Abstract
Theory suggests sexual traits should show heightened condition-dependent expression. This prediction has been tested extensively in experiments where condition has been manipulated through environmental quality. Condition-dependence as a function of genetic quality has, however, only rarely been addressed, despite its central importance in evolutionary theory. To address the effect of genetic quality on expression of sexual and non-sexual traits, we here compare gene expression in Drosophila melanogaster head tissue between flies with intact genomes (high condition) and flies carrying a major deleterious mutation (low condition). We find that sex-biased genes show heightened condition-dependent expression in both sexes, and that expression in low condition males and females regresses towards a more similar expression profile. As predicted, sex-biased expression was more sensitive to condition in males compared to females, but surprisingly female-biased, rather than male-biased, genes show higher sensitivity to condition in both sexes. Our results thus support the fundamental predictions of the theory of condition-dependence when condition is a function of genetic quality.
Keywords: condition-dependence, Drosophila melanogaster, gene expression, genetic quality, sexual dimorphism
1. Background
Investment into reproduction should commonly come with a cost to survival. For males this cost is often direct, since they face increased risk of injury when in combat with other males and death through predation when displaying to females [1,2]. The cost of reproductive investment can also be indirect, since a smaller pool of resources available for somatic maintenance should result in elevated mortality for males as well as females [2,3]. Optimal investment into reproduction should hence depend on the amount of resources an individual has access to and the efficiency by which these can be metabolized, collectively defining the condition of an individual [4,5]. While most traits can be expected to show condition-dependent expression to some extent, theory thus suggests that condition-dependence should be especially pronounced for sexual traits [4,6–8]. Theory further suggests that male sexual traits in general should show higher condition-dependence than female sexual traits, as the marginal benefit from investing into current reproduction is often higher for males than females [1,9].
Heightened condition-dependence of male reproductive traits is the core assumption which the good genes theory of female mate choice relies on. This theory suggests that male condition has a genetic component and that choosy females therefore gain indirect genetic benefits from mating with males expressing exaggerated sexual displays (e.g. [10–12]). Assessing if expression of male sexual traits has a clear connection to genetic quality is thus important both for our understanding of sexual selection and for life-history evolution in general, as well as for testing the main assumption behind the theory of female mate choice which has received the most attention.
Condition-dependence of male sexual display traits has been studied in the past (reviewed in [13,14]). Relying on the assumption that environmental and genetic stressors have a concordant effect on condition, many studies have inferred how genetic quality affects male sexual trait expression when condition was varied through environmental quality instead. These studies support male sexual display traits showing condition-dependent expression in general. The assumption that environmental and genetic quality have similar effects on condition may however not always be true [15], and the relatively few studies which have manipulated condition through genetic quality also show a contradictory picture (reviewed in [13,14,16]).
For logistical reasons, most studies on the condition-dependence of sexual traits have been limited to a restricted set of phenotypes. Many genes do however show sex-biased expression, and in combination with transcriptomics, this provides a unique opportunity to test if condition-dependence of sexual (sex-biased) and non-sexual (unbiased) phenotypes differs on a larger scale. A few studies have taken this approach. Manipulating environmental quality, Wyman et al. [17] detected more sex-biased genes in Drosophila melanogaster flies raised on a normal compared to a low nutrient larval food source. Since gene expression was measured from whole animal bodies, interpretation of results is however potentially complicated by differential allometric scaling of tissues in males and females in different condition [18,19]. Sex-biased gene expression in response to environmental quality has also been studied for various tissues in developing scarab beetles [20–22]. Results from these studies also show that the number of sex-biased genes covary positively with environmental quality in general, but not for all tissues tested. The only study to date that has considered the effect of genetic quality on sex-biased gene expression focused on males alone and found that expression of male-biased genes largely covaried positively with each of two indices of male quality, while female-biased genes covaried negatively with one index [23]. These patterns are consistent with condition-dependent expression as a function of male quality, but since no association between the two indices was found it is unclear if the effect was mediated through condition.
Here, we use a direct manipulation of genetic quality to test if sex-biased genes show heightened condition-dependent expression and if this pattern is stronger in males than females. To address these questions, we study gene expression in D. melanogaster head tissue from male and female flies of either high genetic quality (with intact genomes) or low genetic quality (with genomes carrying one of 11 different heterozygous genomic deletions). Drosophila melanogaster males do not invest in any exaggerated ornaments but compete intensely for mating opportunities and allocate substantial time and effort to locating and vigorously courting females with an elaborate courtship ritual, and females exert mate choice. We study gene expression in heads because they are enriched in neural tissue from where reproductive and sexual behaviours are directed. We predict that male- and female-biased genes should show heightened condition-dependent expression and that this effect should be larger in males.
2. Methods
(a). RNA-sequencing data from individuals of high and low genetic quality
To test if reduced genetic quality results in condition-dependent changes in gene expression, we took advantage of previously published data by Chen & Oliver [24] (GEO accession GSE60571). They performed RNA sequencing on male and female D. melanogaster head tissue from flies carrying haploid deletions (drawn from the DrosDel collection project [25,26]), and deletion-free flies carrying the same genetic background (w1118). From the dataset, we selected samples including autosomal deletions (all on chromosome 3 L, each spanning from 0.33 to 0.83 Mb and including between 22 and 129 genes; electronic supplementary material 1), excluding those from ED4685 because no actual deletion was reported to exist in this line [24]. This left us with data from 11 separate deletion lines, which we took as our low genetic quality treatment, while w1118 samples free from deletions were used as our control. To test the robustness of our main results we also used head samples from the wild-type stock Oregon R as an alternate control (also provided by Chen & Oliver [24]), which gave largely similar results (electronic supplementary material 2) to those reported below. There were three biological replicates for each line and sex, including the control; each replicate consisted of 10 heads pooled together. Our analysis used raw count data as processed by Chen and Oliver, where reads were mapped to FlyBase genome release 5.57 [27] using Tophat v. 2.0.10 [28] and then counted using HT-seq v. 0.5.4p1 [29]. Previous studies on flies carrying heterozygous deletions with sizes comparable to the ones used here have shown that they frequently have an effect on both morphology and egg-to-adult survival [30], and estimates of the selection coefficient against single heterozygous null alleles (comparable to deletions of single genes) show that these average at ≈0.6% in both humans and fruit flies [31]. The deletions studied here should hence have a substantial negative effect on organismal performance.
(b). Data analysis
All analyses were performed in the R statistical environment ([32], v. 3.5.1). We used the DESeq2 package [33] to normalize the raw counts, fit a model using the design formula Sex + Category (where Category is either Deletion or Control), and perform subsequent contrasts. All genes showing an absolute log2 fold change (log2FC) between males and females ≥ 3.5 were removed from subsequent analyses in an effort to remove genes that are sex-limited in expression, following Wyman et al. [17]. For each deletion, genes within the deleted area were excluded from the analyses to avoid confounding ‘cis’ effects on expression with reduced condition. When we identify genes that are differentially expressed in the presence of a deletion, we are therefore considering only genes whose expression is affected indirectly (i.e. in ‘trans’) by the deletion. We classified each gene as sex-biased in the control samples (w1118) if they were differentially expressed between males and females (p < 0.01, FDR corrected). Sex-biased genes were further classified as female-biased (FB) or male-biased (MB) depending on which sex showed higher expression. All other genes were classified as unbiased (UB), excluding from the analyses those classified as lowly expressed by DESeq2. We then repeated this classification separately for each deletion to quantify sex-biased genes under reduced genetic quality. The number of sex-biased genes in each deletion line (FB, MB, and their sum) was compared to the respective class in the control using a χ2 test (FDR corrected p-value), to determine if there was a condition-dependent loss in sex-bias. To test the effect of our significance threshold on the detection of sex-biased expression, we performed the same classification using FDR corrected cut-offs of p < 0.05 and p < 0.001 and found all cut-offs produced qualitatively similar results (table 1, electronic supplementary material 3 and 4). Carrying a major deletion is expected to have genome-wide consequences for gene expression, which potentially reduce canalization of expression and cause more variation between samples. To test for this possibility, we calculated the coefficient of expression variation (CV) for each gene, sex, and deletion. We then fitted a linear mixed-effects model using Sex and Deletion Line as factors and bias class (FB, MB, UB) as random effects, and we compared the CVs between deletions and control using the package emmeans and applying FDR correction for multiple comparisons.
Table 1.
Number of sex-biased genes in the control and each deletion line. Within each bias class, all deletions showed a significant reduction in the number of sex-biased genes compared to controls (padj < 0.001, FDR corrected, except where †padj = 0.018). Between brackets we report the number of genes that are common with the control line (where numbers are not reported, no common genes were found).
| deletion | female-biased genes | male-biased genes | total |
|---|---|---|---|
| control | 208 | 156 | 364 |
| ED210 | 31 | 41 (3) | 72 (3) |
| ED211 | 22 (2) | 21 (3) | 43 (5) |
| ED217 | 7 | 57 (1) | 64 (1) |
| ED225 | 5 | 13 | 18 |
| ED230 | 72 (4) | 75 (2) | 147 (6) |
| ED4287 | 146 (40) | 117† (33) | 263 (73) |
| ED4421 | 15 | 13 | 28 |
| ED4457 | 46 (3) | 39 | 85 (3) |
| ED4475 | 27 (2) | 37 | 64 (2) |
| ED4543 | 3 | 9 | 12 |
| ED4978 | 21 (2) | 19 (2) | 40 (4) |
While our 11 deletions are varied in their position along chromosome arm 3 L and their size in base pairs and genes deleted (electronic supplementary material 1), we hypothesize that they generally reduce genetic quality and thus organismal condition. All deletions should consequently affect a largely shared pool of genes whose expression is condition-dependent. This is in contrast to the genes affected by a given deletion generally being those that are close network neighbours to deleted genes, so that the set of affected genes will largely differ between deletions in a random manner based on deletion location. We used a permutation-based approach to test (i) whether genes were more likely to be differentially expressed (relative to the controls) in multiple deletion lines than expected by chance, (ii) whether genes that lost the sex-bias (either FB or MB) that they had in the controls did so in more different deletion lines than expected by chance, and (iii) whether UB genes in the controls gained sex-bias in more deletion lines than expected by chance. The two first predictions are expected to be true if deletions affect general condition, while the third is not. For each test, the observed distribution was constructed based on how many deletion lines a given gene showed the relevant expression change in. To generate the expectation for each test, we first randomly sampled the same number of genes affected in the relevant way by each deletion from the appropriate underlying gene set—for (i) all genes, for (ii) all sex-biased genes in the controls, and for (iii) all UB genes in the controls—and then generated expected distributions based on how many deletion lines each gene was randomly sampled in. For each of the three tests, this random sampling process was repeated 1000 times. To test if the mean of the observed and random distributions were different in each case, we computed a p-value from the overlap of the observed mean with the distribution of means obtained from the 1000 randomly resampled distributions.
To test whether sex-biased genes show condition-dependence in expression level, we first performed the contrast Deletion/Control for each deletion line and sex. Again, all genes showing an absolute log2 fold change (log2FC) ≥ 3.5 were removed from subsequent analyses. We then fitted a linear mixed-effect model using Bayesian Hamiltonian Markov chain Monte Carlo in the package rstanarm [34], with Sex and Bias (FB, MB, or UB) as fixed effects and Deletion ID as a random effect, using log2FC (Deletion/Control contrast) for each gene separately in each deletion line/sex combination as the response. We used weakly informative normally distributed priors for both the intercept (mean = 0, scale = 10) and coefficients (mean = 0, scale = 2.5), running four chains with 2000 iterations each and discarding the first 1000 as warm-up. Comparisons between classes of genes were performed using the function hypothesis of the brms package [35], with the equivalent of a two-tailed p-value reported. We also repeated this analysis while taking tissue-specificity into account (see electronic supplementary material 7 for details).
We also tested the correlation between the degree of condition-dependence and the degree of sex-bias separately for FB and MB genes in each sex using Pearson's correlation coefficient. Degree of condition-dependence for a gene was calculated by performing the contrast Control/Deletion separately for each deletion line and then taking the mean of the resulting log2FC values, such that genes with positive condition-dependence show reduced expression due to deletions. Sex-bias was calculated for FB genes using the contrast Control Female/Control Male, and for MB genes using the contrast Control Male/Control Female, such that sex-bias was always a positive value.
3. Results
We first used a permutation-based approach to test whether genes of any bias class were more likely to be differentially expressed in multiple deletion lines, relative to the control, than expected by chance. This was predicted to be true if differential expression is primarily due to deletions sharing a common effect on organismal condition rather than having largely unique effects on specific gene networks. We found that the mean number of deletions affecting any given gene was higher in the observed distribution than in the random distribution for each sex (p < 0.001 in both). This pattern was corroborated by sex-biased genes that lost their sex-bias doing so in more deletion lines than expected by chance (p < 0.001 in both sexes), as predicted when genetic quality reduces condition.
Given that deletion lines appear to be in reduced condition, we were able to test our hypothesis that a reduction in condition will result in a net reduction in the sex-bias of the transcriptome. We first classified genes as FB, MB, or UB in the controls and separately in each deletion line, and then compared the number of sex-biased genes in each deletion line to the controls. As predicted, we observed a significant reduction in the number of sex-biased genes in each of the deletion lines (table 1, electronic supplementary material 3). Interestingly, almost all sex-biased genes lost their sex-bias in the deletion lines, while those that were sex-biased in the deletion lines were largely unbiased in the control. According to our permutation analysis, genes independently gained sex-bias in multiple deletion lines more often than expected by chance (p < 0.001 in both sexes), which was not consistent with the expectation that gain of sex-bias would be idiosyncratic with respect to deletion line. However, the difference between random and observed means was extremely small (mean ± s.e.: FB Observed 1.04 ± 0.003, Expected 1.01 ± 0.003; MB Observed 1.03 ± 0.002, Expected 1.01 ± 0.003), with nearly all genes gaining sex-bias doing so in only one deletion line as expected if the process is idiosyncratic rather than related to a change in condition.
When we compared the variability of gene expression between deletion lines and the control (Sex χ2 = 75.1, p < 0.001; Line identity χ2 = 1578.0, p < 0.001), we observed that all 11 deletion lines show higher gene expression variation in females, and in males 5 show increased variation (4 show reduced variation and 2 show no change). These results suggest that our analyses probably overestimated the number of genes that lose and gain sex-bias in the deletion lines, as a relative increase of gene expression variation should produce relatively more false positives and negatives in these lines. To visualize the change in sex-bias between control and deletion lines we plotted the correlation in gene expression between males and females for each line (electronic supplementary material 5). Inspection of these plots in comparison to that of the controls shows that sex-biased genes clearly tend to converge towards an unbiased state. It is also obvious from the analyses reported below that the transcriptome of deletion lines shows a reduction in sex-bias.
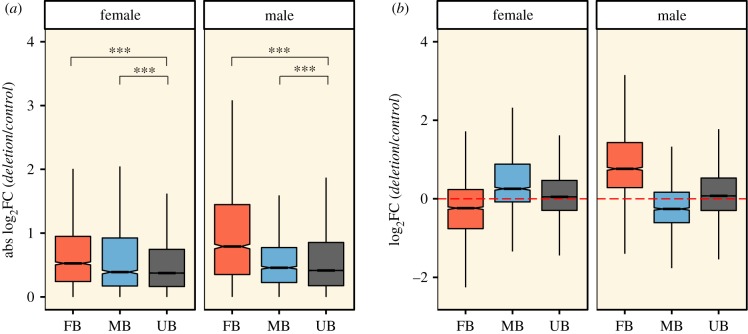
Since we observed a reduction in the number of sex-biased genes under reduced condition, we then tested the magnitude of changes in gene expression level in each sex and bias class (as classified in the control line) as an effect of reduced condition. This allows us to determine whether sex-biased genes show heightened condition-dependent expression versus unbiased genes, and also addresses our second hypothesis, the prediction of a larger condition-dependent effect in males than females. We applied a Bayesian linear modelling framework, where the model coefficients (electronic supplementary material 6) were used to calculate posterior distributions for changes in gene expression that could in turn be used to test our hypotheses. As condition is not expected to have a directional effect on expression of unbiased genes, we first compared the absolute value of expression changes. In both sexes, we observed a larger change in both MB and FB genes than UB genes (both p < 0.001, figure 1a), and thus heightened condition-dependence. Using the change in UB genes as baseline, we observed that sex-biased genes show more heightened condition-dependence in males compared to females (p < 0.001 for both FB and MB genes, figure 1a). We also recorded a greater heightening of condition-dependence for FB genes than MB genes in both sexes (p < 0.001 for both males and females, figure 1a). Taking tissue-specificity of genes into account when analysing condition-dependence generated similar results, with the only exception being that MB and FB genes show similarly heightened condition-dependence in females (electronic supplementary material 7).
Figure 1.
Differential gene expression analysis conducted by contrasting each deletion towards the controls, with genes grouped on the x-axis according to their sex-bias in the controls. Values expressed in (a) absolute log2 fold changes and (b) log2 fold changes. Differences between specific changes in gene expression were tested using a Bayesian linear mixed-model (***p < 0.001). FB, female biased; MB, male biased; UB, unbiased.
Since we expect condition to have a directional effect on expression of sex-biased genes, we tested whether the magnitude of the expected directional effect is elevated in males compared to females, taking the direction of change into account. We again used UB genes as baseline, since these may in part represent a general directional change induced by deletions that is unrelated to condition. Both MB and FB genes in males showed a higher magnitude of change compared to the respective class in females (both p < 0.001, figure 1b). Within each sex we compared the two bias classes and found greater condition-dependence of FB genes than MB genes in both males and females (both p < 0.001, figure 1b). As such, the results when taking the directionality of effects on expression into account mirror those using absolute values in supporting the hypothesis of greater condition-dependence in males.
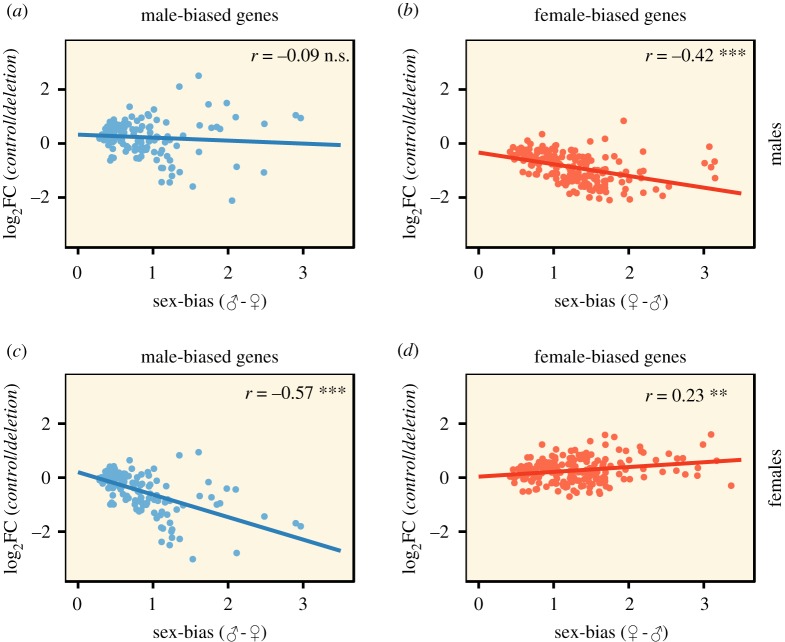
Lastly, we tested for a correlation between degree of condition-dependence and degree of sex-bias for both FB and MB genes separately in males and females (figure 2). In males, the relationship between condition-dependence and sex-bias of MB genes was non-significant (p > 0.05, figure 2a), while the correlation was negative for FB genes (p < 0.001, figure 2b). In females, we observed a negative correlation for MB genes (p < 0.001, figure 2c) and a positive one for FB genes (p < 0.001, figure 2d). As expected, the correlations based on individual deletions show some variation, but are consistent with the general pattern overall, with the exception that in some lines male-biased genes in males show the opposite relationship (electronic supplementary material 8). These results indicate that the more sex-biased MB and FB genes are in females, the more condition-dependent they are, while this is true in general only for FB genes in males. This also suggests that condition-dependent sex-bias in high condition individuals is manifest more strongly in downregulation than upregulation of expression in general.
Figure 2.
Correlation (r = Pearson's correlation coefficient) between degree of condition-dependence (mean log2 fold change value from the contrast Control/Deletion performed separately for all 11 deletion lines) and degree of sex-bias in the controls (calculated separately for male-biased and female-biased genes). Correlations are shown for (a) male-biased genes in males, (b) female-biased genes in males, (c) male-biased genes in females, and (d) female-biased genes in females. Observe that the contrast on the y-axis is reversed relative to figure 1 to make our results easier to compare with those obtained by Wyman et al. [17].
4. Discussion
In this study we use a manipulation of condition through genetic quality in male and female D. melanogaster flies to study the effect on gene expression in head tissue, contrasting sex-biased to unbiased genes. As predicted by theory we find that the effect of reduced condition on sex-biased genes is larger than that on unbiased genes, that it demasculinizes male and defeminizes female expression profiles, and that the effect is more pronounced in males than females. Sex-biased genes thus show heightened condition-dependent expression, which is especially strong in males. Provided that the observed changes in gene expression map proportionally to phenotypic change in sexual and non-sexual phenotypes, these results suggest that male sexual displays should honestly reflect male genetic quality, and that D. melanogaster females should gain indirect genetic benefits from preferentially mating with males expressing exaggerated displays. While we can only draw conclusions with respect to the effect of genetic quality on gene expression in head tissue, we see no a priori reason why our results should not extend to other tissues, but this will need confirmation.
The genic capture hypothesis postulates that genetic variation in male display traits is generated by many small effect loci scattered across the genome, which all affect the focal traits because they each contribute to an individual's condition [4]. Our manipulation of genetic quality involved single major mutations and may therefore appear unfit to test the genic capture hypothesis. Changing the copy number of a small part of the genome does however not only affect expression of genes made haploid, but also genes with which the haploid genes interact with in gene networks (e.g. [36]). This should result in suboptimal expression level of a suite of genes, presumably distinct to each deletion, which collectively should reduce an individual's ability to acquire and metabolize resources and thus its condition. This conclusion is supported by our observation that genes with affected expression are largely shared between independent deletion lines, which is also true for genes which lose their sex-biased expression. Also, the fact that gene expression is more variable between replicates of most deletion lines may reflect reduced condition of flies carrying a deletion, as gene expression in these flies appears to be less canalized.
To our knowledge this study is the first to consider the condition-dependence of sex-biased expression when genetic quality is directly manipulated. We did not have a treatment where condition was manipulated through environmental quality and therefore cannot directly compare the effects of variation in genetic and environmental quality on the sex-biased transcriptome. Regardless, our study generally supports conclusions from earlier contributions exclusively varying environmental quality, both with respect to gene expression [17,20–22] and phenotypic traits (e.g. [37–40]). Our findings also give more weight to studies that have found heightened condition-dependence of display traits when genetic quality has been manipulated (e.g. [41–44]) and when studied in association with male performance traits [23]. If genetic and environmental quality influence condition concordantly, however, requires more research [15].
Surprisingly, we find that female-biased, rather than male-biased, genes show the highest sensitivity to condition in males. Under the assumption that males are exposed to stronger sexual selection, the optimal male phenotype is expected to deviate further than the optimal female phenotype from a hypothetical desexualized base phenotype. Because males and females commonly are defined by the characters they possess, rather than those they lack or have less of, it is easy to assume that males are masculinized primarily through male-biased genes and females feminized through female-biased genes. This assumption was also confirmed in a study of D. melanogaster, where it was shown that the effect of major mutations in sex-biased genes tends to be larger, in terms of viability or sterility, in the sex they are biased towards [45]. Interpreting the effect of changing a gene's expression level is however difficult without detailed knowledge of its function, as this depends on whether the gene acts as an activator or suppressor. In any case our results suggest that condition-dependent masculinization of the transcriptome is primarily achieved through repression of gene expression. This conclusion is further supported by the fact that we detect more female-biased than male-biased genes in head tissue. A slight bias in favour of female-biased genes has also been reported for whole body transcriptomes in D. melanogaster (e.g. [46,47]), indicating that masculinization through suppressed gene expression may also apply to other tissues.
The degree of condition-dependence is predicted to positively covary with the degree of sexual dimorphism [48]. A priori we predicted this relationship to be the strongest for male-biased genes in males, but surprisingly we detect no general association between male-bias and condition-dependence in this sex, while the direction varied between individual deletion lines. This finding contrasts with Wyman et al. [17], who found strong support for highly male-biased genes showing elevated condition-dependence. Our results also contrast with Wyman et al. [17] in that we find a negative correlation between male-biased genes and condition-dependence in females, while they found a positive association. Provided the intersexual genetic correlation with respect to condition-dependent expression of male-biased genes is low, it seems reasonable that females in good condition should suppress expression of male-biased genes in line with our results. If, on the other hand, the intersexual genetic correlation is high and constrains sex-specific evolution [49], strong selection for increased expression in high condition males may cause expression to increase in high condition females also, as observed by Wyman et al. [17]. Since our study was limited to genes sex-biased in head tissue, while Wyman et al. studied gene expression from whole body samples, this may explain why we observe different results. With respect to the association between female-biased expression and condition-dependence, our results align with those of Wyman et al. [17] in both males and females. Condition in both cases was positively associated in females and negatively associated in males, as expected when the sexes are relatively free to modulate gene expression independently. The stronger association between condition and expression of female-biased, rather than male-biased, genes in males observed here gives further support to the conclusion that masculinization of expression in male head tissue is primarily driven by suppression of a specific set of genes.
The number of sex-biased genes reported varies greatly between studies, which probably to a large extent can be explained by experimental power. However, as this study indicates, the number of sex-biased genes detected also depends on genetic quality. In many studies gene expression is measured in inbred individuals. We are not aware of any studies directly comparing the sexual transcriptome of inbred and outbred individuals, but extrapolating from the results reached here it seems plausible that the lower genetic quality of inbred individuals should drastically reduce the sex-bias of many genes (which may also partly explain the general masculinization of asexual stick insects [50]). It is therefore plausible that both the number of sex-biased genes, and the degree to which they show sex-bias, is substantially underestimated in many studies.
5. Conclusion
Theory predicts that investment into reproduction is costly and therefore should show heightened condition-dependence [4,7]. Given that males are often selected to invest more into current reproduction than females [1,9], condition-dependence for male sexual traits should be further heightened. Our results confirm these predictions when condition is a function of genetic quality. Our results thus support the prediction that females should gain indirect genetic benefits from mating with masculinized males, the assumption which the good genes hypothesis for female mate choice is based on. This said it is important to note that condition-dependent expression of male sexual traits should evolve independently of the mechanism that drives females to choose their mate (e.g. good genes, sexy sons, sexual conflict, sensory bias). Any indirect genetic benefits may therefore be reduced by direct (e.g. [51,52]) or indirect (e.g. [53]) costs associated with interacting and mating with preferred males, and the net outcome for females may depend on context [54–56].
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Data accessibility
We used previously published data by Chen & Oliver [24] available at Gene Expression Omnibus (GEO) under the accession number GSE60571. The R code used to run the analyses, the list of genes excluded from each deletion line, and the dataset for the analysis of tissue-specificity are available as electronic supplementary material 9.
Authors' contributions
A.M. conceived the study, analysed the data, and wrote the manuscript; C.M.K. conceived the study, contributed to the analyses, and wrote the manuscript; M.B. conceived the study and contributed to the analyses; U.F. conceived the study and wrote the manuscript. All authors gave final approval for publication.
Competing interests
We have no competing interests.
Funding
A.M. was supported by a fellowship from the Lawski Foundation and the work was financially supported by the Swedish Research Council to U.F.
References
- 1.Trivers R. 1972. Parental investment and sexual selection. Sex. Sel. Descent Man, Aldine Gruyter, New York, pp. 136–179. Chicago, IL: Aldine-Atherton. [Google Scholar]
- 2.Kotiaho JS. 2001. Costs of sexual traits: a mismatch between theoretical considerations and empirical evidence. Biol. Rev. Camb. Philos. Soc. 76, 365–376. ( 10.1017/S1464793101005711) [DOI] [PubMed] [Google Scholar]
- 3.Kirkwood TBL. 1977. Evolution of ageing. Nature 270, 301–304. ( 10.1038/270301a0) [DOI] [PubMed] [Google Scholar]
- 4.Rowe L, Houle D. 1996. The lek paradox and the capture of genetic variance by condition dependent traits. Proc. R. Soc. Lond. B 263, 1415–1421. ( 10.1098/rspb.1996.0207) [DOI] [Google Scholar]
- 5.Hill GE. 2011. Condition-dependent traits as signals of the functionality of vital cellular processes. Ecol. Lett. 14, 625–634. ( 10.1111/j.1461-0248.2011.01622.x) [DOI] [PubMed] [Google Scholar]
- 6.Nur N, Hasson O. 1984. Phenotypic plasticity and the handicap principle. J. Theor. Biol. 110, 275–297. ( 10.1016/S0022-5193(84)80059-4) [DOI] [Google Scholar]
- 7.Houle D. 1998. How should we explain variation in the genetic variance of traits? Genetica 102/103, 241–253. ( 10.1023/A:1017034925212) [DOI] [PubMed] [Google Scholar]
- 8.Andersson M. 1982. Sexual selection, natural selection and quality advertisement. Biol. J. Linn. Soc. 17, 375–393. ( 10.1111/j.1095-8312.1982.tb02028.x) [DOI] [Google Scholar]
- 9.Kokko H. 2001. Fisherian and ‘good genes’ benefits of mate choice: how (not) to distinguish between them. Ecol. Lett. 4, 322–326. ( 10.1046/j.1461-0248.2001.00224.x) [DOI] [Google Scholar]
- 10.Zahavi A. 1975. Mate selection—a selection for a handicap. J. Theor. Biol. 53, 205–214. ( 10.1016/0022-5193(75)90111-3) [DOI] [PubMed] [Google Scholar]
- 11.Grafen A. 1990. Sexual selection unhandicapped by the fisher process. J. Theor. Biol. 144, 473–516. ( 10.1016/S0022-5193(05)80087-6) [DOI] [PubMed] [Google Scholar]
- 12.Iwasa Y, Pomiankowski A, Nee S. 1991. The evolution of costly mate preferences II. The ‘handicap’ principle. Evolution (NY) 45, 1431–1442. ( 10.1111/j.1558-5646.1991.tb02646.x) [DOI] [PubMed] [Google Scholar]
- 13.Cotton S, Fowler K, Pomiankowski A. 2004. Do sexual ornaments demonstrate heightened condition-dependent expression as predicted by the handicap hypothesis? Proc. R. Soc. Lond. B 271, 771–783. ( 10.1098/rspb.2004.2688) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellamy L, Fowler K, Pomiankowski A. 2014. The use of inbreeding to assess the genetic component of condition underlying GEIs in sexual traits. Genotype-by-environment Interact. Sex. Sel., pp. 213–240. Chichester, UK: Wiley-Blackwell.
- 15.Bonduriansky R, Mallet MA, Arbuthnott D, Pawlowsky-Glahn V, Egozcue JJ, Rundle HD. 2015. Differential effects of genetic vs. environmental quality in Drosophila melanogaster suggest multiple forms of condition dependence. Ecol. Lett. 18, 317–326. ( 10.1111/ele.12412) [DOI] [PubMed] [Google Scholar]
- 16.Tomkins JL, Radwan J, Kotiaho JS, Tregenza T. 2004. Genic capture and resolving the lek paradox. Trends Ecol. Evol. 19, 323–328. ( 10.1016/j.tree.2004.03.029) [DOI] [PubMed] [Google Scholar]
- 17.Wyman MJ, Agrawal AF, Rowe L. 2010. Condition-dependence of the sexually dimorphic transcriptome in Drosophila melanogaster. Evolution (NY) 64, 1836–1848. ( 10.1111/j.1558-5646.2009.00938.x) [DOI] [PubMed] [Google Scholar]
- 18.Stewart AD, Pischedda A, Rice WR. 2010. Resolving intralocus sexual conflict: genetic mechanisms and time frame. J. Hered. 101, S94–S99. ( 10.1093/jhered/esq011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mank JE. 2017. The transcriptional architecture of phenotypic dimorphism. Nat. Ecol. Evol. 1, 0006 ( 10.1038/s41559-016-0006) [DOI] [PubMed] [Google Scholar]
- 20.Kijimoto T, Snell-Rood EC, Pespeni MH, Rocha G, Kafadar K, Moczek AP. 2014. The nutritionally responsive transcriptome of the polyphenic beetle Onthophagus taurus and the importance of sexual dimorphism and body region. Proc. R. Soc. B 281, 1–10. ( 10.1098/rspb.2014.2084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ledón-Rettig CC, Moczek AP. 2016. The transcriptomic basis of tissue- and nutrition-dependent sexual dimorphism in the beetle Onthophagus taurus. Ecol. Evol. 6, 1601–1613. ( 10.1002/ece3.1933) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zinna R, Emlen D, Lavine LC, Johns A, Gotoh H, Niimi T, Dworkin I. 2018. Sexual dimorphism and heightened conditional expression in a sexually selected weapon in the Asian rhinoceros beetle. Mol. Ecol. 27, 5049–5072. ( 10.1111/mec.14907) [DOI] [PubMed] [Google Scholar]
- 23.Dean R, Hammer C, Higham V, Dowling DK. 2018. Masculinization of gene expression is associated with male quality in Drosophila melanogaster. Evolution (NY) 72, 2736–2748. ( 10.1111/evo.13618) [DOI] [PubMed] [Google Scholar]
- 24.Chen Z-X, Oliver B. 2015. X chromosome and autosome dosage responses in Drosophila melanogaster heads G3 (Bethesda) 5, 1057–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryder E, et al. 2007. The DrosDel deletion collection: a Drosophila genomewide chromosomal deficiency resource. Genetics 177, 615–629. ( 10.1534/genetics.107.076216) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cook KR, Parks AL, Jacobus LM, Kaufman TC, Matthews KA. 2010. New research resources at the Bloomington Drosophila Stock Center. Fly (Austin) 4, 88–91. ( 10.4161/fly.4.1.11230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.St. Pierre SE, Ponting L, Stefancsik R, McQuilton P. 2014. FlyBase 102—advanced approaches to interrogating FlyBase. Nucleic Acids Res. 42, D780–D788. ( 10.1093/nar/gkt1092) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 ( 10.1186/gb-2013-14-4-r36) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. ( 10.1101/002824) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi KH, Okada Y, Teramura K, Tsujino M. 2011. Deficiency mapping of the genomic regions associated with effects on developmental stability in Drosophila melanogaster. Evolution (NY) 65, 3565–3577. ( 10.1111/j.1558-5646.2011.01400.x) [DOI] [PubMed] [Google Scholar]
- 31.Cassa CA, et al. 2017. Estimating the selective effects of heterozygous protein-truncating variants from human exome data. Nat. Genet. 49, 806–810. ( 10.1038/ng.3831) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.R Core Team. 2017. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 33.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 ( 10.1186/s13059-014-0550-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gabry J, Goodrich B. 2018. rstanarm: Bayesian applied regression modeling via Stan. R Packag. version 2.17.4. http://mc-stan.org/rstanarm.
- 35.Bürkner P-C. 2017. brms: an R package for Bayesian multilevel models using Stan. J. Stat. Softw. 80, 1–28. [Google Scholar]
- 36.Lee H, et al. 2016. Effects of gene dose, chromatin, and network topology on expression in Drosophila melanogaster. PLoS Genet. 12, e1006295 ( 10.1371/journal.pgen.1006295) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.David P, Bjorksten T, Fowler K, Pomiankowski A. 2000. Condition-dependent signalling of genetic variation in stalk-eyed flies. Nature 406, 186–188. ( 10.1038/35018079) [DOI] [PubMed] [Google Scholar]
- 38.Cotton S, Fowler K, Pomiankowski A. 2004. Condition dependence of sexual ornament size and variation in the stalk-eyed fly Cyrtodiopsis dalmanni (Diptera: Diopsidae). Evolution (NY) 58, 1038–1046. ( 10.1111/j.0014-3820.2004.tb00437.x) [DOI] [PubMed] [Google Scholar]
- 39.Bonduriansky R, Rowe L. 2005. Sexual selection, genetic architecture, and the condition dependence of body shape in the sexually dimorphic fly Prochyliza xanthostoma (Piophilidae). Evolution (NY) 59, 138–151. ( 10.1111/j.0014-3820.2005.tb00901.x) [DOI] [PubMed] [Google Scholar]
- 40.Punzalan D, Cooray M, Helen Rodd F, Rowe L. 2008. Condition dependence of sexually dimorphic colouration and longevity in the ambush bug Phymata americana. J. Evol. Biol. 21, 1297–1306. ( 10.1111/j.1420-9101.2008.01571.x) [DOI] [PubMed] [Google Scholar]
- 41.Sheridan L, Pomiankowski A. 1997. Fluctuating asymmetry, spot asymmetry and inbreeding depression in the sexual coloration of male guppy fish. Heredity (Edinb) 79, 515–523. ( 10.1038/hdy.1997.191) [DOI] [Google Scholar]
- 42.Bolund E, Schielzeth H, Forstmeier W. 2010. No heightened condition dependence of zebra finch ornaments—a quantitative genetic approach. J. Evol. Biol. 23, 586–597. ( 10.1111/j.1420-9101.2009.01927.x) [DOI] [PubMed] [Google Scholar]
- 43.Zajitschek SRK, Brooks RC. 2010. Inbreeding depression in male traits and preference for outbred males in Poecilia reticulata. Behav. Ecol. 21, 884–891. ( 10.1093/beheco/arq077) [DOI] [Google Scholar]
- 44.Bellamy L, Chapman N, Fowler K, Pomiankowski A. 2013. Sexual traits are sensitive to genetic stress and predict extinction risk in the stalk-eyed fly, Diasemopsis meigenii. Evolution (NY) 67, 2662–2673. ( 10.1111/evo.12135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Connallon T, Clark AG. 2011. The resolution of sexual antagonism by gene duplication. Genetics 187, 919–937. ( 10.1534/genetics.110.123729) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ayroles JF, et al. 2009. Systems genetics of complex traits in Drosophila melanogaster. Nat. Genet. 41, 299–307. ( 10.1038/ng.332) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Innocenti P, Morrow EH. 2010. The sexually antagonistic genes of Drosophila melanogaster. PLoS Biol. 8, e1000335 ( 10.1371/journal.pbio.1000335) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bonduriansky R. 2007. The genetic architecture of sexual dimorphism: the potential roles of genomic imprinting and condition-dependence. In Sex, size, and gender roles: evolutionary studies of sexual size dimorphism (eds Fairbairn D, Blanckenhorn J, Szekely T), pp. 176–184. Oxford, UK: Oxford University Press. [Google Scholar]
- 49.Griffin RM, Dean R, Grace JL, Ryden P, Friberg U. 2013. The shared genome is a pervasive constraint on the evolution of sex-biased gene expression. Mol. Biol. Evol. 30, 2168–2176. ( 10.1093/molbev/mst121) [DOI] [PubMed] [Google Scholar]
- 50.Parker DJ, Bast J, Jalvingh K, Dumas Z, Robinson-Rechavi M, Schwander T. 2019. Sex-biased gene expression is masculinized in asexual females. bioRxiv, 553172 ( 10.1101/553172) [DOI]
- 51.Friberg U, Arnqvist G. 2003. Fitness effects of female mate choice: preferred males are detrimental for Drosophila melanogaster females. J. Evol. Biol. 16, 797–811. ( 10.1046/j.1420-9101.2003.00597.x) [DOI] [PubMed] [Google Scholar]
- 52.Long TAF, Pischedda A, Stewart AD, Rice WR. 2009. A cost of sexual attractiveness to high-fitness females. PLoS Biol. 7, e1000254 ( 10.1371/journal.pbio.1000254) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pischedda A, Chippindale AK. 2006. Intralocus sexual conflict diminishes the benefits of sexual selection. PLoS Biol. 4, e356 ( 10.1371/journal.pbio.0040356) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Long TAF, Agrawal AF, Rowe L. 2012. The effect of sexual selection on offspring fitness depends on the nature of genetic variation. Curr. Biol. 22, 204–208. ( 10.1016/j.cub.2011.12.020) [DOI] [PubMed] [Google Scholar]
- 55.Yun L, Chen PJ, Kwok KE, Angell CS, Rundle HD, Agrawal AF. 2018. Competition for mates and the improvement of nonsexual fitness. Proc. Natl Acad. Sci. USA 115, 6762–6767. ( 10.1073/pnas.1805435115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Malek HL, Long TAF. 2019. Spatial environmental complexity mediates sexual conflict and sexual selection in Drosophila melanogaster. Ecol. Evol. 9, 2651–2663. ( 10.1002/ece3.4932) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We used previously published data by Chen & Oliver [24] available at Gene Expression Omnibus (GEO) under the accession number GSE60571. The R code used to run the analyses, the list of genes excluded from each deletion line, and the dataset for the analysis of tissue-specificity are available as electronic supplementary material 9.