Abstract
Hydrogels serve as valuable tools for studying cell-extracellular matrix (ECM) interactions in three-dimensional (3D) environments that recapitulate aspects of native ECM. However, the impact of early protein deposition on cell behavior within hydrogels has largely been overlooked. Using a bio-orthogonal labeling technique, we visualized nascent proteins within a day of culture across a range of hydrogels. In two engineered hydrogels of interest in 3D mechanobiology studies – proteolytically degradable covalently crosslinked hyaluronic acid (HA) and dynamic viscoelastic HA hydrogels – mesenchymal stromal cell (MSC) spreading, YAP/TAZ nuclear translocation, and osteogenic differentiation were observed with culture. However, inhibition of cellular adhesion to nascent proteins or reduction in nascent protein remodeling reduced MSC spreading and nuclear translocation of YAP/TAZ, resulting in a shift towards adipogenic differentiation. Our findings emphasize the role of nascent proteins in the cellular perception of engineered materials and have implications for in vitro cell signaling studies and application to tissue repair.
The native extracellular microenvironment provides chemical and physical signals that regulate cell behavior and function1. Synthetic hydrogels have evolved as three-dimensional (3D) culture systems that mimic aspects of physiological cell microenvironments and can be used to explore how cells perceive and respond to these signals2,3, including towards 3D hydrogel design for engineering tissue4. While matrix stiffness is a well-established parameter in mediating cell behavior5, other hydrogel components, such as remodeling through proteolytic degradation6,7 or material stress-relaxation8 are also critical in controlling cell fate. These signals are particularly important for cells in 3D, where dynamic hydrogel reorganization enables cytoskeletal tension, proliferation and differentiation of cells9,10. In addition to these behaviors, cells synthesize and deposit proteins, including extracellular matrix (ECM) proteins11, within hydrogels. However, the influence of the early deposition of these nascent proteins in the pericellular space on cell-hydrogel interactions has largely been overlooked, despite potentially mediating the physical and chemical signals presented to cells.
Within a single tissue, the spatio-temporal presentation and interaction of cells with microenvironmental cues is critical for cell growth and tissue morphogenesis12. For example, at the earliest stages of connective tissue development, cells deposit and interact with a network of ECM in their microenvironment13. This evolving ECM provides critical adhesion cues, mediates cell-cell interactions, and regulates growth factor presentation. As development progresses, the ECM is continuously remodeled, degraded and reassembled by cells to actively shape their surrounding matrix. Thus, this bi-directional signaling is crucial for a range of cell and tissue functions14. Differentiating cells embedded in hydrogels also respond to both mechanical and chemical cues, which define their rate of ECM deposition and retention15–18. However, much of the initial cell-hydrogel interactions are likely lost during the course of cell differentiation as cells secrete and assemble a pericellular matrix that is essential for the progression of tissue maturation19. Indeed, this pericellular matrix was recently reported to influence cell fate within covalently crosslinked hydrogels that restrict cell spreading20; however, there are no reports regarding the mechanoregulatory role of nascent matrix adhesion and remodeling within complex hydrogel environments.
Adhesive interactions of cells and assembled ECM proteins, such as fibronectin and collagen, regulate cellular interactions on 2D substrates, including traction forces21–23; yet, little is known of how these proteins are organized to mediate interactions and mechanotransduction in 3D. Given the importance of ECM as a repository for signals24,25, we hypothesized that early deposition and remodeling of nascent ECM proteins control cell activity and function within 3D hydrogels, overcoming and/or reinforcing cues presented from the material itself. To investigate this, we used metabolic labeling to visualize nascent proteins that undifferentiated human mesenchymal stromal cells (hMSCs) secrete and assemble within various hydrogels, including engineered proteolytically degradable and dynamic viscoelastic hyaluronic acid (HA) hydrogels. These hydrogels are both permissive to cell spreading, through either protease-dependent or protease-independent mechanisms, allowing us to explore the role of adhesion to and remodeling of local nascent ECM on a range of MSC behaviors related to mechanosensing.
Nascent protein deposition occurs early in 3D hydrogels
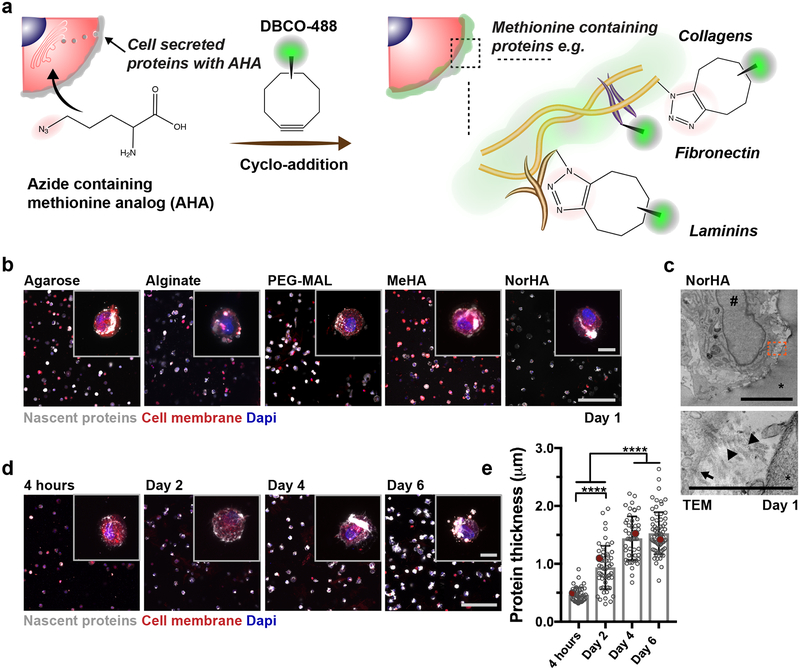
To visualize nascent protein deposition by hMSCs within 3D hydrogels, we adapted a labeling technique where methionine analogs containing azide groups (azidohomoalanine, AHA) are incorporated into proteins as they are synthesized26 and a bio-orthogonal strain-promoted cyclo-addition is then performed with a fluorophore conjugated cyclooctyne (DBCO-488) for visualization (Fig. 1a). The cyclo-addition is performed prior to cell fixation to reduce labeling of intracellular proteins (Supplementary Fig. 1) while maintaining high cell viability (97 ± 2% viability). Thus, this approach allows spatiotemporal visualization of methionine-containing proteins around individual cells15.
Figure 1. Nascent protein deposition by encapsulated hMSCs occurs early, independent of hydrogel type.
a Schematic of nascent extracellular protein labeling. The methionine analog azidohomoalanine (AHA) is added to the culture media and incorporated into nascent proteins (e.g., fibronectin, collagens, laminins). The bio-orthogonal Cu(I)-free strain-promoted cyclo-addition between the azide and DBCO-modified fluorophore (DBCO-488) enables visualization of the nascent proteins. b Representative images of nascent proteins (white) deposited by hMSCs encapsulated in various hydrogels (alginate, agarose, maleimide modified poly(ethylene glycol) (PEG-MAL), methacrylated hyaluronic acid (MeHA), norbornene modified hyaluronic acid (NorHA)), E = ~9 kPa, scale bar 200 μm, inset 20 μm). c Representative transmission electron microscopy (TEM, * hydrogel, # nucleus; scale bar 5 μm left, 1 μm right) image of encapsulated hMSC after 1 day (24 h in culture). Orange box in top image indicates magnification in bottom image (arrow indicates cell membrane and arrowheads show collagen fibrils). d Representative images of nascent proteins (white) deposited by hMSCs encapsulated in non-degradable NorHA hydrogels (9.0 ± 0.7 kPa, mean ± SD, n = 3 independent measurements) and cultured in growth media (supplemented with AHA) up to 6 days (see Supplementary Figure 3 for daily changes up to 14 days, scale bar 200 μm, inset 20 μm). e Quantification of the accumulated nascent protein thickness deposited by hMSCs encapsulated in non-degradable NorHA hydrogels (n = 40 cells (4 hours), 55 cells (Day 2, Day 4) and 70 cells (Day 6) from 3 biologically independent experiments, mean ± SD, **** p ≤ 0.0001, one-way ANOVA with Bonferroni post hoc, red dots indicate measurements for magnified representative images in d).
To illustrate early nascent protein deposition across a range of hydrogel environments, hMSCs were encapsulated within both physically (i.e., agarose, ionically-crosslinked alginate) and covalently (i.e., maleimide poly(ethylene glycol) (PEG-MAL) and methacrylated HA (MeHA) crosslinked via Michael-addition, norbornene modified HA (NorHA) crosslinked via thiol-ene reaction) crosslinked hydrogels, all with ~9 kPa elastic modulus (Fig. 1b). DBCO cyclo-addition labeling after 1 day culture in AHA-supplemented growth media revealed nascent protein deposition across all hydrogels (Fig. 1b). Transmission electron microscopy (TEM) images of hMSCS in NorHA hydrogels confirmed the presence of loosely organized fibrils with a morphology consistent with collagen at the cell-hydrogel interface after 1 day in culture (Fig. 1c) that were not present directly after encapsulation (Supplementary Fig. 2). These findings indicate that nascent protein deposition by hMSCs within the pericellular space occurs rapidly across a range of hydrogel environments.
To further assess the spatio-temporal distribution of nascent proteins using this bio-orthogonal labeling technique, hMSCs were encapsulated within RGD-modified (1 mM) NorHA hydrogels with elastic moduli of 9.0 ± 0.7 kPa and cultured (continuous presence of AHA during entire culture) up to 14 days, consistent with previous studies8,27 (Supplementary Fig. 3). An extensive mesh-like protein structure surrounded the cell body and increased with culture time (Fig. 1d). Additional labeling of the cell membrane confirmed that the proteins were extracellular and permitted quantification of the thickness of cell-produced proteins (Supplementary Fig. 4). The accumulated nascent protein thickness increased over 6 days (Fig. 1e) and was dependent on the initial hydrogel modulus (Supplementary Fig. 3), suggesting that cells accumulate proteins in the pericellular space within environments that restrict cell spreading6. These observations highlight that, even in the absence of differentiation factors, nascent protein deposition occurs early and likely contributes to how cells experience their physical and chemical environment.
Cells adhere to nascent proteins at the cell-hydrogel interface
Previous studies with covalently crosslinked hydrogels indicated that cell spreading in 3D is dependent on the local degradability of the network6,7,27. Thus, we next encapsulated hMSCs in proteolytically degradable HA hydrogels to determine whether nascent protein deposition modulates cell spreading (Fig. 2a). Extracellular nascent proteins were observed during 6 days in culture within degradable NorHA hydrogels (Fig. 2b) and nascent protein thickness and cell spreading aspect ratios increased with culture (Fig. 2c, Supplementary Fig. 5). TEM imaging after 6 days further confirmed that ECM fibrils were still present in close association with the cells and along the cell membrane (Fig. 2d, Supplementary Fig. 6). Encapsulation of hMSCs into degradable NorHA hydrogels of lower (~3 kPa) and higher (~20 kPa) elastic moduli resulted in similar cell spreading and nascent protein deposition; however, the protein thickness was lower for stiffer hydrogels (Supplementary Fig. 5), likely indicating that the distribution and assembly of nascent proteins is mediated through hydrogel crosslinking.
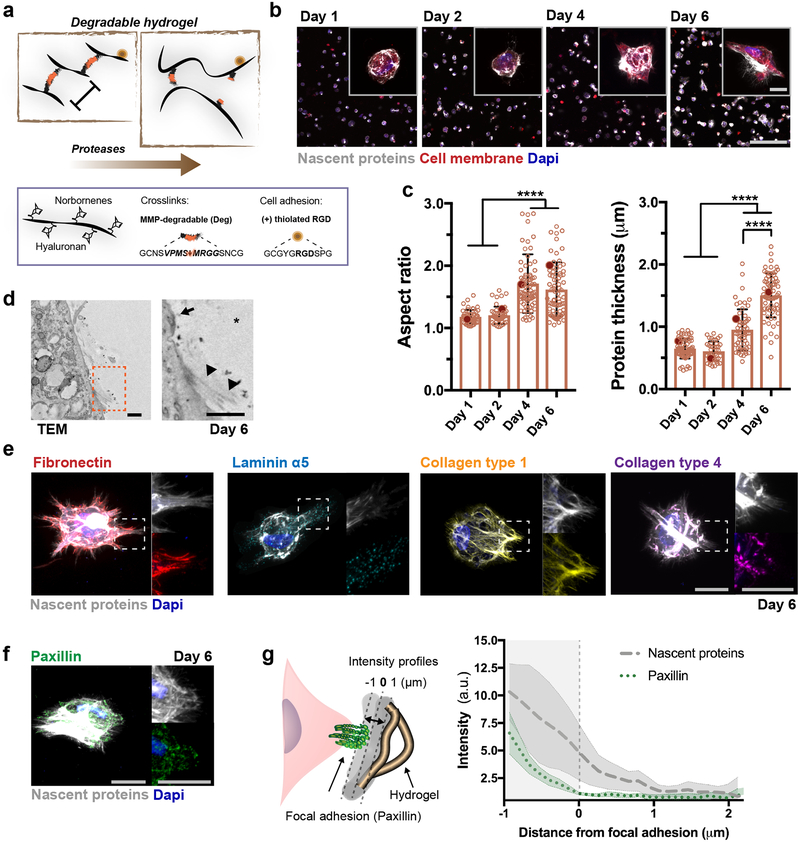
Figure 2. Nascent ECM proteins create an adhesive layer at the cell-hydrogel interface.
a Schematic illustrating norbornene-modified hyaluronic acid (NorHA) hydrogels crosslinked via a thiol-ene reaction with MMP-degradable dithiol peptide crosslinkers and incorporating RGD for adhesion. b Representative images of nascent proteins (white, visualized via fluorescent DBCO labeling) deposited by hMSCs encapsulated in degradable NorHA hydrogels (9.0 ± 0.7 kPa, mean ± SD, n = 3 independent measurements) and cultured in growth media for up to 6 days (see Supplementary Figure 5 for daily changes up to 14 days, scale bar 200 μm, inset 20 μm). c Quantification of the accumulated nascent protein thickness deposited by hMSCs encapsulated in degradable NorHA hydrogels (n = 40 cells (4 hours), 68 cells (day 1, day 2, day 4) and 72 cells (day 6) from 3 biologically independent experiments), mean ± SD, **** p ≤ 0.0001, one-way ANOVA with Bonferroni post hoc, red dots indicate measurements for magnified representative images in b). d Representative transmission electron microscopy (TEM, * hydrogel) images of encapsulated hMSC after 6 days (scale bar 0.5 μm). Orange box in left image indicates magnification in right image (arrow indicates cell membrane and arrowheads show randomly aligned collagen fibrils). e Representative images (magnifications on right) of nascent proteins and fibronectin, laminin α5 and collagen type 1 and type 4 at 6 days (scale bars 20 μm). f Representative image (magnifications of single channels on right) of accumulated nascent proteins and focal adhesions stained for paxillin (scale bars 20 μm). g Schematic (left) illustrating the region used to generate intensity profiles (right) emanating from single FAs (representative image in f), n = 50 adhesions from 10 individual cells (2 biologically independent experiments), lines show median intensity profile, shaded areas demonstrate 95% confidence interval).
We then asked how the spatial pattern of AHA labeled proteins colocalized with specific ECM proteins such as cellular fibronectin, laminin α5, and collagen types 1 and 4. High degrees of structural similarity were observed (Fig. 2e), suggesting that AHA incorporates into nascent proteins of the pericellular matrix. No staining of ECM proteins was observed directly after encapsulation (Supplementary Fig. 7) and we confirmed that serum proteins did not alter cell behavior and adhesion to the hydrogels, as hMSCs cultured in defined serum-free media exhibited similar ECM protein deposition and spreading relative to hMSCs in serum-containing growth media (Supplementary Fig. 8) and there was minimal adhesion of hMSCs on 2D hydrogels (without RGD presentation) cultured in serum-containing media (Supplementary Fig. 9). To investigate whether these changes in the pericellular matrix alter how hMSCs interact and sense the hydrogel environment, we next examined adhesive contacts between cells and extracellular ligands28. Paxillin was used to visualize focal adhesions (FAs) in hMSCs (Fig. 2f). We observed paxillin complexes directly coupled to the actin cytoskeleton (Supplementary Fig. 10), indicating formation of FAs within degradable NorHA hydrogels. Intensity profile quantification across single FAs (paxillin) relative to the location of nascent protein revealed that the localization of single FAs is consistent with adhesion to the deposited ECM (Fig. 2g), with an average protein thickness of 1.2 ± 0.8 μm, consistent with the accumulated protein thickness at day 6 (Fig. 2c). This indicates that hMSCs directly interact with their nascent ECM during adhesion and spreading in degradable hydrogels, and do not rely solely on epitopes presented by the hydrogel.
Nascent protein adhesion influences cell mechanosensing
Having visualized the local accumulation of and cellular adhesion to nascent proteins, we sought to understand whether this adhesion alters cellular behavior within degradable hydrogels. The binding of integrins to specific matrix ligands may actively induce conformational changes in ECM proteins, triggering downstream cell signaling cascades29, such as with the binding of the integrin α2 domain with collagen (GFOGER)30,31 and fibronectin (RGD)22,32. To investigate how this binding regulates nascent protein assembly and cell behavior, we added function-perturbing monoclonal antibodies that selectively block interactions with either secreted collagen (anti Integrin alpha2 (anti-α2)) or human fibronectin (HFN7.1). Soluble RGD peptides (sol RGD) were also used as a competitive inhibitor to integrin-mediated binding33. All inhibitors were administered daily during the 6-day culture period. After 1 day, there were minimal changes in protein thickness with HFN7.1 (5 ug/mL) and sol RGD (0.5 mM) administration and a small increase with anti-α2 (20 ug/mL) treatment (Supplementary Fig. 11a,b). When treatment with the antibodies/peptides was continued for 6 days, hMSC spreading was reduced across all groups when compared to untreated and IgG isotype controls, with minimal changes in nascent protein thickness observed (Fig. 3a, Supplementary Fig. 11c). hMSC spreading depended on the dose of antibodies/peptides at day 6, and concentrations were selected that had minimal impact on cell viability (Supplementary Fig. 11d,e). Although significantly reduced, blocking of collagen or fibronectin interactions did not completely abrogate cell spreading during the 6-day culture period, suggesting that multiple protein binding interactions are involved for cell spreading in 3D hydrogels.
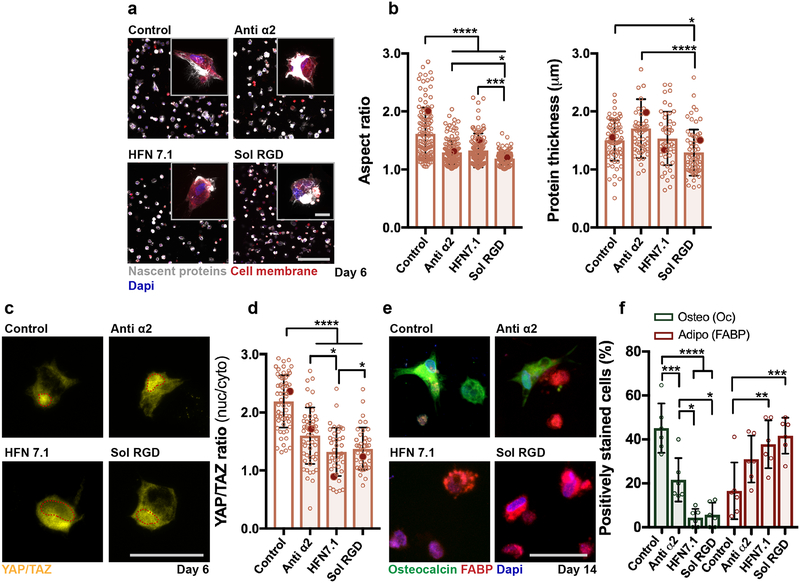
Figure 3. Adhesion to nascent proteins controls hMSC mechanosensing in degradable hydrogels.
a Representative images (scale bar 200 μm, inset 20 μm) of nascent protein deposition in MMP-degradable NorHA hydrogels after 6 days treatment without (control, note same image as in Fig. 2b), with monoclonal antibodies against integrin alpha 2 (anti α2, 20 μg/mL) or human fibronectin (HFN7.1, 5 μg/mL) or with soluble RGD (sol RGD, 0.5 mM). b Quantification of cell aspect ratio and accumulated nascent protein thickness deposited by hMSCs encapsulated in degradable NorHA hydrogels (n = 133 cells (control), n = 155 cells (anti α2), and n = 152 cells (HFN7.1), n = 124 cells (sol RGD) from 2 biologically independent experiments), mean ± SD, **** p ≤ 0.0001, *** p ≤ 0.001, * p ≤ 0.05, one-way ANOVA with Bonferroni post hoc, red dots indicate measurements for magnified images in a). c Representative images and d quantification of nuclear/cytoplasmic (nuc/cyto) YAP/TAZ ratios of hMSCs encapsulated in degradable NorHA hydrogels, cultured for 6 days in adipogenic-osteogenic media (scale bar 50 μm), quantifications: n = 60 cells (control), n = 51 cells (anti α2), and n = 40 cells (HFN7.1), n = 39 cells (sol RGD) from 2 biologically independent experiments), mean ± SD, **** p ≤ 0.0001, * p ≤ 0.05, one-way ANOVA with Bonferroni post hoc, red dots indicate measurements for magnified images in c). e Immunostaining for fatty-acid binding protein (FABP, adipogenic marker) and osteocalcin (Oc, osteogenic marker) after 14 days in adipogenic-osteogenic media (scale bar 50 μm). f Quantification of positively stained cells (percentage,%) towards osteogenesis (Oc positive) and adipogenesis (FABP positive) after 14 days, n = 6 samples from 2 biologically independent experiments), mean ± SD, **** p ≤ 0.0001, *** p ≤ 0.001, ** p ≤ 0.01, * p ≤ 0.05, two-way ANOVA with Bonferroni post hoc).
To assess whether binding to these adhesive domains of collagen and fibronectin alters downstream behaviors of hMSCs, mechanosensitive signaling pathways were analyzed. Yes-associated protein/Transcriptional co-activator (YAP/TAZ) is critical in cellular sensing and transduction of mechanical signals, with enhanced nuclear translocation in response to increased traction/tension27,34,35. After 6 days (culture in bipotential adipogenic-osteogenic media6,8), YAP/TAZ was primarily nuclear for hMSCs. Conversely, YAP/TAZ was more cytoplasmic (reduced nuclear to cytoplasmic intensity (nuc/cyto) ratio) when adhesion to nascent proteins was blocked, particularly with HFN7.1 treatment (Fig. 3c,d). As such, the cell-adhesive domain of secreted fibronectin seems to mediate YAP/TAZ mechanosensitive signaling, resulting from cell-nascent protein sensing.
Given that cell contractility and YAP/TAZ nuclear localization are functionally important in directing MSC fate6,34, changes in downstream osteogenic and adipogenic differentiation were also investigated (Fig. 3e). Trends in nascent protein deposition and spreading were similar for hMSCs cultured in bipotential adipogenic-osteogenic differentiation media to those observed for hMSCs that were cultured in growth media (Supplementary Fig. 12). At 14 days, hMSCs showed primarily osteogenic differentiation as indicated with most cells being positive for osteocalcin (Oc), whereas adipogenesis (positive for fatty-acid binding protein (FABP)) was favored in groups treated with sol RGD, anti-α2 or HFN7.1 (Fig. 3f). Interestingly, anti-α2 treatment reduced osteogenesis to a lesser extent than HFN7.1. Not only does HFN7.1 abrogate cell adhesion to fibronectin, but perturbation of α5β1 and αvβ3 integrin recognition also alters fibronectin conformation and fibril formation36,37, further enhancing its effects. This is also consistent with the observation that sol RGD peptides compete against the cell-adhesive domain of fibronectin for interactions with integrins, and thus similarly reduces hMSC osteogenic differentiation (Fig. 3e,f). These results highlight that, in hydrogels permissive to cell spreading, nascent protein adhesion guides cell behavior and fate by complementing signals supplied by the material itself.
Viscoelastic hydrogels modulate cell spreading in 3D
Recent studies have shown that viscoelastic hydrogels with dynamic crosslinks can significantly impact cell behavior through mechanisms that include local crosslink remodeling and ligand clustering8,38,39, in the absence of proteases. To assess whether nascent proteins mediate these behaviors, we designed a dynamic double network (DN) HA hydrogel system based on both covalent and supramolecular guest-host (GH) crosslinking40. Here, the first network is formed through covalent crosslinking of methacrylated HA (MeHA) and dithiols (DTT) via Michael addition (Fig. 4a). The second network consists of a HA hydrogel that forms through non-covalent guest-host interactions of β-cyclodextrin (CD, host) with adamantane (Ad, guest)41,42. Dynamic hydrogels were formed upon mixing and simultaneous crosslinking with interpenetration of the two networks. Fluorescent labeling and confocal imaging of the MeHA and GH networks demonstrated uniformity and microstructural homogeneity of the networks (Supplementary Fig. 13). Because the properties of each network can be controlled independently, we tailored the viscous and elastic properties of the dynamic hydrogels through alterations of the concentration of either network. At a given MeHA crosslink ratio of 0.3 (ratio of thiols to methacrylates), higher concentrations of the GH network (up to 3 wt%) increased the oscillatory viscous moduli (G”), but did not significantly alter the oscillatory elastic moduli (G’) (Fig. 4b). Conversely, G’ of dynamic hydrogels was altered by varying the covalent crosslink ratio of the MeHA network (0.0–0.4), with only small changes in G” (Fig. 4c). Modulation of viscoelasticity was confirmed by the frequency-response of DN hydrogels with an increase in both G’ and G” at higher frequencies, whereas the MeHA network without the GH network behaved as an elastic hydrogel, with little frequency-dependent response (Supplementary Fig. 14). This is consistent with previous studies that varied hydrogel physical and covalent crosslinking to tune viscoelasticity43.
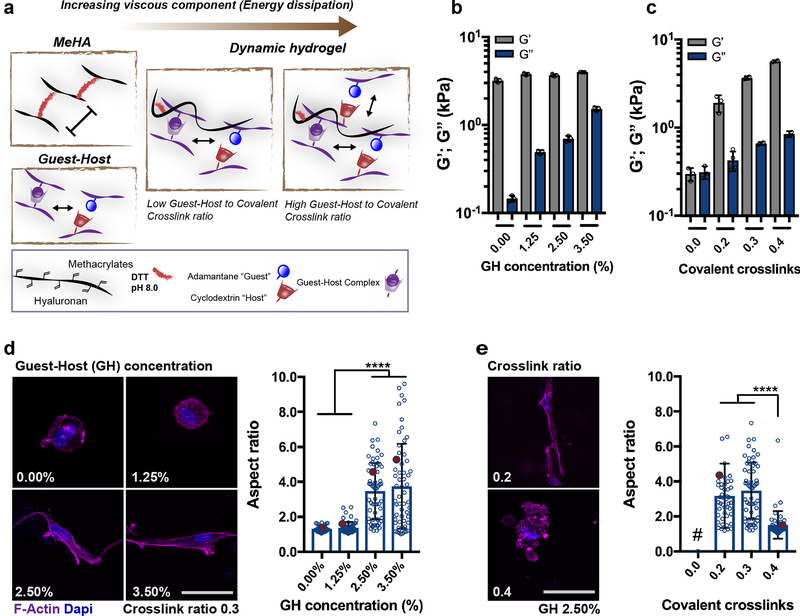
Figure 4. Dynamic hydrogel composition modulates viscoelastic properties and cell spreading.
a Schematic of guest-host double-network (DN) hydrogel formation, from the combination of (i) covalently crosslinked HA hydrogel (MeHA) without RGD peptide and crosslinked with 1,4-dithiothreitol (DTT) and (ii) guest-host (GH) hydrogel assembled through mixing of HA modified with cyclodextrin (host) or adamantane (guest). Rheological measurements (1 Hz, 0.5% strain) of storage modulus (G’, elastic component) and loss modulus (G”, viscous component) for DN hydrogels with b increasing GH polymer concentration at a given covalent crosslink ratio (0.3) and c increasing covalent crosslinking (ratio of thiols to methacrylates) at a given GH polymer concentration (2.50%, n = 3 independent measurements per group, mean ± SD). d Representative images of F-Actin immunostaining and quantification of aspect ratio of hMSCs encapsulated in DN hydrogels (G’ 3.5 ± 0.45 kPa, n = 3 independent measurements per group, mean ± SD) with different GH concentration but same covalent crosslinking ratio (0.3) (scale bar 20 μm), quantifications: n = 34 cells (0.00%), n = 58 cells (1.25%, 2.50%) and n = 56 cells (3.50%) from 2 biologically independent experiments), mean ± SD, **** p ≤ 0.0001, one-way ANOVA with Bonferroni post hoc, red dots indicate measurements for images on the left. e Representative images of F-Actin immunostaining and quantification of aspect ratio of hMSCs encapsulated in DN hydrogels with different covalent crosslinking ratios but same GH polymer concentration (2.50%). Note that image of 0.3 corresponds to 2.50% in d) (scale bar 20 μm), quantifications: n = 46 cells (0.2) and n = 49 (0.4) from 2 biologically independent experiments), mean ± SD, **** p ≤ 0.0001, one-way ANOVA with Bonferroni post hoc, red dots indicate measurements for images on the left, # GH only hydrogels (i.e. no covalent crosslinks) were not stable over 6 days in culture).
Using this material, we showed that encapsulated hMSC behavior was altered by the hydrogel composition and viscoelasticity, as measured through morphological changes at 6 days. When encapsulated within hydrogels of the same G’ (3.6 ± 0.4 kPa, without RGD), but altered viscosity, cell spreading was suppressed within MeHA only (0.00% GH) and DN hydrogels with low GH concentration (1.25% GH) (Fig. 4d); however, cell spreading greatly increased at higher GH concentrations, reaching aspect ratios greater than those observed in the covalent degradable hydrogels. Furthermore, when comparing dynamic hydrogels with increasing G’ (ranging from 1.9 ± 0.2 to 5.7 ± 0.3 kPa), cell spreading decreased as a function of covalent crosslink ratio (Fig. 4e). Note that single GH hydrogels (no covalent crosslinks) were observed to disassemble during the 6-day culture period, which prevented any analysis of cell spreading42. Generally, these findings suggest that increasing the viscous component of dynamic hydrogels influences the ability of hMSCs to spread by physically remodeling their pericellular environment through protease-independent mechanisms, consistent with previous reports8,44.
Nascent protein remodeling alters cell behavior
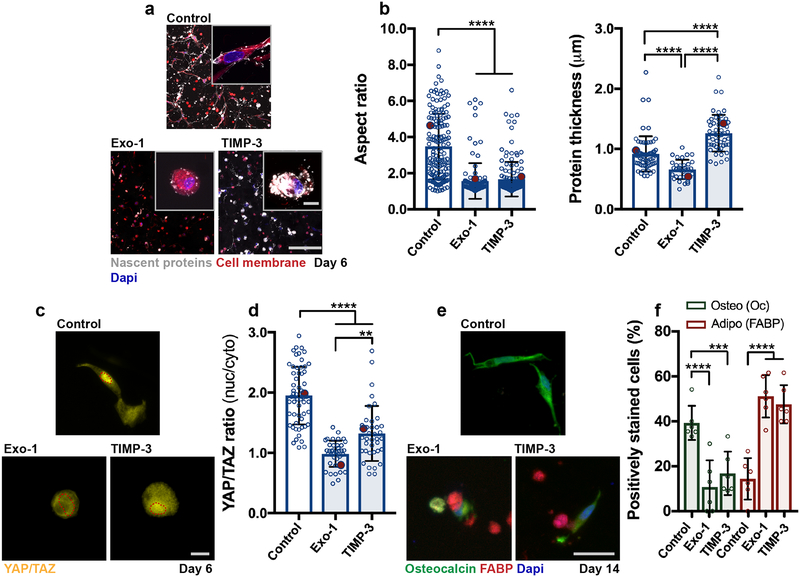
Given that hMSCs were able to spread in dynamic hydrogels without local hydrogel degradation, we used one formulation (GH 2.50%, crosslink ratio 0.3) to investigate how nascent protein deposition and remodeling regulates cell behavior within these viscoelastic environments. At 3 days, encapsulated hMSCs spread in the DN hydrogels with a nascent ECM layer that was evident by 4 hours at the cell-hydrogel interface; RGD did not alter cell spreading (Supplementary Fig. 15). This indicates that the mechanism driving spreading in dynamic hydrogels is not greatly influenced by tethered adhesive ligands; thus, subsequent studies were performed without RGD modification. Cell spreading was then investigated in response to alterations in the nascent proteins. Specifically, 2-(4-Fluorobenzoylamino)-benzoic acid methyl ester (Exo-1) was added to perturb the transport and secretion of extracellular proteins45–47 and TIMP-3 (endogenous tissue inhibitor of metalloproteinase 3) was encapsulated48 with hMSCs to locally limit nascent protein remodeling over the 6 day culture period49 (Fig. 5a). Blocking exocytosis with Exo-1 (120 nM) reduced the nascent protein thickness and cell spreading compared to controls (Fig. 5b), whereas blocking protein remodeling with TIMP-3 (5 nM encapsulated and added daily to media) increased the average nascent protein thickness and reduced hMSC spreading (Fig. 5b). Inhibition of exocytosis may be associated with limitations, including its influence on the secretion of extracellular vesicles and growth factors; though these treatments did not significantly alter 2D YAP/TAZ nuclear localization and contractility relative to controls (Supplementary Fig. 16).
Figure 5. Nascent protein remodeling is required for cell spreading and osteogenesis in dynamic hydrogels.
a Representative images of nascent protein deposition of hMSCs encapsulated in DN hydrogels (G’ 3.6 ± 0.1 kPa, G” 0.7 ± 0.05 kPa, n = 3 independent measurements, mean ± SD) treated with an inhibitor of exocytosis and vesicular trafficking Exo-1 (120 nM) and a recombinant tissue inhibitor of metalloproteinases-3 (TIMP-3, 5 nM encapsulated) during 6 days in growth media (scale bar 200 μm, insets 20 μm). b Quantification of cell aspect ratio and accumulated nascent protein thickness deposited by hMSCs encapsulated in dynamic hydrogels (aspect ratio: n = 161 cells (control), n = 150 cells (TIMP-3), and n = 122 cells (Exo-1), protein thickness: n = 76 cells (control), n = 61 cells (TIMP-3), and n = 55 cells (Exo-1), from 3 biologically independent experiments), mean ± SD, **** p ≤ 0.0001, one-way ANOVA with Bonferroni post hoc, red dots indicate measurements for magnified images in a). c Representative images and d quantification of nuclear/cytoplasmic (nuc/cyto) YAP/TAZ ratios of hMSCs encapsulated in dynamic hydrogels and cultured for 6 days in adipogenic-osteogenic media (scale bar 50 μm, n = 59 cells (Control) and n = 39 cells (TIMP-3, Exo-1), from 2 biologically independent experiments), mean ± SD, **** p ≤ 0.0001, ** p ≤ 0.01, one-way ANOVA with Bonferroni post hoc, red dots indicate measurements for images in c). e Immunostaining for fatty-acid binding protein (FABP, adipogenic marker) and osteocalcin (Oc, osteogenic marker) after 14 days in adipogenic-osteogenic media (scale bar 50 μm). f Quantification of positively stained cells (percentage,%) towards osteogenesis (Oc positive) and adipogenesis (FABP positive) after 14 days, n = 6 samples from 2 biologically independent experiments), mean ± SD, **** p ≤ 0.0001, *** p ≤ 0.001, two-way ANOVA with Bonferroni post hoc).
Noting that cell spreading and matrix remodeling are important for the functional behavior of these cells, we next evaluated if nascent protein deposition and remodeling alters hMSC fate. Upon changing the media to bipotential adipogenic-osteogenic media for 6 days, the same trends in nascent protein deposition and spreading as observed in growth media were maintained (Supplementary Fig. 17). After 6 days of culture, YAP/TAZ nuc/cyto ratios were significantly lower with Exo-1 or TIMP-3 treatment compared to the control group, indicating that these treatments resulted in YAP/TAZ retention in the cytoplasm (Fig. 5c,d). Following 14 days of culture in bipotential media, adipogenesis was significantly increased in both Exo-1 and TIMP-3 treated groups relative to the increased osteogenesis observed in the control group (Fig. 5e,f). This is consistent with the results in degradable hydrogels and previous findings of enhanced spreading and osteogenic commitment as functional outcomes of increased YAP/TAZ nuclear localization8,27, and highlights the essential role of nascent protein adhesion and remodeling in enabling MSC behavior and fate. Similar observations occurred in the absence of differentiation factors, albeit to a lesser extent (Supplementary Fig. 18) and similar trends in cell behavior and nascent protein accumulation were observed across multiple donors (Supplementary Fig. 19). These findings suggest that cell behavior when encapsulated within engineered materials is influenced by nascent protein deposition and subsequent pericellular remodeling at the cell-hydrogel interface.
Outlook
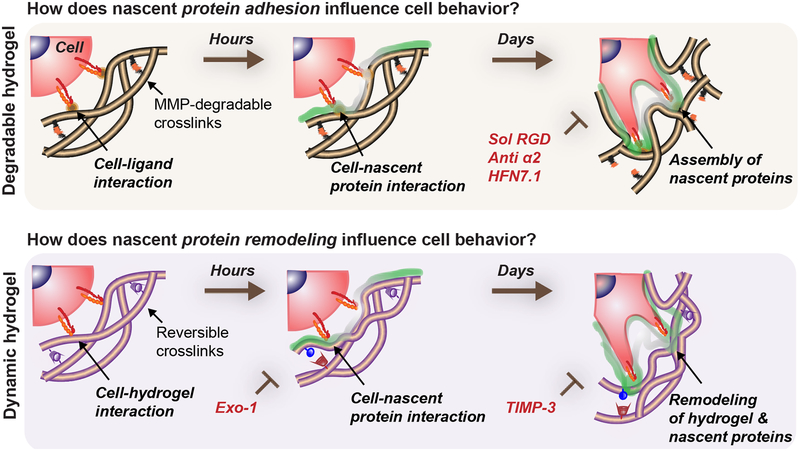
Numerous synthetic hydrogels have been used to investigate how biophysical cues regulate cell behavior in 3D. Often, direct interactions of cells with the engineered microenvironment are implicated in explaining observed phenomena. However, our results indicate that cellular outcomes are not only influenced by the initial engineered interface presented to the cell, but also by adhesion to and remodeling of nascent proteins deposited locally by cells very soon after cultures are initiated. This is often overlooked in the assessment of results, potentially due to the difficulty in analyzing this interface. By adapting a nascent extracellular protein labeling approach, we elucidated insight into the dynamic nature of the cell-hydrogel interface, which plays synergistic effects in directing cell behavior. Specifically, we found that secreted proteins increasingly mask the presentation of signals from the engineered hydrogel, that this process has meaningful consequences as early as day 1, and that the contributions of these nascent proteins persist through differentiation events occurring over several weeks (Fig. 6). Cellular adhesion naturally emerged from focal adhesions interacting with nascent proteins and local matrix remodeling within hydrogels that underwent either proteolytic degradation or dynamic polymer reorganization to permit cell spreading. When specific nascent protein adhesion or remodeling was blocked, the ability of the cell to spread and respond to the material environment was diminished, and this had downstream consequences on YAP/TAZ signaling and differentiation. This work demonstrates that, in many hydrogel systems, nascent proteins shape and define the pericellular microenvironment, supplementing cues from engineered hydrogels.
Figure 6. Nascent protein adhesion and remodeling enhance cell spreading in degradable/dynamic hydrogels.
Cells interact with a 3D hydrogel and presented ligands for a short time period before depositing nascent proteins to form a pericellular matrix. The hydrogel properties determine if encapsulated cells can spread, but adhesion and active remodeling of the nascent proteins are required for spreading. For example, in hydrogels that cells can locally degrade (e.g., protease-sensitive crosslinkers), nascent proteins guide cell behavior as an assembled interfacial layer that cells adhere to. Perturbation of cell-ECM adhesion through inhibiting specific cell-nascent protein interactions (e.g., sol RGD, anti α2, HFN7.1) inhibits spreading and decreases its downstream cellular outcomes (YAP/TAZ nuclear translocation, osteogenic differentiation). Similarly, in dynamic microenvironments (e.g., viscoelastic hydrogels) where spreading is protease-independent, nascent protein deposition and remodeling are needed for mechanosensing (YAP/TAZ nuclear translocation, osteogenic differentiation) and are blocked by inhibiting nascent protein secretion (Exo-1) and remodeling (TIMP-3).
Previous work has studied ECM protein secretion in the context of MSC and progenitor cell differentiation in which hydrogel cues influenced ECM assembly and tissue maturation15,38,50,51. However, the mechanism described herein could be of more general importance in 3D matrices, where nascent proteins could accumulate until the engineered cues are masked, delimiting their direct influence on cell behavior16,52 or formation of organoids53,54. This may also be relevant in the interpretation of cell-matrix events, such as adhesive ligand clustering, local matrix stiffening and cell tractions. The conjugation of specific ECM-binding molecules may further be used to design nascent ECM microenvironments55 and to study the interplay of nascent ECM protein tethering and mechanosensing in 3D hydrogels. Our data show that nascent ECM adhesion and assembly influence how cells sense and interpret microenvironmental cues, but the functional assembly of individual proteins, and how this might be regulated by initial material-presented cues, remains to be elucidated in this complex feedback mechanism.
Methods
Cell isolation, culture, and antibody and small molecule inhibition.
Human MSCs were isolated from fresh unprocessed bone marrow from human donors (Lonza, male 23 years, female 18 years, male 22 years) as previously described56. Briefly, diluted bone marrow (1:4 with PBS) was separated with Ficoll density gradient centrifugation (800 rcf, 20 min). Mononuclear cells were collected from the liquid interface, plated on tissue culture plastic (TCP), cultured in alpha-modified essential medium (α-MEM, 10% FBS, 1% penicillin/streptomycin, 5 ng/mL basic fibroblast growth factor) at 37°C/5% CO2 until 80% confluency of the colonies and stored in liquid nitrogen (95% FBS, 5% dimethyl sulfoxide (DMSO)). All hMSCs were expanded in standard growth media (α-MEM, 10% FBS, 1% penicillin/streptomycin (P/S)) for one passage prior encapsulation, trypsinized for 5 min at 37°C/5% CO2 using TrypLE Express (Invitrogen), washed in α-MEM (serum-free) and resuspended in PBS unless otherwise stated. All hydrogel constructs were cultured in glutamine, methionine and cystine-free high glucose DMEM (Life Technologies) supplemented with 0.201 mM cystine, 100 μg/mL sodium pyruvate, 50 μg/mL ascorbate 2-phosphate, 1% P/S, 10% FBS (Gibco) and 0.1 mM azidohomoalanine (AHA) (“AHA growth media”), and medium was replenished every second day. For differentiation studies, adipogenic/osteogenic supplement (R&D System) was added in a 1:1 ratio to “AHA growth media”, a commonly used culture environment where MSC fate is biased by the engineered hydrogel cues rather than being directed by the media components alone6,8. Media was changed every other day.
Perturbation of nascent protein adhesion was achieved using monoclonal antibodies against collagen (20 μg/mL anti Integrin α2, EMD Millipore MAB1950Z) and human fibronectin (5 μg/mL HFN7.1, Developmental Studies Hybridoma Bank) or soluble RGD peptides (sol RGD, 0.5 mM), and added daily. Notably, the HFN7.1 antibody recognizes human fibronectin and is not reactive with FBS-derived (bovine) fibronectin57. Isotype controls (IgG1 (5 μg/mL and 20 μg/mL R&D System MAB002) were added daily to culture media. Blocking of protein transport and exocytosis was achieved using 120 nM 2-(4-Fluorobenzoylamino)-benzoic acid methyl ester (Exo-1, Sigma Aldrich), and replenished daily. Local inhibition of matrix metalloproteinases was achieved with 5 nM tissue inhibitor of metalloproteinase 3 (recombinant TIMP-3, R&D Systems) encapsulated into dynamic hydrogels, and then added daily to culture media.
Hydrogel preparation.
‘Degradable’: Norbornene-hyaluronic acid (NorHA):
NorHA was synthesized as previously described58. Briefly, tetrabutylammonium salt of sodium hyaluronate (Lifecore, 64 kDa) (HA-TBA) was prepared using Dowex 50W proton exchange resin. Modification of HA-TBA with norbornene groups was performed via esterification with 5-norbornene-2-carboxylic acid (3 equivalent (equiv.)), 4-(dimethylamino)pyridine (DMAP, 1.5 equiv.), and ditert-butyl dicarbonate (Boc2O, 0.4 equiv.) for 20 h at 45 °C (under nitrogen) and dialyzed at room temperature (RT) for 7 days. The degree of modification was 23% by 1H NMR (Supplementary Fig. 20). Non-degradable and MMP-degradable peptides were synthesized with standard solid state methods59. Peptides were cleaved in trifluoroacetic acid, precipitated in ether and purity was confirmed using matrix-assisted laser desorption/ionization (MALDI) (Supplementary Fig. 21)41. Thiolated RGD peptide (GCGYGRGDSPG) was purchased from Genscript. NorHA hydrogels were fabricated by thiol-ene addition crosslinking with ultraviolet (UV) light and the photoinitiator 2-Hydroxy-4′-(2-hydroxyethoxy)-2-methylpropiophenone (Irgacure 2959, Sigma Aldrich). Hydrogel precursor solutions (4 wt% polymer, 0.3 ratio of thiols to norbornenes) were mixed with 3×106 mL−1 hMSCs with or without 1 mM thiolated RGD and UV-polymerized (320–390 nm, Omnicure S1500 UV Spot Cure System, Exfo) between two coverslips for 3 min at 2 mW/cm−2. 5 mm x 5 mm gels were then cut from hydrogel films (ca. 300 μm) and cultured in 48 well plates.
PEG-DA, agarose and alginate:
PEG-DA was synthesized from linear PEG (10 kDa), acrylated through reaction of PEG-OH (Fluka) with acryloyl chloride and trimethylamine in dichloromethane (DCM)60. 1H NMR revealed high degree of end group modifications (Supplementary Fig. 22). For fabrication of PEG-DA hydrogels, 3.5 wt% PEG-DA in PBS containing 0.05 wt% 2-Hydroxy-4’-(2-hydroxyethoxy)-2-methylpropiophenone (I2959) was mixed with 3×106 mL−1 hMSCs and hydrogels were exposed to 5 mW/cm−2 UV light for 5 min. For hMSC encapsulation in 1.5 wt% agarose hydrogels, molten 2-Hydroxyethyl agarose (3 wt%, Sigma Aldrich) was combined with warm cell suspension in a 1:1 ratio and solidified between two coverslips. Alginate constructs were prepared as previously described61. 2.5 wt% alginate (MVG FMC BioPolymer) in 280 μL serum-free “AHA growth media” was rapidly mixed with 70 μL serum-free “AHA growth media” containing 57 mM calcium sulfate and 1.05×106 hMSCs. The solution was pipetted between two coverslips and allowed to gel for 45 min at RT. PEG-DA, agarose and alginate hydrogels were fragmented into 5 mm x 5 mm gels and cultured in 48 well plates.
‘Dynamic’: MeHA and guest-host double-network hydrogels (DN):
HA was modified with methacrylates (MeHA), adamantane (Ad-HA) and β-cyclodextrin (CD-HA) as previously described41. Briefly, MeHA with 86% modification of disaccharides was synthesized by reacting HA with 25 equiv. methacrylic anhydride (MA) at pH 9–10 for 4 hours and repeated without purification (Supplementary Fig. 23). For the preparation of Ad-HA and CD-HA, HA-TBA was dissolved in DMSO and modified by esterification with 1-adamantane acetic acid (3.0 equiv.) via Di-tert-butyl dicarbonate (Boc2O, 0.67 equiv.)/4-(Dimethylamino)pyridine (DMAP, 0.5 equiv.) catalysis or separate amidation with aminated CD (0.8 equiv.)41. Modifications were determined to be 29% for Ad (Supplementary Fig. 24) and 24% for CD (Supplementary Fig. 25). For MeHA hydrogel formation, MeHA was dissolved in serum-free Media 199 (Invitrogen, pH 8.5, supplemented with 2.5 mM tris(2-carboxyethyl)phosphine (TCEP), 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)40. Dithiothreitol (DTT) was added at 20x concentration with a ratio of thiol/methacrylates (crosslink ratio) as specified and an overall MeHA concentration of 3 wt%. The hydrogel solution was pipetted between two coverslips and crosslinked for 30 min at 37 °C/5% CO2. For guest-host double network hydrogels (DN), CD-HA and MeHA were dissolved together in Media199 buffer, and DTT thoroughly mixed with Ad-HA and hMSCs immediately before hydrogel formation such that their combination resulted in the desired concentrations.
Mechanical testing.
Shear rheology:
Hydrogels were formed between the geometry (1° cone angle, 20 mm diameter) and Peltier plate (37°) of a stress-controlled rheometer (TA Instruments, AR2000). Measurements were performed by oscillatory time sweeps (1.0 Hz, 0.5% strain) and frequency sweeps (0.01–100 Hz, 0.5% strain).
Compressive Dynamic Mechanical Analysis:
Hydrogels were cast into 5 mm diameter cylinders and crosslinked prior to compression testing as specified. Samples were preloaded (0.01 N) and compression performed (0.5 N min−1) for determining the elastic moduli (slope from 10–20% strain).
Nascent protein labeling and immunostaining.
For membrane and DBCO-488 (Click Chemistry Tools) labeling, all washing and staining was performed in PBS with 1% bovine serum albumin (BSA, Sigma Aldrich). For normal nascent ECM protein labeling, hydrogels were washed twice, followed by 30 min staining with plasma membrane stain (1:1000 dilution, Molecular Probes) and 40 min incubation in 30 μM DBCO-488 at 37°C/5% CO2. After three washes with PBS, hydrogels were fixed in 10% formalin for 30 min at room temperature (RT) followed by three washes in PBS. For immunostaining, fixed hydrogels were incubated for 2 h at 4°C in permeabilization solution containing 0.2% Triton X-100 in PBS supplemented with 2% BSA, 320 mM sucrose and 6 mM magnesium chloride. For staining of focal adhesions, gels were fixed for 30 min at 37 °C with a microtubule stabilization buffer (1 mM magnesium sulfate (MgSO4), 1 mM ethylene glycol-bis(2-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA), 0.1 M 1,4-piperazinediethanesulfonic acid (PIPES), pH 6.75, 1% Triton X-100, 4% (w/v) poly(ethylene glycol) (PEG, 8000), 2% paraformaldehyde). Prior to immunostaining, hydrogels were blocked in PBS containing 2% BSA. Primary antibodies were diluted in PBS containing 2% BSA and hydrogels stained at 4°C overnight. Antibodies and dilutions included anti-YAP/TAZ (1:200; Santa Cruz sc-101199), anti-fibronectin (1:200; Sigma Aldrich F6140), anti-collagen type 1 (1:200, Abcam ab138492), anti-laminin α5 (1:200, Abcam ab220399), anti-collagen type 4 (1:200, Thermofisher MA1–22148), anti-paxillin (1:200, BD 610052), anti-osteocalcin (1:10, R&D Systems MAB1419) and anti-FABP6 (1:20; R&D System AF3880). For inhibition experiments within degradable hydrogels (including mouse monoclonal antibodies), anti-YAP/TAZ (1:200; Cell Signaling 8418) and anti-osteocalcin (1:50, Bioss bs-4917R-TR) were used. After three PBS washes, secondary antibodies Alexa Flour-488/594/647 IgG H&L (1:200; Abcam ab150113/ab150080/ab150143) were added for 2 h at RT. For F-Actin staining, rhodamine-conjugated phalloidin (1:100; Invitrogen R415) was added alone or with the secondary antibodies and incubated for 2 h at RT. Hydrogels were washed three times followed by DAPI staining (1:1000; Invitrogen D1306, in PBS) for 20 min at RT.
Imaging and quantification.
A Nikon A1R Confocal Microscope was used to acquire z-stack images at 20×0.75 NA (1.23 μm per pixel) and 100×1.4 NA (0.13 μm per pixel). For measuring average local ECM thickness, cell membrane and ECM were determined through binary masks of z-stack images in ImageJ with Otsu’s thresholding. The cell membrane mask was then inverted and superimposed with the ECM mask which generated a mask that encompassed the ECM only. The ImageJ plugin ‘BoneJ’ was used to calculate the local average ECM thickness per slice62. At least five different slices throughout the cell-body were measured for average local thickness to calculate the average thickness per cell. YAP/TAZ nuclear/cytoplasmic ratios were quantified as previously described27. Briefly, binary masks of 3D DAPI and F-Actin image stacks were used to generate nuclear 3D YAP/TAZ images. The ratio of nuclear and cytoplasmic pixel intensities was then calculated and normalized to the nuclear and cytosolic volumes of a cell. Focal adhesion aspect ratios were quantified from maximum intensity z-projections and focal adhesion/nascent protein intensity profiles from single stacks. Evaluation of hMSC differentiation was performed by dividing the number of cells positively stained for OC and FABP, respectively, by the total number of nuclei (identified with DAPI).
Transmission Electron Microscopy (TEM).
Hydrogels for TEM were fixed for 30 min at RT in 2.0% paraformaldehyde, 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4). After three PBS washes, hydrogels were incubated for 1 h at RT in 2.0% osmium tetroxide, washed with deionized water and stained with 2% uranyl acetate. Dehydration was performed with graded ethanol series, and hydrogels were embedded in Embed-812 (Electron Microscopy Sciences). Sections were incubated in Uranyl acetate and lead citrate and imaged with a JEOL 1010 electron microscope including a Hamamatsu digital camera (AMT Advantage image capture software).
2D hydrogels and cell seeding.
NorHA hydrogel films (4 wt% polymer, 0.3 ratio with a thickness of ca. 100 μm) were UV photopolymerized on thiolated coverslips in the presence of 1 mM thiolated RGD58. hMSCs were seeded at 3000 cells cm−2 and cultured in “AHA growth media” for 24 h or 6 days before fixation and subsequent YAP/TAZ and F-actin immunostaining. Culture media was supplemented with inhibitors directly after seeding and replenished daily.
Traction Force Microscopy (TFM).
Traction Force Microscopy images were acquired as previously described52,63. NorHA hydrogel films were prepared as described with 0.2 μm diameter fluorescent beads at 1% vol/vol (Invitrogen F8810). hMSCs were cultured for 18 hours before TFM analysis. Embedded beads and cells were captured in phase contrast and fluorescence using a DeltaVision Deconvolution Microscope (GE Healthcare Life Sciences, Marlborough, MA). Images were acquired prior and after cell lysis with PBS buffer containing 10% SDS (sodium dodecyl sulfate)/1% Triton X-100. ImageJ was used for TFM data analysis, including stack alignment, particle image velocimetry (PIV), Fourier transform traction cytometry (FTTC))64. For FTTC variables, a Poisson’s ratio of 0.45 was assumed and 1e−9 was utilized as regularization parameter.
Statistical analysis and Reproducibility.
GraphPad Prism 7 software was used for all statistical analyses. Statistical comparisons between two experimental groups were performed using two-tailed Student’s t-tests and comparisons among more groups were performed using one-way or two-way analysis of variance (ANOVA) with Bonferroni post hoc testing. Exact p-values and degrees of freedom are reported in Supplementary Table 1. All experiments were repeated as described in the text. For representative immunofluorescence images at least two biological repeats of all experiments were performed with similar results. TEM images were acquired for at least six cells per condition with similar results.
Supplementary Material
Acknowledgments
This work was supported by the Swiss National Foundation through an SNF Early Postdoc Mobility Fellowship (to C.L.), the National Science Foundation (DMR Award 1610525, the Center for Engineering MechanoBiology CMMI: 15–48571), and the National Institutes of Health (R01 EB008722). We are grateful for help from the Penn EMRL Electron Microscopy Core for TEM and the Penn CDB Microscopy Core Facility for TFM, and would like to thank D. Seliktar for providing the PEG-DA, and A. Garcia, M. Davidson, R. Daniels, B. Cosgrove and M. D’Este for helpful conversations.
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
Data Availability Statement
All the data generated or analyzed during this study are included within this article and its Supplementary Information. Additional information is available from the corresponding author on request.
References
- 1.Kim SH, Turnbull J & Guimond S Extracellular matrix and cell signalling: the dynamic cooperation of integrin, proteoglycan and growth factor receptor. J Endocrinol 209, 139–151, (2011). [DOI] [PubMed] [Google Scholar]
- 2.Guvendiren M & Burdick JA Engineering synthetic hydrogel microenvironments to instruct stem cells. Curr Opin Biotechnol 24, 841–846, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tibbitt MW & Anseth KS Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol Bioeng 103, 655–663, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drury JL & Mooney DJ Hydrogels for tissue engineering: scaffold design variables and applications. Biomaterials 24, 4337–4351, (2003). [DOI] [PubMed] [Google Scholar]
- 5.Wells RG The role of matrix stiffness in regulating cell behavior. Hepatology 47, 1394–1400, (2008). [DOI] [PubMed] [Google Scholar]
- 6.Khetan S et al. Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nat Mater 12, 458–465, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schultz KM, Kyburz KA & Anseth KS Measuring dynamic cell-material interactions and remodeling during 3D human mesenchymal stem cell migration in hydrogels. Proc Natl Acad Sci U S A 112, E3757–3764, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaudhuri O et al. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat Mater 15, 326–334, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H & Heilshorn SC Adaptable hydrogel networks with reversible linkages for tissue engineering. Adv Mater 27, 3717–3736, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosales AM & Anseth KS The design of reversible hydrogels to capture extracellular matrix dynamics. Nat Rev Mater 1, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unlu G, Levic DS, Melville DB & Knapik EW Trafficking mechanisms of extracellular matrix macromolecules: insights from vertebrate development and human diseases. Int J Biochem Cell Biol 47, 57–67, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gattazzo F, Urciuolo A & Bonaldo P Extracellular matrix: a dynamic microenvironment for stem cell niche. Biochim Biophys Acta 1840, 2506–2519, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadler KE, Hill A & Canty-Laird EG Collagen fibrillogenesis: fibronectin, integrins, and minor collagens as organizers and nucleators. Curr Opin Cell Biol 20, 495–501, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gjorevski N & Nelson CM Bidirectional extracellular matrix signaling during tissue morphogenesis. Cytokine Growth Factor Rev 20, 459–465, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLeod CM & Mauck RL High fidelity visualization of cell-to-cell variation and temporal dynamics in nascent extracellular matrix formation. Sci Rep 6, 38852, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bian L, Guvendiren M, Mauck RL & Burdick JA Hydrogels that mimic developmentally relevant matrix and N-cadherin interactions enhance MSC chondrogenesis. Proc Natl Acad Sci U S A 110, 10117–10122, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicodemus GD, Skaalure SC & Bryant SJ Gel structure has an impact on pericellular and extracellular matrix deposition, which subsequently alters metabolic activities in chondrocyte-laden PEG hydrogels. Acta Biomater 7, 492–504, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huebsch N et al. Matrix elasticity of void-forming hydrogels controls transplanted-stem-cell-mediated bone formation. Nat Mater 14, 1269–1277, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai R, Nakamoto T, Kawazoe N & Chen G Influence of stepwise chondrogenesis-mimicking 3D extracellular matrix on chondrogenic differentiation of mesenchymal stem cells. Biomaterials 52, 199–207, (2015). [DOI] [PubMed] [Google Scholar]
- 20.Ferreira SA et al. Bi-directional cell-pericellular matrix interactions direct stem cell fate. Nat Commun 9, 4049, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubow KE et al. Mechanical forces regulate the interactions of fibronectin and collagen I in extracellular matrix. Nat Commun 6, 8026, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li B, Moshfegh C, Lin Z, Albuschies J & Vogel V Mesenchymal stem cells exploit extracellular matrix as mechanotransducer. Sci Rep 3, 2425, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott LE, Mair DB, Narang JD, Feleke K & Lemmon CA Fibronectin fibrillogenesis facilitates mechano-dependent cell spreading, force generation, and nuclear size in human embryonic fibroblasts. Integr Biol (Camb) 7, 1454–1465, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daley WP, Peters SB & Larsen M Extracellular matrix dynamics in development and regenerative medicine. J Cell Sci 121, 255–264, (2008). [DOI] [PubMed] [Google Scholar]
- 25.Jansen KA, Atherton P & Ballestrem C Mechanotransduction at the cell-matrix interface. Seminars in Cell & Developmental Biology 71, 75–83, (2017). [DOI] [PubMed] [Google Scholar]
- 26.Dieterich DC et al. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Protoc 2, 532–540, (2007). [DOI] [PubMed] [Google Scholar]
- 27.Caliari SR, Vega SL, Kwon M, Soulas EM & Burdick JA Dimensionality and spreading influence MSC YAP/TAZ signaling in hydrogel environments. Biomaterials 103, 314–323, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doyle AD & Yamada KM Mechanosensing via cell-matrix adhesions in 3D microenvironments. Exp Cell Res 343, 60–66, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hytonen VP & Wehrle-Haller B Protein conformation as a regulator of cell-matrix adhesion. Phys Chem Chem Phys 16, 6342–6357, (2014). [DOI] [PubMed] [Google Scholar]
- 30.Tuckwell D, Calderwood DA, Green LJ & Humphries MJ Integrin alpha 2 I-domain is a binding site for collagens. J Cell Sci 108 (Pt 4), 1629–1637, (1995). [DOI] [PubMed] [Google Scholar]
- 31.Connelly JT, Petrie TA, Garcia AJ & Levenston ME Fibronectin- and collagen-mimetic ligands regulate bone marrow stromal cell chondrogenesis in three-dimensional hydrogels. Eur Cell Mater 22, 168–176; discussion 176–167, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keselowsky BG, Collard DM & Garcia AJ Integrin binding specificity regulates biomaterial surface chemistry effects on cell differentiation. Proc Natl Acad Sci U S A 102, 5953–5957, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Massia SP & Hubbell JA Vascular endothelial cell adhesion and spreading promoted by the peptide REDV of the IIICS region of plasma fibronectin is mediated by integrin alpha 4 beta 1. Journal of Biological Chemistry 267, 14019–14026, (1992). [PubMed] [Google Scholar]
- 34.Dupont S et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183, (2011). [DOI] [PubMed] [Google Scholar]
- 35.Brusatin G, Panciera T, Gandin A, Citron A & Piccolo S Biomaterials and engineered microenvironments to control YAP/TAZ-dependent cell behaviour. Nat Mater, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fogerty FJ, Akiyama SK, Yamada KM & Mosher DF Inhibition of binding of fibronectin to matrix assembly sites by anti-integrin (alpha 5 beta 1) antibodies. The Journal of Cell Biology 111, 699–708, (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDonald JA et al. Fibronectin’s cell-adhesive domain and an amino-terminal matrix assembly domain participate in its assembly into fibroblast pericellular matrix. Journal of Biological Chemistry 262, 2957–2967, (1987). [PubMed] [Google Scholar]
- 38.Lee HP, Gu L, Mooney DJ, Levenston ME & Chaudhuri O Mechanical confinement regulates cartilage matrix formation by chondrocytes. Nat Mater 16, 1243–1251, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cameron AR, Frith JE, Gomez GA, Yap AS & Cooper-White JJ The effect of time-dependent deformation of viscoelastic hydrogels on myogenic induction and Rac1 activity in mesenchymal stem cells. Biomaterials 35, 1857–1868, (2014). [DOI] [PubMed] [Google Scholar]
- 40.Rodell CB, Dusaj NN, Highley CB & Burdick JA Injectable and Cytocompatible Tough Double-Network Hydrogels through Tandem Supramolecular and Covalent Crosslinking. Adv Mater 28, 8419–8424, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loebel C, Rodell CB, Chen MH & Burdick JA Shear-thinning and self-healing hydrogels as injectable therapeutics and for 3D-printing. Nat Protoc 12, 1521–1541, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodell CB, Kaminski AL & Burdick JA Rational design of network properties in guest-host assembled and shear-thinning hyaluronic acid hydrogels. Biomacromolecules 14, 4125–4134, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dooling LJ, Buck ME, Zhang WB & Tirrell DA Programming Molecular Association and Viscoelastic Behavior in Protein Networks. Adv Mater 28, 4651–4657, (2016). [DOI] [PubMed] [Google Scholar]
- 44.McKinnon DD, Domaille DW, Cha JN & Anseth KS Biophysically defined and cytocompatible covalently adaptable networks as viscoelastic 3D cell culture systems. Adv Mater 26, 865–872, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng Y et al. Exo1: a new chemical inhibitor of the exocytic pathway. Proc Natl Acad Sci U S A 100, 6469–6474, (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Kleist L & Haucke V At the Crossroads of Chemistry and Cell Biology: Inhibiting Membrane Traffic by Small Molecules. Traffic 13, 495–504, (2012). [DOI] [PubMed] [Google Scholar]
- 47.Mishev K, Dejonghe W & Russinova E Small Molecules for Dissecting Endomembrane Trafficking: A Cross-Systems View. Chemistry & Biology 20, 475–486, (2013). [DOI] [PubMed] [Google Scholar]
- 48.Purcell BP et al. Injectable and bioresponsive hydrogels for on-demand matrix metalloproteinase inhibition. Nat Mater 13, 653–661, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolf K et al. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol 201, 1069–1084, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sridhar BV et al. Development of a cellularly degradable PEG hydrogel to promote articular cartilage extracellular matrix deposition. Adv Healthc Mater 4, 702–713, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blache U et al. Notch-inducing hydrogels reveal a perivascular switch of mesenchymal stem cell fate. EMBO Rep 19, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cosgrove BD et al. N-cadherin adhesive interactions modulate matrix mechanosensing and fate commitment of mesenchymal stem cells. Nat Mater 15, 1297–1306, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gjorevski N et al. Designer matrices for intestinal stem cell and organoid culture. Nature 539, 560, (2016). [DOI] [PubMed] [Google Scholar]
- 54.Cruz-Acuna R et al. Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat Cell Biol 19, 1326–1335, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hezaveh H et al. Encoding Stem-Cell-Secreted Extracellular Matrix Protein Capture in Two and Three Dimensions Using Protein Binding Peptides. Biomacromolecules 19, 721–730, (2018). [DOI] [PubMed] [Google Scholar]
- 56.Gardner OF, Alini M & Stoddart MJ Mesenchymal Stem Cells Derived from Human Bone Marrow. Methods Mol Biol 1340, 41–52, (2015). [DOI] [PubMed] [Google Scholar]
- 57.Schoen RC, Bentley KL & Klebe RJ Monoclonal antibody against human fibronectin which inhibits cell attachment. Hybridoma 1, 99–108, (1982). [DOI] [PubMed] [Google Scholar]
- 58.Gramlich WM, Kim IL & Burdick JA Synthesis and orthogonal photopatterning of hyaluronic acid hydrogels with thiol-norbornene chemistry. Biomaterials 34, 9803–9811, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wade RJ, Bassin EJ, Rodell CB & Burdick JA Protease-degradable electrospun fibrous hydrogels. Nat Commun 6, 6639, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Almany L & Seliktar D Biosynthetic hydrogel scaffolds made from fibrinogen and polyethylene glycol for 3D cell cultures. Biomaterials 26, 2467–2477, (2005). [DOI] [PubMed] [Google Scholar]
- 61.Bauer A et al. Hydrogel substrate stress-relaxation regulates the spreading and proliferation of mouse myoblasts. Acta Biomater 62, 82–90, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Doube M et al. BoneJ: Free and extensible bone image analysis in ImageJ. Bone 47, 1076–1079, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loebel C et al. Cross-Linking Chemistry of Tyramine-Modified Hyaluronan Hydrogels Alters Mesenchymal Stem Cell Early Attachment and Behavior. Biomacromolecules 18, 855–864, (2017). [DOI] [PubMed] [Google Scholar]
- 64.Tseng Q et al. Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proc Natl Acad Sci U S A 109, 1506–1511, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.