Abstract
Introduction
One of the main problems involved in heart transplantation (HT) is antibody-mediated rejection (AMR). Many aspects of AMR are still unresolved, including its etiology, diagnosis and treatment. In this project, we hypothesize that variants in genes involved in B-cell biology in HT patients can yield diagnostic and prognostic information about AMR.
Methods
Genetic variants in 61 genes related to B-cell biology were analyzed by next generation sequencing in 46 HT patients, 23 with and 23 without AMR.
Results
We identified 3 single nucleotide polymorphisms in ITGA4 gene (c.1845G>A, c.2633A>G, and c.2883C>T) that conformed the haplotype AGT-ITGA4. This haplotype is associated with the development of AMR. Moreover, AMR patients with the haplotype AGT-ITGA4 present lower levels of integrin α-4 in serum samples compared to the reference GAC haplotype in control patients.
Conclusion
We can conclude that polymorphisms in genes related to the biology of B-cells could have an important role in the development of AMR. In fact, the AGT haplotype in ITGA4 gene could potentially increase the risk of AMR.
Introduction
The spectrum of clinical rejection in heart transplantation (HT) involves both arms of the adaptive immune response, the T cell-mediated response leading to cellular rejection (CR), and the humoral response leading to antibody-mediated rejection (AMR) [1]. The humoral arm of the immune response is dominated by B-cells and production of antibodies. Administration of immunosuppressive agents enables more than 90% of heart transplant patients to survive longer than a year. However, conventional immunosuppressive agents are more effective in preventing responses by T cells than responses by B-cells and, therefore, conventional therapies have little impact on AMR [2]. Thus, the treatment of clinical and subclinical AMR remains sub-optimal and new approaches are needed to improve it.
The occurrence and severity of AMR are variable, and genetic polymorphisms that affect the magnitude and nature of the B-cell response are likely to contribute to such phenotypic variation. In fact, several studies investigated the relationship between single nucleotide polymorphisms (SNPs) in genes known to impact B-cell activation/function and antibody effector function [3,4]. In the field of transplantation, most of these studies were performed in kidney transplantation and the strongest evidence available suggests that variants affecting the expression of immunomodulatory cytokines, particularly IL-10, may impact on renal transplant outcomes [4–7]. To our knowledge, no study has thoroughly examined the relationship of variants in genes related to B-cell biology and AMR in HT.
We hypothesize that variants in exon-coding sequences and surrounding area of genes involved in B-cell biology in HT patients can yield diagnostic and prognostic information about AMR. Thus, identifying variants associated with AMR might allow risk stratification of patients and a genetic-based immunosuppression regimen.
Materials and methods
Study design and patients characteristics
This is a retrospective case-control study to evaluate a set of 61 genes related to the biology of B-cells (S1 Fig, S1 Table) and its association with AMR in HT patients from the Advanced Heart Failure and Transplant Unit of the Complejo Hospitalario Universitario de A Coruña (CHUAC). The study was carried out on 46 HT recipients, 23 with and 23 without AMR diagnosis, who underwent the transplantation between June 2000 and November 2016 [8]. Due to the different AMR criteria in the last years, in this study the patients before 2013 (n = 15) were diagnosed as: (1) allograft dysfunction (left ventricular ejection fraction <30% and/or heart failure), (2) no evidence of other causes of allograft dysfunction (acute cellular rejection or CAV), (3) evidence of complement activation on endomyocardial biopsy (C4d and/or C3d staining), and (4) favourable response to therapy addressing AMR (e.g., plasmapheresis, rituximab, steroid boluses). Whereas AMR in patients who underwent HT after 2013 (n = 8) was classified according to International Society for Heart and Lung Transplantation (ISHLT) [9]. The categories for the reporting of AMR are as follows [9]: pAMR 0-negative for pathologic AMR: histopathologic and immunopathologic studies are both negative. pAMR1(H+)-histopathologic AMR alone: histopathologic findings present and immunopathologic findings negative. pAMR1(I+)-immunopathologic AMR alone: histopathologic findings negative and immunopathologic findings positive; that is, CD68+ and/or C4d+ for IHC and C4d+ with or without C3d+ for IF. pAMR2-pathologic AMR: histopathologic and immunopathologic findings are both present. In AMR patients, the inclusion criteria was having at least one positive endomyocardial biopsy (pAMR1 or higher).
The 23 AMR cases were matched to 23 controls by gender, age (±5 years), and follow-up post-transplant. Control patients did not present any distinguishing signs of AMR (pAMR0) or allograft dysfunction. The study protocol was approved by the local Ethics Committee of "Investigación de Galicia" (Reference: 2014/012) and the samples were in the “Colección de muestras para la investigación de insuficiencia cardiaca avanzada y trasplante cardiaco” of the Instituto de Salud Carlos III (C_0000419, 2012/348). All patients gave written informed consent to participate in the study. The study conforms to the ethical guidelines of the 1975 Declaration of Helsinki.
Genetic study
DNA was extracted from clots and blood samples using QIAamp DNA Blood Mini Kit (Qiagen Inc. Hilden, Germany), as previously described [8,10]. In all cases and controls, sequencing was performed on NextSeq500 using TruSight One Sequencing Panel (Illumina, San Diego, CA, USA), which included the selected 61 genes related to the biology of B-cells (S1 Table). To assess quality of the NGS data, we define sensitivity as true positive/(true positive+ false negative), specificity as true negative/(true negative+false positive), and accuracy as (true positive+true negative)/(true positive+false positive+ false negative+true negative). To calculate these parameters, direct sequencing of 8 amplicons, containing at least one variant (S2 Table), was performed as previously described [8,10].
Databases and in silico tools used
The potential effect of SNPs associated with the presence or absence of AMR was predicted using in silico tools as previously described [8]. Moreover, the minor allele frequency (MAF) of the SNPs detected by NGS was extracted from Single Nucleotide Polymorphism Database (dbSNP) and/or The Exome Aggregation Consortium (ExAC).
Localization of variants
Topological placement of the mutations was done using the Swissprot database (http://ca.expasy.org/uniprot/). The Uniprot database provides generally accepted residue ranges corresponding with each domain region and specialized subregion.
Predicting damaging amino acid substitution
Five online tools were used to predict the pathogenicity of the missense variants: SIFT [11] (http://sift.jcvi.org/www/SIFT_seq_submit2.html), Polyphen-2 [12] (http://genetics.bwh.harvard.edu/pph2/), PhDSNP [13] (http://snps.uib.es/phd-snp/phdsnp.html), SNAP2 [14] (https://www.rostlab.org/services/snap/), and MutationTaster [15] (http://www.mutationtaster.org).
Predicting splice site variants
We evaluated the effect of synonymous variants in the vicinity (±6 pb) of the splice site, including splice site modification and splice-enhancing sequences, using 5 tools: GeneSplicer [16] (http://www.cbcb.umd.edu/software/GeneSplicer/), NetGene2 [17] (http://www.cbs.dtu.dk/services/NetGene2/), ESEfinder 3.0 [18] (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi), SSP [19] (http://www.fruitfly.org/seq_tools/splice.html), and HSF [20] (http://www.umd.be/HSF/).
Enzyme-Linked Immunosorbent Assay (ELISA) measurements
Pre-transplant serum samples were used for the determination of functional ITGA4 (n = 16) levels, using a commercially available competitive sandwich ELISA kits (ITGA4 ELISA Kit (Human) (OKEH01078) from Aviva Systems Biology (San Diego, USA)).
ITGA4 measurable concentration range was between 0.156 to 10 ng/mL. Serum samples were measured in duplicate without making any dilution. The absorbance at 450 nm was measured in a spectrophotometer.
Statistical analyses
Genotype frequencies were tested by multiple inheritance models (codominant, dominant, recessive, log-additive) using SNPStats software [21] and the association function of R software. When these models showed a significant association, McNemar’s chi-square test was performed with R. Moreover, a haplotype association analysis for SNPs described in the same gene was also realized with SNPStats and THESIAS program [22] (http://genecanvas.ecgene.net). In all analysis, P-values≤0.05 were considered to indicate statistical significance.
Results
Quality of the NGS data
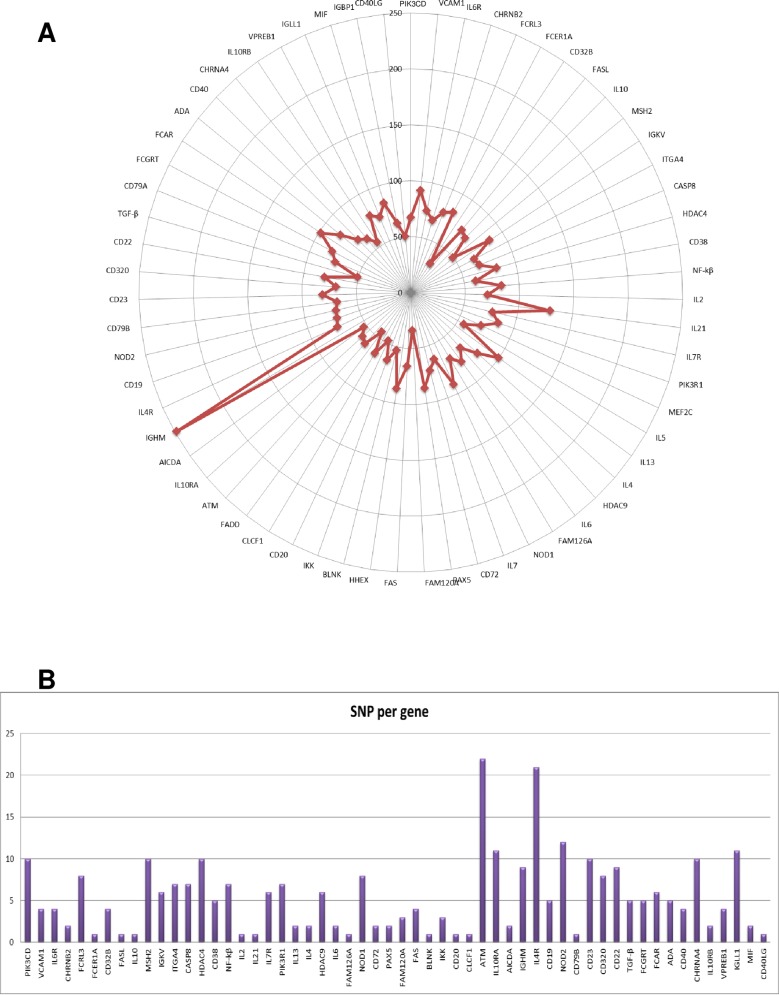
The mean coverage over all the genes related to the biology of B-cells studied was 73.5±3.6 fold (Fig 1A). Base calling accuracy, measured by the Phred quality score (Q score), indicates the probability that a given base is called incorrectly by the sequencer. The runs in the NextSeq500 platform showed a Q score > 30, which means a probability of an incorrect base call of 1 in 1000 reads and a base call accuracy 99.9%, in 80.6±2.6% of reads. The range of bases with Q30 score showed in percentage was 75–90% including the five runs performed.
Fig 1.
A, Sequence coverage. Graph representing the sequence coverage of the genes related to the complement cascade analysed in the study. Over all, mean coverage of the genes studied was 73,5±3,6. B, SNPs distribution per gene. Bar graph showing the distribution of SNPs per gene in AMR (A) and control patients (B).
To determine the accuracy, specificity, and sensitivity of the NGS protocol, we directly sequenced 8 selected amplicons containing at least one variant detected by NGS in 13 samples. From 6094 readable bases in the Sanger sequencing, we observed 6067 true negative calls and 27 true positive calls. No false positive or false negative calls were found, together resulting in a sensitivity, specificity, and accuracy of 100%.
Genotype association to AMR
The AMR patients included in the study, n = 23, and their paired control patients, n = 23, presented the clinical characteristics summarized in Table 1. Fifteen AMR patients diagnosed before 2013 presented the clinical and pathological evidence described in methods section, whereas patients diagnosed after 2013 were classified as: 4 pAMR(I+) and 4 pAMR2.
Table 1. Clinical characteristics of the patients included in the study.
| Variable | AMR (n = 23 patients) | Control (n = 23 patients) | p-value |
|---|---|---|---|
| Age±sd (years) | 50.2±16.5 | 50.0±15.9 | 0.8 |
| Male | 87.0%(20) | 87.0%(20) | 1.0 |
| Female | 13.0%(3) | 13.0%(3) | |
| Primary heart disease | |||
| Dilated cardiomyopathy | 26.1%(6) | 43.5%(10) | 0.4 |
| Ischemic cardiomyopathy | 52.2%(12) | 26.1%(6) | |
| Valvular cardiomyopathy | 8.7%(2) | 13.0%(3) | |
| Others | 13.0%(3) | 17.4%(4) | |
| Follow-up (years) | |||
| <2 years | 17.39%(4) | 4.35%(1) | 0.2 |
| 2–5 years | 30.43%(7) | 17.39%(4) | |
| ≥6 years | 52.17%(12) | 78.26%(18) | |
| Time to AMR diagnosis after transplant (years) | 3.5±0.6 | ||
| pAMR classification (n = 8)* | |||
| pAMR1(I+) | 4 | ||
| pAMR2 | 4 | ||
| Immunosupression | |||
| Steroids | 100%(23) | 100%(23) | 0.9 |
| Mycophenolate mofetil | 87.0%(20) | 95.7%(22) | |
| Basiliximab | 82.6%(19) | 95.7%(22) | |
| Cyclosporine | 56.5%(13) | 69.6%(16) | |
| Tacrolimus | 65.2%(15) | 47.4%(11) | |
| Everolimus | 26.1%(6) | 39.1%(9) | |
| Sandinmune | 20.8%(5) | 8.3%(2) | |
| Daclizumab | 4.4%(1) | 4.4%(1) | |
| Plasmapheresis | 65.2%(15) | 0 | |
| Rituximab | 65.2%(15) | 0 | |
| OKT3 | 12.5%(3) | 0 |
* pAMR classification only available for patients who underwent HT after 2013 (n = 8).
After sequencing the whole coding region and the flanking intronic region of 61 genes, we examined the frequency of 285 polymorphisms found in 23 AMR cases and/or in 23 controls (Fig 1B, S3 Table). We found statistically significant differences, after SNPStats and McNemar analysis, in the genotypes of 3 SNPs in the ITGA4 gene: c.1845G>A, c.2633A>G, and c.2883C>T (Table 2). For c.1845G>ASNP in ITGA4, the greatest significance was achieved for a dominant model where A-mutant alleles in homozygotes or heterozygotes (AA/AG) had increased odds of developing AMR compared to the GG homozygotic wild type [p = 0.002; odds ratio (OR) = 7.4; 95% confidence interval1.9–28.9;Table 2]. Moreover, a higher frequency of the A allele in AMR patients was confirmed in the codominant, recessive, and Log-additive model as well as in the McNemar test. For the second ITGA4 SNP,c.2633A>G, the lowest P-value was obtained after applying the dominant model where G-mutant alleles in homozygotes or heterozygotes (GG/AG) had increased odds of developing AMR compared to the AA homozygotic wild type (p<0.001; OR = 9.7; 95% CI 2,4–39.6;Table 2). Additionally, a higher frequency of the G allele in AMR patients was confirmed in the codominant, recessive, overdominant and Log-additive model as well as in the McNemar test. Finally, for the third ITGA4 SNP,c.2883C>T, the most significant P-value was obtained after applying the dominant model where T-mutant alleles in homozygotes or heterozygotes (TT/CT) had increased odds of developing AMR compared to the CC homozygotic wild type (p = 0.002; OR = 7.4; 95% CI 1.9–28.4; Table 2). In addition, a higher frequency of the T allele in AMR patients was confirmed in the codominant and Log-additive model as well as in the McNemar test.
Table 2. Statistical analysis of 3 variants in ITGA4 gene in control and AMR groups.
| SNP | Model | Genotype | Control | AMR | OR (95% CI) | p-value | AIC | |
|---|---|---|---|---|---|---|---|---|
| c.1845G>A | R (F(x) association) | Codominant | G/G | 14 (60.9%) | 4 (17.4%) | 1.0 | 0.003 | 58.0 |
| G/A | 8 (34.8%) | 12 (52.2%) | 5.2 (1.3–21.9) | |||||
| A/A | 1 (4.3%) | 7 (30.4%) | 24.5 (2.3–262.5) | |||||
| Dominant | G/G | 14 (60.9%) | 4 (17.4%) | 1.0 | 0.002 | 58.2 | ||
| G/A-A/A | 9 (39.1%) | 19 (82.6%) | 7.4 (1.9–28.9) | |||||
| Recessive | G/G-G/A | 22 (95.7%) | 16 (69.6%) | 1.0 | 0.01 | 61.0 | ||
| A/A | 1 (4.3%) | 7 (30.4%) | 9.6 (1.1–86.2) | |||||
| Overdominant | G/G-A/A | 15 (65.2%) | 11 (47.8%) | 1.0 | 0.2 | 66.0 | ||
| G/A | 8 (34.8%) | 12 (52.2%) | 2.0 (0.6–6.7) | |||||
| Log-additive | --- | --- | --- | 5.06 (1.74–14.70) | <0.001 | 56.0 | ||
| McNemar test | McNemar Chi squared equals 5.79 with 1 degrees of freedom | 0.02 | ||||||
| c.2633A>G | R (F(x) association) | Codominant | A/A | 17 (81.0%) | 7 (30.4%) | 1.0 | 0.001 | 53.5 |
| A/G | 4 (19.0%) | 13 (56.5%) | 7.9 (1.9–32.8) | |||||
| G/G | 0 (0%) | 3 (13.0%) | 0.0 | |||||
| Dominant | A/A | 17 (81.0%) | 7 (30.4%) | 1.0 | 0.001 | 53.0 | ||
| A/G-G/G | 4 (19.0%) | 16 (69.6%) | 9.7 (2.4–39.6) | |||||
| Recessive | A/A-A/G | 21 (100%) | 20 (87.0%) | 1.0 | 0.2 | 60.8 | ||
| G/G | 0 (0%) | 3 (13.0%) | 0.0 | |||||
| Overdominant | A/A-G/G | 17 (81.0%) | 10 (43.5%) | 1.0 | 0.009 | 58.1 | ||
| A/G | 4 (19.0%) | 13 (56.5%) | 5.5 (1.4–21.7) | |||||
| Log-additive | --- | --- | --- | 8.6 (2.2–33.3) | 0.001 | 51.7 | ||
| McNemar test | McNemar Chi squared equals 5.26 with 1 degrees of freedom | 0.02 | ||||||
| c.2883C>T | R (F(x) association) | Codominant | C/C | 14 (60.9%) | 4 (17.4%) | 1.0 | 0.006 | 59.6 |
| C/T | 7 (30.4%) | 12 (52.2%) | 6.0 (1.4–25.6) | |||||
| T/T | 2 (8.7%) | 7 (30.4%) | 12.2 (1.8–84.0) | |||||
| Dominant | C/C | 14 (60.9%) | 4 (17.4%) | 1.0 | 0.002 | 58.2 | ||
| C/T-T/T | 9 (39.1%) | 19 (82.6%) | 7.4 (1.9–28.9) | |||||
| Recessive | C/C-C/T | 21 (91.3%) | 16 (69.6%) | 1.0 | 0.06 | 64.1 | ||
| T/T | 2 (8.7%) | 7 (30.4%) | 4.6 (0.8–25.2) | |||||
| Overdominant | C/C-T/T | 16 (69.6%) | 11 (47.8%) | 1.0 | 0.1 | 65.5 | ||
| C/T | 7 (30.4%) | 12 (52.2%) | 2.5 (0.7–8.3) | |||||
| Log-additive | --- | --- | --- | 3.9 (1.5–10.4) | 0.002 | 58.2 | ||
| McNemar test | McNemar Chi squared equals 9.39 with 1 degrees of freedom | 0.002 | ||||||
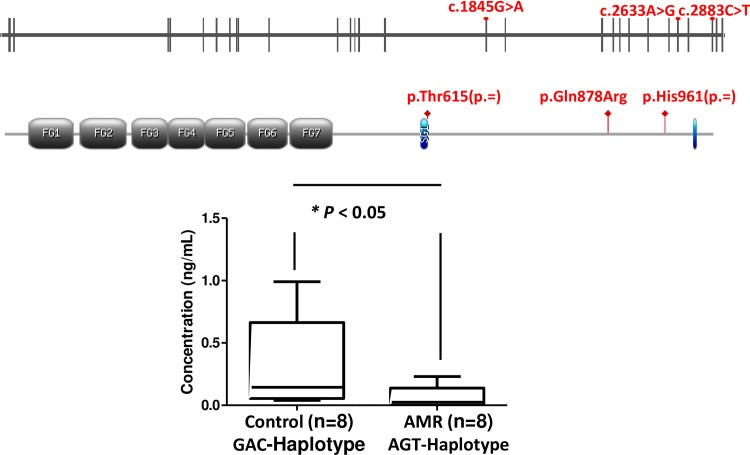
A haplotype association analysis for these 3 SNPs in each subpopulation was then performed (S4 Table). The four observed haplotypes were analyzed: 1.) c.1845G-c.2633A-c.2883C haplotype (GAC); 2.) c.1845A-c.2633G-c.2883T haplotype (AGT), which is a variant with minor allele frequency in the 3 SNPs; 3.) c.1845A-c.2633A-c.2883T haplotype (AAT); and 4.) and c.1845G-c.2633A-c.2883T haplotype (GAT). We found differences in haplotype frequencies to be statistically significant for AGT haplotype (p = 0.005 in SNPstats and p<0.001 in THESIAS) being more frequent in cases (41.3%) than in controls (12.4%). Furthermore, haplotype analysis suggests that the AGT haplotype is associated with the development of AMR with an OR of 11.0 (95% CI 2.3–53.6) and 7.3 (95% CI 2.5–21.3) in SNPstats and THESIAS respectively.
In silico analysis of ITGA4 variants associated with the presence or absence of AMR in genes related to B-cell biology
Although all the variants associated with the presence or absence of AMR in genes related to B-cell biology found in this study were polymorphisms previously known, we used several in silico software in order to predict an impact on the function of the protein (Table 3).
Table 3. ITGA4 variants associated to AMR.
| cDNA | dbSNP | Chr 2 position | Protein | 1kg_maf | Effect | SIFT | POLYPHEN | PhDSNP | MutationTaster | SNAP2 | Grantham score | SSP | HSF | ESEfinder | GENESCAN | NetGene2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| c.1845G>A | rs1143674 | 182374534 | p.Thr615(p=) | 0.3813 (A) | synonymous | - | - | - | - | - | - | New splicing site (0.9) | 43.2 vs 72.2 (+67%) | 6.8 vs 7.3 (+16%) | 7.2 vs 13.1 (+81.5%) | New splicing site (0.8) |
| c.2633A>G | rs1143676 | 182395345 | p.Gln878Arg | 0.2633 (G) | missense | tolerated(0.61) | benign(0.04) | Neutral | Polymorphism(1) | Effect(63%) | 43 | - | - | - | - | - |
| c.2883C>T | rs7562325 | 182399097 | p.His961(p=) | 0.3737 (T) | synonymous | - | - | - | - | - | - | 0.8 vs 0.6 (-30.1%) | 86.6 vs 86.8 (+0.2) | 7.3 vs 8.2 (+12.4%) |
5.0 vs 4.0 (-20%) |
0.6 vs 0.3 (-51.6%) |
The missense polymorphism (p.Gln878Arg-ITGA4) does not seem to alter the function of the protein (Table 3). In the case of synonymous polymorphisms in the vicinity of splice site (Table 3), we used the Houdayer et al criteria of a decrease of at least 20% in the score of the predictions made to consider a possible effect on the splicing process [23]. Only the variant p.His961(p=) in ITGA4 gene was predicted to have a possible impact on the corresponding natural splice site, by 3 out of 5 software used. However, the synonymous variant p.Thr615(p=) could modify the exonic splicing enhancer, and thus, it could have an impact in protein expression.
Serum integrin α-4 levels in AMR transplant patients with ITGA4-AGT haplotype
The median integrin α-4 serum level measured before transplantation in 8 AMR patients that carried the AGT haplotype was 0.02 ng/mL ([IQR] 0.00–0.1 ng/mL) (Fig 2). This value is significant lower (p<0.05) compared to the median integrin α-4 serum level measured before transplantation in 8 patients who did not present with AMR and in whom the analyzed haplotype was not present (0.1 ng/mL; [IQR] 0.05–0.7 ng/mL).
Fig 2. ITGA4 gene.
Schematic structure of the human ITGA4 gene and primary protein domains of ITGA4 showing relative position of the 3 common polymorphism found (c.1845G>A), c.2633A>G), c.2883C>T). ITGA4 serum levels according to ITGA4 haplotype among 8 AMR patients (AGT) and 8 control patients (GAC).
Discussion
The present data represents the first study on the effect of variants on genes involved in the B-cell biology related to AMR in HT patients to date. We have identified three SNPs in ITGA4 gene [p.Thr615(p=), p.Gln878Arg, and p.His961(p=)], which correlates with the development of AMR. These 3 SNPs conform a haplotype: AGT-ITGA4, which could be associated with development of AMR in HT patients. Our data showed that the AGT haplotype in ITGA4 gene in AMR patients present lower levels of integrin α-4 in serum samples compared to the reference GAC haplotype in control patients. In the past decade, efforts to understand how an individual´s genetic can impact on disease susceptibility, severity and responses to treatment have progressed rapidly and the AGT haplotype described in this study could open a new approach to AMR.
Cell adhesion is critical in immune system function, and α4 integrin (also named as CD49d or VLA4), encoded by ITGA4 gene, plays a particularly prominent role in the immune system through its adhesive and signalling functions [24]. These functions include immune cell trafficking, activation of myeloid cells and naïve T and B lymphocytes, differentiation of effector T cells into Th1, Th2, or Th16, immunological synapse and binding of α4 integrin provides costimulatory signals to T cells [25]. Moreover, α4 integrin mediates B-cell localization in the bone marrow and in germinal centers of secondary lymphoid organs. α4 integrin is also involved in survival signalling pathway that inhibits apoptosis of germinal center B-cells through the up-regulation of the antiapoptotic B-cell lymphoma gene family member, Bcl-XL [24]. The prominent role of integrins in the immune system is also demonstrated because they have been implicated in the pathogenesis of several autoimmune and inflammatory diseases, including asthma, multiple sclerosis, contact hypersensitivity, rheumatoid arthritis, and Crohn´s disease [26–29].
In the field of transplantation, most of the studies focus on integrins were done in CR due to their role in T-cells. It has been described that αvβ6, which belong to another family of integrins but with similar functions to α4 integrin, are increased in acute CR of heart allograft and cardiac allograft vasculopathy in humans [30,31]. In two models of rat kidney transplantation, αv integrin antagonist was able to significantly ameliorate the infiltration of CD8+ T cells and macrophages into the graft and inhibit proliferation of mononuclear cells and fibroblasts in the interstitium of kidney allografts [32]. Treatment with anti-α4 antibody in islet allografts prolongs graft survival, suggesting that the α4 adhesion system may be important in antigen-presentation, T cell activation and lymphocyte recruitment to the graft [33]. Moreover, previous studies have demonstrated that anti-α4 integrin antibody treatment prolongs, but not indefinitely, the survival of cardiac allografts in animal models [34,35].
Our study is the first analysis of ITGA4 genotype and α4 integrin expression focus on AMR patients. In our cohort, the haplotype AGT present a higher frequency in patients with AMR and it is associated with lower serum levels of α4 integrin. To explain a possible role of α4 integrin in AMR, we have to take into account that α4 integrin are expressed at higher levels by B-cell precursors than mature B-cells [36]. Thus, we can speculate that our lower serum levels in AMR patients could be due the presence of less B-cell precursors and more mature B-cells and these mature B-cells would secrete antibodies that bind to antigens and activate complement, responsible for the AMR [37]. Moreover, Weinländer et al described that inhibition of endothelial cell spreading and migration by inflammatory cytokines is mediated by human guanylate binding protein-1 (GBP-1) through induction of ITGA4 expression [38]. Thus, a down regulation of ITGA4 gene could have an impact in GBP1 induction, which in turn may result in a higher endothelial cell proliferation and migration during AMR. Furthermore, a down-regulation of the ITGA4 gene has also been described in other diseases, such as Crohn´s disease [39] or the first stages of B-cell chronic lymphocytic leukemia [40]. Interestingly, in the same line as our haplotype decreases the expression of ITGA4, 2 different SNPs (rs7023923 and rs6740847) downregulate ITGA4 expression (Maugeri et al 2011). However, more research must be done to validate this hypothesis and the role of the AGT haplotype in AMR. If so, these results can open a new approach of the prevention of AMR because natalizumab, a humanized monoclonal antibody against the cell adhesion molecule α4-integrin approved by FDA for multiple sclerosis and Crohn's disease, could be a candidate therapy for preventing AMR.
Limitations
The current study presents some limitations including a retrospective design, a relatively small sample size, and an evolving criteria of AMR definition. However, we believe that this study opens a new pathway of understanding that variants in genes related to the B cell biology may influence the development of AMR, a field not yet explored.
Conclusions
The main results of this study are: 1.) AGT haplotype is associated with the development of AMR and 2.) integrin α-4 serum levels in patients carried AGT haplotype are lower that patients carried the reference GAC haplotype. Thus, polymorphisms in the genes related to the biology of B-cells could have an important role in the development of AMR in cardiac transplants. Specifically, the AGT haplotype in ITGA4 gene could increase the risk of AMR. However, the detailed molecular mechanisms on how this haplotype exerts effects in the immune response leading to AMR are unclear and more research must be done to assess its role.
Supporting information
In the bone marrow, development progresses through the pro-B cell and pre-B cell, immature-B cell stages and the most important genes involved are showed. During this differentiation, rearrangements at the immunoglobulin locus result in the generation and surface expression of the pre-B cell receptor and finally a mature B cells that are capable of binding antigen. The 61 genes analysed in this study are showed in bold.
(TIF)
(DOC)
(DOC)
(DOC)
(DOC)
Acknowledgments
We thank Zulaika Grille-Cancela and Paula Blanco-Canosa for their contributions with samples and database records and David Couto-Mallón and Gonzalo Barge-Caballero for the clinical follow-up of the patients.
Abbreviations
- AMR
antibody-mediated rejection
- CR
cellular rejection
- dbSNP
Single Nucleotide Polymorphism Database
- ELISA
Enzyme-Linked Immunosorbent Assay
- ExAC
The Exome Aggregation Consortium
- FN
false-negative
- FP
false-positive
- HT
heart transplantation
- MAF
minor allele frequency
- OR
odds ratio
- SNPs
single nucleotide polymorphisms
- TP
true-positive
- TN
true-negative
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by a grant from Instituto de Salud Carlos III (PI13/02174) and it is part of the research activities of the “Centro de investigación Biomédica en Red Enfermedades Cardiovasculares (CIBERCV)”. Co-financed with FEDER Funds.
References
- 1.Tan CD, Baldwin WM 3rd, Rodriguez ER. Update on cardiac transplantation pathology. Arch Pathol Lab Med. 2007; 131: 1169–1191. 10.1043/1543-2165(2007)131[1169:UOCTP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 2.Platt JL, Tsuji S, Cascalho M. Novel functions of B-cells in transplantation. Curr Opin Organ Transplant. 2011; 16: 61–68. 10.1097/MOT.0b013e328342551c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z, Wang K, Yang H, Han Z, Tao J, Chen H, et al. Associations between HVEM/LIGHT/BTLA/CD160 polymorphisms and the occurrence of antibody-mediate rejection in renal transplant recipients. Oncotarget. 2017; 8: 100079–100094. 10.18632/oncotarget.21941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banham GD, Clatworthy MR. B-cell biomarkers in transplantation—from genes to therapy. Tissue Antigens. 2015; 85: 82–92. 10.1111/tan.12520 [DOI] [PubMed] [Google Scholar]
- 5.Alakulppi NS, Kyllönen LE, Jäntti VT, Matinlauri IH, Partanen J, Salmela KT, et al. Cytokine gene polymorphisms and risks of acute rejection and delayed graft function after kidney transplantation. Transplantation. 2004; 78: 1422–1428. [DOI] [PubMed] [Google Scholar]
- 6.Thakkinstian A, Dmitrienko S, Gerbase-Delima M, McDaniel DO, Inigo P, Chow KM, et al. Association between cytokine gene polymorphisms and outcomes in renal transplantation: a meta-analysis of individual patient data. Nephrol Dial Transplant. 2008; 23: 3017–3023. 10.1093/ndt/gfn185 [DOI] [PubMed] [Google Scholar]
- 7.Khan F, Sar A, Gonul I, Benediktsson H, Doulla J, Yilmaz S, et al. Graft inflammation and histologic indicators of kidney chronic allograft failure: low-expressing interleukin-10 genotypes cannot be ignored. Transplantation. 2010; 90: 630–638. 10.1097/TP.0b013e3181ea391e [DOI] [PubMed] [Google Scholar]
- 8.Marrón-Liñares GM, Núñez L, Crespo-Leiro MG, Barge-Caballero E, Pombo J, Paniagua-Martin MJ, et al. Polymorphisms in genes related to the complement system and antibody mediated cardiac allograft rejection. J Heart Lung Transplant. 2018; 37: 477–485. 10.1016/j.healun.2017.07.006 [DOI] [PubMed] [Google Scholar]
- 9.Berry GJ, Burke MM, Andersen C, Bruneval P, Fedrigo M, Fishbein MC, et al. The 2013 International Society for Heart and Lung Transplantation Working Formulation for the standardization of nomenclature in the pathologic diagnosis ofantibody-mediated rejection in heart transplantation. J Heart Lung Transplant. 2013; 32: 1147–1162. 10.1016/j.healun.2013.08.011 [DOI] [PubMed] [Google Scholar]
- 10.Núñez L, Crespo-Leiro MG, Marrón-Liñares GM, Suarez-Fuentetaja N, Barge-Caballero E, Paniagua-Martín MJ, et al. Analysis of variants in the HCN4 gene and in three single nucleotide polymorphisms of the CYP3A4 gene for association with ivabradine reduction in heart rate: A preliminary report. Cardiol J. 2016; 23: 573–582. 10.5603/CJ.a2016.0050 [DOI] [PubMed] [Google Scholar]
- 11.Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012; 40: W452–457. 10.1093/nar/gks539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010; 7: 248–249. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capriotti E, Calabrese R, Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics. 2006; 22: 2729–2734. 10.1093/bioinformatics/btl423 [DOI] [PubMed] [Google Scholar]
- 14.Bromberg Y & Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007; 35: 3823–3835. 10.1093/nar/gkm238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010; 7: 575–576. 10.1038/nmeth0810-575 [DOI] [PubMed] [Google Scholar]
- 16.Pertea M, Lin X, Salzberg SL. GeneSplicer: A new computational method for splice site prediction. Nucleic Acids Res. 2001; 29: 1185–1190. 10.1093/nar/29.5.1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hebsgaard SM, Korning PG, Tolstrup N, Engelbrecht J, Rouze P, Brunak S. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 1996; 24: 3439–3452. 10.1093/nar/24.17.3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003; 31: 3568–3571. 10.1093/nar/gkg616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved Splice Site Detection in Genie. J Comp Biol. 1997; 4: 311–323. [DOI] [PubMed] [Google Scholar]
- 20.Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acid Research. 2009;37:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006; 22: 1928–1929. 10.1093/bioinformatics/btl268 [DOI] [PubMed] [Google Scholar]
- 22.Tregouet DA, Garelle V. A new JAVA interface implementation of THESIAS: Testing Haplotypes Effects in Association Studies. Bioinformatics. 2007; 23: 1038–1039. 10.1093/bioinformatics/btm058 [DOI] [PubMed] [Google Scholar]
- 23.Houdayer C, Dehainault C, Mattler C, Michaux D, Caux-Moncoutier V, Pagès-Berhouet S, et al. Evaluation of in silico splice tools for decision-making in molecular diagnosis. Hum Mutat. 2008; 29: 975–982. 10.1002/humu.20765 [DOI] [PubMed] [Google Scholar]
- 24.Rose DM, Han J, Ginsberg MH. Alpha4 integrins and the immune response. Immunol Rev. 2002; 186: 118–124. [DOI] [PubMed] [Google Scholar]
- 25.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002; 110: 673–687. 10.1016/s0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- 26.Yednock TA, Cannon C, Fritz LC, Sanchez-Madrid F, Steinman L, Karin N. Prevention of experimental autoimmune encephalomyelitis by antibodies against alpha 4 beta 1 integrin. Nature. 1992; 356: 63–66. 10.1038/356063a0 [DOI] [PubMed] [Google Scholar]
- 27.Abraham WM, Sielczak MW, Ahmed A, Cortes A, Lauredo IT, Kim J, et al. Alpha 4-integrins mediate antigen-induced late bronchial responses and prolonged airway hyperresponsiveness in sheep. J Clin Invest. 1994; 93: 776–787. 10.1172/JCI117032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lobb RR, Hemler ME. The pathophysiologic role of alpha 4 integrins in vivo. J Clin Invest. 1994; 94: 1722–1728. 10.1172/JCI117519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chisholm PL, Williams CA, Lobb RR. Monoclonal antibodies to the integrin alpha-4 subunit inhibit the murine contact hypersensitivity response. Eur J Immunol. 1993; 23: 682–688. 10.1002/eji.1830230317 [DOI] [PubMed] [Google Scholar]
- 30.Yamani MH, Yang J, Masri CS, Ratliff NB, Bond M, Starling RC, et al. Acute cellular rejection following human heart transplantation is associated with increased expression of vitronectin receptor (integrin alphavbeta3). Am J Transplant. 2002; 2: 129–133. [DOI] [PubMed] [Google Scholar]
- 31.Yamani MH, Masri CS, Ratliff NB, Bond M, Starling RC, Tuzcu EM, et al. The role of vitronectin receptor (alphavbeta3) and tissue factor in the pathogenesis of transplant coronary vasculopathy. J Am Coll Cardiol. 2002; 39: 804–810. 10.1016/s0735-1097(01)01823-x [DOI] [PubMed] [Google Scholar]
- 32.Bedke J, Kiss E, Behnes CL, Popovic ZV, Heuser M, Stojanovic T, et al. Anti-inflammatory effects of alphav integrin antagonism in acute kidney allograft rejection. Am J Pathol. 2007; 171: 1127–1139. 10.2353/ajpath.2007.070329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stegall MD, Dean PG, Ninova D, Cohen AJ, Shepard GM, Gup C, et al. Alpha4 integrin in islet allograft rejection. Transplantation. 2001; 71: 1549–1555. [DOI] [PubMed] [Google Scholar]
- 34.Paul LC, Davidoff A, Benediktsson H, Issekutz T. Anti-integrin (LFA-1, VLA-4, and Mac-1) antibody treatment and acute cardiac graft rejection in the rat. Transpl Int. 1996; 9: 420–425. [DOI] [PubMed] [Google Scholar]
- 35.Isobe M, Suzuki J, Yagita H, Okumura K, Yamazaki S, Nagai R, et al. Immunosuppression to cardiac allografts and soluble antigens by anti-vascular cellular adhesion molecule-1 and anti-very late antigen-4 monoclonal antibodies. J Immunol. 1994; 153: 5810–5818. [PubMed] [Google Scholar]
- 36.Ryan DH, Nuccie BL, Abboud CN, Winslow JM. Vascular cell adhesion molecule-1 and the integrin VLA-4 mediate adhesion of human B-cell precursors to cultured bone marrow adherent cells. J Clin Invest. 1991; 88: 995–1004. 10.1172/JCI115403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang R. Donor-Specific Antibodies in Kidney Transplant Recipients. Clin J Am Soc Nephrol. 2018; 13: 182–192. 10.2215/CJN.00700117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weinländer K, Naschberger E, Lehmann MH, Tripal P, Paster W, Stockinger H, et al. Guanylate binding protein-1 inhibits spreading and migration of endothelial cells through induction of integrin alpha4 expression. FASEB J. 2008; 22: 4168–4178. 10.1096/fj.08-107524 [DOI] [PubMed] [Google Scholar]
- 39.Burakoff R, Hande S, Ma J, Banks PA, Friedman S, Makrauer F, et al. Differential regulation of peripheral leukocyte genes in patients with active Crohn's disease and Crohn's disease in remission. J Clin Gastroenterol. 2010; 44: 120–126. 10.1097/MCG.0b013e3181a9ef53 [DOI] [PubMed] [Google Scholar]
- 40.Eksioğlu-Demiralp E, Alpdoğan O, Aktan M, Firatli T, Oztürk A, Budak T, et al. Variable expression of CD49d antigen in B-cell chronic lymphocytic leukemia is related to disease stages. Leukemia. 1996; 10: 1331–1339. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In the bone marrow, development progresses through the pro-B cell and pre-B cell, immature-B cell stages and the most important genes involved are showed. During this differentiation, rearrangements at the immunoglobulin locus result in the generation and surface expression of the pre-B cell receptor and finally a mature B cells that are capable of binding antigen. The 61 genes analysed in this study are showed in bold.
(TIF)
(DOC)
(DOC)
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.