Abstract
Oral mucosa contains a unique transcriptional network that primes oral wounds for rapid resolution in humans. Our previous work identified genes that were consistently upregulated in the oral mucosa and demonstrated that induction of one of the identified genes, transcription factor SOX2, promoted cutaneous wound healing in mice. In this study, we investigated the molecular and cellular mechanisms by which SOX2 accelerates wound healing in skin. RNA-seq analysis showed that SOX2 induced a proliferative and wound-activated phenotype in skin keratinocytes prior to wounding. During wound healing, SOX2 induced proliferation of epithelial and connective tissue cells and promoted angiogenesis. ChIP-assay revealed that SOX2 directly regulates expression of EGFR ligands, resulting in activation of EGFR. In vitro, skin keratinocytes overexpressing SOX2 promoted cell migration via the EGFR/MEK/ERK pathway. We conclude that induction of SOX2 in skin keratinocytes accelerates cutaneous wound healing by promoting keratinocyte migration and proliferation, and enhancement of angiogenesis via the upregulation of EGFR ligands and activation of EGFR/MEK/ERK pathway. Through the identification of putative cutaneous SOX2 targets such as HBEGF, this study opens venues to determine clinical targets for treatment of skin wounds.
INTRODUCTION
Wound healing has four overlapping phases in the repair process: hemostasis, inflammation, proliferation and remodeling (Singer and Clark, 1999). In the proliferation phase, diverse types of cells, including keratinocytes, fibroblasts, endothelial cells, macrophages and leukocytes migrate into the wound area for re-epithelialization, angiogenesis and production of the extracellular matrix (ECM). Growth factors derived from various cells localized in the wounded area are essential for proper and efficient wound healing (Eming et al., 2014). Activation of epidermal growth factor receptor (EGFR) through EGFR ligands, regulates various phases of cutaneous wound healing (Repertinger et al., 2004, Tokumaru et al., 2000). Dysregulation of these interactive processes in diabetic wounds results in delayed wound healing (Brem and Tomic-Canic, 2007, Gurtner et al., 2008, Singer and Clark, 1999).
Oral wound healing has long been considered as an ideal system of wound resolution when compared to skin because oral wounds resolve faster and without scar formation. Several studies determined the differences of the injury responses between oral mucosal and cutaneous wounds, focusing on the inflammation and migrating capacity of keratinocytes (Szpaderska et al., 2003, Turabelidze et al., 2014). We recently reported a longitudinal clinical wound healing study using healthy human subjects where paired comparative gene analysis was done between oral mucosa and skin tissue (Iglesias-Bartolome et al., 2018). This analysis revealed that wound-activated transcriptional networks are present at the basal state in the oral mucosa, priming the epithelium for wound repair. SOX2 and PITX1 were identified as being highly expressed in the oral mucosa when compared to the skin and we demonstrated that they play important functions during wound resolution (Iglesias-Bartolome et al., 2018).
SOX2, a member of the SoxB1 transcription factor family, is an important regulator of stem cell maintenance in epithelial tissues and has been found to promote tumor growth (Arnold et al., 2011, Boumahdi et al., 2014, Siegle et al., 2014). It has also reported that SOX2 is associated with the development of immature tumors, including glioblastomas and teratomas (Garros-Regulez et al., 2016, Phi et al., 2007). In the skin, SOX2 is expressed in cells localized around the bulge region of hair follicles, dermal papilla, Merkel cells, and neural crest precursor cells, but has not been detected in epidermal keratinocytes (Driskell et al., 2009, Johnston et al., 2013). It has been reported that SOX2 has an important role in skin repair (Johnston et al., 2013), and we demonstrated that inducible epidermal-specific expression of SOX2 promoted cutaneous wound healing in mice (Iglesias-Bartolome et al., 2018). However, the detailed cellular and molecular mechanisms of rapid SOX2-mediated wound resolution in skin keratinocytes has not been determined. Herein, we performed in-depth analysis of the molecular and cellular changes induced by SOX2 in skin keratinocytes in the basal state and during wound healing. We identified SOX2-target genes and downstream EGFR/MEK/ERK pathway effectors as essential components in the promotion of cutaneous wound healing.
Results
Epidermal specific SOX2 expression accelerates cutaneous wound healing by promoting re-epithelialization and formation of granulation tissue
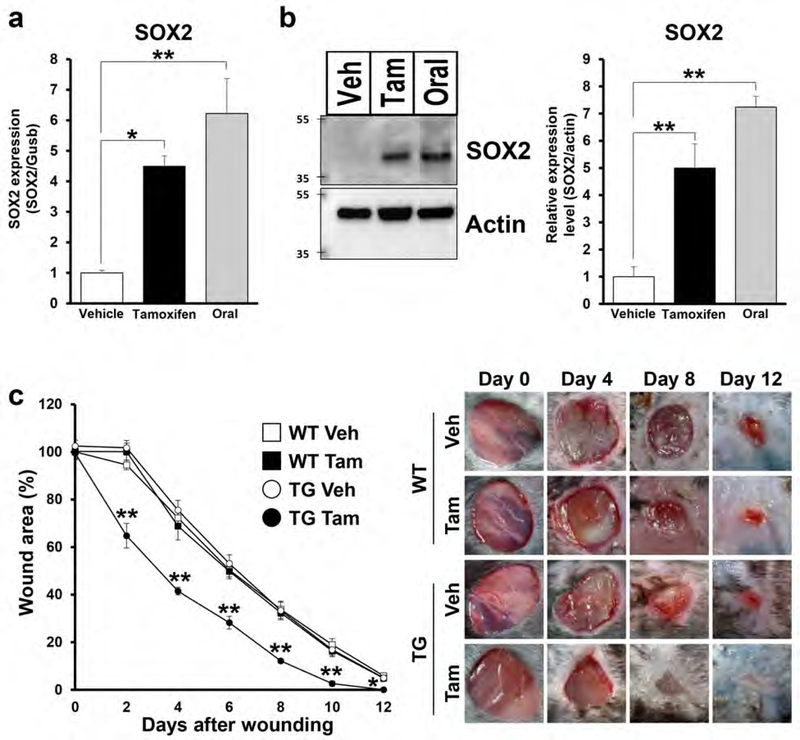
Utilizing a tamoxifen-inducible K14CreERTM/LSL-SOX2 (TG) mouse model (Supplementary Figure S1a), we determined that mRNA and protein levels of SOX2 expression were significantly increased in tamoxifen (Tam) treated TG mice when compared with vehicle (Veh) treated mice, using murine oral mucosa (buccal mucosa and hard palate) as positive controls (Figure 1a, 1b and Supplementary Figure S1b). Skin punch biopsies were obtained from SOX2-overexpressing (TG Tam) and age-matched control mice (TG Veh, Wild type (WT) Tam and WT Veh) to assess the wound healing process every 2 days up to 12 days, when there was almost complete wound closure in TG Tam mice. WT Tam and WT Veh treated mice confirmed that topical application of tamoxifen had no effect on cutaneous wound healing. SOX2-overexpressing mice exhibited significantly accelerated wound closure kinetics when compared with control groups from day 2 to day 12 (Figure 1c). Thickness of the epidermis was significantly increased in SOX2-overexpressing mice when compared to control mice (TG Veh or WT Tam) (Supplementary Figure S1c). The re-epithelialization area was significantly larger in SOX2-overexpressing mice when compared with control (TG Veh) mice at days 2 and 4 after wounding (Supplementary Figure S1d), corroborating our previous work (Iglesias-Bartolome et al., 2018). Furthermore, the granulation tissue area was significantly increased in SOX2-overexpressing mice when compared with control (TG Veh) mice (Supplementary Figure S1e). These results indicate that SOX2 expression in skin keratinocytes accelerates cutaneous wound healing via the promotion of re-epithelialization and formation of granulation tissue.
Figure 1. SOX2+ skin keratinocytes promote re-epithelialization and cutaneous wound healing.
(a) SOX2 mRNA levels at basal state in skin from TG mice treated with vehicle or tamoxifen and oral mucosa. n=4 for each group. (b) Immunoblot showing SOX2 expression levels at basal state in skin from TG mice treated with vehicle or tamoxifen and oral mucosa. Actin was used as a loading control. Right panel shows quantification of SOX2 protein. (c) Percent wound area at each time point relative to the original wound area in WT and TG mice treated with vehicle or tamoxifen. Quantification of the wound areas in n=8 wounds per groups was performed using Image J software. Right panel shows photographs of the wound areas after topical treatment with vehicle or tamoxifen in WT and TG mice at 0, 4, 8, and 12 days after wounding. Data expressed as means ± SEM. **P<0.01, *P<0.05.
Cutaneous SOX2 suppresses epidermal differentiation but induces wound-activated phenotype in basal state
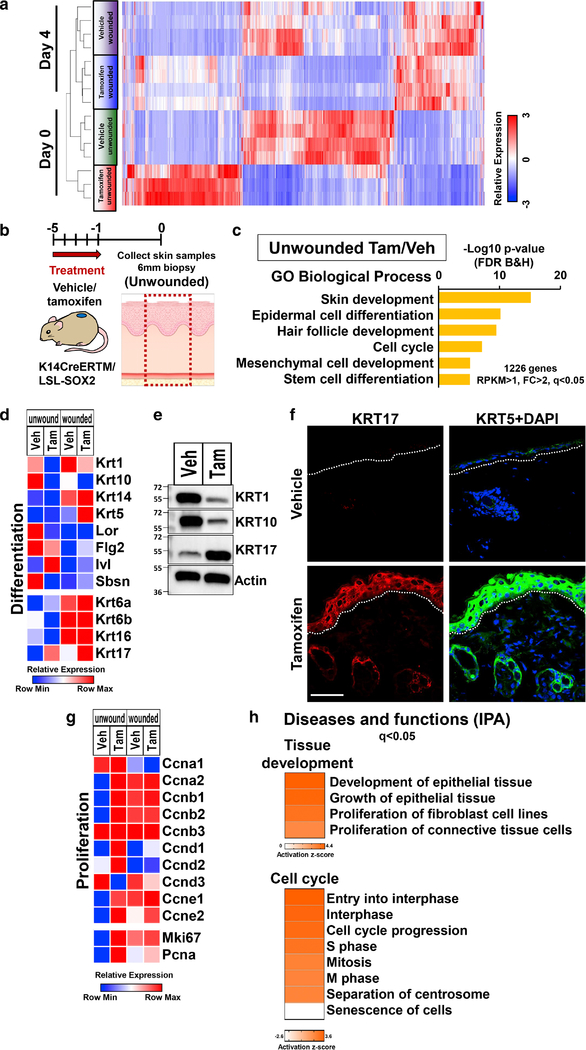
To elucidate the molecular mechanisms underlying the accelerated wound healing in SOX2-overexpressing mice, we performed RNA sequencing (RNA-seq) analysis and compared gene expression between SOX2-overexpressing and control mice at unwounded (day 0) and wounded (day 4 after wounding) timepoints (Figure 2a). Clustering analysis showed that there were large differences in gene expression between SOX2-overexpressing and control mice in the unwounded timepoint. While these differences decreased after wounding, there was clear separation between tamoxifen wounded and vehicle wounded mice in principal component analysis (PCA) (Supplementary Figure S2a).
Figure 2. SOX2 expressing skin keratinocytes induce wound-activated phenotype in basal state.
</p>(a) Unsupervised clustering analysis of RNA-seq gene expression data of the samples for each group in unwounded (day 0) and wounded (day 4) timepoints in TG mice treated with vehicle and tamoxifen. (b) Schematic representation of biopsy site and timecourse of treatment in the unwounded skin. (c) GO biological process terms enriched in datasets of genes differentially regulated by SOX2-overexpressing skin keratinocytes in TG mice in unwounded timepoint. (d) Relative mRNA expression levels of keratinization and epidermal cell differentiation markers throughout the wound healing process. (e) Immunoblot image showing KRT1, KRT10 and KRT17 expression of unwounded skin samples from TG mice treated with vehicle and tamoxifen. Actin was used as a loading control. n=3. (f) Representative images of unwounded skin tissue from TG mice treated with vehicle and tamoxifen stained to detect expression of KRT17 (red), KRT5 (green) and DAPI (blue). Scale bar = 50μm. (g) Relative mRNA expression levels of proliferation markers throughout the wound healing process. (h) IPA analysis of RNA-seq data from unwounded skin samples treated with vehicle and tamoxifen in TG mice: Diseases and functions terms related to tissue development and cell cycle found in differentially regulated genes. Data is expressed as means ± SEM. **P<0.01, *P<0.05.
Next, we focused on differences in gene expression at basal state (Day 0, unwounded) between SOX2-overexpressing and control mice (Figure 2b). Gene Ontology (GO) analysis of the genes upregulated in SOX2-overexpressing mice (unwounded Tam/Veh: RPKM>1, FC>2, q<0.05, 1226 genes) showed enrichment of genes related to skin development, epidermal cell differentiation, hair follicle development and cell cycle (Figure 2c). Expression levels of epidermal differentiation genes (KRT1, KRT10, LOR, SBSN) were suppressed in SOX2-overexpressing mice (Figure 2d, 2e and Supplementary Figure S2b). However, SOX2-overexpressing skin did not result in expression of keratin 13 (KRT13), a well-established oral keratinocyte marker (Supplementary Figure S2c). Keratinocytes at the wound edge switch their phenotype from differentiating to activated, and start expressing KRT6, KRT16 and KRT17 (Freedberg et al., 2001, Wikramanayake et al., 2014). KRT17 is expressed in several skin disorders including psoriasis and tumors (Markey et al., 1992, Yang et al., 2017) and promotes cell proliferation, migration and wound healing (Mazzalupo et al., 2003). Interestingly, SOX2-overexpressing skin showed significantly increased expression of KRT17 in the unwounded timepoint (Figure 2d–2f and Supplementary Figure S2b). It has been reported that SOX2 directly regulates Sonic hedgehog (Shh) in the development of neural stem cells (Favaro et al., 2009). Shh has important roles in the regulation of wound healing (Asai et al., 2006) and in the regulation of KRT17 expression (Callahan et al., 2004). RNA-seq analysis showed that Shh signaling associated genes were upregulated at basal state in SOX2-overexpressing mice (Supplementary Figure S2d). Immunofluorescence staining showed that Shh was highly expressed in SOX2-overexpressing skin (Supplementary Figure S2e).
SOX2 promotes cell proliferation and tumor growth via regulation of cyclin expression (Boumahdi et al., 2014). Transcript levels of most cyclins were upregulated in SOX2-overexpressing mice (Figure 2g). We previously revealed that SOX2-overexpressing skin has a proliferative phenotype at basal state using PCNA staining (Iglesias-Bartolome et al., 2018). We confirmed this result in transcriptome and protein levels with another proliferation maker, Ki-67. Ki-67 and PCNA were also upregulated in SOX2-overexpressing mice (Figure 2g and Supplementary Figure S2f).
When using Ingenuity Pathway Analysis (IPA) on the genes upregulated in SOX2-overexpressing mice in the unwounded timepoint, we found induction of networks associated with development and growth of epithelial tissue and cell cycle progression (Figure 2h) and genes associated with cellular movement (Supplementary Figure S2g).
Taken together, these results indicate that in epidermal keratinocytes, SOX2 suppresses epidermal differentiation while inducing a wound-activated phenotype. This is supported by the increased expression of KRT17 and upregulation of genes associated with cell proliferation and migration in the basal state.
SOX2 in skin keratinocytes promotes proliferation of keratinocytes and angiogenesis during wound healing
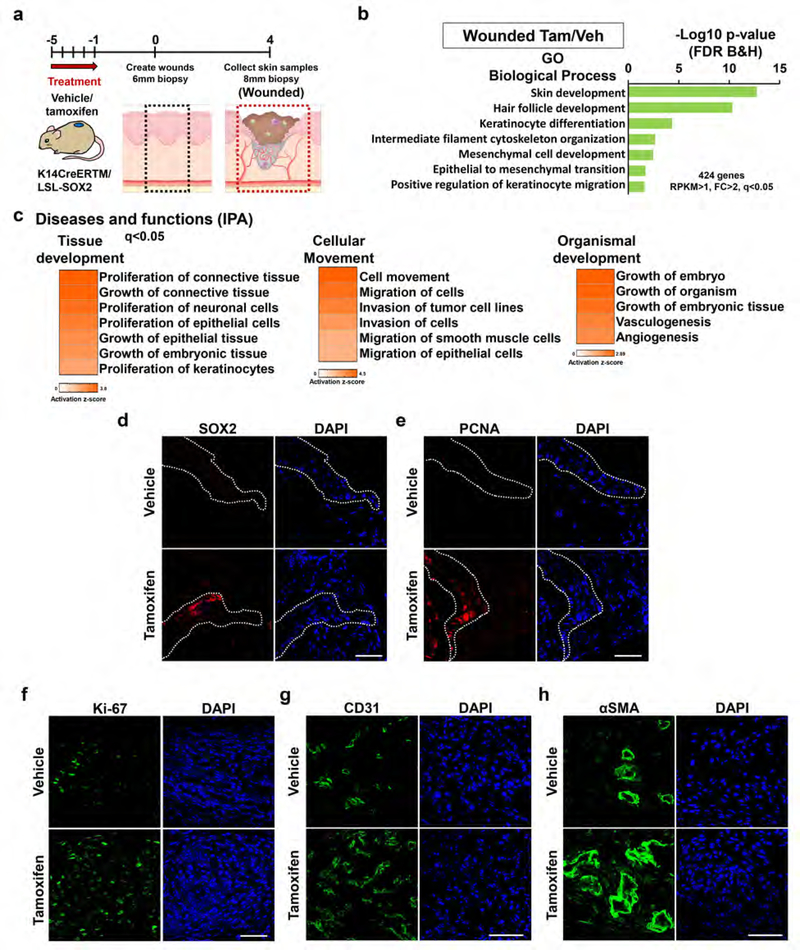
To investigate gene networks regulated by SOX2 during wound healing, we compared the gene expression profiles of SOX2-overexpressing and control mice in the wounded state (Day 4) (Figure 3a). GO analysis of genes significantly upregulated in SOX2-overexpressing mice (wounded Tam/Veh, RPKM>1, FC>2, q<0.05, 424 genes) showed enrichment for processes related to skin development, keratinocyte differentiation, intermediate filament cytoskeleton organization, mesenchymal cell development and positive regulation of keratinocyte migration (Figure 3b). Expression levels of genes described in wound-activated keratinocytes including keratins KRT6, KRT16 and KRT17 were activated in SOX2-overexpressing mice particularly in the wounded timepoint, with the most notable change being for KRT17 (Figure 2d). Diseases and functions analysis via IPA on the upregulated genes in SOX2-overexpressing mice showed that cutaneous SOX2 induced gene networks associated with proliferation of connective tissue and epithelial cells and cell movement during wound healing (Figure 3c and Supplementary Figure S3a). We performed in-depth analysis of migrating tongue and granulation tissue to evaluate the correlation between SOX2 expression and the promotion of re-epithelialization and formation of granulation tissue. Immunofluorescence staining showed that SOX2+ cells were present in the migrating tongue, and a large number of PCNA+ positive keratinocytes were found in the wound area in SOX2-overexpressing mice (Figure 3d and 3e). Ki67, another marker of proliferation, showed upregulation in the RNA-seq analysis (Figure 2g). Furthermore, the numbers of Ki-67 positive cells in the wound area were significantly increased in SOX2-overexpressing mice (Figure 3f and Supplementary Figure S3c). Analysis via IPA also showed induction of gene networks associated with vasculogenesis and angiogenesis in organism development (Figure 3c). It has been reported that SOX2 in tumors promotes angiogenesis through production of pro-angiogenic factors including VEGF, TGFA and EREG (Siegle et al., 2014). In addition to the upregulation of angiogenesis pathways (Figure 3c and Supplementary Figure S3b), we determined that the number of CD31+ endothelial cells and aSMA+ pericytes/myofibroblasts in the wound area were significantly increased in SOX2-overexpressing mice (Figure 3g, 3h and Supplementary Figure S3c). However, SOX2-overexpressing mice did not have increased mRNA levels of members of collagen family (Col1a1, Col1a2, Col3a1) and FN1, and those expression levels correlated with expression levels of TGF-β (Supplementary Figure S3d and S3e).
Figure 3. SOX2 expressing skin keratinocytes promote proliferation of keratinocytes, connective tissue cells and angiogenesis during wound healing.
(a) Schematic representation of biopsy site in the skin at day 4 after wounding (wounded) and timecourse of treatment. (b) GO biological process terms enriched in datasets of genes differentially regulated in SOX2-overexpressing skin keratinocytes of TG mice during wound healing (wounded). (c) IPA analysis of RNA-seq data from wounded skin samples treated with vehicle and tamoxifen in TG mice: Diseases and functions terms related to tissue development, cellular movement and organismal development found in differentially regulated genes. (d-e) Representative images of migratory tongue in TG mice treated with vehicle or tamoxifen at day 4 to show expression of (d) SOX2 and (e) PCNA positive keratinocytes. Scale bar = 50μm. (f-h) Representative images of skin tissue from TG mice treated with vehicle and tamoxifen at day 4 (wounded) stained to show expression of the (f) Ki-67+ cells, (g) endothelial cells (CD31+) and (h) pericytes/myofibroblasts (αSMA+). Scale bar = 50μm.
Neutrophils and macrophages infiltrate the wound area and promote re-epithelialization and angiogenesis via production of chemokines and growth factors (Lamagna et al., 2006). Therefore, we examined the numbers of CD68+ macrophages and myeloperoxidase (MPO)+ neutrophils in the wound area by immunofluorescence staining. However, there was no significant difference between SOX2-overexpressing mice and control mice (Supplementary Figure S3f and S3g).
These results indicate that cutaneous SOX2 enhances cell proliferation in epidermis, as well as connective tissue formation and cell migration. The resulting promotion of re-epithelialization and angiogenesis is potentially directly involved in the accelerated wound healing process.
SOX2-overexpressing mice upregulates EGFR ligands genes
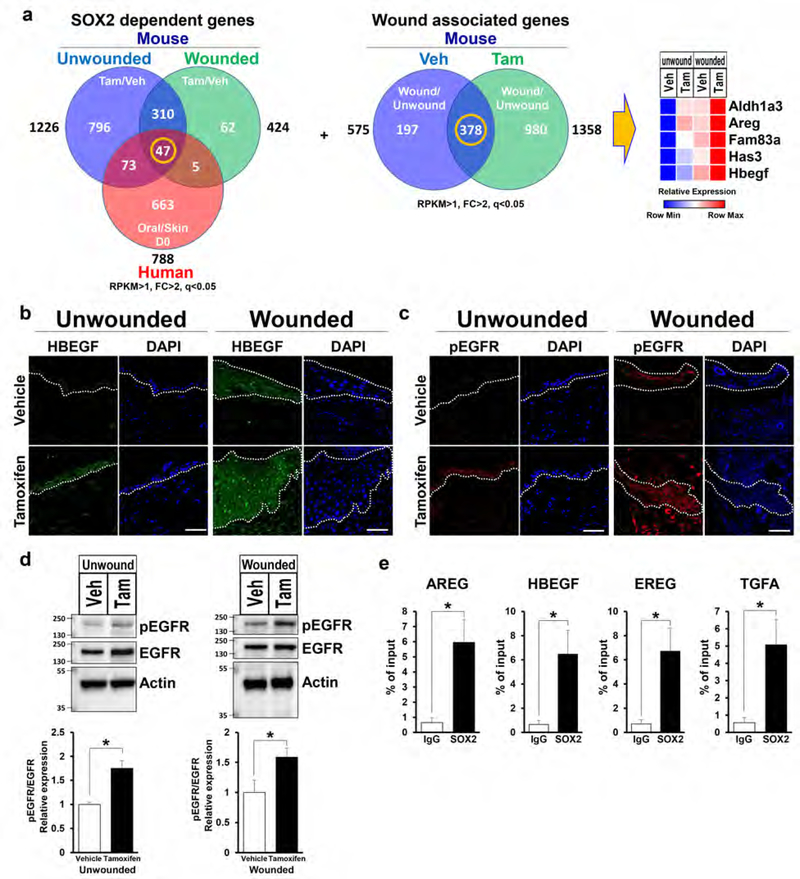
Next, we compared genes from groups (1) unwounded Tam/Veh, (2) wounded Tam/Veh in mice and (3) Oral/Skin (day 0) in human (Iglesias-Bartolome et al., 2018) to determine SOX2 target genes. This analysis identified 47 genes that were upregulated in all three groups (Figure 4a and Supplementary Figure S4a). Consecutively, we performed another analysis comparing genes from groups (4) Veh wounded/unwounded and (5) Tam wounded/unwounded in mice and determined 378 genes commonly upregulated during the wound healing process (Figure 4a). Finally, we cross-referenced these two gene lists and identified 5 SOX2 target genes which potentially are instrumental to accelerated wound healing (Figure 4a). Among these genes, ALDH1A3 encodes an aldehyde dehydrogenase enzyme which is required for conversion of retinol to retinoic acids. It has been reported that topical application of retinoic acid improves cutaneous wound healing (Kitano et al., 2001). HAS3 is involved in the synthesis of the unbranched glycosaminoglycan hyaluronan which is a major constituent of the ECM. Hyaluronan binds to its receptor CD44 present on endothelial cells, triggering the proliferation of endothelial cells and production of angiogenic cytokines (Pardue et al., 2008). Alcian blue staining revealed that hyaluronan was highly elevated in the wound area in SOX2-overexpressing mice (Supplementary Figure S4b).
Figure 4. SOX2 expressing skin keratinocytes induce upregulation of EGFR ligands expression and activation of EGFR.
(a) Venn diagram summarizing comparison of upregulated genes. Left panel shows 1: Unwounded Tam/Veh (blue), 2: Wounded Tam/Veh (green) in mice and 3: Oral/Skin in the basal state (Red) in human. Center panel shows 4: Veh wounded/unwounded (blue) and 5: Tam wounded/unwounded (green). Right panel shows relative mRNA expression levels of 5 genes determined as common genes between 47 genes and 378 genes (center) throughout the wound healing process. (b-c) Representative images of unwounded and wounded skin tissue from TG mice treated with vehicle and tamoxifen stained to show expression of the (b) HBEGF (green), (c) pEGFR (red) and DAPI (blue). Scale bar = 50μm. (d) Immunoblots show EGFR and pEGFR expression in unwounded and wounded skin with vehicle or tamoxifen treated TG mice. Actin was used as a loading control. Bottom panels show quantification of each protein. n=4 for unwounded, n=8 for wounded. (e) qPCR analyses on chromatin samples from epidermis of tamoxifen treated TG mice after immunoprecipitation with anti-SOX2 and IgG control antibodies. n=3. Data expressed as means ± SEM. *P<0.05.
AREG and HBEGF are EGFR ligands which regulate a number of cellular processes such as proliferation, differentiation, migration, and survival via activation of EGFR signaling pathway (Singh et al., 2016). FAM83A activates downstream of EGFR signaling, independent of EGFR activation (Lee et al., 2012). Significantly heightened mRNA levels of AREG and HBEGF expression was confirmed by qPCR in each group, corroborating the trend found in the RNA-seq analysis (Supplementary Figure S4c). Interestingly, other EGFR ligands were also upregulated in SOX2-overexpressing mice when compared with control mice (Supplementary Figure S4d).
Immunofluorescence staining showed that HBEGF was highly expressed in keratinocytes in SOX2-overexpressing mice when compared to control mice in the unwounded and wounded conditions (Figure 4b). Moreover, we found that phospho (p)EGFR was also significantly upregulated in SOX2-overexpressing mice in the unwounded and wounded timepoints (Figure 4c and 4d and Supplementary Figure S4e).
It has been reported that SOX2 increases the expression of EREG and TGFA in hair follicle stem cells and cancer cells (Siegle et al., 2014), which lead us to test if SOX2 directly regulates transcription of EGF ligands in skin keratinocytes by ChIP-assay. Integrative analysis using the RNA-seq dataset and FIMO motif analysis revealed higher enrichment of SOX2 binding to the loci of AREG, HBEGF, EREG and TGFA. This was confirmed by ChIP-qPCR, where the targets showed approximately 9 fold-change when compared with IgG control (Figure 4e). These results suggest that in skin keratinocytes, SOX2 potentially directly regulates the expression of EGFR ligands resulting in the activation of EGFR signaling.
SOX2 in skin keratinocytes promotes proliferation and migration in vitro
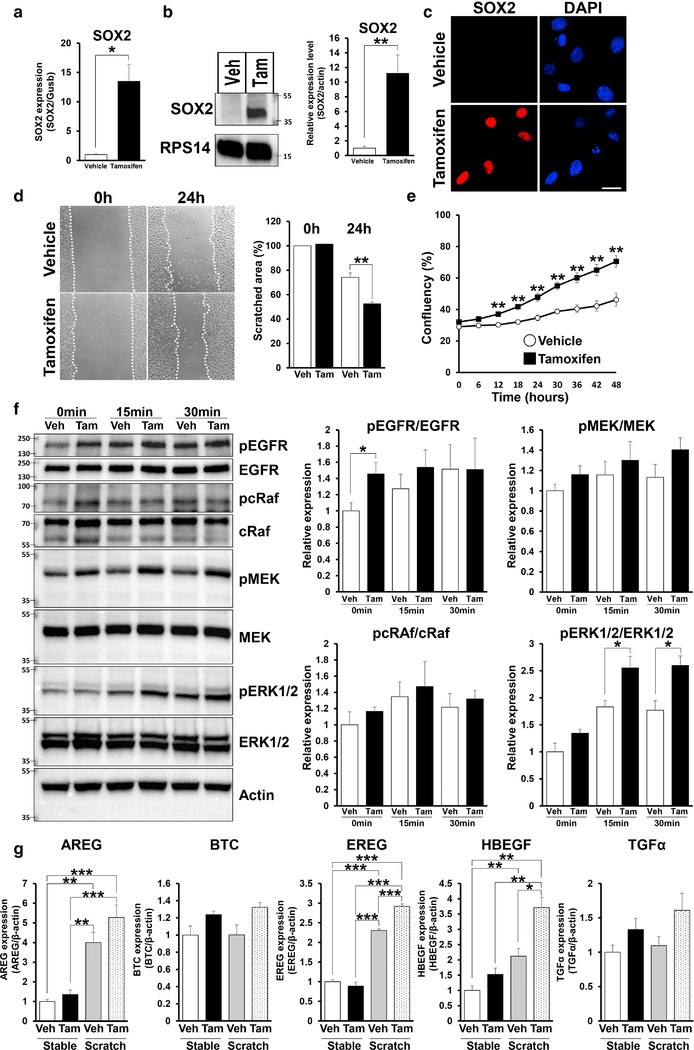
Next, we examined the effect of SOX2 expression in skin keratinocytes in vitro. We confirmed that mRNA and protein levels of SOX2 were significantly upregulated in tamoxifen treated primary keratinocytes derived from TG mice when compared with those treated with vehicle (Figure 5a–c). Scratch assays demonstrated that SOX2 expression in skin primary keratinocytes lead to increased cell migration (Figure 5d), and an increased the proliferative capacity (Figure 5e). We also showed that knockdown of SOX2 expression reverses the enhancement of cell movement in SOX2 expressing mouse keratinocytes (Supplementary Figure S5a and S5b). Next, we examined whether SOX2 affects the EGFR signaling pathway in vitro. Western blot analysis shows that pEGFR was significantly upregulated in SOX2 expressing keratinocytes in the basal state (Figure 5f). Moreover, pERK1/2 was significantly upregulated in SOX2 expressing keratinocytes at 15 and 30 minutes after scratching (Figure 5f).
Figure 5. SOX2+ skin primary keratinocytes promote migration, proliferation and enhance pEGFR via EGFR ligands expression.
(a) mRNA expression of SOX2 in TG mice keratinocytes treated by vehicle and tamoxifen for 48 hours. (b) Immunoblot images showing SOX2 expression in TG mice keratinocytes treated by vehicle or tamoxifen for 48 hours. RPS14 was used as a loading control. Right panel shows quantification of SOX2 protein. (c) Representative images of TG mice keratinocytes treated by vehicle and tamoxifen for 48 hours stained to show expression of SOX2 (red) and DAPI (blue). Scale bar = 20μm. (d) Migrating TG mice keratinocytes treated with vehicle and tamoxifen after scratch. Images were taken at 0 and 24 hours after scratch. Values were determined by relative migrating area in 3 microscopic fields in n=3 per group. (e) Proliferation assays with TG mice keratinocytes treated by vehicle and tamoxifen were quantified every 6 hours for 48 hours, n=4. (f) Immunoblot images showing expression of each protein after scratch (0 minute, 15 minutes and 30 minutes) in TG mice keratinocytes treated with vehicle or tamoxifen. Right panels show quantification of each protein. n=4. (g) Quantification of mRNA levels of expression of EGFR ligands genes, n=4. Data expressed as means ± SEM. ***P<0.001, **P<0.01, *P<0.05.
We also examined SOX2-regulated transcription of EGFR ligands in primary keratinocytes by qPCR. In the basal state, the expression of EGFR ligands was upregulated in SOX2 expressing keratinocytes when compared to control keratinocytes. After scratching, expression of AREG, EREG and HBEGF was significantly upregulated in both control and SOX2 expressing cells when compared to the basal state. In particular, expression of HBEGF and EREG was significantly increased in SOX2 expressing skin keratinocytes when compared with that in control after scratching (Figure 5g). These results indicate that SOX2 potentially modulates EGFR signaling pathway by regulating expression of EGFR ligands, resulting in promotion of cell migration.
Inhibition of EGFR signaling pathway suppresses enhancement of cell migration in SOX2 expressing skin keratinocytes
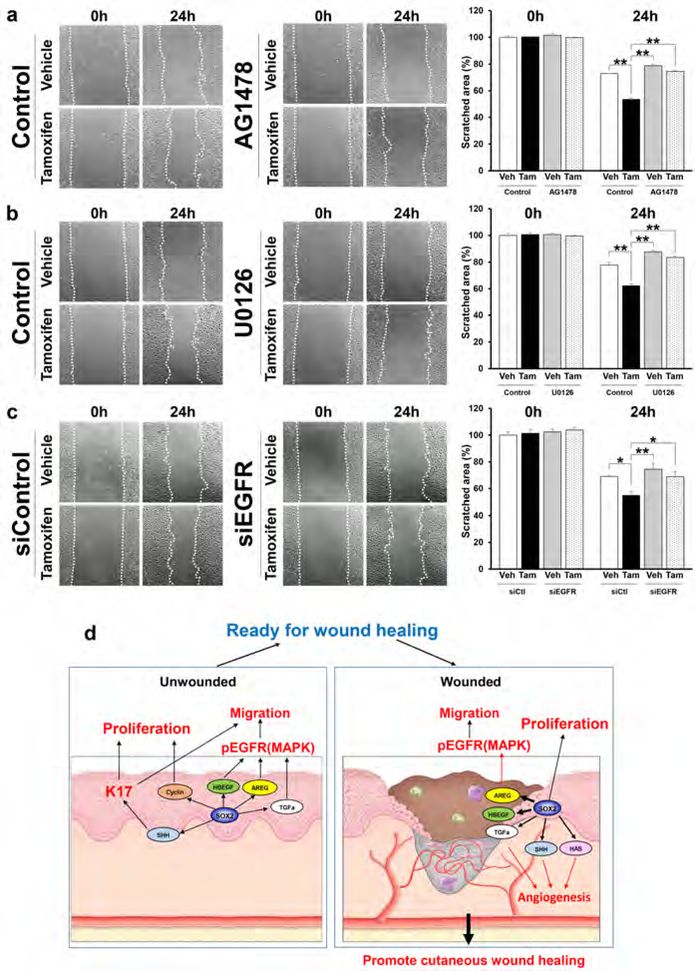
To determine the mechanism through which SOX2 enhances cell motility mediated by activation of the EGFR signaling pathway, we selectively blocked EGFR and ERK activation using an EGFR inhibitor (AG1478), a MEK inhibitor (U0126) and siEGFR (Supplementary Figure S6a). AG1478 (Figure 6a), U0126 (Figure 6b) and siEGFR (Figure 6c) significantly diminished the enhancement of cell migration by SOX2, indicating that SOX2 promotes the migration of keratinocytes via the EGFR/MEK/ERK signaling pathway. Taken together, our results provide mechanistic insight into the roles of SOX2 in epithelial keratinocytes in promoting cutaneous wound healing (Figure 6d).
Figure 6. Inhibition of EGFR/MEK/ERK signaling pathway suppresses cell migration in SOX2 expressing skin keratinocyte.
(a-c) Migrating primary keratinocytes treated with vehicle and tamoxifen in absence or presence of (a) 1uM AG1478, (b) 10μM U0126, and (c) siCtl and siEGFR. Images were taken at 0 and 24 hours after scratch. Values were determined by relative migrating area in 3 microscopic fields in n=3–4 per group. Data expressed as means ± SEM. **P<0.01, *P<0.05. (d) Schematic model summarizing the mechanistic roles of SOX2 in epithelial keratinocyte promote cutaneous wound healing.
DISCUSSION
This study investigates the mechanistic role of SOX2 in epidermal keratinocytes during cutaneous wound healing. We previously reported that the transcription factor SOX2 establishes a network that is efficacious in promoting wound healing (Iglesias-Bartolome et al., 2018). The endogenous expression of SOX2 in epithelia of buccal mucosa and palate confers intrinsic features to these tissues, which translates to accelerated wound healing when compared with skin.
In an effort to determine common shared mechanisms or pathways that accelerate wound healing in oral mucosa and SOX2-overexpressing skin, we utilized an animal model for conditional inducible cutaneous expression of SOX2 and performed gene expression analysis of unwounded and wounded skin, with and without SOX2 expression. We previously demonstrated that the oral mucosa has a unique transcriptional network that prime the epithelium for rapid wound healing in the basal state (Iglesias-Bartolome et al., 2018). This network is associated with psoriasis enriched genes which establish a wound-activated phenotype in the oral mucosa at basal state. Herein, transcriptomic analysis showed that SOX2 in epidermal keratinocyte induced hyperproliferation, wound-activated keratin expression and expression of genes associated with cell migration before wounding, suggesting that SOX2 function within the cutaneous keratinocyte coordinates the expression of a network of genes required for wound healing at basal state.
During the wound healing process, gene analysis revealed that cutaneous SOX2 resulted in increased expression of genes associated with proliferation of connective tissue and epithelium, cell movement and angiogenesis. While it is known that the keratinocytes migrating into the wound area do not have high proliferative capacity, SOX2+ keratinocytes maintain this capacity during migration, therefore potentially contributing to the promotion of re-epithelialization. It has been reported that keratinocytes and fibroblasts interact through cytokines or growth factors during wound healing (Werner et al., 2007), and that angiogenesis is regulated by various keratinocyte-derived growth factors, cytokines and ECM environment (Tonnesen et al., 2000). It has been reported that in fibroblasts, SOX2 regulates the expression of type I collagen using a fibrosis model in lung and skin (Chuang et al., 2018, Liu et al., 2014). However, SOX2 expression in skin keratinocytes did not increase the expression of type I collagen. Our analysis showed significantly increased expression of 29 genes associated with proliferation of connective tissue cells and 40 angiogenesis associated genes, including growth factors, cytokines, ECM and four EGFR ligand (AREG, BTC, HBEGF and TGFA), in SOX2-overexpresing mice during wound healing. There are seven ligands that bind to the EGFR: EGF, TGFA, HBEGF, BTC, AREG, EREG and EPGN (Singh et al., 2016). EGFR signaling plays an exceedingly important role in normal skin integrity and wound healing by regulating re-epithelialization, angiogenesis, migration and proliferation of fibroblasts and inflammation (Bodnar, 2013, Repertinger et al., 2004). This points to these factors being key molecules produced by SOX2+ keratinocytes for rapid wound repair. We present evidence that SOX2+ keratinocytes promote wound healing by regulating both epidermis and dermis through production of growth factors and ECM that induce the promotion of re-epithelialization and formation of granulation tissue via enhancement of proliferation of keratinocytes and fibroblast, cell movement, and angiogenesis in the wounded region.
Recent work has demonstrated that impaired EGFR signaling has been implicated in diabetic wounds (Berlanga-Acosta et al., 2017, Xu and Yu, 2011), and exogenous administration of soluble EGFR ligands, especially HBEGF, accelerated cutaneous wound healing in normal and diabetic mouse models (Johnston et al., 2013, Shirakata et al., 2005). These studies indicate that activation of EGFR signaling by EGFR ligands has the capacity to accelerate wound healing.
Through ChIP assays, we demonstrate that HBEGF, AREG, EREG and TGFA are potential molecular targets of SOX2. We showed that the SOX2-dependent upregulation of EGFR ligands led to enhancement of activation of the EGFR. Utilizing selective inhibition, we also show that enhancement of cell migration in SOX2+ keratinocytes is dependent on EGFR/MEK/ERK signaling.
Taken together, we demonstrate that SOX2 in epidermal keratinocytes altered the cellular phenotype to a wound-activated condition before wounding, allowing skin tissue to more quickly respond to injury, which mimicked the rapid wound resolution in the oral mucosa. After wounding, SOX2 promoted EGFR/MEK/ERK kinase signaling and increased expression of angiogenesis-associated genes resulting in enhancement of re-epithelialization, angiogenesis, and wound healing.
MATERIALS AND METHODS
The detailed protocols and statistical analysis are described in Supplementary Materials and Methods online.
Mice
All animal studies were carried out according to the protocol approved by the Animal Use and Care Committee at the National Institute of Arthritis and Musculoskeletal and Skin Diseases. K14CreERTM/LSL-SOX2 mice were generated by mating KRT14-cre/ERT/20Efu/JK14Cre mice (The Jackson Laboratory) and Rosa26CAG-loxp-stop-loxp-Sox2-IRES-Egfp mice (Lu et al., 2010).
Wound healing assay in vivo
Full-thickness wounds were created and examined as described previously (Zheng et al., 2007). To confirm the effect of SOX2 overexpression on cutaneous wound healing in mice, 5mg/ml of tamoxifen dissolved in ethanol 200μl or same volume of ethanol (as a control) was topically applied to K14CreERTM/LSL-SOX2 and WT mice dorsal skin for 5 consecutive days.
RNA preparation and RNA-seq data analysis
Total RNA from skin tissue and cells was extracted using RNeasy kit (Qiagen) according to the manufacturer’s instructions. mRNA expression profiling and analysis were performed as described in Supplementary Materials and Methods online in the NIAMS Genome Core Facility at the NIH.
Accession numbers
Raw and analyzed RNA-Seq data have been deposited in the Gene Expression Omnibus (GEO) site, accession number GSE118859.
Statistical analysis
P values were calculated using the Student’s t-test (two-sided) or by analysis of one-way ANOVA. Data analysis was done with GraphPad Prism version 7 (GraphPad Software Inc., San Diego, CA). Error bars represent standard errors of the mean and numbers of experiments (n) are as indicated.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the Intramural Research Programs of the National Institute of Arthritis and Musculoskeletal and Skin Diseases (M.I.M ZIA AR041124). Dr. Uchiyama was supported by a Japanese Society for the Promotion of Science Research Fellowship for Japanese Biomedical and Behavioral Researcher at NIH (KAITOKU-NIH). We thank members of the Laboratory of Skin Biology (LSB) and Dr. Ramiro Iglesias-Bartolome for helpful suggestions and discussions. We also thank Gustavo Gutierrez-Cruz, Stefania Dell’Orso and members of the NIAMS Genome Analysis Core Facility and Evelyn Ralston, Aster Kenea and members of the NIAMS Light Imaging Core Facility. We thank Marie-Liesse Asselin-Labat and Mark W. Onaitis (Moores Cancer Center, University California San Diego, La Jolla, CA, USA) for providing the Rosa26CAG-loxp-stop-loxp-Sox2-IRES-Egfp (LSL-SOX2) mice.
Abbreviations:
- ECM
extracellular matrix
- EGFR
epidermal growth factor receptor
- Tam
Tamoxifen
- Veh
Vehicle
- TG
transgenic
- WT
Wild type
- RNA-seq
RNA sequencing
- PCA
principal component analysis
- KRT
keratin
- MPO
myeloperoxidase
Footnotes
CRediT
Conceptualization: A. Uchiyama, J. S. Gutkind, M. Morasso.
Data Curation: A. Uchiyama, S. Brooks, M. Morasso.
Formal Analysis: A. Uchiyama, S. Brooks, M. Morasso.
Funding Acquisition: Intramural Research Programs of the National Institute of Arthritis and Musculoskeletal and Skin Diseases (M.I.M ZIA AR041124), Japanese Society for the Promotion of Science Research Fellowship for Japanese Biomedical and Behavioral Researcher at NIH (KAITOKU-NIH).
Investigation: A. Uchiyama, S. Nayak, R. Graf, M. Cross, K. Hasneen, S. Brooks, M. Morasso.
Methodology: A. Uchiyama, S. Nayak, M. Cross, S. Brooks, M. Morasso.
Project Administration: A. Uchiyama, M. Morasso.
Resources: A. Uchiyama, M. Morasso.
Software: A. Uchiyama, S. Nayak, S. Brooks, M. Morasso.
Supervision: M. Morasso.
Validation: A. Uchiyama, M. Morasso.
Visualization: A. Uchiyama, S. Brooks, M. Morasso.
Writing - Original Draft Preparation: A. Uchiyama, S. Nayak, R. Graf, M. Cross, J.S. Gutkind, M. Morasso.
Writing - Review and Editing: A. Uchiyama, M. Morasso.
CONFLICT OF INTEREST
The authors state no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, et al. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 2011;9(4):317–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai J, Takenaka H, Kusano KF, Ii M, Luedemann C, Curry C, et al. Topical sonic hedgehog gene therapy accelerates wound healing in diabetes by enhancing endothelial progenitor cell-mediated microvascular remodeling. Circulation 2006;113(20):2413–24. [DOI] [PubMed] [Google Scholar]
- Berlanga-Acosta J, Fernandez-Montequin J, Valdes-Perez C, Savigne-Gutierrez W, Mendoza-Mari Y, Garcia-Ojalvo A, et al. Diabetic Foot Ulcers and Epidermal Growth Factor: Revisiting the Local Delivery Route for a Successful Outcome. BioMed research international 2017;2017:2923759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar RJ. Epidermal Growth Factor and Epidermal Growth Factor Receptor: The Yin and Yang in the Treatment of Cutaneous Wounds and Cancer. Adv Wound Care (New Rochelle) 2013;2(1):24–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boumahdi S, Driessens G, Lapouge G, Rorive S, Nassar D, Le Mercier M, et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature 2014;511(7508):246–50. [DOI] [PubMed] [Google Scholar]
- Brem H, Tomic-Canic M. Cellular and molecular basis of wound healing in diabetes. J Clin Invest 2007;117(5):1219–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan CA, Ofstad T, Horng L, Wang JK, Zhen HH, Coulombe PA, et al. MIM/BEG4, a Sonic hedgehog-responsive gene that potentiates Gli-dependent transcription. Genes Dev 2004;18(22):2724–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang HM, Ho LI, Huang MH, Huang KL, Chiou TW, Lin SZ, et al. Non-Canonical Regulation of Type I Collagen through Promoter Binding of SOX2 and Its Contribution to Ameliorating Pulmonary Fibrosis by Butylidenephthalide. Int J Mol Sci 2018;19(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell RR, Giangreco A, Jensen KB, Mulder KW, Watt FM. Sox2-positive dermal papilla cells specify hair follicle type in mammalian epidermis. Development 2009;136(16):2815–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eming SA, Martin P, Tomic-Canic M. Wound repair and regeneration: mechanisms, signaling, and translation. Science translational medicine 2014;6(265):265sr6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro R, Valotta M, Ferri AL, Latorre E, Mariani J, Giachino C, et al. Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat Neurosci 2009;12(10):1248–56. [DOI] [PubMed] [Google Scholar]
- Garros-Regulez L, Garcia I, Carrasco-Garcia E, Lantero A, Aldaz P, Moreno-Cugnon L, et al. Targeting SOX2 as a Therapeutic Strategy in Glioblastoma. Frontiers in oncology 2016;6:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature 2008;453(7193):314–21. [DOI] [PubMed] [Google Scholar]
- Iglesias-Bartolome R, Uchiyama A, Molinolo AA, Abusleme L, Brooks SR, Callejas-Valera JL, et al. Transcriptional signature primes human oral mucosa for rapid wound healing. Science translational medicine 2018;10(451). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AP, Naska S, Jones K, Jinno H, Kaplan DR, Miller FD. Sox2-mediated regulation of adult neural crest precursors and skin repair. Stem Cell Reports 2013;1(1):38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano Y, Yoshimura K, Uchida G, Sato K, Harii K. Pretreatment with topical all-trans-retinoic acid is beneficial for wound healing in genetically diabetic mice. Archives of dermatological research 2001;293(10):515–21. [DOI] [PubMed] [Google Scholar]
- Lamagna C, Aurrand-Lions M, Imhof BA. Dual role of macrophages in tumor growth and angiogenesis. J Leukoc Biol 2006;80(4):705–13. [DOI] [PubMed] [Google Scholar]
- Lee SY, Meier R, Furuta S, Lenburg ME, Kenny PA, Xu R, et al. FAM83A confers EGFR-TKI resistance in breast cancer cells and in mice. J Clin Invest 2012;122(9):3211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Herault Y, Pavlovic G, Leask A. Skin progenitor cells contribute to bleomycin-induced skin fibrosis. Arthritis & rheumatology (Hoboken, NJ) 2014;66(3):707–13. [DOI] [PubMed] [Google Scholar]
- Lu Y, Futtner C, Rock JR, Xu X, Whitworth W, Hogan BL, et al. Evidence that SOX2 overexpression is oncogenic in the lung. PLoS One 2010;5(6):e11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markey AC, Lane EB, Macdonald DM, Leigh IM. Keratin expression in basal cell carcinomas. The British journal of dermatology 1992;126(2):154–60. [DOI] [PubMed] [Google Scholar]
- Mazzalupo S, Wong P, Martin P, Coulombe PA. Role for keratins 6 and 17 during wound closure in embryonic mouse skin. Dev Dyn 2003;226(2):356–65. [DOI] [PubMed] [Google Scholar]
- Pardue EL, Ibrahim S, Ramamurthi A. Role of hyaluronan in angiogenesis and its utility to angiogenic tissue engineering. Organogenesis 2008;4(4):203–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phi JH, Park SH, Paek SH, Kim SK, Lee YJ, Park CK, et al. Expression of Sox2 in mature and immature teratomas of central nervous system. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc 2007;20(7):742–8. [DOI] [PubMed] [Google Scholar]
- Repertinger SK, Campagnaro E, Fuhrman J, El-Abaseri T, Yuspa SH, Hansen LA. EGFR enhances early healing after cutaneous incisional wounding. J Invest Dermatol 2004;123(5):982–9. [DOI] [PubMed] [Google Scholar]
- Shirakata Y, Kimura R, Nanba D, Iwamoto R, Tokumaru S, Morimoto C, et al. Heparin-binding EGF-like growth factor accelerates keratinocyte migration and skin wound healing. J Cell Sci 2005;118(Pt 11):2363–70. [DOI] [PubMed] [Google Scholar]
- Siegle JM, Basin A, Sastre-Perona A, Yonekubo Y, Brown J, Sennett R, et al. SOX2 is a cancer-specific regulator of tumour initiating potential in cutaneous squamous cell carcinoma. Nat Commun 2014;5:4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer AJ, Clark RA. Cutaneous wound healing. The New England journal of medicine 1999;341(10):738–46. [DOI] [PubMed] [Google Scholar]
- Singh B, Carpenter G, Coffey RJ. EGF receptor ligands: recent advances. F1000Res 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpaderska AM, Zuckerman JD, DiPietro LA. Differential injury responses in oral mucosal and cutaneous wounds. Journal of dental research 2003;82(8):621–6. [DOI] [PubMed] [Google Scholar]
- Tokumaru S, Higashiyama S, Endo T, Nakagawa T, Miyagawa J-i, Yamamori K, et al. Ectodomain Shedding of Epidermal Growth Factor Receptor Ligands Is Required for Keratinocyte Migration in Cutaneous Wound Healing. The Journal of Cell Biology 2000;151(2):209–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonnesen MG, Feng X, Clark RA. Angiogenesis in wound healing. The journal of investigative dermatology Symposium proceedings 2000;5(1):40–6. [DOI] [PubMed] [Google Scholar]
- Turabelidze A, Guo S, Chung AY, Chen L, Dai Y, Marucha PT, et al. Intrinsic differences between oral and skin keratinocytes. PLoS One 2014;9(9):e101480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikramanayake TC, Stojadinovic O, Tomic-Canic M. Epidermal Differentiation in Barrier Maintenance and Wound Healing. Adv Wound Care (New Rochelle) 2014;3(3):272–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Yu FS. Impaired epithelial wound healing and EGFR signaling pathways in the corneas of diabetic rats. Investigative ophthalmology & visual science 2011;52(6):3301–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Fan X, Cui T, Dang E, Wang G. Nrf2 Promotes Keratinocyte Proliferation in Psoriasis through Up-Regulation of Keratin 6, Keratin 16, and Keratin 17. J Invest Dermatol 2017;137(10):2168–76. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Watanabe M, Kuraishi T, Hattori S, Kai C, Shibuya M. Chimeric VEGF-ENZ7/PlGF specifically binding to VEGFR-2 accelerates skin wound healing via enhancement of neovascularization. Arterioscler Thromb Vasc Biol 2007;27(3):50311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.