Abstract
Group A Streptococcus (GAS; Streptococcus pyogenes) is a bacterial pathogen for which a commercial vaccine for humans is not available. Employing the advantages of high-throughput DNA sequencing technology to vaccine design, we have analysed 2,083 globally sampled GAS genomes. The global GAS population structure reveals extensive genomic heterogeneity driven by homologous recombination and overlaid with high levels of accessory gene plasticity. We identified the existence of more than 290 clinically associated genomic phylogroups across 22 countries, highlighting challenges in designing vaccines of global utility. To determine vaccine candidate coverage, we investigated all previously described GAS candidate antigens for gene carriage and gene sequence heterogeneity. Only 15 of 28 vaccine antigen candidates were found to have both low naturally occurring sequence variation and high (>99%) coverage across this diverse GAS population. This technological platform for vaccine coverage determination is equally applicable to prospective GAS vaccine antigens identified in future studies.
Introduction
GAS causes >700 million cases per year of superficial diseases such as pharyngitis and impetigo, and >600,000 cases per year of serious invasive infection. Immune sequelae such as acute rheumatic fever (ARF) and acute post-streptococcal glomerulonephritis each account for >400,000 cases per year1,2. As a consequence of ARF, >30 million people live with rheumatic heart disease, involving mitral and/or aortic regurgitation3. GAS ranks within the top 10 infectious disease causes of deaths worldwide1. Despite over 100 years of research, a commercial vaccine has not been developed2. Obstacles that have hindered development of a GAS vaccine include serotype diversity, GAS antigen carriage and variation, and vaccine safety concerns due to the immune sequelae caused by repeated GAS infection2,4. A limited number of Phase I clinical trials have since been conducted, focused primarily on multivalent N-terminal M protein vaccine candidates5,6. Other candidate GAS vaccine antigens that have demonstrated efficacy in preclinical (animal) vaccine studies include the J8 peptide incorporated in the C-terminal repeats of M protein7, and non-M protein candidate vaccine antigens such as the group A carbohydrate8,9 and other surface or secreted proteins (Supplementary Table 1)2,4. While a number of GAS antigens have been selected to avoid autoimmune concerns10,11 or specifically engineered to remove potential autoimmune-involved epitopes7,9, the capacity to investigate issues of serotype diversity, antigen carriage and antigenic variation is impeded by the considerable genetic diversity within the global GAS population12. To address this issue, we have developed a compendium of all GAS vaccine antigen sequences from 2,083 isolates employing high-throughput genomic technology.
Results
GAS population genetics
We have compiled the most geographically and clinically diverse database of GAS genome sequences to date, comprising 2,083 strains, of which 645 isolates are reported for the first time (Supplementary Table 2). Extracting the classical GAS epidemiological and genotypic markers of differentiation from 2,083 genome assemblies, the database constitutes 150 emm types (347 emm sub-types)13, 39 known M-protein clusters14 and 484 multi-locus sequence types (MLSTs)15.
To assess the genome-wide relationships within this global database, we identified the core genome of GAS to be 1,306 coding DNA sequences (CDS), based on an 80% nucleotide sequence coverage threshold and presence in >99% of the 2,083 genomes. To examine signatures of recombination within the core 1,306 genes, we analysed each core gene separately for evidence of mosaicism using the homologous recombination detection tool fastGEAR16. Using this algorithm, we found 890 core genes with a recombinatorial evolutionary history (Supplementary Fig. 1, Supplementary Table 3), leaving 416 non-recombinogenic core genes (Supplementary Table 4) encoded by 266,960 bp of sequence (~15% of a complete GAS genome). This number is likely to be an under-representation of the total levels of GAS core genome recombination based on the limitations in sampling (for example, the potential donor genome not being represented in the collection and/or larger recombination blocks encompassing multiple genes may be missed). A pseudo-core sequence alignment was generated using these 416 core GAS genes. After removal of repeat sequences that can confound read mapping, a total of 30,738 single nucleotide polymorphisms (SNPs) and 23,923 parsimony informative sites were identified within the 266,960 bp pseudo-reference. Phylogenetic analysis of the 416 gene pseudo-core GAS genome identified a deep branching star-like population structure indicative of an early radiation of GAS into distinct lineages (Fig. 1a). While the overall branching topology of the tree is supported by comparing genome-specific and lineage-specific SNPs (Supplementary Fig. 2), low bootstrap support towards the polytomous root of the tree prevents accurate inferences regarding the evolutionary relationships of lineage-specific radiations (Fig 1a). Comparative analyses of the core phylogenetic tree topologies prior (1,306 genes) and post (416 genes) removal of the predicted recombinogenic CDS, did not affect the overall clustering of the isolates at the terminal branches of the tree (Supplementary Fig. 3), indicating that recombination events within the ‘core’ GAS genome have blurred the ancestral evolutionary relationships between GAS lineages, yet have not introduced sufficient homoplasy to disrupt recent evolutionary signals.
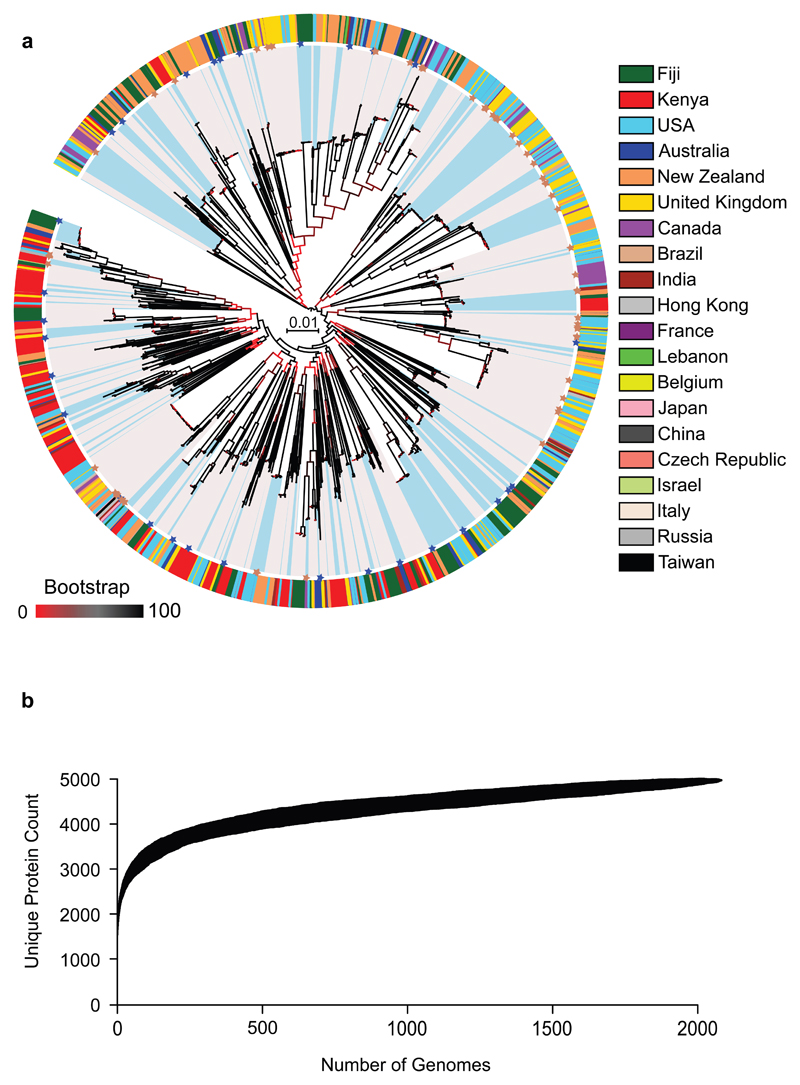
Figure 1. Population structure and pangenome of 2,083 globally distributed GAS strains.
(a) Maximum-likelihood phylogenetic tree of 30,738 SNPs generated from an alignment of 416 core genes. Branch colours indicate bootstrap support according to the legend. Distinct genetic lineages (n = 299) are highlighted in alternating colours (blue and grey) from the tips of the tree. Coloured asterisks refer to the relative position of complete GAS reference genome sequences (existing references are shown in brown; 30 new reference genomes are shown in dark blue). Colour coded around the outside of the phylogenetic tree is the country of isolation for each isolate. (b) Pangenome accumulation curve of 2,083 GAS genomes based on clustering of protein sequence at 70% homology.
Applying the population network approach of PopPUNK17, we identified 299 distinct genetic clusters of evolutionarily related lineages, herein termed phylogroups (Figure 1a, Supplementary Fig. 4, Supplementary Fig. 5a). This clustering approach is derived from core and accessory genetic distances between all 2,083 genomes using optimisation of a clustering network score to find a global distance boundary to define phylogroups (Supplementary Fig. 4a, b), and is designed to be iterative, meaning that new genomes can be added to this database using the same parameters and nomenclature as presented in this study without needing to refit the model. The median nucleotide divergence between phylogroups was 0.47% (range 0.25 – 0.56%), whereas genomes within the same phylogroup differed by a median divergence of 0.01% (range 0 – 0.14%). Of the 299 phylogroups, 206 phylogroups were represented by 2 or more isolates (Supplementary Fig. 4c). Overlaying the geographical origin of the isolates suggests that over half these 206 phylogroups have a diverse geographical distribution (Fig. 1a). The maintenance of so many distinct genetic lineages of GAS not appearing to be restricted by geographical boundaries is suggestive of rapid international spread followed by diversifying selection likely driven through immune selection and/or strain competition between phylogroups. Furthermore, these lineages do not appear to be restricted by clinical association. For example, 172 of the 206 phylogroups (83%) contain at least one clinically defined invasive GAS isolate (Supplementary Fig. 5b). The imbalanced nature of geographical and clinical sampling in this study prevents formal statistical inferences, and such phylogroup informed associations would require representative genomic epidemiological surveillance of the underlying population of GAS worldwide which, to-date, does not exist. Examination of the distribution of the classic GAS molecular epidemiological markers relative to the 206 multi-isolate phylogroups revealed that 179 (87%) carried a single emm sequence type, 140 (68%) carried a single emm sub-type, and 129 (63%) were of a single multi-locus sequence type (Supplementary Fig. 6). Only 3 (1.5%) of the emm sequence types and 55 (27%) of the emm sub-types were unique to a single phylogroup of 2 or more isolates, inferring extensive heterogeneity within GAS emm types. To further investigate these associations, we plotted the pairwise genetic distance of isolates based on common GAS epidemiological markers (emm type, emm sub-type, M-cluster and MLST). Greater than 66% of emm types (84/128 multi-isolate representatives) and 32% of the emm sub-types (65/204 multi-isolate representatives) exceeded the minimal median nucleotide divergence between any two phylogroups (0.25% which equates to 655 SNPs within 416 core genes), showing that many emm types, emm sub-types and M-clusters do not share a close evolutionary history and in many cases represent different genetic lineages (Supplementary Fig. 7). Conversely, <1% of MLST (2/269 multi-isolate representatives) exceeded the minimal median nucleotide divergence between phylogroups, yet MLST was a defining marker in only 27% of phylogroups. Furthermore, 6 of the 7 MLST genes (murI, xpt, gtr, gki, recP, and mutS) had evidence of homologous recombination within their evolutionary history while another MLST gene (yqiL) is not part of the core GAS genome (Supplementary Tables 3 and 4). Additionally, 3 emm18 genomes were also identified to have a deleted xpt gene18, and have been assigned the null allele xpt0 by MLST database curators. While emm-type and MLST have served as important markers for clonal associations within high income settings, our data suggest that emm-type and MLST may have limited capacity for assigning evolutionary relationships within a globally evolving GAS population.
The identification of hundreds of distinct genetic lineages (299 phylogroups) represents a challenge to unravelling the microevolution of dynamically evolving bacterial populations. Indeed, only 32 of the phylogroups identified in this study contain a complete GAS reference genome (n = 68). Furthermore, the vast majority of publicly available GAS reference genomes are of strains and emm-types from North America and Europe, with very few reference types from high-disease burden geographical regions. Moreover, the emm-types circulating in these high-burden settings are often rarely encountered within high-income regions. To enable future research into global and regional GAS population and evolutionary dynamics, 30 geographically and genetically distinct isolates were completely sequenced using the long-read PacBio platform (Supplementary Table 5). Based on our estimated structure of the global GAS population, these reference genomes represent 27 previously unsampled phylogroups (Fig. 1a). These high quality geographically, clinically and evolutionary diverse genomes will act as an important reference tool for new studies into the context of global GAS genome evolution, transmission and disease signatures.
To further assess the relative contribution of recombination on individual phylogroups, we quanitifed the genome-wide rate and fragment length of recombination within 36 of the most highly sampled phylogroups (constituting 1,062 genomes). The microevolution of each lineage was assessed by mapping to a phylogroup specific reference genome and recombination assessed by Gubbins19, a tool previously shown to exhibit high concordance with other recombination detection approaches20. The average number of SNPs observed within the 36 phylogroups was 5,536 SNPs (range 191 to 24,899 SNPs) of which an average 20.5% of SNPs (range of 0.1 to 100%) were found to be vertically inherited within a phylogroup (Supplementary Table 6). Overall the mean ratio of recombination derived mutation versus vertically inherited mutation (r/m) was found to be 4.95 (median of 3.12), and noteably, is significantly greater than 1 (one-sample Wilcoxon test p-value of 7 x 10-7) suggesting that recombination is the primary driver of SNP derived variation in GAS (Supplementary Fig. 8). The average number of recombination events per phylogroup was 58.9 (range 0 to 299) (Supplementary Table 6). Plotting the length of recombination blocks/fragments revealed that the majority of the events were small in length (< 5000 bp) with large events occurring infrequently (Supplementary Fig. 9). The average recombination fragment length in each of the 36 phylogroups was 5,437 bp, ranging from 0 bp (phylogroup 23) to 101,894 bp (phylogroup ‘0’). Removal of recombination events associated with putative mobile genetic elements had a limited effect on the total number of recombination events per phylogroup (Supplementary Fig. 9b), suggesting that hertitable heterogeneity is largely mobile genetic element (MGE) independent. These data highlights that evolution across the core genome of GAS lineages is not uniform and is primarily driven by small homologous recombination events.
Analysis of the variable gene content (defined as protein coding genes present in less than 99% of the 2,083 genomes) across the entire 2,083 genomes identified 3,672 ‘accessory’ genes when homologues were clustered at a conservative 80% amino acid identity using Roary21 (average of 1,717 protein coding genes per genome). Plotting of unique protein counts per new genome added shows that GAS has an ‘open’ pangenome (Fig. 1b), indicating that further genes will continue to be identified as new GAS genomes are sequenced. Annotation of the accessory genome derived from prophage analysis of the draft genome assemblies estimated ~50% of the accessory gene pool of GAS to be phage-related. Plotting of the accessory content relative to the core genome phylogenetic structure of the global population revealed extensive variation both in total overall, and prophage content, within and between GAS core genome lineages (Supplementary Fig. 10), in-line with observations from GAS microevolutionary analyses22–25. Collectively, this high level of heterogeneity both in the context of core genome sequence and accessory gene content provides a unique database for the examination of disease signatures as well as exploring conservation and sequence variation within GAS proteins such as vaccine antigens.
Disease signatures within global GAS database
The lack of correlation between evolutionary lineages and clinical association such as invasive infection, suggests that disease propensity is not restricted to an evolutionary lineage or clone. The interrogation of genomic databases enables an assessment on whether there are common genetic factors over-represented with a clinical phenotype, within a globally disseminated genetically diverse bacterial population. Invasive propensity in GAS has been linked with a number of bacterial genetic factors and regulatory mutations2,26. To ascertain statistical support of gene content, gene polymorphisms or combinations thereof with clinical GAS invasiveness within this global genomic framework, we used the bacterial GWAS method of pyseer27. In this study, we defined invasiveness as those GAS isolated from a normally sterile site (blood, cerebrospinal fluid, bronchopulmonary aspirate) or severe cellulitis with positive GAS culture as invasive (n = 1,048); and those from clinical superfical infections such as throat, skin or urine as non-invasive (n = 896). We included country of origin as a regression covariate, to correct for geographical bias as previously defined26. Through this approach, we identified 184 hits provisionally associated with GAS invasiveness. The underlying population structure was identified as a cofounder, even though correction was applied. The confounding effect caused associations at the same p-value across the entire genome (Supplementary Fig. 11). The top five k-mers which exceeded this confounder threshold include a GAS virulence marker isp (immunogenic secreted protein)28; a LacI family transcriptional regulator; and a hypothetical open reading frame neighbouring the cysteine protease speB (Supplementary Table 7). Further studies are required to ascertain a link between genotype and an invasive phenotype. This analysis demonstrates the utility of the global database for generating new disease insights.
GAS vaccine target variation
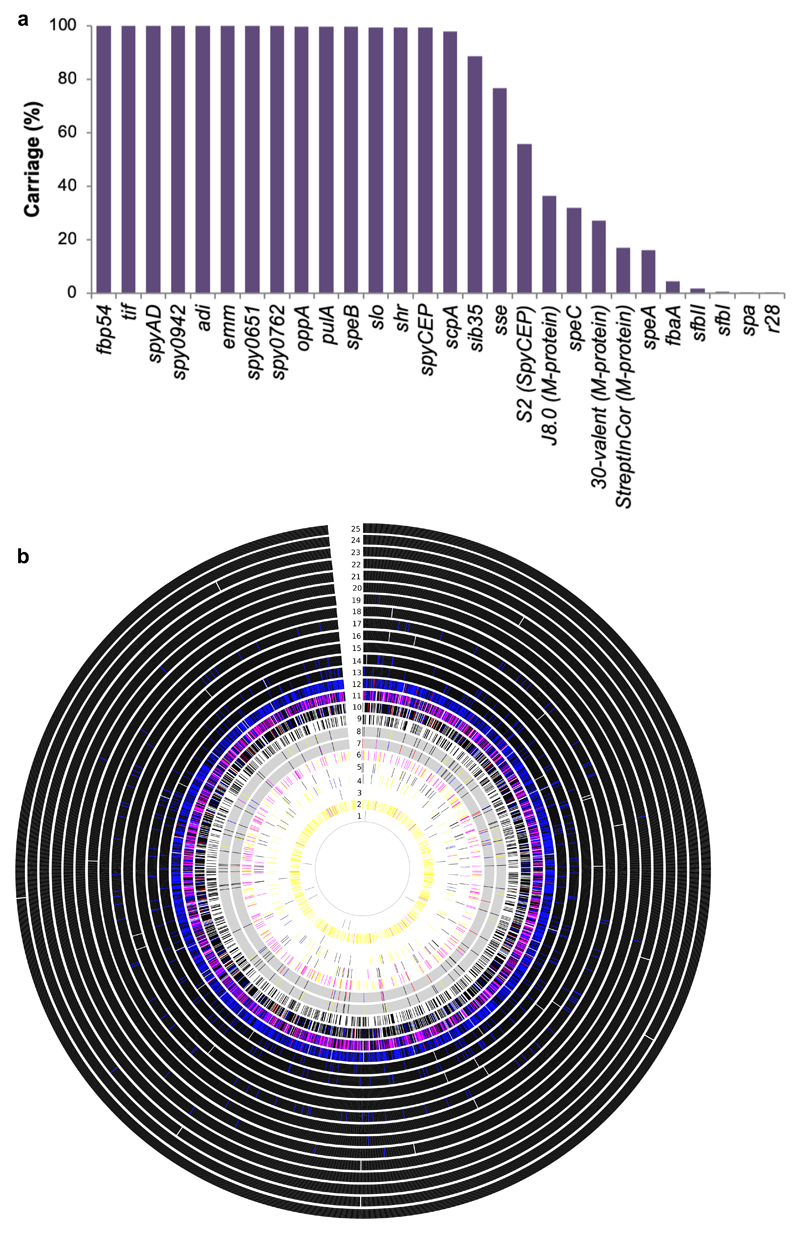
To examine natural variation of proposed GAS vaccine antigens within this genetically diverse GAS population, antigen carriage (gene presence/absence) and amino acid sequence variation of 29 proteinaceous GAS antigens, including 4 peptide fragments, was determined (Supplementary Table 1). The list of identified vaccine antigens analysed in this study have all been shown to convey protection in various murine models (reviewed by Henningham et al. 20124) but little is known about the conservation of these antigens within the global GAS population. Applying a sequence homology-based screening approach to the 2,083 GAS genome assemblies, 13 antigen genes were identified in >99% of isolates (Fig. 2a) at a 70% BlastN cut-off. The group A carbohydrate antigen is derived from a 12 gene biosynthetic cluster (gac) that has displayed protective properties in an animal model9. 2,017 GAS genomes (97%) shared all 12 protein coding genes with high DNA sequence conservation. Some genomes harboured frameshift mutations in several gac genes suggesting that not all 12 genes are critical for GAS survival, commensurate with previous findings on 520 gac loci29.
Figure 2. Antigenic variation within vaccine targets from 2,083 GAS genomes.
(a) Gene carriage (presence/absence) of vaccine antigens. (b) Amino acid sequence variation within 25 protein antigens for each of the 2,083 GAS genomes. Each ring represents a single antigen with protein similarity colour coded according to pairwise BlastP similarity: Black (>98%); Blue (between 95 – 98%); Red (between 90 - 95%); Pink (80 - 90%); Yellow (70 - 80%); Grey (<70%); and White (protein absence). Rings correspond to: 1) R28; 2) Sfb1; 3) Spa; 4) SfbII; 5) FbaA; 6) SpeA; 7) M1 (whole protein); (8) M1 (180bp N-terminal) 9) SpeC; 10) Sse; 11) Sib35; 12) ScpA; 13) SpyCEP; 14) PulA; 15) SLO; 16) Shr; 17) OppA; 18) SpeB; 19) Fbp54; 20) SpyAD; 21) Spy0651; 22) Spy0762; 23) Spy0942; 24) ADI; and 25) TF.
In addition to being omnipresent within the GAS population, an ideal GAS vaccine candidate would exhibit low levels of naturally occurring sequence variation within a genetically diverse dataset. To examine this question, pairwise BlastP cut-off values for 25 protein antigens were calculated. Eighteen antigens exhibited low levels (<2%) of amino-acid sequence variation (Supplementary Fig. 12). When plotted relative to overall carriage within 2,083 genomes, 13 of the 25 antigens were not only carried by >99% of the 2,083 genome sequences but also exhibited low levels of allelic variation (<2% sequence divergence) (Fig. 2b, Supplementary Fig. 12). Furthermore, 11 of these 14 core genome vaccine antigens had signatures of homologous recombination in their evolutionary history (Supplementary Fig. 13). The highest level of sequence heterogeneity in pre-clinical vaccine antigens was observed within the M-protein. Collectively only 33% of genomes had an N-terminal emm sub-type (685 out of 2,083) represented within the 30-valent M-protein vaccine formulation30 (Fig. 2a). We also examined the prevalence of other GAS peptide-based vaccine antigens, namely the C-terminal M-protein sequences of J831 and StreptInCor32; and the S2 peptide from the serine protease SpyCEP33. Carriage of these peptide antigens were assessed at an exact 100% match with the query peptide sequence within the 2,083 GAS genomes. The analyses revealed that 37% of the 2,083 isolates harboured the J8.0 allele of the M-protein; 17% carry the conserved overlapping B and T cell epitope of the StreptInCor M-protein vaccine candidate; and 56% of isolates encode the S2 peptide from SpyCEP protein. Further interrogation of known J8 sequence variants within the multi-copy M- and M-like C-repeat sequences represented in the 2,083 genome assemblies identified J8.12 (79%) and J8.40 (76%) as the most frequently encountered variants (Supplementary Fig. 14).
The characterisation of core gene products under different selection pressures may be used to identify potential novel vaccine antigen targets. Using the ratio of non-synonymous to synonymous codon subsititutions (dN/dS ratio) of each of the non-recombinogenic 416 genes, we identified that the average dN/dS ratio across the core GAS genome was greater than expected under a neutrality ratio of 1 (1.16), consitituing 49% of core genes (205 out of 416), suggestive of an overall positive selection across the GAS genome (Supplementary Table 4). Of the 3 ‘non-recombinogenic’ core vaccine targets analysed in this study, the streptococcal hemoprotein receptor (Shr) had signatures of positive selection (dN/dS 1.22) while the hypothetical membrane associated protein Spy0762 and the nucleoside-binding protein Spy0942 both exhibited signatures of purifying selection with dN/dS ratios of 0.57 and 0.66 respectively (Supplementary Table 4).
Antigenic heterogeneity within GAS vaccine antigens
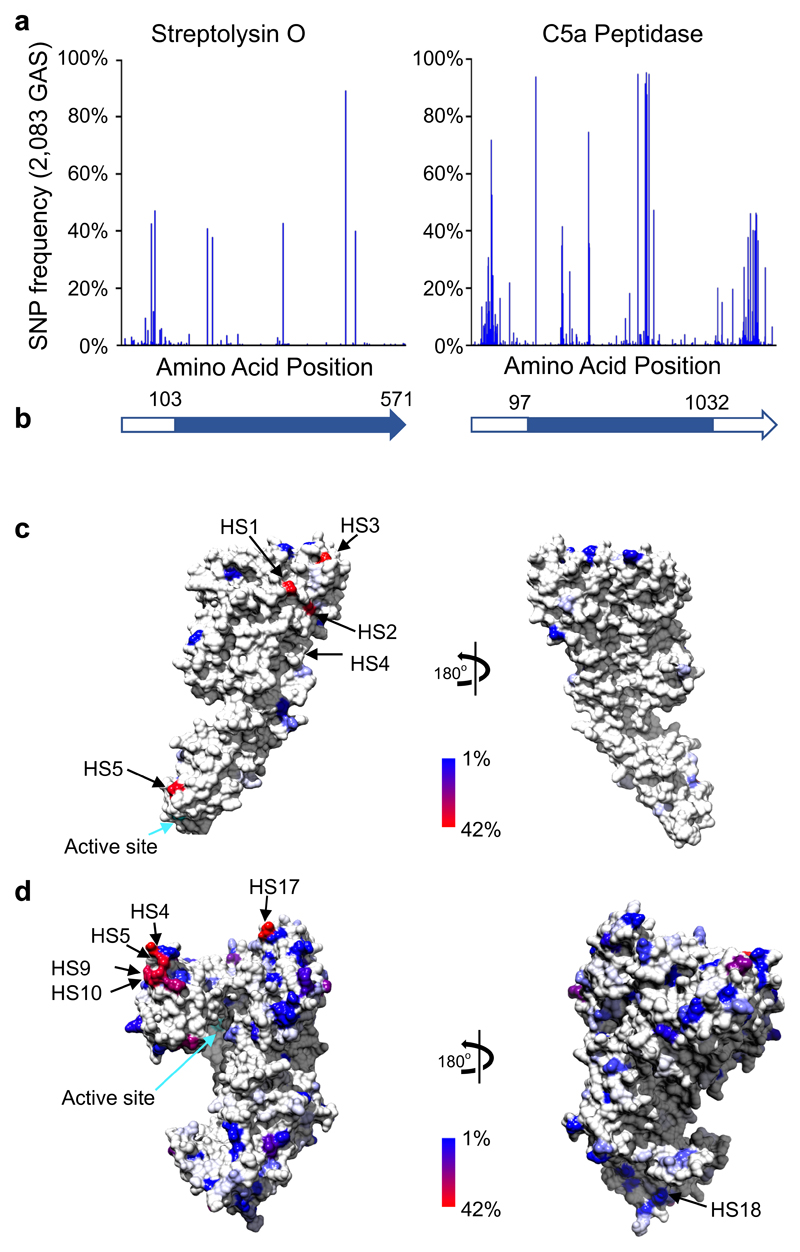
The ascertainment of antigenic variation within genome sequence databases allows such data to be overlaid onto protein structures, yielding important insight regarding potential sites of structural plasticity or immunodominance, which in turn can be used to inform vaccine design through identification of invariant surface regions and/or structurally constrained domains or subdomains. Two crystal structures are publicly available for GAS proteins that fulfil the criteria of global vaccine antigen coverage as defined in this study (>98% carriage and <2% amino acid sequence variation): Streptolysin O34 and C5a peptidase35. Identification of polymorphism location and polymorphism frequency within the 2,083 GAS genomes for the Streptolysin O (Fig. 3a, Supplementary Table 8) and C5a peptidase (Fig. 3b, Supplementary Table 9) proteins were determined. Using this data, we derived the consensus amino acid sequence for each protein. We then modelled the consensus sequence and population derived polymorphisms onto the corresponding crystal structures of the mature Streptolysin O protein (amino acids 103-501, Fig. 3b, c)34 and C5a peptidase (amino acids 97-1032; Fig. 3b, d)35. Using data extracted from the 2,083 genomes, further examination of amino acid heterogeneity present within the mature Streptolysin O protein revealed 5 sequence diversity hotspots (Fig. 3c). All hotspot polymorphisms were bimorphic in nature indicating restrictions in Streptolysin O plasticity (Supplementary Table 10). In comparison, we identified 20 sequence diversity hotspots within the mature C5a peptidase protein of which half were bimorphic (Fig. 3a, Supplementary Table 11), indicating more plasticity can be accommodated within the C5a peptidase than Streptolysin O. To ascertain the functional consequence of the most common protein variations, we examined mutational sensitivity and structural integrity of these amino acids variants using Phyre236 and the SuSPect platform37. All substitutions in both Streptolysin O and C5a peptidase were at locations where it was predicted that a change in the amino acid would not likely impact protein structure or activity (Supplementary Tables 10 and 11). To further examine selective pressures within these antigens, we assessed the selective constraints at each codon position. We found that 10.5% (60/571) of amino acid residues had higher diversity at first and second codon positions than at third codon positions for Streptolysin O and 16.5% (170/1032) for C5a peptidase, indicating that these sites are undergoing positive selection (Supplementary Tables 8 and 9). Of the diversity hotspots, 40% (2/5) of the Streptolysin O sites and 60% (12/20) of the C5a peptidase sites demonstrated signatures of positive selection. These data may reflect immune selection and/or the amount of plasticity that can be encompassed without compromising protein function.
Figure 3. Global amino acid variation mapped onto the protein crystal structure of the mature GAS Streptolysin O34 and C5a peptidase35.
(a) Frequency of amino acid variations within 2,083 genomes. (b) Schematic of the Streptolysin O and C5a peptidase open reading frame representing the location of amino acids within the mature enzymes (blue block). Model of the consensus sequence of the Streptolysin O (c) and C5a peptidase (d) mature enzymes. Plotted against the structure is the amino acid variation frequency within the 2,083 GAS genomes as represented in the colour gradient from 1% variable (blue) to 42% variable (red); invariant sites are coloured in light grey. Position of the top 5 most variable surface hotspots (“HS”) are annotated (as defined in Supplementary Tables 10 and 11). Active sites for each enzyme are indicated (cyan arrow).
Discussion
There is a strong case for the development of a safe and efficacious GAS vaccine1,2. One of several hurdles to be addressed in the development of a GAS vaccine suitable for worldwide use is the extensive genetic diversity of the global GAS population. To address issues of vaccine antigen gene carriage within the global GAS population and the extensive variation of antigen amino acid sequences between isolates, we have developed a platform for the interrogation of candidate antigens at unprecedented resolution. We have demonstrated that GAS is a genetically diverse species containing a large dispensable gene pool. Within the core or ‘conserved’ genome we have identified extensive evidence of recombination that will initiate future research into the biology and underlying drivers of such dynamic evolution. This diversity also has consequences for vaccine induced evolutionary sweeps of bacterial populations and subsequent emergence of vaccine escape clones, as has been observed in serotype-specific Streptococcus pneumoniae 38 and Bordetella pertussis 39 vaccination programs. Our findings identify that selection pressures are variable across the core GAS genome and proposed vaccine candidates, likely reflective of distinct and ongoing evolutionary adaptation. Collectively, within an evolving global bacterial pathogen such as GAS, we have identified that a number of proposed pre-clinical GAS vaccine antigens fulfil the criteria for a global vaccine. It is tempting to speculate that multi-antigenic formulations would provide an ideal approach against a rapidly evolving pathogen as well as increasing global coverage. Indeed, the incorporation of additional antigens to existing serotype-specific approaches in GAS enhances theoretical vaccine coverage40 (Supplementary Table 12).
We reveal that the global population structure of GAS is one of extensive genetic diversity, likely to be reflective of rapid international spread of genetically diverse lineages driven by diversifying selection from the immune system and/or competition between lineages. This may lead to negative frequency-dependant selection as has been proposed for other human bacterial pathogens such as S. pneumoniae and E. coli 41,42. Recombination has previously been identified to be high in GAS43,44 and at a genome-wide population level, our findings suggest a major role for homologous recombination of small DNA fragments in driving the evolutionary dynamics of GAS, indicating that evolution of GAS lineages is more likely to arise by recombination rather than by mutation43. All GAS lineages do not evolve at the same rate and this is likely to have key, yet undefined, biological significance. Similar impact and rates of homologous recombination have been observed in other bacterial pathogens such as S. pneumoniae 45 and Legionella pneumophila 46. A comparison of the relative rates of recombination versus mutation, based on whole-genome and gene-restricted MLST approaches, places S. pyogenes with other highly recombinogenic species such as K. pneumoniae and S. pneumoniae (Table 1).
Table 1. Comparative ratio of nucleotide changes resulting from recombination relative to point mutation (r/m) in selected bacterial pathogens.
| Species | r/m ratio (genome-wide) |
r/m ratio (MLST-derived) * |
References |
|---|---|---|---|
|
Streptococcus
pyogenes |
4.95 | 17.2 | This Study and Enright et al. 15 |
|
Streptococcus
pneumoniae |
6.36 | 23.1 | Chaguza et al. 53 and Hanage et al. 54 |
|
Staphylococcus
aureus |
0.6 | 0.1 | Driebe et al. 55 and Enright et al. 56 |
|
Legionella
pneumophilla |
47.8 | 0.9 | David et al. 46and Coscolla and Gonzalez-Candelas 57 |
|
Klebsiella
pneumoniae |
4.75 | 0.3 | Diancourt et al. 58and Wyres et al. 59 |
Multi-locus Sequence Type (MLST) allele-derived r/m ratios as defined by Vos and Didelot44.
The generation of high quality, well-curated reference genomes acts as a landmark for understanding the evolutionary context of a species, especially given the high levels of genetic diversity encountered in bacterial populations such as GAS and the contrasting epidemiology of infection observed between high-income countries and lower socioeconomic regions of the world which accounts for the overwhelming burden of GAS disease. The availability of new GAS reference genomes will enable targeted evolutionary and pathobiological studies of this genetically diverse pathogen. The 30 new GAS reference genomes reveal that despite an open pangenome where accessory gene content varies significantly across the population and recombination appears frequent, the overall size of the GAS genome remains at a steady state. Only recently have plasmids been characterised within the GAS genome19,47,48. We have identified a further 5 small plasmids in GAS ranging in size from 2,645 bp to 6,485 bp, harbouring bacteriocin-like genetic markers that are suggested to play a role in inter-bacterial inhibition49. In the context of vaccination, the availability of a globally representative reference database will provide a platform for examining the effect of future vaccination programs38,39.
Modelling of population based antigenic variation against protein crystal structures enables the identification of residues that may be under functional or structural constraints, or alternatively, selection pressure. This population-derived sequence approach could be assessed alongside immunological studies to define protective epitopes. Such information can be incorporated into further refinement of vaccine antigens such as peptide-based approaches that factor in naturally occurring population heterogeneity, enabling the targeting of immunogenic epitopes within antigens that are less amenable to variation.
This platform for population genomics-informed vaccine design is equally applicable to all known GAS antigens and those that remain to be defined. Thus, informed selection of putative vaccine antigens for human trial evaluation will now be possible, allowing identification of highly conserved antigens or combinations of antigens that ensure complete vaccine coverage across GAS emm types from differing geographic regions. For example, GAS vaccine antigens such as SLO, SpyCEP, ADI, TF and C5a peptidase, found here to be highly conserved across geographic regions, protect against multiple GAS emm types in animal models10,50,51. An approach similar to that used in this study would also be applicable to other pathogens that exhibit high levels of global strain diversity.
Online Methods
Bacterial isolates
The global collection of 2,083 Streptococcus pyogenes isolates examined in this study included short read genome sequence data from population-based studies that we have generated within Kenya59 and Fiji26, and other disease specific population-based studies of invasive GAS from Canada60, USA18 and the United Kingdom61,62 that was available as of 1st July 2018. We selected a small subset of isolates from published microevolution (outbreak) studies to avoid biasing the collection on single genetically related lineages. Sixty-eight GAS reference genomes and publicly available draft genomes from Lebanon63 were also included. To increase genomic representation from regions endemic for GAS infection and other under-sampled geographical regions, we collected a further 271 isolates from Australia, 279 isolates from New Zealand, 50 isolates from Brazil, 45 isolates from India and 7 isolates from Belgium. The rationale underpinning isolate selection was difference in epidemiological markers (emm type), anatomical site of isolation (skin, throat, blood) and clinical presentation, all key factors in GAS vaccine design. Metadata pertaining to the database of isolates are provided in Supplementary Table 2.
Genome sequencing and assembly
Genomic DNA was extracted and paired-end multiplex libraries were created and sequenced using the Illumina Hi-seq 2500 platform at the read-length between 75 to 125 bp (Wellcome Trust Sanger Institute, UK). Draft genome sequences were generated using an iterative Velvet-based assembly pipeline with secondary read mapping validation64 or using SKESA v2.3.065 with default parameters. Gene predictions and annotations were generated using PROKKA66 and streptococcal RefSeq specific databases64. Annotations pertaining to the mga locus (including emm and emm-like genes) were manually curated using in-house databases due to ambiguity when using pipeline procedures. The assembly pipeline generated assemblies of an average length of 1,791,171 bp (range 1,641,039 bp – 1,986,343) and an N50 of 252,789 bp (range 2,276 – 1,953,601 bp). On average, 1,711 coding sequences were identified per draft genome (range 1,495 – 1,976 coding sequences [CDS]). All draft genome assemblies are publicly available through GenBank. Accession numbers are listed in Supplementary Table 2.
Sequence mapping
To examine the genetic relationship of the 2,083 GAS genome sequences, we employed a single reference based mapping approach using sub-sampled Illumina fastqs at an estimated coverage of 75x. Published reference and draft genome datasets accessed from public databases were each shredded into an estimated 75x coverage of paired-end 100 bp reads using SAMtools wgsim. Sequence reads were mapped to the M1 GAS reference genome MGAS5005 (GenBank accession number CP000017)67 with BWA MEM (version 0.7.16) and read depth calculated with SAMtools (version 1.6) with a Phred quality score ≥20. Single nucleotide polymorphism (SNPs) with a Phred quality score ≥30 were identified in each isolate using SAMtools pileup with a minimum coverage of 10x. Core genes were defined as a minimum 80% of the MGAS5005 reference gene with a minimum 10x coverage. Using this approach, we identified 1,306 MGAS5005 genes with 99% carriage in 2,083 genomes. A core SNP genome alignment of 171,273 SNPs was generated by concatenating the SNPs located within the 1,306 core genes, giving a total of 1,201,767 bp. SNPs residing within repeat regions (minimum length of 20 nucleotides) and mobile genetic elements are considered evolutionary confounders and were identified as previously described68 or identified using PHASTER69. SNPs within these regions were excised from the core alignment, reducing the length from 1,201,767 bp to 1,197,326 bp and the SNP count from 171,273 to 170,653. Therefore, a total of 170,653 SNPs were aligned for phylogenetic analysis of the 1,306 ‘core’ genome (Supplementary Fig. 3).
Recombination detection
To examine evidence of recombination within the core GAS genome, FastGEAR16 was run on 1,306 individual gene alignments, comprising all 2,083 GAS strains included in the study. This method infers population structure for each alignment allowing for detection of lineages that have ancestral and recent recombinations between them. Default parameters were used with a minimum threshold of 4 bp applied for recombination length. A total of 890 genes had signatures of recombination and were excluded from evolutionary analyses. The remaining 416 genes were concatenated, corresponding to 268,003 bp of sequence. SNPs residing within repeat regions were removed as described above, resulting in 266,960 bp of sequence used as a best estimate for the global GAS population structure.
For intra-phylogroup recombination analyses, 36 most highly represented PopPunk phylogroups were chosen to investigate the influence of recombination (1,062 isolates). For each phylogroup, core genome alignments were performed using Snippy v4.3.5 (https://github.com/tseemann/snippy), against a reference strain within each phylogroup (Supplementary Table 6), maximum likelihood trees were inferred using IQtree v1.6.570, which were used as inputs for the recombination detection tool Gubbins v.2.3.419. Gubbins was run with maximum number of iterations of 20 with the minimum number of 5 SNPs to identify a recombination block, with a window size of 100 to 10,000 bp, with any taxa with more than 25% gaps filtered from the analysis. Recombinogenic blocks that overlapped with predicted mobile genetic elements (MGEs) in the reference genome were discarded. Phage regions were determined using PHASTER69 and integrative conjugative elements (ICE) were determined by manual inspection of reference genomes based on similarity of blast hits from known ICE. Recombination versus vertically inferited mutation (r/m) ratios for each lineage were calculated as the average r/m including all isolates within the phylogroup. For the species values of r/m was determined by the average across all 36 phylogroups (Table 1).
Phylogenetic analysis
Maximum-likelihood trees were generated for the 416 and 1,306 core genome alignments using IQ-tree v1.6.570. The generalized time-reversible nucleotide substitution with gamma correction for site-specific rate variation was performed with 100 bootstrap random resampling’s of the alignment data to support for maximum-likelihood bipartitions. For figure generation, phylogenetic trees and associated metadata were collated using the web portal, Interactive Tree of Life71.
Population genomics and cluster designation
To define evolutionary related clusters (phylogroups) in the population we used PopPUNK (Population Partitioning Using Nucleotide K-mers), which has previously been shown to give high quality clusters in a subset of S. pyogenes isolates included in this study17. We used k-mers between 15 and 29 nucleotides long in steps of two to calculate core and accessory distances between all pairs of isolates (Supplementary Fig. 4a). We clustered these distances first with the default two-component Bayesian Gaussian Mixture Model, then used the 'refine fit' mode to move the boundary of this fit such that the network was highly transitive and sparse, obtaining a network score (ns) of 0.980 (Supplementary Fig. 4b, 4c). To increase the utility of the GAS population clusters defined here, we created a database so that others can assign sample clusters using the same model and nomenclature as we present here. To do this we used PopPUNK to extract one sample per clique in the network, giving a reduced size query database containing 359 sequences. This database can be accessed at https://doi.org/10.6084/m9.figshare.6931439.v1 and contains an example command for database query and future expansion. The PopPUNK cluster designation (“phylogroup”) for each of 2,083 genomes have been added to Supplementary Table 2 and to the Microreact72 interative web application (https://microreact.org/project/5DEFpeck4).
Nucleotide divergence was derived by calculating the pairwise hamming distance from the 416 core genome alignment (266,960 bp). For pairwise hamming distance plots based on epdemiological markers (Supplementary Fig. 7), a reference genome was assigned for each marker based on the most representative distance within each type (minimum combined hamming distance) from the 416 core genome alignment.
Pangenome analysis
The pangenome was defined using Roary v3.11.221 without splitting paralogs and with clustering at 80%. Accessory genome was defined as the pan less the core, totalling 3,672 genes. Identification of prophage CDS within each of the 2,083 genomes was performed using PHASTER69. Clustering with CD-HIT-EST73 at ≤90% nucleotide homology resulted in 1,438 gene clusters. 584 core genes and 1,567 accessory genes hit these phage regions with blastn v2.3.0+ with a 90% nucleotide cut-off over 90% of the gene length. These data were then processed to generate a binary gene content matrix in which the presence of a gene is defined as >90% coverage to a corresponding phage gene cluster.
Vaccine antigen screening pipeline
To examine naturally occurring antigenic variation of proposed GAS vaccine targets within this genetically diverse GAS population, carriage of 29 vaccine antigens (Supplementary Table 1) and the group A carbohydrate biosynthesis loci was determined. The list of vaccine antigens screened have been shown to convey a significant level of protection in murine models4, but less is known about the conservation of these antigens within a global context. The presence of vaccine antigen genes was determined by BlastN analysis of 2,083 genome assemblies based on a 70% nucleotide cut-off over 70% of the gene length. Nucleotide sequences of whole and N-terminal regions of the M-protein were extracted using publicly available databases to account for known higher levels of allelic variation. This data was then converted into a binary gene content matrix in which gene presence was defined as >70% homology across a minimum 70% of the query gene length. Allelic variation was examined by plotting tBlastN (or BlastN for group A carbohydrate genes) scores relevant to the query reference sequence. To facilitate future studies assessing vaccine antigen carriage and sequence variation within GAS genome sequences, we have generated a bioinformatics pipeline for assessing antigenic variation from genome assemblies. This script, as used in this study, is available at https://github.com/shimbalama/screen_assembly and requires a query sequence (such as a vaccine antigen) and will run BlastN, tBlastN or BlastP at a user defined cut-off generating numerous outputs and plots as represented in this study (see Fig. 3a, Supplementary Fig. 13 and Supplementary Tables 8 and 9). Futhermore, this screening approach is applicable to any pathogen where genome assemblies are supplied.
Streptolysin O and C5a peptidase surface variation
Protein sequences of streptolysin O and C5a peptidase were chosen for further analyses as well characterised crystal structures exist for each of these GAS antigens. A protein alignment corresponding to the published crystalised structures of streptolysin O (amino acid residues 103 – 571, Protein Data Bank [PDB] accession number 4HSC34) and C5a peptidase (amino acid residues 97 - 1032, PDB accession number 3EIF35) was generated. Using this data, we derived the consensus amino acid sequence for each protein as defined by the most common amino acid identified within the global GAS genome database and modelled the consensus against the mature crystal structures. Amino acid polymorphic sites were converted into a binary matix and presented as a percentage of 2,083 genomes in Fig. 3. Visualisation of polymorphic sites on the crystal structure was determined using Chimera (version 1.11.2)74. Mutational sensitivity and structural integrity analyses was performed using Phyre236 that incorporates the SuSPect platform37.
Signatures of molecular adaptation
We investigated molecular signatures of selective constraints in all non-recombinogenic core genes (n = 416) by fitting a codon model to each of the individual genes and estimating the ratio of synonymous to nonsynonymous substitutions, dN/dS (also known as ω). Recombinogenic core genes (n = 890), as identified by fastGEAR, were excluded from analyses as such evolutionary processes invalidate phylogenetic codon model fitting. For each gene alignment, ambiguous codon sites were first excluded, before fitting the M0 codon model in CODEML, part of the PAML v4.0 package75. This model estimates a global dN/dS which allows for straight-forward comparison between genes. For the Streptolysin O and C5a peptidase protein coding genes we conducted more detailed analyses, by assessing selective constraints across codon sites. To do this we counted the number synonymous and nonsynonymous substitutions in each codon position, to obtain a similar quantity to the dN/dS value above76. Although this method does not explicitly use a codon model, it is scalable for the large number of samples used here. Despite the objective of this study being centered around global diversity, our database does contain sample bias in the context of clinical and geographical sampling, and the selection analyses should be interpreted carefully, as they may not represent current global selective trends.
Generation of 30 new GAS reference genomes
The vast majority of publicly available completely sequenced reference genomes are of emm-types from North America and Europe and very few are of emm-types from high-disease burden geographical regions. To facilitate the expansion of studies within the highest disease burden regions, 30 isolates were completely sequenced using long-read sequencing technology. Long-read sequences were obtained using the Pacific Biosciences RS II platform from a single molecule real-time (SMRT) cell as described previously77. Briefly, genome sequences were assembled using the SMRTpipe version v2.1.0 using the Hierarchical Genome Assembly Process (HGAP.2) and Quiver for post-assembly consensus validation. Secondary validation of the assemblies was performed using the Canu assembler78. To correct long-read sequence errors, primarily around homopolymeric regions, Illumina short read sequences from each of the 30 genomes were mapped using BWA MEM v0.7.16. Single contigs were achieved for all genomes and associated plasmids where present, with an average coverage depth of 80x. Genomes were annotated using the same pipeline as for the Illumina draft genomes64 with putative prophage regions defined using the PHASTER server69. The average size of these new reference genomes was 1,810,671 bp (ranging from 1,701,466 bp to 1,950,606) with 5 strains containing circular plasmids ranging from 2,645 bp to 6,485 bp in size (Supplementary Table 5).
Genome-Wide Association of GAS Invasiveness
To identify genomic signatures within the global GAS population overrepresented with severe GAS infection (‘invasive’) we ran pyseer27 on 1,944 samples (1,048 defined as invasive) using the linear mixed model. A total of 87M k-mers between 9 and 100 bases long were counted using fsm-lite. We only tested common k-mers, those with a minor allele frequency >1% (of which 18M were counted in our dataset). We created a kinship matrix from our recombination-free core phylogenetic tree of 2,083 genomes (416 genes, Figure 1a). The country of isolation was used as a covariate in pyseer’s model to account for geographical signal as defined previously26. All k-mers were mapped to the MGAS5005 GAS reference genome using bwa and visualised with R. We used a Bonferroni correction to adjust the significance threshold passed the number of unique patterns tested, which gave 9.4x10-7 for a 0.05 family-wise error rate. 184 k-mers were significantly associated with severe infection.
Supplementary Material
Acknowledgments
This work was supported by the National Health and Medical Research Council (NHMRC) project and program grants for Protein Glycan Interactions in Infectious Diseases and Cellular Microbiology; an Australian and New Zealand joint initiative, the Coalition to Accelerate New Vaccines Against Streptococcus (CANVAS); and The Wellcome Trust, UK. For part of this study, MRD was supported by a NHMRC postdoctoral training fellowship (635250) and AM was a GENDRIVAX fellow funded by European Union's Seventh Framework Programme FP7/2007-2013/ under REA grant agreement n°251522. We acknowledge the assistance of the sequencing and pathogen informatics core teams at the Wellcome Trust Sanger Institute. We acknowledge and thank the database curators of the S. pyogenes MLST and emm databases (especially Prof. Debra Bessen). We dedicate this work to the memory of our friend and colleague Prof. Gusharan Singh Chhatwal.
Footnotes
Author Contributions
MRD, GD and MJW conceived the project. MRD, AM, JAL, JALees, SD, PRS, MTGH, SYCT, PMG, ACS, JAB, GSC, SDB, RAS, TL, JDF, NJM, JRC, ACS, JP, AS, DAW, BJC and MJW designed experiments. MRD, LM, JAL, JALees, SD, AM, RJT, KAW, SRH, TRH, HRF, RSLAT, OB, AJC, RB, PNS, NJM and DAW performed experimental protocols. MRD, LM, JAL, JALees, SD, DJP, AM, PRS, NJM, GD and MJW analysed experimental results. MRD and MJW wrote the manuscript and all authors reviewed the manuscript.
Competing Interests Statement
AS is an employee of the GSK group of companies having a commercial interest in GAS vaccine development. The company had no influence over study design.
Data Availability
Illumina sequence reads and draft genome assemblies were deposited into the European Nucleotide Archive under the accession numbers specified in Supplementary Table 2. Genbank accession numbers for the 30 new GAS reference genomes are provided in Supplementary Table 5. To facilitate community accessibility and interrogation of the data presented in this study, the phylogenetic (Fig 1a), PopPUNK phylogroup designations, and associated metadata components have been uploaded to the interactive web interface Microreact72 (https://microreact.org/project/5DEFpeck4). The PopPUNK database for assigning new genomes is available at https://doi.org/10.6084/m9.figshare.6931439.v1.
References
- 1.Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5:685–94. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 2.Walker MJ, et al. Disease manifestations and pathogenic mechanisms of group A Streptococcus . Clin Microbiol Rev. 2014;27:264–301. doi: 10.1128/CMR.00101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watkins DA, et al. Global, regional, and national burden of rheumatic heart disease, 1990-2015. N Engl J Med. 2017;377:713–722. doi: 10.1056/NEJMoa1603693. [DOI] [PubMed] [Google Scholar]
- 4.Henningham A, Gillen CM, Walker MJ. Group A streptococcal vaccine candidates: potential for the development of a human vaccine. Curr Top Microbiol Immunol. 2013;368:207–42. doi: 10.1007/82_2012_284. [DOI] [PubMed] [Google Scholar]
- 5.Kotloff KL, et al. Safety and immunogenicity of a recombinant multivalent group A streptococcal vaccine in healthy adults: phase 1 trial. JAMA. 2004;292:709–15. doi: 10.1001/jama.292.6.709. [DOI] [PubMed] [Google Scholar]
- 6.McNeil SA, et al. Safety and immunogenicity of 26-valent group A Streptococcus vaccine in healthy adult volunteers. Clin Infect Dis. 2005;41:1114–22. doi: 10.1086/444458. [DOI] [PubMed] [Google Scholar]
- 7.Brandt ER, et al. New multi-determinant strategy for a group A streptococcal vaccine designed for the Australian Aboriginal population. Nat Med. 2000;6:455–9. doi: 10.1038/74719. [DOI] [PubMed] [Google Scholar]
- 8.Sabharwal H, et al. Group A Streptococcus (GAS) carbohydrate as an immunogen for protection against GAS infection. J Infect Dis. 2006;193:129–35. doi: 10.1086/498618. [DOI] [PubMed] [Google Scholar]
- 9.van Sorge NM, et al. The classical lancefield antigen of group A Streptococcus is a virulence determinant with implications for vaccine design. Cell Host Microbe. 2014;15:729–740. doi: 10.1016/j.chom.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henningham A, et al. Conserved anchorless surface proteins as group A streptococcal vaccine candidates. J Mol Med (Berl) 2012;90:1197–207. doi: 10.1007/s00109-012-0897-9. [DOI] [PubMed] [Google Scholar]
- 11.Valentin-Weigand P, Talay SR, Kaufhold A, Timmis KN, Chhatwal GS. The fibronectin binding domain of the Sfb protein adhesin of Streptococcus pyogenes occurs in many group A streptococci and does not cross-react with heart myosin. Microb Pathog. 1994;17:111–20. doi: 10.1006/mpat.1994.1057. [DOI] [PubMed] [Google Scholar]
- 12.Steer AC, Law I, Matatolu L, Beall BW, Carapetis JR. Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect Dis. 2009;9:611–6. doi: 10.1016/S1473-3099(09)70178-1. [DOI] [PubMed] [Google Scholar]
- 13.Beall B, Facklam R, Thompson T. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J Clin Microbiol. 1996;34:953–8. doi: 10.1128/jcm.34.4.953-958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanderson-Smith M, et al. A systematic and functional classification of Streptococcus pyogenes that serves as a new tool for molecular typing and vaccine development. J Infect Dis. 2014;210:1325–38. doi: 10.1093/infdis/jiu260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enright MC, Spratt BG, Kalia A, Cross JH, Bessen DE. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect Immun. 2001;69:2416–27. doi: 10.1128/IAI.69.4.2416-2427.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mostowy R, et al. Efficient inference of recent and ancestral recombination within bacterial populations. Mol Biol Evol. 2017;34:1167–1182. doi: 10.1093/molbev/msx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lees JA, et al. Fast and flexible bacterial genomic epidemiology with PopPUNK. Genome Research. 2019;29:1–14. doi: 10.1101/gr.241455.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chochua S, et al. Population and whole genome sequence based characterization of invasive group A streptococci recovered in the United States during 2015. MBio. 2017;8 doi: 10.1128/mBio.01422-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Croucher NJ, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marttinen P, et al. Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res. 2012;40:e6. doi: 10.1093/nar/gkr928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Page AJ, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beres SB, et al. Genome-wide molecular dissection of serotype M3 group A Streptococcus strains causing two epidemics of invasive infections. Proc Natl Acad Sci U S A. 2004;101:11833–8. doi: 10.1073/pnas.0404163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nasser W, et al. Evolutionary pathway to increased virulence and epidemic group A Streptococcus disease derived from 3,615 genome sequences. Proc Natl Acad Sci U S A. 2014;111:E1768–76. doi: 10.1073/pnas.1403138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turner CE, et al. Emergence of a new highly successful acapsular group A Streptococcus clade of genotype emm89 in the United Kingdom. MBio. 2015;6:e00622. doi: 10.1128/mBio.00622-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.You Y, et al. Scarlet fever epidemic in China caused by Streptococcus pyogenes serotype M12: Epidemiologic and molecular analysis. EBioMedicine. 2018 doi: 10.1016/j.ebiom.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lees JA, et al. Sequence element enrichment analysis to determine the genetic basis of bacterial phenotypes. Nat Commun. 2016;7 doi: 10.1038/ncomms12797. 12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lees JA, Galardini M, Bentley SD, Weiser JN, Corander J. pyseer: a comprehensive tool for microbial pangenome-wide association studies. Bioinformatics. 2018;34:4310–4312. doi: 10.1093/bioinformatics/bty539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McIver KS, Subbarao S, Kellner EM, Heath AS, Scott JR. Identification of isp, a locus encoding an immunogenic secreted protein conserved among group A streptococci. Infect Immun. 1996;64:2548–55. doi: 10.1128/iai.64.7.2548-2555.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henningham A, et al. Virulence role of the GlcNAc side chain of the Lancefield cell wall carbohydrate antigen in non-M1-serotype group A Streptococcus . MBio. 2018;9 doi: 10.1128/mBio.02294-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dale JB, Penfound TA, Chiang EY, Walton WJ. New 30-valent M protein-based vaccine evokes cross-opsonic antibodies against non-vaccine serotypes of group A streptococci. Vaccine. 2011;29:8175–8. doi: 10.1016/j.vaccine.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Batzloff MR, et al. Protection against group A Streptococcus by immunization with J8-diphtheria toxoid: contribution of J8- and diphtheria toxoid-specific antibodies to protection. J Infect Dis. 2003;187:1598–608. doi: 10.1086/374800. [DOI] [PubMed] [Google Scholar]
- 32.Guilherme L, et al. Towards a vaccine against rheumatic fever. Clin Dev Immunol. 2006;13:125–32. doi: 10.1080/17402520600877026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pandey M, et al. Combinatorial synthetic peptide vaccine strategy protects against hypervirulent CovR/S mutant streptococci. J Immunol. 2016;196:3364–74. doi: 10.4049/jimmunol.1501994. [DOI] [PubMed] [Google Scholar]
- 34.Feil SC, Ascher DB, Kuiper MJ, Tweten RK, Parker MW. Structural studies of Streptococcus pyogenes streptolysin O provide insights into the early steps of membrane penetration. J Mol Biol. 2014;426:785–92. doi: 10.1016/j.jmb.2013.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kagawa TF, et al. Model for substrate interactions in C5a peptidase from Streptococcus pyogenes: A 1.9 A crystal structure of the active form of ScpA. J Mol Biol. 2009;386:754–72. doi: 10.1016/j.jmb.2008.12.074. [DOI] [PubMed] [Google Scholar]
- 36.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–58. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yates CM, Filippis I, Kelley LA, Sternberg MJ. SuSPect: enhanced prediction of single amino acid variant (SAV) phenotype using network features. J Mol Biol. 2014;426:2692–701. doi: 10.1016/j.jmb.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Croucher NJ, et al. Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat Genet. 2013;45:656–63. doi: 10.1038/ng.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bart MJ, et al. Global population structure and evolution of Bordetella pertussis and their relationship with vaccination. MBio. 2014;5:e01074. doi: 10.1128/mBio.01074-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Courtney HS, et al. Trivalent M-related protein as a component of next generation group A streptococcal vaccines. Clin Exp Vaccine Res. 2017;6:45–49. doi: 10.7774/cevr.2017.6.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corander J, et al. Frequency-dependent selection in vaccine-associated pneumococcal population dynamics. Nat Ecol Evol. 2017;1:1950–1960. doi: 10.1038/s41559-017-0337-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McNally A, et al. Signatures of negative frequency dependent selection in colonisation factors and the evolution of a multi-drug resistant lineage of Escherichia coli . bioRxiv. 2018 400374. [Google Scholar]
- 43.Bao YJ, Shapiro BJ, Lee SW, Ploplis VA, Castellino FJ. Phenotypic differentiation of Streptococcus pyogenes populations is induced by recombination-driven gene-specific sweeps. Sci Rep. 2016;6 doi: 10.1038/srep36644. 36644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vos M, Didelot X. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 2009;3:199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- 45.Chewapreecha C, et al. Dense genomic sampling identifies highways of pneumococcal recombination. Nat Genet. 2014;46:305–309. doi: 10.1038/ng.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.David S, et al. Dynamics and impact of homologous recombination on the evolution of Legionella pneumophila . PLoS Genet. 2017;13:e1006855. doi: 10.1371/journal.pgen.1006855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bergmann R, Nerlich A, Chhatwal GS, Nitsche-Schmitz DP. Distribution of small native plasmids in Streptococcus pyogenes in India. Int J Med Microbiol. 2014;304:370–8. doi: 10.1016/j.ijmm.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 48.Woodbury RL, et al. Plasmid-Borne erm(T) from invasive, macrolide-resistant Streptococcus pyogenes strains. Antimicrob Agents Chemother. 2008;52:1140–3. doi: 10.1128/AAC.01352-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wescombe PA, Heng NC, Burton JP, Chilcott CN, Tagg JR. Streptococcal bacteriocins and the case for Streptococcus salivarius as model oral probiotics. Future Microbiol. 2009;4:819–35. doi: 10.2217/fmb.09.61. [DOI] [PubMed] [Google Scholar]
- 50.Bensi G, et al. Multi high-throughput approach for highly selective identification of vaccine candidates: the group A Streptococcus case. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.015693. M111 015693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ji Y, Carlson B, Kondagunta A, Cleary PP. Intranasal immunization with C5a peptidase prevents nasopharyngeal colonization of mice by the group A Streptococcus . Infect Immun. 1997;65:2080–7. doi: 10.1128/iai.65.6.2080-2087.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chaguza C, et al. Recombination in Streptococcus pneumoniae lineages increase with carriage duration and size of the polysaccharide capsule. MBio. 2016;7 doi: 10.1128/mBio.01053-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hanage WP, et al. Using multilocus sequence data to define the pneumococcus. J Bacteriol. 2005;187:6223–30. doi: 10.1128/JB.187.17.6223-6230.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Driebe EM, et al. Using whole genome analysis to examine recombination across diverse sequence types of Staphylococcus aureus . PLoS One. 2015;10:e0130955. doi: 10.1371/journal.pone.0130955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus . J Clin Microbiol. 2000;38:1008–15. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coscolla M, Gonzalez-Candelas F. Population structure and recombination in environmental isolates of Legionella pneumophila . Environ Microbiol. 2007;9:643–56. doi: 10.1111/j.1462-2920.2006.01184.x. [DOI] [PubMed] [Google Scholar]
- 57.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43:4178–82. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wyres KL, et al. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae . bioRxiv. 2018 doi: 10.1371/journal.pgen.1008114. 414235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seale AC, et al. Invasive group A Streptococcus infection among children, rural Kenya. Emerg Infect Dis. 2016;22:224–32. doi: 10.3201/eid2202.151358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Athey TB, et al. Deriving group A Streptococcus typing information from short-read whole-genome sequencing data. J Clin Microbiol. 2014;52:1871–6. doi: 10.1128/JCM.00029-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chalker V, et al. Genome analysis following a national increase in scarlet fever in England 2014. BMC Genomics. 2017;18:224. doi: 10.1186/s12864-017-3603-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kapatai G, Coelho J, Platt S, Chalker VJ. Whole genome sequencing of group A Streptococcus: development and evaluation of an automated pipeline for emmgene typing. PeerJ. 2017;5:e3226. doi: 10.7717/peerj.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ibrahim J, et al. Genome analysis of Streptococcus pyogenes associated with pharyngitis and skin infections. PLoS One. 2016;11:e0168177. doi: 10.1371/journal.pone.0168177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Page AJ, et al. Robust high-throughput prokaryote de novo assembly and improvement pipeline for Illumina data. Microb Genom. 2016;2:e000083. doi: 10.1099/mgen.0.000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Souvorov A, Agarwala R, Lipman DJ. SKESA: strategic k-mer extension for scrupulous assemblies. Genome Biol. 2018;19:153. doi: 10.1186/s13059-018-1540-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 67.Sumby P, et al. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J Infect Dis. 2005;192:771–82. doi: 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- 68.He M, et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile . Nat Genet. 2013;45:109–13. doi: 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arndt D, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–5. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Argimon S, et al. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom. 2016;2:e000093. doi: 10.1099/mgen.0.000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–2. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 75.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–91. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 76.Weyrich LS, et al. Neanderthal behaviour, diet, and disease inferred from ancient DNA in dental calculus. Nature. 2017;544:357–361. doi: 10.1038/nature21674. [DOI] [PubMed] [Google Scholar]
- 77.Davies MR, et al. Emergence of scarlet fever Streptococcus pyogenes emm12 clones in Hong Kong is associated with toxin acquisition and multidrug resistance. Nat Genet. 2015;47:84–7. doi: 10.1038/ng.3147. [DOI] [PubMed] [Google Scholar]
- 78.Koren S, et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Illumina sequence reads and draft genome assemblies were deposited into the European Nucleotide Archive under the accession numbers specified in Supplementary Table 2. Genbank accession numbers for the 30 new GAS reference genomes are provided in Supplementary Table 5. To facilitate community accessibility and interrogation of the data presented in this study, the phylogenetic (Fig 1a), PopPUNK phylogroup designations, and associated metadata components have been uploaded to the interactive web interface Microreact72 (https://microreact.org/project/5DEFpeck4). The PopPUNK database for assigning new genomes is available at https://doi.org/10.6084/m9.figshare.6931439.v1.