Abstract
Functional validation of candidate genes involved in adaptation and speciation remains challenging. We here exemplify the utility of a method quantifying individual mRNA transcripts in revealing the molecular basis of divergence in feather pigment synthesis during early-stage speciation in crows. Using a padlock probe assay combined with rolling circle amplification, we quantified cell type specific gene expression in the histological context of growing feather follicles. Expression of Tyrosinase Related Protein 1 (TYRP1), Solute Carrier Family 45 member 2 (SLC45A2) and Hematopoietic Prostaglandin D Synthase (HPGDS) was melanocyte-limited and significantly reduced in follicles from hooded crow explaining the substantially lower eumelanin content in grey vs. black feathers. The central upstream Melanocyte Inducing Transcription Factor (MITF) only showed differential expression specific to melanocytes - a feature not captured by bulk RNA-seq. Overall, this study provides insight into the molecular basis of an evolutionary young transition in pigment synthesis, and demonstrates the power of histologically explicit, statistically substantiated single-cell gene expression quantification for functional genetic inference in natural populations.
Keywords: feather, melanogenesis, speciation, single-cell gene expression, MITF, HPGDS
Introduction
Feathers are a key evolutionary innovation of the epidermis arising in the dinosaur lineage basal to the avian clade (Godefroit et al., 2014; Xu et al., 2001). Millions of years of evolution have modified the development of epidermal appendages from scales to the complex morphological structure of a feather (Figure 1). Its basic bauplan has been repeatedly expanded to a variety of feather types ranging from downy feathers for insulation to pennaceous wing feathers enabling flight (Prum and Dyck, 2003; Widelitz et al., 2003; Yu et al., 2002). By integrating light-absorbing, chemical compounds modulating feather coloration, the avian plumage further provides ample opportunity for inter- and intraspecific signaling (Hill and McGraw, 2006). Mediating between the natural and social environment, feather coloration constitutes an important component of natural and sexual selection with implications for adaptation and speciation (Cuthill et al., 2017; Hubbard et al., 2010). Coloration differences arising within short evolutionary timescales within and among populations are thus believed to play an active role in the initial steps of species divergence (Hugall and Stuart-Fox, 2012; Price, 2008).
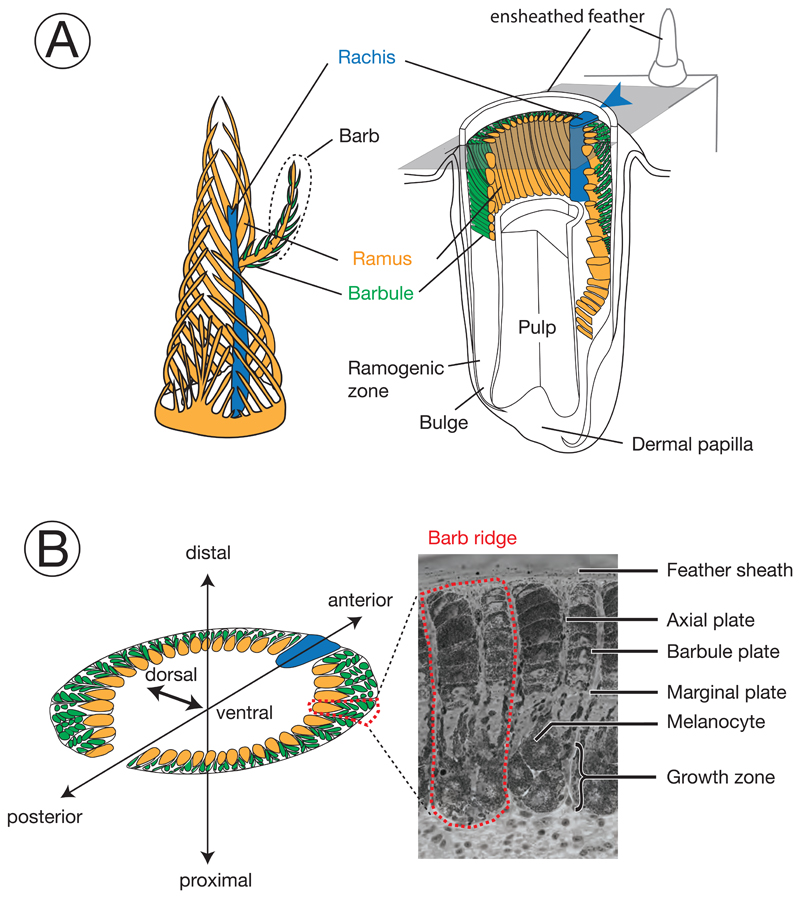
Figure 1. Schematic diagram and terminology of feather follicle morphology.
The feather is a complex epidermal organ of cellular origin. Its basic bauplan follows a hierarchical structure of ramification into barbs and barbules. It develops within follicles, an epidermal modification derived from invagination into the dermis (Chen et al., 2015; d’Ischia et al., 2013). Cells fulfilling different functions in the mature barbs are derived from stem cells at the collar bulge of the basal layer (Yue et al., 2005). Developing progenitors of these cells migrate and gradually differentiate to form mature cells in barbs, including basal cells, marginal plate cells, axial plate cells, and barbulous plate cells (Prum, 1999). Development of the main feather progresses along the proximal-distal axis with helical displacement of barb loci. Barb loci emerge on the posterior midline and are gradually displaced towards the anterior midline where they fuse with the rachis. For more information on the feather morphology and development consult (Lucas and Stettenheim, 1972; Yu et al., 2004).
Colors define homologous structures in the respective panels; arrowheads indicating the rachis facilitate orientation in the remaining figures. A) left: Schematic, three-dimensional representation of a growing pennaceous feather with the afterfeather shown in front. right: Partial section a feather follicle. The cutting plane in grey shows the position of cross sections in panel B. B) left: Schematic cross section of a feather follicle with three-dimensional axes for orientation. right: Example of a cross-section from a growing, ensheathed carrion crow feather displaying two barb ridges.
The chemical compounds contributing to the vibrant colors of birds include metabolized ketocarotenoids producing yellow-orange color and porphyrins resulting in bright pink, yellow, red, or green (Shawkey et al., 2006). Iridescence is mediated by refraction of light on feather microstructures often involving an ordered array of melanin granules within a keratin substrate (Shawkey et al., 2006). In this study, we focus on genesis and deposition of melanin polymers into the growing feather setting the basis for black, reddish, or brown color space (Hill and McGraw, 2006). While the molecular pathways of pigment synthesis and color patterning across the body are well characterized in mammals and insects (Kronforst et al., 2012; Silvers, 2012), inference in birds is usually indirect and often assumed to follow the general pathways characterized in other vertebrates (Hill and McGraw, 2006) (but see Lopes et al., 2016; Mundy et al., 2016). Moreover, functional investigation is generally restricted to domesticated model species such as chicken (Yu, 2002; Yu et al., 2004), duck (Li et al., 2012), or quail (Niwa et al., 2002; Shiojiri et al., 1996). Recently, however, high-throughput sequencing of genomes and transcriptomes has opened coloration genetic research to variation segregating in natural populations (San-Jose and Roulin, 2017). Genome-wide association mapping approaches yield initial candidate genes contributing to variation in pigmentation within and among populations (Küpper et al., 2016; Vijay et al., 2016). Differential gene expression analysis between color morphs using bulk mRNA sequencing provides an independent axis to screen for genes potentially involved in color mediating pathways (Du et al., 2017; Vijay et al., 2016). Yet, while useful to generate initial hypotheses, bulk mRNA sequencing integrates over a vast number of cells and cell types, and falls short of mapping gene expression relevant to the variance in pigmentation. Therefore, cell type specific approaches measuring gene expression in the chromatophores themselves are required for functional characterization of candidate genes.
We here present a powerful approach allowing detailed quantification of transcript abundance in single cells. The basic approach is not limited to color genetics and can be used for any cell type and phenotype of interest. We focus on melanogenesis using an avian model of color-mediated early-stage speciation in European crows. Black carrion crows Corvus (corone) corone (CC) and grey-coated hooded crows Corvus (corone) cornix (HC) hybridize along narrow contact zones in Europe and Asia that have likely formed in the Holocene (Mayr, 1942; Vijay et al., 2016). The characteristic differences in the amount of melanin deposited in the plumage (Figure 2A) have been shown to be associated with assortative mating (Randler, 2007) and social marginalization of minority phenotypes (Saino & Scatizzi, 1991). These behaviors linked to plumage pigmentation patterns are believed to act in concert to reduce the amount of crossbreeding (Brodin & Haas, 2006, 2009; Londei, 2013). Yet, this evidence of behavioural isolation by phenotype contrasts with near-identical genomes making the crow system a suitable model to investigate the genetic underpinnings of divergence in color patterns and their role during the early stages of speciation (Knijff, 2014; Pennisi, 2014; Wolf and Ellegren, 2017). Previous population genetic investigations suggest that only few, confined regions of the genome associated with color divergence appear to be under divergent selection resisting gene flow across the hybrid zone. Most genomic regions unrelated to pigmentation introgress more freely across the hybrid zone (Poelstra et al., 2014). This finding was corroborated by subsequent functional work. Transcriptome analyses across several tissues revealed a limited number of differentially expressed genes between taxa. Differences were almost exclusively restricted to skin tissue with active feather follicles maturing into grey or black feathers. Among these differentially expressed genes, genes involved in melanogenesis were strongly enriched and predominantly down-regulated in the grey coated hooded crows (Poelstra et al., 2014, 2015). Together with population genomic scans for candidate genes that may causally be involved in divergence of the pigmentation pattern these results suggested divergence of one or a few upstream melanogenesis genes. Half of the genes, including TYR, TYRP1, SLC45A2, SLC45A4, RAB38, EDNRB2, MC1R, NDP, AXIN2, HPGDS, MLANA, OCA2, and RAS-GRF1, involved in this metabolic pathway regulate, or are modulated by, the key transcription factor MITF (Poelstra et al., 2014, 2015) and have partly been suggested to modulate pigmentation in other species (Cuthill et al., 2017; Hoekstra, 2006; Hubbard et al., 2010; Manceau et al., 2010; Vickrey et al., 2018). Yet, despite generalized differential expression throughout the melanogensis pathway, MITF assuming a central regulatory role in mammals showed no evidence of differential expression in crow feather (Poelstra et al., 2015).
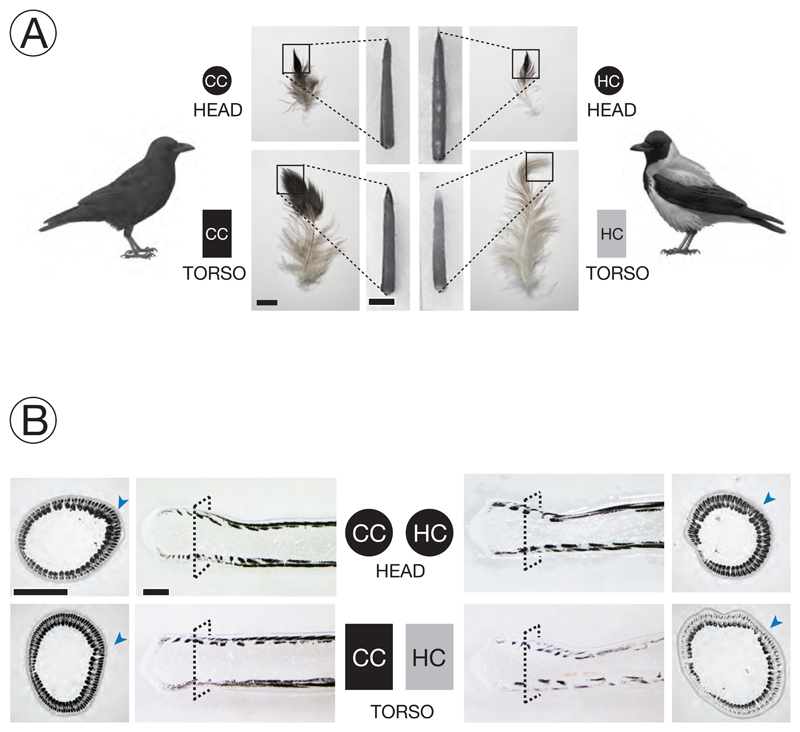
Figure 2. Experimental setup and phenotypic classification.
A) Melanin-based plumage pigmentation differs between of all-black carrion crows (CC) and grey-coated hooded crows (HC). Growing feather follicles were sampled in the head region (circle) and on the ventral part of the torso (rectangle). Photographs adjacent to the schematic representation of birds show an example of semiplume, pennaceous body feather (bar = 1 cm) from torso or head for each taxon. Squares define the area of the mature feather represented by the ensheathed feather follicle (bar = 2 mm) which forms the raw material of the experiment. Lighter pigmentation of mature feathers of HC torso is already visible in ensheathed feather follicles. Symbol color imitates the pigmentation of mature feather tips. [Bird drawings courtesy of Dan Zetterström].
B) Bright-field images from sections of ensheathed feather follicles. The dashed square on the longitudinal sections defines the corresponding position of the cross section at 1000 μm above the dermal papilla. The arrowhead in blue indicates the location of the rachis (see Figure 1). Dark areas represent regions where light-absorbing eumelanin is accumulated. Follicles from black-fathered regions show relatively higher eumelanin content in barb ridges than those sampled from the grey torso of hooded crows; bar = 0.5 mm.
Building upon information derived from bulk mRNAseq, we here characterize the molecular basis of (divergence in) avian melanin pigmentation at single cell resolution. We first examined whether phenotypic and anatomical characteristics may contribute to explain the striking color contrast. We characterized melanocyte maturation, melanosome transport and the formation of barbed ridges in the native histological context of growing feather follicle of regenerating torso feathers (grey in HC, black in CC) and head feathers (black in both taxa) from both taxa. We then quantified gene expression patterns of candidate genes both along the longitudinal axis of a feather and cross sections to reveal spatial-temporal expression patterns within and between taxa.
We quantified gene expression for a set of four candidate genes that based on previous work 1) were involved in melanogenesis, 2) covered a large range of expression levels and 3) were suggested to play a role in pigmentation divergence (see Poelstra et al., 2015 and references therein). Gene targets included down-stream ‘effector’ genes SLC45A2 and TYRP1. SLC45A2 is a solute transporter of the basic melanin unit L-Tyrosine from the cytosol of melanocytes into the melanosome. The melanosomal TYRP1 enzyme catalyzes tyrosine metabolism and is thus directly involved in melanin synthesis. TYRP1 is predicted to be under control of MITF (D’Mello et al., 2016), and is well characterized in coloration of mammal epidermis (Box et al., 1998) and skin appendages (Bourgeois et al., 2016; Du et al., 2017). HPGDS encodes the PGDS protein that may indirectly interact with MITF, but has generally not been well characterized in the context of melanogensis (Borovansky and Riley, 2011; Vachtenheim and Borovansky, 2010). Based on bulk mRNA sequencing of skin tissue of torso including regrowing feathers, SLC45A2, TYRP1 and HPGDS showed significantly higher levels of steady state mRNA in carrion crows (Poelstra et al., 2015). The regulatory core gene MITF, however, was not differentially expressed. This may be due to a different role in avian regulatory networks or due to ubiquitous expression in cell types other than melanocytes diluting the effect. To quantify mRNA transcripts in the cell type of origin we developed a histologically explicit padlock-probe assay coupled with rolling circle amplification allowing (RCA) (Weibrecht et al., 2013) to partition variation in gene expression among taxa into its biologically relevant components.
Overall, this study provides detailed insight into the molecular mechanisms involved in feather melanization and gene regulatory evolution. It also demonstrates the power of the quantitative padlock assay allowing for statistical inference on gene expression differences in a cell type specific context that can readily be transferred to other systems.
Material and Methods
Taxonomic considerations
The taxonomic status of the Corvus (corone) ssp. species complex is contentious with some authors treating populations differing in plumage coloration as separate species (Parkin et al., 2003). European all-black carrion crows and grey-coated hooded crows, however, have near identical genomes arguing against species status (Poelstra et al., 2014). Hence, until formal revision of taxonomic status recent work has treated carrion [C. (corone) corone] and hooded crows [C. (corone) cornix] with uncertainty regarding taxonomic status (Dutoit et al., 2017; Poelstra et al., 2013; Vijay et al., 2016). Given that for most part of the genome - including the target genes under investigation - genetic differentiation approaches null, taxa can essentially be treated as color morphs segregating within a population for the purpose of functional genetic work.
Population sampling
Crow hatchlings were obtained directly from the nest at an age of about three weeks. Hooded crows [C. (corone) cornix] were sampled in the area around Uppsala, Sweden (59°52'N, 17°38'E), carrion crows [C. (corone) corone] were obtained from the area of Konstanz, Germany (47°45N', 9°10'E) (Supplementary Table S1). To avoid any confounding effects of relatedness, only a single individual was selected from each nest. After transfer of carrion crows to Sweden by airplane, all crows were hand-raised indoors at Tovetorp fieldstation, Sweden (58°56'55"N, 17° 8'49"E). When starting to feed by themselves they were released to large roofed outdoor enclosures (6.5 x 4.8 x 3.5 m) specifically constructed for the purpose. Carrion and hooded crows were raised and maintained under common garden conditions in groups of maximum six individuals separated by sub-species and sex.
Feather sampling
For the purpose of the study, feather follicles needed to be sampled in a comparable, developmental stage with active expression of genes involved in the melanogenesis pathway. We therefore synchronized feather regrowth by plucking feathers on defined 3x3 cm patches on the torso (black in carrion crow, grey in hooded crow) and head (black in both subspecies) exploiting the natural disposition for molting during the months of July-September (Blotzheim et al., 1993). Previous work has shown that regrown feathers are identical in color and shape to the original feathers (Poelstra et al., 2014). We restricted sample collection to the molting seasons in August of 2015 and 2016, corresponding to an age of 1 and 2 years, respectively (Supplementary Table S1). This excludes any potential ontogenetic changes in coloration sometimes occurring at the juvenile molt 4-6 weeks after fledging (Blotzheim et al., 1993). We then allowed feathers to regrow for 10 to13 days, at which stage the previously plucked areas of skin contained densely spaced feather follicles still ensheathed within the shaft. Regenerating feather follicles ranging from 0.8 to 1.0 centimeters were collected in a stage of early development prior to apoptosis and keratinization. Immediately after retrieval follicles were fixed in phosphate buffered saline (PBS) solution containing 4 % paraformaldehyde (AH Diagnostics, Sweden). This material forms the basis of the subsequent histological examination and mRNA quantification (Weibrecht et al., 2013).
Histology of melanocytes and characteristics of melanosomes
Feather follicles pretreated with paraformaldehyde (PFA) as described above were dehydrated in an ethanol serial dilution of 70%, 95%, and 99.9%, and then embedded in Technovit 7100 (EMS, USA). Serial plastic cross-sections of 3 μm in thickness were obtained using a microtome (MICROM HM 360, ThermoFisher Scientific Inc., USA), and subsequently stained with hematoxylin (Merck & Co. Inc., USA) and eosin (BDH Chemicals, UK). To identify the morphologic features of melanosomes, PFA-fixed feathers were dissected and squeezed, and then wet-mounted with PBS. Bright-field histological or whole-mounted images were taken using a Leica Imager M2 equipped with a Hamamatsu digital camera C1440.
Quantification of targeted mRNA molecules in a single-cell context
LNA primer and padlock probe design
The abundance of mRNA transcripts was quantified in situ for four gene targets, HPGDS (Hematopoietic Prostaglandin D Synthase), SLC45A2 (Solute Carrier Family 45 member2), TYRP1 (Tyrosinase-Related Protein 1), and MITF (Melanocyte Inducing Transcription Factor) acting in different parts of the melanogenesis pathway. For in situ quantification we followed Weibrecht et al. (2013) using padlock probes in combination with rolling circle amplification (RCA) (see Supplementary Figure S1) on cryosections of PFA-fixed feather follicles which were infiltrated with a serial gradient of 15% and 30% sucrose in PBS, and then embedded in OTC Cryomount medium (Histolab AB, Sweden). The actin beta (ACTB), which shows no expression differences between taxa across several tissues including feather follicles (Poelstra et al., 2015), was included as the internal reference to control for technical variation in staining efficiency among sections.
First, appropriate sites for Locked Nucleic Acid (LNA™) primers initiating cDNA synthesis and for padlock probes hybridizing to the cDNA were designed on gene model of the Corvus (corone) cornix reference assembly ASM738373v2 available at NCBI (https://www.ncbi.nlm.nih.gov/) for five targeted genes, including TYRP1 (XM_010392933), HPGDS (4 isoforms: XM_010411872, XM_010411873, XM_010411874, XM_010411875), SLC45A2 (2 isoforms: XM_010395148, XM_019283413), MITF (14 isoforms: XM_010390220, XM_010390221, XM_010390223, XM_010390224, XM_010390225, XM_19280403, XM_19280404, XM_19280405, XM_19280406, XM_19280407, XM_19280409, XM_19280410, XM_19280411, XM_19280412), and ACTB (XM_010392369). Where several transcript isoforms were annotated, constitutively expressed exons were targeted (see Table 2 for primer and padlock probe design). To guarantee the same specificity across individuals, and importantly across taxa, primers were targeted at monomorphic regions devoid of genetic variation segregating within and between taxa (Poelstra et al., 2014). Using the Padlock Design Assistant version 1.6 (Weibrecht et al., 2013), we first identified a unique, 150-bp DNA fragment of roughly 50% GC content, that was suitable for both LNA primers and padlock probes. Blast searches at a statistical significance level of e-4 and e-9 of the resulting candidate sequences of LNA primers and padlock probes against the Corvus (corone) cornix genome (reference ASM738373v2), respectively, confirmed unique matching. For the best in situ spatial signaling of RCA, the 3’-end of a LNA primer were designed to overlap with the hybridization site of the padlock probe by at least 6 bp (Weibrecht et al., 2013).
Table 2.
Sequences of Locked Nucleic Acid (LNA™) primers and padlock probes for targeted pigmentation and internal control mRNAs. Lowercase letters in LNA primer sequences indicate the position of the locked nucleic acid modification. In the of padlock probes or detection oligonucleotides, sequence in blue is the hybridization site against targeted cDNA derived from corresponding mRNA, sequence in red indicates the hybridization site for corresponding detection oligonucleotide, and sequence in black is the extra fragment to stabilize padlock probe structure. Fluorescent dyes for detection oligonucleotides: FITC = Fluorescein isothiocyanate, Cy3 = Cyanines 3, Cy5 = Cyanines 5
| Primer or Probe | Sequence |
|---|---|
| LNA Primers | |
| LNA-primer ACTB | 5’/CaCcTgAaCcTcTcATTGCCA |
| LNA-primer TYRP1 | 5’/AaGaCtTcGaAcTgCTGGGTC |
| LNA-primer MITF | 5’/GtTtGgAtCcCaTtTTGGCAG |
| LNA-primer HPGDS | 5’/TgCcAgGtCaGgTtTACAGGA |
| LNA-primer SLC45A2 | 5’/AgCaGcCaAtCaGtGCCACT |
| Padlock probes | |
| Padlock-probe ACTB | 5’/Phos/TGAACTCCCTGATGGCAGTGAATGCGAGTCCGTCTGACAGTCCCAGTGTACCCACCCTGGAGAAGAGCTA |
| Padlock-probe TYRP1 | 5'/Phos/GTGATCCTTCAGGAAACTAGTGCTGGATGATCGTCCGACAGTCCCAGTGTACCCACACAGTAGAAGGGTACA |
| Padlock-probe MITF | 5’/Phos/CAGTGTTCCTGCCAATATCTGCTTATGTCGCCCGGGACAGTCCCAGTGTACCCACACCAGCTGGCACTTA |
| Padlock-probe HPGDS | 5'/Phos/GACACTTCTGTCCTGCCTCAATGCACATGTTTGGCTCCGACAGTCCCAGTGTACCCACGGATGTGTGCAGTAC |
| Padlock-probe SLC45A2 | 5’/Phos/TCAGCATCCACAGTGAGCGATCTGCGAGACCGTATGACAGTCCCAGTGTACCCACAAACCGTGATGCCGTCACCATGATT |
| Detection oligonucleotide | |
| Detection oligonucleotide ACTB | 5’/FITC/ AAAAAACAGTGAATGCGAGTCCGTCTUUUU |
| Detection oligonucleotide TYPR1 | 5'-/Cy5/CCACGCCTAGTGCTGGATGATCGTCC |
| Detection oligonucleotide MITF | 5’/Cy3/GTATCTGCTTATGTCGCCCG |
| Detection oligonucleotide HPGDS | 5’/Cy5/CCTCAATGCACATGTTTGGCUCC |
| Detection oligonucleotide SLC45A2 | 5’/Cy3/AGCGATCTGCGAGACCGTAT |
Pre-treatment of histological section
We performed in situ padlock probe with RCA on sections of feather follicles to obtain visualize the targeted mRNA molecules in the spatial-temporal context of the growing feather. All reactions were done on Superfrost Plus slides (Thermo Scifentific co. USA) with secure-seal hybridization chambers (Life technologies co. USA). Sections of 10 μm in the thickness were obtained with the assistance of cryofilms (type 3C, 16UF, Section-lab co. Japan) and were mounted on slides with histology mounting medium (Pertex HistoLab, Sweden). All sections were permeabilized in 0.1 M HCl with 20 mg/ml pepsin (Sigma-Aldrich co. USA) at 37 °C for 1 minute, followed by washes of DEPC-PBS twice. Hereafter, all washes were done by wash buffer of DEPC-PBS-tween 20 (0.05 %) twice between each reaction.
In situ reverse transcription
After washing twice, we added reverse transcription mixture containing 1 μM of LNA primer of each targeted mRNA (Actin-B, HPGDS, MITF, SLC45A2, TYRP1) with 20 U/μl RevertAid H minus reverse transcriptase (ThermoFisher Scientific co. USA), 1 U/μ1 Ribolock ribonuclease inhibitor (ThermoFisher Scientific co. USA), 0.5 mM dNTP, and 0.2 μg/μl BSA (BioNordika AB, Sweden) at 37 °C for 3 hours. After washing twice, sections were post-fixed in freshly prepared 4 % PFA in DEPC-PBS for 45 minutes incubated at room temperature. Preparing the section for padlock probe hybridization and ligation, sections were treated with 2 % glycine in DEPC-PBS at room temperature for 20 minutes.
RNA degradation, hybridization and ligation of padlock probe
We then eliminated RNA molecules for padlock probe circularization by adding the mixtures of 0.4 U/μl RNase H (ThermoFisher Scientific co. USA), 1U/μl Ribolock RNase inhibitor, 0.5 U/μl Tth Ligase (Genecraft co, Germany), 0.05 M KCl, 0.2 ug/μl BSA, and 20 % (V/V) formamide (ThermoFisher Scientific co. USA), plus 0.1 uM/μ1 of padlock probes of each targeted genes, at 37 °C for 30 minutes and followed by the other incubation of 45 minutes at 45 °C.
In situ rolling circle amplification and fluorescent-labeled oligonucleotide hybridization
After washing, the RCA reaction took place at 37 °C for 12 hours with the presence of 1 U/μl Phi29 polymerase (ThermoFisher Scientific co. USA), 1 U/μl Ribolock RNase inhibitor, 0.2 ug/μl BSA, and 5% (V/V) glycerol. Detection oligonucleotide hybridization was performed in two serial staining reactions. Staining mixture containing 1x SSC, 20 % (V/V) formamide, plus 0.1 μM of each fluorescent-labeled detection oligonucleotide (see Table 2) at 37 °C for 30 minutes. After imaging for the first staining, detection oligonucleotides were dehybridized and washed with distilled water for 5 minutes three times, followed by the incubation of 30 minutes at 50 °C. Slides were checked to confirm absence of any trace of fluorescent oligonucleotide before the subsequent staining.
Quantification of mRNA abundance along the proximal-distal axis of a feather
In situ mRNA abundance was quantified by counting independent, fluorescent signals on a Leica fluorescence microscope Imager M2 equipped with a Hamamatsu digital camera C11440 and corresponding filter setting. Fluorescent dyes were chosen such that mRNA abundance of three different genes could be investigated simultaneously on the same histological section. The first combination of detection oligonucleotides included the FITC fluorophore (filter excitation: 450 - 490 nm, filter emission: 500 - 550 nm) for the ACTB gene, the Cy3 fluorophore (filter excitation: 538 - 562 nm, filter emission: 570 - 640 nm) for SLC45A2 and the Cy5 fluorophore (filter excitation: 625 - 655 nm, filter emission: 665 - 715 nm) for HPGDS. The second set of detection oligonucleotides consisted of ACTB labeled with the FITC, MITF labeled with the Cy3, and TYRP1 labeled with the Cy5 (Table 2). To investigate gene expression for both sets on the same histological section, padlock probes of the first gene set were stripped off with 20 % formamide at 50 °C for 30 minutes. Absence of any remaining fluorescence residues was confirmed prior to hybridization with detection oligonucleotides of the second gene set.
To capture all RCA in situ signals across the 10 μm cryosection, each section was scanned at different focal planes resulting in z-stacks of images. Prior to quantification each z-stack of images was algorithmically merged into a best-focal-point image using ImageJ ver. 1.51h (Schindelin et al., 2012) with the Stack Focuser plugin (Umorin, 2006), optimally representing all RCA signals present in the sample. To ensure comparability between images taken separately for the control, dye set 1 and dye set 2, all images for the same section were aligned using a feature based alignment algorithm (Hast et al., 2013) in Matlab release 2016b. For some special cases, the alignment needed to be performed manually. This was done by using the ImageJ plugin Align Image by Line ROI (Schindelin et al., 2012). Quantification of signal position, intensity and cumulative number of RCA signals was automated using the Cellprofiler ver. 2.4.0rc1 pipeline (Carpenter et al., 2006). The basic steps of the pipeline were segmentation of the feather outline, detection of fluorescent signals, and detection of autofluorescence. The outline of the feather was extracted by enhancing the edges followed by morphological operations. The fluorescent signals were segmented using an adaptive local thresholding method based on ellipse fit (Ranefall et al., 2016). Regions with high autofluorescence were detected on images acquired before staining, and used to mask out the detected signals. All scripts are available in the supplement scripts and codes.
Defining melanocyte boundaries
The abovementioned approach measures abundance of mRNA molecules for the entire image including cell types other than melanocytes. To specifically quantify mRNA abundance in melanocytes, we determined cell boundaries in cross sections using protein immunostaining against the TYPR1 protein. The TYRP1 protein is suited as a melanocyte-specific marker for the following reasons: (i) based on UniProtKB gene ontology data it is functionally limited to melanogenesis where it serves as a central downstream regulator of melanogenesis (humans: http://www.uniprot.org/uniprot/P17643, mouse: …/P07147, chicken: …/O57405) (“UniProt,” 2017); (ii) in crows, TYRP1 transcripts are abundant only in skin with feather follicles (mean read count forebrain/liver/gonads/black feathered skin: 11/12/40/21121) and (iii) the protein product is only detectable in melanocytes within feather follicles (Poelstra et al., 2014).
Immunostaining was performed with a primary antibody against human TRP1 at 1:500 dilution (ab83774, Abcam Co., USA) and secondary goat-anti-rabbit IgG H&L antibody (ab150077, Alexa Fluor 488, Abcam Co., USA) at 1:200 dilution. In addition, TYRP1 and ACTB mRNAs were visualized using the padlock-RCA approach described above, and cell nuclei were stained using 1 μg/μl Hoechst 33342 (ThermoFisher Scientific Inc., USA; filter excitation: 335 - 383 nm, filter emission: 420 - 470 nm). Combining the information from cell identity, melanocyte specific mRNA and protein expression (TYRP1) and ubiquitous mRNA signals (ACTB) melanocyte boundaries could be localized (Figure 3). Cell boundaries were determined on cross sections at 1000 μm distal to the dermal papilla for a subset of three follicle samples of carrion crows (Supplementary Figure S2C). Since the inferred cell body boundaries consistently coincided with the melanin cluster at the ventral end of the barbed ridges, melanin patterns were thereafter used to extrapolate melanocyte boundaries in all other sections (Supplementary Figure S2D).
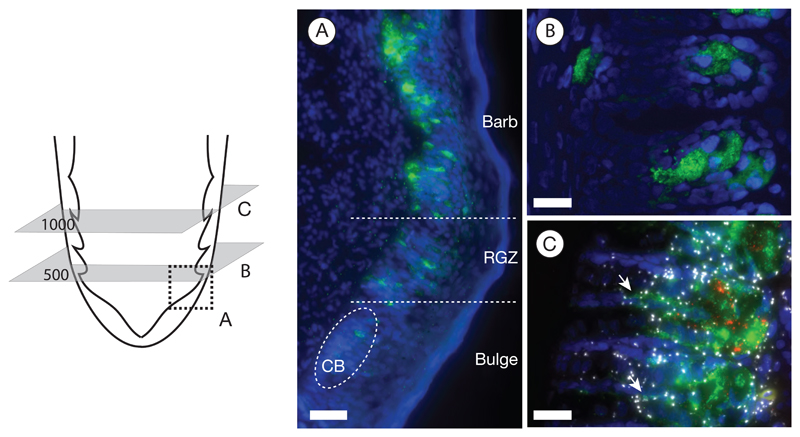
Figure 3. Melanocyte characterization.
The schematic of the proximal end of a feather follicle on the left side depicts the cutting planes for the images to the right. Melanocytes are visualized by immunostaining against the TYRP1 protein (in green); nuclei were stained with Hoechst 33342 (in blue).
A) The longitudinal section represented by the area of the dashed square illustrates the emergence of melanocytes in the collared bulge (CB) and their subsequent maturation in the ramogenic zone (RGZ) with increasing accumulation of the TYRP1 protein; bar = 50 μm.
B) Cross section showing barb ridges at an early developmental stage at 500 μm above the dermal papilla. The TYRP1 protein begins accumulating in a tubular cell body located in the ventral growth zone; bar = 20 μm.
C) Protein immunostaining combined with in situ mRNA padlock probe detection on a cross section at 1000 μm above the dermal papilla. At this developmental stage, melanocytes appear fully mature: the TYRP1 protein (in green) is predominantly accumulated in a spherical cell body sending dendritic cytoplasm into barbules plates (white arrow). Signals of rolling circle amplification (RCA) of in situ mRNA padlock probing tagging individual TYRP1 mRNA transcripts (shown in red) are confined to the cell body. In contrast to melanocyte specific gene expression of TYRP1, mRNA of the internal control gene ACTB (in white) is ubiquitously expressed across other cell types; bar = 20 μm.
Statistical comparison of transcript abundance
A major goal of this study was to quantify variation of in situ gene expression in melanocytes with respect to taxon (carrion/hooded crow), body parts (head/torso) and color (black/grey). To identify homologous regions for cross sections matched by developmental stage we first quantified transcript abundance along the feather using the padlock-RCA approach on longitudinal sections of the follicle (Figure 4). Two suitable regions were identified: a region of early differentiation at 500 μm distal of the dermal papilla, and a mature, differentiated region at 1000 μm where (i) expression levels of the central transcription factor MITF peaked and (ii) the lowly expressed SLC45A2 and HPGDS genes started saturating. Moreover, we found that 1000 μm constitutes a compromise between maximal gene expression levels and density of the melanin attenuating the signal of RCA products. At each region five serial cross cryosections of 10 μm in thickness were sampled covering together approximately at least one whole melanocyte. For each section, accumulated RCA signals within melanocytes of five barbs located next to the rachis were measured and summed across the serial sections for each barb ridge as described above (Supplementary Figures 2B & D). The sum of RCA signals of one barb ridge reflects the number of detected mRNA transcripts of an average of 3 (range: 2-4) melanocytes present per barb.
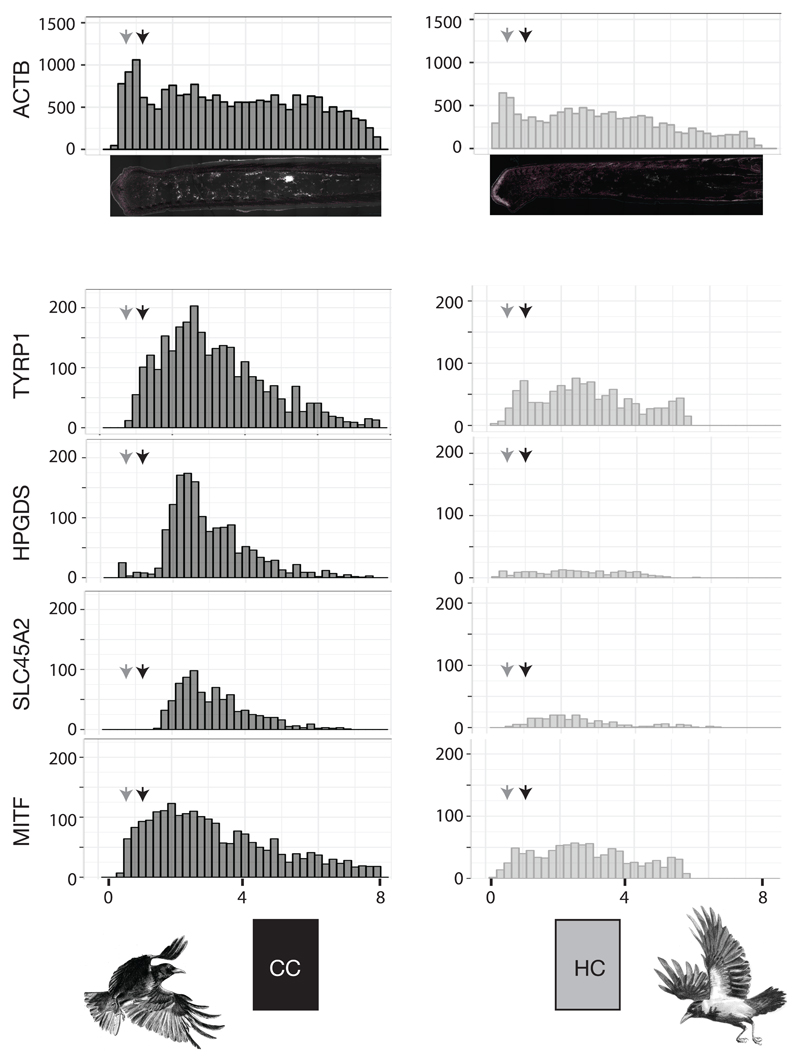
Figure 4. Longitudinal gene expression patterns of targeted mRNA transcripts in feathers from torso.
Histogram summarizing mRNA transcript abundance along the proximal-distal axis of feather follicles sampled from black torso of carrion crows (CC, left column) and grey torso of hooded crows (HC, right column). Each bin represents the raw counts of targeted RCA signals from mRNA transcripts of each candidate gene. Grey and black arrows indicate the position of cross sections at 500 μm and 1,000 μmabove the dermal papilla, respectively. (x-axis is the distance from the proximal end in mm. y-axis is accumulated raw RCS signal counts).
To control for technical differences in staining efficiency among and within sections, the abundance of each target gene was normalized relative to the ubiquitously expressed ACTB gene (an independent ACTB reference was used for each dye set, see above). Statistically, this can be modeled using the binomial distribution where RCA signal counts of the target gene K are expressed as a function of the total number of counts N of both target gene and ACTB. The probability of obtaining K successes is given by The probability of success p can here be thought of as the expression of the target gene relative to the ACTB control. The response variable was accordingly coded in matrix format containing RCA counts of the candidate gene alongside the control (~success, failure). A generalized linear model with binomial error structure was then used to assess significant differences in normalized gene expression of the target gene (parameter p) relative to the additive effects of the explanatory variables taxon (carrion/hooded crow), body area (head/torso) and color (black/grey). Overdispersion of the models arising due to variation not accounted for by any of these explanatory factors was taken into account by fitting a random factor for each data point separately resulting in a mixed effect model with basic structure: response ~ intercept + explanatory 1 + explanatory 2 + … + (1|observation). The observation-level random effect, where each data point receives a unique level, models the extra-binomial variation present in the data (Harrison, 2014). Statistical models were run for each target gene separately using the R package lme4 of R version 3.4.1. (Bates et al., 2015; R Development Core Team, 2008).
The resulting set of candidate models including all possible, additive combinations of explanatory factors was compared using the Akaike Information Criterion (AIC). To control for possible effects of melanocyte maturity, the position of the cross-section was included in all models as a fixed factor (500 μm vs. 1000 μm). Following Burnham and Anderson (2002) models differing by ΔAIC ≤ 2 were considering to be supported by the data. Akaike’s weight wAIC was used to assess the relative importance of each explanatory variable.
Quantifying melanocyte-specificity of gene expression
To assess melanocyte-specificity of gene expression we quantified total mRNA abundance in five barb ridges for a total of 115 cross sections at both 500 μm and 1000 μm of each individual (torso HC, CC and head CC, HC) (Supplementary Figure S2A). In addition, we quantified the number of in situ tagged mRNA molecules residing exclusively in an area reflecting melanocyte boundaries delimited by TYRP1 protein immunostaining (Supplementary Figure S2C). Subtracting the latter from the total number of counts in barb ridges yields the number of transcripts expressed in cell types other than melanocytes (Supplementary Figure S2D). Normalizing both melanocyte-specific and unspecific counts by the respective area in the cross section provides a relative measure of transcript density, δmel and δother respectively. Since the expectation of a ratio of two random variables is generally not equal to the ratio of the expectation of the two random variables (Hejmans, 1999) we summed across all counts and normalized by the cumulative volume to obtain a single measure across sections for follicles from each of the eight combinations (CC/HC, torso/head, 500/1000 μm). Assuming similar levels of signal detectability in both regions allows defining a standardized measure of melanocyte-specificity as the Smel= δmel / (δmel + δother). A value of 0.5 indicates equal density in both areas and hence ubiquitous gene expression independent of cell type. A value of 0.5> Smel ≥1 indicates an excess of expression in melanocytes reaching 1 if expression were fully limited to melanocytes. Values of 0≤ Smel<0.5 indicate a depletion of gene expression in melanocytes relative to other cell types. The relationship of cell type specificity with taxon (CC/HC), body region (torso/head), melanocyte maturity (500/1000 μm) and candidate gene was statistically assessed with an analysis of variance in R version 3.4.1. (Bates et al., 2015; R Development Core Team, 2008). A post-hoc Tukey test was used to assess statistical significance between individual genes.
Results
Morphological and anatomical characterization
Pennaceous wing and tail feathers, as well as body feathers covering the head and chest appear black in both carrion and hooded crows. Body feathers of hooded crows are light grey in contrast to all-black carrion crows (Figure 2A). In both taxa semiplume body feathers are not uniformly pigmented. The intensity of eumelanin pigmentation increases from the proximal end, which is dominated by plumulacous barbs, to the fully pigmented, pennacoues tip of the feather. Feathers from hooded crow torso, however, are overall lighter, but show the same pigmentation gradient along the feather (Figure 2A). As the basis for all subsequent analyses we collected feather follicles from head (black in both taxa) and the ventral side of the torso (black in CC, grey in HC) at an early stage of development when still ensheathed by a protective outer epidermal layer (Figures 1A & 2A). As feathers grow from proximal to distal, our sample represents the visible tip of the feather reflecting the natural contrast in plumage coloration between taxa in the torso region (Figure 2A).
Both longitudinal sections and cross-sections illustrate that differences in coloration are a direct consequence of taxon-specific variation in the level of melanization (Figure 2B). In both black and grey feathers melanin was deposited in rod-shaped melanosomes aggregating into higher-order granular structures (Supplementary Figure S3). While sections of black feathers (head CC & HC, torso CC) were saturated with melanin granules locally absorbing all light, grey feathers of hooded crow torso were more sparsely pigmented (Figure 2B, Supplementary Figures S4). Independent of the overall intensity, melanin was not uniformly distributed within barb ridges. Cross-sections illustrate that melanosomes were restricted to barbule plate cells separated by a melanin-free axial plate. Melanin was further concentrated at the dorsal end of barb ridges flanking the feather sheath, and ventrally in the growth zone adjacent to the ramus. The ‘melanin desert’ in between corresponds to rows of approximately three to four keratinocytes. In grey feathers, melanin levels appeared to be more depleted dorsally relative to the ventral growth zone of the barb ridge coinciding with the location of mature melanocytes (Figures 1B & 2B, Supplementary Figure S4).
Melanin is synthesized in melanocytes that are derived from activated progenitor cells at the very base of the follicle. Melanocyte maturation followed feather development in proximal-distal direction in both taxa: developing melanocytes first appeared in the collar bulge and were sparsely distributed in the basal layer of the ramogenic zone until about 500 μm from the distal end of the follicle (Figure 3A). At this stage most melanocytes were rod- to spindle-shaped (Figure 3B). At 1000 μm where barb ridges are fully formed, melanocytes were fully differentiated and spherical in shape (Supplementary Movie S1) measuring on average 32.8 ± 6.4 μm in diameter (n = 16). Mature melanocytes were situated in the ventral growth zone of a barb ridge and developed dendrite-like outgrowth of cytoplasm passing through cellular space in the axial plate and depositing melanosomes into keratinocytes of the barbule plates (Figures 1B & 3C, Supplementary Movie S2). Dendrite outgrowth appeared to be bounded by the marginal plate and axial plate, and thus limited to within single barb ridges. Melanocyte development was accompanied by melanin deposition appearing in the middle-upper region bulge, the collar bulge, with a significant increase in the ramogenic zone (Figure 3A, Supplementary Figure S2A). In addition to the proximal-distal axis of feather maturation, the development of barbs also followed the anterior-posterior maturation pattern. Anterior barb ridges close to the rachis were developmentally more advanced than posterior barbs (Figure 2B, Supplementary Figures S2B & S4) as expected by the relative age of rami (Figure 1A). Qualitatively, taxa did not differ in any of the morphological features considered above.
Cell type specificity of gene expression
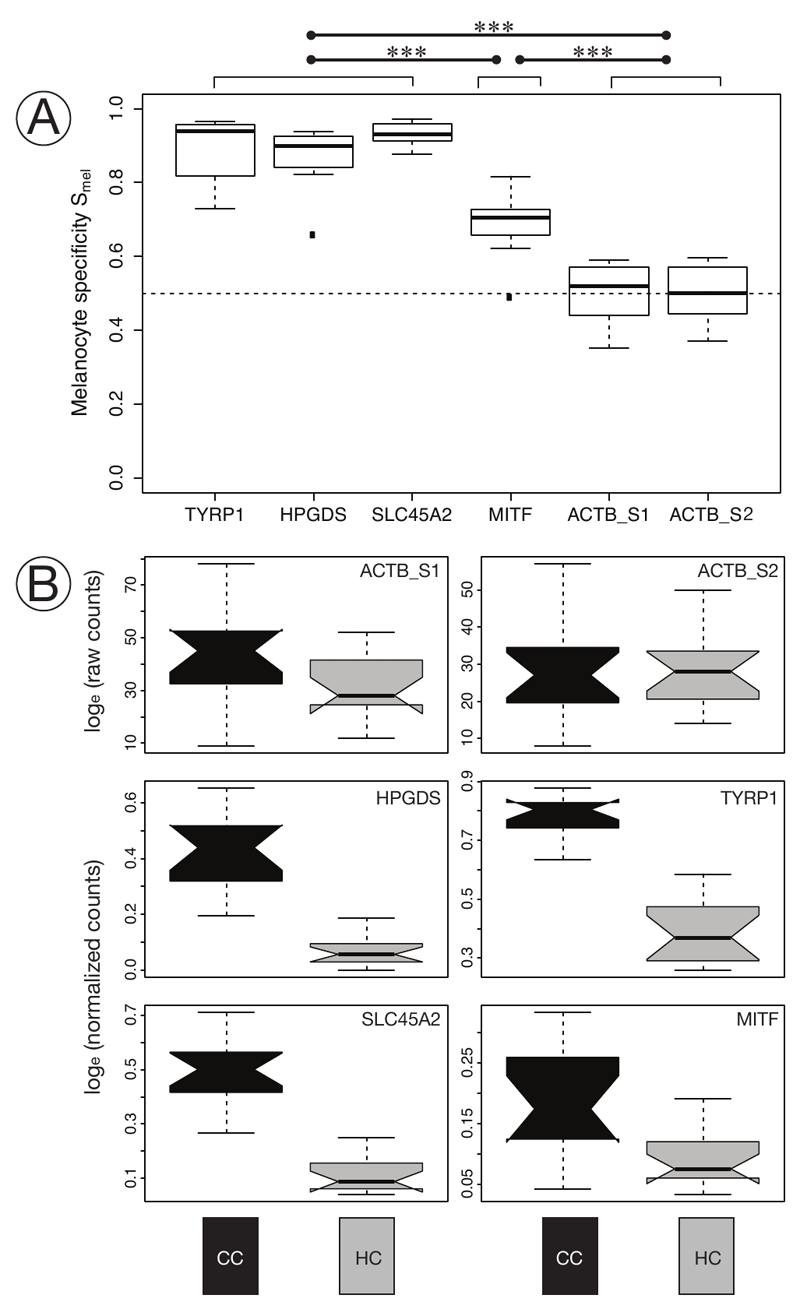
To assess the functional role of these gene targets in avian melanogenesis we first quantified melanocyte-specificity of gene expression. TYRP1 mRNA transcripts almost exclusively accumulated in the spherical melanocyte cell body with only rare traces in dendrites, and were lacking altogether in other cell types (Figure 3C & 5). Transcripts of HPGDS and SLC45A2 were also limited to the barb ridge growth zone likewise supporting melanocyte specificity (Figure 5). Expression of MITF and the ACTB control gene, however, was not restricted to melanocytes. Transcripts of both genes were detected in different cell types throughout the follicle in pulp, bulge and the ramogenic zone. In mature barb ridges transcripts were broadly distributed throughout including barbule, marginal and axial plates, and even the feather sheath (Figures 3C & 5). This qualitative assessment was corroborated by a quantitative measure of melanocyte-specificity Smel (see Methods; a value of 0.5 indicates ubiquitous gene expression independent of cell type, larger values reflect increasing melanocyte specificity reaching 1 if expression is fully limited to melanocytes). Cell type specificity was comparable among taxa (ANOVA: F(1,39)=1.77, p>0.05), body region (F(1,39)=3.77, p>.05) and melanocyte maturity (F(1,39)=0.14, p>0.05), but differed substantially between genes (F(5, 39)=46.64, p<0.01). The internal control gene ACTB showed no expression bias reflecting ubiquitous expression across all cells in the histological section of the feather follicle (median Smel ACTB_S1: 0.52, ACTB_S2: 0.50; with ACTB_S1 reflecting ACTB counts from first staining coupled with SLC45A2 and HPGDS and ACTB_S2 from the second staining of the same section coupled with MITF and TYRP1). On the contrary, TYRP1, HPGDS and SLC45A2 were strongly biased towards expression in melanocytes (median Smel 0.94, 0.90, 0.93 respectively). MITF showed only a slight bias towards melanocytes with a specificity index of 0.71 (Figure 6A). In summary, this provides evidence for melanocyte-specificity of TYRP1, HPGDS and SLC45A2, melanocyte affinity for MITF and ubiquitous expression of the ACTB control.
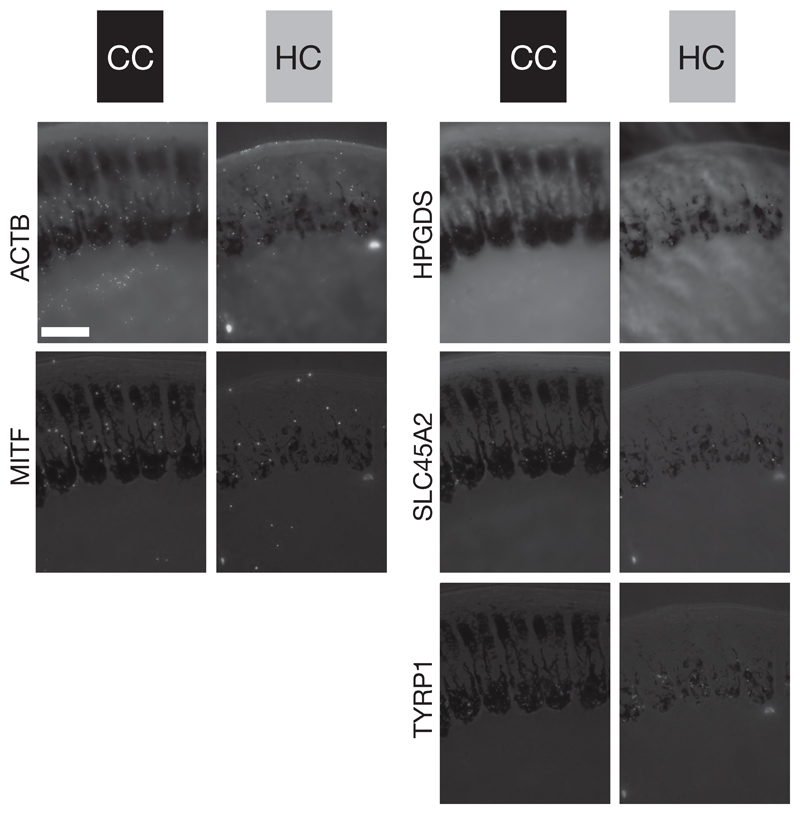
Figure 5. Five-way in situ gene expression in an explicit histological context.
Illustration of in situ stained mRNA transcripts of five genes on the same histological cross section of carrion crow (CC black) and hooded crow (HC grey) follicles sampled from torso at 1000 μm above the dermal papilla. For orientation, these cross sections reflect the schematic shown in Figure 1B. Bright-field images are superimposed for orientation and direct comparison among genes. The images represent one of five serial sections used for statistical analyses. ACTB and MITF (left column) are ubiquitously expressed in all cell types of a follicle. In contrast, HPGDS, SLC45A2 and TYRP1 (right column) are restricted to melanocyte cell bodies; bar = 50 μm.
Figure 6. Specificity of gene expression and differentiation by taxon and pigmentation intensity.
A) Boxplot of the specificity index of targeted genes. TYRP1, HPGDS, and SLC45A2 are near-exclusively expressed in melanocytes and do not in specificity (post-hoc Tukey test), p>0.05) for all comparisons). MITF transcripts are significantly less specific to melanocytes, but include a variety of other cell types. The ACTB control is ubiquitously expressed. Highly significant comparisons among (p<0.001, post-hoc Tukey test) are indicated among groups of genes with a triple asterisk. (n = 8).
B) Notched boxplot of raw counts (ACTB gene) or normalized in situ RCA signals of targeted mRNA transcripts (HPGDS, SLC45A2, TYRP1, MITF) in melanocytes at 1000 μm above the dermal papilla of carrion crow (CC) and hooded crow (HC) torso feathers. Gene expression of all four genes is significantly higher in melanocytes of carrion crow (CC black), compared to hooded crows (HC grey) (see Table 1). For gene expression of head see Supplementary Figure S5. (n = 30)
Gene expression transition along the proximal-distal axis of feather
We quantified the level of gene expression along the proximal-distal axis of the feather follicle (Figure 4). Absolute abundance of transcripts differed, as expected, among genes and by follicle origin (black CC torso vs. grey HC torso). For the internal control gene ACTB transcript abundance was high throughout. Regardless of taxon, body region or pigmentation intensity, gene expression started at the very proximal end in the bulge region, peaked in the ramogenic zone at around 500 μm, was then stably maintained and after several millimeters slowly declined. In the highly pigmented follicles of CC torso, gene expression profiles of the melanocyte-specific TYRP1 gene differed markedly from the pattern of the ubiquitously expressed ACTB gene. mRNA abundance of this core melanosomal enzyme closely mirrored melanocyte maturation as inferred from morphology and melanin deposition (see above). The first signals of TYRP1 transcripts were detected in earliest developing melanocytes in the collared bulge. Signals quickly increased in concert with melanocyte maturation in the ramogenic zone. Expression peaked at around 1 mm from the base where spherical melanocytes were fully differentiated. Beyond this region expression gradually leveled off indicating reduced activity of melanogenesis. With the exception of near-absent expression at early stages of melanocyte maturation in the ramogenic zone, the other melanocyte-specific genes HPGDS and SLC45A2 had overall lower expression levels, but otherwise mirrored the mRNA abundance pattern of TYRP1 transcripts. Expression profiles of the broadly expressed MITF gene were similar to TYRP1 with a steep onset of expression in the ramogenic zone, but with a somewhat earlier peak of expression at around 1 mm from the base of the follicle. In follicles sampled from grey torso in HC, expression patterns were generally similar to those in black torso feather despite reduced signal counts of each gene. Expression profiles in follicles sampled from head were similar in shape, but showed lower abundance for HPGDS, SLC45A2 and MITF.
Comparison of gene expression
We formally tested whether abundance of mRNA transcripts within melanocytes differed among taxa (CC vs. HC), body region (head vs. torso) and feather pigmentation (black vs. grey). mRNA transcripts were quantified in the cell body of melanocytes for all four target genes and ACTB as internal control. Generalized linear mixed effect models using statistical model selection based on the Akaike Information Criterion (AIC) suggested an effect of taxon and pigmentation for TYRP1, HPGDS and MITF, and an additional effect of body region for SLC45A2 (Table 1). Overall, the parameter with the by far strongest influence was feather pigmentation (wAIC = 1.00 for all genes) followed by taxon (wAIC ranging from 0.57 for TYRP1 to 0.83 for MITF). Gene expression was slightly higher in head of hooded crows (black in both species), but substantially lower in follicles sampled from the grey torso region (Figure 6B, Supplementary Figure S5). These results were independent of melanocyte maturity both at 500 μm vs. 1000 μm above the dermal papilla.
Table 1.
Summary of the statistical models exploring the effect of taxon (carrion vs. hooded crow), body region (head vs. torso) and color (black vs. grey) on normalized gene expression for each of the four target genes seperately. upper part: Factors included in each model are indicated by ‘x’ (e.g. M7: gene expression ~ intercept + taxon + bodyregion + color + |random effect of each section). Akaike’s information criterion (AIC) and difference to the best model (ΔAIC) are shown. The best model is shown in bold on dark grey background; other models with support (ΔAIC≤2) are highlighted in light grey. lower part: Akaike’s weights (wAIC) indicating the relative importance of a factor are shown for each gene.
| Intercept | Taxon | Body region | Color | HPGDS | SLC45A2 | TYRP1 | MITF | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AIC | ΔAIC | AIC | ΔAIC | AIC | ΔAIC | AIC | ΔAIC | |||||
| M0 | x | 723.13 | 97.20 | 757.73 | 89.85 | 884.62 | 115.13 | 559.95 | 34.30 | |||
| M1 | x | x | 686.90 | 60.97 | 732.16 | 64.29 | 852.75 | 83.25 | 554.80 | 29.15 | ||
| M2 | x | x | 689.39 | 63.45 | 745.91 | 78.04 | 849.98 | 80.48 | 549.29 | 23.64 | ||
| M3 | x | x | 627.74 | 1.80 | 671.44 | 3.57 | 770.82 | 1.32 | 529.72 | 4.07 | ||
| M4 | x | x | x | 625.93 | 0.00 | 672.47 | 4.59 | 769.50 | 0.00 | 525.65 | 0.00 | |
| M5 | x | x | x | 629.65 | 3.71 | 671.17 | 3.30 | 770.57 | 1.07 | 529.52 | 3.88 | |
| M6 | x | x | x | 671.53 | 45.59 | 724.02 | 56.14 | 819.84 | 50.34 | 543.82 | 18.17 | |
| M7 | x | x | x | x | 626.97 | 1.03 | 667.87 | 0.00 | 771.13 | 1.64 | 527.65 | 2.00 |
| wAIC | Taxon | 0.74 | 0.75 | 0.57 | 0.83 |
| Body region | 0.35 | 0.82 | 0.40 | 0.31 | |
| Color | 1.00 | 1.00 | 1.00 | 1.00 |
Discussion
The study of animal coloration has made an important contribution to our understanding of the genetic basis underlying inheritance and evolution (Hoekstra, 2006; Hubbard et al., 2010). Long confined to traditional model organisms such as mouse or fruit fly, high-throughput nano-sequencing technology now allows investigating the genetic basis of color variation across a broad array of organisms (San-Jose and Roulin, 2017). Yet, functional genetic studies remain difficult for the vast majority of species that are not amenable to assays such as gene knock-outs, RNA interference or CRISPR-Cas9 modification that are standard for laboratory models (Wolf and Ellegren, 2017).
Functional validation of genes by padlock probe combined with rolling circle amplification
We here suggest a versatile approach for functional validation of genetic signals from genome scans or association mapping applicable to a wide variety of natural systems. In contrast to bulk mRNA sequencing which integrates over diverse cell populations, in situ padlock probes are a flexible tool to visualize mRNA transcripts of any gene of interest in an explicit histological context (Larsson et al., 2010). Combined with rolling circle amplification, single mRNA molecules are imaged as bright, round-shaped fluorescent signals that can readily be quantified using automated image analyses. In contrast to conventional in situ hybridization relying on relative fluorescence intensity, the approach chosen here results in numeric count data. This property allows for flexible statistical analyses of complex treatment design. Moreover, referencing counts to a control gene known to be unaffected by the contrast in question (housekeeping gene) controls for methodological variation in staining efficiency and allows integrating data across multiple sections into one statistical analysis. Using an evolutionary model for incipient speciation characterized by a striking polymorphism in feather pigmentation, we demonstrate the utility of the approach to characterize gene expression relevant to natural phenotypic variation.
Morphological considerations
In a first step, we characterized the morphological features of black carrion and hooded crow feathers as the basis for subsequent quantification of gene expression at cellular resolution in a histological context. Independent of taxon (CC vs HC), body region (head vs. torso) and feather coloration (black vs. gray), the investigated crow feathers followed the common blueprint of feather development, including the hierarchical structure of follicle development with emerging barbs and barbules (Chen et al., 2015; d’Ischia et al., 2013), cellular development (Yue et al., 2005) and cell differentiation (Prum, 1999) (Lucas and Stettenheim, 1972; Yu et al., 2004). All cellular features associated with melanin production, such as melanocytes with dendritic outgrowth and localized melanin deposition in the growth zone and barbule plates, were qualitatively comparable in follicles of both grey and black feathers. The only marked difference consisted in the increased eumelanin deposition in black feathers. This corroborates the assumption that plumage differences between carrion and hooded crows are merely due to differences in melanin production resulting from differential gene expression, and are not caused by any defect of melanocyte development (Poelstra et al., 2014). This set the stage to examine melanocyte-specific gene expression using the padlock assay.
Single mRNA transcript counts are overall well correlated with RNA-seq based quantification
Padlock probes for the detection of mRNA were recently developed (Larsson et al., 2010) and have so far only been used in a few studies in a biomedical context (El-Heliebi et al., 2017; Johansson et al., 2016; Ke et al., 2013). Following published guidelines (Weibrecht et al., 2013) we designed probes for four different targets and one control gene and applied them to analyze expression profiles in the complex tissue of nascent feather follicles. Given the added difficulty of working with keratizined, water repelling feather material suggests that the method is applicable across a wide variety of tissues. Relative estimates of steady-state mRNA concentration as measured by FPKM (fragments per kilobase per million reads) via bulk mRNAseq in skin tissue containing re-growing feather follicles differed by more than tenfold among candidate genes. SLC45A2 and MITF were most lowly expressed, followed by HPGDS with intermediate levels of expression and highest values for TYPR1 (see Fig. 4 in Poelstra et al., 2015). The normalized mRNA counts derived here using the padlock/RCA assay were highly correlated with the FPKM based estimates in all cells (R2=0.92, p<0.001), and to a lower extent when exclusively considering melanocyte-specific transcripts (R2=0.79, p<0.001). The correlation was largely unaffected by body region and taxon. This suggests that the method is not only suited to compare expression by treatment separately for each gene, but also to relate absolute transcript numbers from different genes normalized by the same control.
Cell type specific quantification of the ubiquitously expressed MITF transcripts reveals taxon-specific expression
Poelstra et al. (2015) found widespread differential expression in the melanogenesis pathway including genes such as SLC45A2 or TYRP1 which are well known to act downstream in the melanin production pathway (Goding, 2000). MITF, however, which assumes a central regulatory function in melanogenesis by integrating signals from several interacting pathways (WNT, cAMP-dependent, MAPK) was not differentially expressed. Using the padlock/RCA approach this conflict could be resolved. Developing a measure of cell type specificity, we could show that transcripts of SLC45A2 and TYRP1 were near-exclusively restricted to melanocytes where they were further enriched in the cell body relative to dendrites. MITF transcripts, on the contrary, were expressed more broadly. Considering melanocyte-specific transcripts alone, MITF expression followed the expected pattern of significant upregulation in black carrion crow feathers (Poelstra et al., 2015). The lack of differential expression in MITF using bulk mRNA sequencing was thus not due to a different functional role in avian regulatory networks, but reflects signal dilution by other cell types.
MITF has been shown to modulate cell-cycle progression altering melanocyte survival and turnover and to regulate the amount of melanin produced in melanocytes (Goding, 2000; Levy et al., 2006). Our data lend support the latter function. Pigmentation differences were exclusively due to change in the concentration of eumelanin contained in rod-shaped granules in both taxa, and there was no evidence for differences in melanocyte shape or development. Moreover, while absolute transcript numbers differed significantly between grey hooded crow feather follicles and black carrion crow follicles, longitudinal expression patterns did not. In both taxa MITF transcripts were detected at the very onset of melanocyte development and preceded expression of TYPR1 and SLC45A2. This is consistent with a role of MITF in early melanocyte development and in regulation of downstream elements such as TYPR1 and SLC45A2 being transcribed at a more advanced stage of melanocyte maturation. Altogether, this suggests a role of MITF in regulating differential melanin concentration among taxa, and more generally, corroborates a central role of MITF in regulating melanogenesis in a non mammalian system.
A functional role for the HPGDS gene in melanogenesis
Using population genomic screens in combination with bulk mRNA sequencing it had been suggested in the crow system that MITF may itself be regulated by upstream elements that have diverged during the course of evolution (Poelstra et al., 2014, 2015). These include CACNG calcium channel genes regulating AMPA receptors known to influence expression of MITF and genes less known for their role in melanogenesis such as Norrie-disease protein (NDP) (Poelstra et al., 2015; Vickrey et al., 2018). Another candidate from both population genetic and bulk mRNA studies was the Hematopoietic Prostaglandin D Synthase gene, HPGDS, a gene that is generally associated with gluthathione metabolism with no clear bearing on melanogenesis (Masoodi et al., 2010; Takeda et al., 2006). Yet, it has been suggested that HPGDS may interact with MITF through SOX9 or by its calcium ion binding functionality (Poelstra et al., 2014, 2015 and references therein). The observation of the present study that HPGDS transcripts were limited to melanocytes lends further support to a potential role in melanogenesis. Moreover, just as MITF, it was expressed at an early stage of melanocyte maturation which is consistent with the proposition that it may be involved in regulatory feed-back with MITF (Shimanuki et al., 2012; Takeda et al., 2006). Additionally, in human cell lines, it had been shown that prostaglandins positively enhanced tyrosinase activity (Abdel-Malek et al., 1987). Thus, increased expression level of HPGDS is likely to contribute to elevated melanin production in black as compared to grey feathers.
Conclusion
This study introduces padlock probe assays as a versatile tool to validate functional inference from bulk transcriptomic analyses at single-cell resolution. Using this approach, we statistically substantiated differential gene expression of the central upstream regulator MITF in melanocytes of feather follicles collected from all-black carrion and grey-coated hooded crow under common garden conditions. We were able to visualize gene expression in an explicit histological context and provide a quantitative measure of cell type specificity in gene expression for a set of candidate genes at a resolution inaccessible to bulk mRNA sequencing. The results from this study corroborated that plumage divergence in the crow system is not due to any defect on melanocyte development and differentiation during feather regeneration. Much rather, the findings are consistent with the prediction that few, melanocyte specific upstream regulators differentially modulate melanin production between taxa through the central hub gene MITF. Overall, this is consistent with the idea that population divergence may arise quickly through modification of few genes conveying prezygotic isolation.
Supplementary Material
Acknowledgements
We express our gratitude to Sven Jakobsson for providing the infrastructure for animal husbandry at Tovetorp research station. We would also like to thank Christen Bossu, Jelmer Poelstra and Matthias Weissensteiner for their contribution in obtaining samples. Kristaps Solokovskis, Thomas Giegold, Nils Andbjer, TamaraVolkmer, Barbara Martinschitsch and Luisa Sontheimer were of invaluable support for raising and maintaining the captive crow population. Martin Wikelski, Inge Müller and additional staff from the Max-Planck-Institute for Ornithology in Radolfzell facilitated sampling in Germany and transport to Sweden. Margareta Mattson and Helena Malmikumpu provided help with sectioning. Finally, we would like to thank Dirk Metzler for discussing statistical approaches.
Funding
Funding was provided by the European Research Council (ERCStG-336536 FuncSpecGen to JW), the Swedish Research Council Vetenskapsrådet (621-2013-4510 to JW), Knut and Alice Wallenberg Foundation (to JW) and Tovetorp fieldstation through Stockholm University.
Footnotes
Competing Interests
We declare no competing interests.
Author Contributions
CW and JW conceived of the study and together with OS designed the experiments with input from AK in primer design and PR in image analysis. During a pilot phase RM developed parts of the methodological set up. JW collected the samples and together with JB was responsible for animal husbandry. CW and JW wrote the manuscript with input from all other authors.
Ethics Approval
Permission for sampling of wild carrion crows in Germany was granted by Regierungspräsidium Freiburg (Aktenzeichen 55-8852.15/05). Import into Sweden was registered with Veterinäramt Konstanz (Bescheinigungsnummer INTRA.DE.2014.0047502) and Jordbruksverket (Diarienummer 6.6.18-3037/14). Sampling permission in Sweden was granted by Naturvårdsverket (Dnr: NV-03432-14) and Jordbruksverket (Diarienummer 27-14). Animal husbandry and experimentation was authorized by Jordbruksverket (Diarienummer 5.2.18-3065/13, Diarienummer 27-14) and ethically approved by the European Research Council (ERCStG-336536).
Dataset and Scripts
Data used and software scripts was deposited on the Figshare (DOI: 10.6084/m9.figshare.7635791).
References
- Abdel-Malek ZA, Swope VB, Amornsiripanitch N, Nordlund JJ. In Vitro Modulation of Proliferation and Melanization of S91 Melanoma Cells by Prostaglandins. Cancer Res. 1987;47:3141–3146. [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67:1–48. [Google Scholar]
- Blotzheim G, von Bauer KM, Bezzel E. Passeriformes (4. Teil): Corvidae – Sturnidae Rabenvögel, Starenvögel. AULA; Wiesbaden: 1993. [Google Scholar]
- Borovansky J, Riley PA. Melanins and Melanosomes: Biosynthesis, Structure, Physiological and Pathological Functions. John Wiley & Sons; 2011. [Google Scholar]
- Bourgeois YXC, Bertrand JAM, Delahaie B, Cornuault J, Duval T, Milá B, Thébaud C. Candidate Gene Analysis Suggests Untapped Genetic Complexity in Melanin-Based Pigmentation in Birds. J Hered. 2016;107:327–335. doi: 10.1093/jhered/esw017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box NF, Wyeth JR, Mayne CJ, O’Gorman LE, Martin NG, Sturm RA. Complete sequence and polymorphism study of the human TYRP1 gene encoding tyrosinase-related protein 1. Mamm Genome Off J Int Mamm Genome Soc. 1998;9:50–53. doi: 10.1007/s003359900678. [DOI] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR. Model Selection and Multimodel Inference: a Practical Information-Theoretic Approach. Springer Verlag; 2002. [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-F, Foley J, Tang P-C, Li A, Jiang TX, Wu P, Widelitz RB, Chuong CM. Development, regeneration, and evolution of feathers. Annu Rev Anim Biosci. 2015;3:169–195. doi: 10.1146/annurev-animal-022513-114127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthill IC, Allen WL, Arbuckle K, Caspers B, Chaplin G, Hauber ME, Hill GE, Jablonski NG, Jiggins CD, Kelber A, et al. The biology of color. Science. 2017;357 doi: 10.1126/science.aan0221. eaan0221. [DOI] [PubMed] [Google Scholar]
- d’Ischia M, Wakamatsu K, Napolitano A, Briganti S, Garcia-Borron J-C, Kovacs D, Meredith P, Pezzella A, Picardo M, Sarna T, et al. Melanins and melanogenesis: methods, standards, protocols. Pigment Cell Melanoma Res. 2013;26:616–633. doi: 10.1111/pcmr.12121. [DOI] [PubMed] [Google Scholar]
- D’Mello SAN, Finlay GJ, Baguley BC, Askarian-Amiri ME. Signaling pathways in melanogenesis. Int J Mol Sci. 2016;17:1144. doi: 10.3390/ijms17071144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Huang K, Zhao J, Song X, Xing X, Wu Q, Zhang L, Xu C. Comparative transcriptome analysis of raccoon dog skin to determine melanin content in hair and melanin distribution in skin. Sci Rep. 2017;7 doi: 10.1038/srep40903. 40903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutoit L, Vijay N, Mugal CF, Bossu CM, Burri R, Wolf JBW, Ellegren H. Covariation in levels of nucleotide diversity in homologous regions of the avian genome long after completion of lineage sorting. Proc R Soc B. 2017;284 doi: 10.1098/rspb.2016.2756. 20162756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Heliebi A, Kashofer K, Fuchs J, Jahn SW, Viertler C, Matak A, Sedlmayr P, Hoefler G. Visualization of tumor heterogeneity by in situ padlock probe technology in colorectal cancer. Histochem Cell Biol. 2017;148:105–115. doi: 10.1007/s00418-017-1557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godefroit P, Sinitsa SM, Dhouailly D, Bolotsky YL, Sizov AV, McNamara ME, Benton MJ, Spagna P. A Jurassic ornithischian dinosaur from Siberia with both feathers and scales. Science. 2014;345:451–455. doi: 10.1126/science.1253351. [DOI] [PubMed] [Google Scholar]
- Goding CR. Mitf from neural crest to melanoma: signal transduction and transcription in the melanocyte lineage. Genes Dev. 2000;14:1712–1728. [PubMed] [Google Scholar]
- Harrison XA. Using observation-level random effects to model overdispersion in count data in ecology and evolution. PeerJ. 2014;2:e616. doi: 10.7717/peerj.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hast A, Nysjö J, Marchetti A. Optimal RANSAC - Towards a repeatable algorithm for finding the optimal set. J WSCG. 2013;21:21–30. [Google Scholar]
- Hejmans R. When does the expectation of a ratio equal the ratio of the expectations? Stat Pap. 1999;40:107–115. [Google Scholar]
- Hill GE, McGraw KJ. Bird Coloration: Function and Evolution. Harvard University Press; 2006. [Google Scholar]
- Hoekstra HE. Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity. 2006;97:222–234. doi: 10.1038/sj.hdy.6800861. [DOI] [PubMed] [Google Scholar]
- Hubbard JK, Uy JAC, Hauber ME, Hoekstra HE, Safran RJ. Vertebrate pigmentation: from underlying genes to adaptive function. Trends Genet. 2010;26:231–239. doi: 10.1016/j.tig.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Hugall AF, Stuart-Fox D. Accelerated speciation in colour-polymorphic birds. Nature. 2012;485:631–634. doi: 10.1038/nature11050. [DOI] [PubMed] [Google Scholar]
- Johansson MM, Lundin E, Qian X, Mirzazadeh M, Halvardson J, Darj E, Feuk L, Nilsson M, Jazin E. Spatial sexual dimorphism of X and Y homolog gene expression in the human central nervous system during early male development. Biol Sex Differ. 2016;7:5. doi: 10.1186/s13293-015-0056-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke R, Mignardi M, Pacureanu A, Svedlund J, Botling J, Wählby C, Nilsson M. In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods. 2013;10:857–860. doi: 10.1038/nmeth.2563. [DOI] [PubMed] [Google Scholar]
- de Knijff P. How carrion and hooded crows defeat Linnaeus’s curse. Science. 2014;344:1345–1346. doi: 10.1126/science.1255744. [DOI] [PubMed] [Google Scholar]
- Kronforst MR, Barsh GS, Kopp A, Mallet J, Monteiro A, Mullen SP, Protas M, Rosenblum EB, Schneider CJ, Hoekstra HE. Unraveling the thread of nature’s tapestry: the genetics of diversity and convergence in animal pigmentation. Pigment Cell Melanoma Res. 2012;25:411–433. doi: 10.1111/j.1755-148X.2012.01014.x. [DOI] [PubMed] [Google Scholar]
- Küpper C, Stocks M, Risse JE, dos Remedios N, Farrell LL, McRae SB, Morgan TC, Karlionova N, Pinchuk P, Verkuil YI, et al. A supergene determines highly divergent male reproductive morphs in the ruff. Nat Genet. 2016;48:79–83. doi: 10.1038/ng.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson C, Grundberg I, Söderberg O, Nilsson M. In situ detection and genotyping of individual mRNA molecules. Nat Methods. 2010;7:395–397. doi: 10.1038/nmeth.1448. [DOI] [PubMed] [Google Scholar]
- Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Li S, Wang C, Yu W, Zhao S, Gong Y. Identification of genes related to white and black plumage formation by RNA-Seq from white and black feather bulbs in ducks. Plos One. 2012;7:e36592. doi: 10.1371/journal.pone.0036592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes RJ, Johnson JD, Toomey MB, Ferreira MS, Araujo PM, Melo-Ferreira J, Andersson L, Hill GE, Corbo JC, Carneiro M. Genetic basis for red coloration in birds. Curr Biol. 2016;26:1427–1434. doi: 10.1016/j.cub.2016.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas AM, Stettenheim PR. Avian Anatomy - Integument. Washington, DC: U.S. Department of Agriculture; 1972. [Google Scholar]
- Manceau M, Domingues VS, Linnen CR, Rosenblum EB, Hoekstra HE. Convergence in pigmentation at multiple levels: mutations, genes and function. Philos Trans R Soc B Biol Sci. 2010;365:2439–2450. doi: 10.1098/rstb.2010.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoodi M, Nicolaou A, Gledhill K, Rhodes LE, Tobin DJ, Thody AJ. Prostaglandin D2 production in FM55 melanoma cells is regulated by α-melanocyte-stimulating hormone and is not related to melanin production. Exp Dermatol. 2010;19:751–753. doi: 10.1111/j.1600-0625.2010.01098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E. Systematics and the Origin of Species. New York: Columbia University Press; 1942. [Google Scholar]
- Mundy NI, Stapley J, Bennison C, Tucker R, Twyman H, Kim K-W, Burke T, Birkhead TR, Andersson S, Slate J. Red carotenoid coloration in the zebra finch Is controlled by a cytochrome P450 gene cluster. Curr Biol. 2016;26:1435–1440. doi: 10.1016/j.cub.2016.04.047. [DOI] [PubMed] [Google Scholar]
- Niwa T, Mochii M, Nakamura A, Shiojiri N. Plumage pigmentation and expression of its regulatory genes during quail development - histochemical analysis using Bh (black at hatch) mutants. Mech Dev. 2002;118:139–146. doi: 10.1016/s0925-4773(02)00256-3. [DOI] [PubMed] [Google Scholar]
- Parkin DT, Collinson M, Helbig AJ, Knox AG, Sangster G. The taxonomic status of carrion and hooded Crows. Br Birds. 2003;96:274–290. [Google Scholar]
- Pennisi E. Disputed islands. Science. 2014;345:611–613. doi: 10.1126/science.345.6197.611. [DOI] [PubMed] [Google Scholar]
- Poelstra JW, Ellegren H, Wolf JBW. An extensive candidate gene approach to speciation: diversity, divergence and linkage disequilibrium in candidate pigmentation genes across the European crow hybrid zone. Heredity. 2013;111:467–473. doi: 10.1038/hdy.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelstra JW, Vijay N, Bossu CM, Lantz H, Ryll B, Müller I, Baglione V, Unneberg P, Wikelski M, Grabherr MG, et al. The genomic landscape underlying phenotypic integrity in the face of gene flow in crows. Science. 2014;344:1410–1414. doi: 10.1126/science.1253226. [DOI] [PubMed] [Google Scholar]
- Poelstra JW, Vijay N, Hoeppner MP, Wolf JBW. Transcriptomics of colour patterning and coloration shifts in crows. Mol Ecol. 2015;24:4617–4628. doi: 10.1111/mec.13353. [DOI] [PubMed] [Google Scholar]
- Price T. Speciation in Birds. Greenwood Village, Colorado, U.S.A: Roberts & Company Publishers; 2008. [Google Scholar]
- Prum RO. Development and evolutionary origin of feathers. J Exp Zool. 1999;285:291–306. [PubMed] [Google Scholar]
- Prum RO, Dyck J. A hierarchical model of plumage: morphology, development, and evolution. J Exp Zool Part B-Mol Dev Evol. 2003;298B:73–90. doi: 10.1002/jez.b.27. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- Randler C. Assortative mating of carrion Corvus corone and hooded crows C. cornix in the hybrid zone in eastern Germany. Ardea. 2007;95:143–150. [Google Scholar]
- Ranefall P, Sadanandan SK, Wählby C. Fast adaptive local thresholding based on ellipse fit. 2016 IEEE 13th International Symposium on Biomedical Imaging; Prague, Czech Republic. IEEE; 2016. pp. 205–208. [Google Scholar]
- San-Jose LM, Roulin A. Genomics of coloration in natural animal populations. Phil Trans R Soc B. 2017;372 doi: 10.1098/rstb.2016.0337. 20160337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shawkey MD, Hauber ME, Estep LK, Hill GE. Evolutionary transitions and mechanisms of matte and iridescent plumage coloration in grackles and allies (Icteridae) J R Soc Interface. 2006;3:777–786. doi: 10.1098/rsif.2006.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimanuki M, Takeda K, Kawaguchi M, Suzuki T, Shibahara S. Lipocalin-type prostaglandin D synthase as a marker for the proliferative potential of melanocyte-lineage cells in the human skin. J Dermatol. 2012;39:699–704. doi: 10.1111/j.1346-8138.2011.01485.x. [DOI] [PubMed] [Google Scholar]
- Shiojiri N, Oguri Y, Satoh H, Nakamura A. Distribution of melanocytes in feather germs of a plumage mutant Bh (Black at Hatch) embryo of Japanese quail (Coturnix coturnix japonica) Zool Sci Jpn. 1996;13:719–723. [Google Scholar]
- Silvers WK. Coat Colors of Mice: a Model for Mammalian Gene Action and Interaction. New York: Springer New York; 2012. [Google Scholar]
- Takeda K, Yokoyama S, Aburatani H, Masuda T, Han F, Yoshizawa M, Yamaki N, Yamamoto H, Eguchi N, Urade Y, et al. Lipocalin-type prostaglandin D synthase as a melanocyte marker regulated by MITF. Biochem Biophys Res Commun. 2006;339:1098–1106. doi: 10.1016/j.bbrc.2005.11.125. [DOI] [PubMed] [Google Scholar]
- Umorin M. The plugin in imageJ. Stack Focuser. 2006 https://imagej.nih.gov/ij/plugins/stack-focuser.html.
- Vachtenheim J, Borovansky J. “Transcription physiology” of pigment formation in melanocytes: central role of MITF. Exp Dermatol. 2010;19:617–627. doi: 10.1111/j.1600-0625.2009.01053.x. [DOI] [PubMed] [Google Scholar]
- Vickrey AI, Bruders R, Kronenberg Z, Mackey E, Bohlender RJ, Maclary ET, Maynez R, Osborne EJ, Johnson KP, Huff CD, et al. Introgression of regulatory alleles and a missense coding mutation drive plumage pattern diversity in the rock pigeon. eLife. 2018;7 doi: 10.7554/eLife.34803. e34803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay N, Bossu CM, Poelstra JW, Weissensteiner MH, Suh A, Kryukov AP, Wolf JBW. Evolution of heterogeneous genome differentiation across multiple contact zones in a crow species complex. Nat Commun. 2016;7 doi: 10.1038/ncomms13195. 13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibrecht I, Lundin E, Kiflemariam S, Mignardi M, Grundberg I, Larsson C, Koos B, Nilsson M, Söderberg O. In situ detection of individual mRNA molecules and protein complexes or post-translational modifications using padlock probes combined with the in situ proximity ligation assay. Nat Protoc. 2013;8:355–372. doi: 10.1038/nprot.2013.006. [DOI] [PubMed] [Google Scholar]
- Widelitz RB, Jiang TX, Yu MK, Shen T, Shen JY, Wu P, Yu ZC, Chuong CM. Molecular biology of feather morphogenesis: A testable model for evo-devo research. J Exp Zool Part B-Mol Dev Evol. 2003;298B:109–122. doi: 10.1002/jez.b.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JBW, Ellegren H. Making sense of genomic islands of differentiation in light of speciation. Nat Rev Genet. 2017;18:87–100. doi: 10.1038/nrg.2016.133. [DOI] [PubMed] [Google Scholar]
- Xu X, Zhou Z, Prum RO. Branched integumental structures in Sinornithosaurus and the origin of feathers. Nature. 2001;410:200–204. doi: 10.1038/35065589. [DOI] [PubMed] [Google Scholar]
- Yu HS. Melanocyte destruction and repigmentation in vitiligo: A model for nerve cell damage and regrowth. J Biomed Sci. 2002;9:564–573. doi: 10.1159/000067283. [DOI] [PubMed] [Google Scholar]
- Yu M, Wu P, Widelitz RB, Chuong C-M. The morphogenesis of feathers. Nature. 2002;420:308–312. doi: 10.1038/nature01196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu MK, Yue ZC, Wu P, Wu DY, Mayer JA, Medina M, Widelitz RB, Jiang TX, Chuong CM. The developmental biology of feather follicles. Int J Dev Biol. 2004;48:181–191. doi: 10.1387/ijdb.031776my. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Jiang T-XX, Widelitz RB, Chuong C-MM. Mapping stem cell activities in the feather follicle. Nature. 2005;438:1026–1029. doi: 10.1038/nature04222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.