Abstract
Traits can evolve rapidly through changes in gene expression or protein-coding sequences. However, these forms of genetic variation can be correlated and changes to one can influence the other. As a result, we might expect traits lacking differential expression to preferentially evolve through changes in protein sequences or morphological adaptation. Given the lack of differential expression across the distribution of sidewinder rattlesnakes (Crotalus cerastes), we tested this hypothesis by comparing the coding regions of genes expressed in the venom gland transcriptomes and fang morphology. We calculated Tajima's D and FST across four populations comparing toxin and nontoxin loci. Overall, we found little evidence of directional selection or differentiation between populations, suggesting that changes to protein sequences do not underlie the evolution of sidewinder venom or that toxins are under extremely variant selection pressures. Although low-expression toxins do not have higher sequence divergence between populations, they do have more standing variation on which selection can act. Additionally, we found significant differences in fang length among populations. The lack of differential expression and sequence divergence suggests sidewinders—given their generalist diet, moderate gene flow and environmental variation—are under stabilizing selection which functions to maintain a generalist phenotype. Overall, we demonstrate the importance of examining the relationship between gene expression and protein-coding changes to understand the evolution of complex traits.
Keywords: balancing selection, stabilizing selection, genetic differentiation, SNPs, ecological specialization theory, transcriptomics
1. Background
Traits involved in antagonistic coevolutionary interactions can evolve rapidly under strong selective pressures [1–3]. Selection can act through changes in gene expression or through mutational changes to the genes themselves, altering the structure and function of the proteins underlying the trait. These forms of genetic variation on which selection can act can be highly correlated: high-expression genes tend to show lower rates of coding sequence evolution due to their importance in producing the focal phenotype [4]. Therefore, to understand the evolution of traits at the heart of coevolutionary interactions, differences in both coding sequence evolution and gene expression should be examined.
Snake venom is a rapidly evolving polygenic trait in which significant changes to both gene expression and protein-coding sequences have been observed [5–7]. Snake venom exhibits considerable variation at both intra- and interspecific levels, which is largely believed to be the result of local adaptation to either prey or predator resistance, potentially resulting in coevolutionary arms races [7–11]. Importantly, despite being polygenic, venom is the result of a relatively straightforward pathway connecting the expression of venom genes to the venom proteins and ultimately the venom phenotype [12,13]. This linear genotype to phenotype pathway allows variation at both levels (i.e. expression and sequence evolution) to directly affect venom functionality, making it simple to simultaneously study the effects of selection on both protein-coding sequences and expression of toxins. Positive selection is frequently detected in toxin genes in a variety of venomous snake species [5,6,14,15]. Frequent toxin gene duplication has facilitated neo- and sub-functionalization and led to the formation of large toxin gene families [5,15,16]. However, recent research at the intraspecific level suggests that snake venom evolution might be driven by changes in toxin gene expression (i.e. toxin quantity) rather than coding sequence evolution (i.e. toxin function) [7,17–20]. The architecture of venom allows expression variation to occur without significant deleterious effects to other genes (i.e. pleiotropy), allowing for rapid evolutionary adaptation to coevolutionary interactions [21–23]. Toxin expression differences are known in several venomous snake systems. For example, mainland and insular populations of eastern diamondback rattlesnakes (Crotalus adamanteus) display differential expression that results in local adaptation to the resistance phenotype of sympatric prey [7]. High-expression toxins are considered vital for venom functionality (i.e. general prey killing) and therefore evolve under strong purifying selection across populations [7,24,25]. Meanwhile, low-expression toxins can exhibit prey-specific functions over relatively short time scales, and therefore have higher expression variation and differentiation in response to changes in prey resistance [7,24,25].
Undoubtedly, selection on both toxin function and quantity have influenced the evolution of snake venom, as evidenced by the high variability exhibited by numerous species [26–28]. However, most tests for positive selection have examined multiple species rather than populations of a single species. Similarly, some species have similar venom expression profiles across large geographical scales, displaying little evidence of differential expression [17,29]. The lack of differential expression suggests that the venoms of these species are under different selection pressures than those exhibiting high intraspecific expression variation. For example, eastern coral snakes (Micrurus fulvius) lack variation in toxin expression across their distribution, and although there is evidence of positive selection on toxin sequences when compared to other coral snake species, no study has examined intraspecific sequence variability [17,30]. Examining the coding sequences of toxins in species that lack differential expression patterns can highlight the role of selection on protein-coding regions to test if there is a relationship between toxin expression and sequence divergence, and how variation in one form of genetic variation (or lack thereof) can influence the other.
The sidewinder rattlesnakes (Crotalus cerastes) is a small (approx. 40 cm) rattlesnake native to warm deserts of the southwestern United States and northwestern Mexico (figure 1) [31,32]. Hofmann et al. [29] used venom-gland transcriptomics to test for evidence of differential expression between subspecies, mitochondrial lineages and lineages inferred from loci in the transcriptome. Unlike the extreme differences found in venom expression profiles of some sympatric rattlesnake species—including Mojave rattlesnakes, C. scutulatus [33,34] and southern Pacific Rattlesnakes, C. helleri [35,36]—few genes were differentially expressed between lineages of sidewinders, and overall venom expression was similar across their range [29]. The lack of differential expression among toxin genes could be a product of a relatively flat adaptive landscape and stabilizing selection functioning to maintain a generalist venom arsenal [37–39]. Sidewinders prey on a variety of small mammals and lizards, and there is no evidence of geographical variation in their diet, supporting this hypothesis [40,41]. Alternatively, if there is a relationship between toxin expression and sequence divergence, changes to sidewinder venom could instead occur via changes to the protein sequences as they respond to an adaptive landscape. A single case of neurotoxicity in sidewinder venom [42], together with data showing that expression and sequence evolution are highly correlated [4], favour this hypothesis.
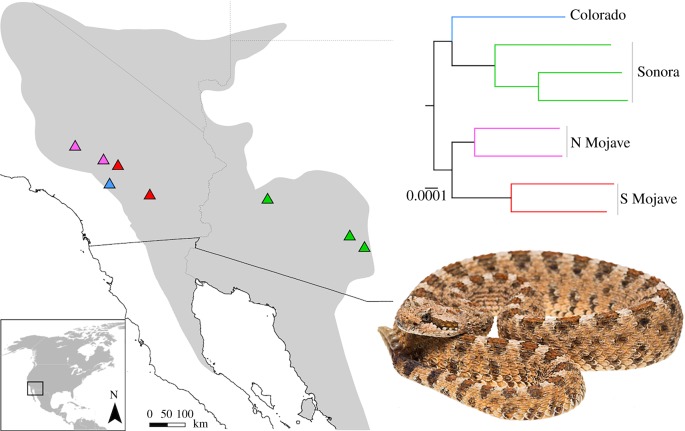
Figure 1.
Distribution of sidewinder rattlesnakes (Crotalus cerastes), and localities of samples used herein. Sample colours correspond to lineage assignments based on phylogenetic analyses of 1508 nontoxin loci (approx. 1.4 million bp) [28] (upper right). Inset photo of C. cerastes by Travis Fisher. (Online version in colour.)
Here, we investigate the role sequence divergence has played in driving the evolution of sidewinder rattlesnake venom. Specifically, because sidewinder rattlesnakes lack considerable differential expression of toxin genes, we hypothesize that their venoms have diverged in protein sequences. Secondarily, because high-expression genes are constrained by selection to ensure translational efficiency and production of the focal phenotype, they tend to have lower rates of sequence evolution [43]. Therefore, we hypothesize that sequence divergence occurs mainly in low-expression proteins. To test these hypotheses, we use recently characterized venom gland transcriptomes and compare several metrics of toxin sequence divergence to a larger set of nontoxin genes among lineages of sidewinder rattlesnakes. If positive selection and changes in toxin function underlie the evolution of sidewinder venom, then we expect toxins will have significantly higher rates of coding sequence evolution than nontoxins, with nonsynonymous mutations being highly differentiated between populations. Alternatively, if sidewinders have a generalist venom phenotype evolving via stabilizing selection, then we expect toxins to have lower or similar rates of coding sequence evolution to nontoxins. In addition to examining the venom phenotype, we use museum specimens to test for differences in fang morphology between lineages to fully investigate the potential for local adaptation in this integrated feeding phenotype.
2. Methods
(a). Variant calling
To examine toxin sequence evolution in sidewinder rattlesnakes, we used samples and venom-gland transcriptomes generated by [29]. First, to assess the role of sequence variation on venom evolution, we called single-nucleotide polymorphisms (SNPs) using a combination of BWA-MEM, Picard and GATK (electronic supplementary material, S1).
(b). Sequence evolution
To summarize the variant data, we determined whether each SNP was synonymous or nonsynonymous, calculated the number of SNPs per kilobase, calculated nucleotide diversity (π), and tested for differences between toxins and nontoxin loci (electronic supplementary material, S1).
We tested for evidence of selection by calculating Tajima's D and Weir and Cockerham's FST across all individuals and populations, and then tested for differences in means estimates between toxins and nontoxins (electronic supplementary material, S1). To account for differences in sample sizes between toxins and nontoxins, we randomly sub-sampled nontoxins to toxin sample size and performed 1000 bootstrap replicates of the regression analyses. The proportion of the bootstraps (b) which detected significance (p < 0.05) is reported alongside the results from the full dataset.
To look for specific toxins under selection, we generated a null distribution from the nontoxin transcripts and looked for toxin outliers outside the 95th percentile. Additionally, to test if low-expression toxins were under stronger positive or balancing selection pressures, we used linear regression to correlate average toxin expression [29] to estimates of Tajima's D and FST. We tested positive and negative values of Tajima's D separately to account for potential differences in expression related to alternate selection pressures.
Lastly, we tested for differentiation between lineages using pairwise FST calculated for each locus between the four phylogeographic lineages (i.e. Colorado, Sonora, North Mojave, South Mojave), and analyses were performed as above (electronic supplementary material, S1). To maximize our ability to detect evidence of sequence differentiation, we combined lineages into larger clades and re-performed FST analyses. Larger clades were based on the phylogeny recovered by [29] or on geography (electronic supplementary material, S1).
(c). Fang morphology
To characterize variation in fang length among sidewinder lineages, we measured formalin-preserved specimens housed at the Museum of Vertebrate Zoology at Berkeley (MVZ), the California Academy of Sciences (CAS) and the University of Illinois Museum of Natural History (UIMNH). Specimens were assigned to lineages based on geography, with geographically ambiguous samples removed prior to analysis. Sampling included specimens from the North Mojave (n = 9), South Mojave (n = 12), Colorado (n = 19) and Sonora (n = 14) lineages. The left and right fangs were measured from tip to base using digital callipers, and the average of the left and right measurements was used for statistical analysis. To control for the effect of individual snake body size on fang length, we also measured the head length of each snake. We measured head length using the ‘straight’ tool in ImageJ v. 1.52a [44] using dorsal photographs of the head taken over a 0.5 cm grid. By dividing fang length by head length, we obtained a ratio describing fang size relative to total head size. We tested for an effect of sidewinder lineage on the relative fang lengths using a one-way ANOVA, and compared individual lineages with Tukey's HSD test.
3. Results
(a). Sequence variation
The majority of toxin and nontoxin transcripts had one or more SNPs (table 1). However, toxins maintained a significantly higher number of SNPs per kb (R2 = 0.01, p < 0.01, b = 0.93) and there was a significant association between the type of mutation and whether the transcript was a toxin or nontoxin; toxins showed significantly more nonsynonymous variants (X2 = 175.31, d.f. = 1, p < 0.01; table 1) and maintained significantly higher values of nucleotide diversity (π; R2 < 0.01, p = 0.04, b = 0.20; table 1) than nontoxins.
Table 1.
Sequence variation results between toxins and nontoxins including the number of transcripts found to have SNPs, the number of SNPs by mutation type, SNPs per kilobase (kb) and nucleotide diversity (π).
| transcripts | SNPs | synonymous | nonsynonymous | SNPs/kb | π | |
|---|---|---|---|---|---|---|
| nontoxin | 1777 (90%) | 9423 | 6893 (73%) | 2530 (27%) | 5.47 ± 3.43 | 0.26 ± 0.09 |
| toxin | 58 (94%) | 392 | 166 (42%) | 226 (58%) | 7.69 ± 4.20 | 0.28 ± 0.10 |
(b). Signatures of selection
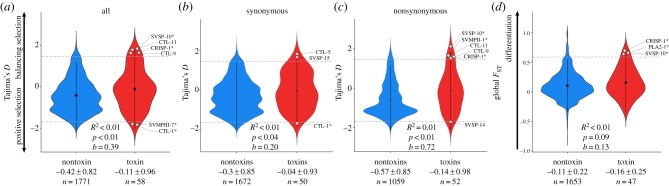
To test for signatures of selection, we first calculated Tajima's D across all eight individuals. On average, estimates of Tajima's D for nontoxin transcripts were found to be significantly different from zero (, t = −21.53, d.f. = 1770, p < 0.01; figure 2a). However, estimates of Tajima's D for toxins were not significantly different from zero (, t = −0.87, d.f. = 57, p = 0.39), suggesting toxins are evolving along the mutation–drift equilibrium with little evidence of selection or that toxins undergo highly variable selection pressures (figure 2a). Estimates of Tajima's D between toxins and nontoxins were significantly different (R2 < 0.01, p < 0.01, b = 0.39), providing further evidence that toxins and nontoxins are evolving differently with nontoxins—unexpectedly—evolving under stronger positive selection (figure 2a).
Figure 2.
Violin plots of Tajima's D for (a) all mutations, and separately for (b) synonymous mutations and (c) nonsynonymous mutations. (d) Weir and Cockerham's FST estimates from nontoxin and toxin transcripts of all eight sidewinders (Crotalus cerastes). Average nontoxin estimates were significantly different than toxins for estimates of Tajima's D, but were not significantly different for estimates of FST. Both metrics had low bootstrap support for significance, with the majority of the signal for Tajima's D coming from nonsynonymous mutations. Nontoxins were used as a null distribution to identify toxin transcripts that might be evolving under strong positive or balancing selection. Toxins were under significant selection if they were located outside the 95th percentile of nontoxin transcripts indicated by the dotted grey line and white diamonds. Asterisks next to toxins indicate that the toxin was proteomically verified by [28]. (Online version in colour.)
We then calculated Tajima's D separately for nonsynonymous and synonymous mutations. We found that, as expected, nonsynonymous mutations are under stronger positive selection than synonymous mutations (figure 2b,c). However, given that toxins have a mean estimate closer to zero for both types of mutations, toxins are either not evolving under strong selection pressures or are under more variable selection pressures which neutralizes the signal in the mean value. Additionally, nonsynonymous mutations appear to be driving the signal of outliers detected in the analysis across all SNPs.
Although we found no evidence of selection across all toxin loci, we did not expect all toxins to be evolving equally. Therefore, we used the nontoxins as a null distribution to identify individual toxin outliers that might be evolving under positive or balancing selection. Based on the sample size of toxins used in our outlier analyses, we would expect to identify between two and three outliers by chance (5%). In this analysis of Tajima's D, we identified six toxin transcripts that fell outside the 95th percentile generated from the nontoxin distribution (figure 2a). Only CTL-1 and SVMPIII-7 were found to be under significant positive selection, while CRISP-1, CTL-9, CTL-11 and SVSP-10 were found to be under significant balancing selection (electronic supplementary material, table S1).
Next, we calculated FST across all lineages. Differentiation in toxins relative to nontoxins was used to represent signatures of divergent selection among lineages. On average, estimates of FST for nontoxins were not significantly different than toxins (R2 < 0.01, p = 0.09, b = 0.13) with toxins maintaining a slightly higher average (0.16 ± 0.25) than nontoxins (0.10 ± 0.22; figure 2d). Using the nontoxins as a null distribution, we identified three toxin transcripts that fell outside the 95th percentile: CRISP-1, PLA2-1 and SVSP-10 (figure 2d; electronic supplementary material, table S1). Only CRISP-1 and SVSP-10 were shared between Tajima's D and FST analyses (electronic supplementary material, table S1).
(c). Effect of toxin expression on sequence evolution
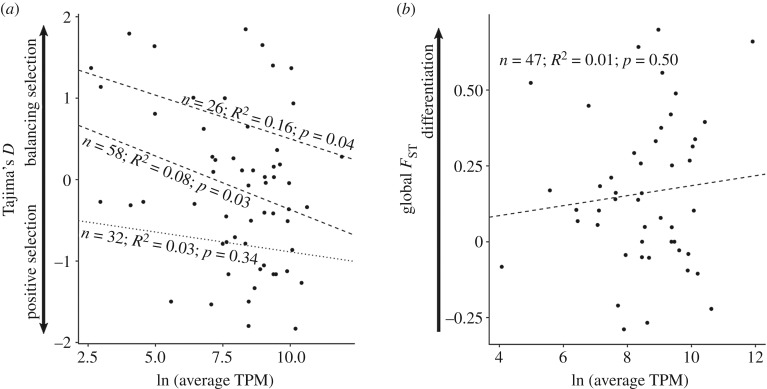
To test our prediction that toxin sequence divergence was inversely related to expression, we used average expression level of toxins estimated by transcripts per million reads (TPM) [29]. First, we performed a linear regression of average TPM and Tajima's D and found that low-expression toxins are more likely to be evolving via balancing selection (n = 58, R2 = 0.08, p = 0.03; figure 3a). Given that positive and negative values of Tajima's D suggest alternate forms of selection (balancing and positive selection, respectively), we tested positive and negative values of Tajima's D separately for trends with expression. The trend toward balancing selection for low-expression transcripts remained significant when analysing only positive values of Tajima's D (n = 26, R2 = 0.16, p = 0.04; figure 3a). However, expression level did not predict the strength of positive selection as there was not a significant trend when analysing only negative values of Tajima's D (n = 32, R2 = 0.03, p = 0.34; figure 3a). Similarly, toxins found to be differentially expressed by Hofmann et al. [29] did not have significantly different values of Tajima's D than toxins not differentially expressed (n = 58, R2 < 0.01, p = 0.46). High-expression toxins are constrained by selection to ensure translational efficiency and produce the focal phenotype [43]. Our results support that low-expression toxins have higher sequence variation and evolve via balancing selection.
Figure 3.
Relationship of (a) Tajima's D and (b) Weir and Cockerham's FST with the average toxin expression calculated by log-transformed transcripts per million (TPM) in [28]. Tajima's D was analysed in three ways: we analysed all toxins together (middle line), and then separately analysed values greater than (upper line) and less than (lower line) zero. Tajima's D shows a significant correlation when considering all toxins or toxins found to have a positive Tajima's D value.
We then performed regressions of average TPM with global FST and found that this approximation of sequence evolution was not correlated to expression (FST: n = 47, R2 = 0.01, p = 0.50; figure 3b). Similarly, toxins found to be differentially expressed by Hofmann et al. [29] did not have significantly different values of FST (n = 47, R2 = 0.05, p = 0.15). These results suggest that toxin expression and sequence divergence are not correlated, rejecting our hypothesis that low-expression toxins have higher sequence divergence.
(d). Population and toxin differentiation
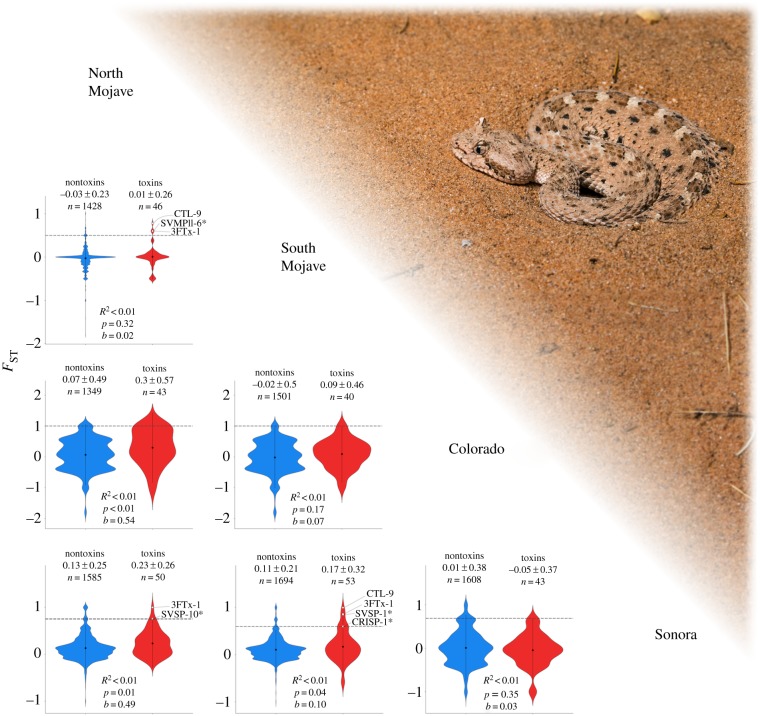
We estimated FST between phylogeographic lineages using the nontoxin transcripts to assess population differentiation, and calculated FST between toxin transcripts to determine if toxins maintained higher levels of sequence divergence. Overall, there is little genetic differentiation within Crotalus cerastes even between the most geographically distant lineages (i.e. North Mojave versus Sonora; figures 1 and 4). The mean nontoxin FST estimate between the North Mojave and Sonoran lineages (FST = 0.13) indicates only moderate differentiation with all other pairwise comparisons having little to no genetic differentiation (figure 4). Mean estimates of FST between phylogeographic lineages indicate that toxins and nontoxins exhibit similar patterns of divergence with geographically distant populations maintaining higher differentiation of both toxins and nontoxins (figure 4). However, Mantel tests based on the average geographical point of our samples for each population found no correlation between geographical distance and FST for either toxins (r = −0.04, p = 0.46) or nontoxins (r = 0.63, p = 0.12). Mantel tests based on the average branch length between populations similarly found nonsignificance (nontoxins: r = 0.45, p = 0.17; toxins: r = 0.15, p = 0.33). Overall, toxins tended to have higher genetic differentiation than nontoxins (figure 4), but this difference was only significant when comparing the North Mojave and Sonora lineages, the North Mojave and Colorado lineages, and South Mojave and Sonora lineages (p < 0.05; figure 4).
Figure 4.
Violin plots showing the pairwise Weir and Cockerham's FST estimates for toxins and nontoxins between the four lineages identified by [28]. Nontoxins were used as a null distribution to identify toxin transcripts that might be evolving under strong selection. Toxins were under significant selection if they were located outside the 95th percentile of nontoxin transcripts indicated by the dotted grey line and white diamonds. Asterisks next to toxins indicate that the toxin was proteomically verified by [28]. Photo of C. cerastes by Travis Fisher. (Online version in colour.)
We used nontoxins as a null distribution in each population comparison to determine which toxins had significant sequence divergence; toxins outside of the 95th percentile were considered significantly divergent. No toxin transcripts exceeded the 95th percentile of the null distribution between the Colorado lineage and either Mojave lineage because the 95th percentile encompassed the maximum value (FST = 1) (figure 4). Nonetheless, a total of nine toxin alleles were found to be fixed between these populations (electronic supplementary material, table S1). The remaining four comparisons recovered several significantly differentiated toxins (figure 4). A total of six unique toxins were found to be significantly divergent between lineage comparisons (figure 4). 3FTx-1 and CTL-9 were the only two toxins found to be significantly divergent in multiple lineage comparisons (figure 4). The comparison of the South Mojave and Sonoran lineage individuals recovered the largest number of significant toxin differentiation with four toxins: 3FTx-1, CRISP-1, CTL-9 and SVSP-1 (figure 4).
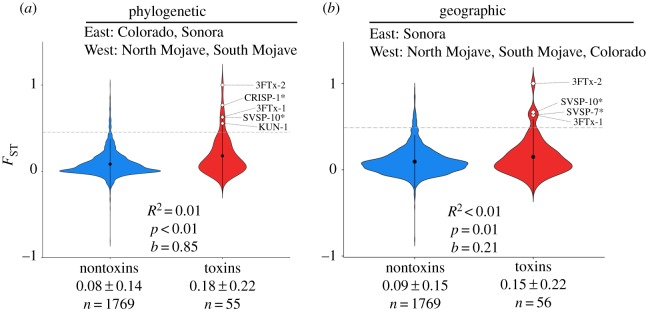
Lastly, to maximize our ability to detect evidence of sequence differentiation, we combined populations based on the phylogeny recovered by Hofmann et al. [29] and based on geography. Based on both phylogeny and geography, respectively, we found that toxin and nontoxin estimates were significantly differentiated (R2 = 0.01, p < 0.01, b = 0.85; R2 < 0.01, p = 0.01, b = 0.21) with toxins maintaining a higher differentiation (FST = 0.18; FST = 0.15) than nontoxins (FST = 0.08; FST = 0.09; figure 5). Based on the phylogeny, five toxins exhibited significant differentiation (figure 5a), while geography resulted in only four toxins (figure 5b). 3FTx-1, 3FTx-2 and SVSP-10 were shared between these two analyses, but CRISP-1 was unique to the phylogenetic grouping, and SVSP-7 was unique to the geographical grouping.
Figure 5.
Violin plots and mean estimates of pairwise Weir and Cockerham's FST between phylogenetic and geographical East and West clades. Nontoxins were used as a null distribution to identify toxin transcripts that might be evolving under strong selection. Toxins were under significant selection if they were located outside the 95th percentile of nontoxin transcripts indicated by the dotted grey line and white diamonds. Asterisks next to toxins indicate that the toxin was proteomically verified by [28]. (Online version in colour.)
(e). Selection on individual toxins
We identified several toxins under significant selection pressures, yet few were shared across tests (electronic supplementary material, table S1). Additionally, further examination of the alleles for these loci found little evidence of allele fixation for a given population (electronic supplementary material, table S2). Toxins 3FTx-1, CRISP-1, CTL-9 and SVSP-10 were significant across multiple tests (electronic supplementary material, table S1). However, it is important to note that neither 3FTx-1 nor CTL-9 were verified proteomically [29], and SNPs in 3FTx-1 induced nonsense mutations (electronic supplementary material, table S2). CRISP-1 and SVSP-10 are only moderately expressed in the transcriptome, but displayed some phylogenetic patterns with regard to the SNPs present (electronic supplementary material, table S2). For example, CRISP-1 contained two fixed differences—one synonymous and one nonsynonymous—between the East and West clades (electronic supplementary material, table S2). Although the former probably does not affect venom function, the latter substitution should. The CRISP proteins are expressed widely across venomous snakes, but their function is not well-understood, as they have demonstrated a variety of biological effects [45,46]. Although CRISP-1 is only moderately expressed in the venom, the nonsynonymous SNP might impact the folding of the protein and function of the venom in each population's respective environment. Beyond these few examples, however, we found little evidence of sequence divergence or changes in toxin function involved in the evolution of sidewinder rattlesnake venom.
(f). Fang morphology
We used head-size-adjusted relative fang lengths to test for variation in morphology of the venom delivery system among sidewinder lineages. Relative fang length varied significantly among lineages (F3,50 = 5.0, p = 0.004, electronic supplementary material, figure S1A). The North Mojave lineage had significantly smaller fangs than both the Colorado (p = 0.01) and Sonora lineages (p < 0.01), while all other pairwise comparisons of mean relative fang length were not significantly different (p > 0.1; electronic supplementary material, figure S1A). Notably, lineages that differed significantly in their fang morphology were also those with the highest FST values for toxin sequences (electronic supplementary material, figure S1B). In fact, toxin sequence differentiation and fang morphology appear to be positively correlated (electronic supplementary material, figure S1B). Although this trend is not significant (R2 = 0.57, p = 0.08), the correlation is significantly different than that of nontoxins (Anova: p = 0.02; electronic supplementary material, figure S1B).
4. Discussion
Venom is often involved in antagonistic interactions, leading to rapid evolutionary responses via changes in gene expression (i.e. toxin quantity) or through mutational changes to the protein sequences themselves (i.e. toxin function). Both toxin quantity and sequence evolution influence the overall evolution of venom in snakes, but these forms of genetic variation might be correlated [4,47]. In this study, we used sidewinder rattlesnakes (Crotalus cerastes) as a model to investigate the role sequence divergence and changes in toxin function play in the evolution of a venom with little differential expression. We hypothesized that toxin expression and sequence divergence are inversely related, expecting to find high sequence divergence across populations.
Instead, we found little evidence of positive or balancing selection acting on toxin loci when compared to nontoxins in the venom-gland transcriptomes of sidewinders. On average, toxins displayed weaker signals of positive selection than nontoxins, and estimates of Tajima's D were not significantly different from zero suggesting either a lack of selection or diverse selection pressures acting on toxins. Similarly, the global toxin FST estimate was not significantly different than the nontoxin FST. Importantly, although significance was detected (p < 0.05) in some analyses of average values, standard deviations between toxins and nontoxins overlapped heavily and the effect size (R2) was ubiquitously low. These results suggest weak differences in Tajima's D and FST metrics between toxins and nontoxins. Additionally, detection of significance was probably a result of the discrepancy in sample size between toxins and nontoxins since bootstrap resampling generally showed low support (figures 2 and 4).
As we did not expect all toxins to evolve similarly, we tested for outliers based on the 95th percentile of nontoxins. Given our sample size of toxins, we expected to find two to three outliers by chance (5%). We detected three or more toxin outliers under positive and balancing selection suggesting that toxins are under diverse selection pressures. In our analysis of average differences between toxins and nontoxins, these outliers probably provided strong conflicting signals of selection which might have influenced our results—particularly Tajima's D. Therefore, using average values might not have provided informative differences between toxins and nontoxins in some tests. Looking for outliers at the tails of the distributions probably provided more informative results for detecting selection. Nonetheless, few toxin outliers were shared across tests which further supports that toxins are under diverse selection pressures. Better statistical analyses could be done with a larger sample size to examine venom variation across the entire distribution.
We hypothesized that low-expression toxins would have greater sequence divergence as a result of positive selection or relaxed purifying selection. Although we did not find evidence of a relationship between expression and positive selection, we did find a significant relationship between expression level and balancing selection. Low-expression toxins displayed evidence of balancing selection, suggesting higher standing sequence variation. Margres et al. [25] found that low-expression toxins have more standing expression variation between populations than high-expression toxins because high-expression toxins are constrained by selection to provide generic killing functions. Together, our results suggest that low-expression toxins tend to be more variable at the population level. This variation, on which selection can now act, probably contributes more heavily to the evolution of venom than high-expression toxins, at least intraspecifically.
Our investigation of relative fang length among Sidewinder lineages revealed variation in fang size concordant with the highest amounts of divergence in toxin sequences. These results conform to previous analyses in Mojave rattlesnakes (C. scutulatus) and eastern diamondback rattlesnakes (C. adamanteus) that demonstrated fang length variation to be associated with population-level variation in venom composition [18,28]. Thus, our work adds to a growing body of evidence suggesting that fang length and venom composition are part of a functionally integrated phenotype for capturing prey. Margres et al. [18] hypothesized that longer fang length might be associated with higher expression of myotoxins. As myotoxins cause hind-limb paralysis in mammals, longer fangs might increase envenomation efficiency. In support of this hypothesis, the population of sidewinders with the longest fangs (Sonora) also exhibited highest myotoxin expression [29].
Overall, our results strongly suggest that sidewinder rattlesnake toxins are not under strong positive selection pressures and are not highly differentiated between populations. Therefore, changes in toxin function do not appear to underlie the evolution of sidewinder venom, contrary to our primary hypothesis. Instead, nontoxins and toxins appear to evolve at similar rates, suggesting that sidewinders have evolved a generalist venom arsenal as a result of stabilizing selection. In support of this result, Margres et al. [39] found that—for generalist species—venom only diverges in the absence of gene flow and that spatial environmental variation favours generalist phenotypes via stabilizing selection, consistent with ecological specialization theory [37].
Sidewinders prey on lizards and small mammals, and dietary studies have shown a cosmopolitan diet lacking geographical variation across the species range [40,41]. Sidewinders also have a large distribution across multiple deserts or environmental conditions further supporting ecological specialization theory [37,39]. Additionally, sidewinders exhibit moderate to high gene flow between populations and across the variable environments they inhabit (figure 4). Gene flow across environments can swamp signals of adaptation to local conditions and might increase the complexity of venom as populations share alleles, particularly for low-expression toxins. Sidewinder venom is composed of 62 putative toxins, the majority of which are moderately to highly expressed across its distribution [29]. This complexity might have evolved in response to a wide variety of ecological conditions and to facilitate a generalist diet.
Taken together, our results support the maintenance of a generalist venom arsenal in sidewinder rattlesnakes. Unlike sympatric species such as Mojave rattlesnakes, which appear to evolve under strong diversifying selection [28,33], the generalist venom of sidewinder rattlesnakes probably evolves along a relatively flat adaptive landscape void of significant differential expression and sequence divergence. In this case, the mild individual variation we observe in both expression and sequence divergence is probably the result of balancing selection and this variation is favoured by stabilizing selection to maintain a generalist venom phenotype [37–39]. Selection to maintain a generalist diet—rather than specialize—probably stabilizes sidewinder venom composition, despite populations diverging an estimated 1.5 Myr ago [48]. In comparison, the Mojave and Sonoran populations of Mojave rattlesnakes which show significantly different venom profiles are estimated to have diverged approximately 0.5–1 Myr ago [28,33,49]. Most venomous snakes demonstrate evidence of strong positive selection in the form of changes to gene expression or to the underlying coding sequence [5–7]. However, we have found no evidence of strong differentiation in either expression or sequence divergence in sidewinder rattlesnakes. Although snake venom often evolves rapidly due to antagonistic coevolutionary interactions with prey resistance, stabilizing selection and gene flow can alternatively function to produce generalist phenotypes that allow species to span a variety of environmental conditions and feed on a similarly generalist diet.
5. Conclusion
Evidence of positive selection is nearly ubiquitous in venom, and few systems exist where significant changes to either the coding sequence or gene expression are not occurring. Here we investigated sequence divergence in the venom of sidewinder rattlesnakes (Crotalus cerastes). Surprisingly, coding sequence and gene expression divergence appear to contribute very little to the evolution of sidewinder venom. Overall, the venom appears remarkably similar across the distribution sampled in the United States and—given its generalist diet, moderate to high gene flow and environmental variation—is probably under the influence of stabilizing selection which functions to maintain a generalist phenotype, rather than involved in strong antagonistic coevolutionary interactions.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Carol Spencer at MVZ, Lauren Scheinberg at CAS, and Chris Phillips and Dan Wylie at UIMNH for access to specimens. Clemson University provided computational resources on their Palmetto Cluster.
Data accessibility
Raw Illumina sequences and transcriptome assemblies are available in Genbank under accession numbers SRA: SRR6768682-SRR6768689; TSA: GGMJ00000000.
Authors' contributions
Conceptualization: R.M.R., E.P.H., D.R.R., C.L.P.; sampling: J.L.S., R.M.R.; sequencing: J.L.S., A.J.M.; data processing and analysis: R.M.R., E.P.H., J.L.S., A.J.M., M.L.H., D.R.R.; writing: R.M.R., E.P.H., M.J.M., M.L.H.; review and editing: all authors.
Competing interests
We declare no competing interests.
Funding
National Science Foundation to D.R.R. (DEB 1145987 and DEB 1638902), C.L.P. (DUE 1161228, DEB 1822417 and DEB 1638879) and M.L.H. (Postdoctoral Research Fellowship in Biology 1711141). Funding was also provided by Clemson University (C.L.P.), PrairieBiotic Research Inc. (J.L.S.), Sigma Xi Grants-in-aid-of-research (J.L.S.), the SnakeDays Research Grant (J.L.S.), the Southwestern Association of Naturalists Howard McCarley Research Grant (J.L.S. and A.J.M.), and the Theodore Roosevelt Memorial Fund through the American Museum of Natural History (J.L.S. and A.J.M.).
References
- 1.Van Valen L. 1974. Molecular evolution as predicted by natural selection. J. Mol. Evol. 3, 89–101. ( 10.1007/BF01796554) [DOI] [PubMed] [Google Scholar]
- 2.Paterson S, et al. 2010. Antagonistic coevolution accelerates molecular evolution. Nature 464, 275–278. ( 10.1038/nature08798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chouteau M, Arias M, Joron M. 2016. Warning signals are under positive frequency-dependent selection in nature. Proc. Natl Acad. Sci. USA 113, 2164–2169. ( 10.1073/pnas.1519216113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Yang J-R. 2015. Determinants of the rate of protein sequence evolution. Nat. Rev. Genet. 16, 409–420. ( 10.1038/nrg3950) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG. 2013. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol. Evol. 28, 219–229. ( 10.1016/j.tree.2012.10.020) [DOI] [PubMed] [Google Scholar]
- 6.Nakashima K-I, et al. 1995. Accelerated evolution in the protein-coding regions is universal in crotalinae snake venom gland phospholipase A2 isozyme genes. Proc. Natl Acad. Sci. USA 92, 5605–5609. ( 10.1073/pnas.92.12.5605) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Margres MJ, Wray KP, Hassinger ATB, Ward MJ, McGivern JJ, Moriarty Lemmon E, Lemmon AR, Rokyta DR. 2017. Quantity, not quality: rapid adaptation in a polygenic trait proceeded exclusively through expression differentiation. Mol. Biol. Evol. 34, 3099–3110. ( 10.1093/molbev/msx231) [DOI] [PubMed] [Google Scholar]
- 8.Daltry JC, Wüster W, Thorpe RS. 1996. Diet and snake venom evolution. Nature 379, 537–540. ( 10.1038/379537a0) [DOI] [PubMed] [Google Scholar]
- 9.Chippaux J-P, Williams V, White J. 1991. Snake venom variability: methods of study, results and interpretation. Toxicon 29, 1279–1303. ( 10.1016/0041-0101(91)90116-9) [DOI] [PubMed] [Google Scholar]
- 10.Jansa SA, Voss RS. 2011. Adaptive evolution of the venom-targeted vWF protein in opossums that eat pitvipers. PLoS ONE 6, e20997 ( 10.1371/journal.pone.0020997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holding ML, Biardi JE, Gibbs HL. 2016. Coevolution of venom function and venom resistance in a rattlesnake predator and its squirrel prey. Proc. R. Soc. B 283, 20152841 ( 10.1098/rspb.2015.2841) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casewell NR, et al. 2014. Medically important differences in snake venom composition are dictated by distinct postgenomic mechanisms. Proc. Natl Acad. Sci. USA 111, 9205–9210. ( 10.1073/pnas.1405484111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Margres MJ, McGivern JJ, Wray KP, Seavy M, Calvin K, Rokyta DR. 2014. Linking the transcriptome and proteome to characterize the venom of the Eastern Diamondback Rattlesnake (Crotalus adamanteus). J. Proteomics 96, 145–158. ( 10.1016/j.jprot.2013.11.001) [DOI] [PubMed] [Google Scholar]
- 14.Gibbs HL, Rossiter W. 2008. Rapid evolution by positive selection and gene gain and loss: PLA2 venom genes in closely related Sistrurus rattlesnakes with divergent diets. J. Mol. Evol. 66, 151–166. ( 10.1007/s00239-008-9067-7) [DOI] [PubMed] [Google Scholar]
- 15.Casewell NR, Wagstaff SC, Harrison RA, Renjifo C, Wüster W. 2011. Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Mol. Biol. Evol. 28, 2637–2649. ( 10.1093/molbev/msr091) [DOI] [PubMed] [Google Scholar]
- 16.Lynch VJ. 2007. Inventing an arsenal: adaptive evolution and neofunctionalization of snake venom phospholipase A2 genes. BMC Evol. Biol. 7, 2 ( 10.1186/1471-2148-7-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Margres MJ, McGivern JJ, Seavy M, Wray KP, Facente J, Rokyta DR. 2015. Contrasting modes and tempos of venom expression evolution in two snake species. Genetics 199, 165–176. ( 10.1534/genetics.114.172437) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Margres MJ, Wray KP, Seavy M, McGivern JJ, Sanader D, Rokyta DR. 2015. Phenotypic integration in the feeding system of the Eastern Diamondback Rattlesnake (Crotalus adamanteus). Mol. Ecol. 24, 3405–3420. ( 10.1111/mec.13240) [DOI] [PubMed] [Google Scholar]
- 19.Rokyta DR, Margres MJ, Ward MJ, Sanchez EE. 2017. The genetics of venom ontogeny in the Eastern Diamondback Rattlesnake (Crotalus adamanteus). PeerJ 5, e3249 ( 10.7717/peerj.3249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan KY, Tan CH, Chanhome L, Tan NH. 2017. Comparative venom gland transcriptomics of Naja kaouthia (monocled cobra) from Malaysia and Thailand: elucidating geographical venom variation and insights into sequence novelty. PeerJ 5, e3142 ( 10.7717/peerj.3142) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraser DJ, Weir LK, Bernatchez L, Hansen MM, Taylor EB. 2011. Extent and scale of local adaptation in salmonid fishes: review and meta-analysis. Heredity (Edinb) 106, 404–420. ( 10.1038/hdy.2010.167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romero IG, Ruvinsky I, Gilad Y. 2012. Comparative studies of gene expression and the evolution of gene regulation. Nat. Rev. Genet. 13, 505–516. ( 10.1038/nrg3229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fraser HB. 2013. Gene expression drives local adaptation in humans. Genome Res. 23, 1089–1096. ( 10.1101/gr.152710.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibbs HL, Sanz L, Calvete JJ. 2009. Snake population venomics: proteomics-based analyses of individual variation reveals significant gene regulation effects on venom protein expression in Sistrurus rattlesnakes. J. Mol. Evol. 68, 113–125. ( 10.1007/s00239-008-9186-1) [DOI] [PubMed] [Google Scholar]
- 25.Margres MJ, Wray KP, Seavy M, McGivern JJ, Herrera ND, Rokyta DR. 2016. Expression differentiation is constrained to low-expression proteins over ecological timescales. Genetics 202, 273–283. ( 10.1534/genetics.115.180547) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holding ML, Margres MJ, Rokyta DR, Gibbs HL. 2018. Local prey community composition and genetic distance predict venom divergence among populations of the Northern Pacific Rattlesnake (Crotalus oreganus). J. Evol. Biol. 31, 1513–1528. ( 10.1111/jeb.13347) [DOI] [PubMed] [Google Scholar]
- 27.Mackessy SP. 2008. Venom composition in rattlesnakes: trends and biological significance. In The biology of rattlesnakes (eds Hayes WK, Beaman KR, Cardwell MD, Bush S.), pp. 495–510. Loma Linda, CA: Loma Linda University Press. [Google Scholar]
- 28.Strickland JL, et al. 2018. Evidence for divergent patterns of local selection driving venom variation in Mojave Rattlesnakes (Crotalus scutulatus). Sci. Rep. 8, 17622 ( 10.1038/s41598-018-35810-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hofmann EP, Rautsaw RM, Strickland JL, Holding ML, Hogan MP, Mason AJ, Rokyta DR, Parkinson CL. 2018. Comparative venom-gland transcriptomics and venom proteomics of four Sidewinder Rattlesnake (Crotalus cerastes) lineages reveal little differential expression despite individual variation. Sci. Rep. 8, 15534 ( 10.1038/s41598-018-33943-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Margres MJ, Aronow K, Loyacano J, Rokyta DR. 2013. The venom-gland transcriptome of the eastern coral snake (Micrurus fulvius) reveals high venom complexity in the intragenomic evolution of venoms. BMC Genomics 14, 531 ( 10.1186/1471-2164-14-531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klauber LM. 1972. Rattlesnakes: their habits, life histories, and influence on mankind. Vol. 1, 2nd edn Los Angeles, CA: University of California Press. [Google Scholar]
- 32.Ernst CH, Ernst EM. 2012. Venomous Reptiles of the United States, Canada, and Northern Mexico. Vol. 2: Crotalus. Baltimore, MD: The John Hopkins University Press. [Google Scholar]
- 33.Strickland JL, Mason AJ, Rokyta DR, Parkinson CL. 2018. Phenotypic variation in Mojave Rattlesnake (Crotalus scutulatus) venom is driven by four toxin families. Toxins (Basel) 10, 1–23. ( 10.3390/toxins10040135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glenn JL, Straight R. 1978. Mojave rattlesnake Crotalus scutulatus scutulatus venom: variation in toxicity with geographical origin. Toxicon 16, 81–84. ( 10.1016/0041-0101(78)90065-X) [DOI] [PubMed] [Google Scholar]
- 35.French WJ, Hayes WK, Bush SP, Cardwell MD, Bader JO, Rael ED. 2004. Mojave toxin in venom of Crotalus helleri (Southern Pacific Rattlesnake): molecular and geographic characterization. Toxicon 44, 781–791. ( 10.1016/j.toxicon.2004.08.008) [DOI] [PubMed] [Google Scholar]
- 36.Sunagar K, et al. 2014. Intraspecific venom variation in the medically significant Southern Pacific Rattlesnake (Crotalus oreganus helleri): biodiscovery, clinical and evolutionary implications. J. Proteomics 99, 68–83. ( 10.1016/j.jprot.2014.01.013) [DOI] [PubMed] [Google Scholar]
- 37.Futuyma DJ, Moreno G. 1988. The evolution of ecological specialization. Annu. Rev. Ecol. Syst. 19, 207–233. ( 10.1146/annurev.es.19.110188.001231) [DOI] [Google Scholar]
- 38.Aird SD, Arora J, Barua A, Qiu L, Terada K, Mikheyev AS. 2017. Population genomic analysis of a pitviper reveals microevolutionary forces underlying venom chemistry. Genome Biol. Evol. 9, 2640–2649. ( 10.1093/gbe/evx199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Margres MJ, Patton A, Wray KP, Hassinger ATB, Ward MJ, Lemmon EM, Lemmon AR, Rokyta DR. 2019. Tipping the scales: the migration–selection balance leans toward selection in snake venoms. Mol. Biol. Evol. 36, 271–282. ( 10.1093/molbev/msy207) [DOI] [PubMed] [Google Scholar]
- 40.Webber MM, Glaudas X, Rodríguez-Robles JA. 2012. Do Sidewinder Rattlesnakes (Crotalus cerastes, Viperidae) cease feeding during the breeding season? Copeia 2012, 100–105. ( 10.1643/CP-10-181) [DOI] [Google Scholar]
- 41.Webber MM, Jezkova T, Rodríguez-Robles JA. 2016. Feeding ecology of Sidewinder Rattlesnakes, Crotalus cerastes (Viperidae). Herpetologica 72, 324–330. ( 10.1655/Herpetologica-D-15-00031.1) [DOI] [Google Scholar]
- 42.Bosak AR, Ruha A-M, Graeme KA. 2014. A case of neurotoxicity following envenomation by the Sidewinder Rattlesnake, Crotalus cerastes. J. Med. Toxicol. 10, 229–231. ( 10.1007/s13181-013-0373-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. 2005. Why highly expressed proteins evolve slowly. Proc. Natl Acad. Sci. USA 102, 14 338–14 343. ( 10.1073/pnas.0504070102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. ( 10.1038/nmeth.2089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fry BG. 2005. From genome to ‘venome’: molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Res. 15, 403–420. ( 10.1101/gr.3228405) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mackessy SP. 2010. Handbook of venoms and toxins of reptiles. Boca Raton, FL: CRC Press. [Google Scholar]
- 47.Pál C, Papp B, Hurst LD. 2001. Highly expressed genes in yeast evolve slowly. Genetics 158, 927–931. ( 10.1016/j.jelectrocard.2006.10.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Douglas ME, Douglas MR, Schuett GW, Porras LW. 2006. Evolution of rattlesnakes (Viperidae; Crotalus) in the warm deserts of western North America shaped by Neogene vicariance and Quaternary climate change. Mol. Ecol. 15, 3353–3374. ( 10.1111/j.1365-294X.2006.03007.x) [DOI] [PubMed] [Google Scholar]
- 49.Schield DR, et al. 2018. Cryptic genetic diversity, population structure, and gene flow in the Mojave rattlesnake (Crotalus scutulatus). Mol. Phylogenet. Evol. 127, 669–681. ( 10.1016/j.ympev.2018.06.013) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw Illumina sequences and transcriptome assemblies are available in Genbank under accession numbers SRA: SRR6768682-SRR6768689; TSA: GGMJ00000000.