Abstract
BMP/SMAD signaling is a crucial regulator of intestinal differentiation1–4. However, the molecular underpinnings of the BMP pathway in this context are unknown. Here, we characterize the mechanism by which BMP/SMAD signaling drives enterocyte differentiation. We establish that the transcription factor HNF4A acts redundantly with an intestine-restricted HNF4 paralog, HNF4G, to activate enhancer chromatin and upregulate the majority of transcripts enriched in the differentiated epithelium; cells fail to differentiate upon double knockout of both HNF4 paralogs. Furthermore, we show that SMAD4 and HNF4 function via a reinforcing feed-forward loop, activating each other’s expression and co-binding to regulatory elements of differentiation genes. This feed-forward regulatory module promotes and stabilizes enterocyte cell identity; disruption of the HNF4-SMAD4 module results in loss of enterocyte fate in favor of progenitor and secretory cell lineages. This intersection of signaling and transcriptional control provides a framework to understand regenerative tissue homeostasis, particularly in tissues with inherent cellular plasticity5.
Cellular identity arises from the combined activity of pioneer factors, which provide access to tissue-specific regulatory elements, and tissue-restricted transcription factors, which assemble at these accessible elements to control tissue-specific gene expression6. To define regulators of intestinal homeostasis, we employed ATAC-seq to identify chromatin-accessible regions in the intestinal duodenal epithelium that are inaccessible in other tissues7 (Supplementary Fig. 1a). We applied DNA-binding motif analysis to identify transcription factors operating at these 4,575 intestine-specific regions (Supplementary Fig. 1a and Supplementary Table 1) and found that the HNF4A/G (hepatocyte nuclear factor 4 alpha/gamma) motif was the top-scoring factor, suggesting that HNF4 is a fundamental regulator of intestine-specific gene expression. HNF4A has been implicated prominently in the human genetics of Inflammatory Bowel Disease8 and Colorectal Cancer9,10. ChIP-seq studies have shown HNF4 can bind to differentiation genes; paradoxically, HNF4A is largely dispensable in the intestinal epithelium in mouse models11–13. We hypothesized that the extent and magnitude of HNF4A’s functions have been masked by genetic redundancy with the paralog HNF4G, and further analysis7,14 shows highly selective Hnf4γ expression in the intestine (Supplementary Fig. 1b–d). HNF4A and HNF4G also exhibited an overlapping immunofluorescence profile in the intestine (Fig. 1a). Using paralog-specific antibodies (Supplementary Fig. 2a), we compared HNF4A and HNF4G binding to the intestinal epithelial genome using ChIP-seq. Strikingly similar binding profiles were observed (Fig. 1b and Supplementary Fig. 2b), with the vast majority of binding sites (98.7%) identified as common between HNF4A and HNF4G (7,193 sites, FDR < 0.05, DiffBind15, Fig. 1c and Supplementary Fig. 2c). HOMER analysis of HNF4A and HNF4G ChIP-seq-defined binding regions (Supplementary Fig. 2d) were enriched with nearly identical DNA-binding motifs, remarkably similar to previously defined HNF4 binding sequence preferences16. Taken together, the similar expression and DNA-binding profiles of the HNF4 paralogs justified further exploration of their possible genetic redundancy.
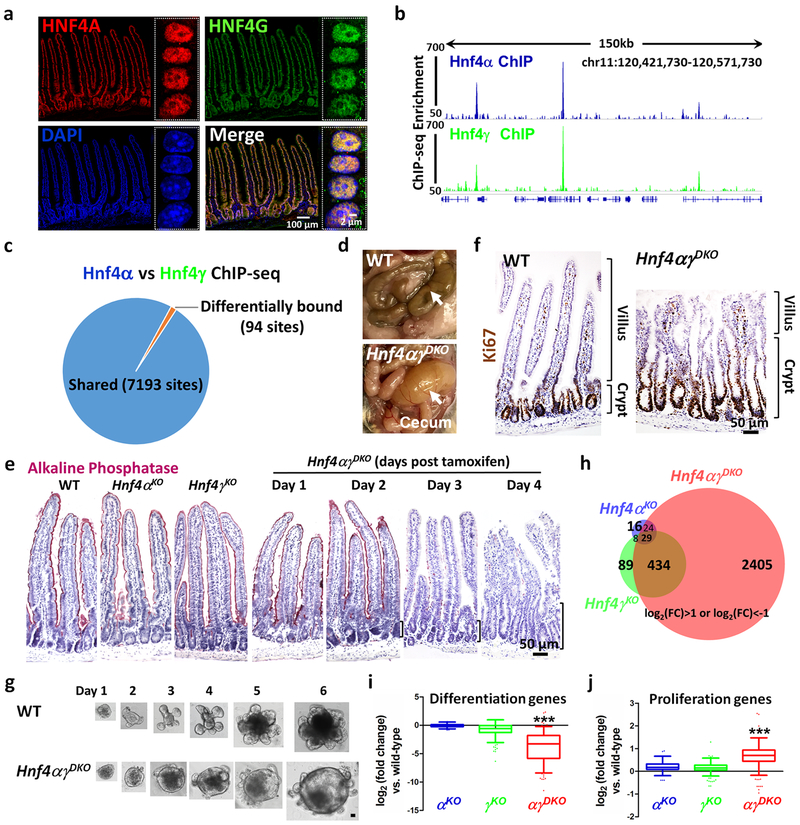
Figure 1. HNF4A and HNF4G are redundantly required to drive intestinal differentiation.
a, Co-localization of HNF4A and HNF4G in intestinal epithelial cells by immunofluorescence confocal microscopy (n = 3 biologically independent mice). The high-resolution images are of villus epithelial cell nuclei as shown in the boxed insets. ChIP-seq reveals strikingly similar binding profiles of HNF4A and HNF4G in the duodenal epithelial cells. b, Example of ChIP-seq tracks. c, DiffBind analysis (FDR < 0.05, n = 2 biologically independent mice) shows that 7,193 sites are shared and only 94 sites are differentially bound by HNF4A and HNF4G. Statistical tests were embedded in DiffBind. d, Necropsy reveals a distended and fluid-filled intestine in the Hnf4αγDKO mouse after 4 days of tamoxifen injection (n = 10 independent experiments). e, Alkaline Phosphatase staining (differentiation marker, pink color, n = 4 biologically independent mice). f, Immunostaining of Ki67 (proliferation marker, brown nuclei). Brackets show elongating crypts in the double mutants (n = 4 biologically independent mice). The proliferation zone is defined by the distribution of Ki67-positive cells. g, In contrast to WT, Hnf4αγDKO organoids show a spherical morphology and fewer lumenal contents, consistent with a failure to differentiate (n = 6 independent experiments). Scale bar, 50 μm. h, Only Hnf4αγDKO mice show a striking alteration of gene expression as evidenced by RNA-seq (n = 3 biologically independent mice). Venn diagram shows the numbers of genes in HNF4 single and double mutants with log2 fold-change > 1 or < −1, FDR < 0.05. Statistical tests were embedded in Cuffdiff. Differentiation genes (i) are reduced while proliferation genes (j) are elevated in the Hnf4αγDKO mice. The middle line represents the median; whiskers represent the 10th and 90th percentile. Post-hoc Dunn’s test was applied following a Kruskal-Wallis test at P < 0.001 ***; n = 3 biologically independent mice).
While previous reports have indicated that Hnf4γ mutants are viable17,18, analysis of intestines lacking HNF4G is incomplete. We generated Hnf4γKO mice via CRISPR-mediated editing (Supplementary Fig. 3a–c). Animals developed normally without HNF4G, were fertile, and showed no gross abnormalities in the histopathology or transcriptome of the intestine (Supplementary Fig. 3d–f). The relatively modest phenotype observed upon loss of either HNF4A11–13,19 or HNF4G indicated that these factors could function redundantly. We therefore generated double mutant Villin-CreERT2; Hnf4αf/f; Hnf4γCrispr/Crispr mice. Hnf4αγDKO adults showed rapid weight loss upon tamoxifen treatment (Supplementary Fig. 4a) and had to be euthanized within 5 days, exhibiting fluid-filled intestines, indicative of intestinal malfunction (Fig. 1d). Expression of differentiation markers, such as alkaline phosphatase and KRT20, indicated that differentiation was severely disrupted in the Hnf4αγDKO mutants (Fig. 1e and Supplementary Fig. 4b,c). Dramatic expansion of the proliferation zone (Fig. 1f and Supplementary Fig. 4d,e) suggested that homeostatic balance was lost in the Hnf4αγDKO mice, with expansion of the progenitor cell zone at the expense of cellular differentiation. Organoids derived from the Hnf4αγDKO mutants grew in a manner consistent with an expansion of proliferative progenitors and a deficit of differentiated cells, with less debris in their lumens (Fig. 1g). Together, these findings reveal redundant functions between HNF4 paralogs in driving intestinal differentiation.
The molecular consequences of HNF4 redundancy were appreciated through transcriptome analysis. Over 2,892 genes were significantly altered in the Hnf4αγDKO epithelium, whereas HNF4G or HNF4A loss yielded only 560 or 77 differentially expressed genes, respectively (FDR < 0.05, |log2(fold change)|>1) (Fig. 1h and Supplementary Fig. 4f). Genes downregulated in the Hnf4αγDKO mutants were enriched for ontologies associated with enterocyte functions in digestive metabolism, enterocyte morphology, and immune signaling (Supplementary Fig. 4g). The number of gene expression changes and their magnitude in the double mutant (Fig. 1h and Supplementary Fig. 4f), versus relatively subtle differences in the single mutants (Supplementary Fig. 5), suggest largely redundant functions between HNF4 factors. Hnf4αγDKO epithelium exhibits a robust shift in the transcriptome away from differentiated cell transcripts20 and towards proliferating cell transcripts20 (Fig. 1i,j, Supplementary Fig. 4h and Supplementary Table 2).
Nearly 45% of genes with reduced expression in the Hnf4αγDKO intestines showed binding of HNF4 within 20 kb, which implies that HNF4 factors directly trans-activate these genes. Conversely, genes upregulated upon loss of both HNF4 paralogs showed no significant enrichment of binding by HNF4 factors (Fig. 2a,b), suggesting that HNF4 binds and activates enterocyte differentiation genes and indirectly suppresses crypt/proliferation (examples illustrated in Supplementary Fig. 4i). Association between HNF4 binding and linked gene expression diminished with distance (Fig. 2c), and the frequency of HNF4 binding events at a gene correlated to the magnitude of linked gene expression changes (Fig. 2d). Expression of HNF4 bound genes showed greater changes in transcript levels than unbound genes in Hnf4αγDKO (Fig. 2e). GO analysis revealed that HNF4 direct-target genes (1,036 genes with log2 FC < −1 and FDR < 0.05, within 30 kb of HNF4 binding sites) function in processes typical of differentiated enterocytes, whereas downregulated genes lacking HNF4 binding sites were more enriched in immune-related functions, suggesting an indirect role for HNF4 factors (Fig. 2f). HNF4-binding regions also exhibited properties of transcriptional enhancers, including ChIP-seq enrichment for chromatin features and co-regulators typically found at enhancers (Supplementary Fig. 6a,b). To explore how HNF4 factors were activating differentiation genes, we profiled enhancer chromatin structures in control and Hnf4αγDKO mutants using H3K27ac-MNase-ChIP-seq. HNF4 binding was most abundant at regions losing H3K27ac signal in the Hnf4αγDKO (9,832 sites, 24.0% of which bind HNF4), whereas regions gaining H3K27ac signal showed the least HNF4 binding (9,974 sites, 2.4% of which bind HNF4, Fig. 2g). This suggests that HNF4 factors function to maintain active enhancer chromatin but not to directly repress chromatin. H3K27ac signal was observed at nucleosomes flanking HNF4 binding sites in control mice, and this pattern was disrupted upon loss of HNF4 factors (Fig. 2h,i). Conversely, H3K27ac levels at promoters were unaffected in the Hnf4αγDKO mutants (Supplementary Fig. 6c). These findings indicate that HNF4 drives differentiation by activating gene expression in the intestinal epithelium through binding to distal enhancer regions and maintaining enhancer chromatin activity.
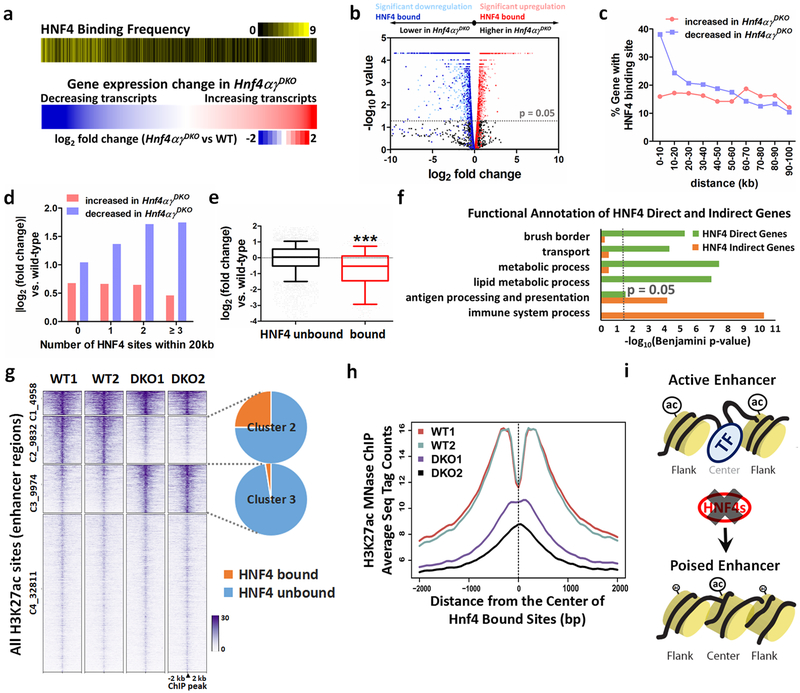
Figure 2. HNF4 binding is required to activate enhancer chromatin and stimulate genes required for intestinal differentiation.
HNF4 factors function primarily to activate rather than repress transcription, as evidenced by (a) heatmaps (Kolmogorov-Smirnov test, one-sided for positive and negative enrichment scores, P < 0.001). The lower heatmap displays a log2 fold-change in gene expression (n = 3 biologically independent mice). The upper heatmap shows the corresponding frequency (yellow shading) of HNF4 binding sites located within 20 kb of the TSSs of the corresponding genes in 10-gene binned groups displayed in the expression heatmap. b, Volcano plot of gene expression changes (n = 3 biologically independent mice). Significant genes were called via Cuffdiff. c, Distribution of TSS distances in 10-kb windows from the nearest HNF4 binding sites. Genes with an FDR < 0.05 are determined as either decreased or increased in the Hnf4αγDKO duodenum compared to the controls (n = 3 biologically independent mice). Downregulated genes are more likely to have nearby HNF4 binding sites than upregulated genes. d, Genes that decrease in HNF4-deficient epithelium are more likely to harbor multiple HNF4 binding events than genes that show increased expression (n = 3 biologically independent mice). e, HNF4 bound genes are significantly downregulated in Hnf4αγDKO compared to unbound genes. The middle line represents the median; whiskers represent the 10th and 90th percentile (Mann-Whitney test, two-sided at P < 0.001 ***; n = 3 biologically independent mice). f, Functional annotation of HNF4 direct and indirect genes (log2 fold-change < −1, FDR < 0.05) by DAVID. Direct genes: HNF4 bound genes (genes within 30 kb of annotated TSSs nearby HNF4 binding sites). Indirect genes: HNF4 unbound genes. Benjamini P values were calculated using DAVID. g, MNase-ChIP-seq profiles (k-means = 4, MACS P ≤ 10−5) of H3K27ac. Statistical tests were embedded in the MACS package. h, Aggregate plots of H3K27ac ChIP-Seq signal are centered on the 5,952 enhancer sites that are bound by HNF4, and loss of HNF4 paralogs prominently reduces H3K27ac signal. i, Model of HNF4-control of active chromatin structures in the intestine.
BMP/SMAD signaling is well-recognized as a driver of intestinal differentiation1–4, though its molecular mechanisms are unclear. Given HNF4’s potent role in driving the differentiation program (Figs. 1,2 and Supplementary Fig. 7), we wondered whether HNF4 and BMP/SMAD could be working in concert. Highly similar transcriptome changes occurred upon loss of the BMP-effector transcription factor SMAD4 and in our Hnf4αγDKO model, suggesting that these factors could be working in the same pathway (Fig. 3a). Interestingly, transcripts of SMAD factors and BMP ligands were dramatically reduced upon HNF4 loss in the epithelium (FDR < 0.001, Fig. 3b). Phosphorylated SMAD1/5/9 and total protein level of SMAD4 and SMAD5 were also diminished in the nucleus of villus epithelial cells upon Hnf4αγDKO (Fig. 3c–e). Consistent with direct activation by HNF4, active chromatin was lost at HNF4-bound regions of Smad4, Smad5, and Bmp1 loci upon Hnf4αγDKO (Fig. 3f and Supplementary Fig. 8a). These findings suggest that HNF4 directly activates BMP/SMAD signaling.
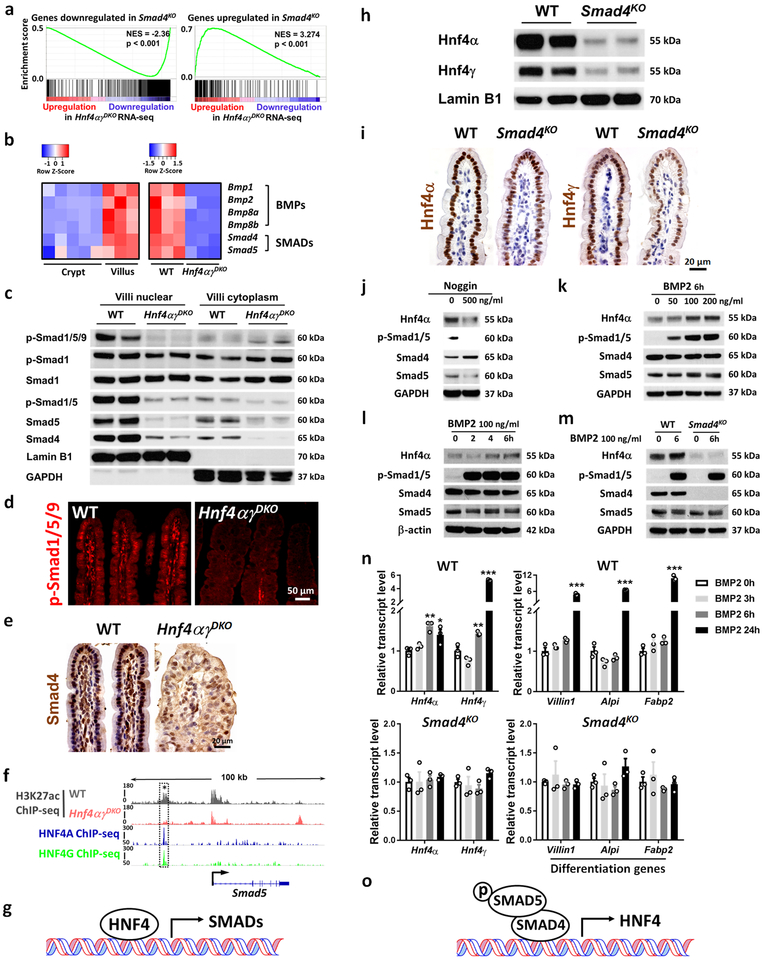
Figure 3. HNF4 and BMP/SMAD reinforce each other’s expression.
a-g, HNF4 binds and activates core components of the BMP/SMAD signaling pathways. a, GSEA reveals that genes downregulated or upregulated upon SMAD4 loss strongly correlate with genes of reduced expression or increased expression in Hnf4αγDKO, respectively (Kolmogorov-Smirnov test, one-sided for positive and negative enrichment scores, P < 0.001, n = 3 biologically independent mice). NES: normalized enrichment score. Heatmap (b) displays that RNA-seq expression levels of BMPs/SMADs are highly expressed in villus compared to the crypt and are significantly reduced in the Hnf4αγDKO intestinal epithelium compared to the WT littermate controls (n = 3 biologically independent mice, FDR < 0.001). Statistical tests were embedded in Cuffdiff. This is also evidenced by (c) western blot (n = 2 independent experiments and 2 biologically independent mice for each experiment). Immunofluorescent staining of p-SMAD1/5/9 (d) and immunohistochemistry staining of SMAD4 (e) also show reduced levels upon HNF4 loss (representative of 3 biologically independent mice). f, ChIP-seq tracks (n = 2 biologically independent mice) show that HNF4 factors bind to Smad5 and loss of HNF4 results in reduced H3K27ac signal (see dashed rectangles). Reciprocally, HNF4 levels are dependent upon BMP/SMAD4. h-o, BMP/SMAD signaling promotes HNF4 expression. Western blot (n = 2 independent experiments and 2 biologically independent mice for each experiment) (h) and immunostaining (n = 3 biologically independent mice) (i) show that SMAD4 knockout (4 days after tamoxifen injection) can cause significant downregulation of nuclear protein levels of Hnf4α and Hnf4γ in the villi. Western blot of the primary WT intestinal organoids in presence of (j) Noggin treatment (BMP antagonist, 72 h, n = 2 independent experiments), (k) 6 h treatment of different dosages of BMP2 (n = 3 independent experiments), and (l) different durations of BMP2 treatment (n = 3 independent experiments). m, BMP2 can induce Hnf4α expression in the primary WT intestinal organoids, but this effect is abolished in the absence of Smad4 (n = 2 independent experiments). Uncropped western blots are shown in Supplementary Fig. 11. qRT-PCR (n) shows that BMP2 (100 ng/ml) promotes Hnf4 and differentiation gene expression in the primary WT intestinal organoids but not in the Smad4KO organoids. All the qPCR data are presented as mean ± SEM (n = 3 independent organoid cultures). Transcript levels relative to BMP 0 h, and statistical comparisons were performed using one-way ANOVA followed by Dunnett’s post test at P < 0.001 ***, P < 0.01 ** and P < 0.05 *. All the primary organoids were harvested at Day 6 after seeding.
Positive feedback mechanisms can stabilize cellular lineage decisions21, and we investigated whether BMP signaling reciprocally stimulates HNF4 expression. Reduced Hnf4α and Hnf4γ transcript and protein levels (Fig. 3h,i and Supplementary Fig. 8b) were found in the villi of Smad4KO mice. In primary intestinal organoids, HNF4A protein levels were suppressed upon addition of the BMP antagonist Noggin, but elevated by BMP2 treatment (Fig. 3j–l). HNF4G expression was also responsive to BMP2 treatment (Supplementary Fig. 8c,d), consistent with redundancy of Hnf4α and Hnf4γ. BMP signaling suppressed stem cell genes (Supplementary Fig. 8e) as previously reported22, but also promoted enterocyte differentiation genes (Fig. 3n). BMP2-mediated activation of HNF4 protein and transcripts were dependent upon SMAD4, as the responses were not observed in Smad4KO tissues (Fig. 3m,n). Hnf4a expression was elevated as early as 6 h after treatment with BMP2 ligand, while enterocyte differentiation markers were not elevated until 24 h (Fig. 3n and Supplementary Fig. 8f), suggesting that Hnf4a is an early target of BMP signaling.
These data indicate a feed-forward differentiation mechanism in which HNF4 transcription factors and BMP/SMAD signaling reciprocally activate each other’s expression in intestinal villi (Fig. 3g,o). Such a regulatory relationship could then impart a differentiation program by activating intermediary differentiation factors or by directly activating a larger cadre of pro-differentiation transcripts. We explored the downstream activities of these two factors using ChIP-seq and RNA-seq in both mouse and human model systems. ChIP-seq of SMAD4 and HNF4A23 in Caco-2 cells indicated a broad regulatory partnership, with over 18,000 genomic regions commonly targeted by both HNF4A and SMAD4 factors (47% of SMAD4 binding sites at MACS P ≤ 10−5, Fig. 4a). Canonical SMAD4 and HNF4A/G motifs were enriched at these regions (Fig. 4b), suggesting that many of these regions were co-occupied by SMAD4 and HNF4. HNF4A and SMAD4 shared regulatory regions present at the HNF4A (Fig. 4c) and SMAD5 (Fig. 4d) loci, consistent with the reciprocal activation between HNF4 and SMAD4 seen in mice. Co-localized binding of HNF4A and SMAD4 was observed at a number of enterocyte-specific genes (Fig. 4e). Overlapping expression (Supplementary Fig. 8g) and co-binding of genomic enhancers of HNF4 and SMAD4 were also observed in ChIP-seq on mouse intestinal epithelium (Fig. 4f–h). At the transcriptome-level, the 541 genes that were significantly downregulated both in the Smad4KO and Hnf4αγDKO mice (Fig. 4i and Supplementary Fig. 8h) were associated with enterocyte functions such as lipid metabolism, microvillus, and absorption. Conversely, genes downregulated only in Hnf4αγDKO and not in Smad4KO were more associated with generalized ontologies of cytoplasm and Golgi apparatus (Fig. 4j and Supplementary Table 3). These findings suggest that the convergence of SMAD4 and HNF4 is critical to achieve a robust, enterocyte-specific gene expression program.
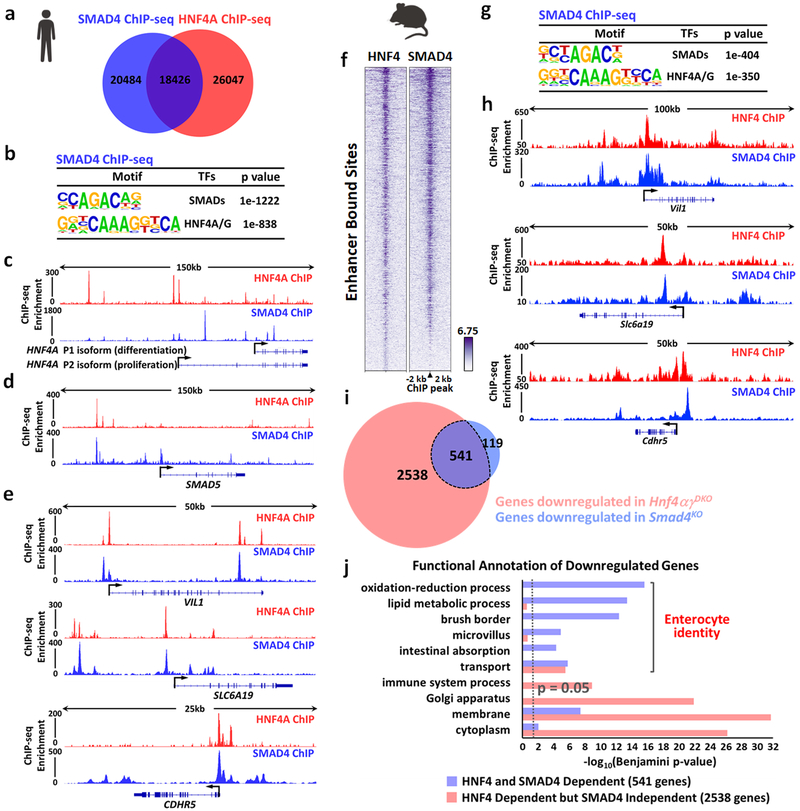
Figure 4. HNF4 and SMAD4 co-bind genomic enhancers, activating enterocyte-identity genes.
One replicate of human Caco-2 cell line and two replicates of biologically independent mouse tissues were used in ChIP-seq analysis. Statistical tests were embedded in MACS and HOMER. ChIP-seq in Caco-2 cells reveals that SMAD4 and HNF4A co-bind genomic regions, as evidenced by (a) Venn diagram (MACS P ≤ 10−5) and (b) HOMER motif analysis (MACS P ≤ 10−10). HNF4A and SMAD4 bind to gene loci of (c) HNF4A, (d) SMAD5 and (e) enterocyte differentiation genes (VIL1, SLC6A19 and CDHR5) in Caco-2 cells. ChIP-seq in mouse intestinal epithelium also reveals that SMAD4 and HNF4 co-bind genomic regions (n = 2 biologically independent mice each). f, Heatmap shows a similar binding pattern of SMAD4 (MACS P ≤ 10−5) and HNF4 (MACS P ≤ 10−3) at their enhancer bound sites. g, HOMER motif analysis of mouse SMAD4 ChIP-seq (MACS P ≤ 10−5, enhancer sites). h, HNF4 and SMAD4 co-bind to enterocyte differentiation genes (Vil1, Slc6a19, and Cdhr5) in mouse intestinal epithelium. i, Venn diagram shows highly overlapping target genes with significant downregulation (FDR < 0.05) in the intestine of Hnf4αγDKO (red circle) and genes with significant downregulation (FDR < 0.05) in Smad4KO (blue circle). Significant genes were called via Cuffdiff. (n = 3 biologically independent mice). j, Enterocyte-related functions are enriched among HNF4-SMAD4 co-dependent genes (common downregulated genes, 541 genes, purple bar) compared to genes only downregulated in Hnf4αγDKO but not in Smad4KO (2,538 genes, red bar). This suggests the combined functions of HNF4 and SMAD4 are specifically driving the transcriptome towards enterocyte identity. Benjamini P values were calculated using DAVID.
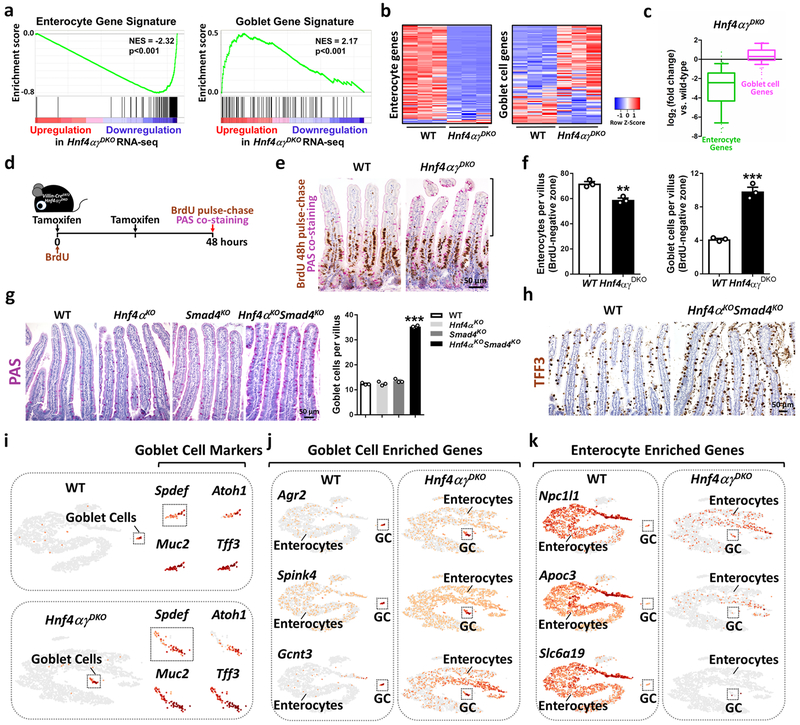
Our findings identify a reinforcing regulatory loop between the transcription factors HNF4 and SMAD4. Loss of this regulatory loop could impair enterocyte differentiation and/or destabilize enterocyte identity. Indeed, we observed a bias towards crypt cell structures and gene expression upon loss of HNF4 factors (Fig. 1). Additionally, a clear shift in the transcriptome occurred away from enterocyte transcripts24 and towards secretory goblet cell transcripts24 upon HNF4 loss (Fig. 5a–c and Supplementary Table 2), and goblet cell numbers increased in Hnf4αγDKO (Supplementary Fig. 9a–c). To help discern whether HNF4 is required for enterocyte differentiation and/or fate stability, we monitored villus-resident enterocytes in Villin-CreERT2; Hnf4αγDKO and their littermate controls. We labeled proliferating cells using BrdU at the time of tamoxifen-induced genetic changes, and monitored enterocyte fate at 48 h post-treatment (Fig. 5d). In this assay, we assume all cells above the BrdU-positive zone represent cells that were post-mitotic and had already migrated out of the crypt at the time of co-treatment with BrdU and tamoxifen to induce the loss of HNF4. These post-mitotic cells in the Hnf4αγDKO mice were more likely to exhibit features of secretory goblet cells than littermate controls (Fig. 5e). One possible explanation for the increased numbers of goblet cells could be that enterocytes are converting to goblet cells in the absence of HNF4 factors. Alternatively, the increase in goblet cell numbers could be attributed to conversion of other cell populations in the villus, or rapid migration of non-dividing goblet progenitor crypt cells towards villus tips. However, given that there appears to be a loss of enterocyte numbers above the BrdU-positive zone that is proportional to the increased number of goblet cells gained (Fig. 5f), and no significant difference in the number of goblet cells in the crypts (Supplementary Fig. 9d), we favor the interpretation that cells are undergoing an enterocyte-to-goblet conversion. Consistent with a co-regulatory relationship between HNF4 and SMAD4, combined loss of SMAD4 and HNF4A also led to accumulation of goblet cells (Fig. 5g,h and Supplementary Fig. 9e).
Figure 5. Disruption of an HNF4-SMAD4 regulatory loop compromises enterocyte identity.
a, GSEA of RNA-seq data (n = 3 biologically independent mice) reveals that the transcriptome moves away from enterocyte transcripts and towards secretory goblet cell transcripts upon HNF4-loss (Kolmogorov-Smirnov test, one-sided for positive and negative enrichment scores, P <0.001). Heatmaps (b) and boxplots (c) show that enterocyte signature transcripts are reduced, while goblet cell signature transcripts are elevated in Hnf4αγDKO. The middle line represents the median; whiskers represent the 10th and 90th percentile (Mann-Whitney test, two-sided, P <0.001, n = 3 biologically independent mice). d, Schematic of BrdU pulse-chase and PAS co-staining experiment. e, Co-staining of BrdU (48 h pulse-chase) and PAS. f, Enterocytes (PAS negative) and goblet cells (PAS positive) were counted within the BrdU negative zone of 50 villi per mouse. The data is presented as mean ± SEM (n = 3 biologically independent mice, Student’s t test, two-sided at P < 0.001 *** and P < 0.01 **). g, Mice were injected with tamoxifen for 4 consecutive days, and harvested 10 days after the first injection. Goblet cells were counted on 30 villi per mouse (WT and single mutants: n = 3 biologically independent mice; double mutants: n = 4 biologically independent mice, one-way ANOVA followed by Dunnett’s post test at P < 0.001***). h, Goblet cell markers are elevated in mutants as evidenced by immunohistochemistry staining of TFF3 (n = 3 biologically independent mice). i-k, Single-cell RNA-seq on dissociated villus epithelium. Canonical goblet cell markers were used to identify goblet cell clusters (i) in WT and mutants. Compared to WT, goblet-cell-enriched genes are increased (j) whereas enterocyte-enriched genes are decreased (k) in the enterocyte clusters of mutants (GC: Goblet Cells), suggesting compromised enterocyte identity. Estimated number of cells in each experiment was WT, n = 4,100; Hnf4αγDKO, n = 4,200.
The coincident increase in goblet cells and decrease in enterocytes within the post-mitotic zone of the villi suggested that enterocytes could be converting towards a goblet cell identity (Fig. 5a–h). To further explore this possibility, single cell RNA-seq was applied to control and mutant villus epithelial cells. Goblet cells were distinguished from enterocytes by canonical goblet cell markers (Spdef, Atoh1, Muc2, and Tff3), and were clearly identified in the dissociated villus cells of WT, Hnf4αγDKO and Hnf4αKOSmad4KO, respectively (Fig. 5i and Supplementary Fig. 9f). Goblet-cell-enriched genes24, such as Agr2, Spink4, Gcnt3, and S100a6, were highly expressed in the goblet cell clusters of WT, Hnf4αγDKO and Hnf4αKOSmad4KO, but were also expressed in the enterocyte clusters of mutants (Fig. 5j and Supplementary Fig. 9g). Conversely, enterocyte-enriched genes24,25, such as Npc1l1, Apoc3, Slc6a19, and Lct, were reduced in the enterocyte clusters of Hnf4αγDKO and Hnf4αKOSmad4KO compared to WT (Fig. 5k and Supplementary Fig. 9h). Thus the majority of enterocytes in these mutant models exhibit a mixed expression profile, featuring ectopic levels of goblet-cell-enriched transcripts and diminished levels of enterocyte-enriched transcripts. Taken together, these data suggest that the BMP/SMAD signaling pathway promotes differentiation via HNF4 transcription factors, and together, they stabilize enterocyte identity via a positive regulatory loop (Supplementary Fig. 10). Disruption of the HNF4-SMAD4 feed-forward circuit results in loss of enterocyte fate in favor of progenitor (Fig. 1) and goblet cell (Fig. 5) identities. Thus, the reinforcing regulatory module identified in this work may be especially important for enterocyte differentiation and lineage stability in a tissue with an underlying permissive chromatin environment5,26,27. The HNF4-SMAD4 regulatory model also predicts hurdles that must be overcome in order for enterocytes to exit their differentiated state and contribute to regenerative or oncogenic programs.
Methods
Mice
The Hnf4γCrispr lines were generated by the genome editing facility (Rutgers Cancer Institute of New Jersey), using the guide RNA: acaagaagaagtacctacgagg. The two resulting strains had identical 4-bp deletions in the 4th exon, and no phenotypic differences between the two lines, which were subsequently crossed together for later studies. The Villin-CreERT2 transgene28, Hnf4αf/f29, Hnf4γCrispr/Crispr and Smad4f/f30alleles were integrated to generate the conditional compound-mutants and controls. Both HNF4α isoforms (promoters P1 and P2) include the floxed exons29 utilized in our model. Experimental mice (8–12 weeks old) were treated with tamoxifen (Sigma T5648) at 50 mg/kg/day by intraperitoneal injection. Histologic analysis, western blot and qPCR were done after 4 consecutive days of tamoxifen treatment. For RNA-seq analysis, mice were harvested after 2 or 3 consecutive days of tamoxifen treatment. For ChIP-seq and micrococcal nuclease (MNase)-ChIP, mice were analyzed after 3 consecutive days of tamoxifen treatment. For the BrdU pulse-chase experiment, mice were injected with 1 mg BrdU and 100 mg/kg tamoxifen on the first day, 50 mg/kg tamoxifen on the second day, and were harvested 48 h after the BrdU injection. For the flow cytometry and single cell RNA-seq experiments, Hnf4αγDKO mice were injected with 100 mg/kg tamoxifen on the first day, 50 mg/kg tamoxifen on the second day, and were harvested at 48 h and 42 h after the initial tamoxifen injection, respectively. For the histologic analysis and single cell RNA-seq experiments of Hnf4αKOSmad4KO, mice were harvested 10 days after the initial tamoxifen treatment. All mouse protocols and experiments were approved by the Rutgers Institutional Animal Care and Use Committee, and we complied with all relevant ethical regulations. All samples were collected between 12:00 and 14:00 to avoid circadian variability.
Histology and Immunostaining
Intestinal tissues were fixed overnight in 4% paraformaldehyde at 4°C, washed with PBS, and dehydrated through ascending alcohols prior to paraffin embedding. 5-μm thick paraffin sections were used for immunohistochemistry and immunofluorescence staining using standard procedures. To mark enterocytes, alkaline phosphatase activity was detected using the AP Staining Kit II (Stemgent). Periodic Acid-Schiff (PAS) staining was used to detect goblet cells, and slides were incubated in 0.5% periodic acid and stained with Schiff’s Reagent (J612171, Alfa Aesar). Co-labeling of goblet cells and BrdUlabel-retaining cells was performed by BrdU immunohistochemistry and followed by PAS staining. Immunohistochemistry was performed using primary antibodies against HNF4A (Santa Cruz sc-6556 X, 1:2,000), HNF4G (Santa Cruz sc-6558 X, 1:2,000), Ki67 (Abcam ab16667, 1:300), BrdU (Bio-Rad MCA2060, 1:500), Keratin 20 (Cell Signaling 13063, 1:2,500), SMAD4 (Santa Cruz sc-7966, 1:500), MUC2 (Santa Cruz sc-15334, 1:300) and TFF3 (LifeSpan BioSciences, LS-C662731, 1:300). After employing secondary antibody and the Vectastain ABC HRP Kit (Vector Labs), slides were developed using 0.05% DAB (Amresco 0430) and 0.015% hydrogen peroxide in 0.1 M Tris, and then counterstained with hematoxylin. The slides were mounted and viewed on a Nikon Eclipse E800 microscope. Images were photographed with a Retiga 1300 CCD (QImaging) camera and QCapture imaging software. A Zeiss Axiovert 200M fluorescence microscope was used for imaging the immunofluorescence staining of p-SMAD1/5/9 (Cell Signaling 13820, 1:200) by using a Retiga-SRV CCD (QImaging). Leica TCS SP8 Spectral Confocal System with Lightning Confocal Super Resolution was used for imaging the immunofluorescence staining of HNF4A (Abcam ab41898, 1:100) and HNF4G (Santa Cruz sc-6558 X, 1:100). A Zeiss LSM 510 Meta confocal microscope was used for imaging the immunofluorescence staining of HNF4A (Abcam ab41898, 1:100) and SMAD4 (Cell signaling 46535, 1:100). ImageJ and Adobe Photoshop were used to adjust contrast and brightness. When adjustments were made, they were applied uniformly for comparative images.
Intestinal epithelium/villi/crypt isolation and single-cell dissociation
Freshly harvested duodenum was flushed with cold PBS, opened longitudinally, cut into 1-cm pieces, and then rotated in 3 mM EDTA in PBS at 4°C. The tissue was then vigorously shaken to release the epithelium from underlying muscular tissue, and the supernatant was collected as the whole epithelium fraction; villi were collected from the top of a 70-μm cell strainer while crypts passed through. Cells were pelleted by centrifugation at 170 g at 4 °C and then washed by cold PBS. Cell pellets were used for RNA extraction, ChIP, or protein extraction for Western blot as described in later sections. To dissociate single cells for flow cytometry and single-cell RNA-seq, isolated duodenal villi were further rotated with 5 U/ml dispase (Stem Cell 07913) and 200 U/ml of DNase I (Sigma D4513) at 37°C for 30 min, and were then washed twice with 1% BSA/PBS, and filtered with a 40-μm cell strainer.
Flow cytometry
To determine the percentage of goblet cells in the duodenal villi of Hnf4αγDKO mice (48 h after tamoxifen injection) and their littermate controls, villus cells were isolated and dissociated as described above. Cells were blocked with TruStain FcX™ anti-mouse CD16/32 antibody (1 μg per 106 cells, BioLegend 101319) for 10 min on ice to prevent non-specific labeling, and were then labeled with Alexa Fluor 647-conjugated anti-CD24 antibody (1:200, BioLegend 101818) and Atto 488-conjugated Ulex europaeus lectin (5 μg/ml, Sigma 19337) on ice for 20 min. Dead cells were eliminated using 0.5 μg/ml DAPI and goblet cells (UEA-1+CD24−)31,32 were detected with Beckman Coulter Gallios Flow Cytometer. Kaluza analysis 2.1 software was used for flow cytometry data analysis.
RNA-seq and data analysis
Epithelial cells were isolated from mouse duodenum, processed for RNA extraction using Trizol (Invitrogen) according to the manufacturer’s instructions, and sequenced using Illumina’s TruSeq RNA Library Prep kit v2. Raw sequencing reads (fastq) were quality checked using fastQC (v0.11.3) and were further aligned to mouse (mm9) genomes using Tophat2 (v2.1.0) to generate bam files. Cuffquant (v2.2.1) was used to generate cxb files from bam files. Cuffnorm (v2.2.1) was performed to calculate FPKM values using quartile normalization. Cuffdiff33 (v2.2.1) was applied to identify differentially expressed genes between the control and the mutants using quartile normalization and per-condition dispersion. Genes with FPKM > 1 were used for further analysis. Cuffdiff was used to generate the pre-ranked gene list. Gene set enrichment analysis (GSEA v3.0) was performed on the pre-ranked gene list as described34. Heatmapper35 was used to display relative transcript levels of genes of interest by using normalized FPKM values from Cuffnorm. Gene ontology analysis was performed with DAVID (v6.8).
Single-cell RNA-seq and data analysis
Single-cell RNA libraries were generated using the Chromium Single Cell 3’ v2 kit (10X Genomics, CA, USA). Briefly, each sample of dissociated duodenal villi was incubated with Dead Cell Removal MicroBeads (Miltenyi, USA) and passed through a magnetic column, removing the majority of dead cells/debris. The remaining live cells were counted using a trypan blue exclusion count assay with a hemocytometer and were diluted for loading onto the Chromium Controller. Loading was performed to target capture of ~3,000 gel beads per sample for downstream analysis, and samples were processed through the Chromium Controller following the standard manufacturer’s specifications. The sequencing library was validated on the Agilent TapeStation (Agilent Technologies, Palo Alto, CA, USA), and quantified with Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA) and quantitative PCR (Applied Biosystems, Carlsbad, CA, USA). The sequencing library was clustered on three lanes of flowcell. After clustering, the flowcells were loaded on the Illumina HiSeq instrument according to manufacturer’s instructions. The samples were sequenced following 10X Genomics single-cell RNA-sequencing protocols. Image analysis and base calling were conducted by the HiSeq Control Software (HCS v3.4.0). Raw sequence data (.bcl files) generated from Illumina HiSeq was converted into fastq files and de-multiplexed using the 10X Genomics’ cellranger (v2.1.1) mkfastq command. Subsequent UMI and cell barcode de-convolution was performed using 10X Genomics’ cellranger count command to generate the final digital gene expression matrices, t-SNE plot, and cloupe files. Single-cell RNA library preparations, sequencing reactions, and initial bioinformatics analysis were conducted at GENEWIZ, LLC. (South Plainfield, NJ, USA). Loupe Cell Browser 2.0.0 was used for downstream analysis of goblet cell and enterocyte enriched genes in each genotype. Plots showing individual gene expression intensity are based on a log2 scale of the gene expression; the plot of wild type cells was reflected over the vertical axis to position the goblet cell cluster for better comparison with knockout samples.
Chromatin Immunoprecipitation (ChIP)
Standard procedures were used for ChIP, as described36, with minor modifications on mouse duodenal epithelial cells. For HNF4A and HNF4G ChIP of mouse duodenal epithelium, cells were isolated as described above and then cross-linked in 1% formaldehyde (Sigma F8775) for 10 min at 4°C and then for 35 or 50 min at room temperature. For SMAD4 ChIP of mouse duodenal epithelium, cells were cross-linked in 2 mM DSG (Thermo 20593) for 45 min, and then further cross-linked in 1% formaldehyde for 20 min at room temperature. Cells were washed with ice-cold PBS, resuspended with lysis buffer (1% SDS, 10 mM EDTA pH 8.0, 50 mM Tris pH 8.0, and protease inhibitor cocktails), and further incubated at room temperature for 10 min. Cell lysates were sonicated using a Diagenode Bioruptor to generate 200-bp to 500-bp fragments, as determined by agarose gel electrophoresis. The supernatant of lysates were diluted in binding buffer (20 mM Tris pH 8.0, 2 mM EDTA pH 8.0, 150 mM NaCl, 1% Triton X-100, and protease inhibitor cocktails) and incubated with HNF4A (6 μg, Santa Cruz sc-6556 X, lot B1015), HNF4G (6 μg, Santa Cruz sc-6558 X, lot F0310) or SMAD4 (20 μl, Cell signaling 46535, lot 2) antibody coupled to Dynabeads (Invitrogen) at 4°C overnight. The final concentrations of SDS in the sonicates for mouse HNF4 and SMAD4 ChIP are 0.125% and 0.167%, respectively. The immunoprecipitates were washed 5 times and rotated at 4°C with RIPA buffer (50 mM HEPES pH 7.6, 1 mM EDTA pH 8.0, 0.7% Na-deoxycholate, 1% NP-40, and 0.5 M LiCl) and a quick wash of TE buffer (0.1 mM EDTA pH 8.0 and 10 mM Tris pH 8.0). To recover the DNA, the samples were incubated overnight in reverse cross-linking buffer (1% SDS and 0.1 M NaHCO3) at 65 °C. The DNA was purified by QIAquick PCR Purification Kit (QIAGEN) and quantified with Picogreen (Life Technologies). ChIP enrichment was confirmed with qPCR for putative targets prior to library preparation.
For MNase-ChIP, mouse duodenal epithelial cells were resuspended in digestion buffer (50 mM Tris-HCl pH7.6, 1 mM CaCl2, 0.2% Triton X-100, 5 mM Na butyrate, and protease inhibitor cocktails) and treated with micrococcal nuclease (MNase, Sigma N3755) at 0.67 U/ml for 5 min at 37°C. The reaction was terminated by adding 5 mM EDTA in 10 mM Tris, pH7.6, and samples were dialyzed in chromatin RIPA buffer (10 mM Tris, pH7.6, 1 mM EDTA, 0.1% SDS, 0.1% Na-deoxycholate, and 1% Triton X-100) before the overnight IP at 4°C with Histone H3K27ac (Abcam ab4729, lot GR184332–2) antibody. IP material was subsequently handled as described above for conventional ChIP of sonicated genomic DNA (a final concentration of 0.13% SDS in the sonicates).
To perform SMAD4 ChIP in Caco-2 cells, lentiviral vectors were produced by transfecting HEK293-FT cells with the viral packaging constructs (VSVG and Delta 8.2) and plasmids with expression constructs. Caco-2 cells were then transduced serially with lentiviral vectors carrying avi-tagged SMAD4 expression constructs (pReceiver-Lv108, GeneCopoeia) with puromycin resistance cassette and BirA expression plasmid with hygromycin B (Addgene plasmid 29649). Caco-2 cells were cultured in DMEM (Gibco 11995–065), containing 10% FBS (Gibco 26140–095) and 1% penicillin and streptomycin (Invitrogen 15140–122). Cells stably expressing both avi-tagged SMAD4 and BirA were selected with medium containing 2 μg/ml puromycin and 0.4 mg/ml Hygromycin B, respectively. Cells were fed 24 hours prior to harvest at approximately 95% confluency. Each 100 μl cell pellet was incubated with 1 ml of cross linking solution containing an equal amount of DSG (Thermo 20593), DSS (Thermo 21655) and EGS (Thermo 21565), creating a final concentration of 2 mM NHS ester (reactive groups). Cells were rocked for 20 min at room temperature, after which fresh formaldehyde was added to make a concentration of 1.22% formaldehyde. The cells were then rocked for an additional 20 min. The cross-linking reaction was stopped with 125 mM glycine. The cells were then washed with PBS, and nuclei were extracted using a Dounce homogenizer and Farnham lysis buffer (5 mM PIPES, pH 8.0, 85 mM KCl, 0.5% NP-40, and protease inhibitor cocktails). IP material was subsequently handled as described above for conventional ChIP of sonicated genomic DNA (a final concentration of 0.2% SDS in the sonicates). The diluted sonicates were then incubated with pre-blocked (0.5% BSA/PBS) Streptavidin beads (Invitrogen) overnight at 4°C. The beads were serially washed in low salt (0.1% SDS, 0.1% Sodium Deoxycholate, 1% Triton X-100, 150 mM NaCl, 1 mM EDTA, and 20 mM HEPES, pH 8.0), high-salt (0.1% SDS, 0.1% Sodium Deoxycholate, 1% Triton X-100, 500 mM NaCl, 1 mM EDTA, and 20 mM HEPES, pH 8.0), LiCl buffer (250 mM LiCl, 0.5% Sodium Deoxycholate, 0.5% NP-40, 1mM EDTA, and 20 mM HEPES, pH 8.0), and a final wash buffer (1 mM EDTA and 20 mM HEPES, pH 8.0). The beads were then incubated at 65 °C for 6 h in reverse cross linking buffer (0.1 mM NaHCO3 and 1% SDS).
ATAC-seq
Duodenal villi were treated with pre-warmed 0.25% Trypsin for 8 min at 37°C on a vortex station (speed set between 6–7), neutralized with 10% FBS, and passed through a 40-μm cell strainer to obtain single cells. 50,000 cells were used for ATAC-seq as described previously37,38 with slight modifications. Briefly, cells were centrifuged at 500 g for 5 min at 4°C and resuspended in ice-cold lysis buffer (10 mM Tris, pH 7.4, 10 mM NaCl, 3 mM MgCl2, and 0.1% NP-40). Cells were then centrifuged at 500 g for 10 min at 4°C. The isolated nuclear pellets were incubated with a 50-μl reaction of Nextera Tn5 Transposase (Illumina FC-121–1030) for 30 min at 37°C. The transposed chromatin was purified with QIAquick PCR Purification Kit (QIAGEN), and PCR was amplified with high-fidelity 2× PCR Master Mix (New England Biolabs M0541). The optimum number of additional cycles was based on 1/3 of the maximum fluorescent intensity. The PCR amplified libraries were purified. Fragment size was selected using Pippin Prep and sequenced on Illumina NextSeq 550.
ChIP-seq, ATAC-seq, and data analysis
For ChIP-seq, 5 ng each of ChIP or input DNA was used to prepare ChIP-seq libraries using Rubicon Genomics ThruPLEX DNA-seq Kit (Illumina). Fragment size was selected using Pippin Prep and sequenced on Illumina HiSeq 2500 or NextSeq 550. FastQC (v0.11.3) was used to check the quality of raw sequencing reads (fastq), and bowtie2 (v2.2.6) was used to align the sequences to mouse (mm9) or human (hg19) genomes and generate bam files. Deeptools bamCoverage39 (v2.4.2, duplicate reads ignored, RPKM normalized and extended reads) was used to generate bigwig files from bam files. BigWigMerge (v2) was used to merge the bigwig files of different replicates. The Integrative Genomics Viewer40 (IGV 2.4.13) was used to visualize normalized bigwig tracks. Model-based Analysis of ChIP-Seq41 (MACS 1.4.1) was used for peak calling and to generate bed files from aligned reads. The shiftsize parameter used in MACS was based on the fragment size of Pippin Prep. HNF4A and HNF4G ChIP-seq of mouse duodenal epithelium are at a P value of 10−3; SMAD4 ChIP-seq of mouse duodenal epithelium is at a P value of 10−5; H3K27ac MNase-ChIP-seq and ATAC-seq of mouse duodenal epithelium or villi are at a P value of 10−5; HNF4A and SMAD4 ChIP-seq of Caco-2 cells are at a P value of 10−5 or 10−10. BEDTools42 (v2.17.0) was used to merge, intersect or subtract the intervals of bed files. k-means clustering heatmaps of ChIP-seq were created with Haystack43 (v0.4.0) quantile normalized bigwigs using computeMatrix and plotHeatmap from deeptools39 (v2.4.2). Genomic regions of desired k-means clusters were extracted from bed files generated by plotHeatmap. DiffBind15,44 (v1.16.3) was used to identify differential signals of HNF4A and HNF4G ChIP-seq, and 0.05 was used as the cutoff for significance. Subsequent analysis of HNF4 binding sites consists of all peaks at MACS P value ≤ 10−3 in each of the 4 wild-type replicates merged into a set of 7,287 binding regions. Promoters were defined as bed regions within 2 kb of the transcription start sites (TSSs) of RefSeq genes, whereas enhancers were defined by excluding promoters and used for subsequent analysis. SitePro45 (v1.0.2) was used to compare the average signal profiles of H3K27ac between WT littermate controls and Hnf4αγDKO around HNF4 bound sites. The Peak2gene/BETA-minus function (v1.0.2) in Cistrome tools46 was used to identify genes within 10 kb, 20 kb, or 30 kb of HNF4 binding sites, and gene ontology (GO) term analysis (DAVID v6.8)47 was used for functional annotation of those genes. Homer findMotifsGenome.pl48 (v4.8.3, homer de novo Results) was used to call transcription factor motifs enriched at peaks. All analysis was done with publicly available algorithms and additional details are available from the corresponding author upon request.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
cDNA was synthesized from total RNA with Oligo(dT)20 primers using SuperScript III First-Strand Synthesis SuperMix (Invitrogen). qRT-PCR was performed to measure changes in mRNA expression using Applied Biosystems 7900HT Sequence Detection System. The sequences of the primers used are available upon request. Briefly, the qRT-PCR was carried out using Power SYBR® Green PCR Master Mix, and the amplification conditions were as follows: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15s and 60°C for 1 min. Hypoxanthine-guanine phosphoribosyl transferase 1 (Hprt1) was used as an internal control. The 2−ΔΔCt method was applied to calculate the fold change of relative transcript level.
Protein extraction and Western blot
The protein of duodenal epithelium or intestinal organoids was extracted with RIPA buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% Na-deoxycholate, 0.1% SDS, protease inhibitor cocktails, and phosphatase inhibitors). The nuclear protein extraction of duodenal villi was performed as described49. 4 cycles and 2 cycles of bioruptor sonication (30s on and 30s off) were used before and after rotating cells in lysis buffer at 4°C for 30 min, respectively. Protein concentration was determined by Pierce BCA Protein Assay Kit (Thermo). Immunodetection was performed using specific antibodies against HNF4A (Santa Cruz sc-6556 X, 1:1,000), HNF4G (Santa Cruz sc-6558 X, 1:1,000), Lamin B1 (Abcam ab16048, 1:1000), GAPDH (Santa Cruz sc-25778, 1:5,000), β-actin (Abcam ab8227, 1:5,000) and SMAD 1/5/9 Antibody Sampler Kit (Cell signaling 12656, 1:1,000).
Organoid culture
Primary crypt-derived organoids were isolated from duodenal epithelium and cultured in Cultrex® reduced growth factor basement membrane matrix, Type R1 (Trevigen) according to established methods50. The organoids were treated with 1 μM tamoxifen dissolved in ethanol for 12 h. Vehicle-treated organoids served as a control. Tamoxifen was added into culture medium of organoids on Day 2 after seeding. Organoids were imaged using a Zeiss Axiovert 200M inverted microscope with a Retiga-SRV CCD (QImaging). For BMP2 and Noggin treatment experiments, recombinant BMP2 (R&D 355-BM) and Noggin (Peprotech 250–38) were prepared according to the supplier’s instructions, and vehicle controls were used in organoid culture. The organoids were harvested on Day 6 for subsequent RNA or protein extraction. QIAGEN RNeasy Micro Kit was used to extract RNA from organoids.
Statistical analysis
The data are presented as mean ± SEM, and statistical comparisons were performed using one-way ANOVA followed by Dunnett’s post test with the GraphPad Prism version 7.02 or two-sided Student’s t test at P < 0.001 ***, P < 0.01 ** or P < 0.05 *. The exact P values are shown in Supplementary Table 4. Mann-Whitney test and Kruskal-Wallis test (followed by post-hoc Dunn’s test) were used as part of RNA-seq analysis. Bioinformatics-related statistical analysis was done with the embedded statistics in each package, including DiffBind15, HOMER48, Cuffdiff33, GSEA34,51 and DAVID47. P < 0.05 (95% confidence interval) was considered statistically significant.
Reporting Summary
Further information on research design is available in the Life Sciences Reporting Summary linked to this article.
Data availability
All RNA-seq, ChIP-seq and ATAC-seq data of this study have been deposited in GEO (GSE112946). The following datasets from GEO were reanalyzed with our sequencing data: the accession numbers for the transcriptome of Smad4KO and HNF4A ChIP in CaCo-2 cells from our previous studies are GSE10217152 and GSE2343623, respectively. GSE513367 and GSE3602514 were used to analyze chromatin accessibility and RNA-seq data across different tissues. GSE5791953 and GSE9872454 were used to mark active chromatin. GSE53545, GSE7076655, and GSE10217152 were used to perform RNA-seq analysis of villus-enriched genes and crypt-enriched genes.
Supplementary Material
Acknowledgements
This research was funded by a grant from the NIH (R01CA190558) to M.P.V. The Verzi laboratory is also supported by the Intestinal Stem Cell Consortium funded by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health under grant number U01 DK103141. L.C. was supported by New Jersey Commission on Cancer Research grant (DFHS18PPC051). N.H.T., S.L., R.P.V. and A.P. were supported by MacMillan Summer Undergraduate Research Fellowships. R. Shivdasani, R. Hart, T. Nakamura, and B. Nickels provided helpful discussions. The research was supported by the Genome Editing shared resource of Rutgers Cancer Institute of New Jersey (P30CA072720), next-generation sequencing services of RUCDR, flow cytometry/cell sorting core facility at EOHSI, and imaging core facility of Human Genetics Institute of New Jersey.
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Davis H et al. Aberrant epithelial GREM1 expression initiates colonic tumorigenesis from cells outside the stem cell niche. Nat Med 21, 62–70 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haramis AP et al. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science 303, 1684–6 (2004). [DOI] [PubMed] [Google Scholar]
- 3.He XC et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat Genet 36, 1117–21 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Auclair BA, Benoit YD, Rivard N, Mishina Y & Perreault N Bone morphogenetic protein signaling is essential for terminal differentiation of the intestinal secretory cell lineage. Gastroenterology 133, 887–96 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Kim TH et al. Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature 506, 511–5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwafuchi-Doi M & Zaret KS Cell fate control by pioneer transcription factors. Development 143, 1833–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vierstra J et al. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science 346, 1007–12 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Consortium, U.I.G. et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet 41, 1330–4 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang B et al. Proteogenomic characterization of human colon and rectal cancer. Nature 513, 382–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Babeu JP, Darsigny M, Lussier CR & Boudreau F Hepatocyte nuclear factor 4alpha contributes to an intestinal epithelial phenotype in vitro and plays a partial role in mouse intestinal epithelium differentiation. Am J Physiol Gastrointest Liver Physiol 297, G124–34 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Cattin AL et al. Hepatocyte nuclear factor 4alpha, a key factor for homeostasis, cell architecture, and barrier function of the adult intestinal epithelium. Mol Cell Biol 29, 6294–308 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.San Roman AK, Aronson BE, Krasinski SD, Shivdasani RA & Verzi MP Transcription factors GATA4 and HNF4A control distinct aspects of intestinal homeostasis in conjunction with transcription factor CDX2. J Biol Chem 290, 1850–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin S et al. Comparison of the transcriptional landscapes between human and mouse tissues. Proc Natl Acad Sci U S A 111, 17224–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stark R & Brown GD DiffBind: Differential Binding Analysis of ChIP-Seq Peak Data. Bioconductor, Available online at: http://bioconductor.org/packages/release/bioc/html/DiffBind.html (2011). [Google Scholar]
- 16.Fang B, Mane-Padros D, Bolotin E, Jiang T & Sladek FM Identification of a binding motif specific to HNF4 by comparative analysis of multiple nuclear receptors. Nucleic Acids Res 40, 5343–56 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerdin AK et al. Phenotypic screening of hepatocyte nuclear factor (HNF) 4-gamma receptor knockout mice. Biochem Biophys Res Commun 349, 825–32 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Baraille F et al. Glucose Tolerance Is Improved in Mice Invalidated for the Nuclear Receptor HNF-4gamma: A Critical Role for Enteroendocrine Cell Lineage. Diabetes 64, 2744–56 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Verzi MP, Shin H, San Roman AK, Liu XS & Shivdasani RA Intestinal master transcription factor CDX2 controls chromatin access for partner transcription factor binding. Mol Cell Biol 33, 281–92 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merlos-Suarez A et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 8, 511–24 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Boyer LA et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–56 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi Z et al. BMP restricts stemness of intestinal Lgr5(+) stem cells by directly suppressing their signature genes. Nat Commun 8, 13824 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verzi MP et al. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev Cell 19, 713–26 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haber AL et al. A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moor AE et al. Spatial Reconstruction of Single Enterocytes Uncovers Broad Zonation along the Intestinal Villus Axis. Cell 175, 1156–1167 e15 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Yousefi M, Li L & Lengner CJ Hierarchy and Plasticity in the Intestinal Stem Cell Compartment. Trends Cell Biol 27, 753–764 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tetteh PW et al. Replacement of Lost Lgr5-Positive Stem Cells through Plasticity of Their Enterocyte-Lineage Daughters. Cell Stem Cell 18, 203–13 (2016). [DOI] [PubMed] [Google Scholar]
- 28.el Marjou F et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–93 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Hayhurst GP, Lee YH, Lambert G, Ward JM & Gonzalez FJ Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol 21, 1393–403 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang X, Li C, Herrera PL & Deng CX Generation of Smad4/Dpc4 conditional knockout mice. Genesis 32, 80–1 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Wong VW et al. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat Cell Biol 14, 401–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jadhav U et al. Dynamic Reorganization of Chromatin Accessibility Signatures during Dedifferentiation of Secretory Precursors into Lgr5+ Intestinal Stem Cells. Cell Stem Cell 21, 65–77 e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7, 562–78 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–50 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Babicki S et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res 44, W147–53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perekatt AO et al. YY1 is indispensable for Lgr5+ intestinal stem cell renewal. Proc Natl Acad Sci U S A 111, 7695–700 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buenrostro JD, Wu B, Chang HY & Greenleaf WJ ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21 29 1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramirez F et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson JT et al. Integrative genomics viewer. Nat Biotechnol 29, 24–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quinlan AR BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr Protoc Bioinformatics 47, 11 12 1–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pinello L, Farouni R & Yuan GC Haystack: systematic analysis of the variation of epigenetic states and cell-type specific regulatory elements. Bioinformatics (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ross-Innes CS et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–93 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shin H, Liu T, Manrai AK & Liu XS CEAS: cis-regulatory element annotation system. Bioinformatics 25, 2605–6 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Liu T et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol 12, R83 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang da W, Sherman BT & Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–89 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shaked H, Guma M & Karin M Analysis of NF-kappaB activation in mouse intestinal epithelial cells. Methods Mol Biol 1280, 593–606 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Sato T et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–5 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Tamayo P, Steinhardt G, Liberzon A & Mesirov JP The limitations of simple gene set enrichment analysis assuming gene independence. Stat Methods Med Res 25, 472–87 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perekatt AO et al. SMAD4 suppresses WNT-driven de-differentiation and oncogenesis in the differentiated gut epithelium. Cancer Res (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Camp JG et al. Microbiota modulate transcription in the intestinal epithelium without remodeling the accessible chromatin landscape. Genome Res 24, 1504–16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saxena M et al. Transcription factor-dependent ‘anti-repressive’ mammalian enhancers exclude H3K27me3 from extended genomic domains. Genes Dev 31, 2391–2404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.San Roman AK, Tovaglieri A, Breault DT & Shivdasani RA Distinct Processes and Transcriptional Targets Underlie CDX2 Requirements in Intestinal Stem Cells and Differentiated Villus Cells. Stem Cell Reports 5, 673–681 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq, ChIP-seq and ATAC-seq data of this study have been deposited in GEO (GSE112946). The following datasets from GEO were reanalyzed with our sequencing data: the accession numbers for the transcriptome of Smad4KO and HNF4A ChIP in CaCo-2 cells from our previous studies are GSE10217152 and GSE2343623, respectively. GSE513367 and GSE3602514 were used to analyze chromatin accessibility and RNA-seq data across different tissues. GSE5791953 and GSE9872454 were used to mark active chromatin. GSE53545, GSE7076655, and GSE10217152 were used to perform RNA-seq analysis of villus-enriched genes and crypt-enriched genes.