Abstract
Developmental silencing of fetal globins serves as both a paradigm of spatiotemporal gene regulation and an opportunity for β-hemoglobinopathy therapeutic intervention. The NuRD chromatin complex participates in γ-globin repression. Here we use pooled CRISPR screening to comprehensively disrupt NuRD protein coding sequences in human adult erythroid precursors. We find essential for fetal hemoglobin (HbF) control a nonredundant subcomplex of NuRD protein family paralogs, whose composition we corroborate by affinity chromatography and proximity labeling mass spectrometry proteomics. Mapping top functional guide RNAs identifies key protein interfaces where in-frame alleles result in loss-of-function due to destabilization or altered function of subunits. We ascertain mutations of CHD4 that dissociate its requirement for cell fitness from HbF repression in both primary human erythroid precursors and transgenic mice. Finally we demonstrate that sequestering CHD4 from NuRD phenocopies these mutations. This work indicates a generalizable approach to discover protein complex features amenable to rational biochemical targeting.
Editorial summary:
Comprehensive CRISPR mutagenesis targeting all members of the NuRD complex identifies a specific sub-complex required for fetal globin silencing and informs a rational targeting strategy for elevating globin levels while avoiding cytotoxicity.
Severe hemoglobinopathies resulting from mutations of the adult β-globin gene (HBB) including sickle cell disease (SCD) and β-thalassemia affect millions worldwide1,2. Derepression of the fetal γ-globin genes (HBG1/HBG2) resulting in fetal hemoglobin (HbF, α2γ2) induction holds great potential to ameliorate the pathophysiology of SCD and β-thalassemia3-7,8,9. γ-globin silencing is an active process dependent on interactions between DNA binding factors and chromatin readers, writers and erasers acting within multiprotein nuclear complexes. The nucleosome remodeling and deacetylase (NuRD) complex is a chromatin modifier composed of six different protein family subunits10,11. Each subunit family is comprised of multiple paralog proteins that can variably combine to form NuRD multiprotein complexes. The catalytic subunits are the ATP-dependent nucleosome remodelers CHD3 and CHD4 and the histone deacetylases HDAC1 and HDAC2. Other members include the methyl CpG binding proteins MBD2 and MBD3 and structural subunits MTA1, MTA2, and MTA3, GATAD2A and GATAD2B, and RBBP4 and RBBP7. Various NuRD members have been implicated in developmental silencing of HbF. NuRD members occupy embryonic and fetal globin genes12,13. Knockdown of MBD2 induces the expression of γ-globin in human β-globin locus transgenic mice14 and in human CD34+ HSPC derived adult erythroid cells15,16. Conditional knockout of Chd4 results in de-repression of γ-globin in β-YAC transgenic mice and cultured murine chemical inducer of dimerization (CID) hematopoietic cells17. Knockdown of CHD4 in primary human erythroid cells results in robust increase in γ-globin expression16,18. A coiled-coil protein interaction between MBD2 and GATAD2A is necessary for γ-globin gene repression and could be a potential target for molecular interruption15. Genetic knockdown or chemical inhibition of HDAC1 and HDAC2 induces HbF in adult erythroid progenitors19-21. Initially discovered by GWAS as a locus associated with HbF level22,23, the transcriptional repressor BCL11A has been validated as a critical negative regulator of γ-globin expression24-32. Biochemical studies have revealed that BCL11A physically interacts with NuRD complex subunits including CHD3/4, HDAC1/2, MTA1/2/3, RBBP4/7, MBD316. More recently ZBTB7A has been reported as a γ-globin repressor33. ZBTB7A confers its repressive activity nonredundantly with BCL11A, yet also physically interacts with NuRD subunits including MTA2, HDAC1/2, GATAD2B. Together these data provide the impetus to define the mechanisms through which NuRD represses HbF and to identify possible molecular targets for pharmacotherapy (also see Supplementary Note).
Here we investigated the coding sequences within the NuRD complex associated with HbF repression by using CRISPR-Cas9 dense mutagenesis in human umbilical cord blood-derived erythroid progenitor (HUDEP-2) adult-stage erythroid cells. Taking into account cellular fitness as a counter-screen, we nominated potential NuRD target regions for therapeutic de-repression of HbF that escape cellular toxicity, validated their effects in primary human cells and transgenic mice, and developed a rational therapeutic strategy for HbF induction to phenocopy potent mutations.
Results
CRISPR dense in situ mutagenesis reveals NuRD complex members essential for HbF repression
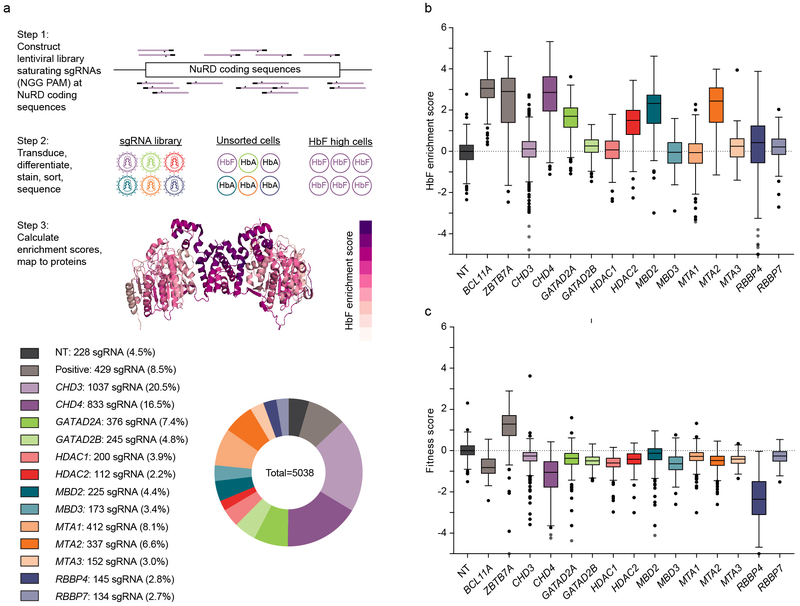
We hypothesized that CRISPR-Cas9 dense in situ mutagenesis could reveal critical NuRD sequences at which in-frame alleles result in loss-of-function. We compared HbF enrichment scores among the different NuRD subunits (Fig. 1a, also see Supplementary Note). As expected, sgRNAs targeting positive control genes BCL11A and ZBTB7A showed robust HbF enrichment as compared to nontargeting (NT) sgRNAs (Fig. 1b). We defined hit genes, i.e. those with biological phenotype, as those at which at least 75% of the sgRNAs exceeded the median NT sgRNA score34. We discovered that among the 13 NuRD subunit genes, only 5 genes, CHD4, GATAD2A, HDAC2, MBD2, and MTA2, were required for HbF repression while perturbations of the coding sequences of the other 8 NuRD genes, CHD3, GATAD2B, HDAC1, MBD3, MTA1, MTA3, RBBP4 and RBBP7, did not substantially affect HbF repression (Fig. 1b). Notably within 5 paralogous gene families (CHD, GATAD2, HDAC, MBD, MTA), only one member was required for HbF repression, whereas the other gene members were dispensable. For example, HDAC2 was required for HbF repression while HDAC1 was not, MTA2 was required for HbF repression while MTA1 and MTA3 were not, and so forth. This observation suggested that a subcomplex of NuRD defined by constituent paralogous family members was required for γ-globin repression.
Fig. 1: Dense mutagenesis of NuRD genes by CRISPR-Cas9 pooled screening.
a, Upper panel shows overview of the CRISPR-Cas9 pooled lentiviral screen. All NGG PAM restricted sgRNAs targeting 13 NuRD genes plus BCL11A and ZBTB7A targeting and nontargeting sgRNAs synthesized by oligonucleotide array and cloned as pool to lentivirus. HUDEP-2 cells were transduced (n=3), selected, and subject to erythroid differentiation culture. Genomic DNA was isolated from unsorted and HbF+ cells. Integrated sgRNA libraries were amplified and deep sequenced to determine sgRNA representation. HbF enrichment scores calculated by comparing sgRNA abundance in HbF-high to unsorted cells, fitness score calculated by comparing sgRNA abundance in unsorted cells to library. Enrichment scores were mapped to linear and 3D protein maps. As an example, HbF enrichment scores from MTA2 and HDAC2 mapped to dimer of PDB ID 5ICN. Lower panel shows sgRNA library distribution. b, c, HbF enrichment (b) and fitness (c) scores for each sgRNA (sample size per lower panel a) targeting NuRD genes and controls. Boxplot shows median, 25th and 75th percentiles with whiskers and outliers per Tukey method.
We observed that almost all gene targeting sgRNAs had a small negative effect on fitness score as compared to nontargeting sgRNAs (Fig. 1c). We speculated that this was related to the known modest cellular fitness impact of Cas9-mediated DNA damage response35-37. Previous pooled CRISPR screens have shown modest negative fitness impact of neutral gene targeting sgRNAs as compared to nontargeting sgRNAs, consistent with these results38. We only observed two NuRD members with obvious negative fitness scores beyond this modest shared effect, namely CHD4 and RBBP4 (Fig. 1c, also see Supplementary Note). By comparing HbF enrichment scores and fitness scores, we distinguished four functional classes of NuRD genes: GATAD2A, HDAC2, MBD2, MTA2 as essential for HbF repression but not cell fitness, CHD4 as essential both for HbF repression and cell fitness, RBBP4 as essential for cell fitness, and CHD3, GATAD2B, HDAC1, MBD3, MTA1, MTA3, RBBP7 as neither essential for HbF repression nor for cell fitness. For each of the six NuRD constituent paralogous families, just one member of each family was required for HbF repression or cellular fitness.
Proteomic analysis corroborates the HbF repressive NuRD subcomplex
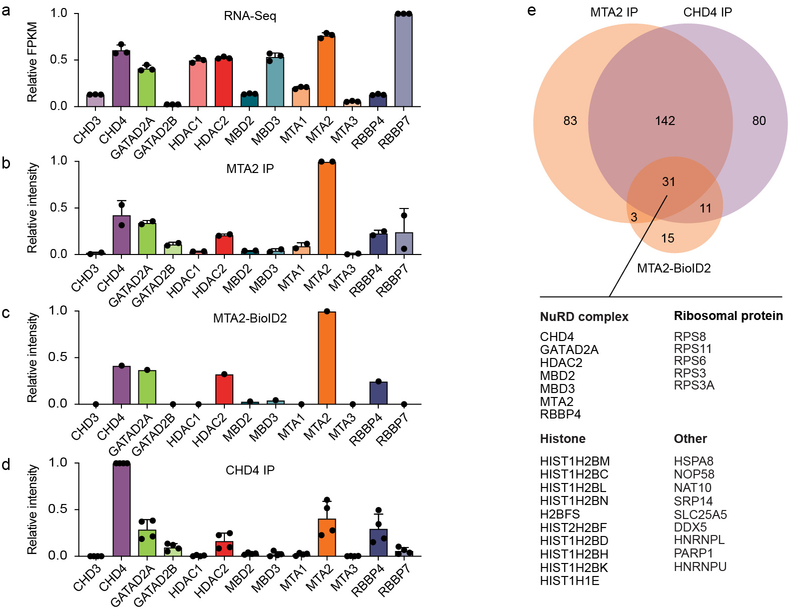
We analyzed RNA-seq data generated from HUDEP-2 cells for all the NuRD genes to compare the expression of each paralog to its functional requirement (Fig. 2a). We found that mRNA expression levels of CHD4, GATAD2A, and MTA2 were significantly higher than their respective paralogs (CHD3, GATAD2B, MTA1 and MTA3) suggesting that higher gene expression could account for the paralog-specific dependence. However HDAC2 and HDAC1 shared similar mRNA expression levels, MBD3 showed higher mRNA expression as compared to MBD2, and RBBP7 showed higher expression as compared to RBBP4.
Fig. 2: NuRD subcomplex expression.
a, Quantification of NuRD gene expression in HUDEP-2 cells by RNA-seq. b-d, Relative abundance of NuRD subunits by LFQ mass spectrometry following immunoprecipitation of MTA2 from HUDEP-2 cells (b), following streptavidin pull-down from MTA2-BioID2 HUDEP-2 cells (c), and following immunoprecipitation of CHD4 from HUDEP-2 (d). Protein intensities expressed as fraction of bait intensity. Error bars in a, b and d represent mean and standard deviation of 3, 2, and 4 independent experiments respectively. e, Venn diagram showing MTA2 IP, MTA2-BioID2 and CHD4 IP proteins as detected by mass spectrometry following affinity purification. The specific interactions in these samples were identified by testing protein LFQ intensity greater than controls with a corrected p-value less than 0.2. Table shows identity of 31 proteins, including 7 NuRD proteins, detected by all three methods.
Protein abundance is not necessarily predicted by mRNA abundance39-41. To further evaluate the paralog-specific composition of the NuRD protein complex, we performed affinity purification of proteins physically associated with MTA2 and CHD4 using immunoprecipitation and label-free quantification by high-resolution mass spectrometry (IP-MS, Supplementary Fig. 2a). To orthogonally test the identity of the erythroid NuRD subcomplex members, we evaluated the proteins physically neighboring MTA2 in its intracellular milieu by proximity labeling (Supplementary Fig. 2b-c). Overall we found strong agreement of all 3 methods in detecting interacting NuRD subunit members (Fig. 2b-e, Supplementary Fig. 2d-f, Supplementary Table 1, Supplementary Note). We found 31 proteins at the intersection of all 3 MS experiments (Fig. 2e). With respect to NuRD, the shared proteins included 7 NuRD subunits, preferentially identifying the NuRD paralogs found to be functional by CRISPR screening. Specifically, 6 of the proteomics identified NuRD members (MTA2, RBBP4, CHD4, GATAD2A, HDAC2, MBD2) were found to be critical by CRISPR screening, whereas only MBD3 was found to be dispensable for HbF repression and cellular fitness (also see Supplementary Note).
Functional maps of NuRD subunits essential for HbF repression
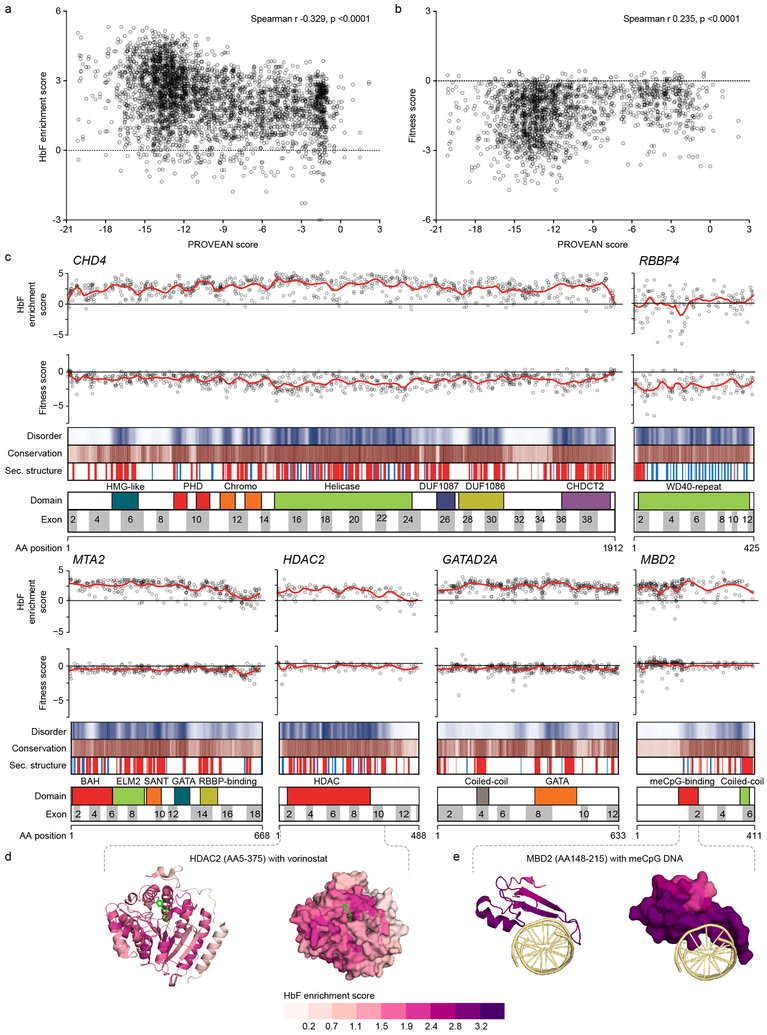
We hypothesized that sgRNAs targeting critical regions of functional NuRD subunits would show heightened enrichment scores due to an increased likelihood of loss-of-function in-frame alleles36,42. To test this, we compared the functional scores against various protein-level sequence annotations, including evolutionary conservation, protein disorder, domain identity and secondary structure (Supplementary Table 2). Indeed, we observed an inverse relationship between PROVEAN conservation score and HbF enrichment (Spearman r −0.329, p <0.0001) indicating that targeting more conserved positions within NuRD hit genes is more likely to result in HbF derepression (Fig. 3a). Similarly, a correlation between PROVEAN score and cellular fitness score (Fig. 3b) showed that the sgRNAs targeting more conserved amino acid residues exerted greater fitness cost (Spearman r 0.235, p <0.0001). These results support the hypothesis that CRISPR-Cas9 mediated comprehensive in situ mutagenesis can identify critical NuRD complex protein coding sequences (also see Supplementary Fig. 3 and Supplementrary Note).
Fig. 3: Maps of functional NuRD subcomplex.
a and b, Scatter plots of HbF enrichment (a) and fitness (b) scores compared to PROVEAN conservation scores for individual sgRNAs for hit genes (n = 3765 sgRNAs for a and 1955 for b). Spearman r and p-values shown. Each dot represents sgRNA average score from three independent experiments. c, Linear maps of hit NuRD genes. HbF enrichment scores and fitness scores are shown for each sgRNA as dots, with LOESS regression line in red. Disorder scores are shown as heatmap from white to blue from maximal disorder to maximal order. Evolutionary conservation PROVEAN scores are shown as heatmap from white to brown from minimal to maximal conservation. Secondary structure predictions are shown with helix in red and sheet in blue. d and e, Structures colored based on LOESS regression HbF enrichment scores for (d) human HDAC2 (PDB ID 4LXZ) in complex with competitive inhibitor vorinostat (in green) and (e) human MBD2 in complex with methylated DNA (in yellow) (PDB ID 6CNQ). Left panels depict cartoon models, right panels surface models.
To visualize amino acid residues that result in maximal HbF derepression and minimal cellular toxicity upon disruption, we generated linear maps of HbF enrichment and fitness scores for all sgRNAs at the NuRD hit genes (Fig. 3c). We plotted individual sgRNA scores as well as a smoothed score based on local polynomial regression. In addition, we mapped the PROVEAN conservation scores, disorder scores, secondary structure predictions, and domain annotations. We observed visually apparent examples of functional scores correlating with each of these annotations. For example, at CHD4 we found heightened HbF enrichment scores at conserved, ordered sequences with secondary structure predictions, as compared to neighboring sequences. This pattern was evident around characterized domains including the catalytic ATP-dependent helicase domain, chromatin-interacting regions including two PHD fingers and two chromodomains, an N-terminal HMG-like domain, two domains of unknown function (DUF1087 and DUF1086) and a C-terminal domain CHDCT2 (Fig. 3c). Similarly, we found heightened scores at the N-terminal regions of MTA2 comprising various domains critical for NuRD complex formation43, at the catalytic histone deacetylase domain of HDAC2, and at the MBD2 domain that binds methyl-CpG DNA44,45. The NuRD regions identified as most critical for HbF repression by dense mutagenesis included those involved in intra-subunit interactions. For example, a heterodimeric interaction of a coiled-coil region of GATAD2A (AA137–179) with a coiled-coil region of MBD2 (AA360–393) is required for γ-globin repression15. Elevated HbF enrichment scores were observed for sgRNAs targeting the GATAD2A coiled-coil region as compared to flanking sequences (Fig. 3c). The sgRNA HbF enrichment score maps did not identify functional sequences within the NuRD subunits CHD3, MTA1, MTA3, MBD3, GATAD2B, HDAC1, and RBBP7 (Supplementary Fig. 4).
We compared the high-resolution functional scores based on Cas9-mediated comprehensive in situ mutagenesis to available protein structures. We colored these structures by overlaying the regression HbF enrichment scores onto each amino acid. We found that the functional scores marked binding interfaces as particularly vulnerable to disruption (also see Supplementary Note). For example, recoloring with HbF enrichment scores the structure of HDAC2 in complex with the HDAC inhibitor vorinostat (PDB ID 4LXZ)46 suggested that residues adjacent to the vorinostat binding site had heightened HbF enrichment scores as compared to neighboring residues (Fig. 3d). The interface of MDB2 abutting meCpG DNA (PDB ID 6CNQ)45 demonstrated elevated scores as compared to the non-DNA facing region of MDB2, suggesting that sgRNAs that disrupt meCpG binding are especially potent in terms of HbF induction (Fig. 3e).
Identification of critical residues of MTA2 required for HbF repression
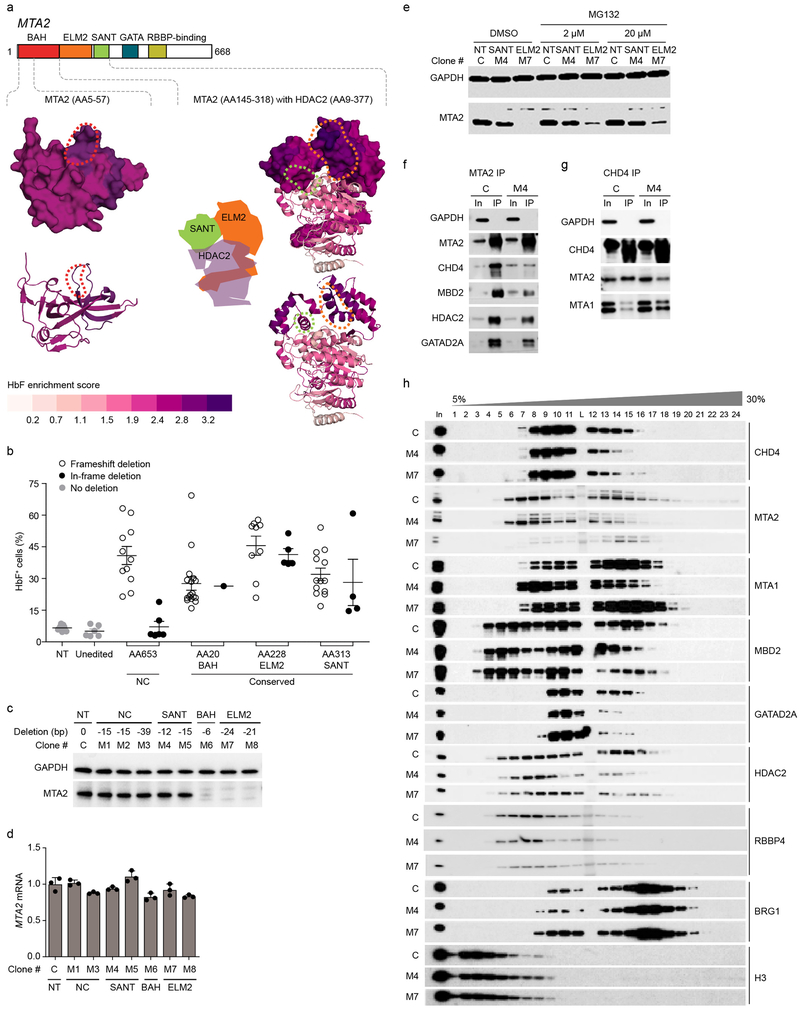
MTA subunits are considered the scaffolds onto which the NuRD complex assembles47. Dense mutagenesis identified multiple regions of heightened HbF enrichment scores at MTA2 overlapping its conserved, ordered domains (Fig. 3c), including the BAH domain implicated in chromatin interaction48, the ELM2 and SANT domains involved in MTA homodimerization and HDAC recruitment49, a GATA-like zinc finger (ZF) domain, and RBBP4 interacting region11. To test the hypothesis that the residues with heightened functional CRISPR scores reveal positions at which in-frame deletions are associated with loss-of-function, we characterized clones with defined mutations of the MTA2 NuRD scaffold subunit (see Supplementary Note). Clones with frameshift mutations showed derepression of HbF in a manner that was independent of whether the targeted amino acid resided in a domain or at NC sequences (Fig. 4a-b). In-frame deletions did not result in elevated HbF levels when targeted to the NC position with modest HbF enrichment scores, but in contrast, clones with in-frame deletions at positions of heightened HbF enrichment score within the BAH, ELM2 and SANT domains demonstrated derepression of HbF, indicating loss-of-function of MTA2. These results support the hypothesis that heightened functional CRISPR scores mark regions where in-frame deletions result in loss-of-function alleles.
Fig. 4: In-frame deletions disrupt MTA2 function.
a, Structures colored based on LOESS regression HbF enrichment scores for MTA2 and HDAC2. MTA2 BAH domain (PDB ID 1W4S, left) and MTA2 ELM2-SANT domain with HDAC2 (PDB ID 5ICN, right) with aligned HbF enrichment scores mapped. Top panels depict surface models for MTA2, bottom panels cartoon models. Dotted lines indicate the position of residues deleted in the hemizygous in-frame deletion clones at BAH (red), ELM2 (orange) and SANT (green) domains. b, Each dot indicates fraction of HbF+ cells from hemizygous MTA2 frameshift or in-frame deletion, non-deletion (unedited), or nontargeted (NT) HUDEP-2 clones. Error bars represent mean of three independent experiments and SEM. c, Immunoblot (representative of three independent experiments) of MTA2 from nuclear lysates of hemizygous in-frame deletion clones shows some clones lose and others preserve MTA2 levels. d, MTA2 mRNA expression in hemizygous in-frame deletion clones by RT-qPCR. Error bars represent mean and SD of three replicates. e, MTA2 level by immunoblot following exposure of HUDEP-2 MTA2+/− hemizygous in-frame deletion clones to four hours of MG132 proteasome inhibitor at indicated concentrations. f, Immunoprecipitation (IP) of MTA2 followed by immunoblot shows interaction with NuRD subunits. g, IP of CHD4 followed by immunoblot shows interaction with NuRD subunits. h, Glycerol gradient density sedimentation analyses on nuclear extracts from control and M4 SANT domain and M7 ELM2 domain hemizygous in-frame deletion HUDEP-2 clones. Each experiment (e-h) was repeated at least three times with similar results.
We evaluated individual in-frame deletion clones (Supplementary Fig. 5c) to examine the biochemical basis for HbF induction following mutation of MTA2. We observed two classes of loss-of-function clones, one in which MTA2 protein was lost, and another in which MTA2 protein level was preserved but function was impaired (Fig. 4c-e and Supplementary Fig. 5d). Immunoprecipitation by anti-MTA2 or anti-CHD4 followed by immunoblot with NuRD subunits showed that the MTA2 in-frame deletion in clone M4 resulted in reduced capacity for NuRD complex formation (Fig. 4f-g and Supplementary Fig. 5e-f). Reduced interaction of MTA2 with CHD4, MBD2, HDAC2, and GATAD2A was observed in this clone. To assess the composition of the NuRD complex, we performed glycerol gradient density sedimentation analysis of nuclear extracts50, comparing control and MTA2 M4 and M7 mutant clones (Fig. 4h). In control cells we observed cosedimentation of CHD4, MTA2, GATAD2A, MBD2, HDAC2, and RBBP4 as the NuRD subcomplex as well as in various lower molecular weight fractions, suggesting the presence of free proteins, intermediate subcomplexes (such as the so-called NuDe complex lacking CHD3/CHD4)51, and alternative complexes. In clone M4 we found MTA2 was expressed yet relatively depleted from higher molecular weight fractions, consistent with reduced incorporation of MTA2 to the NuRD complex. Concomitantly we observed decreased incorporation of CHD4, GATAD2A, MBD2, HDAC2, and RBBP4 to the NuRD complex. In contrast, in clone M7 we observed the absence of MTA2, consistent with its destabilization, along with reduced incorporation of CHD4 and GATAD2A into the NuRD complex. These results suggested that impairment of the recruitment of CHD4 to NuRD could be a final common pathway of NuRD functional disruption. Together these results showed two modes of action of in-frame loss-of-function deletions of MTA2. One mode was loss of MTA2, associated with formation of aberrant complexes. A second mode was mutations that preserve the levels of MTA2 but disrupt its function in supporting the assembly of NuRD.
Decoupling CHD4 roles in HbF repression and cellular fitness
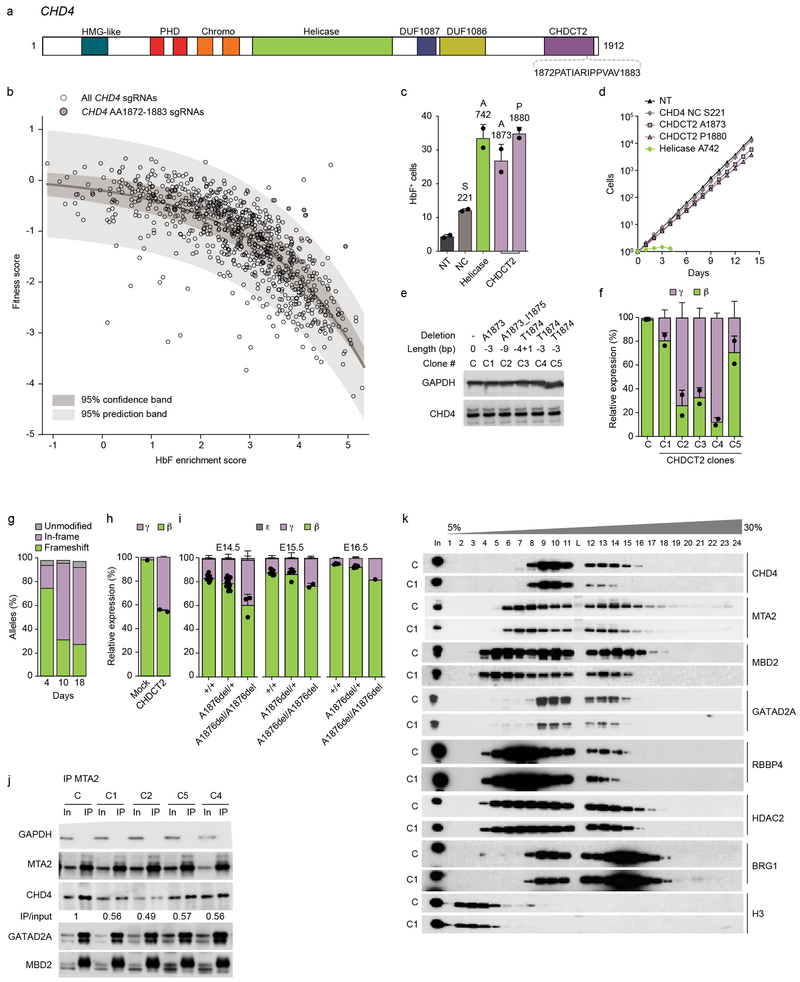
CHD4 possessed the highest median HbF enrichment score of the NuRD subunits (Fig. 1b). The HbF enrichment scores of sgRNAs targeting CHD4 were of similar magnitude as those targeting the critical HbF repressors BCL11A and ZBTB7A. However, unlike the other HbF repressing NuRD hit genes, CHD4 also demonstrated negative fitness scores (Fig. 1c). We observed a strong inverse correlation between HbF enrichment scores and fitness scores (Spearman r −0.784, p<0.0001), indicating that targeting most positions of CHD4 was associated with a similar magnitude of HbF induction and fitness cost. However, we observed a small group of sgRNAs with high HbF enrichment, yet relatively modest fitness scores within the CHDCT2 domain of CHD4 (Fig. 5a-b). We found the interval from AA1872–1883 to be highly significant (p=2.93 ×10−11) as including outliers from the dataset. Using a sliding window to test all contiguous 12 AA segments within CHD4, we did not find any other significant nonoverlapping outlier intervals. We introduced individual sgRNAs within CHD4 CHDCT2 AA1872-AA1883 to test if the resultant mutations could uncouple HbF derepression from negative cellular fitness. We tested two C-terminal targeting sgRNAs around A1873 and P1880 as well as sgRNAs targeting the helicase domain around A742 and a relatively NC, non-domain region of CHD4 around S221. Indeed, the CHDCT2 targeting sgRNAs showed high HbF induction approaching that of the helicase targeting sgRNA, and substantially greater than the sgRNA targeting the NC region (Fig. 5c-e). The sgRNAs targeting the CHD4 helicase domain showed substantial cellular toxicity, with almost no viable cells observed after five days of culture. In contrast, the CHDCT2 targeting sgRNAs had minimal impact on cellular expansion throughout 14 days in culture, similar to the nontargeting or CHD4 NC targeting sgRNAs (Fig. 5d).
Fig. 5: Targeting CHD4 CHDCT2 uncouples HbF induction from cytotoxicity.
a, CHD4 domains. b, Dots indicate HbF enrichment and fitness scores of all 833 sgRNAs targeting CHD4. 95% confidence and prediction bands after non-linear least squares curve fitting. Purple dots show sgRNAs targetting AA1872-1883, the only nonoverlapping contiguous 12 AA segment of CHD4 significantly enriched in outliers from prediction band (p = 2.93 ×10−11, Fisher’s exact test, two-sided). sgRNA scores average of three experiments. c, Fraction HbF+ cells (%) following sgRNA targeting CHD4 NC, helicase, CHDCT2 domain or nontargeting (NT) (n = 2, error bars represent mean and SD). d, HUDEP-2 expansion following sgRNA targeting CHD4 S22, A742, A1873, P1880 and NT (average from three experiments). e, Immunoblot from CHD4 hemizygous in-frame deletion clones. Representative of three experiments. f, β- and γ-globin RT-qPCR in CHD4 hemizygous in-frame deletion clones (mean and SD from n = 2). g, CD34+ HSPCs electroporated with SpCas9:sgRNA targeting CHDCT2 A1873 with indels quantified by amplicon sequencing after 4, 10, or 18 day erythroid culture. h, β- and γ-globin expression in human erythroid precursors (mean and SD from n = 2). i, β-, ε- and γ-globin expression in fetal livers from E14.5, E15.5 and E16.5 Chd4A1876del/+;β-YAC+ × Chd4A1876del/+;β-YAC+ embryos (mean and SD, sample size per Supplementary Fig. 7d). j, IP MTA2 followed by NuRD subunit immunoblot. Quantification of CHD4 by IP/input ratio. k, Density sedimentation analyses of CHDCT2 domain hemizygous in-frame deletion HUDEP-2 clone. Each experiment (j-k) was repeated at least three times with similar results.
We evaluated the effect of CHD4 CHDCT2 mutation on HbF repression in primary erythroid cells. We electroporated CD34+ hematopoietic stem and progenitor cells (HSPCs) with SpCas9 protein and sgRNA ribonucleoprotein complex (RNP). We performed 18 day erythroid differentiation culture, and observed similar terminal erythroid maturation by cell expansion and immunophenotype (Supplementary Fig. 6g). We compared gene editing efficiency at three time points, at the beginning, middle, and end of erythroid differentiation culture (days 4, 10, and 18). We evaluated the indels by deep sequencing of amplicons. We found that frameshift mutations were lost during the course of erythropoiesis (CHD4 frameshift allele frequency from 72.5% to 26.2% from day 4 to day 18) while in-frame deletions were gained (from 19.0% to 63.5%) (Fig. 5g). These results indicated that in-frame mutations at CHD4 CHDCT2 around A1873 selectively escaped negative fitness in primary erythroid precursors, similar to findings in HUDEP-2 immortalized erythroid precursors. Next we evaluated globin gene expression at the end of erythroid differentiation culture. In comparison to mock edited cells, erythroid cells edited at CHD4 CHDCT2 demonstrated elevation of γ-globin from 2.1% to 44.8% of total β-like globin, HbF+ cells from 28.4% to 67.7%, and HbF from 4.9 to 16.0% by HPLC (Fig. 5h and Supplementary Fig. 6e-f). These results validate the comprehensive mutagenesis screen and demonstrate that in-frame mutations at AA1872–1883 of CHD4 induce HbF with minimal impact on cell fitness in human primary erythroid cells. In addition, we generated mice homozygous for in-frame deletion at Chd4 A1876 (orthologous to human CHD4 A1873) and observed impaired developmental silencing of transgenic human γ-globin (Fig. 5i, Supplementary Fig. 7, Supplementary Note).
Rational targeting of CHD4
We conducted biochemical studies to investigate the mechanism whereby in-frame deletions at CHD4 CHDCT2 impact the NuRD complex (see Supplementary Note). In each of the four CHD4 CHDCT2 in-frame deletion clones tested, we observed reduced immunoprecipitation of CHD4 despite similar pull-down of MTA2 itself and other NuRD members, including GATAD2A and MBD2 (Fig. 5j). These results suggested a reduced ability of mutant CHD4 to interact with the NuRD complex. To further investigate the impact of CHDCT2 mutant CHD4 on NuRD complex assembly, we performed fractionation of nuclear extracts by glycerol gradient density sedimentation. We observed decreased incorporation of mutant CHD4 to the NuRD complex along with modest reduction of MTA2, GATAD2A, MBD2, HDAC2, and RBBP4 incorporation (Fig. 5k). Taken together, these results suggest that in-frame deletions at CHD4 CHDCT2 domain around AA1872–1883 impair the interaction of CHD4 with NuRD.
A recent study demonstrated that the previously poorly characterized CHDCT2 domain acts to recruit CHD4 to NuRD by binding to GATAD2 factors GATAD2A and GATAD2B43. A C-terminal segment of GATAD2B was sufficient to pulldown the CHD4 CHDCT2 domain in a rabbit reticulocyte lysate transcription/translation system. Since bridging interactions by endogenous NuRD subunits are minimal under these conditions, the results suggest a direct interaction between CHDCT2 and GATAD2 factors. In addition to the heightened HbF enrichment scores around the N-terminal coiled-coil domain of GATAD2A implicated in binding to MBD2, we observed another cluster of sgRNAs with heightened HbF enrichment scores at GATAD2A between AA335–486, a C-terminal region encompassing a C2C2-type GATA ZF (Fig. 3c). These GATAD2A sequences, including the GATA ZF, are homologous to sequences within the GATAD2B segment shown to bind to CHD4 CHDCT243.
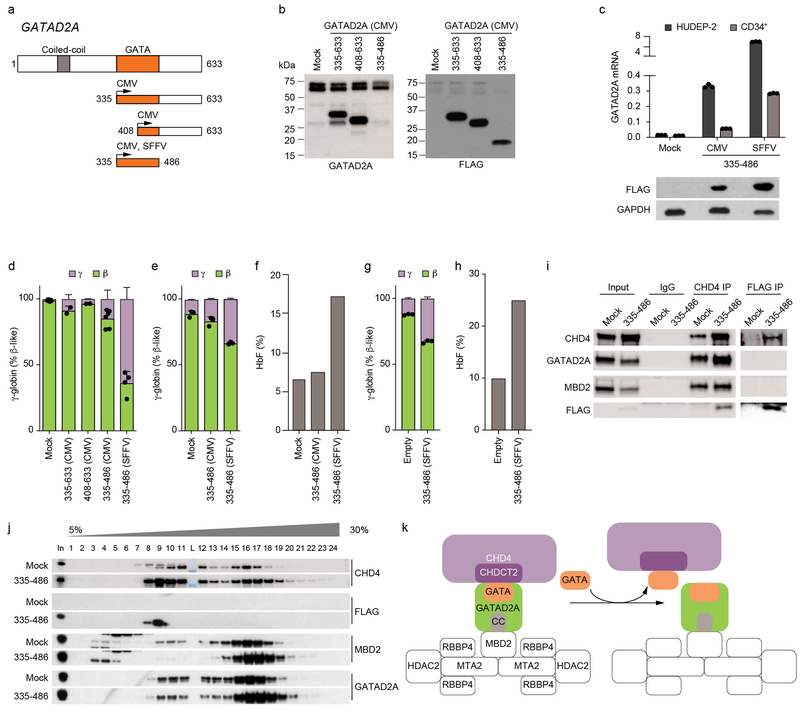
We hypothesized that this C-terminal region of GATAD2A could contribute to HbF repression by binding to CHD4 CHDCT2. Therefore, we reasoned that ectopic expression of this segment might competitively bind to CHD4 CHDCT2, displace CHD4 from NuRD, and mimic CHDCT2 mutations that result in HbF derepression. We expressed several FLAG epitope tagged constructs of GATAD2A in HUDEP-2 cells, including the entire C-terminal segment (AA335–633), a C-terminal segment which includes a truncation of the GATA ZF domain (AA408–633), or a segment encompassing the entire GATA ZF corresponding to the region with heightened HbF enrichment scores (AA335–486) (Fig. 6a). Immunoblot analysis with both GATAD2A and FLAG antibodies suggested that protein levels of the truncated GATAD2A were no greater than that of endogenous GATAD2A (Fig. 6b). Overexpression of these constructs in HUDEP-2 cells led to increased expression of γ-globin, with the greatest effect observed in cells expressing the GATAD2A AA335–486 segment encompassing the GATA ZF domain (Fig. 6d). To test CHD4-GATAD2A disruption in primary human erythroid precursors, we transduced and subjected to erythroid differentiation CD34+ HSPCs with the GATAD2A AA335–486 vector. Enforced GATAD2A expression was lower as compared to HUDEP-2 cells, although relative expression increased under the control of the more potent SFFV enhancer/promoter52 (Fig. 6c).
Fig. 6: GATAD2A ZF sequesters CHD4 from NuRD.
a, Domain structure of GATAD2A and truncation mutants. b, Protein levels of truncated GATAD2A segments under control of CMV promoter in HUDEP-2 cells. c, Expression of GATAD2A mRNA by RT-qPCR (both endogenous and ectopic GATAD2A) in non-transduced HUDEP-2 cells or CD34+ HSPC derived human erythroid precursors (HEPs) with GATAD2A AA335-486 segment driven by CMV or SFFV promoter (top) or protein expression levels of GATAD2A AA335-486 segment driven by CMV or SFFV promoter in HUDEP-2 cells (bottom). Error bars show mean and SD, n = 3. d, γ and β-globin expression by RT-qPCR in HUDEP-2 cells expressing GATAD2A truncation mutants. Error bars show mean and SD, n = 2-7. e, γ and β-globin expression by RT-qPCR in HEPs expressing GATAD2A AA335-486 segment. Error bars show mean and SD, n = 3. f, HbF level by HPLC from HEPs expressing GATAD2A AA335-486 segment. g-h, γ and β-globin expression by RT-qPCR (g) and HbF level by HPLC (h) in HEPs from a SCD patient expressing GATAD2A AA335-486 segment driven by SFFV promoter. Error bars show mean and SD, n = 3 technical replicates. i, CHD4 IP or FLAG IP followed by immunoblot in HUDEP-2 cells expressing GATAD2A AA335-486 segment driven by SFFV promoter. j, Density sedimentation analyses on nuclear extracts from HUDEP-2 cells expressing GATAD2A AA335-486 driven by SFFV promoter. Each experiment (b, i, j) repeated at least three times with similar results k, Schematic model, sequestering CHD4 from NuRD by disruption of GATAD2A-CHDCT2 interaction.
We observed dose-dependent increases in γ-globin expression with higher expression of GATAD2A 335–486 segment in both HUDEP-2 and CD34+ HSPC derived primary erythroid precursors (Fig. 6d-e). Although we observed impaired cell expansion with extremely high level expression of GATAD2A 335–486 in HUDEP-2 cells as driven by SFFV promoter, we observed only slight impact on expansion of primary human erythroid precursors, consistent with relatively more modest expression of the GATAD2A 335–486 segment (Supplementary Fig. 9a-b). The maturation of primary human erythroid precursors by immunophenotype, enucleation frequency, or morphology was unchanged with the GATAD2A 335–486 expression, while γ-globin increased from 10.8 to 33.6% of total β-like globin, F-cells from 22.1 to 37.4%, and HbF level from 6.5 to 17.1% (Fig. 6e-f and Supplementary Fig. 9b-c). These data suggest that expression of a GATAD2A segment could selectively counteract HbF silencing with minimal impact on the proliferation or maturation of erythroid cells. We observed similar γ-globin and HbF induction upon expression of the GATAD2A 335–486 segment in primary human erythroid precursors from a SCD patient (Fig. 6 g-h).
To assess potential interaction of truncated GATAD2A with CHD4, we performed immunoprecipitation. Pulldown of GATAD2A 335–486 with anti-FLAG enriched CHD4 but not endogenous GATAD2A or MBD2, suggesting that this GATAD2A segment sequesters CHD4 from NuRD. Reciprocal pulldown with CHD4 antibody enriched the FLAG-tagged GATAD2A AA335–486 segment (Fig. 6i and Supplementary Fig. 9d). To evaluate the NuRD complex, we performed glycerol gradient sedimentation in cells expressing GATAD2A AA335–486. The truncated GATAD2A segment co-sedimented with low molecular weight forms of CHD4 but not the NuRD complex. We observed increased abundance of low molecular weight forms of CHD4 in the presence of the GATAD2A AA335–486 segment (Fig. 6j and Supplementary Fig. 9e). In contrast the distribution of other endogenous members of NuRD, such as GATAD2A and MBD2, did not shift, indicating that GATAD2A 335–486 sequesters CHD4 from NuRD, leaving NuRD otherwise intact. Together, these data suggest that rational targeting of the CHD4 CHDCT2-GATAD2A interaction may disrupt CHD4 recruitment to NuRD and elevate HbF levels while sparing cytotoxicity (Fig. 6k).
Discussion
Here we used comprehensive CRISPR mutagenesis to reveal the NuRD coding sequences required for HbF repression. We found that a NuRD subcomplex including just one functional paralog per protein family is required. These results suggest that the HbF-repressive functional NuRD complex is nonredundant and that targeting key subunits might provide biological specificity. We performed both affinity purification and proximity labeling proteomics of the MTA2 and CHD4 NuRD subcomplexes. The results were highly congruent with the CRISPR mutagenesis, identifying GATAD2A, HDAC2, MBD2, and RBBP4 to be members of the functional subcomplex along with MTA2 and CHD4. Future experiments will be required to determine how much of the HbF repressive effect depends on direct recruitment of the NuRD subcomplex to the γ-globin genes.
Over the past decade specific DNA binding transcriptional repressors such as BCL11A and ZBTB7A have been identified as crucial for HbF silencing. These discoveries have suggested novel gene therapies based on viral transduction or gene editing of mobilized HSCs, manipulated ex vivo and then autologously re-engrafted53. However given their complexity and cost, these gene therapies are unlikely to scale to the massive global unmet need for β-hemoglobinopathy therapeutics. Small molecule approaches are imperative, yet rational targeting of gene regulation remains challenging. By performing dense mutagenesis in situ we aimed to nominate specific NuRD positions as possible therapeutic targets while avoiding excess cellular toxicity. The results of this screen suggest numerous vulnerable regions within the HbF repressive NuRD subcomplex. For example, at MTA2 we find that disturbing protein structure at some positions destabilizes the protein and at other positions leads to impaired function. Either approach (destabilizing or competitive binding) might be phenocopied by appropriate rational design.
At CHD4, we identified in-frame deletions at the CHDCT2 domain uncoupling HbF repression from cellular toxicity. The CHDCT2 domain of CHD4 has recently been shown to recruit CHD4 to NuRD by binding to GATAD2A. We speculate that the NuRD independent effects of CHD4 may be less reliant on these sequences. We demonstrate that a fragment of GATAD2A able to bind to CHD4 sequesters CHD4 from NuRD, and leads to HbF derepression with minimal toxicity. Improved biochemical and structural understanding of the GATAD2A-CHD4 interaction may afford opportunities for small molecule targeting of this complex. More broadly this study demonstrates how dense mutagenesis correlating protein sequences with function offers a means to achieve specificity when targeting ubiquitous chromatin regulatory complexes (see Supplementary Note). As the resolution and throughput of genome editing perturbation range advance, comprehensive mutagenesis of protein complexes in situ could be readily adapted to the study of many disease-relevant cellular processes.
Methods
HUDEP-2 cell culture.
HUDEP-2 cells54 were cultured as described previously55 (also see Supplementary Note).
Human primary erythroid cell culture.
Human CD34+ HSPCs from mobilized peripheral blood of deidentified healthy donors were obtained from Fred Hutchinson Cancer Research Center, Seattle, Washington. CD34+ HSPCs were expanded and differentiated as described previously55. SpCas9 and sgRNA were delivered by electroporation of ribonucleoprotein complex (RNP)56 (see Supplementary Note, Supplementary Methods).
Design and synthesis of human NuRD lentiviral sgRNA libraries.
Every 20-mer sequence upstream of NGG PAM on sense and antisense strands was identified for human NuRD gene coding regions (Fig. 1a). The sgRNA oligos were synthesized as previously described55,57-59. Synthesized oliogs were amplified using PCR. A second PCR was performed to remove barcodes and insert vector homology. Subsequently oligos were cloned into LentiGuide-Puro (Addgene plasmid 52963)58 using Gibson assembly. Sufficient colonies were isolated for ~1800 colonies per individual sgRNA within the library. Plasmid library was deep sequenced to confirm the representation of sgRNAs. To produce lentivirus, HEK293T cells were cultured with Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 2% penicillin-streptomycin in 15 cm tissue culture dishes. HEK293T cells were transfected at 70–80% confluence in 14 ml of media with 13.3 μg pf psPAX2, 6.7 μg VSV-G and 20 μg of the lentiviral construct plasmid of interest using 180 μg of linear polyethylenimine. 24 hours after transfection, media was changed. Lentiviral supernatant was collected at 72 hours post-transfection and subsequently concentrated by ultracentrifugation (24000 RPM for four hour at 4°C with Beckman Coulter SW 32 Ti rotor).
Tiled pooled CRISPR-Cas9 screen.
LentiCas9-Blast (Addgene plasmid 52962)58 was used to transduce HUDEP-2 cells. Transduced cells were selected with 10 μg ml−1 blasticidin. Cas9 activity was confirmed using the pXPR-011 (Addgene plasmid 59702) GFP reporter assay as described previously60. HUDEP-2 cells with stable SpCas9 expression were transduced at low multiplicity of infection with the human sgRNA lentiviral pool while in expansion phase. An average multiplicity of infection of 0.183 (+/− 0.004) with mimimum representation of 1036 +/− 48 cells per guide RNA across the three biological replicates was achieved (Supplementary Fig. 1c). These results indicate that the majority of transduced cells in the screen only contained a single integrant. 10 μg ml−1 blasticidin and 1 μg ml−1 puromycin were added 24h after transduction to select for lentiviral library integrants in cells with stable SpCas9 expression. Cells were cultured in expansion media for four days followed by differentiation media for additional eight days. Cells were washed with PBS and intracellular staining was performed by fixing cells with 0.05% glutaraldehyde (grade II) for 10 minutes at room temperature. Cells were centrifuged for 5 minutes at 600 × g and then re-suspended in 0.1% Triton X-100 for 5 minutes at room temperature for permeabilization. Triton X-100 was diluted with PBS with 0.1% BSA and then centrifuged at 600 × g for 15 minutes. Cells were stained with anti-human antibody for HbF conjugated with FITC for 20 minutes in the dark and slow shaking. Cells were washed to remove unbound antibody before sort. 0.2 μg HbF antibody was used per 5 million fixed and permeabilized cells. Control cells exposed to non-targeting sgRNA sample and BCL11A exon 2 were used as negative and positive controls respectively to establish flow cytometry conditions. The population of cells with top 10% expression of HbF was FACS sorted. Library preparation and deep sequencing was performed as previously described57. Briefly, genomic DNA was extracted using Qiagen Blood and Tissue kit. Herculase PCR reaction using LentiGuide-Puro specific primers (Supplementary Table 3) was performed as follows: Herculase II reaction buffer (1X), forward and reverse primers (0.5 μM each), dimethyle sulfoxide (DMSO) (8%), deoxynucleotide triphosphates (dNTPs) (0.25 mM each), Herculase II Fusion DNA Polymerase (0.5 μL) using the following cycling conditions: 95°C for 2 minutes, 20 cycles of 95°C for 15s, 60°C for 20s, 72 for 30s; 72°C for 5 minutes. Multiple reactions of no more than 2.5 μg each were used to amplify from ~25 μg of gDNA per pool. Samples were subject to second PCR using handle-specific primers57 to add adaptors and indexes to each sample using the following conditions: Heculase II reaction buffer (1X), forward and reverse primers (0.5 M each) dNTPs (0.25 mM each), Herculase II Fusion DNA Polymerase (0.5 μL) with following conditions: 95°C for 2 minutes, 20 cycles of 95°C for 15s, 60°C for 20s, 72 for 30s; 72°C for 5 minutes. PCR products were run on an agarose gel and the band of expected size was gel purified. Illumina Hiseq paired end sequencing was performed.
sgRNA count and mapping.
Raw read counts from library, unsorted, and high HbF sample groups were used to calculate the log2 fold change in sgRNA abundance with DESeq261. These log2 fold changes were normalized to the median nontargeting sgRNA log2 fold change. sgRNAs were then mapped to the protein by mapping the predicted double strand break site to the two amino acids nearest the double strand break by using genomic coordinates. Lastly, scores for amino acids with no assigned sgRNA were interpolated via LOESS regression, using known sgRNA scores and location as input. The scores for each amino acid were then mapped onto structures publicly available in the Protein Data Bank. The sequence of the protein structure was aligned to the sequence of the protein isoform to which the sgRNA were originally mapped using Biopythons pairwise2 module62 (local alignment with Blosum62 matrix, opening gap cost −10, extension −0.5). Scores from sgRNA mapping to the same amino acid were averaged. Protein structures were recolored in PyMOL (The PyMOL Molecular Graphics System, Version 2.0, Schrödinger, LLC) based on aligned scores. For visual clarity, the scores were divided into 17 bins.
Generation of hemizygous deletion clones.
Hemizygous cellular clones carrying in-frame or frameshift mutations were generated in two steps. In the first step, one allele of MTA2 or CHD4 was deleted using a pair of sgRNA targeting outside of the coding region (Supplementary Fig. 5a, 6a). In the second step, sgRNA was introduced targeting the region of interest in the second allele of gene of interest (see Supplementary Note).
Cloning of individual sgRNAs.
LentiGuide-Puro (Addgene plasmid 52963) was digested with BsmBI in 1X buffer 3.1 at 37°C (New England Biolabs) for linearization. One unit of TSAP thermosensitive alkaline phosphatase (Promega) was added for 1 hour at 37°C to dephosphorylate the linearized lentiGuide and then TSAP was heat inactivated at 74°C for 15 minutes. Linearized and dephosphorylated lentiGuide was run on an agarose gel and gel purified. SgRNA-specifying oligos were phosphorylated and annealed using the following conditions: sgRNA sequence oligo (100 μM); sgRNA reverse complement oligo (100 μM); T4 ligation buffer (1X); and T4 polynucleotide kinase (5 units) with the following temperature conditions (New England Biolabs): 37°C for 30 minutes; 95°C for 5 minutes; and then ramp down to 25°C at 5°C min−1. Annealed oligos were ligated into lentiGuide in a 1:3 ratio (vector: insert) using T4 ligation buffer (1X) and T4 DNA ligase (750 units). Plasmids were verified by sequencing using a U6F promoter forward primer (Supplementary Table 3).
Coimmunoprecipitation.
30 million HUDEP-2 cells were centrifuged (500 × g for 5 minutes) and washed twice with PBS, then lysed in ice cold lysis buffer (50 mM Tris‐HCl, pH 7.5, 150 mM NaCl, 1% NP‐40 + freshly added protease inhibitor (Sigma) for 20 minutes; later cells were centrifuged at 800 × g for 5 minutes at 4°C. Pellet was re-suspended in ice cold lysis buffer (5 million cells/ml buffer) and passed through 26g syringe 5 times. Lysates were centrifuged at 4°C and 800 × g for 8 minutes. Subsequently, pellet (nuclei) was re-suspended in 1ml IP buffer (150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM b-glycerolphosphate, 1 mM Na3VO4, and 1 mM Pefabloc (Roche Applied Science)) and placed on ice for 30 minutes, sonicated using Bioruptor (Diagenode). Lysates were centrifuged at 16000g for 5 minutes. Supernatants were incubated with 5–10 μg of the antibodies overnight. Dynabeads protein G were used to collect target protein-antibody immune complexes. Following washing, bound proteins were eluted by Laemmli Sample Buffer freshly supplemented with 1mM DTT (BioRad).
Immunoblot analysis.
Protein expression, IP efficiency and analysis of NuRD interacting partners were done using SDS-PAGE/ immunoblotting using 4–20% TGX™ Precast Protein Gels (BioRad) with standard procedures. Details of the antibodies used are provided in Supplementary Table 4.
Liquid chromatography nanoelectrospray ionization mass spectrometry.
IP products eluted by Laemmli Sample Buffer were resolved by SDS-PAGE and visualized using silver staining. Gel slices were excised, and destained using 30 mM potassium hexa-cyanoferrate (III) /100 mM sodium thiosulfate. Cysteine residues were reduced (10 mM DTT, 56°C, 60 minutes) and alkylated (55 mM iodoacetamide, 25°C, 45 minutes) prior to in-gel overnight proteolysis at 37°C using 375 ng sequencing grade modified porcine trypsin (Promega, Madison, WI) per gel slice, in 50 mM ammonium bicarbonate pH8.4. Peptides were eluted from gel slabs by 70% acidified acetonitrile and lyophilized using a vacuum centrifuge. Peptide pellets were resuspended in 0.1% aqueous formic acid and 15% of the solution was analyzed. The liquid chromatography system (Ekspert NanoLC 425, Eksigent, Redwood City, CA) consisted in a vented trap-elute architecture63 coupled to an Orbitrap Fusion mass spectrometer (Thermo, San Jose, CA) via a nanoelectrospray ion source (New Objective). Peptides were resolved using a 30 cm × 75 μm internal diameter column, packed with ReproSil-Pur C18-AQ 1.9 μm particles (Dr. Maisch, Ammerbuch-Entringen, Germany) over 90 minutes using a 5–40% acetonitrile gradient in water at 250 nl/minute. The protein abundance was quantified based on LFQ intensity (sum of the integrated area of the extracted ion chromatogram (XIC) for each peptide assigned to the protein)64 (see Supplementary Note Methods)
HbF expression and globin gene expression.
1×106 HUDEP-2 cells were stained and anlyzed for HbF expression by intracellular flow-cytometry as described previously55 HbF expression was also measured by HPLC using D-10 hemoglobin analyzer (Bio-Rad). Globin gene expression was assesed using RT-qPCR (Supplementary Note and Supplementary Table 3).
Cellular expansion.
HUDEP-2 cells were seeded in triplicate at 100,000 cells per well in 6-well plates and cultured in the presence of blasticidin and puromycin as appropriate. Viable cells were counted using Trypan Blue exclusion by Countess II FL Automated Cell Counter (ThermoFisher).
Immunocytochemistry and quantification of DNA damage biomarkers γH2AX and 53BP1.
24 hours after transduction, cells were selected with puromycin. After an additional 48 hours of culture, cells were attached on poly-L-lysine coated glass cover slips (15,000 cells per cover slip) for 30 minutes. Subsequently, cells were fixed with 4% paraformaldehyde and immunostained for DNA damage repair markers γH2AX and 53BP1. Antibody labeling was visualized using Alexa Fluor secondary antibodies. To visualize nuclei cells were counterstained with DAPI. Quantitative analysis of γH2AX foci was performed by using Zeiss (Axioskpe2) fluorescent microscope equipped with a Leica DFC300FX camera and the Leica microsystem LAS program. Pictures at 20X and 40X magnification were made of five areas in the culture dish. Mean fluorescent intensity of foci in each cell in the image were measured using ImageJ software. Similarly, pyknotic nuclei were counted using DAPI stain and particle counter plugin of ImageJ software.
Etoposide sensitivity assay.
HUDEP-2 cell hemizygous CHD4 clones with mutations in CHDCT2 region and control were seeded at 0.5 × 106 in 24-well plates and treated with either DMSO or etoposide (100, 300, 1000 nM) for 24 hours. At the end of the treatment period, cells were stained with Annexin V and DAPI, and the cell viability was measured using flow cytometry.
Cloning and expression of BioID2 in HUDEP-2 cells.
Humanized BioID2 sequence together with GGGGS13 linker was amplified from Addgene plasmid ID #7422465 by introducing the BsrG1 and BamHI cloning sites. The fragment was cloned in lentiCas9-Blast back bone (Addgene plasmid #52962) carrying dCas9 (dCas9 sequence from Addgene plasmid #47948) sequence between BamHI and BsiWI cloning sites. Together plasmid was initially reconstructed as lenti-dCas9-linkerGGGGS13-BioID2A-Blast with BioID2-linker and dCas9 sequence as one ORF. MTA2 sequence was obtained from Harvard Plasmid Data Base (ID: HsCD00337978) and PCR amplified using following primers with complementary overhangs (see primer sequences in Supplementary Table 3. Subsequently, lenti-dCas9-linkerGGGGS13-BioID2A-Blast was digested with BsiWI-HF and BamHI-HF. PCR amplified MTA2 sequence and backbone from BsiWI-HF/BamHI-HF digestion of lenti-dCas9-linkerGGGGS13-BioID2A-Blast were subjected to Gibson Assembly. In parallel, the oligos encoding Kozak, start codon, FLAG and NLS (SV40) sequences and BsiWI/BamHI compatible ends (Supplementary Table 3) were annealed and ligated into BamHI/BsiWI digested lenti-dCas9-linkerGGGGS13-BioID2A-Blast vector to produce a control plasmid.
To produce lentivirus HEK293T cells were transfected with 13.3 μg of psPAX2, 6.7 μg VSV-G and 20 μg of the lentiviral construct plasmid (MTA2-BioID2/NLS-BioID2) using 180 μg of linear polyethylenimine. Lentiviral supernatant was collected after 72 hours post-transfection and subsequently concentrated by ultracentrifugation. MTA2 biallelic deleted HUDEP-2 cells (MTA2−/−) were transduced with 5 μl of concentrated lentivirus. Following 24 hours of transduction, cells were exposed to 10 μg ml-1 blasticidin in HUDEP-2 expansion media. Blasticidin resistant colonies were expanded and tested for MTA2-BioID2 expression and function using immunoblot and intracellular HbF staining respectively.
Protein complex gradient sedimentation.
Glycerol sedimentation assay was performed according to Wang et al50 with modifications. Briefly, pellets of 40 million HUDEP-2 cells were lysed on ice for 10 min in Buffer A (10 mM Tris-Cl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1 mM EDTA, 0.5% NP-40, 1 mM DTT, and 1X complete protease inhibitor cocktail ((Roche)). Nuclei were sedimented by centrifugation at 1000xg for 5 min at 4C. Nuclei were then resuspended in Buffer B (10mM HEPES pH 7.6, 3 mM MgCl2, 100 mM KCl, 0.1 mM EDTA, 10% glycerol, 1 mM DTT, and 1X complete protease inhibitor cocktail), and were lysed by addition of ammonium sulfate to a final concentration of 0.3 M for 30 min on ice. Soluble nuclear proteins were collected from supernatant after ultracentrifugation at 40,000 rpm for 10 min at 4C (SW 41-Ti, swinging-bucket), and precipitated with 0.3 g/ml ammonium sulfate for 30 min on ice. Protein precipitate was pelleted by centrifugation at 17,000xg for 20 min at 4C, and resuspended in HEMG buffer (25 mM HEPES pH 7.9, 0.1 mM EDTA, 12.5 mM MgCl2, 100 mM KCl, 1 mM DTT, and 1X complete protease inhibitor cocktail). Resuspended nuclear extract (1 mg in 500 μl HEMG buffer) was overlaid onto a 12 ml 5–30% continuous glycerol gradient (prepared in buffer containing 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Igepal CA-630, 0.5% Na-deoxycholate, 0.1% SDS, 1 mM DTT) prepared in 14 ml 14×95 mm polyallomer centrifuge tube (Beckman Coulter, 331374). Tubes were centrifuged at 40,000 rpm at 4C for 16h in a SW 40-Ti swinging-bucket rotor. Fractions (0.5 ml) were collected from top to bottom of the tube, and were analyzed using 26-well 4–20% Criterion TGX gels (Biorad) and subsequent immunoblotting.
Expression of GATAD2A constructs.
Human GATAD2A ORF was purchased from GenScript (Clone ID: OHu17401D, RefSeq: NM_017660.3) and was PCR amplified and cloned between EcoRI and BamHI restriction sites of a pLVX-Puro vector (Clontech) using Gibson assembly with primers provided in Supplementary Table 3. To generate SFFV promoter driven GATAD2A 335–486 construct, CMV promoter of pLVX-puro was replaced with SFFV promoter (amplified from pHR-SFFV-dCas9-BFP-KRAB, Addgene #46911). Briefly, pLVX-puro GATAD2A 335–486 was digested with ClaI and EcoRI, and the large vector fragment was gel purified for further Gibson assembly with PCR amplified SFFV promoter. The sequence integrity of all the cloned products were verified by Sanger sequencing. All the cloned GATAD2A constructs contain a SV-40 NLS (PKKKRKV) and a FLAG-tag (DYKDDDDK) at the N-terminus. Wild type HUDEP-2 cells were transduced with lentivirus carrying GATAD2A constructs for 24 hours, followed by selection with puromycin (1 μg/ml).
RNA sequencing:
SpCas9 expressing HUDEP-2 cells in expansion media were transduced in parallel with sgRNAs targeting CHD4 and MTA2 and controls. 24 hours following transduction, media was replaced with fresh expansion media containing 1 μg ml−1 puromycin. After 48 hours of transduction, a small fraction of puromycin selected cells was used to extract genomic DNA. PCR was performed using extracted genomic DNA around the sgRNA cut site, PCR amplified product was purified using DNA purification kit and used for Sanger sequencing and subsequent TIDE analysis. Each sgRNA’s indel formation efficiency was measured more than 80% by TIDE. Four days following transduction cells were lysed and RNA was extracted using RNeasy Mini Kit (Qiagen). Libraries were synthesized using Illumina TruSeq Stranded mRNA sample preparation kits from 500 ng of purified total RNA according to the manufacturer’s protocol. The final dsDNA libraries were sequenced on Illumina NextSeq500 with single-end 75bp reads by the Dana-Farber Cancer Institute Molecular Biology Core Facilities (also see Supplementary Methods).
Generation of Chd4A1876del mice
Animal experiments were performed under protocols approved by the Boston Children’s Hospital Animal Care and Use Committee (15–12-3065R). To generate Chd4A1876del mice, a sgRNA targeting the terminal domain on Chd4 was designed and synthesized. Two overlapping oligonucleotides carrying the T7 RNA polymerase promoter, the target sequence and the sgRNA scaffold were annealed and used as template for in vitro transcription (MEGAshortscript T7 Transcription Kit, Ambion). In vitro transcribed RNA was purified using Nucaway Spin Columns66,67. A single strand donor DNA template including a 3 bp deletion and a silent mutation to prevent re-cleavage of edited alleles was obtained from IDT (PAGE Ultramer, 60 bp). Primers, sgRNA and DNA donor template sequences are listed in Supplemental Table 1. Prior to microinjection, 100 ng/μl SpCas9 protein (PNA Bio), 100 ng/μl sgRNA and 200 ng/μl DNA donor template were mixed in sterile RNase-free microinjection buffer (1 mM Tris–HCl pH 7.5; 0.1 mM EDTA pH 7.5), centrifuged for 30 min at 14,000g at 4°C and kept on ice until use66 One-cell-stage mouse embryos (C57BL/6NCrl) were isolated, microinjected, and transferred into CD1 foster mothers. Founder mice were screened by PCR followed by Sanger sequencing. Mosaicism was assessed by targeted amplicon sequencing. Sequence analysis was performed using CRISPResso68. To monitor fetal hemoglobin repression in vivo, we used mice transgenic for the human β-globin gene cluster on a yeast artificial chromosome (β-YAC)36. Mice heterozygous for the Chd4A1876del allele and carrying one copy of the human β-globin gene cluster were bred; embryos were collected from staged pregnancies at E14.5, E15.5 and E16.5 stage. Fetal livers were dissected and mechanically dissociated in PBS, and used for genotyping and globin expression analyses.
Data Availability
The data that support the findings of this study are available from the corresponding author upon request. Data and analysis are included in the article and Supplementary Note. Mass spectrometery raw data is accessable from proteomecentral under accession number PXD009793, http://proteomecentral.proteomexchange.org/cgi/GetDataset.
Next-generation sequencing (NGS) data (RNA-seq and CRISPR screen) are available from NCBI SRA portal under accession No. PRJNA496556, https://www.ncbi.nlm.nih.gov/sra
Code Availability
For analysis and visualization of functional readout from tiled pooled CRISPR screen, we used a custom computational pipeline available at https://gitlab.com/bauerlab/crispro.
Supplementary Material
Acknowledgements
We thank X. Wang, C. Brendel, E.C. Smith, A. Gutierrez, G.D. Ginder and D.C. Williams for useful discussions and J. Desimini for graphical assistance. V.A.C.S. was supported by an Individual Travel Grant (ITG) from Radboud University. D.S.V. was supported by the Cooley’s Anemia Foundation. L.M.K.D. was supported by NHLBI (5T32HL007574–36) and a Burroughs Wellcome Fund Postdoctoral Enrichment Grant (PDEP 1015098). S.A.W. and K.L. were supported by NIAID (R01AI117839) and NIGMS (R01GM115911). L.P. was supported by a National Human Genome Research Institute (NHGRI) Career Development Award (R00HG008399). T. M. was supported by NIH (5R01DK111455) and JSPS Grant-in-Aid for Scientific Research A (17H01567). A.K. is the Damon Runyon-Richard Lumsden Foundation Clinical Investigator and acknowledges support of the St. Baldrick’s Arceci Innovation Award, and NCI R01 CA204396 and P30 CA008748. Generation of mouse model was supported by a NIDDK Cooperative Centers of Excellence in Hematology (CCEH) award (U54DK110805) to S.H.O. S.H.O. was supported by Doris Duke Charitable Foundation, and is an Investigator of the Howard Hughes Medical Institute. D.E.B. was supported by NIDDK (K08DK093705, R03DK109232), NHLBI (DP2OD022716, P01HL032262), the Doris Duke Charitable Foundation, Burroughs Wellcome Fund, the American Society of Hematology, and an Epigenetics Seed Grant from Harvard Medical School.
Footnotes
Competing Interests Statement
The authors declare no competing interests.
References (main text)
- 1.Modell B & Darlison M Global epidemiology of haemoglobin disorders and derived service indicators. Bull World Health Organ 86, 480–7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Piel FB et al. Global epidemiology of sickle haemoglobin in neonates: a contemporary geostatistical model-based map and population estimates. Lancet 381, 142–51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darling RC, Smith CA, Asmussen E & Cohen FM Some Properties of Human Fetal and Maternal Blood. J Clin Invest 20, 739–47 (1941). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schroeder WA, Shelton JR, Shelton JB & Cormick J The Amino Acid Sequence of the Alpha Chain of Human Fetal Hemoglobin. Biochemistry 2, 1353–7 (1963). [DOI] [PubMed] [Google Scholar]
- 5.Blau CA & Stamatoyannopoulos G Hemoglobin switching and its clinical implications. Curr Opin Hematol 1, 136–42 (1994). [PubMed] [Google Scholar]
- 6.Vinjamur DS, Bauer DE & Orkin SH Recent progress in understanding and manipulating haemoglobin switching for the haemoglobinopathies. Br J Haematol (2017). [DOI] [PubMed] [Google Scholar]
- 7.Bauer DE, Brendel C & Fitzhugh CD Curative approaches for sickle cell disease: A review of allogeneic and autologous strategies. Blood Cells Mol Dis 67, 155–168 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Nuinoon M et al. A genome-wide association identified the common genetic variants influence disease severity in beta0-thalassemia/hemoglobin E. Hum Genet 127, 303–14 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Lettre G et al. DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc Natl Acad Sci U S A 105, 11869–74 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torchy MP, Hamiche A & Klaholz BP Structure and function insights into the NuRD chromatin remodeling complex. Cell Mol Life Sci 72, 2491–507 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Millard CJ et al. The structure of the core NuRD repression complex provides insights into its interaction with chromatin. Elife 5, e13941 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kransdorf EP et al. MBD2 is a critical component of a methyl cytosine-binding protein complex isolated from primary erythroid cells. Blood 108, 2836–45 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harju-Baker S, Costa FC, Fedosyuk H, Neades R & Peterson KR Silencing of Agamma-globin gene expression during adult definitive erythropoiesis mediated by GATA-1-FOG-1-Mi2 complex binding at the −566 GATA site. Mol Cell Biol 28, 3101–13 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rupon JW, Wang SZ, Gaensler K, Lloyd J & Ginder GD Methyl binding domain protein 2 mediates gamma-globin gene silencing in adult human betaYAC transgenic mice. Proc Natl Acad Sci U S A 103, 6617–22 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gnanapragasam MN et al. p66Alpha-MBD2 coiled-coil interaction and recruitment of Mi-2 are critical for globin gene silencing by the MBD2-NuRD complex. Proc Natl Acad Sci U S A 108, 7487–92 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu J et al. Corepressor-dependent silencing of fetal hemoglobin expression by BCL11A. Proc Natl Acad Sci U S A 110, 6518–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costa FC, Fedosyuk H, Chazelle AM, Neades RY & Peterson KR Mi2beta is required for gamma-globin gene silencing: temporal assembly of a GATA-1-FOG-1-Mi2 repressor complex in beta-YAC transgenic mice. PLoS Genet 8, e1003155 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amaya M et al. Mi2beta-mediated silencing of the fetal gamma-globin gene in adult erythroid cells. Blood 121, 3493–501 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradner JE et al. Chemical genetic strategy identifies histone deacetylase 1 (HDAC1) and HDAC2 as therapeutic targets in sickle cell disease. Proc Natl Acad Sci U S A 107, 12617–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esrick EB, McConkey M, Lin K, Frisbee A & Ebert BL Inactivation of HDAC1 or HDAC2 induces gamma globin expression without altering cell cycle or proliferation. Am J Hematol 90, 624–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shearstone JR et al. Chemical Inhibition of Histone Deacetylases 1 and 2 Induces Fetal Hemoglobin through Activation of GATA2. PLoS One 11, e0153767 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uda M et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci U S A 105, 1620–5 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menzel S et al. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat Genet 39, 1197–9 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Bauer DE et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 342, 253–7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu N et al. Direct Promoter Repression by BCL11A Controls the Fetal to Adult Hemoglobin Switch. Cell 173, 430–442 e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sankaran VG et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science 322, 1839–42 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Thein SL et al. Intergenic variants of HBS1L-MYB are responsible for a major quantitative trait locus on chromosome 6q23 influencing fetal hemoglobin levels in adults. Proc Natl Acad Sci U S A 104, 11346–51 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sankaran VG et al. Developmental and species-divergent globin switching are driven by BCL11A. Nature 460, 1093–7 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sankaran VG et al. A functional element necessary for fetal hemoglobin silencing. N Engl J Med 365, 807–14 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu J et al. Transcriptional silencing of {gamma}-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev 24, 783–98 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yi Z et al. Sox6 directly silences epsilon globin expression in definitive erythropoiesis. PLoS Genet 2, e14 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu J et al. Correction of sickle cell disease in adult mice by interference with fetal hemoglobin silencing. Science 334, 993–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masuda T et al. Transcription factors LRF and BCL11A independently repress expression of fetal hemoglobin. Science 351, 285–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schoonenberg VAC et al. CRISPRO: identification of functional protein coding sequences based on genome editing dense mutagenesis. Genome Biol 19, 169 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aguirre AJ et al. Genomic Copy Number Dictates a Gene-Independent Cell Response to CRISPR/Cas9 Targeting. Cancer Discov 6, 914–29 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munoz DM et al. CRISPR Screens Provide a Comprehensive Assessment of Cancer Vulnerabilities but Generate False-Positive Hits for Highly Amplified Genomic Regions. Cancer Discov 6, 900–13 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Morgens DW, Deans RM, Li A & Bassik MC Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat Biotechnol 34, 634–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morgens DW et al. Genome-scale measurement of off-target activity using Cas9 toxicity in high-throughput screens. Nat Commun 8, 15178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laurent JM et al. Protein abundances are more conserved than mRNA abundances across diverse taxa. Proteomics 10, 4209–12 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vogel C & Marcotte EM Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 13, 227–32 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gautier EF et al. Comprehensive Proteomic Analysis of Human Erythropoiesis. Cell Rep 16, 1470–1484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi J et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol 33, 661–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torrado M et al. Refinement of the subunit interaction network within the nucleosome remodelling and deacetylase (NuRD) complex. FEBS J 284, 4216–4232 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scarsdale JN, Webb HD, Ginder GD & Williams DC Jr. Solution structure and dynamic analysis of chicken MBD2 methyl binding domain bound to a target-methylated DNA sequence. Nucleic Acids Res 39, 6741–52 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu K et al. Structural basis for the ability of MBD domains to bind methyl-CG and TG sites in DNA. J Biol Chem 293, 7344–7354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lauffer BE et al. Histone deacetylase (HDAC) inhibitor kinetic rate constants correlate with cellular histone acetylation but not transcription and cell viability. J Biol Chem 288, 26926–43 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar R & Wang RA Structure, expression and functions of MTA genes. Gene 582, 112–21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang N & Xu RM Structure and function of the BAH domain in chromatin biology. Crit Rev Biochem Mol Biol 48, 211–21 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Ding Z, Gillespie LL & Paterno GD Human MI-ER1 alpha and beta function as transcriptional repressors by recruitment of histone deacetylase 1 to their conserved ELM2 domain. Mol Cell Biol 23, 250–8 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X et al. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat Genet 49, 289–295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Low JK et al. CHD4 Is a Peripheral Component of the Nucleosome Remodeling and Deacetylase Complex. J Biol Chem 291, 15853–66 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Demaison C et al. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of imunodeficiency] virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum Gene Ther 13, 803–13 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Esrick EB & Bauer DE Genetic therapies for sickle cell disease. Semin Hematol 55, 76–86 (2018). [DOI] [PubMed] [Google Scholar]
Methods-only references
- 54.Kurita R et al. Establishment of immortalized human erythroid progenitor cell lines able to produce enucleated red blood cells. PLoS One 8, e59890 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Canver MC et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 527, 192–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu Y et al. Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat Med (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shalem O et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanjana NE, Shalem O & Zhang F Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen S et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160, 1246–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Doench JG et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol 32, 1262–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cock PJ et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–3 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cifani P & Kentsis A High Sensitivity Quantitative Proteomics Using Automated Multidimensional Nano-flow Chromatography and Accumulated Ion Monitoring on Quadrupole-Orbitrap-Linear Ion Trap Mass Spectrometer. Mol Cell Proteomics 16, 2006–2016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cox J et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13, 2513–26 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim DI et al. An improved smaller biotin ligase for BioID proximity labeling. Mol Biol Cell 27, 1188–96 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seruggia D, Fernandez A, Cantero M, Pelczar P & Montoliu L Functional validation of mouse tyrosinase non-coding regulatory DNA elements by CRISPR-Cas9-mediated mutagenesis. Nucleic Acids Res 43, 4855–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harms DW et al. Mouse Genome Editing Using the CRISPR/Cas System. Curr Protoc Hum Genet 83, 15 7 1–27 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pinello L et al. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat Biotechnol 34, 695–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request. Data and analysis are included in the article and Supplementary Note. Mass spectrometery raw data is accessable from proteomecentral under accession number PXD009793, http://proteomecentral.proteomexchange.org/cgi/GetDataset.
Next-generation sequencing (NGS) data (RNA-seq and CRISPR screen) are available from NCBI SRA portal under accession No. PRJNA496556, https://www.ncbi.nlm.nih.gov/sra