PVE of 34.85%. Except for the chromosome 1 QTL (qLC-II_1), all of them were identified in a single MP, DP-1 (S1 Table). Similarly, QTL analysis under zero fertilizer dose revealed that a total of five main-effect QTLs were detected for the traits of PH-I, TN-I and III, and LC-II. Three QTLs (qTN-III_3, qPH-I_4, and qTN-I_5) were detected on chromosomes 3, 4, and 5 and they explained PV of 17.02%, 19.17%, and 28.44% at stages I and III of fertilizer application. Another two QTLs (qLC-II_8 and qLC-II_11) were associated with a single trait (LC) at first stages of fertilizer application. QTLs on chromosome 3, 4, and 5 were contributed by the negative additive effect from donor alleles, whereas QTLs on chromosomes 8 and 11 were contributed by the positive additive effect from the WTR-1 allele.
Exploring putative QTLs for candidate genes for future breeding programs
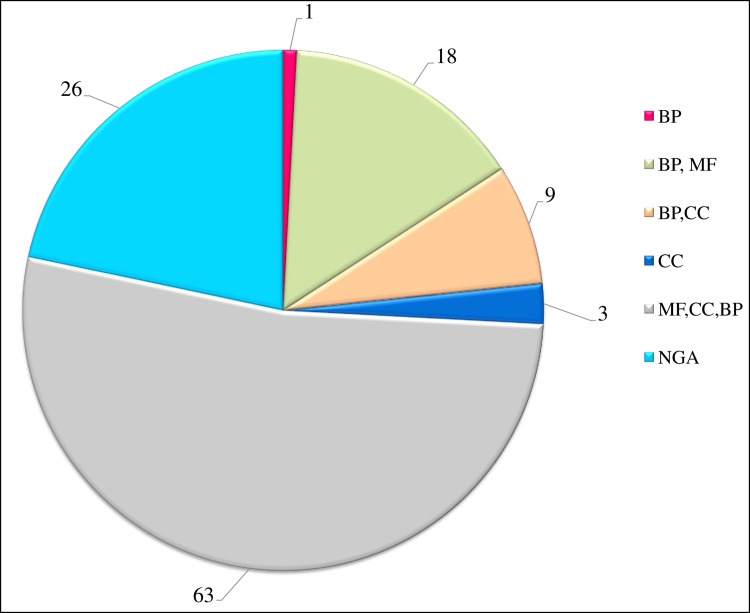
The putative QTLs detected from the extreme threshold parameters were further explored to showcase the possible candidate genes within the loci to understand the molecular and physiological mechanisms underlying the traits conferring NPK deficiency tolerance. A total of 19 M-QTLs were located on all chromosomes, except on chromosome 7. Further, the possible candidate genes within the M-QTLs were filtered based on the SNP flanking marker positions with a threshold score of ≤1 Mb interval regions of M-QTLs (Table 6). The lowest number (3) and highest number (25) of candidate genes were associated with a single trait (TN at stages III and IV) in the MP DP-1. A total of 120 genes were identified (S2 Table), of which 78.33% (94 genes) were functionally annotated while 21.66% (26 genes) were reported as hypothetical, retrotransposon proteins, and expressed proteins. The 94 functionally annotated genes were classified into six biological functions related to cellular component (CC), biological process (BP), and molecular function (MF) (Fig 5). Out of these 94 genes, 63 candidate genes were involved in MF, CC, and BP, followed by 18 genes for BP and MF, 9 genes for BP and CC, 3 genes for CC, and a single gene for BP.
Table 6. Putative QTLs (≤1 Mb interval regions) at DGI stages for favorable agronomic traits.
| Cross combination | Fertilizer dose (% NPK) |
QTL designation | Chromosome | Left marker | Right marker | Size of the QTL (kb) | LOD score | Additive effect | R2 (%) | No. of candidate genes detected within the locus |
|---|---|---|---|---|---|---|---|---|---|---|
| HAN × WTR-1 | 80% | qPH-III_2 | 2 | S2_29072519 | S2_28879120 | 193.40 | 3.65 | 2.86 | 18.12 | 15 |
| HAN × WTR-1 | 0% | qLC-I_2 | 2 | S2_30451119 | S2_30564762 | 113.60 | 3.04 | 0.64 | 10.79 | 12 |
| HAN × WTR-1 | 80% | qPH-I_5 | 5 | S5_17801786 | S5_17996757 | 195.10 | 3.59 | 1.21 | 15.61 | 16 |
| HAN × WTR-1 | 80% | qTN-III_5 | 5 | S5_17859957 | S5_17801758 | 58.20 | 26.77 | 3.62 | 28.98 | 3 |
| HAN × WTR-1 | 0% | qTN-IV_5 | 5 | S5_17465069 | S5_17801758 | 336.70 | 25.70 | 4.72 | 5.85 | 25 |
| CH448 × WTR-1 | 0% | qLC-II_8 | 8 | S8_8206216 | S8_8918154 | 711.90 | 5.26 | 1.37 | 22.85 | 30 |
| HAN × WTR-1 | 100% | qTN-I_9 | 9 | S9_20844013 | S9_20779092 | 64.92 | 3.11 | -0.65 | 13.00 | 10 |
| HAN × WTR-1 | 80% | qTN-III_12 | 12 | S12_6585982 | S12_5905028 | 681.11 | 3.10 | -1.00 | 1.89 | 9 |
Fig 5.
Functional classification of annotated genes (MF = Molecular function; CC = Cellular component; BP = Biological process; NGO = No gene ontology classification).
Discussion
For the ever-increasing global population and to meet food demand, the development of rice varieties with higher grain yield is essential [80]. The application of the key nutrients N, P, and K either organically or through chemical fertilizer plays a foremost role in increasing yield and sustaining soil fertility [81]. With the recent trends in crop breeding, higher rice productivity has been successfully attained by applying high fertilizer doses [9,35,82,83]. However, without knowledge of the correct stage, timing, and dose of fertilizer application, any assurance for increasing yield cannot be determined. The excessive use of fertilizer is a major contributor to increasing soil, water, and environmental pollution, along with farm operation costs [9,18,33,84]. China is leading in per hectare usage of fertilizer (300 to 350 kg ha-1 N in Jiangsu Province), and this amount is around four times higher than the average world fertilizer consumption for rice production [18,33]. Hence, it is crucial to undertake a systematic breeding program involving identifying genotypes with higher input use efficiency and genomic regions derived from these lines to improve elite varieties to assure sustainable crop production. This would not only reduce environmental and human hazards but would also improve the livelihood of farmers by reducing farm operational costs along with a higher expected grain yield. Hence, in the present study, we have developed MPs generated from three different donors (HAN, CH448, and Z413) and evaluated them under three different fertilizer doses, 100% (222.23 kg ha-1 N), 80% (177.77 kg ha-1 N), and 0% (0 kg ha-1 N), to study the genetic variation for low input use efficiency, especially for N, P, and K. Further, we identified the genomic regions influencing FATs at DGI stages under different fertilizer doses while dissecting the molecular genetic information.
Dissecting molecular genetics of nutrient deficiency tolerance in rice
ANOVA carried out using different MPs for FATs at different stages indicated the existence of significant variation among all the MPs for the target traits. As expected, all the genotypes, including parents of the MPs, showed a higher mean performance for all the AFTs at 100% NPK, followed by 80% and 0% NPK. This trend observed mainly because of the higher doses of the key nutrients N, P, and K, which are proven to enhance rice yield. Several researchers also reported the similar effect of key nutrients on plant growth and yield-attributed traits [85–88]. The variance components of FATs indicated that the segregation pattern at different DGI stages under different fertilizer doses varied from normal to skewed distribution (Table 2). This clearly confirmed the influence of few or many genes with a cumulative and additive effect, which is difficult to dissect using traditional low-resolution genotyping platforms. Hence, to understand the molecular genetic basis of the traits that influence the genomic regions conferring FATs at different DGI stages under varied fertilizer doses, a high-resolution and informative genotyping platform was employed. Numerous high-quality SNPs retrieved from the advanced genotyping tool tGBS were used for the molecular mapping of the key traits. A total of 19 M-QTLs were identified from three different MPs for the three FATs at four DGI stages under three different fertilizer doses. Out of these QTLs, 13 QTLs from DP-1, 4 QTLs from DP-4, and 2 QTLs from DP-7 explained PV ranging from 1.89% to 28.98%, 7.74% to 28.44%, and 30.62% to 34.85%, respectively. With the extreme threshold parameters, eight M-QTLs were considered as putative QTLs possess ≤1 Mb QTL regions, whereas the remaining 11 QTLs possess ≥1 Mb genetic regions of QTLs (Table 6). The putative QTLs qPH-III_2, qLC-I_2, qPH-I_5, qTN-III_5, qTN-IV_5, qLC-II_8, qTN-I_9, and qTN-III_12 suggested that these are closely associated with the respective traits (PH, TN, and LC) at DGI stages of fertilizer application. However, the majority of these QTLs on chromosomes 2, 5, and 8 were attributed to WTR-1 allele, whereas the QTLs on chromosomes 9 and 12 by HAN.
Potentiality of the putative QTLs and promising ILs
The mean performance of all the individuals of the MPs showed higher values than their parents for all the FATs (Table 2; S1 Fig). This indicates a more positive response by ILs to fertilizer application than their parents, even to suboptimal and zero fertilizer doses. The main reason for the improved performance of the ILs is the existence of transgressive segregation for all the FATs at all DGI stages under different fertilizer doses. The QTL qTN-I_9 detected under 100% NPK was found to be responsible for the transgressive segregation of the ILs for the trait TN for the mapping population derived from DP-1. Similarly, QTLs qPH-III_2, qPH-I_5, qTN-III_5, and qTN-III_12 contributed to the superior performance of ILs from the MPs derived from DP-1. Most importantly, three QTLs, qLC-I_2, qTN-IV_5, and qLC-II_8, detected on chromosomes 2, 5, and 8 at 0% NPK fertilizer dose, contributed solely to the better phenotypic performance of the ILs in MPs DP-1 and DP-4. This finding has an immense application in MAS through the introgression of the same QTLs in the background of elite lines to give them more NuUE under zero fertilizer conditions. To date, very few rice cultivars with tolerance of low inputs have been identified by using different doses of individual N, P, and K fertilizers reported from various phenotypic screening methodologies [13,89–91]. However, in the present study, we could identify several ILs that showed transgressive segregation even under suboptimal and zero fertilizer application, especially for LC. The genotypes of the mapping populations DP-1 (LC-I and LC-II ≥24.10%, LC-III ≥80.83%, LC-IV ≥27.50%), DP-4 (LC-I and LC-II ≥23.80%, LC-III ≥76.10%, LC-IV ≥23.80%), and DP-7 (LC-I ≥60.01%, LC-II ≥34.52%, LC-III ≥80.01%, LC-IV ≥36.31%) have significantly exceeded the performance of the parents and checks. However, a total of 11 ILs, two ILs from DP-1 (GSR-IR2-1-R11-L1-L2, GSR-IR2-1-R12-L1-R2), four ILs (GSR-IR2-4-L7-Y1-L2, GSR-IR2-4-R4-S1-L2, GSR-IR2-4-Y10-L1-Y2, and GSR-IR2-4-Y12-L1-Y2) from DP-4, and five ILs (GSR-IR2-7-L4-SU1-Y2, GSR-IR2-7-L8-SU2-R2, GSR-IR2-7-R13-SU1-L2, GSR-IR2-7-Y11-SU3-R2, and GSR-IR2-7-Y11-SU3-Y2) from DP-7, have been commonly identified as promising ILs that show constitutively increased LC at DGI stages. These ILs, being highly responsive to fertilizer application, could be an imperative source of NuUE traits for future breeding programs and also an excellent source for the genetic dissection of tolerance of low inputs in rice. These potential ILs are highly useful for medium to low input marginal farmers who may not incur much of their cost in buying fertilizer for rice cultivation. Therefore, the area and production of rice would increase, which could further help in ensuring food security and farmers’ livelihood. This is one of the remarkable findings and applications of the present study and could be attributed to the contribution of the putative QTLs detected from the potential genetic backgrounds.
Promising QTLs and comparison with previous QTLs related to deficiency of N, P, and K fertilizers
The rice Gramene database (http://archive.gramene.org), QTL Genome Viewer (http://qtaro.abr.affrc.go.jp), and previous reports from comprehensive literature surveys showed hundreds of QTLs associated with morphological, physiological, and biochemical traits that influence individual nutrient fertilizer deficiency in rice [10,41,52,92]. Among the 19 M-QTLs detected in the present study, 14 M-QTLs were detected in the same genomic region that was previously reported. These QTLs were significantly associated with more than 20 traits reported under N, P, and K deficiency tolerance, and were located on all chromosomes except chromosomes 7 and 8. The remaining five QTLs were found to be novel (Table 7). Among the 14 QTLs associated with FATs under DGI stages, nine QTLs were related to PH-I, LC-I, and LC-II, and four stages to TN on seven different chromosomes (2, 3, 4, 5, 6, 8, and 11) under deficiency of complete fertilizer (0 kg ha‒1). Six QTLs were related to PH-I, PH-III, PH-IV, TN-III, and LC-II on five chromosomes (1, 2, 3, 5, and 12) under suboptimal fertilizer dose (177.77 kg ha-1 N), and four QTLs were related to TN-I, TN-II, TN-III, and LC-II on four chromosomes (1, 9, 10, and 12) under the recommended dose of fertilizer, 100% (222.23 kg ha‒1 N). Some of these QTLs were co-localized with low N, P, and K tolerance on chromosomes 2, 4, 6, 10, 11, and 12. These QTL clusters might play an important role in NuUE in rice, and their related SNP markers could be useful for MAS in low input breeding programs.
Table 7. M-QTLs sharing similar genomic regions with previously reported QTLs.
| S. no. | FDa (% NPK) | QTL designation | QTL region (Mb) | Common region shared with earlier reported QTLs | |
|---|---|---|---|---|---|
| QTLs | Reference | ||||
| 1 | 0% | qLC-I_2 | 29.07–28.88 | qRPH ¥, qRSDW¥, and qRKUP¥ | [68] |
| 2 | 0% | qTN-III_3 | 27.66–29.5 | qDWR_3#, qNL_3.1#,qNL_3.2#, qaNUE_3#, qSP_3b#, qGD_3a#, and qFGp_3a# | [58,93] |
| 3 | 0% | qTN-II_4 | 31.28–32.42 | qRRN_4*, OsPTR5*, and OsPTR6* | [98] |
| 4 | 0% | qPH-I_4 | 15.43–21.82 | qSB_4# and qRRW_4* | [57,67] |
| 5 | 0% | qTN-IV_5 | 17.8–18 | qDWR_5#, agNUE_5#,qDWS#, qDM#, qFW#, qTN#, and qaNUE# | [93] |
| 6 | 0% | qTN-I_5 | 17.80–17.86 | qDWR_5# and qagNUE_5# | [93] |
| 7 | 0% | qTN-II_6 | 2.37–10.28 | qSP-6a,b#, qNCm6-12#, qNCD-6#, qPUP*, qRRDW_6.1*, qRSDW_6.2*, qRTDW_6.3*, qRPUC_6.4*, qRRL_6.4*, and qRWRSR_6.6* | [53,58,64,67,94] |
| 8 | 0% | qLC-II_8 | 8.21–8.92 | Novel | - |
| 9 | 0% | qLC-II_11 | 3.07–19.65 | qRRDW_11*, qPNP-11.1*, qRWRSR*, and qRRDW-11_1# | [65,67,99] |
| 10 | 80% | qLC-II_1 | 6.15–24.88 | qPN1.7*, qSPF* | [67,98] |
| 11 | 80% | qPH-III_2 | 30.45–30.56 | Novel | - |
| 12 | 80% | qPH-IV_3 | 0.24–1.24 | Novel | - |
| 13 | 80% | qPH-I_5 | 17.53–18.57 | Novel | - |
| 14 | 80% | qTN-III_5 | 17.47–17.80 | Novel | - |
| 15 | 80% | qTN-III_12 | 0.85–2.97 | qRTHK_12_1# and qRDW_12# | [99,100] |
| 16 | 100% | qLC-II_1 | 29.88–33.6 | qRRDW_1_1#, and qSRL_1_2# | [99,101] |
| 17 | 100% | qTN-I_9 | 20.78–20.84 | qDRW_9_5# | [100] |
| 18 | 100% | qTN-II_10 | 4.70–14.56 | qNAA_10# and qPUP* | [53,102] |
| 19 | 100% | qPH-III_12 | 5.91–6.59 | Novel | - |
aFertilizer dose
bPercentage of phenotypic variation explained by each QTL
#QTLs for nitrogen deficiency
*QTLs for phosphorus deficiency
¥QTLs for potassium deficiency in rice.
By comparing with the previously reported QTLs, most of the QTLs were sharing common genetic regions at DGI stages in all populations. The QTLs were associated with other multiple QTLs, ranging from one to 11 under N, P, and K deficiency tolerance (Table 7). In the zero percentage of fertilizer condition, a total of three putative QTLs related to relative PH (cm) (qRPH), relative shoot dry weight (g/p) (qRSDW), and relative potassium uptake (mg/p) (qRKUP) shared the same genomic region with qLC-I on chromosome 2. Among the three QTLs, qRPH had the highest phenotypic variation of 14.70%, with an LOD score of 4.04, identified by using 123 DH lines under low-K stress conditions [68]. On chromosome 3, six QTLs related to N concentration in leaf sheaths plus stems (qNS%_3.1), N concentration in leaf blades (qNL%_3.2), dry weight of roots (qDWR_3.3), grain density per panicle (qGD_3a), number of filled grains per panicle (qFGP_3a), and number of spikelets per panicle (qSP_3b) shared the same genetic region with qTN-III_3 at 29.07–28.88 Mb [58,93]. These QTLs were responsible for nitrogen use efficiency (NUE) and were also associated with yield-attributed traits under low nitrogen rates. Two genetic regions on chromosome 4 at 15.43–21.82 Mb and 31.28–32.42 Mb region of two QTLs (qPH-I_4 and qTH-II_4) were shared with three QTLs related to shoot and root growth traits under low nitrogen and phosphorus tolerance and another two of them were related to phosphorus transporters [57,67]. By using the F9-10 generation of 169 recombinant inbred lines (RILs) derived from a cross between IR64 (O. sativa L. var. indica), and Azucena (O. sativa L. var. japonica), Thi et al. [93] identified 44 QTLs for 15 agronomic and NUE-related traits on all chromosomes, except chromosome 9. Among these traits, seven NUE-related traits such as agronomic NUE, number of tillers, total fresh matter, dry weight of roots, total dry matter, dry weight of sheaths plus stems, and dry weight of leaf blades were shared with the current study for two QTLs (qTN-I_5 and qTN-IV_5) on chromosome 5 at 17.80–17.86 Mb region and 17.80–18 Mb region, respectively. However, the highest number of QTLs (11 QTLs) for low nitrogen and phosphorus tolerance were shared with qTN-II_6 at the 2.37–10.28 Mb region on chromosome 6. Earlier, under low nitrogen conditions, Hu et al. [94] identified three QTLs for nitrogen content in shoots by using 116 DH populations, which have been developed through anther culture of F1 hybrids from indica rice variety Taichung Native 1 (TN1) and japonica rice variety ChunJiang 06 (CJ06). One of the main-effect QTLs, qNCm6-12 (nitrogen content in plant shoots at mature stage), is significantly associated with the present QTL related to TN at stage II of fertilizer application. These QTLs mapped between RM527 and RM3 and explained PV of 9.73%. In addition, Feng et al. [58] identified 28 QTLs for yield-attributed traits on seven chromosomes under low N conditions. Two QTLs (qPL-6a and qSP-6a) for panicle length and number of spikelets per panicle explained PV of 15.58% and 6.40% from the analysis of 138 F14 RILs, which were derived from a super hybrid rice (Xieyou 9308) in China. Another QTL related to absorbed NUE (qaNUE) was also located in the same position [93]. The remaining seven QTLs were associated with low P tolerance QTLs. Of these, qPUP_6 was associated with P uptake [95], four QTLs (qRRDW, qRSDW, qRTDW, and qRPUC) for root traits and P uptake in rice [64], and two QTLs (qRRL, and qRWRSR) for root growth and weight traits [67]. Interestingly, to date, there are no reported QTLs matched to/shared with the currently identified major QTL qLC-II_8 (PVE of 22.85%) at the 8.21–8.92 Mb region on chromosome 8, which indicates that these QTLs are novel loci controlling for LC. However, Shimizu et al. [62], Wang et al. [96], and Tong et al. [97] reported 11 QTLs associated with LC on chromosomes 1, 2, 3, 4, 7, and 12 under low N and P conditions. As compared to the above studies, the current study identified four QTLs for LC on chromosomes 8 and 11 that are novel genetic regions under zero fertilizer conditions. The individual QTLs for LC had PVE ranging from 10.79% to 30.62%.The alleles from WTR-1 were in the direction of increasing the LC. These results indicate that these DGI stages of fertilizer application and cluster QTLs were significantly associated with low N, P, and K tolerances in rice.
Under suboptimal doses of fertilizer, a total of six QTLs (qLC-II_1, qPH-III_2, qPH-IV_3, qPH-I_5, qTN-III_5, and qTN-III_12) were associated with LC, TN, and PH located on five different chromosomes (1, 2, 3, 5, and 12), explaining an average PV of 19.09%. The smallest genetic region of qPH-III_2 (0.11 Mb) was located on chromosome 2, and qPH-IV_3 (1 Mb) was located on chromosome 3. Two QTLs, qPH-I_5 (1.04 Mb) and qTN-III_5 (0.33 Mb), were located on chromosome 5, and those are not shared with any reported QTLs. Therefore, these QTLs were considered as novel loci for DGI stages of fertilizer application. The remaining two QTLs, qLC-II_1 on chromosome 1 and qTN-III_12 on chromosome 12, at 6.15–24.88 Mb region and 0.85–2.97 Mb region, were shared. A total of three QTLs were related to low N and two QTLs to low P tolerance traits. Three QTLs (qTNCS_1b, qSY_1b, and qGD_1a) under low N on chromosome 1 shared the same genetic region with LC-II_1. The first two QTLs for total nitrogen content of shoot and straw yield were noticed by Cho et al. [54] by using 166 F8 lines derived from a cross between a Korean tongil type of rice, variety Dasanbyeo, and a Chinese japonica variety, TR22183. Similarly, Feng et al. [58] identified three QTLs for grain density (GD) per panicle by using 138 F14 RILs, under low N. One of the QTLs, qGD_1a (6.04% PVE) located on chromosome 1, was shared with qLC-II_1. Two other QTLs (qRTDW_1 and qRSDW_1) were shared with qLC-II_1 on chromosome 1. These two QTLs were associated with low P tolerance. Lastly, chromosome 12 at the 0.85–2.97 Mb region showed the presence of two QTLs (qRTHK_12_1 and qRDW_12) for root thickness [100] and root dry weight [99] by using RILs under low N conditions. On the other hand, for the recommended dose of fertilizer, four QTLs (qLC-II_1, qTN-I_9, qTN-II_10 and qPH-III_12) located on four different chromosomes (1, 9, 10, and 12) had considerable association with earlier reports of several QTLs related to low P and N tolerance in rice.
For yield-attributed and NUE traits related to QTLs under low N conditions, Lian et al. [99] and Dai et al. [102] reported a total of 15 main-effect QTLs for root and shoot weight and 13 QTLs distributed on all 12 chromosomes. Of these, two QTLs, for relative root weight, qRRW_1a [99], and seminal root length, SRL_1.2 [101], on chromosome 1 shared a common genetic region with qLC-II_1. Similarly, a single QTL, qDRW-9.5 [100], on chromosome 9 was shared with qTN-I_9 under low N conditions. The genetic region of 4.70–14.56 Mb associated with qTN-II_10 is shared with two QTLs: for nitrogen accumulation amount, qNAA_10 [102], and phosphorus uptake, qPUP_10 [53]. Lastly, chromosome 12 at the 5.91–6.59 Mb region is associated with qPH-III_12 (PVE of 23.06%), which is not shared with any previous reports of QTLs and is considered to be a novel locus. In summary, among all the shared genetic regions, QTLs detected on chromosomes 1, 4, 5, 10 and 11 confer N and P deficiency tolerance, those on chromosomes 3 and 4 confer N deficiency tolerance, those on chromosome 4 confer P deficiency tolerance, and those on chromosome 2 confer K deficiency tolerance in rice. The clusters of QTLs detected under different doses of fertilizer at DGI stages and novel QTLs are valuable genetic resources to identify useful genes and introgression in elite lines to improve rice in NuUE.
Putative candidate genes and functions
The development of highly nutrient-responsive genotypes for low nutrient application is imperative during this era when the area and production of rice are under threat. MAS is one of the main strategies for accelerating the process of developing lines with higher NuUE. Effective MAS using major-effect QTLs mainly depends on the highest co-segregation of the markers and trait. Understanding of the molecular mechanisms and gene structure of the loci detected increases the efficiency of MAS. Hence, in the present study, we explored the candidate genes within the major-effect loci interval region (≤1 Mb) and their predicted molecular and physiological functions. A total of eight putative QTLs detected on five different chromosomes (2, 5, 8, 9, and 12) were explored for the candidate gene analysis survey (Table 7). A total of 120 genes were identified in the promising QTL interval regions. Each QTL has a variable number of underlying associated genes ranging from 3 genes (58.2 kb) on chromosome 3 to 30 genes (711.90 kb) on chromosome 8. In silico analysis revealed that 78.33% of the genes are functionally annotated while the remaining genes are expressed and hypothetical proteins. The list of all possible candidate loci and their functions is detailed in S2 Table and Fig 5. These genes were functionally related to numerous physiological and molecular functions such as photosynthetic rate, synthesis of chlorophyll precursors, phosphate transporters (peptide transporter), zinc transporters, growth stimulators of auxin-responsive genes, abscisic acid (ABA) signaling pathways, and activation of various transcription factors, all of which were found within the promising M-QTL interval regions.
A QTL (qPH-II_2) located at 28.88–29.07 Mb on chromosome 2 associated with PH at stage II of fertilizer application was neighboring the candidate gene Os02g46140, which encodes for F-box protein domain involved in several biological functions, is related to phytohormone signaling pathways, and regulates various developmental processes as a part of the abiotic stress response mechanism [103]. Another candidate gene, Os02g46090, is associated with calcium signaling pathways. Several families of calcium sensors have been reported and they have played major roles in nitrogen metabolism and abiotic and biotic stress response mechanisms [104]. Recently, researchers identified a novel mutant of the calcium-dependent protein-kinase-encoding gene, esl4, which is significantly expressed in roots, shoots, and enzyme activity of several genes related to nitrogen metabolism [105]. Interestingly, two candidate genes, a tonoplast-localized low-affinity nitrate transporter, OsNPF7.2 [106], and a transcriptional regulator, GROWTH-REGULATOR FACTOR 4 (OsGRF4), along with a growth inhibitor of DELLA proteins [107], are located in the same genetic region on chromosome 2. The hotspot region of the root-specific transporter plays a vital role in intracellular allocation of nitrate in roots, especially in sclerenchyma, cortex, and stele, and ultimately leads to influences on plant growth. The interaction of GRF4 and DELLA proteins is involved in physiological activities to regulate multiple genes for carbon and nitrogen metabolism, plays a significant role in the homeostatic coordination of nitrogen metabolism, and also increases shoot biomass and PH [107,108]. Overexpression and mutant analysis of OsNPF7.2 showed that the significant expression of OsNPF7.2 caused an increase in TN by regulating the cytokinin and strigolactone pathway [109]. Another QTL, qLC-I_2 at the 30.45–30.56 Mb region, was found to be located on chromosome 2, adjacent to another candidate gene, OsGS1;1 (cytosolic glutamine synthatase), which regulates nitrogen metabolic pathways and also influences plant growth and development [110,111]. The GS1 proteins have a major role in generating glutamine, which is the primary form of remobilized nitrogen during natural senescence in leaves for long-distance transport [112]. In addition to that, qLC-I_2 is adjacent to the diacylglycerolacetyl transferase (OsDGAT) (Os02g48350) gene and interacts with OsTCP19 in regulating seedling establishment by modulating stress signaling molecules and abscisic acid pathways in various abiotic stresses [113].
Three QTLs, qPH-I, qTN-III, and qTN-IV (17.47–18.00 Mb), were found to be located on chromosome 5 adjacent to Os05g30970, and they encode for copine-like protein. It is mainly involved in the nutritional role of glutamate for aiding in seedling establishment under nitrogen deficiency [114]. In the same position are three other possible putative candidate loci: Os05g30870 (OsRLCK185), for diverse roles in plant growth and development and stress responses [115]; and Os05g30240, encoding for pentatricopeptide-repeat protein for upregulation in salt stress conditions [116]. Hence, to overcome nutrient deficiency, balanced mineral nutrients are essential for optimal plant growth and development. Each of these genes/loci played a critical role in various physiological and molecular responses such as cytokinins negatively regulating Pi (phosphate) starvation and also regulating metabolic changes under low N [117,118], and phytohormones such as auxins and seconday metabolites that are involved in the maintenance of homeostasis, root hair development, and signaling pathways under low NPK tolerance mechansim [119,120]. Similarly, phytohormones, ABA, jasmonates (JA), and their associated biosynthetic genes at vegetative and reproductive stages regulate various signaling pathways, leading to adaptation to nutrient deficiency through the root architecture, and maintaining N, P, and K homeostasis [121–123]. The in silico expression analysis was performed for the candidate genes, Os02g46140, Os02g46090, and Os02g48350 on chromosome 2 and Os05g30970, Os05g30870, and Os05g30240 on chromosome 5 located within M-QTL regions using RiceXpro [124]. The expression analysis indicated wide differential expression pattern of these candidate genes (S2 Fig). The high level of expression was observed in ovary development for the two candidate genes, Os02g46090 and Os05g30240, whereas for the other genes high level of expression was noticed in spikelet hull (lemma and palea) during the early stages of seed development and higher expression during roots and panicles development. Hence, these candidate genes within the M-QTL interval region can be considered as the promising putative candidate genes. However, further validation and developing functional markers are necessary for their effective application in breeding programs.
Conclusions
Genetic dissection of low N, P, and K tolerance at DGI stages of fertilizer application is an important target area for modern breeding programs in rice. Hence, in the present study, we identified the genomic regions that confer low N, P, and K tolerance for favorable agronomic traits at different stages of fertilizer application and selected ILs possessing the highest tolerance of low NPK using three different MPs derived from three donors (HAN, CH448, and Z413). ANOVA and descriptive statistics indicated the existence of significant genetic variation among the FATs and the predominance of non-Mendelian inheritance. To identify the genetic factors that influence these quantitatively inherited traits, QTL analysis was carried out by using the precisely estimated phenotypic values and high-quality SNPs derived from the tGBS genotyping platform. A total of 19 M-QTLs on different chromosomes were detected. Among these M-QTLs, eight QTLs were considered as putative QTLs, with the smallest locus size (≤1 Mb) contributing the highest toward trait expression. Among the putative QTLs, qTN-I_92, qTN-III_5, qLC-I_2, and qLC-II_8 detected under 100%, 80%, and 0% NPK were found to be responsible for transgressive segregation of the ILs for the trait. Notably, qLC-I_2, qTN-IV_5, and qLC-II_8 detected at zero fertilizer application showed higher performance for LC under 0% of NPK fertilizer. These QTLs not only help in building a tolerance of low N, P, and K nutrient simultaneously but also improve genotypes to make them highly responsive to lower nutrient application. This is one of the remarkable achievements of the current study, which helps low-input and marginal farmers to cultivate rice without incurring high fertilizer cost and to eventually ensure food security and sustainable agriculture. On the other hand, in silico functional annotation of candidate genes within the putative QTLs indicated that two and five candidate genes found to confer tolerance of low N, P, and K and related to several physiological and metabolic pathways were also found to be involved in abiotic stress tolerance. However, additional investigation is needed for further confirmation to examine the potential physiological and molecular mechanisms. These studies can help in understanding the underlying complex genetic interactions involved in nutrient use efficiency and the identification of more efficient breeding materials containing these genetic factors. Furthermore, the detected genomic regions related to stage-specific tolerance of low fertilizer doses and promising ILs can be useful for MAS and future breeding programs for low-input conditions.
Supporting information
(XLSX)
(XLSX)
(TIFF)
(TIFF)
Acknowledgments
The authors are grateful to the internal reviewers from the International Rice Research Institute for their valuable comments and suggestions that helped to considerably improve the quality of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
All the authors would like to thank and acknowledge the Bill & Melinda Gates Foundation (BMGF) for providing the research grant to ZL for the Green Super Rice Project under ID OPP1130530. We would also like to thank the Department of Agriculture (DA) of the Philippines for providing funds to JA under the Next-Gen project. We would also like to thank the Department of Science and Technology (DOST)-Philippine Council for Agriculture, Aquatic and Natural Resources Research and Development (PCCARRD), Philippines, for providing funds to JA through the University of the Philippines, Los Baños (UPLB), under the improved resource use efficient (iRUE) rice project. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Godfray HC, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, et al. Food security: the challenge of feeding 9 billion people. Science. 2010; 327:812–818. 10.1126/science.1185383 [DOI] [PubMed] [Google Scholar]
- 2.Tester M, Langridge P. Breeding technologies to increase crop production in a changing world. Science. 2010; 327:818–822. 10.1126/science.1183700 [DOI] [PubMed] [Google Scholar]
- 3.Tilman D, Cassman KG, Matson PA, Naylor R, Polasky S. Agricultural sustainability and intensive production practices. Nature. 2002; 418:671–677. 10.1038/nature01014 [DOI] [PubMed] [Google Scholar]
- 4.Xu Y. Envirotyping for deciphering environmental impacts on crop plants. Theor Appl Genet. 2016; 129:653–673. 10.1007/s00122-016-2691-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tadele Z. Raising crop productivity in Africa through intensification. Agronomy. 2017; 7:22. [Google Scholar]
- 6.Lopez-Ridaura S, Frelat R, Van Wijk MT, Valbuena D, Krupnik TJ, Jat ML. Climate smart agriculture, farm household typologies and food security: An ex-ante assessment from Eastern India. Agric Syst. 2018; 159:57–68. 10.1016/j.agsy.2017.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kesavan PC, Swaminathan MS. Strategies and models for agricultural sustainability in developing Asian countries. Philos Trans R Soc B Biol Sci. 2007; 363(1492):877–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimaano NG, Ali J, Anumalla M, Cruz PC, Aurora M, Diaz MG, et al. Novel quantitative trait loci for weed competitive ability traits using the early generation of backcross rice populations. Prepints. 2018; 1–30. [Google Scholar]
- 9.Rahman KM, Zhang D. Effects of fertilizer broadcasting on the excessive use of inorganic fertilizers and environmental sustainability. Sustainability. 2018; 10(3):759. [Google Scholar]
- 10.Ali J, Jewel ZA, Mahender A, Anandan A, Hernandez J, Li Z. Molecular genetics and breeding for nutrient use efficiency in rice. Int J Mol Sci. 2018; 19:1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahender A, Swamy B, Anandan A, Ali J. Tolerance of iron-deficient and -toxic soil conditions in rice. Plants. 2019; 8:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma SB, Sayyed RZ, Trivedi MH, Gobi TA. Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. Springerplus; 2013; 2:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wissuwa M, Kretzschmar T, Rose TJ. From promise to application: root traits for enhanced nutrient capture in rice breeding. J Exp Bot. 2016; 67:3605–3615. 10.1093/jxb/erw061 [DOI] [PubMed] [Google Scholar]
- 14.Kulcheski FR, Côrrea R, Gomes IA, de Lima JC, Margis R. NPK macronutrients and microRNA homeostasis. Front Plant Sci. 2015; 6:451 10.3389/fpls.2015.00451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fageria NK. The use of nutrients in crop plants. Boca Raton, FL: CRC Press, 2009. Available: 10.1201/9781420075113 [DOI] [Google Scholar]
- 16.Li G, Huang G, Li H, van Ittersum MK, Leffelaar PA, Zhang F. Identifying potential strategies in the key sectors of China’s food chain to implement sustainable phosphorus management: a review. Nutr Cycl Agroecosyst. 2016; 104:341–359. [Google Scholar]
- 17.Herrera JM, Rubio G, Häner LL, Delgado JA, Lucho-Constantino CA, Islas-Valdez S, et al. Emerging and established technologies to increase nitrogen use efficiency of cereals. Agronomy. 2016; 6:25. [Google Scholar]
- 18.Sui B, Feng X, Tian G, Hu X, Shen Q, Guo S. Optimizing nitrogen supply increases rice yield and nitrogen use efficiency by regulating yield formation factors. Field Crops Res. 2013; 150:99–107. [Google Scholar]
- 19.Guo J, Hu X, Gao L, Xie K, Ling N, Shen Q, et al. The rice production practices of high yield and high nitrogen use efficiency in Jiangsu, China. Sci Rep. 2017; 7:2101 10.1038/s41598-017-02338-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L, Lin L, Cai G, Sun Y, Huang T, Wang K, et al. Identification of nitrogen, phosphorus, and potassium deficiencies in rice based on static scanning technology and hierarchical identification method. PLoS One. 2014; 9:e113200 10.1371/journal.pone.0113200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng B, Chen K, Cui Y, Wu Z, Zheng T, Zhu Y, et al. Genetic dissection and simultaneous improvement of drought and low nitrogen tolerances by designed QTL pyramiding in rice. Front Plant Sci. 2018; 9:306 10.3389/fpls.2018.00306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rashid MM, Ahmed N, Jahan M, Islam KS, Nansen C, Willers JL, et al. Higher fertilizer inputs increase fitness traits of brown planthopper in rice. Sci Rep. 2017; 7:4719 10.1038/s41598-017-05023-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang F, Cui Z, Chen X, Ju X, Shen J, Chen Q, et al. Integrated nutrient management for food security and environmental quality in China. Adv Agron. 2012. pp. 1–40. [Google Scholar]
- 24.Liu X, Zhang Y, Han W, Tang A, Shen J, Cui Z, et al. Enhanced nitrogen deposition over China. Nature. 2013; 494:459–62. 10.1038/nature11917 [DOI] [PubMed] [Google Scholar]
- 25.Smith LED, Siciliano G. A comprehensive review of constraints to improved management of fertilizers in China and mitigation of diffuse water pollution from agriculture. Agric Ecosyst Environ. 2015; 209:15–25. [Google Scholar]
- 26.Huang M, Jiang P, Shan S, Gao W, Ma G, Zou Y, et al. Higher yields of hybrid rice do not depend on nitrogen fertilization under moderate to high soil fertility conditions. Rice. 2017; 10:43 10.1186/s12284-017-0182-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Q. Strategies for developing Green Super Rice. Proc Natl Acad Sci. 2007; 104:16402–16409. 10.1073/pnas.0708013104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali J, Xu JL, Gao YM, Fontanilla MA, Li ZK. Green super rice (GSR) technology: an innovative breeding strategy-achievements & advances In. Jogloy S, ed. Proc. 12th Inter. Cong. SABRAO; January 13–16, 2012, Chiang Mai, Thailand, 16–17. [Google Scholar]
- 29.Chauhan BS, Opeña J, Ali J. Response of 10 elite “Green Super Rice” genotypes to weed infestation in aerobic rice systems. Plant Prod Sci. Crop Science Society of Japan; 2015; 18:228–233. [Google Scholar]
- 30.Yorobe JM Jr, Ali J, Pede VO, Rejesus RM, Velarde OP, Wang H, et al. Yield and income effects of rice varieties with tolerance of multiple abiotic stresses: The case of green super rice (GSR) and flooding in the Philippines. Agric Econ (United Kingdom). Wiley Online Library; 2016; 47:261–271. [Google Scholar]
- 31.Li T, Ali J, Marcaida M, Angeles O, Franje NJ, Revilleza JE, et al. Combining limited multiple environment trials data with crop modeling to identify widely adaptable rice varieties. PLoS One. Public Library of Science; 2016; 11:e0164456 10.1371/journal.pone.0164456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dobermann A, Witt C, Dawe D, Abdulrachman S, Gines HC, Nagarajan R, et al. Site-specific nutrient management for intensive rice cropping systems in Asia. Field Crops Res. 2002; 74:37–66. [Google Scholar]
- 33.Peng S, Buresh RJ, Huang J, Yang J, Zou Y, Zhong X, et al. Strategies for overcoming low agronomic nitrogen use efficiency in irrigated rice systems in China. Field Crops Res. 2006; 96:37–47. [Google Scholar]
- 34.Peng XL, Liu YY, Luo SG, Fan LC, Song TX, Guo YW. Effects of site-specific nitrogen management on yield and dry matter accumulation of rice from cold areas of northeastern China. Agric Sci China. 2007; 6:715–723. [Google Scholar]
- 35.Liu C, Liu Y, Li Z, Zhang G, Chen F. A novel way to establish fertilization recommendations based on agronomic efficiency and a sustainable yield index for rice crops. Sci Rep. 2017; 7:1001 10.1038/s41598-017-01143-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Li J. Branching in rice. Curr Opin Plant Biol. 2011; 14:94–99. 10.1016/j.pbi.2010.11.002 [DOI] [PubMed] [Google Scholar]
- 37.Liang W, Shang F, Lin Q, Lou C, Zhang J. Tillering and panicle branching genes in rice. Gene. 2014; 537:1–5. 10.1016/j.gene.2013.11.058 [DOI] [PubMed] [Google Scholar]
- 38.Koumoto T, Shimada H, Kusano H, She KC, Iwamoto M, Takano M. Rice monoculm mutation moc2, which inhibits outgrowth of the second tillers, is ascribed to lack of a fructose-1, 6-bisphosphatase. Plant Biotechnol. 2013; 30:47–56. [Google Scholar]
- 39.Wang Q, Nian J, Xie X, Yu H, Zhang J, Bai J, et al. Genetic variations in ARE1 mediate grain yield by modulating nitrogen utilization in rice. Nat Commun. 2018; 9:735 10.1038/s41467-017-02781-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar A, Dixit S, Ram T, Yadaw RB, Mishra KK, Mandal NP. Breeding high-yielding drought-tolerant rice: genetic variations and conventional and molecular approaches. J Exp Bot. 2014; 65:6265–6278. 10.1093/jxb/eru363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahender A, Anandan A, Pradhan SK, Singh ON. Traits-related QTLs and genes and their potential applications in rice improvement under low phosphorus condition. Arch Agron Soil Sci. 2018; 64. [Google Scholar]
- 42.Shabir G, Aslam K, Khan A, Shahid M, Manzoor H, Noreen S, et al. Rice molecular markers and genetic mapping: Current status and prospects. J Integr Agric. 2017; 16:1879–1891. [Google Scholar]
- 43.Chen H, Xie W, He H, Yu H, Chen W, Li J, et al. A high-density SNP genotyping array for rice biology and molecular breeding. Mol Plant. 2014; 7:541–553. 10.1093/mp/sst135 [DOI] [PubMed] [Google Scholar]
- 44.Lau WC, Rafii MY, Ismail MR, Puteh A, Latif MA, Ramli A. Review of functional markers for improving cooking, eating, and the nutritional qualities of rice. Front Plant Sci. 2015; 6:832 10.3389/fpls.2015.00832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y, Li P, Yang Z, Xu C. Genetic mapping of quantitative trait loci in crops. Crop J. 2017; 5:175–184. [Google Scholar]
- 46.Kumar S, Banks TW, Cloutier S. SNP discovery through next-generation sequencing and its applications. Int J Plant Genomics. 2012; Article ID 831460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He J, Zhao X, Laroche A, Lu ZX, Liu H, Li Z. Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front Plant Sci. 2014; 5:484 10.3389/fpls.2014.00484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parida SK, Mukerji M, Singh AK, Singh NK, Mohapatra T. SNPs in stress-responsive rice genes: validation, genotyping, functional relevance and population structure. BMC Genomics. 2012; 13:426 10.1186/1471-2164-13-426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu QH, Chen YX, Zhou SH, Fu L, Chen JJ, Xiao Y, et al. High-density genetic linkage map construction and QTL mapping of grain shape and size in the wheat population Yanda1817 x Beinong6. PLoS One. 2015; 10:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Yu K, Li H, et al. High-density SNP map construction and QTL identification for the apetalous character in Brassica napus L. Front Plant Sci. 2015; 6:1164 10.3389/fpls.2015.01164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramkumar G, Sivaranjani AKP, Pandey MK, Sakthivel K, Shobha Rani N, Sudarshan I, et al. Development of a PCR-based SNP marker system for effective selection of kernel length and kernel elongation in rice. Mol Breed. 2010; 26:735–740. [Google Scholar]
- 52.Vinod KK, Heuer S. Approaches towards nitrogen- and phosphorus-efficient rice. AoB Plants. 2012; 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wissuwa M, Yano M, Ae N. Mapping of QTLs for phosphorus-deficiency tolerance in rice (Oryza sativa L.). Theor Appl Genet. 1998; 97:777–783. [Google Scholar]
- 54.Cho YI, Jiang W, Chin J-H, Piao Z, Cho YG, McCouch SR, et al. Identification of QTLs associated with physiological nitrogen use efficiency in rice. Mol Cells. 2007; 23:72–79. [PubMed] [Google Scholar]
- 55.Wei D, Cui K, Pan J, Ye G, Xiang J, Nie L, et al. Genetic dissection of grain nitrogen use efficiency and grain yield and their relationship in rice. Field Crops Res. 2011; 124:340–346. [Google Scholar]
- 56.Wei D, Cui K, Ye G, Pan J, Xiang J, Huang J, et al. QTL mapping for nitrogen-use efficiency and nitrogen-deficiency tolerance traits in rice. Plant Soil. 2012; 359:281–295. [Google Scholar]
- 57.Ogawa S, Valencia MO, Lorieux M, Arbelaez JD, McCouch S, Ishitani M, et al. Identification of QTLs associated with agronomic performance under nitrogen-deficient conditions using chromosome segment substitution lines of a wild rice relative, Oryza rufipogon. Acta Physiol Plant. 2016; 38:103. [Google Scholar]
- 58.Yue F, Zhai RR, Lin ZC, Cao LY, Wei XH, Cheng SH. Quantitative trait locus analysis for rice yield traits under two nitrogen levels. Rice Sci. 2015; 22:108–115. [Google Scholar]
- 59.Zhou Y, Tao Y, Tang D, Wang J, Zhong J, Wang YY, et al. Identification of QTL associated with nitrogen uptake and nitrogen use efficiency using high throughput genotyped CSSLs in rice (Oryza sativa L.). Front Plant Sci. 2017; 8:1166 10.3389/fpls.2017.01166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang K, Cui K, Liu G, Xie W, Yu H, Pan J, et al. Identification of quantitative trait loci for phosphorus use efficiency traits in rice using a high density SNP map. BMC Genet. 2014; 15:155 10.1186/s12863-014-0155-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ni JJ, Wu P, Senadhira D, Huang N. Mapping QTLs for phosphorus deficiency tolerance in rice (Oryza sativa L.). Theor Appl Genet. 1998; 97:1361–1369. [Google Scholar]
- 62.Shimizu A, Yanagihara S, Kawasaki S, Ikehashi H. Phosphorus deficiency-induced root elongation and its QTL in rice (Oryza sativa L.). Theor Appl Genet. 2004; 109:1361–1368. 10.1007/s00122-004-1751-4 [DOI] [PubMed] [Google Scholar]
- 63.Lang NT, Buu BC. Mapping QTLs for phosphorus deficiency tolerance in rice. Omonrice. 2006; 14:1–9. [Google Scholar]
- 64.Ming F, Zheng XW, Mi GH, He P, Zhu LH, Zhang FS. Identification of quantitative trait loci affecting tolerance to low phosphorus in rice (Oryza sativa L.). Chinese Sci Bull. 2000; 45:520–525. [Google Scholar]
- 65.Li J, Xie Y, Dai A, Liu L, Li Z. Root and shoot traits responses to phosphorus deficiency and QTL analysis at seedling stage using introgression lines of rice. J Genet Genomics. 2009; 36:173–183. 10.1016/S1673-8527(08)60104-6 [DOI] [PubMed] [Google Scholar]
- 66.Chin JH, Lu X, Haefele SM, Gamuyao R, Ismail A, Wissuwa M, et al. Development and application of gene-based markers for the major rice QTL Phosphorus uptake 1. Theor Appl Genet. 2010; 120:1073–1086. 10.1007/s00122-009-1235-7 [DOI] [PubMed] [Google Scholar]
- 67.Xi C, Ren J, Zhao XQ, Ding ZS, Jing ZH, Wang C, et al. Genetic dissection of low phosphorus tolerance related traits using selected introgression lines in rice. Rice Sci. 2015; 22:264–274. [Google Scholar]
- 68.Wu P, Ni JJ, Luo AC. QTLs underlying rice tolerance to low-potassium stress in rice seedlings. Crop Sci. 1998; 38:1458–1462. [Google Scholar]
- 69.Wang Y, Wu WH. Genetic approaches for improvement of the crop potassium acquisition and utilization efficiency. Curr Opin Plant Biol. 2015; 25:46–52. 10.1016/j.pbi.2015.04.007 [DOI] [PubMed] [Google Scholar]
- 70.Wang Y, Lu J, Ren T, Hussain S, Guo C, Wang S, et al. Effects of nitrogen and tiller type on grain yield and physiological responses in rice. AoB Plants. 2017; 9(2):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou W, Lv T, Yang Z, Wang T, Fu Y, Chen Y, et al. Morphophysiological mechanism of rice yield increase in response to optimized nitrogen management. Sci Rep. 2017; 7:17226 10.1038/s41598-017-17491-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sasaki T. Breeding green super rice (GSR) varieties for sustainable rice cultivation Li Z, Chinese Academy of Agricultural Sciences, China; and Ali J., International Rice Research Institute, Philippines. Achieving sustainable cultivation of rice. Volume 1 Burleigh Dodds Science Publishing; 2017. pp. 131–152. [Google Scholar]
- 73.Ali J, Aslam UM, Tariq R, Murugaiyan V, Schnable PS, Li D, et al. Exploiting the genomic diversity of rice (Oryza sativa L.): SNP-typing in 11 early-backcross introgression-breeding populations. Front Plant Sci. 2018; 9:849 10.3389/fpls.2018.00849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ali J, Xu JL, Gao YM, Fontanillo MA, Marfori CM, Li Z. Innovative Green Super Rice (GSR) molecular breeding strategy: achievements and advances. Philipp J Crop Sci. 2012; 37:49. [Google Scholar]
- 75.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009; 84:210–223. 10.1016/j.ajhg.2009.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, et al. TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One. 2014; 9:e90346 10.1371/journal.pone.0090346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meng L, Li H, Zhang L, Wang J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015; 3:269–283. [Google Scholar]
- 78.Kosambi DD. The estimation of map distances from recombination values. Ann Eugenics 12:172–175. [Google Scholar]
- 79.R Core Team (2013). R: A Language and Environment for Statistical Computing. Version 3.0.1. R Foundation for Statistical Computing, Vienna: Available at http://www.R‐project.org. [Google Scholar]
- 80.Ray DK, Mueller ND, West PC, Foley JA. Yield trends are insufficient to double global crop production by 2050. PLoS One. 2013; 8:e66428 10.1371/journal.pone.0066428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yadav RL, Dwivedi BS, Prasad K, Tomar OK, Shurpali NJ, Pandey PS. Yield trends, and changes in soil organic-C and available NPK in a long-term rice–wheat system under integrated use of manures and fertilisers. Field Crops Res. 2000; 68:219–246. [Google Scholar]
- 82.Mulvaney RL, Khan SA, Ellsworth TR. Synthetic nitrogen fertilizers deplete soil nitrogen: a global dilemma for sustainable cereal production. J Environ Qual. 2009; 38:2295–2314. 10.2134/jeq2008.0527 [DOI] [PubMed] [Google Scholar]
- 83.Lu C, Tian H. Global nitrogen and phosphorus fertilizer use for agriculture production in the past half century: shifted hot spots and nutrient imbalance. Earth Syst Sci Data. 2017; 9:181–192. [Google Scholar]
- 84.Tilman D, Balzer C, Hill J, Befort BL. Global food demand and the sustainable intensification of agriculture. Proc Natl Acad Sci. 2011; 108:20260–20264. 10.1073/pnas.1116437108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Djomo SH, Mbong GA, Malla DK, Suh C. Effect of different doses of NPK fertilizer on the growth and yield of rice in Ndop, North West of Cameroon. African J Agric Res. 2017; 12:1244–1252. [Google Scholar]
- 86.Dongarwar UR, Patke N, Dongarwar LN, Kashiwar SR. Influence of different fertilizer doses on growth, yield and economics of direct seeded rice in Eastern Vidharbha Zone of Maharashtra, India. Int J Curr Microbiol App Sci. 2018; 7:3837–3845. [Google Scholar]
- 87.Zhang Q, Wang G. Studies on nutrient uptake of rice and characteristics of soil microorganisms in a long-term fertilization experiment for irrigated rice. J Zhejiang Univ Sci B. 2005; 6:147–154. 10.1631/jzus.2005.B0147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yousaf M, Li J, Lu J, Ren T, Cong R, Fahad S, et al. Effects of fertilization on crop production and nutrient-supplying capacity under rice-oilseed rape rotation system. Sci Rep. 2017; 7:1270 10.1038/s41598-017-01412-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang Y, Tan L, Zhu Z, Yuan L, Xie D, Sun C. TOND1 confers tolerance to nitrogen deficiency in rice. Plant J. 2015; 81:367–376. 10.1111/tpj.12736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Saito K, Linquist B, Atlin GN, Phanthaboon K, Shiraiwa T, Horie T. Response of traditional and improved upland rice cultivars to N and P fertilizer in northern Laos. Field Crops Res. 2006; 96:216–223. [Google Scholar]
- 91.Mohammed N. Exploring Rice Genetic Resources to Improve Nutrient Use Efficiency. PhD thesis, University of York; 2018. Available: http://etheses.whiterose.ac.uk/20489/.
- 92.Elanchezhian R, Krishnapriya V, Pandey R, Rao AS, Abrol YP. Physiological and molecular approaches for improving phosphorus uptake efficiency of crops. Curr Sci. 2015; 108:1271–1279. [Google Scholar]
- 93.Hạnh NT, Cường PV, Pierre B. Rice nitrogen use efficiency: genetic dissection. J Sci Devel. 2013; 11:814–825. Available: http://www1.vnua.edu.vn/tapchi/Upload/27102013-07.%20Nguyen%20Thi%20Thuy%20Hanh%20814-825.pdf [Google Scholar]
- 94.Hu SK, Zeng DL, Shi SY, Ye WJ, Dong GJ, Zhu L, et al. QTL analysis of nitrogen content of plant shoot under two nitrogen conditions in rice (Oryza sativa L.). Aust J Crop Sci. 2012; 6:1737. [Google Scholar]
- 95.Wissuwa M, Yano M, Ae N. Mapping of QTLs for phosphorus-deficiency tolerance in rice (Oryza sativa L.). Theor Appl Genet. 1998; 87:777–783. [Google Scholar]
- 96.Wang B, Lan T, Wu W, Li W. Mapping of QTLs controlling chlorophyll content in rice. Yi Chuan Xue Bao. 2003; 30(12):1127–1132. [PubMed] [Google Scholar]
- 97.Tong HH, Mei HW, Yu XQ, Xu XY, Li MS, Zhang SQ, et al. Identification of related QTLs at late developmental stage in rice (Oryza sativa L.) under two nitrogen levels. Acta Genet Sin. 2006; 33:458–467. 10.1016/S0379-4172(06)60073-5 [DOI] [PubMed] [Google Scholar]
- 98.Ouyang J, Cai Z, Xia K, Wang Y, Duan J, Zhang M. Identification and analysis of eight peptide transporter homologs in rice. Plant Sci. 2010; 179:374–382. [Google Scholar]
- 99.Lian X, Xing Y, Yan H, Xu C, Li X, Zhang Q. QTLs for low nitrogen tolerance at seedling stage identified using a recombinant inbred line population derived from an elite rice hybrid. Theor Appl Genet. 2005; 112:85–96. 10.1007/s00122-005-0108-y [DOI] [PubMed] [Google Scholar]
- 100.MacMillan K, Emrich K, Piepho H-P, Mullins CE, Price AH. Assessing the importance of genotype× environment interaction for root traits in rice using a mapping population II: conventional QTL analysis. Theor Appl Genet. 2006; 113:953–964. 10.1007/s00122-006-0357-4 [DOI] [PubMed] [Google Scholar]
- 101.Obara M, Tamura W, Ebitani T, Yano M, Sato T, Yamaya T. Fine-mapping of qRL6.1, a major QTL for root length of rice seedlings grown under a wide range of NH4+ concentrations in hydroponic conditions. Theor Appl Genet. 2010; 121:535–547. 10.1007/s00122-010-1328-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dai GJ, Cheng SH, Hua ZT, Zhang ML, Jiang HB, Feng Y, et al. Mapping quantitative trait loci for nitrogen uptake and utilization efficiency in rice (Oryza sativa L.) at different nitrogen fertilizer levels. Genet Mol Res. 2015; 14:10404–10414. 10.4238/2015.September.8.1 [DOI] [PubMed] [Google Scholar]
- 103.Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, et al. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 2007; 143:1467–1483. 10.1104/pp.106.091900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.DeFalco TA, Bender KW, Snedden WA. Breaking the code: Ca2+ sensors in plant signalling. Biochem J. 2010; 425:27–40. [DOI] [PubMed] [Google Scholar]
- 105.Xing Y, Guo S, Chen X, Du D, Liu M, Xiao Y, et al. Nitrogen metabolism is affected in the nitrogen-deficient rice mutant esl4 with a calcium-dependent protein kinase gene mutation. Plant Cell Physiol. 2018; 59:2512–2525. 10.1093/pcp/pcy169 [DOI] [PubMed] [Google Scholar]
- 106.Hu R, Qiu D, Chen Y, Miller AJ, Fan X, Pan X, et al. Knock-down of a tonoplast localized low-affinity nitrate transporter OsNPF7.2 affects rice growth under high nitrate supply. Front Plant Sci. 2016; 7:1529 10.3389/fpls.2016.01529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li S, Tian Y, Wu K, Ye Y, Yu J, Zhang J, et al. Modulating plant growth–metabolism coordination for sustainable agriculture. Nature. 2018; 560:595 10.1038/s41586-018-0415-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Serrano-Mislata A, Bencivenga S, Bush M, Schiessl K, Boden S, Sablowski R. DELLA genes restrict inflorescence meristem function independently of plant height. Nat Plants. 2017; 3:749 10.1038/s41477-017-0003-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang J, Lu K, Nie H, Zeng Q, Wu B, Qian J, et al. Rice nitrate transporter OsNPF7.2 positively regulates tiller number and grain yield. Rice. 2018; 11:12 10.1186/s12284-018-0205-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tabuchi M, Sugiyama K, Ishiyama K, Inoue E, Sato T, Takahashi H, et al. Severe reduction in growth rate and grain filling of rice mutants lacking OsGS1;1, a cytosolic glutamine synthetase1; 1. Plant J. 2005; 42:641–651. 10.1111/j.1365-313X.2005.02406.x [DOI] [PubMed] [Google Scholar]
- 111.Huang Y, Guo Y, Liu Y, Zhang F, Wang Z, Wang H, et al. 9-cis-epoxycarotenoid dioxygenase 3 regulates plant growth and enhances multi-abiotic stress tolerance in rice. Front Plant Sci. 2018; 9:162 10.3389/fpls.2018.00162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yamaya T, Kusano M. Evidence supporting distinct functions of three cytosolic glutamine synthetases and two NADH-glutamate synthases in rice. J Exp Bot. 2014; 65:5519–5525. 10.1093/jxb/eru103 [DOI] [PubMed] [Google Scholar]
- 113.Mukhopadhyay P, Tyagi AK. OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Sci Rep. 2015; 5:9998 10.1038/srep09998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kan CC, Chung TY, Wu HY, Juo YA, Hsieh MH. Exogenous glutamate rapidly induces the expression of genes involved in metabolism and defense responses in rice roots. BMC Genomics. 2017; 18:186 10.1186/s12864-017-3588-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vij S, Giri J, Dansana PK, Kapoor S, Tyagi AK. The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: organization, phylogenetic relationship, and expression during development and stress. Mol Plant. 2008; 1:732–750. 10.1093/mp/ssn047 [DOI] [PubMed] [Google Scholar]
- 116.Chen G, Zou Y, Hu J, Ding Y. Genome-wide analysis of the rice PPR gene family and their expression profiles under different stress treatments. BMC Genomics. 2018; 19:720 10.1186/s12864-018-5088-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Martín AC, Del Pozo JC, Iglesias J, Rubio V, Solano R, De La Peña A, et al. Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J. 2000; 24:559–567. [DOI] [PubMed] [Google Scholar]
- 118.Sakakibara H. Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol. 2006; 57:431–449. 10.1146/annurev.arplant.57.032905.105231 [DOI] [PubMed] [Google Scholar]
- 119.Ashley MK, Grant M, Grabov A. Plant responses to potassium deficiencies: a role for potassium transport proteins. J Exp Bot. 2005; 57:425–436. 10.1093/jxb/erj034 [DOI] [PubMed] [Google Scholar]
- 120.Zhang Z, Li Q, Li Z, Staswick PE, Wang M, Zhu Y, et al. Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol. 2007; 145:450–464. 10.1104/pp.107.106021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lynch SR. Why nutritional iron deficiency persists as a worldwide problem. J Nutr. 2011; 141:763–768. [DOI] [PubMed] [Google Scholar]
- 122.Yang C, Li D, Mao D, Liu X, Ji C, Li X, et al. Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ. 2013; 2207–2218. 10.1111/pce.12130 [DOI] [PubMed] [Google Scholar]
- 123.Singh A, Jha SK, Bagri J, Pandey GK. ABA inducible rice protein phosphatase 2C confers ABA insensitivity and abiotic stress tolerance in Arabidopsis. PLoS One. 2015; 10:e0125168 10.1371/journal.pone.0125168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sato Y, Antonio BA, Namiki N, Takehisa H, Minami H, Kamatsuki K, et al. RiceXPro: a platform for monitoring gene expression in japonica rice grown under natural field conditions. Nucleic Acids Res. 2010; 39:1141–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(TIFF)
(TIFF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.