Abstract
Non-vesicular lipid transport between bilayers at membrane contact sites plays important physiological roles. Mechanistic insight into the action of lipid transport proteins localized at these sites (bridge/tunnel versus shuttle models) requires a determination of the distance between bilayers at which this transport can occur. Here, we developed DNA-origami nanostructures to organize size-defined liposomes at precise distances and used them to study lipid transfer by the SMP domain of E-Syt1. Pairs of DNA-ring-templated donor and acceptor liposomes were docked through DNA pillars, which determined their distance. The SMP domain was anchored to donor liposomes via an unstructured linker and lipid transfer was assessed via a FRET-based assay. We show that lipid transfer can occur over distances that exceed the length of SMP dimer, compatible with a shuttle model. The DNA nanostructures developed here can be adapted to study other processes occurring where two membranes are closely apposed to each other.
Introduction
In eukaryotic cells, close appositions between the membranes of two different organelles mediated by protein tethers play a variety of functions in the control of organelle homeostasis, including lipid transport between the two adjacent bilayers via lipid-transport modules1–4. Examples of such modules are SMP (Synaptotagmin-like Mitochondrial lipid binding Protein) domains, which represent a branch within the TULIP (TUbular LIPid-binding proteins) domain superfamily5–13. SMP domains are found in proteins that also function as tethers between the endoplasmic reticulum (ER) and other membranes12,14–17, such as the three extended synaptotagmins (E-Syt1, E-Syt2 and E-Syt3), named tricalbin in yeast18–20. The E-Syts/tricalbins are anchored to the ER membrane via N-terminal hydrophobic hairpins, which are followed by the SMP domain and by variable numbers of C2 domains that mediate contacts of the ER membrane with the plasma membrane in trans20–23.
The SMP domain of the E-Syts dimerizes in an anti-parallel fashion to form a 90-Å elongated structure comprising a deep groove lined with hydrophobic residues that run from one end to the other of the module. This groove harbors glycerophospholipids, without selectivity for specific head groups10. A role of the SMP domain of the E-Syts in glycerolipid transport between bilayers has been supported by cell-free liposomes- and FRET-based assays and by evidence for a Ca2+-dependent role of E-Syt1 in the control of plasma membrane lipid homeostasis in response to acute perturbations24–26. However, how the SMP domain transfers lipids between two membranes remains unclear. Two models have been proposed10. In one model, the SMP domain directly bridges the two bilayers so that lipids can enter and exit it at its tips, and slide along the hydrophobic channel (tunnel model). In the second model, the SMP domain shuttles between the two bilayers, extracting lipids from one and delivering them to the other (shuttle model).
The tunnel model implies that transport can only occur when the two bilayers are separated by a distance that does not exceed the length of the SMP dimer (9 nm). The shuttle model, in contrast, is compatible with greater distances, limited only by the length of the predicted unstructured linker that connects the SMP domain to the bilayer of the ER. Analysis of ER-plasma membrane contact sites by cryo-electron tomography (cryo-ET) in mammalian cells showed that the ER-plasma membrane distance at contact sites can be variable and in the 10–30 nm range22, which seems too far for the dimeric SMP domain to span it. However, small, puncta-like closer appositions of the two bilayers (<10 nm), which would be compatible for the tunnel model, can occasionally be seen22,27. Moreover, direct proof that E-Syts’ SMP domains can transfer lipids over a distance greater than their length is still lacking. Commonly used in vitro lipid transfer assays do not have the ability to control the distance between membranes. The goal of this study is to develop a method to assess the property of the SMP domain of the E-Syts, or of any other lipid-transport proteins or modules, to transfer lipids between bilayers over well-defined distances.
DNA origami is a technique that generates self-assembled and programmable DNA nanostructures28–30. Recently, it was reported that rationally designed DNA structures represent a versatile toolbox to grow, mold and fuse liposomes in a precise and deterministic way31–38. Here we have harnessed the engineering power of DNA in nanoscale to develop a distance-dependent lipid transfer assay using DNA-origami-organized liposomes. The DNA nanostructure consists of two DNA rings that guide the formation of liposomes and a DNA pillar that keeps the two rings, and thus the two liposomes, at a predefined distance. This methodology offers a powerful solution to control spacing of lipid bilayers at a nanoscale distance and holds great potential for creating a generic platform for studies of protein-mediated lipid transfer or of other protein functions at sites where two membranes are closely apposed to each other. Our data provide evidence that SMP domains can transfer lipids over distances that exceed their length, compatible with a shuttle model of lipid transfer.
Results
A DNA-origami nanostructure to control liposome distance
In the commonly used liposome-based assays of protein-mediated lipid transport, the protein of interest is added to a suspension of donor and acceptor liposomes. In most cases, however, efficient transport requires tethering of the two bilayers, mimicking conditions in living cells, where much of lipid transport occurs at membrane contact sites3,12,24–26,39,40. Tethering can be achieved by protein domains that flank the lipid transport module or by an additional protein added to the mixture. Protein-mediated liposome tethering, however, leads to aggregation and does not allow controlling the distance between bilayers. Given the limitation of this system, we explored the possibility of using DNA nanotechnology (specifically DNA origami) to develop an aggregation-independent lipid transport assay, in which the distance between liposomes could be tightly controlled. To this aim, we capitalized on a recently reported method to generate small liposomes within DNA rings33,34 and further developed this method to obtain pairs of such liposomes with a well-defined distance between them.
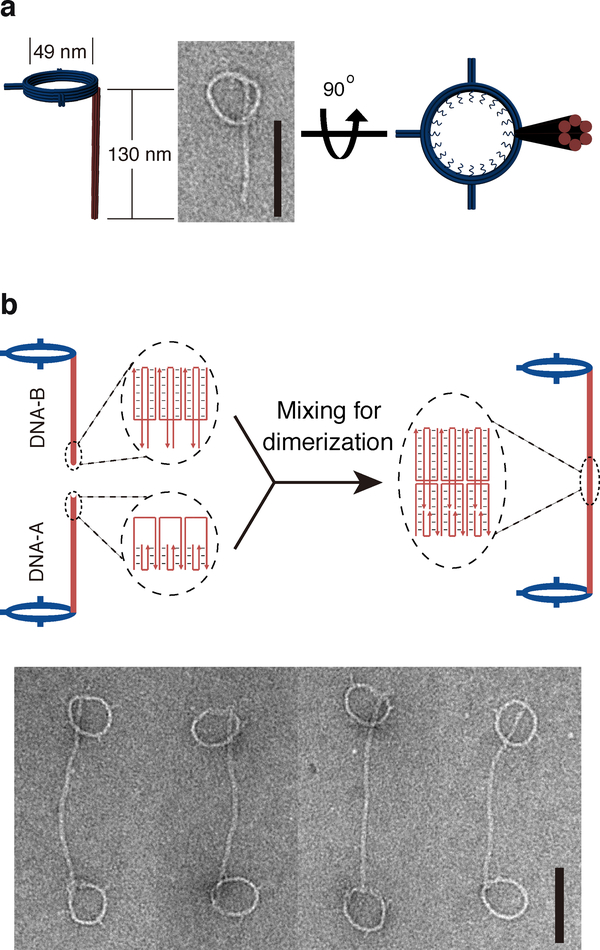
We first constructed a DNA-based scaffold capable of holding two DNA rings at a precisely defined distance. Briefly, we designed DNA rings for templating size-defined liposomes as described previously33,34. A ring (inner diameter ≈ 49 nm) consisted of a bundle of eight curved DNA helices with two of the inner helices lined by a total of 32 single-stranded (ss) oligonucleotide handles, which can hybridize to lipidated oligonucleotide anti-handles to nucleate the formation of liposomes (Fig. 1a and Supplementary Fig. 1). Additionally, each ring was connected to a six-helix-bundle DNA rod (130, 46, or 23 nm long) equipped with a “sticky end” composed of 3-nt vacancy (DNA-A) or overhang (DNA-B) on each helix (Fig. 1b). These DNA nanostructures were folded by thermal annealing, purified by rate-zonal centrifugation41, and examined by negative-staining transmission electron microscopy (TEM), which confirmed the formation of rod-bearing rings (Fig. 1a). Next, these structures, whose rods have complementary sticky ends (DNA-A and DNA-B), were incubated for 90 min at room temperature (RT) to form dumbbell-like dimers, where two rings are connected by a pillar, as confirmed by TEM and agarose gel electrophoresis (Fig. 1b and Supplementary Fig. 2).
Figure 1. DNA nanostructures designed for this study.
a, Cartoon models and a negative-staining TEM image of monomeric DNA nanostructure comprising a ring connected to a rod. b, Schematics of DNA nanostructure dimerization. Diagrams in the dashed ovals illustrate the dimerization mediated by DNA rods with complementary sticky ends. Negative-stain TEM images of DNA-origami dimers (two rings joint by a pillar) are shown at the bottom. Scale bars: 100 nm. The experiments were repeated independently three times with similar results.
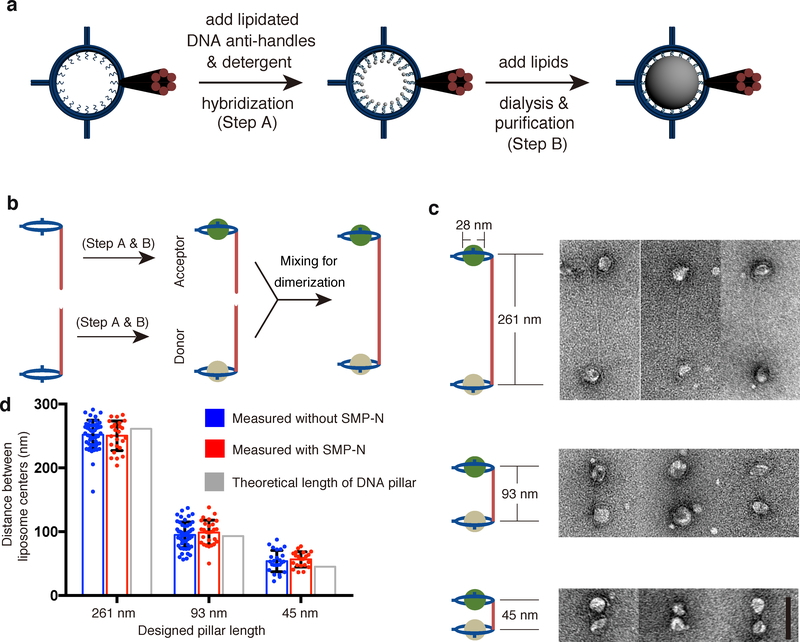
To produce dimeric DNA nanostructures harboring liposomes of different lipid compositions, monomeric DNA rings were labeled with phospholipids through inner handle/anti-handle hybridization, incubated with additional lipids and detergent, and dialyzed against detergent-free buffer solutions to allow liposome formation within the DNA rings (Fig. 2a)33,34. Two sets of DNA-ring-templated liposomes with different lipid compositions were generated and purified by iodixanol-gradient centrifugation. One set, termed “donor” liposomes (formed on DNA rings carrying DNA-A-labeled rods), contained DOPC, DGS-NTA(Ni), and a pair of fluorescence resonance energy transfer (FRET) dye-labeled lipids: NBD-PE and rhodamine-PE. The other set, termed “acceptor” liposomes (formed on DNA rings carrying DNA-B-labeled rods), contained DOPC, POPS, and PI(4,5)P2 (see Online Methods for lipid acronyms and molar ratios). DNA-ring-templated donor and acceptor liposomes were then mixed to allow dimerization via DNA-A/DNA-B hybridization (Fig. 2b). Negative-staining TEM showed liposome-containing DNA-origami dimers (Fig. 2c and Supplementary Fig. 3) with center-to-center distance of the two liposomes consistent with the theoretical length of the DNA pillar. For example, using DNA rings with 130-nm rods, we obtained a liposome center-to-center distance of 252 ± 23 nm (Fig. 2d), confirming that the dimerized DNA pillars (~261 nm) were rigid enough to keep the two liposomes at the predicted distance. As the DNA-ring-templated liposomes were homogenous in size and on average ~28 nm in diameter34, the closest distance between their membranes was on average ~224 nm. Similarly, when the donor and accepter liposomes were separated by 93-nm and 45-nm DNA pillars (Fig. 2c), TEM revealed liposome center-to-center distances of 95 ± 20 nm and 54 ± 17 nm, respectively. These values were again in good agreement with the design (Fig. 2d) and translated to membrane separations of circa 67 nm and 26 nm, respectively. We next used these donor/acceptor liposome pairs with controlled bilayer distances for lipid transfer assays.
Figure 2. DNA-directed liposome formation and assembly.
a, Schematics of DNA-ring-templated liposome formation. b, Schematics of the donor and acceptor liposome heterodimer assembly. c, Cartoon models and TEM images of DNA-organized liposome dimers separated by DNA pillars with different lengths (261, 93, and 45 nm). d, Dot plots showing center-to-center distance of DNA-tethered liposomes, with or without SMP-N, measured from negative-stain TEM images compared with theoretical lengths of DNA pillars. Mean and standard deviations (error bars) are shown (n=50, 25, 50, 27, 25, and 24 liposome pairs). Scale bars: 100 nm.
Lipid transfer between DNA-origami templated liposomes
As a premise to the use of dimeric DNA nanostructures to investigate the impact of bilayer distance on lipid transport by the SMP domain of E-Syt1, we first assessed whether presence of DNA rings affected the property of the SMP domain to transfer lipids between them. In this system, a small portion of the liposome surface is in contact with the negatively charged DNA nanostructure, and this could impact the properties of the bilayer. Furthermore, DNA-origami-templated liposomes are generally maintained in a buffer that differs from those typically used for lipid transport assays24,25 as it contains 10-mM Mg2+ (rather than 0-mM Mg2+) and 400-mM KCl (rather than 100-mM NaCl/KCl).
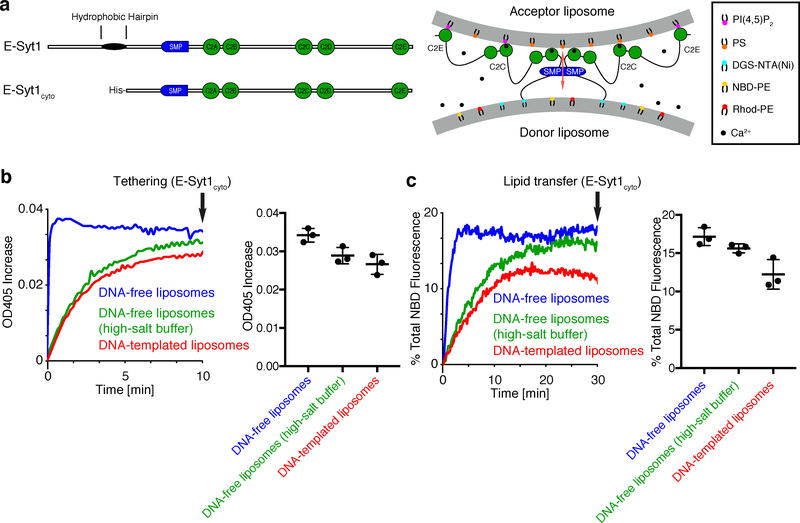
To address the potential impact of such parameters in our assay, lipid transfer with either DNA-free liposomes or DNA-templated liposomes was compared using the entire cytosolic region of E-Syt1. E-Syt1 with an N-terminal His-tag (E-Syt1cyto, Supplementary Fig. 4) to mediate binding to DGS-NTA(Ni) was added to a mixture of DNA-free or DNA-templated donor [containing DGS-NTA(Ni)] and acceptor liposomes (Fig. 3a). Next, Ca2+ was also added as it was required both to enable liposome tethering by the C2 domains of E-Syt1 and to relieve their inhibitory function on lipid transport24–26. Thus, in this system, E-Syt1cyto acted both as the tether (His-tag and C2 domains) and as the lipid transport protein (SMP domain). As expected, addition of E-Syt1cyto resulted in an increase in turbidity of the liposome mixture, which was revealed by optical density reading at 405 nm (turbidity) (Fig. 3b), reflecting E-Syt1cyto - dependent aggregation of liposomes into large particles. Importantly, lipid transport was observed under all conditions, as the fluorescence of NBD, which was quenched by rhodamine in the donor liposomes, increased due to dequenching, revealing dilution of NBD-PE and rhodamine-PE as they were transferred to acceptor liposomes. However, tethering and lipid transfer between DNA-free liposomes occurred with slower kinetics when the standard buffer was replaced by the high-salt buffer (Fig. 3b and c). Additionally, in the high salt buffer, DNA-ring-templated liposomes showed slightly lower tethering and lipid transfer activities than DNA-free liposomes (Fig. 3b and c). This slight difference in lipid transport cannot be attributed to the smaller size of DNA-ring-templated liposomes (28 nm in diameter) relative to DNA-free liposomes (large unilamellar vesicles, LUV) as control experiments using DNA-free liposomes of different size (sonicated small unilamellar vesicles, SUVs; 100- and 400-nm LUVs) showed that in fact lipid transport was slightly faster with the SUV (~30 nm) liposomes (Supplementary Fig. 5a, see below for the constructs used in this assay). Thus, both high-salt and DNA nanostructures partially reduce the activity of E-Syt1cyto. We conclude that in spite of the slower transfer achieved by DNA-templated liposomes, these results validate the use of our experimental system for investigations of lipid transport between bilayers.
Figure 3. E-Syt1cyto clusters and mediates lipid transfer between DNA-origami-templated liposomes.
a, Domain structures of E-Syt1 and E-Syt1cyto (left) and schematic representation of E-Syt1cyto-mediated liposome tethering and lipid transfer in the presence of Ca2+ (right). b, (Left) Representative time courses of tethering of donor and acceptor liposomes in the presence of E-Syt1cyto and Ca2+, as assessed by the increase in turbidity (OD at 405 nm). (Right) Dot plots showing OD405 levels after incubation for 10 min (arrow in the left panel). c, (Left) Representative time courses of lipid transfer between donor and acceptor liposomes in the presence of E-Syt1cyto and 100 μM Ca2+ at RT as assessed by the dequenching of NBD-PE fluorescence. (Right) Dot plots showing quantification of NBD fluorescence at the end of the incubation (arrow in the left panel). Mean and standard deviations (error bars) are shown (n=3 independent experiments).
Validation of shuttle mechanism of lipid transfer
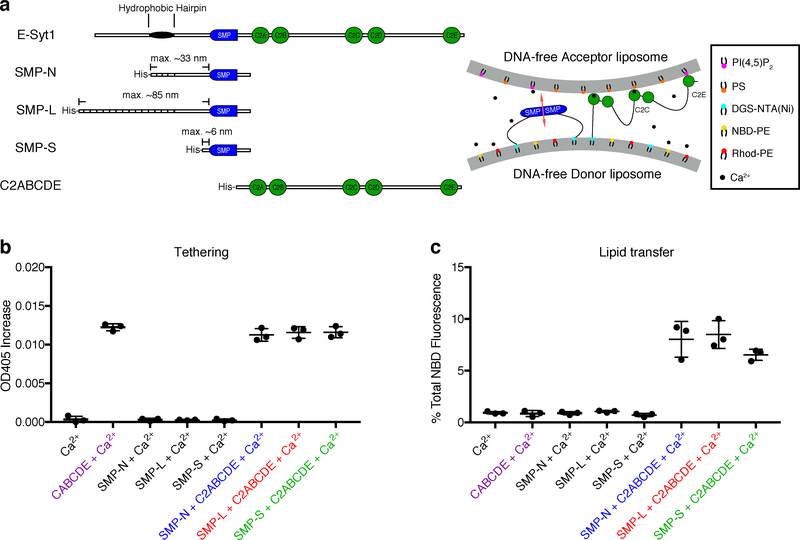
To assess whether SMP domains can transport lipids by shuttling between the two liposomes within DNA-organized liposome dimers, we generated constructs comprising the SMP domain of E-Syt1 and upstream “linker” sequences of different length. These constructs, which were preceded by a His-tag, were devoid of the entire downstream C2 domain-containing region of the protein to prevent E-Syt1-dependent liposome aggregation, thus limiting our interrogation to lipid transport within individual DNA-templated liposome dimers (Fig. 4a). In one construct, SMP-native (SMP-N, Fig. 4a and Supplementary Fig. 4), the upstream linker region consisted of the 47 amino acid (a.a.) sequence (predicted to be unfolded) that connects the SMP domain to the hydrophobic hairpin region in wild-type E-Syt1, plus 35 a.a. derived from the vector. In another construct, SMP-long (SMP-L, Fig. 4a and Supplementary Fig. 4), the native a.a. sequence was replaced by a concatemer of 4 such sequences, resulting in a 214 a.a. linker. We used a concatemer of the native sequence for this longer construct to minimize the chance that this sequence could negatively impact the function of the SMP domain. In a third construct, SMP-short (SMP-S, Fig. 4a and Supplementary Fig. 4), the linker region comprised the SMP domain flanked at its N-terminus only by the 16 a.a. derived from the vector.
Figure 4. The SMP domain of E-Syt1 alone does not cluster liposomes but transfers lipids between protein-tethered liposomes.
a, Domain structures of E-Syt1 constructs (left) and schematic representation of SMP-mediated lipid transfer between liposomes tethered by the C2ABCDE of E-Syt1 in the presence of Ca2+ (right). b, Dot plots showing liposomes aggregation (or the lack thereof), due to tethering of donor and acceptor liposomes in the presence of various E-Syt1 constructs and 100 μM Ca2+ at RT, as assessed by the increase in turbidity (OD405) levels after incubation for 10 min (arrow in Supplementary Fig. 5b). c, Dot plots showing lipid transfer between donor and acceptor liposomes in the presence of E-Syt1 constructs and 100 μM Ca2+ at RT, as assessed by the increase in NBD-PE fluorescence at the end of the incubation (arrow in Supplementary Fig. 5c). Mean and standard deviations (error bars) are shown (n=3 independent experiments).
Prior to performing lipid transfer studies using these constructs with DNA-templated liposome dimers, it was important to confirm that all three constructs did not tether liposomes, thus ensuring that liposomes were kept in close proximity only with the help of DNA nanostructures, and not by liposome aggregation. A turbidity assay confirmed that this was indeed the case, whereas the same assay performed with the additional C2ABCDE fragment of E-Syt1 (Fig. 4a and Supplementary Fig. 4) resulted in major turbidity increases, reflecting robust aggregation (Fig. 4b and Supplementary Fig. 5b). As expected, SMP domain-dependent lipid transfer, which was detected by fluorescence increase, occurred only when liposomes were tethered by the C2ABCDE fragment (Fig. 4c and Supplementary Fig. 5c). Furthermore, similar lipid transfer activities were observed with all three constructs, confirming that the different linkers had negligible influence on the function of the SMP domain.
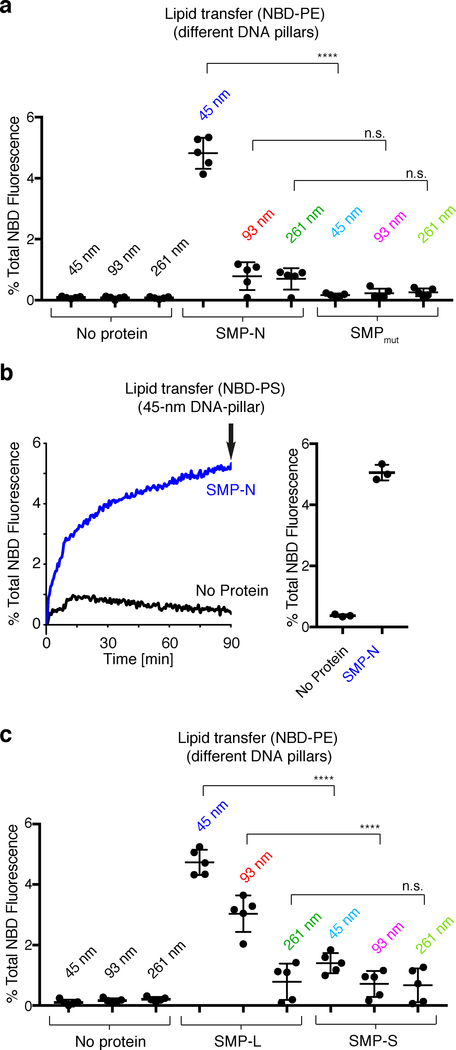
We next examined SMP-N-mediated lipid transfer using DNA-templated liposome dimers with a pillar length of 45 nm. In such dimers, the average distance between the bilayers of the two liposomes is estimated to be ~26 nm (see above). SMP-N (~33 nm when fully stretched, note that the thickness of the SMP domain is 2.5 or 4.5 nm depending on its orientation to the membrane) was first added to the monomeric DNA-templated donor liposomes at a protein to lipid molar ratio of 1:250. 20% DGS-NTA(Ni) was used in the donor liposomes to allow efficient binding of the protein to the membrane (Supplementary Fig. 6). Subsequently, these liposomes were mixed with the monomeric DNA-templated acceptor liposomes for 90 min to allow dimer formation and lipid transport (Supplementary Fig. 7a). Presence of the protein did not affect dimerization efficiency or the distance between the two liposomes (Fig. 2d and Supplementary Fig. 8). However, such presence resulted in a robust increase in fluorescence, signaling the occurrence of lipid transfer (Fig. 5a and Supplementary Fig. 7b). Consistent with the reported property of the E-Syts to harbor and/or transfer glycerolipids between membranes without major selectivity for specific head groups10, a similar increase in fluorescence was observed when NBD-PE in the donor liposomes was replaced by NBD-PS (Fig. 5b). In contrast, the mutant SMP-N carrying V169W and L308W, which was reported to impair lipidtransfer activity of E-Syt125, showed no NBD fluorescence increase (Fig. 5a and Supplementary Fig. 7b).
Figure 5. SMP mediates lipid transfer between the two liposomes within DNA-organized liposomes dimers.
a, Dot plots showing lipid transfer between donor and acceptor liposomes separated by 45-nm, 93-nm, and 261-nm DNA pillars in the absence or presence of the SMP-N or SMPmut at RT, as assessed by the increase in the NBD-PE fluorescence at the end of the incubation (arrow in Supplementary Fig. 7b). b, Same as in (a), but using 45-nm DNA pillars and donor liposomes in which NBD-PE was replaced by NBD-PS. Representative time courses are on the left and dot plots showing the NBD fluorescence at the end of the incubation (arrow in the left panel) are on the right. c, Same as in (a), but transport was mediated by SMP constructs with two different polypeptide linkers (SMP-L, 214 a.a. and SMP-S, 16 a.a.). Mean and standard deviations (error bars) are shown [n=3 independent experiments in (b), n=5 independent experiments in (a) and (c)]. Bonferroni’s multiple comparisons test. **** P<0.0001; n.s., not significant.
Next we tested SMP-S and SMP-L with dimeric DNA nanostructures comprising 45-, 93- and 261-nm DNA pillars (and thus bilayer distances of circa 26, 67 and 224 nm, see above). Similar to SMP-N, SMP-L, with a linker predicted to span 85 nm when fully stretched (Fig. 4a), showed robust lipid transfer between liposomes separated by the 45-nm pillars (Fig. 5c and Supplementary Fig. 7c). More importantly, lipid transfer activity of SMP-L was observed when liposomes were separated by the 93-nm pillars (bilayer distance approximately 67 nm), but not by the 261-nm pillars (bilayer distance approximately 224 nm) (Fig. 5c and Supplementary Fig. 7c). In contrast, shortening the linker region of the SMP domain to a maximum length of ~6 nm (SMP-S, Fig. 4a) dramatically reduced the lipid transfer activity on all liposome dimers, even with the bilayers placed as close as ~26 nm (Fig. 5c and Supplementary Fig. 7c). This defect was not due to an impairment of the SMP domain per se, as SMP-S was functional on liposomes tethered by the C2ABCDE construct (Fig. 4 and Supplementary Fig. 5).
To confirm that the properties shown here are not unique of the SMP domain of E-Syt1, we tested in the same DNA-origami-based lipid transport assay the SMP domain of TMEM24, which is similar in structure and length to the SMP domain of of E-Syt112. TMEM24 is an ER-plasma membrane tethering protein and lipid transporter enriched in neuroendocrine cells and neurons12,42. We generated and purified the SMP of TMEM24 (SMPTMEM24) with a His-tagged N-terminal linker region composed of 78 a.a. (~31 nm when fully stretched, Supplementary Fig. 9a). Consistent with the results obtained with SMP-N (Fig. 5a and Supplementary Fig. 7b), SMPTMEM24, in the presence of DNA-ring-templated liposome pairs, transported lipids between liposomes separated by the 45-nm DNA pillars, although the bilayer distance (~26 nm) far exceeds the length of the SMP domain. However, it did not transport lipids between liposome dimers separated by 93- or 261-nm pillars, where the bilayer distances far exceed the length of the linker (Supplementary Fig. 9b).
Collectively, these findings further indicate that exchange of lipids occurs within DNA-organized liposome dimers and validate the suitability of our nano-engineered system to study distance-dependent lipid transport between bilayers. Importantly, our data support the hypothesis that SMP domains can act as shuttles in transporting lipids between apposed bilayers (Fig. 6).
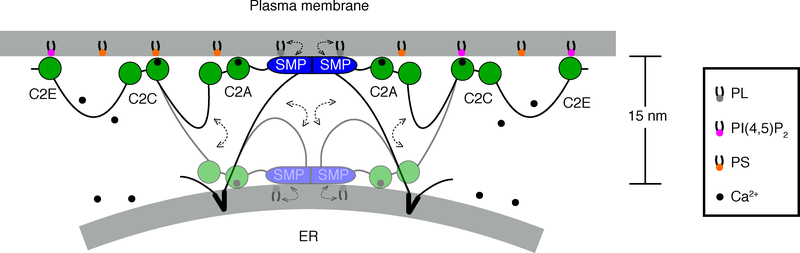
Figure 6. Model for E-Syt1-mediated lipid transfer.
The interaction of E-Syt1 with the plasma membrane mediated by its C2C and C2E domain allows E-Syt1 to function as a tether at ER-plasma membrane contact sites. Both these interactions are triggered by elevation of cytosolic Ca2+.20,22–25,35 The SMP domain shuttles between the two membranes to transfer lipids. The Ca2+-binding C2A domain, which is tightly conjoined with the C2B domain, is theorized to move together with SMP domain. Ca2+ ions are shown as small black circles. Double-headed dashed arrows indicate the movement of SMP domain together with the C2A-C2B domain module.
Discussion
DNA nanotechnology has allowed for the construction of precisely engineered lipid bilayer structures that are amenable to biochemical assays (e.g. FRET-based analyses), thus providing a unique opportunity to study membrane dynamics quantitatively and systematically32. In this work, we joined two DNA-ring-templated liposomes with distinct lipid compositions via a rigid DNA pillar. Unlike liposomes tethered by proteins, which cluster into tight aggregates, here the two closely apposed lipid bilayers can be maintained at customized distances with nanoscale precision by programmable DNA nanostructures. Thus, the DNA-origami-directed membrane engineering technique offers a solution to generate membrane appositions in vitro with controllable membrane proximity.
Here we have used DNA-origami-organized liposomes to assess whether the SMP domain of E-Syt1 can transfer lipids at a distance that exceeds its length. In our experimental system, membrane tethering via C2 domains was replaced by membrane tethering via DNA nanostructures, allowing to control liposome separation distance in a pre-defined, non-aggregating way. An important issue of these studies was the demonstration that the FRET signal changes observed in our assay reflected lipid transfer between pairs of liposomes within dimeric DNA nanostructures. This was found to be the case as only minimal FRET signal changes occurred with untethered liposomes by SMP domain. Note that the average distance of untethered liposomes in suspension under the conditions of our assay — liposome size used and lipid concentration (0.1–0.5 mM) used — is estimated to be around 403–709 nm43.
We show that upon anchoring to the donor liposomes via unstructured linker regions of different length, the SMP domain of E-Syt1 can transfer lipids within the dimeric structures at distances that greatly exceed the length of dimeric SMP domain (~9 nm) but are not longer than the length of those linker regions. The shuttle model is consistent with the hydrophilic nature of the two tips of the SMP domain and with the lack of evidence for the property of the SMP domain to form tetramers or longer adducts to bind bilayers and bridge them10. The polypeptide sequence that links the SMP domain of E-Syt1 to its intramembrane hydrophobic hairpin (47 a.a.) is predicted to be unfolded. With the maximal theoretical length approaching 19 nm, this linker is long enough to allow the SMP domain to shuttle lipids over the inter-membrane distance typically found between the ER and the plasma membrane (~15 nm) in cells expressing E-Syt1 under high cytosolic Ca2+, where recruitment of E-Syt1 to the plasma membrane and its lipid transfer functions are stimulated20–25.
A shuttling of the SMP domain of E-Syt1 over an approximately 15 nm distance, however, raises the conundrum of how such shuttling can be compatible with the presence at the C-terminal side of the SMP domain of a closely adjacent C2 domain (C2A domain) with membrane binding properties18,24. The C2A domains of the E-Syts bind membranes in a Ca2+-dependent way18,24,44 and were proposed to bind the plasma membrane45. However, the length of the unstructured linker that separates it from the SMP domain is too short (~ 8 nm when fully stretched) to span the ER-plasma membrane distance when the SMP domain is in close proximity of the ER during its shuttling. Thus, we propose that in living cells the tandem arranged C2A-C2B domains10,45 move together with the SMP domain between the two membranes during lipid transport (Fig. 6). In support of this possibility, membrane binding by the C2A domain is not selective for negatively charged bilayers, thus making plausible its binding to both bilayers18,24. Its proximity to the SMP domain may help orient the SMP domain at the membrane interface for lipid extraction/delivery. Clearly, elucidating the dynamics of E-Syt-mediated lipid transfer warrants further investigation in the future. Outstanding questions include how and at what pace the SMP domain extracts lipids from a bilayer and delivers them to the target bilayer. A combination of our platform with single-molecule experiments could shed light on these questions.
Our current study with the SMP domain of E-Syt1 validates the concept of using DNA-origami-guided membrane structures to modulate and study lipid transport. In theory, this system can be adapted to study other lipid transport proteins as well as enzymatic reactions thought to involve “in trans” interactions of two membranes46,47. Modification of the nanostructures used here will enable the generation of different membrane curvature and/or topology. An important design consideration is the rigidity of the DNA nanostructures that host and organize membranes. While the six-helix-bundle DNA rods (persistence length ≈ 1–3 μm)48,49 appear to be sufficiently stiff for the purpose this study (maximum inter-membrane distance < 300 nm), reinforced DNA structures can be constructed50 when studying proteins that function across longer range or exert mechanical forces on membranes. Finally, it may be possible to use these nanostructures to aid the structural analysis of tethering proteins by cryo-EM.
Online Methods
Chemicals
Chemicals were from the following sources: Isopropyl-β-D-thiogalactoside (IPTG) (AmericanBio), His60 Ni Superflow Resin (Takara), tris (2-carboxyethyl) phosphine (TCEP) (Thermo), Iodixanol solution (60%, w/v) (Cosmo Bio USA), Octyl β-D-glucopyranoside (OG) (EMD Millipore), n-dodecyl-β-D-maltopyranoside (DDM) (Avanti Polar Lipids). All DNA oligonucleotides were purchased from Integrated DNA Technologies. All lipids were obtained from Avanti Polar Lipids: 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC); 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS); L-α-phosphatidylinositol- 4,5-bisphosphate [PI(4,5)P2]; 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2–1,3-benzoxadiazol-4-yl) (NBD-PE); 1,2-dioleoyl-sn-glycero-3-phospho-L-serine-N-(7-nitro-2–1,3-benzoxadiazol-4-yl) (NBD-PS); 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (Rhod-PE); 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl) iminodiacetic acid) succinyl] [DGS-NTA(Ni)]; 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[4-(p-maleimidophenyl)butyramide] (MPB-PE).
Plasmids
The plasmids encoding E-Syt1cyto and C2ABCDE were previously described24.
The region coding for the SMP domain of E-Syt1 plus its upstream sequence up to the hydrophobic hairpin (a.a. 93–327 of E-Syt1, referred to here as SMP-N) or SMP domain of TMEM24 (a.a. 34–260) was cloned using NotI and XhoI sites into the pET-28a vector (Novagen). SMP-S (a.a. 135–327 of E-Syt1) was cloned using NdeI and NotI sites into the pET-28a vector. SMP-L including 3× a.a. 93–136 and 93–327 was cloned into the pET-28a vector using BamHI, SacI, SalI, NotI and XhoI sites, respectively. L308W/V169W mutations were introduced in SMP-N by site-directed mutagenesis25.
Protein Expression and Purification
Soluble fragments of human E-Syt1 or TMEM24 were expressed in Expi293 cells or BL21 (DE3) RIL Codon Plus (Agilent) E. coli cells and purified as described previously24. Briefly, cells were harvested and lysed in buffer A [25 mM Tris-HCl, pH 8.0, 300 mM NaCl, 10 mM imidazole, 1x complete EDTA-free protease inhibitor cocktail (Roche), 0.5 mM TCEP] by three freeze–thawing cycles using liquid nitrogen (for Expi293 cells) or by sonication (for bacteria). The suspension was clarified by centrifugation at 17,000 × g for 30 min at 4 °C and the protein was purified from the supernatant by a Ni-NTA column. After elution, the protein was further purified by gel filtration. Fractions containing E-Syt1 or TMEM24 fragments were pooled and concentrated.
Liposome Preparation
DNA-free donor liposomes were composed as follows: 87:1.5:1.5:10 mole percent of DOPC: NBD-PE: Rhod-PE: DGS-NTA(Ni) or 87:1.5:1.5:10 mole percent of DOPC: NBD-PS: Rhod-PE: DGS-NTA(Ni). DNA-free acceptor liposomes were composed as follows: 85:10:5 mole percent DOPC: POPS: PI(4,5)P2. Liposome preparation was performed as previously described24. Briefly, lipid mixtures were dried onto a film. Lipid films were then hydrated with buffer A. DNA-free liposomes were generated by ten freeze-thaw cycles in liquid N2 and 37 °C water bath. LUVs were formed by extrusion through polycarbonate filters with a pore size of 50, 100 or 400 nm (Avanti Polar Lipids). SUVs were formed by sonication for 20 min using a probe tip sonicator (Virtis Virsonic).
DNA Origami Design and Preparation
Monomeric DNA-origami cages were designed using caDNAno (cadnano.org). Inner and outer handle sequences (21-nt) were generated by NUPACK (nupack.org) and manually added to the 3’-ends of the appropriate staple strands. DNA scaffold strands (8064-nt) were produced using E. coli and M13-derived bacteriophages28,30. The DNA cage monomers were assembled from a scaffold strand (20 nM) and a pool of staple strands (140 nM each) in buffer B (5 mM Tris-HCl, pH 8.0, 12 mM MgCl2) using a 36-hour thermal annealing program. Correctly assembled DNA cages were purified via rate-zonal centrifugation in glycerol gradients as described previously34.
Preparation of lipidated DNA anti-handles
The lipid-DNA conjugate was prepared as previously described with slight changes34. Briefly, 1 mM 5’-thiol labeled DNA oligonucleotides were treated with 20 mM TCEP in buffer C (25 mM HEPES, pH 7.4, 140 mM KCl) for 30 min. 900 μM pre-treated thiol-modified DNA oligonucleotides were reacted with 3.3 mM pre-dried MPB-PE and 6.7 mM pre-dried donor lipid mixtures [77% DOPC, 20% DGS-NTA(Ni), 1.5% Rhod-PE, 1.5% NBD-PE] or pre-dried acceptor lipid mixtures [85% DOPC, 10% POPS, 5% PI(4,5)P2] in buffer C containing 2% OG at RT for 30 min. This reaction mixture was then dialyzed in 7 Kd molecular weight cut-off (MWCO) cassette against buffer C overnight in order to incorporate lipidated DNA molecules into liposomes. The conjugation products were separated from unconjugated DNA via isopycnic centrifugation in iodixanol gradients and later analyzed by SDS-PAGE. Fractions containing DNA-lipid conjugates were combined.
Preparation of DNA-Origami-Organized Liposomes
DNA-origami-organized liposomes were prepared as previously described with slight changes34. Briefly, 20 nM assembled DNA nanostructures were first labeled with 1280 nM lipidated anti-handles buffer D (25 mM HEPES, pH 7.4, 400 mM KCl, 10 mM MgCl2) containing 1% OG at 37°C for 1 h. To form DNA-origami-organized liposomes, 0.6 mM rehydrated donor or acceptor lipid mixtures were added to 10 nM lipid-labeled DNA nanostructures. The solution was diluted to 500 μL in buffer D with 1% OG, gently shaken for 30 min at RT, put into a 7 kD MWCO dialysis cassette, and dialyzed against buffer D overnight. The dialyzed solutions were subjected to centrifugation in iodixanol gradients. A quasi-linear gradient containing 2.4 mL of 6%–26% (w/v) iodixanol was loaded on top of 2 mL dialyzed sample containing 30% iodixanol. Gradients were then centrifuged at 50,000 rpm for 5 h at 4°C. Fractions were analyzed by 1.5% agarose gel with 0.05% SDS and those containing DNA-origami-organized liposomes were combined.
Electron Microscopy
To prepare negatively stained DNA nanostructures or DNA-origami-organized liposomes with or without proteins, a drop of sample (5 μL) was deposited on a glow discharged formvar/carbon coated copper grid and incubated for 1–3 min at RT. Fluid was then removed by blotting with a filter paper. The grid was immediately stained for 3 min with 2% (w/v) uranyl formate. Grids were examined using a JEOL JEM-1400 Plus microscope equipped with a LaB6 filament (acceleration voltage: 80 kV). Images were acquired by an Advanced Microscopy Technologies bottom-mount 4k×3k CCD camera.
Analysis of DNA Nanostructures Dimerization Efficiency
5 μL of DNA nanostructures or DNA-origami-organized liposomes with or without proteins were incubated with 0.2 μL 1 mM 3’-Cy3 labeled DNA oligonucleotides at 37 °C for 30 min. The samples were then analyzed by electrophoresis in 1.5% agarose gels with 0.05% SDS and 10 mM MgCl2. Fluorescence of DNA bands was visualized using a Typhoon FLA 9500 scanner (GE Healthcare) and analyzed using ImageJ (NIH).
Lipid Transfer Assays Using DNA-Free Liposomes
All in vitro lipid transfer assays with soluble proteins were performed as previously described24. Briefly, reactions were performed in 50 μL volumes with a final lipid concentration of 0.5 mM, with donor and acceptor liposomes added at a 1:1 ratio. Reactions were initiated by the addition of proteins to the liposome mixtures (protein: lipid ratio 1: 1000) in a 96-well plate (Corning). The fluorescence intensity of NBD was monitored with an excitation of 460 nm and emission of 538 nm every 10 sec over 30 min at RT by using SpectraMax M5 Microplate Reader (Molecular Devices). All data were corrected by setting the data point at 0 min to zero, and subtracting the baseline values obtained in the absence of proteins. The data were expressed as a percentage of the maximum fluorescence, determined after adding 10 μL of 2.5% DDM to the reactions after 30 min. All experiments were repeated 3 times and a representative trace is shown. Bars represent average fluorescence values ± standard deviation from the three-pooled readings.
Lipid Transfer Assays Using DNA-Origami-Organized Liposomes
Lipid transfer assays using DNA-origami-organized liposomes were performed as for the lipid transfer assays using DNA-free liposomes, with the exception that 1 μM SMP domain was preincubated with 100 μM DNA-ring-templated donor liposomes for 5 min at RT. Reactions were initiated by the addition of 100 μM DNA-ring-templated acceptor liposomes in a 96-well plate (Corning). The DDM detergent was added to stop the reactions after 90 min. All experiments were repeated 3 or 5times and a representative trace is shown. Bars represent average fluorescence values ± standard deviation from the three- or five-pooled readings.
Liposome Tethering Assays
Liposome tethering assays with soluble proteins were performed as previously described24. Briefly, the reactions were initiated by the addition of proteins to the mixture of donor and acceptor liposomes (1:1 ratio) in a 96-well plate (Corning) using SpectraMax M5 Microplate Reader (Molecular Devices). The absorbance at 405 nm was measured to assess turbidity. Data were expressed as absolute absorbance values subtracted by the absorbance prior to protein addition. All experiments were repeated 3 times. Bars represent average 405 nm absorbance values from the three-pooled readings.
Liposome Sedimentation Assays
SMP-N was added to 100 μL 250 μM DNA-ring-templated liposomes comtaining 0%, 5%, 10%, and 20% DGS-NTA(Ni), respectively, at a final concentration of 2.5 μM. After incubation at RT for 1 h, liposomes were pelleted by centrifugation at 165,000 × g for 30 min at 4 °C. Corresponding amounts of the supernatants and pellets were analyzed by SDS-PAGE and the Coomassie-stained gels were analyzed using Image J (NIH).
Statistical Analysis
Comparisons of data were carried out by t-test with Bonferroni corrections for multiple comparisons as appropriate with Prism 7 (GraphPad software).
Data Availability Statement
All raw data are available from the corresponding authors upon reasonable request.
Supplementary Material
Acknowledgments
We thank Yiying Cai for discussion. This work was supported by NIH grants NS036251 and DA018343, the HHMI and the Kavli Foundation to P.D.C., an NIH Director’s New Innovator Award (GM114830) and a Yale University faculty startup fund to C.L., a Human Frontier Science Program Long-term Fellowship to X.B., and by an Agency for Science, Technology and Research Graduate Scholarship (Singapore) to Q.X..
Footnotes
Competing Financial Interests Statement
The authors declare no conflict of interest.
References
- 1.Saheki Y & De Camilli P Endoplasmic Reticulum-Plasma Membrane Contact Sites. Annu Rev Biochem 86, 659–684 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Wu H, Carvalho P & Voeltz GK Here, there, and everywhere: The importance of ER membrane contact sites. Science 361, eaan5835 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong LH, Gatta AT & Levine TP Lipid transfer proteins: the lipid commute via shuttles, bridges and tubes. Nat Rev Mol Cell Biol 20, 85–101 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Stefan CJ, Manford AG & Emr SD ER-PM connections: sites of information transfer and inter-organelle communication. Curr Opin Cell Biol 25, 434–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.AhYoung AP et al. Conserved SMP domains of the ERMES complex bind phospholipids and mediate tether assembly. Proc Natl Acad Sci U S A 112, E3179–88 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeong H, Park J, Jun Y & Lee C Crystal structures of Mmm1 and Mdm12-Mmm1 reveal mechanistic insight into phospholipid trafficking at ER-mitochondria contact sites. Proc Natl Acad Sci U S A 114, E9502–E9511 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawano S et al. Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES. J Cell Biol 217, 959–974 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kopec KO, Alva V & Lupas AN Homology of SMP domains to the TULIP superfamily of lipid-binding proteins provides a structural basis for lipid exchange between ER and mitochondria. Bioinformatics 26, 1927–31 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee I & Hong W Diverse membrane-associated proteins contain a novel SMP domain. FASEB J 20, 202–6 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Schauder CM et al. Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature 510, 552–5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong LH & Levine TP Tubular lipid binding proteins (TULIPs) growing everywhere. Biochim Biophys Acta Mol Cell Res 1864, 1439–1449 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lees JA et al. Lipid transport by TMEM24 at ER-plasma membrane contacts regulates pulsatile insulin secretion. Science 355, eaah6171 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alva V & Lupas AN The TULIP superfamily of eukaryotic lipid-binding proteins as a mediator of lipid sensing and transport. Biochim Biophys Acta 1861, 913–23 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Kornmann B et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325, 477–81 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu LK, Choudhary V, Toulmay A & Prinz WA An inducible ER-Golgi tether facilitates ceramide transport to alleviate lipotoxicity. J Cell Biol 216, 131–147 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toulmay A & Prinz WA A conserved membrane-binding domain targets proteins to organelle contact sites. J Cell Sci 125, 49–58 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirabayashi Y et al. ER-mitochondria tethering by PDZD8 regulates Ca(2+) dynamics in mammalian neurons. Science 358, 623–630 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Min SW, Chang WP & Sudhof TC E-Syts, a family of membranous Ca2+-sensor proteins with multiple C2 domains. Proc Natl Acad Sci U S A 104, 3823–8 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manford AG, Stefan CJ, Yuan HL, Macgurn JA & Emr SD ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev Cell 23, 1129–40 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Giordano F et al. PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell 153, 1494–509 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang CL et al. Feedback regulation of receptor-induced Ca2+ signaling mediated by E-Syt1 and Nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Rep 5, 813–25 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Fernandez-Busnadiego R, Saheki Y & De Camilli P Three-dimensional architecture of extended synaptotagmin-mediated endoplasmic reticulum-plasma membrane contact sites. Proc Natl Acad Sci U S A 112, E2004–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Idevall-Hagren O, Lu A, Xie B & De Camilli P Triggered Ca2+ influx is required for extended synaptotagmin 1-induced ER-plasma membrane tethering. EMBO J 34, 2291–305 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bian X, Saheki Y & De Camilli P Ca(2+) releases E-Syt1 autoinhibition to couple ER-plasma membrane tethering with lipid transport. EMBO J 37, 219–234 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saheki Y et al. Control of plasma membrane lipid homeostasis by the extended synaptotagmins. Nat Cell Biol 18, 504–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu H et al. Extended synaptotagmins are Ca2+-dependent lipid transfer proteins at membrane contact sites. Proc Natl Acad Sci U S A 113, 4362–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orci L et al. From the Cover: STIM1-induced precortical and cortical subdomains of the endoplasmic reticulum. Proc Natl Acad Sci U S A 106, 19358–62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Douglas SM et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothemund PW Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Dietz H, Douglas SM & Shih WM Folding DNA into twisted and curved nanoscale shapes. Science 325, 725–30 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grome MW, Zhang Z, Pincet F & Lin C Vesicle Tubulation with Self-Assembling DNA Nanosprings. Angew Chem Int Ed Engl 57, 5330–5334 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu W et al. A Programmable DNA Origami Platform to Organize SNAREs for Membrane Fusion. J Am Chem Soc 138, 4439–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y et al. Self-assembly of size-controlled liposomes on DNA nanotemplates. Nat Chem 8, 476–83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z, Yang Y, Pincet F, Llaguno MC & Lin C Placing and shaping liposomes with reconfigurable DNA nanocages. Nat Chem 9, 653–659 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan YH, van Lengerich B & Boxer SG Effects of linker sequences on vesicle fusion mediated by lipid-anchored DNA oligonucleotides. Proc Natl Acad Sci U S A 106, 979–84 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franquelim HG, Khmelinskaia A, Sobczak JP, Dietz H & Schwille P Membrane sculpting by curved DNA origami scaffolds. Nat Commun 9, 811 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perrault SD & Shih WM Virus-inspired membrane encapsulation of DNA nanostructures to achieve in vivo stability. ACS Nano 8, 5132–40 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beales PA & Vanderlick TK Specific binding of different vesicle populations by the hybridization of membrane-anchored DNA. J Phys Chem A 111, 12372–80 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Kumar N et al. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J Cell Biol 217, 3625–3639 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valverde DP et al. ATG2 transports lipids to promote autophagosome biogenesis. J Cell Biol, jcb.201811139 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin C, Perrault SD, Kwak M, Graf F & Shih WM Purification of DNA-origami nanostructures by rate-zonal centrifugation. Nucleic Acids Res 41, e40 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun EW et al. Lipid transporter TMEM24/C2CD2L is a Ca(2+)-regulated component of ER-plasma membrane contacts in mammalian neurons. Proc Natl Acad Sci U S A 116, 5775–5784 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu W, Wang J, Rothman JE & Pincet F Accelerating SNARE-Mediated Membrane Fusion by DNA-Lipid Tethers. Angew Chem Int Ed Engl 54, 14388–92 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma L et al. Single-molecule force spectroscopy of protein-membrane interactions. Elife 6, pii: e30493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu J et al. Structure and Ca(2)(+)-binding properties of the tandem C(2) domains of E-Syt2. Structure 22, 269–80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stefan CJ et al. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell 144, 389–401 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Eden ER, White IJ, Tsapara A & Futter CE Membrane contacts between endosomes and ER provide sites for PTP1B-epidermal growth factor receptor interaction. Nat Cell Biol 12, 267–72 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Kauert DJ, Kurth T, Liedl T & Seidel R Direct mechanical measurements reveal the material properties of three-dimensional DNA origami. Nano Lett 11, 5558–63 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Liedl T, Hogberg B, Tytell J, Ingber DE & Shih WM Self-assembly of three-dimensional prestressed tensegrity structures from DNA. Nat Nanotechnol 5, 520–4 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castro CE, Su HJ, Marras AE, Zhou L & Johnson J Mechanical design of DNA nanostructures. Nanoscale 7, 5913–21 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data are available from the corresponding authors upon reasonable request.