Abstract
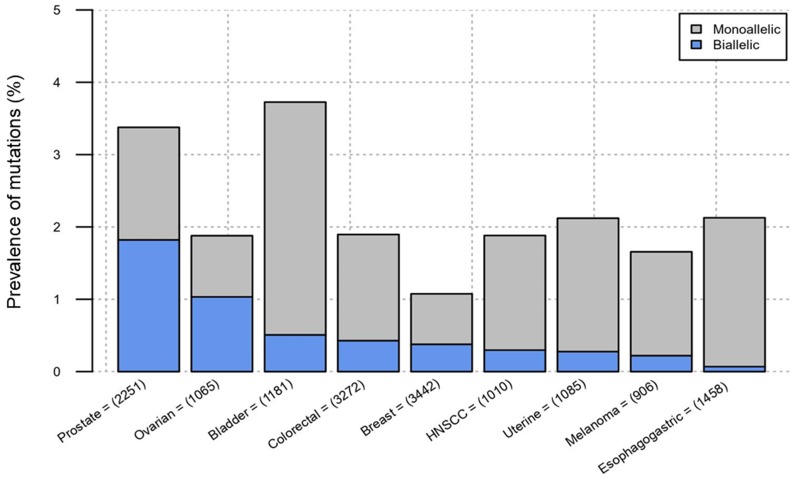
Inactivating CDK12 alterations have been reported in ovarian and prostate cancers and may have therapeutic implications; however, the prevalence of these mutations across other cancer types is unknown. We searched the cBioPortal and GENIE Project (public release v4.1) databases for cancer types with > 200 sequenced cases, that included patients with metastatic disease, and in which the occurrence of at least monoallelic CDK12 alterations was > 1%. The prevalence of at least monoallelic CDK12 mutations was highest in bladder cancer (3.7%); followed by prostate (3.4%), esophago-gastric (2.1%) and uterine cancers (2.1%). Biallelic CDK12 inactivation was highest in prostate cancer (1.8%), followed by ovarian (1.0%) and bladder cancers (0.5%). These results are the first (to our knowledge) to estimate the prevalence of monoallelic and biallelic CDK12 mutations across multiple cancer types encompassing over 15,000 cases.
Keywords: prostate cancer, CDK12, genetics, immunotherapy, biomarkers
INTRODUCTION
Inactivating CDK12 alterations have been reported in ovarian and prostate cancers; however, the prevalence of these mutations across all cancer types is unknown [1]. While CDK12 was initially thought to be involved in homologous-recombination DNA repair, emerging data suggest a unique role of this gene in DNA replication-associated repair. To this end, it has been suggested that inactivating CDK12 mutations lead to widespread focal genomic duplications that generate gene fusion-induced neoantigens and favorable responses to immune-checkpoint blockade therapy using PD-1 inhibitors [2]. Given this potentially actionable molecular subtype, we sought to determine the prevalence of monoallelic and biallelic CDK12 alterations across tumor types.
RESULTS
Datasets (in cBioPortal and GENIE) from prostate, breast, colorectal, bladder, ovarian, uterine, head-and-neck squamous cell carcinoma, melanoma, and esophago-gastric cancers were included (Table 1); other tumor types did not reach a 1% frequency of CDK12 alterations. The prevalence of at least monoallelic CDK12 mutations was highest in bladder cancer (3.7%); followed by prostate (3.4%), esophago-gastric (2.1%) and uterine cancers (2.1%). Biallelic CDK12 inactivation was highest in prostate cancer (1.8%), followed by ovarian (1.0%) and bladder cancers (0.5%) (Figure 1).
Table 1. Datasets publically available from cBioPortal and GENIE Project that were used, by disease group, with overall sample size.
| Disease | Dataset | Sample Size | Total |
|---|---|---|---|
| Bladder | BLCA_TCGA_PAN_CAN_ATLAS_2018 | 408 | 1,181 |
| DFCI-ONCOPANEL-3 | 69 | ||
| MSK-IMPACT341 | 95 | ||
| MSK-IMPACT410 | 326 | ||
| MSK-IMPACT468 | 143 | ||
| UTUC_MSKCC_2013 | 84 | ||
| VICC-01-T5A | 3 | ||
| VICC-01-T7 | 53 | ||
| Breast | BRCA_IGR_2015 | 216 | 3,442 |
| BRCA_MBCPROJECT_WAGLE_2017 | 157 | ||
| DFCI-ONCOPANEL-3 | 304 | ||
| MSK-IMPACT341 | 410 | ||
| MSK-IMPACT410 | 1,021 | ||
| MSK-IMPACT468 | 1,076 | ||
| VICC-01-T5A | 87 | ||
| VICC-01-T7 | 171 | ||
| Colorectal | CRC_MSK_2018 | 1,134 | 3,272 |
| DFCI-ONCOPANEL-3 | 351 | ||
| MSK-IMPACT341 | 209 | ||
| MSK-IMPACT410 | 906 | ||
| MSK-IMPACT468 | 465 | ||
| VICC-01-T5A | 47 | ||
| VICC-01-T7 | 160 | ||
| Esophagogastric | DFCI-ONCOPANEL-3 | 146 | 1,458 |
| EGC_MSK_2017 | 341 | ||
| ESCA_TCGA_PAN_CAN_ATLAS_2018 | 182 | ||
| MSK-IMPACT341 | 122 | ||
| MSK-IMPACT410 | 216 | ||
| MSK-IMPACT468 | 106 | ||
| STES_TCGA_PUB | 288 | ||
| VICC-01-T5A | 11 | ||
| VICC-01-T7 | 46 | ||
| HNSCC | DFCI-ONCOPANEL-3 | 83 | 1, 010 |
| HNC_MSKCC_2016 | 151 | ||
| HNSC_TCGA_PAN_CAN_ATLAS_2018 | 517 | ||
| MSK-IMPACT341 | 37 | ||
| MSK-IMPACT410 | 132 | ||
| MSK-IMPACT468 | 75 | ||
| VICC-01-T5A | 6 | ||
| VICC-01-T7 | 9 | ||
| Melanoma | 906 | ||
| MSK-IMPACT341 | 64 | ||
| MSK-IMPACT410 | 364 | ||
| MSK-IMPACT468 | 214 | ||
| VICC-01-T7 | 140 | ||
| VICC-01-T5A | 37 | ||
| Ovarian | DFCI-ONCOPANEL-3 | 125 | 1,065 |
| MSK-IMPACT341 | 88 | ||
| MSK-IMPACT410 | 139 | ||
| MSK-IMPACT468 | 155 | ||
| OV_TCGA_PUB | 489 | ||
| VICC-01-T5A | 31 | ||
| VICC-01-T7 | 38 | ||
| Prostate | DFCI-ONCOPANEL-3 | 97 | 2,251 |
| MSK-IMPACT341 | 153 | ||
| MSK-IMPACT410 | 569 | ||
| MSK-IMPACT468 | 377 | ||
| PRAD_FHCRC | 149 | ||
| PRAD_MICH | 61 | ||
| PRAD_MSKCC | 194 | ||
| PRAD_SU2C_2015 | 150 | ||
| PRAD_MSKCC_2017 | 501 | ||
| Uterine | DFCI-ONCOPANEL-3 | 120 | 1,085 |
| MSK-IMPACT341 | 119 | ||
| MSK-IMPACT410 | 258 | ||
| MSK-IMPACT468 | 326 | ||
| UCEC_TCGA_PUB | 232 | ||
| VICC-01-T7 | 19 | ||
| VICC-01-T5A | 11 |
Figure 1. Prevalence of CDK12 mutations across 9 cancer types.
DISCUSSION
In the era of precision oncology, inactivation of CDK12 may represent a new molecular subtype with therapeutic implications [6], although the pan-cancer prevalence of this genomic alteration was previously unknown. These results are the first (to our knowledge) to estimate the prevalence of monoallelic and biallelic CDK12 mutations across nine cancer types encompassing >15,000 cases. This is important as CDK12 alterations may be implicated in favorable responses to immune checkpoint inhibition, with biallelic alterations theoretically expected to respond better than monoallelic alterations. Prospective clinical trials (e.g. NCT03570619) are now needed to adequately assess this therapeutic hypothesis, and our data could be useful in the design of such trials.
Our results are limited to data that were publicly available. In addition, genotyping and mutation calling are sensitive to several factors, e.g. quality of the sample, sequencing depth and platform, and the pipeline used. Additionally, datasets from the GENIE Project revealed overall lower CDK12 mutation rates than datasets retrieved from cBioPortal. The reason for this is unclear but may include different pipelines with different sensitivity and specificity, artifacts due to DNA damage in sample preparation found in the capture-panels used in the GENIE Project, and differing sample quality (all samples from the GENIE Project were formalin-fixed paraffin-embedded while most from cBioPortal were fresh-frozen samples) [3–5]. Because of this, we hypothesize that our reported prevalences are likely underestimates of the true frequency of these mutations. Nevertheless, our analysis suggests that there are at least nine cancer types with a CDK12 mutation prevalence between 1-4%, hopefully prompting further exploration of immunotherapy approaches using a basket-trial design. Given the recent FDA-approval of larotrectinib for NTRK-altered cancers regardless of histologic type, we envision a similar mode of clinical exploration for CDK12-altered tumors.
METHODS
We searched the cBioPortal [3,4] and GENIE Project (public release v4.1) [5] databases for cancer types with ≥200 sequenced cases, that included patients with metastatic disease, and in which the prevalence of at least monoallelic CDK12 alterations was ≥1%. Analyses were restricted to datasets containing both CDK12 mutation and copy-number alteration (CNA) data using hybridization-capture panels from Dana-Farber Cancer Institute, Memorial Sloan-Kettering Cancer Center and Vanderbilt-Ingram Cancer Center. CDK12 mutations were considered inactivating (i.e. resulting in loss-of-function) in the case of homozygous loss, genomic rearrangements, frameshift or nonsense protein-truncating mutations, splice-site mutations, or missense mutations within the kinase domain. Monoallelic alterations were defined as at least one protein-truncating CDK12 variant; biallelic alterations were defined as a protein-truncating variant plus a second protein-truncating variant, a kinase domain missense variant, or loss-of-heterozygosity of the wild-type CDK12 allele. All analyses were performed in R.
Footnotes
CONFLICTS OF INTEREST
E.S.A. is a paid consultant/advisor to Janssen, Astellas, Sanofi, Dendreon, Medivation, Bristol Myers Squibb, AstraZeneca, Clovis, and Merck; he has received research funding to his institution from Janssen, Johnson & Johnson, Sanofi, Dendreon, Genentech, Novartis, Tokai, Bristol Myers-Squibb, AstraZeneca, Clovis, and Merck; and he is the co-inventor of a biomarker technology that has been licensed to Qiagen.
C.H.M. has previously received research funding from the Conquer Cancer Foundation (Bristol Myers-Squibb), travel support from Dava Oncology, and is a paid consultant to McGraw-Hill.
All other authors report no financial disclosures.
FUNDING
L.M and E.L.I. received support from National Institute of Health R01CA200859.
E.S.A. and C.H.M. are partially supported by National Institutes of Health Cancer Center Support Grant & P30 CA006973.
E. S. A. is partially supported by National Institutes of Health grant R01CA185297 and Department of Defense grant W81XWH-16-PCRP-CCRSA.
REFERENCES
- 1.Chilà R, Guffanti F, Damia G. Role and therapeutic potential of CDK12 in human cancers. Cancer Treat Rev. 2016;50:83–88. doi: 10.1016/j.ctrv.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Wu YM, Cieślik M, Lonigro RJ, Vats P, Reimers MA, Cao X, Ning Y, Wang L, Kunju LP, de Sarkar N, Heath EI, Chou J, Feng FY, et al. PCF/SU2C International Prostate Cancer Dream Team Inactivation of CDK12 Delineates a Distinct Immunogenic Class of Advanced Prostate Cancer. Cell. 2018;173:1770–1782.e14. doi: 10.1016/j.cell.2018.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–04. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.AACR Project GENIE Consortium AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017;7:818–31. doi: 10.1158/2159-8290.CD-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antonarakis ES. Cyclin-Dependent Kinase 12, Immunity, and Prostate Cancer. N Engl J Med. 2018;379:1087–89. doi: 10.1056/NEJMcibr1808772. [DOI] [PubMed] [Google Scholar]