Abstract
Mechanical sensitization is one of the most difficult clinical pain problems to treat. However, the molecular and genetic bases of mechanical nociception are unclear. Here we develop a Drosophila model of mechanical nociception to investigate the ion channels and signaling pathways that regulate mechanical nociception. We fabricated von Frey filaments that span the subthreshold to high noxious range for Drosophila larvae. Using these, we discovered that pressure (force/area), rather than force per se, is the main determinant of aversive rolling responses to noxious mechanical stimuli. We demonstrated that the RTK PDGF/VEGF receptor (Pvr) and its ligands (Pvfs 2 and 3) are required for mechanical nociception and normal dendritic branching. Pvr is expressed and functions in class IV sensory neurons, whereas Pvf2 and Pvf3 are produced by multiple tissues. Constitutive overexpression of Pvr and its ligands or inducible overexpression of Pvr led to mechanical hypersensitivity that could be partially separated from morphological effects. Genetic analyses revealed that the Piezo and Pain ion channels are required for mechanical hypersensitivity observed upon ectopic activation of Pvr signaling. PDGF, but not VEGF, peptides caused mechanical hypersensitivity in rats. Pharmacological inhibition of VEGF receptor Type 2 (VEGFR-2) signaling attenuated mechanical nociception in rats, suggesting a conserved role for PDGF and VEGFR-2 signaling in regulating mechanical nociception. VEGFR-2 inhibition also attenuated morphine analgesic tolerance in rats. Our results reveal that a conserved RTK signaling pathway regulates baseline mechanical nociception in flies and rats.
SIGNIFICANCE STATEMENT Hypersensitivity to touch is poorly understood and extremely difficult to treat. Using a refined Drosophila model of mechanical nociception, we discovered a conserved VEGF-related receptor tyrosine kinase signaling pathway that regulates mechanical nociception in flies. Importantly, pharmacological inhibition of VEGF receptor Type 2 signaling in rats causes analgesia and blocks opioid tolerance. We have thus established a robust, genetically tractable system for the rapid identification and functional analysis of conserved genes underlying mechanical pain sensitivity.
Keywords: Drosophila, opioid tolerance, pain, PDGF/PDGFR, TRP ion channels, VEGF/VEGFR
Introduction
Animals must discriminate gentle touch from noxious mechanical stimuli capable of damaging tissue. High-threshold noxious mechanical stimuli elicit specific escape behaviors to avoid potential damage. These behaviors are initiated by mechanosensitive peripheral nociceptors (Dubin and Patapoutian, 2010). Diverse behavioral assays have been used to study mechanical nociception. One approach used extensively in humans (Levin et al., 1978), rodents (Bonin et al., 2014), worms (Chalfie et al., 2014), and flies (Zhong et al., 2010; Kim et al., 2012) is von Frey filaments, which are small fibers that produce a defined force when applied to the skin (Levin et al., 1978). Studies in Drosophila larvae have found that application of similar forces to the skin produced variable behavioral responses (Zhong et al., 2010; Kim et al., 2012; Guo et al., 2014), implying that applied force may not be the variable that determines the behavioral response.
Drosophila Type II multidendritic (md) sensory neurons are structurally and functionally similar to vertebrate nociceptors. These neurons have nonmyelinated endings and display elaborate peripheral dendritic projections that contact nearly every larval epidermal cell (Singhania and Grueber, 2014). There are four classes of Type II multidendritic sensory neurons (I-IV) defined by peripheral arbor complexity (Singhania and Grueber, 2014). Class III neurons respond to gentle touch (Tsubouchi et al., 2012; Yan et al., 2013), whereas class IV neurons transduce noxious mechanical stimuli (Hwang et al., 2007). Noxious mechanical stimuli elicit an aversive rolling behavior in Drosophila larvae distinct from normal locomotion and gentle touch responses (Im and Galko, 2012). Nociceptive responses are assumed to be efforts by an organism to avoid potential tissue damage. However, it is not known whether all nociceptive responses correlate with epidermal tissue damage, and conversely, whether stimuli can also induce tissue damage without producing nociceptive behavioral responses.
Ion channels are important sensors of mechanical stimuli (Eijkelkamp et al., 2013). In Drosophila, transient-receptor potential A family members Pain and Trpa1 mediate behavioral responses to high-threshold mechanical stimuli (Tracey et al., 2003; Zhong et al., 2012). Piezo-like channels (Coste et al., 2010) mediate touch responses (Faucherre et al., 2013; Ranade et al., 2014) and mechanical allodynia (Murthy et al., 2018; Szczot et al., 2018) in vertebrates. The Drosophila Piezo gene is expressed in nociceptors and is required for mechanical nociception (Kim et al., 2012). However, the upstream signaling pathways that activate different ion channels that mediate mechanical nociception in Drosophila are not known.
In vertebrates, activation of the platelet-derived growth factor receptor (PDGFR) regulates mechanical sensitivity (Narita et al., 2005; Masuda et al., 2009). PDGFR signaling also selectively mediates opioid tolerance (Wang et al., 2012). In addition, VEGFR signaling is involved in the genesis of cancer-associated pain (Selvaraj et al., 2015; Yang et al., 2018). Drosophila have a cognate receptor, Pvr, which is related to both PDGFR and VEGFR (Heino et al., 2001). Pvr and its three ligands (Pvf1, Pvf2, and Pvf3) mediate multiple physiological processes, including wound-induced cell migration (Wu et al., 2009), hemocyte proliferation (Munier et al., 2002), and cellular development and differentiation (Mondal et al., 2014). The role of Pvr in nociception in the fly has not been studied previously, and no previous work has determined whether VEGFRs modulate opioid tolerance.
Here, using larval-specific von Frey filaments of our own design, we characterize the larval dose–response to mechanical stimuli. We show that applied pressure (force/area), rather than force, is the critical determinant of behavioral responsiveness. We demonstrate that Pvr and two of its ligands (Pvf2 and Pvf3) regulate mechanical nociception. Genetic interaction analysis reveals that Piezo and Pain channels are required to modulate mechanical nociceptive responsiveness after Pvr activation. We establish that mechanical nociception in vertebrates is regulated by PDGF ligands and VEGFR signaling, suggesting a novel interaction between these related signaling systems in the regulation of mechanical sensitivity. We also demonstrate that VEGFR inhibition causes analgesia and blocks opioid tolerance, and that the analgesia induced by VEGFR inhibition is opioid-mediated.
Materials and Methods
Drosophila stocks and genetics
All fly stocks used in this work were maintained at 18°C. Crosses to generate larval progeny were performed on regular cornmeal food at 25°C.
The following mutants were used: PvrC02859 is a hypomorphic allele and Pvrc02195 herein referred to as Pvrnull (Cho et al., 2002). Pvf1EP1624, here referred to as Pvf1null, is a null allele (Cho et al., 2002). Pvf2C06947 (Cho et al., 2002) and Pvf309531 (BL17577) are hypomorphic alleles, here referred to as Pvf2hypo and Pvf3hypo, respectively. Pvf2MI00770 (BL32696) and Pvf3M04168 (BL37270) are MiMIC lines that disrupt Pvf2 and Pvf3. Pvf2-3 is a targeted deletion that removes the coding region of Pvf3 and decreases Pvf2 expression (Parsons and Foley, 2013). Ion channel deficiencies: Piezo Df (2L)Exel7034 (BL7807); Pain Df (2R)BSC602 (BL25435). Ion channels mutants: PiezoKO (Kim et al., 2012) is a deletion of piezo; pain70 is a deletion allele of pain (Im et al., 2015).
The GAL4/UAS system was used to drive tissue-specific gene expression of transgenes under UAS control (Brand and Perrimon, 1993). The following Gal4 drivers were used: embryonic and larval epidermis, e22c-Gal4 (Lawrence et al., 1995); larval epidermis, A58-Gal4 (Galko and Krasnow, 2004); gut, Myo1a-Gal4 (Jiang and Edgar, 2009); embryonic and larval hemocytes, hmlΔ-Gal4 (Sinenko and Mathey-Prevot, 2004); somatic muscle, Dmef2-Gal4 (Zars et al., 2000); fatbody, fatbody-Gal4 (Wu et al., 2009); class IV multidendritic nociceptive sensory neurons, ppk1.9-Gal4 (Ainsley et al., 2003); daughterless-Gal4 (da-Gal4) (Wodarz et al., 1995) was used for ubiquitous expression. Temporal control of the Gal4/UAS system was performed using tubP-gal80ts under permissive temperature (McGuire et al., 2004). e22c-Gal4, UAS-DsRed2Nuc(21), FasIII-GFP (Wang et al., 2015) was used to label the larval epidermis.
UAS-RNAi lines from Vienna Drosophila Research Center (Dietzl et al., 2007) (stock numbers indicated below in parentheses) were as follows: painRNAi#1 (v39477); PvrRNAi#2 (v13502); PvrRNAi#3 (v43459), and PvrRNAi#4 (v105353); Pvf2RNAi#2 (v7628) and Pvf2RNAi#4 (v7626); Pvf3RNAi#3 (v37933) and Pvf3RNAi#4 (v37935); PiezoRNAi#1 (v102440), PiezoRNAi#2 (v2796), PiezoRNAi#3 (v105132). UAS-RNAi lines from the TRiP (Ni et al., 2011) Bloomington collection were as follows: painRNAi#2 (BL31510), painRNAi#3 (BL31511), painRNAi#4 (BL51835); mCherryRNAi (BL35785) (RNAi targeting mCherry under UAS control) and UAS-LuciferaseRNAi (BL31603) were used as irrelevant control genes (not present in the Drosophila genome) for behavior and immunostaining experiments.
UAS-RNAi lines from NIG-Fly (http://www.shigen.nig.ac.jp/fly/nigfly/index.jsp) were as follows: PvrRNAi#1 (8222R-3), Pvf2RNAi#1 (formerly NIG13780), Pvf2RNAi#3 (13780R-1), Pvf2RNAi#5 (13780R-2), Pvf3RNAi#1 (13781R-1), and Pvf3RNAi#2 (13781R-3). UAS transgenes used: UAS-Pvf1 (McDonald et al., 2003), Pvf2d02444 (a UAS-containing EP allele of Pvf2 that allows overexpression of this ligand in the presence of a Gal4 driver) (Cho et al., 2002), and UAS-Pvf3 (Rosin et al., 2004) were used to overexpress Pvf1, Pvf2, and Pvf3, respectively. UAS-Pvr and UAS-PvrCA (Duchek et al., 2001) were used to overexpress the WT and the constitutively active forms of Pvr, respectively. UAS-PvrDN (Duchek et al., 2001) was used to overexpress the dominant negative form of Pvr. UAS-Pain (Al-Anzi et al., 2006) and UAS-Piezo (BL58773) were used to overexpress these ion channels.
Recombination between transgenes/alleles was genetically and molecularly confirmed. The oligonucleotide primers used to confirm the recombination of pain70, UAS-Pvr and piezo, UAS-Pvr are available on request.
Mechanical probes and mechanical nociception assays
We custom-built mechanical probes similar to von Frey filaments. Each probe was made by gluing (Aleene's All-Purpose Wood Glue) nitinol filaments (Mailin) of specified lengths and diameters to a graspable holder (Stained Craft Sticks, Loew Cornell) (see Fig. 1A). The probes were calibrated to apply a measured amount of force (in grams) when they bend against a scale (Table 1). The mass (m; recorded in grams) was then converted to force in millinewtons (mN) using the formula f = ma where the measured mass was multiplied by gravitational acceleration (9.8 m/s2). Finally, the measured force (in mN) was converted to pressure (force/area; in kilopascals [kPa]) by dividing the force by the surface area of the smoothened tip of the filament (Table 1).
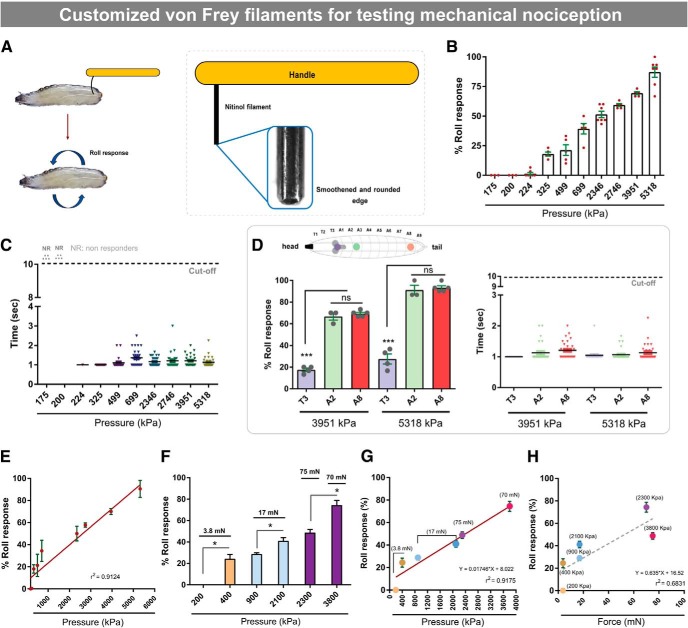
Figure 1.
Development of a refined mechanical nociception assay using von Frey-like filaments in Drosophila larvae. A, Schematic of mechanical probe design and assay. B, Dose–response of aversive rolling versus increasing mechanical pressure. C, Latencies of aversive rolling upon application of the indicated mechanical pressures. D, Aversive rolling to mechanical probes as a function of the anatomical location stimulated. Diagram represents the approximate locations of thoracic segment 3 (T3) and abdominal segments 2 (A2) and 8 (A8). Both percentage response (left graph) and response latencies (right graph) are shown. E, Linear regression model relating mechanical pressure to nociceptive behavior. F, Nociceptive behavioral responses to mechanical probes exerting approximately equivalent force but differing pressures. G, Linear regression model relating applied pressure to nociceptive responses. H, Linear regression model relating applied force to nociceptive responses. Error bars indicate mean ± SEM. Two-tailed unpaired t test (F) and one-way ANOVA with post hoc Tukey's test (D) were used for statistical analysis: *p < 0.05; ***p < 0.001, ns, not significant.
Table 1.
Calibration of the different mechanical probesa
| Measurement no. | Mechanical probes (P) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | P10 | |
| 1 | 0.14 | 0.16 | 0.16 | 0.27 | 0.40 | 0.55 | 1.80 | 4.92 | 7.00 | 17.36 |
| 2 | 0.14 | 0.16 | 0.18 | 0.27 | 0.40 | 0.55 | 1.90 | 5.10 | 7.20 | 17.40 |
| 3 | 0.14 | 0.16 | 0.19 | 0.27 | 0.40 | 0.58 | 1.92 | 4.94 | 7.22 | 16.46 |
| 4 | 0.13 | 0.15 | 0.19 | 0.26 | 0.41 | 0.57 | 1.87 | 4.86 | 7.20 | 17.00 |
| 5 | 0.13 | 0.16 | 0.18 | 0.26 | 0.39 | 0.55 | 1.90 | 5.00 | 6.99 | 16.90 |
| 6 | 0.14 | 0.16 | 0.18 | 0.26 | 0.40 | 0.57 | 1.88 | 4.81 | 7.20 | 16.90 |
| 7 | 0.14 | 0.15 | 0.18 | 0.27 | 0.40 | 0.56 | 1.89 | 4.99 | 7.01 | 17.40 |
| 8 | 0.13 | 0.15 | 0.17 | 0.26 | 0.40 | 0.57 | 1.85 | 4.95 | 7.04 | 17.42 |
| 9 | 0.14 | 0.16 | 0.17 | 0.26 | 0.39 | 0.58 | 1.92 | 4.91 | 6.99 | 16.53 |
| 10 | 0.14 | 0.16 | 0.18 | 0.26 | 0.40 | 0.55 | 1.91 | 5.05 | 7.33 | 17.00 |
| Average mass (g) | 0.14 | 0.16 | 0.18 | 0.26 | 0.40 | 0.56 | 1.88 | 4.95 | 7.12 | 17.04 |
| SD | 0.005 | 0.005 | 0.009 | 0.005 | 0.006 | 0.013 | 0.037 | 0.086 | 0.125 | 0.357 |
| Force (mN) | 1.37 | 1.57 | 1.76 | 2.55 | 3.92 | 5.49 | 18.42 | 48.51 | 69.78 | 166.99 |
| Pressure (kPa) | 175 | 200 | 224 | 325 | 499 | 699 | 2346 | 2746 | 3951 | 5318 |
aCalibration and forces/pressures produced by the different mechanical probes. Each mechanical probe was measured 10 times, and the average mass (in grams) is reported. The forces are converted to millinewtons (mN), and to pressure in kilopascals (kPa) based upon the diameter of the wire tip. The wire diameters of the different mechanical probes were as follows: P1–P7 = 0.01 cm; P8–P9 = 0.015 cm; P10 = 0.02 cm.
To assess behavioral responses in the nociceptive assay, each mechanical probe was applied to the posterior dorsal side of the larva (abdominal segment A8) until the filament bent, typically 1–2 s. A complete roll of 360 degrees along the body axis within 10 s of probe bending was considered a positive response. Other responses, such as fast crawling, attempting to turn, and wiggling, were categorized as negative responses. A 200 kPa probe was used as a subthreshold standard, and a 2346 kPa probe was used as a mid-range noxious probe. Other probes were used where indicated. The experimenter was blind to the genotype of the larvae being tested. Three independent experiments, each using 30 larvae, were performed unless otherwise indicated.
Immunofluorescence
To evaluate epidermal tissue damage induced by mechanical probes, third instar epidermal reporter larvae (e22c-Gal4, UAS-DsRed2Nuc(21), FasIII-GFP) were etherized (Ethyl Ether Anhydrous, Fisher Scientific, catalog #E138-500), dissected in ice-cold PBS, and then fixed for 1 h in 4% PFA. After several washes in PBS-Tx (1× PBS with 0.3% Triton X-100), all samples were mounted in Vectashield (Vector Laboratories, catalog #H-1000).
For isolated class IV sensory neuron immunostaining, the experimental procedure was as detailed previously (Im et al., 2015). Briefly, third instar larvae expressing ppk-Gal4>UAS-mCD8-GFP were cut open in Schneider's medium (where gut and fat body were then removed from the carcass by vortexing) and washed three times in 1 ml of Rinaldini solution. The cleaned carcasses were incubated to 0.5 mg/ml Collagenase I solution (Sigma-Aldrich, catalog #C9722) for 1 h at room temperature, washed in Schneider's medium (Thermo Fisher Scientific, catalog #21720-024), and then mechanically dissociated by repeated pipetting for 10 min. The dissociated tissue was then filtered through a 40 μm cell strainer (Corning, product # 431750), and cells were incubated with anti-mCD8a antibody-conjugated magnetic beads (eBioscience, catalog #13-0081-82) on ice for 1 h followed by washes. Finally, isolated class IV sensory neurons were plated on concanavalin A (Sigma-Aldrich, catalog #C5275) coated coverslips and immunostained with primary rabbit anti-Pvr antibody (1:500 dilution in PBS-Tx; gift from Jocelyn McDonald, Kansas University) (McDonald et al., 2003) at 4°C overnight. The secondary antibody was applied for 2 h at room temperature and was a Cy3-conjugated goat antiserum (1:500 dilution in PBS-Tx) against rabbit IgG (Jackson ImmunoResearch Laboratories, catalog #111-165-003). Nuclei were identified by DAPI (Sigma-Aldrich, catalog #D9542) staining (1× dilution in PBS-Tx, 10 min incubation followed by washing in PBS-Tx). All stained samples were mounted in Vectashield (Vector Laboratories, catalog #H-1000).
Confocal microscopy and stereomicroscope
Larvae were imaged on an Olympus FV1000 Confocal microscope, and Fluoview software was used to obtain the images. The following confocal microscope lasers were used: wavelengths 405 (DAPI), 488 (GFP), and 543 (far red fluorescent protein). Images were captured at a resolution of 1024 × 1024 pixels for tissue damage experiments using a 20× numerical aperture (NA) 0.85 oil objective lens at 1× zoom; for isolated sensory neurons, images were captured at the same resolution using a 60×, NA 1.4 oil objective lens at 4× zoom. A Z-series stack, step size of 1.5 μm, was collected and processed into a single Z projection. Identical settings for laser intensity and other image capture parameters were applied for comparison of staining in the control and experimental groups. A MZ16FA stereomicroscope (Leica Microsystems) with Planapo 1.6× objective was used to image scab formation. All figures were assembled with Photoshop CS6 and Illustrator CS6 (Adobe).
Quantitation of tissue damage
To measure the area of epidermal tissue damage, a maximum intensity projection Z-stack image was created for each wounded/probed larval whole mount. The area of the gap (if any) in the epidermal membrane marker (FasciclinIII-GFP) was measured using the Fluoview software.
Quantitation of Pvr expression in Class IV neurons
To measure Pvr levels in class IV sensory neurons, total fluorescence measurements were performed using Fiji software on maximum intensity projection Z-stack images (McCloy et al., 2014) of Pvr-stained neurons.
Quantitation of Class IV neuronal morphology
Drosophila third instar larvae (ppk-Gal4>UAS-mCD8-GFP with or without Pvr, Pvf2, or Pvf3 allele combinations; see Fig. 4) were etherized, immobilized, gently immersed in 1:5 (v/v) diethyl ether to halocarbon oil (Sigma-Aldrich, catalog #H8898), and covered with 22 × 50 mm glass coverslips (Das et al., 2017). Images of Class IV multidendritic sensory neurons (right hemi-segment A5 or A6) were captured using an Olympus Fluoview confocal microscope at a resolution of 1024 × 1024 pixels using a 20× dry objective (1.4× zoom, NA 0.7), at a step size of 1.5 μm. Images obtained from maximum intensity projection of the Z stacks were manually curated as necessary (Ferreira et al., 2014; Das et al., 2017) to eliminate nonspecific autofluorescence. Image processing was performed in Fiji. Background image noise was eliminated by 3D median filtering (typical radius = 1–3) (Ferreira et al., 2014). The pixel intensity for each image was globally thresholded in Fiji/ImageJ. The dendritic morphology of each image was skeletonized using the Skeletonize3D plugin in Fiji/ImageJ followed by use of the Analyze Skeleton Fiji/ImageJ plugin (Das et al., 2017). The resulting neurometric data (total dendritic length and total dendritic branches) were analyzed and plotted in GraphPad. The Sholl analysis plugin in Fiji/ImageJ (Ferreira et al., 2014) was used to quantify the distribution of branches within a dendritic arbor and determine the critical value (or peak number of intersections of the arbor tree with arbitrary concentric circles [centered on the cell body] of increasing radius). The corresponding critical radius (the distance from the cell soma at which the critical value is observed) was also measured (Das et al., 2017). All images were assembled using Photoshop CS6 and Illustrator CS6 (Adobe). Total dendritic length and branch numbers were expressed as mean ± SEM. The Sholl profile for each neuron was fitted to an eighth-degree polynomial, from which the critical value (the maximum of the polynomial function) and critical radius (the distance at which the critical value occurs) were determined.
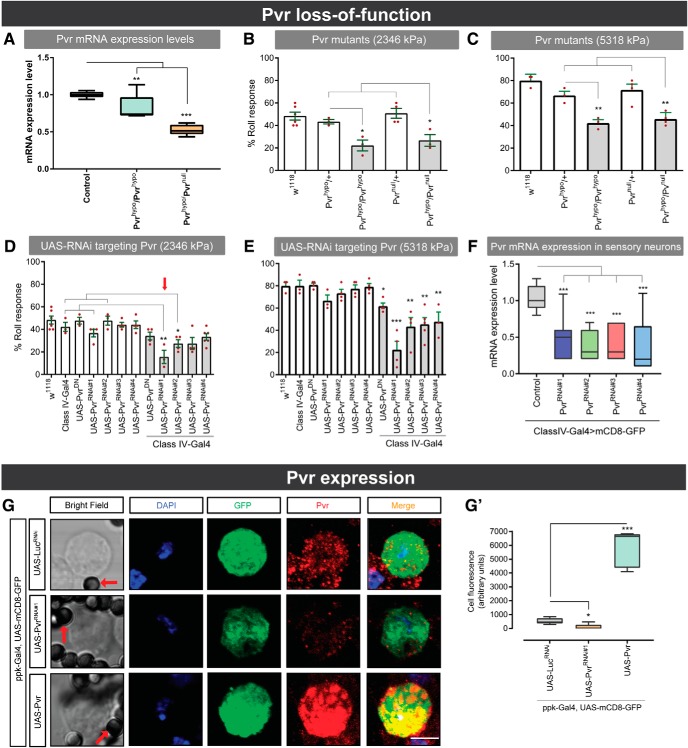
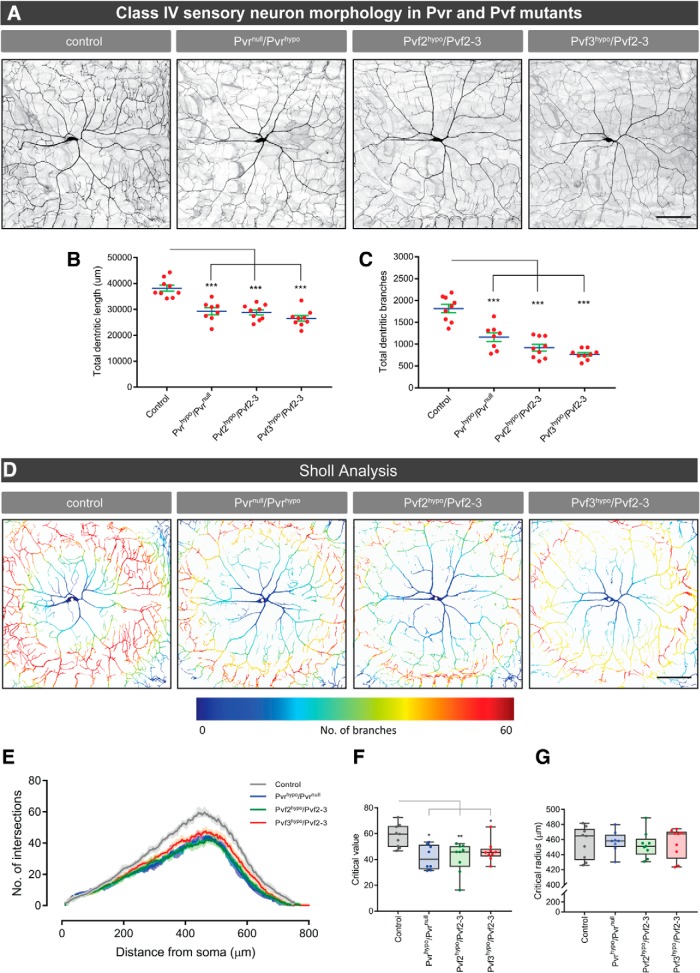
Figure 4.
Pvr is expressed in Class IV sensory neurons and is required for mechanical nociception. A, Pvr mRNA expression in Pvr mutants and control larvae. B, C, Behavioral responses to noxious mechanical pressures of 2346 kPa (B) and 5318 kPa (C) of indicated Pvr mutants and associated controls. D, E, Responses to noxious mechanical pressures of 2346 kPa (D) and 5318 kPa (E) of relevant genetic controls and larvae expressing UAS-PvrRNAi in class IV multidendritic sensory neurons. F, Pvr mRNA expression levels in relevant control larvae and larvae expressing UAS-PvrRNAi transgenes in class IV sensory neurons. G, Pvr expression in isolated class IV multidendritic sensory neurons expressing GFP. Red arrow indicates magnetic beads. G′, Cell fluorescence quantification. Box plots of total fluorescence represent median (horizontal line). Whiskers represent the 2.5th and 97.5th percentiles. C, F, Box plots represent median (horizontal line). Whiskers represent the 10th and 90th percentiles. Data are mean ± SEM. Statistical analysis was performed using two-tailed unpaired t test, one-way ANOVA with Dunnett's and Tukey's post hoc tests: *p < 0.05; **p < 0.01; ***p < 0.001.
Measuring mRNA levels from whole larvae and Class IV neurons
Total RNA from 50 larvae was extracted using Trizol (Invitrogen, catalog #15596018). RNA quantity and quality were determined by measuring absorbance at 260 and 260 nm/280 nm (ratio of 1.8–2.1), using NanoDrop technology (NANODROP 1000 spectrophotometer, Thermo Fisher Scientific). Then RNA samples treated with DNase I (New England BioLabs, catalog #M0303S) at 37°C for 10 min were purified using the RNeasy Mini Kit (QIAGEN, catalog #74104). cDNA synthesis was performed using the High Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific, catalog #4387406). The cDNA samples were then treated with RNase H (New England BioLabs, catalog #M0297S) for 20 min at 37°C, and purified using the QIAquick PCR Purification Kit (QIAGEN, catalog #28104). qPCR products from this cDNA template were measured using the curve of the SYBR Green method (Thermo Fisher Scientific, catalog #4367659) and an Applied Biosystems QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific). GAPDH was used as reference gene: GAPDH (forward), TAAATTCGACTCGACTCACGGT; GAPDH (reverse), CTCCACCAACATACTCGGCTC amplified a product of 151 bp. The primers used to amplify Pvr were Pvr (forward), GGATGAGAATCGCATATTTGCCT; Pvr (reverse), ACTTTCGTTTTGCTTTAGCACCT, which amplified a 205 bp product spanning two exons common to all 13 isoforms of Pvr. Reaction conditions were as follows: 10 min at 95°C followed by 35 cycles of 15 s at 95°C and 1 min at 64°C. Three samples were taken for each gene, and the qRT-PCR was repeated three times for each sample. The ddCt, 2−ΔΔCT method was used to calculate real-time PCR results.
The gene expression analysis of Pvr-mRNA levels in isolated class IV sensory neurons was performed by TaqMan Gene Expression Assay (Thermo Fisher Scientific, catalog #4369016). Briefly, 15 class IV sensory neurons (ppk-Gal4>UAS-mCD8-GFP) for each genotype were collected using FACS. Single Cell-to-CT qRT-PCR Kit (Thermo Fisher Scientific, catalog #4458237) was used for sample preparation, reverse transcription, preamplification, and qPCR. The predesigned TaqMan Assay, probe and primer sets, used for Pvr (TaqMan Assay ID: Dm01803625_m1) covered all the Pvr transcripts variants. Rpl32 was used as gene of reference, TaqMan Assay ID: Dm02151827_g1.
Vertebrate methods
Animals.
Male Sprague Dawley rats (150–200 g, Envigo, catalog #002) were housed in groups of 2 and were maintained on a 12 h light/dark cycle with ad libitum access to food and water. Rats habituated to the colony room for 1 week before experimental manipulations. All protocols were approved by the University of Pittsburgh Institutional Animal Care and Use Committee.
Drug administration.
Peptides: Carrier-free PDGF and VEGF peptides were obtained from R&D Systems (PDGF-AA catalog #1055-AA; PDGF-BB catalog #520-BB/CF; PDGF-CC catalog #1687-CC/CF; VEGF-A catalog #564-RV/CF; VEGF-B catalog #2595-VE/CF; VEGF-C catalog #9199-VC/CF; VEGF-D catalog #469-VD/CF). They were reconstituted as directed by the manufacturer and stored at −80°C until used. Inhibitors: VEGFR-1, ZM306416 (ChemCruz Biochemicals, catalog #HY-13785); VEGFR-2, cabozantinib (LC Laboratories, catalog #C-9000); and VEGFR-3, SAR 131675 (APEX Bio, catalog #B2301). Drugs were dissolved in DMSO (Thermo Fisher Scientific, catalog #D128-500; 100 mg/ml) before dilution to working concentration. Morphine (catalog #M8777) and naloxone (catalog #PHR1802) were obtained from Sigma Aldrich. All drugs and peptides were dissolved to their working concentration in a solution of 10% β-cyclodextrin sulfobutyl ether (Captisol, CyDex, catalog #RC-0C7-100) and 0.9% saline, which was used as vehicle in all vertebrate experiments. Drugs were administered daily via lumbar puncture (20 μl/injection) as previously described (Xu et al., 2006).
Nociceptive testing.
Mechanical sensitivity was measured using the Randall-Selitto paw-pressure test (Analgesy-meter, Ugo Basile). A linearly increasing pressure was applied to the hindpaw of rats. The weight (in grams) eliciting a paw withdrawal was defined as the mechanical nociceptive threshold. A maximum of 250 g was used as a cutoff to avoid tissue damage. Animals were handled for 4 d, and baselines were taken 1 d before the beginning of the injections. Nociceptive testing was performed daily 40 min after intrathecal injection.
Statistical analysis
All statistical analyses were performed using Prism version 7 (GraphPad Software). Drosophila data were tested for normal distribution using Kolmogorov–Smirnov tests, then were analyzed using either the two-tailed unpaired Student's t test or one-way ANOVA followed by Tukey's or Dunnett's post hoc tests. Rodent behavioral assays were analyzed using two-way ANOVA followed by Sidak or Dunnett's post hoc tests.
Results
A new tool for assessing mechanical nociception in Drosophila larvae
To evaluate mechanical nociception in Drosophila larvae, we modified an established assay (Zhong et al., 2010; Kim et al., 2012; Gorczyca et al., 2014; Guo et al., 2014) with the goal of generating a stimulus–response curve extending from subthreshold mechanical stimuli that did not produce a behavioral response through stimuli that produced the stereotypical rolling response in nearly all larvae. We constructed mechanical probes of defined length and diameter from nitinol wire. These are similar to the von Frey filaments used for testing mechanical stimuli in humans and other vertebrates (Bonin et al., 2014). Each probe was smoothed at the tip and attached to a handle (Fig. 1A) before calibration (Table 1) and conversion of measured force values to pressure based on the tip area (for details, see experimental procedures and Table 1).
To assess behavior, each probe was pressed against the larva until bending, exerting a constant force and pressure. A response of a 360 degree roll along the body axis was considered aversive (Fig. 1A; Movie 1). This response differed from normal locomotion and light touch responses (Movie 2). Pressures <200 kPa did not elicit rolling responses (Fig. 1B). The prevalence of observed aversive rolling steadily increased with applied pressure from 224 to 5318 kPa (Fig. 1B). When we measured response latency (time to rolling after probe bending), we found that all probes induced aversive rolling between 1 and 2 s after probe application (Fig. 1C). To determine whether responsiveness differed along the larval body axis, we stimulated thoracic (T3) or abdominal (A2 and A8) segments with noxious probes (Fig. 1D). Neither percentage of responders nor latency (Fig. 1D) differed between A2 and A8. Most larvae failed to respond when these probes, 3951 kPa (one-way ANOVA, F(2,9) = 242.9, p < 0.0001, Tukey's post hoc test for T3 vs A2 comparison: p < 0.0001; for T3 vs A8 comparison: p < 0.0001) and 5318 kPa (one-way ANOVA, F(2,9) = 116.1, p < 0.0001, Tukey's post hoc test for T3 vs A2 comparison: p < 0.0001; for T3 vs A8 comparison: p < 0.0001), were applied to the T3 segment (Fig. 1D). A8 was chosen for our standard assay because it was procedurally easier to stimulate this segment.
Aversive response to noxious mechanical stimulation.

Normal larval locomotion.

Studies of mechanical nociception in Drosophila larvae have generated variable behavioral responses from probes delivering similar forces (Zhong et al., 2010; Kim et al., 2012; Guo et al., 2014; Hu et al., 2017). We observed that the relationship between pressure and behavioral responsiveness was linear (Fig. 1E). To test whether force or pressure was the main determinant of behavioral responsiveness, we used probe pairs that generated nearly identical forces but differing pressures. Behavioral responses differed markedly between probes generating identical forces (Fig. 1F). For one pair of 3.8 mN probes, one generating 200 kPa of pressure produced no aversive responses, whereas the other probe did (400 kPa; 24% responders) (t test, p < 0.001). Two 17 mN probes generating different pressures also produced differing behavioral responses (28% vs 41% responders; Fig. 1F,G) (t test, p = 0.0003). At higher intensities, a probe of slightly lower force generating higher pressure (70 mN, 3800 kPa) produced more rolling behavior than a probe of higher force generating less pressure (75 mN, 2300 kPa) (t test, p < 0.001; Fig. 1F,G). Thus, the relationship between applied force and behavioral responsiveness is not linear (Fig. 1H). In sum, our results suggest that the most robust and reliable way to assess larval mechanical nociception is by plotting applied pressure (force/area of probe tip) versus behavioral responsiveness.
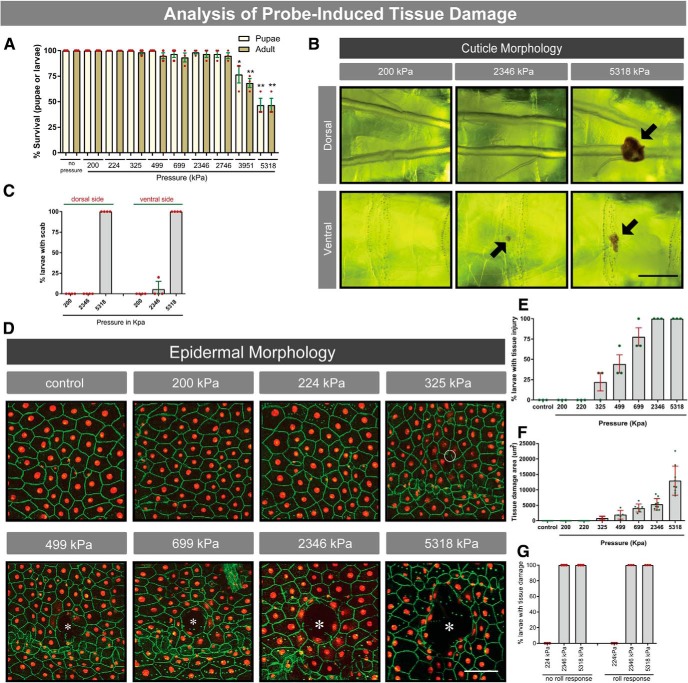
A determinant of whether a particular mechanical stimulus is noxious includes whether the stimulus impacts survival and/or causes tissue damage. We determined whether application of increasing mechanical pressure to larvae affected survival to the pupal or adult stage. Pressures <2746 kPa did not reduce survival (Fig. 2A). However, probing once with a 3951 kPa probe or a 5318 kPa probe reduced survival by 30% (t test, pupae p = 0.0488; t test, adult p = 0.0020) and 50% (t test, pupae p = 0.0013; t test, adult p = 0.0013), respectively (Fig. 2A). Pressures >2346 kPa induced visible cuticular scabs (Fig. 2B) and gaps in the epidermal sheet (Fig. 2D,E). The percentage of larvae exhibiting scabs (Fig. 2C) and epidermal disruption (Fig. 2E) increased with increasing pressure, as did the area of tissue damage (Fig. 2F). As expected, a subthreshold probe (200 kPa) did not induce visible tissue damage. Interestingly, our data showed that even larvae that did not show a behavioral response to 2346 kPa pressure (∼50% of larvae) suffered tissue damage (Fig. 2G), indicating that stimuli that do not elicit aversive rolling can cause epidermal damage.
Figure 2.
Survival and tissue damage analysis induced by mechanical stimulation. A, Larval survival in response to increasing pressure. Yellow bars represent pupae. Brown bars represent adults. B, Live whole mounts of third instar larvae probed with the indicated pressures at dorsal segment A8 and viewed by light microscopy. Black arrows indicate melanized scabs. Scale bar, 250 μm. C, Percentage of larvae developing melanized scabs on the dorsal and ventral sides. D, Dissected whole mounts of third instar larvae with fluorescently labeled epidermis probed at dorsal segment A8 with indicated pressures. Red represents epidermal nuclei. Green represents epidermal cell membranes. Dark areas (white asterisks) represent epidermal gaps. White circle represents broken epidermal membrane. Scale bar, 100 μm. Percentage of open wounds (E) and area of epidermal damage (F) after mechanical stimulation (E,F; n = 9 animals for each condition). G, Percentage of larvae that showed tissue damage, probed at dorsal segment A8 with the indicated pressures, that did or did not exhibit a nociceptive behavioral response. Error bars indicate mean ± SEM. Two-tailed unpaired t test was used for statistical analysis: *p < 0.05; **p < 0.01.
The PDGF/VEGF-like ligands Pvf2 and Pvf3 mediate mechanical nociception
Because PDGFRs and VEGFRs have been implicated in mechanical nociception in vertebrates (Masuda et al., 2009; Hulse et al., 2014), we hypothesized that PDGF/VEGF-related ligands might affect mechanical nociception in flies. The Drosophila genome encodes three such ligands: Pvf1, Pvf2, and Pvf3 (Heino et al., 2001; Cho et al., 2002). Larvae homozygous for a Pvf1 null allele (Cho et al., 2002) showed mechanical nociception responses equivalent to relevant controls (Fig. 3A). By contrast, larvae homozygous or transheterozygous for two alleles of both Pvf2 and Pvf3 showed substantially attenuated mechanical nociception (Fig. 3A) (t test, Pvf2 homozygous vs Pvf2/+, p = 0.0014; t test, Pvf3 homozygous vs Pvf3/+, p = 0.0017; t test, Pvf3MI04168 homozygous vs Pvf3MI04168/+, p = 0.046; one-way ANOVA, Pvf2 heterozygous, F(2,6) = 25.95, p = 0.0011, Tukey's post hoc test for Pvf2hypo/Pvf2MI00770 vs Pvf2hypo/+ comparison: p = 0.0014; for Pvf2hypo/Pvf2MI00770 vs Pvf2MI00770/+ comparison: p = 0.0028; one-way ANOVA, Pvf3 heterozygous, F(2,6) = 9.972, p = 0.0124, Tukey's post hoc test for Pvf3hypo/Pvf3MI04168 vs Pvf3hypo/+ comparison: p = 0.0126; for Pvf3hypo/Pvf3MI04168 vs Pvf3MI04168/+ comparison: p = 0.0383). The Pvf2MI00770 mimic allele had a mild phenotype when homozygous but led to a transheterozygous phenotype significantly more severe than either Pvf2 allele (Pvf2MI00770 or Pvf2hypo) alone. We also tested larvae-bearing allele combinations targeting both Pvf2 and Pvf3 (Parsons and Foley, 2013), as these genes are chromosomally adjacent. Larvae carrying hypomorphic alleles of Pvf2 or Pvf3 over a genomic deletion that disrupts both Pvf2 and Pvf3 (Pvf2-3) displayed attenuated mechanical nociceptive responses (one-way ANOVA, F(2,8) = 43.68, p < 0.0001, Tukey's post hoc test for Pvf2hypo/Pvf2-3 vs Pvf2hypo/+ comparison: p < 0.0001; Pvf2hypo/Pvf2-3 vs Pvf2-3/+ comparison: p < 0.0001; for Pvf3hypo/Pvf2-3 vs Pvf3hypo/+ comparison: p < 0.0001; for Pvf3hypo/Pvf2-3 vs Pvf2–3/+ comparison: p < 0.0001). Comparison between larvae carrying the hypomorphic alleles of Pvf2 or Pvf3 and the Pvf2MI00770 or Pvf3MI04168 mimic alleles also showed a decreased mechanical nociceptive response (one-way ANOVA, F(2,8) = 11.25, p = 0.0047, Tukey's post hoc test for Pvf2MI00770/Pvf3MI04168 vs Pvf2MI00770/+ comparison: p = 0.0088; for Pvf2MI00770/Pvf3MI04168 vs Pvf3MI04168/+ comparison: p = 0.0133; one-way ANOVA, F(2,7) = 18.51, p = 0.0016, Tukey's post hoc for Pvf2hypo/Pvf3MI04168 vs Pvf2hypo/+ comparison: p = 0.002; for Pvf2hypo/Pvf3MI04168 vs Pvf3MI04168/+ comparison: p = 0.0062; one-way ANOVA, F(2,6) = 17, p = 0.0034, Tukey's post hoc test for Pvf3hyp.o./Pvf2MI00770 vs Pvf3hypo/+ comparison: p = 0.0039; for Pvf3hypo/Pvf2MI00770 vs Pvf2MI00770/+ comparison: p = 0.0090). The strongest defects were observed with larvae transheterozygous for various hypomorphic alleles of Pvf2/Pvf3 (one-way ANOVA, F(2,6) = 27.31, p = 0.0010, Tukey's post hoc test for Pvf2hypo/Pvf3hypo vs Pvf2hypo/+ comparison: p = 0.0019; for Pvf2hypo/Pvf3hypo vs Pvf3hypo/+ comparison: p = 0.0015) (Fig. 3A). These results indicate that Pvf2 and Pvf3 are required for mechanical nociception and suggest that they may work in concert.
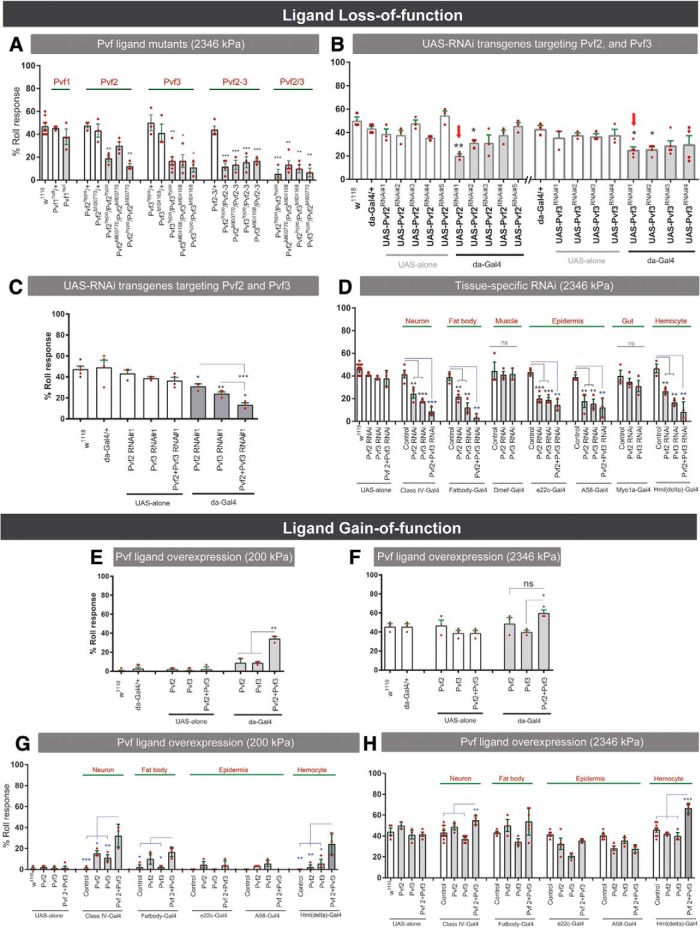
Figure 3.
The Pvr-ligands Pvf2 and Pvf3 are required for mechanical nociception. A, Responses to noxious mechanical pressure (2346 kPa) of the indicated Pvf alleles and respective controls. B, Responses to noxious mechanical pressure (2346 kPa) of relevant controls and UAS-RNAi transgenes targeting either Pvf2 or Pvf3. Red arrows indicate the strongest RNAi transgenes. C, Responses to noxious mechanical pressure (2346 kPa) of relevant controls and UAS-RNAi transgenes targeting both Pvf2 and Pvf3 ligands in all tissues. D, Responses to noxious mechanical pressure (2346 kPa) of relevant controls and larvae expressing UAS-Pvf2RNAi#3 or UAS-Pvf3RNAi#4 in the indicated tissues. E, F, Response to non-noxious (200 kPa) (E) and noxious (2346 kPa) (F) mechanical pressure of relevant controls and larvae ubiquitously overexpressing Pvf2 and Pvf3. G, H, Behavioral responses of larvae overexpressing Pvf ligands in the indicated combinations and indicated tissues to 200 kPa (G) and 2346 kPa (H) mechanical probes. Controls: w1118 or tissue-specific Gal4 driver or UAS transgene alone. Data are mean ± SEM. Statistical analysis was performed using two-tailed unpaired t test, and one-way ANOVA with post hoc Tukey's test: *p < 0.05; **p < 0.01; ***p < 0.001, ns, not significant.
To determine the tissue source(s) of functional Pvf2 and Pvf3, we expressed UAS-RNAi transgenes targeting them in specific tissues and evaluated mechanical nociception in progeny larvae. Ubiquitous expression of select UAS-RNAi transgenes targeting Pvf2 or Pvf3 via the ubiquitously expressed da-Gal4 significantly attenuated mechanical nociception and identified the strongest UAS-RNAi transgene targeting each ligand (t test, p = 0.0132; t test, p = 0.048) (Fig. 3B). Consistent with our mutant analysis, targeting Pvf2 and Pvf3 together led to a stronger defect in mechanical nociception (one-way ANOVA, F(2,6) = 49.05, p = 0.0002, Tukey's post hoc test for da-Gal4>Pvf2RNAi#1+Pvf3RNAi#1 vs Pvf2RNAi#1/+ comparison: p = 0.0002; for da-Gal4>Pvf2RNAi#1+Pvf3RNAi#1 vs Pvf3RNAi#1 comparison: p = 0.0004) (Fig. 3C). We used the strongest UAS-RNAi transgenes to survey which tissues might produce active ligands. Targeting Pvf2 or Pvf3 in fat body (t test, fatbody-Gal4>Pvf2RNAi vs fatbody-Gal4/+, p = 0.0046; t test, fatbody-Gal4>Pvf3RNAi vs fatbody-Gal4/+, p = 0.0058), class IV nociceptive sensory neurons (t test, classIV-Gal4>Pvf2RNAi vs classIV-Gal4/+, p = 0.0229; t test, classIV-Gal4>Pvf3RNAi vs classIV-Gal4/+, p = 0.0024), epidermis (t test, e22c-Gal4>Pvf2RNAi vs e22c-Gal4/+, p = 0.0008; t test, e22c-Gal4>Pvf3RNAi vs e2cc-Gal4/+, p = 0.0011; t test, A58-Gal4>Pvf2RNAi vs A58-Gal4/+, p = 0.0238; t test, A58-Gal4>Pvf3RNAi vs A58-Gal4/+, p = 0.0069), and hemocytes (t test, Hml(delta)-Gal4>Pvf2RNAi vs Hml(delta)-Gal4/+, p = 0.0096; Hml(delta)-Gal4> Gal4>Pvf3RNAi vs Hml(delta)-Gal4/+, p = 0.0022) (Fig. 3D) led to defects in mechanical nociception. This distributed nature of the requirement for Pvf ligands suggested that both ligands might function within the same tissue. When we targeted Pvf2 and Pvf3 together in the same tissue, we observed a stronger mechanical nociception defect in all of the tissues tested (t test, fatbody-Gal4>Pvf2RNAi + Pvf3RNAi vs fatbody-Gal4/+, p = 0.0005; t test, classIV-Gal4>Pvf2RNAi + Pvf3RNAi vs classIV-Gal4/+, p = 0.0008; t test, Hml(delta)-Gal4>Pvf2RNAi + Pvf3RNAi vs Hml(delta)-Gal4/+, p = 0.0015; t test, e22c-Gal4>Pvf2RNAi + Pvf3RNAi vs e22c-Gal4/+, p = 0.0012; t test, A58-Gal4> Pvf2RNAi vs A58-Gal4/+, p = 0.0074) (Fig. 3D). These data suggest that Pvf2 and Pvf3 may have additive effects on mechanical nociception.
We further tested the possibility that Pvf2 and Pvf3 work together using a gain-of-function approach. Ubiquitous overexpression of Pvf2, Pvf3, or both ligands simultaneously induced “genetic” mechanical allodynia (one-way ANOVA, F(2,6) = 25.32, p = 0.0012, Tukey's post hoc test for da-Gal4>Pvf2+Pvf3 vs da-Gal4>Pvf2 comparison: p = 0.0020; for da-Gal4>Pvf2+Pvf3 vs da-Gal4>Pvf3 comparison: p = 0.0020) (Fig. 3E) and mechanical hyperalgesia (one-way ANOVA, F(2,6) = 5.712, p = 0.0408, Tukey's post hoc test for da-Gal4>Pvf2+Pvf3 vs da-Gal4>Pvf2 comparison: p = 0.1108; for da-Gal4>Pvf2+Pvf3 vs da-Gal4>Pvf3 comparison: p = 0.0180) (Fig. 3F), which was more pronounced when the ligands were coexpressed. Genetically induced mechanical allodynia was more pronounced in most tissues when both ligands were coexpressed than when they were expressed singly (Fig. 3G) (neuron: one-way ANOVA, F(3,9) = 16.16, p = 0.0006, Tukey's post hoc test for classIV-Gal4>Pvf2+Pvf3 vs classIV-Gal4/+ comparison: p = 0.0002; for classIV-Gal4>Pvf2+Pvf3 vs classIV-Gal4>Pvf2 comparison: p = 0.0190; for classIV-Gal4>Pvf2+Pvf3 vs classIV-Gal4>Pvf3 comparison: p = 0.0049; fatbody: one-way ANOVA, F(3,9) = 5.904, p = 0.0165, Tukey's post hoc test for fatbody-Gal4>Pvf2+Pvf3 vs fatbody-Gal4/+ comparison: p = 0.0166; for fatbody-Gal4>Pvf2+Pvf3 vs fatbody-Gal4>Pvf3 comparison: p = 0.0164; hemocyte: one-way ANOVA, F(3,8) = 8.955, p = 0.0062, Tukey's post hoc test for Hml(delta)-Gal4>Pvf2+Pvf3 vs Hml(delta)-Gal4/+ comparison: p = 0.0043; for Hml(delta)-Gal4>Pvf2+Pvf3 vs Hml(delta)-Gal4>Pvf2 comparison: p = 0.0075; for Hml(delta)-Gal4>Pvf2+Pvf3 vs Hml(delta)-Gal4>Pvf3 comparison: p = 0.0190). Mechanical hyperalgesia was most pronounced when both ligands were overexpressed in neurons or hemocytes (neuron: one-way ANOVA, F(3,19) = 9.746, p = 0.0004, Tukey's post hoc test for classIV-Gal4>Pvf2+Pvf3 vs classIV-Gal4/+ comparison: p = 0.0042; for classIV-Gal4>Pvf2+Pvf3 vs classIV-Gal4>Pvf3 comparison: p = 0.0002; hemocyte: F(3,14) = 18.88, p < 0.0001, Tukey's post hoc test for Hml(delta)-Gal4>Pvf2+Pvf3 vs Hml(delta)-Gal4/+ comparison: p = 0.0001; for Hml(delta)-Gal4>Pvf2+Pvf3 vs Hml(delta)-Gal4>Pvf2 comparison: p = 0.0001; for Hml(delta)-Gal4>Pvf2+Pvf3 vs Hml(delta)-Gal4>Pvf3 comparison: p = 0.0001) (Fig. 3H).
Pvr, a PDGFR/VEGFR-like receptor tyrosine kinase, mediates mechanical nociception
Loss of Pvf ligands attenuated responsiveness to noxious mechanical stimulation, whereas overexpression induced mechanical hypersensitivity (Fig. 3). This suggested that the Pvr RTK might also be required for mechanical nociception. We therefore assayed mechanical nociceptive responses in Pvr mutants. Larvae homozygous for hypomorphic Pvr alleles and transheterozygous for hypomorphic/null alleles of Pvr, all of which showed reduced Pvr mRNA (one-way ANOVA, F(2,30) = 66.40, p < 0.0001, Dunnett's post hoc test for Pvrhypo/Pvrhypo vs control (w1118) comparison: p = 0.0009; for Pvrhypo/Pvrnull vs control (w1118) comparison: p = 0.0001) (Fig. 4A), exhibited decreased responsiveness to a noxious 2346 kPa probe (t test, Pvrhypo/Pvrhypo vs w1118, p = 0.030; one-way ANOVA, F(2,7) = 8.67, p = 0.0128, Tukey's post hoc test for Pvrhypo/Pvrnull vs Pvrhypo/+ comparison: p = 0.0734; for Pvrhypo/Pvrnull vs Pvrnull/+ comparison: p = 0.0107) (Fig. 4B) and 5318 kPa probe (t test, Pvrhypo/Pvrhypo vs w1118, p = 0.0072; one-way ANOVA, F(2,8) = 11.63, p = 0.0043, Tukey's post hoc test for Pvrhypo/Pvrnull vs Pvrhypo/+ comparison: p = 0.0218; Pvrhypo/Pvrnull vs Pvrnull/+ comparison: p = 0.0045) (Fig. 4C). Thus, Pvr is required for full responsiveness to noxious mechanical stimuli.
We next determined whether Pvr was expressed in and functions in class IV multidendritic nociceptive sensory neurons (Hwang et al., 2007; Zhong et al., 2010; Kim et al., 2012). Expressing Pvr-specific UAS-DN or UAS-RNAi transgenes in class IV sensory neurons diminished responsiveness to a noxious 2346 kPa (PvrRNAi#1: one-way ANOVA, F(2,7) = 9.721, p = 0.0095, Tukey's post hoc test for classIV-Gal4>PvrRNAi#1 vs classIV-Gal4/+ comparison: p = 0.0079; for classIV-Gal4>PvrRNAi#1 vs PvrRNAi#1/+ comparison: p = 0.0179; PvrRNAi#2: one-way ANOVA, F(2,7) = 8.23, p = 0.0145, Tukey's post hoc test for classIV-Gal4>PvrRNAi#2 vs classIV-Gal4/+ comparison: p = 0.0466; for classIV-Gal4>PvrRNAi#2 vs PvrRNAi#2/+ comparison: p = 0.0113) (Fig. 4D) and 5318 kPa probe (PvrDN: one-way ANOVA, F(2,8) = 12.62, p = 0.0034, Tukey's post hoc test for classIV-Gal4>PvrDN vs classIV-Gal4/+ comparison: p = 0.0070; for classIV-Gal4>PvrDN vs PvrDN/+ comparison: p = 0.0035; PvrRNAi#1: one-way ANOVA, F(2,7) = 22.44, p = 0.0009, Tukey's post hoc test for classIV-Gal4>PvrRNAi#1 vs classIV-Gal4/+ comparison: p = 0.0008; for classIV-Gal4>PvrRNAi#1 vs PvrRNAi#1/+ comparison: p = 0.0035; PvrRNAi#2: one-way ANOVA, F(2,7) = 8.23, p = 0.0145, Tukey's post hoc test for classIV-Gal4>PvrRNAi#2 vs classIV-Gal4/+ comparison: p = 0.0466; for classIV-Gal4>PvrRNAi#2 vs PvrRNAi#2/+ comparison: p = 0.0113; PvrRNAi#3: one-way ANOVA, F(2,9) = 14.26, p = 0.0016, Tukey's post hoc test for classIV-Gal4>PvrRNAi#3 vs classIV-Gal4/+ comparison: p = 0.0028; for classIV-Gal4>PvrRNAi#3 vs PvrRNAi#3/+ comparison: p = 0.0029; PvrRNAi#4: one-way ANOVA, F(2,7) = 10.52, p = 0.0078, Tukey's post hoc test for classIV-Gal4>PvrRNAi#4 vs classIV-Gal4/+ comparison: p = 0.0102; for classIV-Gal4>PvrRNAi#4 vs PvrRNAi#4/+ comparison: p = 0.0083) (Fig. 4E). All of the RNAi transgenes tested substantially reduced Pvr mRNA levels within class IV sensory neurons (Fig. 4F) (one-way ANOVA, F(4,40) = 10.84, p < 0.0001, Dunnett's post hoc test for classIV-Gal4>mCD8-GFP>PvrRNAi#1 vs classIV-Gal4>mCD8-GFP/+ comparison: p = 0.0002; for classIV-Gal4>mCD8-GFP>PvrRNAi#2 vs classIV-Gal4>mCD8-GFP/+ comparison: p = 0.0001; for classIV-Gal4>mCD8-GFP>PvrRNAi#3 vs classIV-Gal4>mCD8-GFP/+ comparison: p = 0.0001; for classIV-Gal4>mCD8-GFP>PvrRNAi#4 vs classIV-Gal4>mCD8-GFP/+ comparison: p = 0.0001). The behavioral and mRNA expression results suggest that Pvr is expressed in class IV multidendritic neurons. Harvesting and plating class IV nociceptive sensory neurons revealed that Pvr was present, and its levels could be reduced by expression of UAS-PvrRNAi (t test, ppk-Gal4, mCD8-GFP>PvrRNAi#1 vs ppk-Gal4, mCD8-GFP>LucRNAi#, p = 0.0032) or enhanced by expression of UAS-Pvr (t test, ppk-Gal4, mCD8-GFP>Pvr vs ppk-Gal4, mCD8-GFP>LucRNAi#, p = 0.0004) (Fig. 4G–G′).
Pvr and Pvf2/Pvf3 alter dendritic branching of class IV sensory neurons
Because Pvr is expressed in class IV sensory neurons (Fig. 4G) and Pvr/Pvf mutants display defective mechanical nociception (Figs. 3A, 4A,B), it seemed possible that developmental alterations in the neurons could underlie these effects. Therefore, we examined dendritic branching of class IV mechanical nociceptors in Pvr, Pvf2, and Pvf3 mutants. Although class IV multidendritic neurons are present in the normal number and location, Pvr, Pvf2, and Pvf3 mutants showed reduced total dendritic length (one-way ANOVA, F(3,30) = 17.64, p < 0.0001, Dunnett's post hoc test for ppk-Gal4-mCD8-GFP, Pvrhypo/Pvrnull vs control (ppk-Gal4-mCD8-GFP) comparison: p < 0.0001; for ppk-Gal4-mCD8-GFP, Pvf2hypo/Pvf2–3 vs control comparison: p < 0.0001; for ppk-Gal4-mCD8-GFP, Pvf3hypo/Pvf2-3 vs control comparison: p < 0.0001) (Fig. 5A,B) and dendritic branching compared with controls (one-way ANOVA, F(3,29) = 14.03, p < 0.0001, Dunnett's post hoc test for ppk-Gal4-mCD8-GFP, Pvrhypo/Pvrnull vs control (ppk-Gal4-mCD8-GFP) comparison: p < 0.0001; for ppk-Gal4-mCD8-GFP, Pvf2hypo/Pvf2–3 vs control comparison: p = 0.0003; for ppk-Gal4-mCD8-GFP, Pvf3hypo/Pvf2-3 vs control comparison: p < 0.0001) (Fig. 5C). For all numerical values of branch length, number, critical value, and critical radius, see Table 2. A measure of neuronal arbor complexity of these mutants (Sholl analysis) (Ferreira et al., 2014) demonstrated a decrease in the critical value (for maximum number of crossings; see Experimental procedures) in all three mutant genotypes (Pvr, 27%; Pvf2, 29%; Pvf3, 21%; Fig. 5D–F) (one-way ANOVA, F(3,31) = 5.776, p = 0.0029, Dunnett's post hoc test for ppk-Gal4-mCD8-GFP, Pvrhypo/Pvrnull vs control (ppk-Gal4-mCD8-GFP) comparison: p = 0.0036; for ppk-Gal4-mCD8-GFP, Pvf2hypo/Pvf2–3 vs control comparison: p = 0.0030; for ppk-Gal4-mCD8-GFP, Pvf3hypo/Pvf2-3 vs control comparison: p < 0.0331). The critical radius (see Experimental procedures) was similar between mutants and controls (Fig. 5G). This morphological analysis indicates that Pvr signaling in class IV sensory neurons alters both dendritic length and branching.
Figure 5.
Neuronal morphology of class IV multidendritic neurons in Pvr and Pvf mutants. A, class IV multidendritic sensory neurons labeled via Ppk1.9-Gal4, UAS-mCD8-GFP in controls and Pvr and Pvf mutants. B, Quantitation of total dendritic length. C, Quantitation of dendritic branch number in controls and Pvr and Pvf mutants. D, Sholl analysis of Pvr and Pvf mutants. Representative class IV sensory neurons of controls and Pvr and Pvf mutants with a heatmap of dendritic branch density. Blue represents low. Red represents high. E, Graphic Sholl profiles of class IV sensory neurons in controls and Pvr and Pvf mutants. Dendritic intersections of individual neurons were pooled for each genotype, and the average density profile was fitted to a polynomial function. F, G, Critical values (maximum number of dendritic crossings) (F) and critical radius (the distance at which the highest number of intersections occur) (G) of neurons by genotype. D–G, n = 8 or 9 neurons. B, C, Data are mean ± SEM. F, G, Box plots represent median. Whiskers represent the 10th and 90th percentiles. All quantitation based upon n = 8 or 9 neurons total per genotype. One-way ANOVA followed by Dunnett's post hoc test was used for statistical analysis: *p < 0.05; **p < 0.01; ***p < 0.001. Scale bar, 100 μm.
Table 2.
Morphometric properties of class IV sensory neuronsa
| Control | Pvrhypo/Pvrnull | Pv2hypo/Pvf2-3 | Pv3hypo/Pvf2-3 | |
|---|---|---|---|---|
| Total length (mm) | 38.2 ± 3.5 | 29.1 ± 4.1 | 28.9 ± 2.6 | 26.5 ± 3.3 |
| Total branches | 1745.1 ± 351.3 | 1148 ± 307.3 | 917.6 ± 237.9 | 766.7 ± 119.7 |
| Critical value | 58.4 ± 8.8 | 42.7 ± 9.8 | 41.3 ± 12.5 | 46.2 ± 8.31 |
| Critical radius | 455.9 ± 21.1 | 457.1 ± 15.6 | 452.8 ± 18.4 | 455.1 ± 19.9 |
aThe total dendritic length, branch numbers, and Sholl profiles (critical value and critical radius) of class IV sensory neuron are expressed as mean ± SEM.
Constitutive versus inducible activation of Pvr separates nociceptive signaling effects from developmental morphology
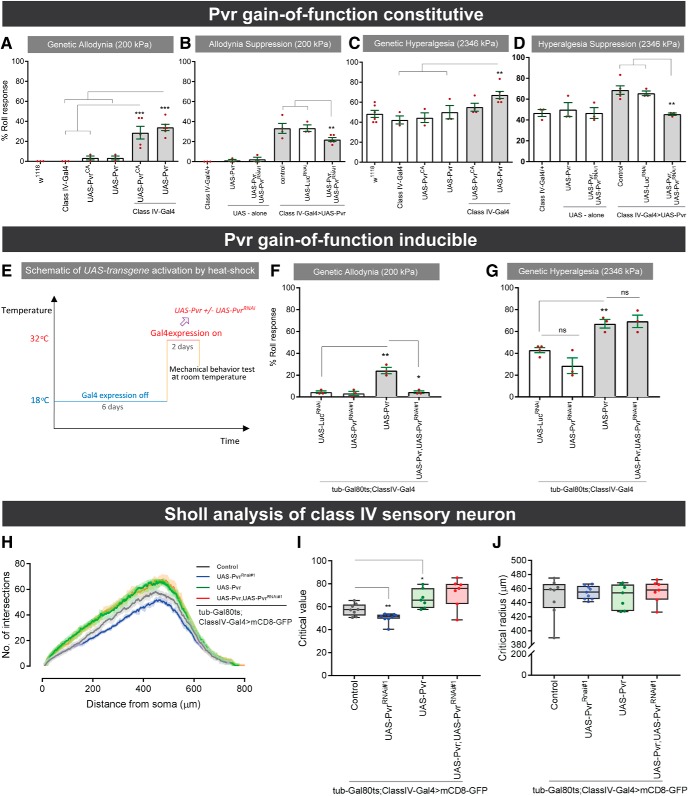
We next investigated whether Pvr overexpression or hyperactivation in class IV sensory neurons could also induce hypersensitivity. Both overexpression (one-way ANOVA, F(2,9) = 43.69, p < 0.0001, Tukey's post hoc test for classIV-Gal>Pvr vs classIV-Gal4/+ comparison: p < 0.0001; for classIV-Gal>Pvr vs Pvr/+ comparison: p < 0.0001) and hyperactivation (one-way ANOVA, F(2,8) = 9.612, p = 0.0075, Tukey's post hoc test for class-Gal4>PvrCA vs classIV-Gal4/+ comparison: p = 0.0090; classIV-Gal>Pvr vs PvrCA/+ comparison: p = 0.0170) of Pvr caused mechanical allodynia (Fig. 6A), whereas only overexpression induced mechanical hyperalgesia (one-way ANOVA, F(2,9) = 8.579, p = 0.00082, Tukey's post hoc test for classIV-Gal4>Pvr vs classIV-Gal4/+ comparison: p = 0.0069; for classIV-Gal4>Pvr vs Pvr/+ comparison: p = 0.0454) (Fig. 6C). Both types of Pvr-induced hypersensitivity could be attenuated by reducing Pvr function in class IV sensory neurons (for mechanical allodynia, see Fig. 6B; one-way ANOVA, F(2,9) = 8.309, p = 0.0090, Tukey's post hoc test for classIV-Gal4>Pvr, PvrRNAi#1 vs control (classIV-Gal4/+) comparison: p = 0.00112; for classIV-Gal4>Pvr,PvrRNAi#1 vs classIV-Gal4>LuciferaseRNAi (LucRNAi) comparison: p = 0.0176) and Figure 6D for mechanical hyperalgesia (one-way ANOVA, F(2,8) = 11.81, p = 0.0041, Tukey's post hoc test for classIV-Gal4>Pvr, PvrRNAi#1 vs control (classIV-Gal4/+) comparison: p = 0.0028; for classIV-Gal4>Pvr,PvrRNAi#1 vs classIV-Gal4>LucRNAi comparison: p = 0.0120). Coexpression of an irrelevant UAS-RNAi transgene, UAS-LuciferaseRNAi (UAS-lucRNAi) in class IV neurons did not attenuate ectopic mechanical allodynia or hyperalgesia (Fig. 3E,G), suggesting that the observed attenuation is not caused by titration of the Gal4/UAS system.
Figure 6.
Behavioral and morphological analysis of constitutive and inducible Pvr overexpression and loss of function. A, Mechanical allodynia induced by constitutive overexpression of UAS-Pvr in class IV multidendritic sensory neurons. B, Partial suppression of Pvr-induced mechanical allodynia by coexpression of UAS-PvrRNAi#1. C, Mechanical hyperalgesia induced by constitutive overexpression of UAS-Pvr in class IV multidendritic sensory neurons. D, Suppression of Pvr-induced mechanical allodynia by coexpression of UAS-PvrRNAi#1 but not UAS-LucRNAi. E, Schematic approach to UAS-transgene activation by temporally induced heat shock. Switching to the permissive temperature (32°C) relieves inhibition of Gal4 and permits transgene expression. F, G, Mechanical allodynia (F) and mechanical hyperalgesia (G) induced by overexpression of UAS-Pvr ± UAS-PvrRNAi in class IV multidendritic sensory neurons. H, Sholl profiles of class IV sensory neurons after heat shock-induced expression of UAS-Pvr ± UAS-PvrRNAi. I, Critical values and critical radii (J) of neurons by genotype. H–J, n = 7–9 neurons. A–D, F, G, Data are mean ± SEM. I, J, Box plots represent median. Whiskers represent the 10th and 90th percentiles. A–D, One-way ANOVA followed by Dunnett's post hoc test was used for statistical analyses. F, G, I, J, Two-tailed t tests were used for statistical analyses: *p < 0.05; **p < 0.01; ***p < 0.001, ns, not significant.
Constitutive Pvr overexpression in class IV sensory neurons caused mechanical hypersensitivity, whereas loss of Pvr function (Fig. 5) caused mild dendritic branching defects and reduced mechanical nociception. To determine whether these effects on nociceptive thresholds could be separated from effects upon developmental morphology, we performed conditional expression of relevant Pvr variants (RNAi and overexpression) using the tub-Gal80ts system (McGuire et al., 2004) and a heat shock regimen (Fig. 6E). A 2 d induction of Pvr expression in class IV multidendritic neurons induced mechanical allodynia (Fig. 6F; t test, p = 0.0031) and hyperalgesia (Fig. 6G; t test, p = 0.0025). Allodynia, but not hyperalgesia, was attenuated by coexpression of a UAS-PvrRNAi transgene (Fig. 6F,G, respectively). At the morphological level, conditional reduction of Pvr in class IV multidendritic neurons showed a decrease of the critical value (t test, p = 0.0071), but not the critical radius (t test, p = 0.5074) (Fig. 6H–J). Overexpression of Pvr caused a slight increase in the critical value that was not reduced by coexpression of UAS-PvrRNAi (Fig. 6I). Given that the behavioral effects of conditional Pvr overexpression ± Pvr reduction do not directly correlate with induced morphological changes, it appears that the changes in nociceptive thresholds may be separable from effects on neuronal morphology.
The Painless and Piezo ion channels interact genetically with Pvr to regulate mechanical nociception
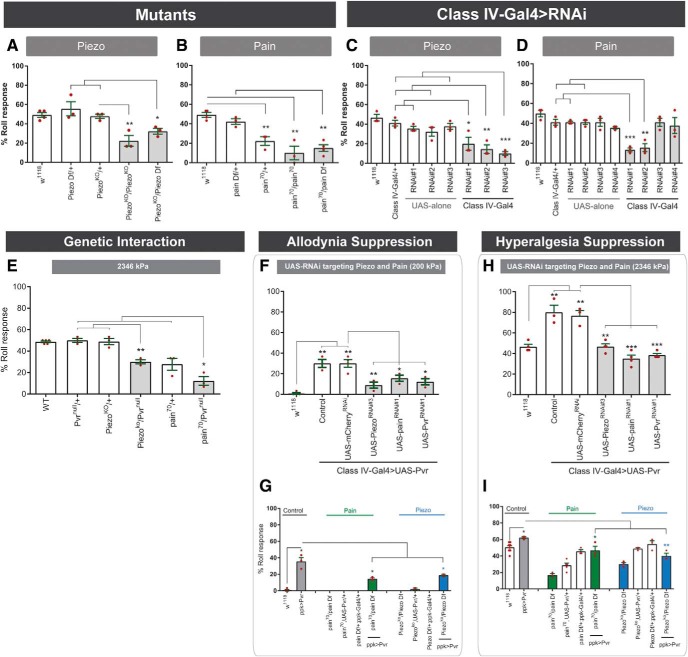
We next asked whether piezo and TRP channels play a role in mechanical nociception. As reported previously (Tracey et al., 2003; Kim et al., 2012), mutants for Piezo and painless (pain), a TRP channel, showed defects in mechanical nociception (t test, Piezoko/Piezoko vs Piezoko/+, p = 0.0129; t test, pain70/+ vs w1118, p = 0.0024; t test, pain70/pain70 vs w1118, p = 0.0018) (Fig. 7A,B). Transheterozygotes of each mutant allele over a corresponding deficiency for Piezo and pain70 also led to defects in baseline mechanical nociception (Piezo: one-way ANOVA, F(2,6) = 6.356, p = 0.0330, Tukey's post hoc test for Piezoko/Piezo Df vs Piezo Df/+ comparison: p = 0.0226; pain: F(2,7) = 14.59, p = 0.0032, Tukey's post hoc test for pain70/pain Df vs pain Df/+ comparison: p = 0.0027) (Fig. 7A,B). Interestingly, larvae heterozygous for pain70 were haploinsufficient for mechanical nociception (Fig. 7B). To determine whether these channels functioned in class IV nociceptive sensory neurons, we used ppk1.9-Gal4 to express gene-specific UAS-RNAi transgenes targeting Piezo and pain. RNAi transgenes targeting each channel significantly attenuated mechanical nociception (PiezoRNAi: one-way ANOVA, F(2,6) = 6.212, p = 0.0345, Tukey's post hoc test for classIV-Gal4>PiezoRNAi#1 vs classIV-Gal4/+ comparison: p = 0.0334; PiezoRNAi#2: one-way ANOVA, F(2,6) = 11.49, p = 0.0089, Tukey's post hoc test for classIV-Gal4>PiezoRNAi#2 vs classIV-Gal4/+ comparison: p = 00079; for classIV-Gal4>PiezoRNAi#2 vs PiezoRNAi#2/+ comparison: p = 0.0459; PiezoRNAi#3: one-way-ANOVA, F(2,6) = 41.90, p = 0.0003, Tukey's post hoc test for classIV-Gal4>PiezoRNAi#3 vs classIV-Gal4/+ comparison: p = 0.0004; for classIV-Gal4>PiezoRNAi#3 vs PiezoRNAi#3/+ comparison: p = 0.0007; painRNAi#1: one-way ANOVA, F(2,6) = 56.74, p = 0.0001, Tukey's post hoc test for classIV-Gal4>painRNAi#1 vs classIV-Gal4/+ comparison: p = 0.0002; for classIV-Gal4>painRNAi#1 vs painRNAi#1/+ comparison: p = 0.0002; painRNAi#2: one-way ANOVA: F(2,6) = 22.20, p = 0.0017, Tukey's post hoc test for classIV-Gal4>painRNAi#2 vs classIV-Gal4/+ comparison: p = 0.0028; for classIV-Gal4>painRNAi#2 vs painRNAi#2/+ comparison: p = 0.0029) (Fig. 7C,D). These results confirm that Piezo and the Trp channel pain are required in class IV multidendritic neurons for mechanical nociception.
Figure 7.
Pvr-induced mechanical hypersensitivity is mediated by Piezo and Painless in Class IV sensory neurons. A, B, Quantitation of responses to noxious mechanical pressure (2346 kPa) of control larvae and indicated mutants and deficiencies for Piezo (A) and pain (B). C, D, Quantitation of responses to noxious mechanical pressure (2346 kPa) of relevant controls and ppk1.9-Gal4 expressing UAS-PiezoRNAi (C) and UAS-painRNAi (D). E, Genetic interaction between Pvr and piezo or Pvr and pain. F, H, Suppression of Pvr-induced mechanical allodynia (F) and hyperalgesia (H) by coexpression of either UAS-PiezoRNAi or UAS-painRNAi in class IV multidendritic sensory neurons. G, I, Suppression of Pvr-induced genetic mechanical allodynia (G) and hyperalgesia (I) in piezo and pain mutant larvae. Data are mean ± SEM. Two-tailed unpaired t test and one-way ANOVA with post hoc Tukey's test were used for statistical analyses: *p < 0.05; **p < 0.01; ***p < 0.001.
Our data implicate Pvr signaling and two ion channels (Pain and Piezo) in mechanical nociception. Therefore, to determine whether Pvr interacted genetically with piezo and pain, we combined loss-of-function alleles of each gene to create double-heterozygous larvae carrying alleles of Pvr/Piezo and Pvr/pain. Both combinations showed reduced mechanical nociception compared with single alleles (one-way ANOVA, F(2,6) = 23.79, p = 0.0014, Tukey's post hoc test for Piezoko/Pvrnull vs Pvrnull/+ comparison: p = 0.0015: for Piezoko/Pvrnull vs Piezoko/+ comparison: p = 0.0021; F(2,4) = 26.44, p = 0.0049, Tukey's post hoc test for Pain70/Pvrnull vs Pvrnull/+ comparison: p = 0.0036, respectively) (Fig. 7E), indicating genetic interactions.
We next determined whether coexpression of the strongest RNAi transgenes targeting Piezo or pain could suppress Pvr-induced genetic mechanical hypersensitivity. As expected, coexpression of UAS-PvrRNAi#1 rescued Pvr-induced genetic mechanical allodynia (t test, classIV-Gal4>Pvr + PvrRNAi#1 vs classIV-Gal4>Pvr + mCherry, p = 0.0154) (Fig. 7F) and hyperalgesia (t test, classIV-Gal4>Pvr + PvrRNAi#1 vs classIV-Gal4>Pvr + mCherry, p = 0.0004) (Fig. 7H). UAS-RNAi transgenes targeting Piezo or pain also attenuated Pvr-induced mechanical allodynia (t test, classIV-Gal4>Pvr + PiezoRNAi#3 vs classIV-Gal4>Pvr + mCherry, p = 0.0078; t test, classIV-Gal4>Pvr + painRNAi#1 vs classIV-Gal4>Pvr + mCherry, p = 0.0326, respectively) (Fig. 7F) and mechanical hyperalgesia (t test, classIV-Gal4>Pvr + PiezoRNAi#3 vs classIV-Gal4>Pvr + mCherry, p = 0.0024; t test, classIV-Gal4>Pvr + painRNAi#1 vs classIV-Gal4>Pvr + mCherry, p = 0.0009, respectively) (Fig. 7H). Coexpression of an independent exogenous UAS-RNAi transgene targeting mCherry did not suppress the Pvr-induced genetic mechanical allodynia, or hyperalgesia (Fig. 7F,H), indicating that suppression of Pvr-induced genetic mechanical hypersensitivity was not due to titration of the Gal4/UAS system. As predicted by the RNAi results, Pvr-induced genetic mechanical allodynia (Fig. 7G) and hyperalgesia (Fig. 7I) were also attenuated in pain (t test, pain70/pain Df, ppk-Gal4>Pvr vs ppk-Gal4>Pvr, p = 0.0480; t test, pain70/pain Df, ppk-Gal4>Pvr vs ppk-Gal4>Pvr, p = 0.0410, respectively) and Piezo mutant larvae (t test, Piezoko/Piezo Df, ppk-Gal4>Pvr vs ppk-Gal4>Pvr, p = 0.0281; t test, Piezoko/Piezo Df, ppk-Gal4>Pvr vs ppk-Gal4>Pvr, p = 0.0032). Together, our results indicate that Pvr signaling mediates mechanical nociception and hypersensitivity, at least in part through Piezo and pain in class IV sensory neurons.
Vertebrate PDGFR and VEGFR signaling modulates mechanical nociception
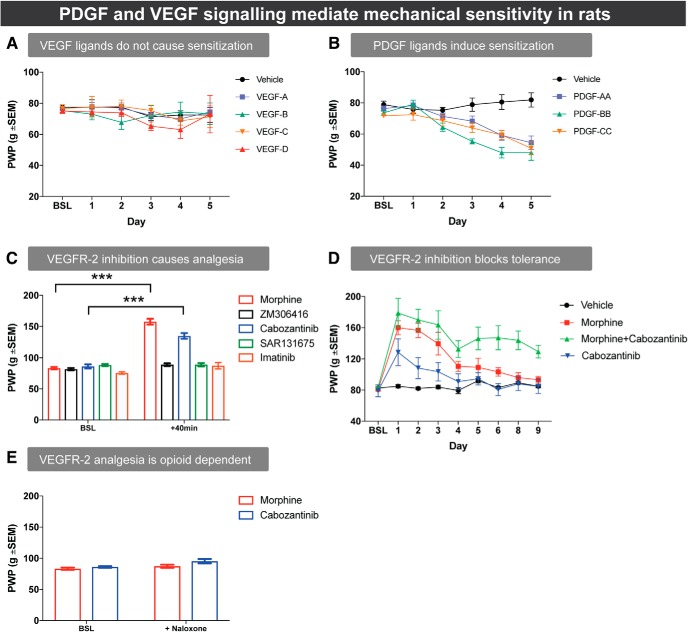
Given that Pvf 2 and Pvf 3 overexpression caused mechanical sensitization in the fly, we determined whether VEGFR and PDGFR ligands could induce mechanical hypersensitivity in rats. Using an analogous assay in vertebrates, we found that none of the VEGFR ligands caused mechanical sensitization (Fig. 8A; two-way ANOVA; interaction: F(20,125) = 0.5838, p = 0.9174, not significant; time: F(5,120) = 2.587, p = 0.0293; treatment: F(4,24) = 0.349, p = 0.8421). However, consistent with previous results (Narita et al., 2005; Masuda et al., 2009), we found that PDGF AA, BB, and CC induced mechanical hypersensitivity (Fig. 8B; two-way ANOVA; interaction: F(15,100) = 5.8, p < 0.0001; treatment: F(3,20) = 15.94, p < 0.0001; day: F(5,100) = 25.12, p < 0.001; Dunnett's multiple-comparison test: vehicle vs PDGF-AA: p = 0.0020; vehicle vs PDGF-BB: p = 0.0001; vehicle vs PDGF-CC: p = 0.0001).
Figure 8.
PDGFR and VEGFR modulate mechanical sensitivity in rats. A, Effect of daily intrathecal injections of VEGF peptides (10 pmol) on the mechanical threshold. B, Effect of daily intrathecal injections of PDGF peptides (10 pmol) on the mechanical threshold. C, Effect of acute injections with either morphine (0.6 nmol) or 10 μg of the VEGFR1 inhibitor ZM306416, the VEGFR-2 inhibitor cabozantinib, the VEGFR3 inhibitor SAR131675, or the PDGFR inhibitor imatinib on mechanical threshold. D, Effect of cabozantinib on the development of morphine analgesic tolerance. E, Effect of naloxone (10 mg/kg s.c.) on morphine-mediated (0.6 nmol i.t.) or cabozantinib-mediated (10 μg, i.t.) acute analgesia. For all experiments, mechanical sensitivity was measured 40 min after injections using the Randall–Sellitto test. Data are mean ± SEM. ***p < 0.001. n = 6 rats/group. BSL, Baseline.
Our previous findings showed that PDGFR inhibition in vertebrates did not cause analgesia (Wang et al., 2012; Donica et al., 2014). However, Drosophila Pvr loss of function decreases nociception. Therefore, we investigated whether inhibition of the phylogenetically related VEGFR could induce analgesia in rats. We treated rats with selective inhibitors targeting VEGFR-1 (Antczak et al., 2012; Wu et al., 2017), VEGFR-2 (Yakes et al., 2011), or VEGFR-3 (Alam et al., 2012). Cabozantinib, a VEGFR-2 inhibitor, caused mechanical analgesia. In contrast, inhibition of VEGFR-1 (ZM306416), VEGFR-3 (SAR 131675), or PDGFR (imatinib) did not cause analgesia (Fig. 8C; two-way ANOVA; interaction: F(4,25) = 81.7, p < 0.0001; treatment: F(4,25) = 40.89, p < 0.0001; time: F(1,25) = 328.8, p < 0.0001; Sidak's multiple-comparison test: baseline vs + 40 min: morphine, p < 0.0001; cabozantinib, p < 0.0001).
Our previous discovery that PDGFR inhibition eliminated morphine analgesic tolerance (Wang et al., 2012) prompted us to test whether VEGFR signaling could also modulate tolerance. Rats treated with morphine and the VEGFR-2 inhibitor cabozantinib demonstrated a marked inhibition of analgesic tolerance (Fig. 8D; two-way ANOVA; interaction: F(24,160) = 4.486, p < 0.0001; treatment: F(3,20) = 13.7, p < 0.0001; day: F(8,160) = 23.95, p < 0.0001; Dunnett's multiple-comparison test, morphine vs morphine + cabozantinib: day 5, p = 0.0205; day 6, p = 0.0047; day 8, p = 0.0018; day 9, p = 0.0252). It is possible that the tolerance inhibition by cabozantinib could be due to its analgesic effect. However, two lines of evidence argue against this possibility. First, the initial analgesic effect of morphine was not changed by coadministration with cabozantinib on day 1 (Fig. 8D; day 1, Dunnett's multiple-comparison test, morphine vs morphine + cabozantinib, p = 0.9943, not significant), suggesting that cabozantinib did not augment the acute analgesic effect of morphine. Second, tolerance to the analgesic effect of cabozantinib developed over time (Fig. 8D). This development of tolerance led us to investigate whether the analgesic effects of cabozantinib were opioid-mediated. Remarkably, naloxone treatment completely eliminated the acute analgesic effect of cabozantinib (Fig. 8E; two-way ANOVA; interaction: F(1,9) = 1.414, p = 0.264, not significant; treatment: F(1,9) = 8.561, p = 0.0169; time: F(1,9) = 4.565, p = 0.614). In sum, our vertebrate data suggest that mechanical nociception mediated by Pvr and its ligands in Drosophila is mediated in rats by PDGF ligands and VEGFR-2. We also established a role for VEGFR-2 signaling in opioid tolerance.
Discussion
Our work establishes a role for Pvr signaling in modulating mechanical nociception in Drosophila and shows that this role is conserved in mammals using the homologous vertebrate receptors, PDGFR and VEGFR. Using custom-made metal filaments, we generated a full behavioral dose–response curve to mechanical stimulation and found that pressure, rather than force, is linearly related to behavioral responsiveness in Drosophila larvae. Genetically, we showed that Pvr and two of its ligands (Pvf2 and Pvf3) determine baseline responsiveness to mechanical nociceptive stimuli. Pvr signaling appears to modulate nociception either by controlling the degree of arborization of class IV multidendritic neurons and/or by regulating Pain and Piezo ion channel function.
Many assays of mechanical nociception use mechanical probes that produce a defined amount of force and induce an aversive behavioral response when applied to the skin (Levin et al., 1978; Zhong et al., 2010; Bonin et al., 2014; Chalfie et al., 2014). Our finding that pressure correlates more reliably with aversive behavioral responsiveness than force in Drosophila larvae could explain the variability in behavioral responses reported by previous investigators (Zhong et al., 2010; Kim et al., 2012; Guo et al., 2014). Drosophila larvae are small (∼5 mm long and 0.7 mm wide at the stage tested), and it is possible that pressure is a more relevant variable for animals when the potentially noxious stimulus affects a relatively large percentage of the animal's body surface area. Alternatively, it is possible that pressure is also an operant variable in larger vertebrates, although most studies have reported only force (Dellon et al., 1993; Cunha et al., 2004). The relevance of pressure versus force may be worth revisiting in other animal models. In our study, noxious pressures (320–5300 kPa) produced gradually increasing damage to the local epidermis and overlying cuticle. The extent of damage was correlated with aversive behavior. Intriguingly, epidermal damage can be observed even when there is no aversive response, suggesting that even non-noxious stimuli can be damaging to the organism.
We found that Pvr and its ligands, Pvf2 and Pvf3, determine the mechanical nociceptive baseline in Drosophila. Some of these effects are likely developmental, as Pvr mutants exhibit reduced dendritic arbor complexity. However, developmental effects may not be necessary for sensitization, as acute hyperactivation of Pvr causes mechanical hypersensitivity. Conditional induction of Pvr revealed a separability of behavior and morphology that reinforces the idea that morphological changes may not be the sole driver of behavioral output. Functional Pvf2 and Pvf3 are produced by multiple tissues (fat body, hemocyte, epidermis, and neurons), suggesting that the mechanical nociceptive threshold may be set by diverse physiological stimuli modulating the ambient levels of these ligands in multiple tissues. Pvf2 and Pvf3 act more efficiently together, as knocking them both out or overexpressing both of them produced a stronger mechanical nociceptive defect or hypersensitivity, respectively. RTKs have not previously been implicated as a modulator of baseline mechanical nociception in Drosophila, although Neuropeptide F and its G-protein-coupled receptor do act in a similar manner (Hu et al., 2017).
In agreement with previous reports, our findings suggest that Piezo, a mechanically activated channel, and Pain, a transient-receptor potential A family member, function in larval class IV nociceptive sensory neurons to mediate mechanical nociception (Tracey et al., 2003; Kim et al., 2012). One interesting question is why nociceptors contain multiple ion channels that can respond to noxious mechanical stimulation. One possibility is that each channel responds distinctly to pressure-induced membrane deformation (Lewis and Grandl, 2015; Cox et al., 2016). Another possibility is that TRP and other channels form different complexes that mediate mechanical nociception (Staruschenko et al., 2010; Ma et al., 2011). In sum, modulation of mechanosensation by multiple effectors could function to tailor response sensitivity and reduce response ambiguity (Ohyama et al., 2015). It is also possible that having multiple redundant channels could serve as a “fail-safe” mechanism to ensure that signals critical in detecting sensory stimuli are reliably transmitted (Tsubouchi et al., 2012; Turner et al., 2016).
While the signaling mechanisms underlying mechanosensation remain elusive in vertebrates (Peirs and Seal, 2016), our findings suggest that vertebrate homologs of RTKs would be likely candidates. A striking result of our study is that the role of Pvr in mechanical nociception is conserved in the rat by the VEGFR and PDGF mammalian orthologs. Consistent with previous studies (Masuda et al., 2009; Wang et al., 2012), we found that PDGF ligands caused mechanical sensitization while PDGFR inhibition did not cause analgesia. Other recent studies have noted VEGFR expression on peripheral nerves and effects on nociception with peripherally administered VEGF ligands (Selvaraj et al., 2015; Yang et al., 2018). While we did not see effects of intrathecally administered VEGF peptides, inhibition of VEGFR-2 raised the mechanical threshold in rats. Our results suggest the intriguing possibility that crosstalk between PDGF peptides and VEGF receptors could modulate mechanical sensitivity. PDGF peptides can bind with extremely high affinity to the VEGFR-2, and computational predictions suggest that PDGF peptides may account for a large percentage of VEGFR-2 binding in vivo (Mamer et al., 2017). Also, the Flt3 RTK can alter mechanical sensitivity in a TRP channel-dependent manner (Rivat et al., 2018), further supporting the concept that RTKs may regulate mechanical nociception. Further studies are indicated to better understand how RTKs, piezo, and TRP channels might interact to regulate mechanical sensitivity in vertebrates.
We also made the intriguing observation that VEGFR-2 signaling blocks opioid tolerance. We previously showed that hedgehog signaling regulated thermal nociceptive sensitivity in the fly, and also modulated opioid tolerance in the rat (Babcock et al., 2011). While there is no known opioid receptor or peptide in the fly, our findings suggest that signals regulating nociceptive sensitivity in the fly may also modulate opioid analgesic signaling in vertebrates. VEGFR-2 inhibition of tolerance is consistent with previous work by our group and others indicating that blocking activation of the phylogenetically related RTKs PDGF receptor-β (Wang et al., 2012), fibroblast growth factor receptor (Fujita-Hamabe et al., 2011), and ephrin B receptor (Liu et al., 2011) inhibits morphine tolerance. These emerging findings suggest that RTK inhibitors could eventually be of therapeutic importance in pain treatment. Our discovery that analgesia induced by the VEGFR-2 inhibitor cabozatinib is opioid-mediated suggests a complex and previously undescribed relationship between opioid receptor and VEGFR-2 signaling systems.
In conclusion, we have refined a genetically tractable model for studying mechanical nociception and used the power of Drosophila genetics to identify novel conserved mediators of mechanical nociception: Pvr and its ligands. This regulatory system is conserved in the rat, as PDGF ligands and VEGFR-2 modulate mechanical thresholds. VEGFR-2 antagonism also blocks opioid tolerance in vertebrates. Together, our results demonstrate the power of Drosophila genetics to quickly identify and screen novel therapeutic targets of potential clinical importance.
Footnotes
H.B.G. was supported by National Institute on Drug Abuse. M.J.G. was supported by National Institute on Neurological Diseases and Stroke and National Institute of General Medical Sciences, and a University of Texas M. D. Anderson Clark Fellowship. C.-R.T. was supported by American Heart Association Predoctoral Fellowship 16PRE30880004. H.N.T. was supported by a National Institutes of Health Predoctoral Kirchstein National Research Service Award from National Institute of Neurological Disorders and Stroke F31 NS083306 and a Marilyn and Frederick R. Lummis, Jr. MD Fellowship. Dr. Paulucci-Holthauzen at the Basic Science Research Building Microscopy Facility provided training and support, and the National Institutes of Health shared Instrumentation Grant 1S10OD024976-01 supporting the confocal microscope. The Flow Cytometry & Cell Sorting Core Laboratory at M. D. Anderson was funded by National Cancer Institute Cancer Center Support Grant P30CA16672. The Vienna Drosophila RNAi Center, the Bloomington Drosophila Stock Center, and the Japanese NIG-Fly stock center provided Drosophila stocks. We thank the M.J.G. and H.B.G. laboratories for comments on the manuscript; Ardem Patapoutian for piezoKO; Edan Foley for Pvf2-3; Jocelyn McDonald for anti-Pvr; and Thomas Wang for developing prototype von Frey filaments.
The authors declare no competing financial interests.
References
- Ainsley JA, Pettus JM, Bosenko D, Gerstein CE, Zinkevich N, Anderson MG, Adams CM, Welsh MJ, Johnson WA (2003) Enhanced locomotion caused by loss of the Drosophila DEG/ENaC protein Pickpocket1. Curr Biol 13:1557–1563. 10.1016/S0960-9822(03)00596-7 [DOI] [PubMed] [Google Scholar]
- Alam A, Blanc I, Gueguen-Dorbes G, Duclos O, Bonnin J, Barron P, Laplace MC, Morin G, Gaujarengues F, Dol F, Hérault JP, Schaeffer P, Savi P, Bono F (2012) SAR131675, a potent and selective VEGFR-3-TK inhibitor with antilymphangiogenic, antitumoral, and antimetastatic activities. Mol Cancer Ther 11:1637–1649. 10.1158/1535-7163.MCT-11-0866-T [DOI] [PubMed] [Google Scholar]
- Al-Anzi B, Tracey WD Jr, Benzer S (2006) Response of Drosophila to wasabi is mediated by painless, the fly homolog of mammalian TRPA1/ANKTM1. Curr Biol 16:1034–1040. 10.1016/j.cub.2006.04.002 [DOI] [PubMed] [Google Scholar]
- Antczak C, Mahida JP, Bhinder B, Calder PA, Djaballah H (2012) A high-content biosensor-based screen identifies cell-permeable activators and inhibitors of EGFR function: implications in drug discovery. J Biomol Screen 17:885–899. 10.1177/1087057112446174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock DT, Shi S, Jo J, Shaw M, Gutstein HB, Galko MJ (2011) Hedgehog signaling regulates nociceptive sensitization. Curr Biol 21:1525–1533. 10.1016/j.cub.2011.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin RP, Bories C, De Koninck Y (2014) A simplified up-down method (SUDO) for measuring mechanical nociception in rodents using von Frey filaments. Mol Pain 10:26. 10.1186/1744-8069-10-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401–415. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Hart AC, Rankin CH, Goodman MB (2014) Assaying mechanosensation. WormBook 1.172.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho NK, Keyes L, Johnson E, Heller J, Ryner L, Karim F, Krasnow MA (2002) Developmental control of blood cell migration by the Drosophila VEGF pathway. Cell 108:865–876. 10.1016/S0092-8674(02)00676-1 [DOI] [PubMed] [Google Scholar]
- Coste B, Mathur J, Schmidt M, Earley TJ, Ranade S, Petrus MJ, Dubin AE, Patapoutian A (2010) Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 330:55–60. 10.1126/science.1193270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox CD, Bae C, Ziegler L, Hartley S, Nikolova-Krstevski V, Rohde PR, Ng CA, Sachs F, Gottlieb PA, Martinac B (2016) Removal of the mechanoprotective influence of the cytoskeleton reveals PIEZO1 is gated by bilayer tension. Nat Commun 7:10366. 10.1038/ncomms10366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha TM, Verri WA Jr, Vivancos GG, Moreira IF, Reis S, Parada CA, Cunha FQ, Ferreira SH (2004) An electronic pressure-meter nociception paw test for mice. Braz J Med Biol Res 37:401–407. 10.1590/S0100-879X2004000300018 [DOI] [PubMed] [Google Scholar]
- Das R, Bhattacharjee S, Patel AA, Harris JM, Bhattacharya S, Letcher JM, Clark SG, Nanda S, Iyer EP, Ascoli GA, Cox DN (2017) Dendritic cytoskeletal architecture is modulated by combinatorial transcriptional regulation in Drosophila melanogaster. Genetics 207:1401–1421. 10.1534/genetics.117.300393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellon ES, Crone S, Mouery R, Dellon AL (1993) Comparison of the Semmes-Weinstein monofilaments with the pressure-specifying sensory device. Restor Neurol Neurosci 5:323–326. 10.3233/RNN-1993-55602 [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448:151–156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Donica CL, Cui Y, Shi S, Gutstein HB (2014) Platelet-derived growth factor receptor-beta antagonism restores morphine analgesic potency against neuropathic pain. PLoS One 9:e97105. 10.1371/journal.pone.0097105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin AE, Patapoutian A (2010) Nociceptors: the sensors of the pain pathway. J Clin Invest 120:3760–3772. 10.1172/JCI42843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchek P, Somogyi K, Jékely G, Beccari S, Rørth P (2001) Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell 107:17–26. 10.1016/S0092-8674(01)00502-5 [DOI] [PubMed] [Google Scholar]
- Eijkelkamp N, Quick K, Wood JN (2013) Transient receptor potential channels and mechanosensation. Annu Rev Neurosci 36:519–546. 10.1146/annurev-neuro-062012-170412 [DOI] [PubMed] [Google Scholar]
- Faucherre A, Nargeot J, Mangoni ME, Jopling C (2013) piezo2b regulates vertebrate light touch response. J Neurosci 33:17089–17094. 10.1523/JNEUROSCI.0522-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira TA, Blackman AV, Oyrer J, Jayabal S, Chung AJ, Watt AJ, Sjöström PJ, van Meyel DJ (2014) Neuronal morphometry directly from bitmap images. Nat Methods 11:982–984. 10.1038/nmeth.3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita-Hamabe W, Nakamoto K, Tokuyama S (2011) Involvement of NCAM and FGF receptor signaling in the development of analgesic tolerance to morphine. Eur J Pharmacol 672:77–82. 10.1016/j.ejphar.2011.04.029 [DOI] [PubMed] [Google Scholar]
- Galko MJ, Krasnow MA (2004) Cellular and genetic analysis of wound healing in Drosophila larvae. PLoS Biol 2:E239. 10.1371/journal.pbio.0020239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorczyca DA, Younger S, Meltzer S, Kim SE, Cheng L, Song W, Lee HY, Jan LY, Jan YN (2014) Identification of Ppk26, a DEG/ENaC channel functioning with Ppk1 in a mutually dependent manner to guide locomotion behavior in Drosophila. Cell Rep 9:1446–1458. 10.1016/j.celrep.2014.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Wang Y, Wang Q, Wang Z (2014) The role of PPK26 in Drosophila larval mechanical nociception. Cell Rep 9:1183–1190. 10.1016/j.celrep.2014.10.020 [DOI] [PubMed] [Google Scholar]
- Heino TI, Kärpänen T, Wahlström G, Pulkkinen M, Eriksson U, Alitalo K, Roos C (2001) The Drosophila VEGF receptor homolog is expressed in hemocytes. Mech Dev 109:69–77. 10.1016/S0925-4773(01)00510-X [DOI] [PubMed] [Google Scholar]
- Hu C, Petersen M, Hoyer N, Spitzweck B, Tenedini F, Wang D, Gruschka A, Burchardt LS, Szpotowicz E, Schweizer M, Guntur AR, Yang CH, Soba P (2017) Sensory integration and neuromodulatory feedback facilitate Drosophila mechanonociceptive behavior. Nat Neurosci 20:1085–1095. 10.1038/nn.4580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulse RP, Beazley-Long N, Hua J, Kennedy H, Prager J, Bevan H, Qiu Y, Fernandes ES, Gammons MV, Ballmer-Hofer K, Gittenberger de Groot AC, Churchill AJ, Harper SJ, Brain SD, Bates DO, Donaldson LF (2014) Regulation of alternative VEGF-A mRNA splicing is a therapeutic target for analgesia. Neurobiol Dis 71:245–259. 10.1016/j.nbd.2014.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang RY, Zhong L, Xu Y, Johnson T, Zhang F, Deisseroth K, Tracey WD (2007) Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr Biol 17:2105–2116. 10.1016/j.cub.2007.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SH, Galko MJ (2012) Pokes, sunburn, and hot sauce: Drosophila as an emerging model for the biology of nociception. Dev Dyn 241:16–26. 10.1002/dvdy.22737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SH, Takle K, Jo J, Babcock DT, Ma Z, Xiang Y, Galko MJ (2015) Tachykinin acts upstream of autocrine Hedgehog signaling during nociceptive sensitization in Drosophila. Elife 4:e10735. 10.7554/eLife.10735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Edgar BA (2009) EGFR signaling regulates the proliferation of Drosophila adult midgut progenitors. Development 136:483–493. 10.1242/dev.026955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SE, Coste B, Chadha A, Cook B, Patapoutian A (2012) The role of Drosophila piezo in mechanical nociception. Nature 483:209–212. 10.1038/nature10801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence PA, Bodmer R, Vincent JP (1995) Segmental patterning of heart precursors in Drosophila. Development 121:4303–4308. [DOI] [PubMed] [Google Scholar]
- Levin S, Pearsall G, Ruderman RJ (1978) von Frey's method of measuring pressure sensibility in the hand: an engineering analysis of the Weinstein-Semmes pressure aesthesiometer. J Hand Surg 3:211–216. 10.1016/S0363-5023(78)80084-7 [DOI] [PubMed] [Google Scholar]
- Lewis AH, Grandl J (2015) Mechanical sensitivity of Piezo1 ion channels can be tuned by cellular membrane tension. Elife 4:e12088. 10.7554/eLife.12088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Liu WT, Liu YP, Dong HL, Henkemeyer M, Xiong LZ, Song XJ (2011) Blocking EphB1 receptor forward signaling in spinal cord relieves bone cancer pain and rescues analgesic effect of morphine treatment in rodents. Cancer Res 71:4392–4402. 10.1158/0008-5472.CAN-10-3870 [DOI] [PubMed] [Google Scholar]
- Ma X, Nilius B, Wong JW, Huang Y, Yao X (2011) Electrophysiological properties of heteromeric TRPV4–C1 channels. Biochim Biophys Acta 1808:2789–2797. 10.1016/j.bbamem.2011.07.049 [DOI] [PubMed] [Google Scholar]
- Mamer SB, Chen S, Weddell JC, Palasz A, Wittenkeller A, Kumar M, Imoukhuede PI (2017) Discovery of high-affinity PDGF-VEGFR interactions: redefining RTK dynamics. Sci Rep 7:16439. 10.1038/s41598-017-16610-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda J, Tsuda M, Tozaki-Saitoh H, Inoue K (2009) Intrathecal delivery of PDGF produces tactile allodynia through its receptors in spinal microglia. Mol Pain 5:23. 10.1186/1744-8069-5-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A (2014) Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle 13:1400–1412. 10.4161/cc.28401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JA, Pinheiro EM, Montell DJ (2003) PVF1, a PDGF/VEGF homolog, is sufficient to guide border cells and interacts genetically with taiman. Development 130:3469–3478. 10.1242/dev.00574 [DOI] [PubMed] [Google Scholar]
- McGuire SE, Mao Z, Davis RL (2004) Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE 2004:pl6. 10.1126/stke.2202004pI6 [DOI] [PubMed] [Google Scholar]
- Mondal BC, Shim J, Evans CJ, Banerjee U (2014) Pvr expression regulators in equilibrium signal control and maintenance of Drosophila blood progenitors. Elife 3:e03626. 10.7554/eLife.03626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munier AI, Doucet D, Perrodou E, Zachary D, Meister M, Hoffmann JA, Janeway CA Jr, Lagueux M (2002) PVF2, a PDGF/VEGF-like growth factor, induces hemocyte proliferation in Drosophila larvae. EMBO Rep 3:1195–1200. 10.1093/embo-reports/kvf242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy SE, Loud MC, Daou I, Marshall KL, Schwaller F, Kühnemund J, Francisco AG, Keenan WT, Dubin AE, Lewin GR, Patapoutian A (2018) The mechanosensitive ion channel Piezo2 mediates sensitivity to mechanical pain in mice. Sci Transl Med 10:eaat9897. 10.1126/scitranslmed.aat9897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Usui A, Narita M, Niikura K, Nozaki H, Khotib J, Nagumo Y, Yajima Y, Suzuki T (2005) Protease-activated receptor-1 and platelet-derived growth factor in spinal cord neurons are implicated in neuropathic pain after nerve injury. J Neurosci 25:10000–10009. 10.1523/JNEUROSCI.2507-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, Binari R, Booker M, Brennecke J, Perkins LA, Hannon GJ, Perrimon N (2011) A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods 8:405–407. 10.1038/nmeth.1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama T, Schneider-Mizell CM, Fetter RD, Aleman JV, Franconville R, Rivera-Alba M, Mensh BD, Branson KM, Simpson JH, Truman JW, Cardona A, Zlatic M (2015) A multilevel multimodal circuit enhances action selection in Drosophila. Nature 520:633–639. 10.1038/nature14297 [DOI] [PubMed] [Google Scholar]
- Parsons B, Foley E (2013) The Drosophila platelet-derived growth factor and vascular endothelial growth factor-receptor related (Pvr) protein ligands Pvf2 and Pvf3 control hemocyte viability and invasive migration. J Biol Chem 288:20173–20183. 10.1074/jbc.M113.483818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirs C, Seal RP (2016) Neural circuits for pain: recent advances and current views. Science 354:578–584. 10.1126/science.aaf8933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranade SS, Woo SH, Dubin AE, Moshourab RA, Wetzel C, Petrus M, Mathur J, Bégay V, Coste B, Mainquist J, Wilson AJ, Francisco AG, Reddy K, Qiu Z, Wood JN, Lewin GR, Patapoutian A (2014) Piezo2 is the major transducer of mechanical forces for touch sensation in mice. Nature 516:121–125. 10.1038/nature13980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivat C, Sar C, Mechaly I, Leyris JP, Diouloufet L, Sonrier C, Philipson Y, Lucas O, Mallié S, Jouvenel A, Tassou A, Haton H, Venteo S, Pin JP, Trinquet E, Charrier-Savournin F, Mezghrani A, Joly W, Mion J, Schmitt M, et al. (2018) Inhibition of neuronal FLT3 receptor tyrosine kinase alleviates peripheral neuropathic pain in mice. Nat Commun 9:1042. 10.1038/s41467-018-03496-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin D, Schejter E, Volk T, Shilo BZ (2004) Apical accumulation of the Drosophila PDGF/VEGF receptor ligands provides a mechanism for triggering localized actin polymerization. Development 131:1939–1948. 10.1242/dev.01101 [DOI] [PubMed] [Google Scholar]
- Selvaraj D, Gangadharan V, Michalski CW, Kurejova M, Stösser S, Srivastava K, Schweizerhof M, Waltenberger J, Ferrara N, Heppenstall P, Shibuya M, Augustin HG, Kuner R (2015) A functional role for VEGFR1 expressed in peripheral sensory neurons in cancer pain. Cancer cell 27:780–796. 10.1016/j.ccell.2015.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinenko SA, Mathey-Prevot B (2004) Increased expression of Drosophila tetraspanin, Tsp68C, suppresses the abnormal proliferation of ytr-deficient and Ras/Raf-activated hemocytes. Oncogene 23:9120–9128. 10.1038/sj.onc.1208156 [DOI] [PubMed] [Google Scholar]
- Singhania A, Grueber WB (2014) Development of the embryonic and larval peripheral nervous system of Drosophila. Wiley Interdiscip Rev Dev Biol 3:193–210. 10.1002/wdev.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staruschenko A, Jeske NA, Akopian AN (2010) Contribution of TRPV1-TRPA1 interaction to the single channel properties of the TRPA1 channel. J Biol Chem 285:15167–15177. 10.1074/jbc.M110.106153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczot M, Liljencrantz J, Ghitani N, Barik A, Lam R, Thompson JH, Bharucha-Goebel D, Saade D, Necaise A, Donkervoort S, Foley AR, Gordon T, Case L, Bushnell MC, Bönnemann CG, Chesler AT (2018) PIEZO2 mediates injury-induced tactile pain in mice and humans. Sci Transl Med 10:eaat9892. 10.1126/scitranslmed.aat9892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey WD Jr, Wilson RI, Laurent G, Benzer S (2003) painless, a Drosophila gene essential for nociception. Cell 113:261–273. 10.1016/S0092-8674(03)00272-1 [DOI] [PubMed] [Google Scholar]
- Tsubouchi A, Caldwell JC, Tracey WD (2012) Dendritic filopodia, Ripped Pocket, NOMPC, and NMDARs contribute to the sense of touch in Drosophila larvae. Curr Biol 22:2124–2134. 10.1016/j.cub.2012.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner HN, Armengol K, Patel AA, Himmel NJ, Sullivan L, Iyer SC, Bhattacharya S, Iyer EP, Landry C, Galko MJ, Cox DN (2016) The TRP channels Pkd2, NompC, and Trpm act in cold-sensing neurons to mediate unique aversive behaviors to noxious cold in Drosophila. Curr Biol 26:3116–3128. 10.1016/j.cub.2016.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Barker K, Shi S, Diaz M, Mo B, Gutstein HB (2012) Blockade of PDGFR-beta activation eliminates morphine analgesic tolerance. Nat Med 18:385–387. 10.1038/nm.2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Antunes M, Anderson AE, Kadrmas JL, Jacinto A, Galko MJ (2015) Integrin adhesions suppress syncytium formation in the Drosophila larval epidermis. Curr Biol 25:2215–2227. 10.1016/j.cub.2015.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A, Hinz U, Engelbert M, Knust E (1995) Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell 82:67–76. 10.1016/0092-8674(95)90053-5 [DOI] [PubMed] [Google Scholar]
- Wu WB, Xu YY, Cheng WW, Yuan B, Zhao JR, Wang YL, Zhang HJ (2017) Decreased PGF may contribute to trophoblast dysfunction in fetal growth restriction. Reproduction 154:219–229. 10.1530/REP-17-0253 [DOI] [PubMed] [Google Scholar]
- Wu Y, Brock AR, Wang Y, Fujitani K, Ueda R, Galko MJ (2009) A blood-borne PDGF/VEGF-like ligand initiates wound-induced epidermal cell migration in Drosophila larvae. Curr Biol 19:1473–1477. 10.1016/j.cub.2009.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JJ, Walla BC, Diaz MF, Fuller GN, Gutstein HB (2006) Intermittent lumbar puncture in rats: a novel method for the experimental study of opioid tolerance. Anesth Analg 103:714–720. 10.1213/01.ane.0000226100.46866.ea [DOI] [PubMed] [Google Scholar]
- Yakes FM, Chen J, Tan J, Yamaguchi K, Shi Y, Yu P, Qian F, Chu F, Bentzien F, Cancilla B, Orf J, You A, Laird AD, Engst S, Lee L, Lesch J, Chou YC, Joly AH (2011) Cabozantinib (XL184), a novel MET and VEGFR-2 inhibitor, simultaneously suppresses metastasis, angiogenesis, and tumor growth. Mol Cancer Ther 10:2298–2308. 10.1158/1535-7163.MCT-11-0264 [DOI] [PubMed] [Google Scholar]
- Yan Z, Zhang W, He Y, Gorczyca D, Xiang Y, Cheng LE, Meltzer S, Jan LY, Jan YN (2013) Drosophila NOMPC is a mechanotransduction channel subunit for gentle-touch sensation. Nature 493:221–225. 10.1038/nature11685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Li S, Jin ZR, Jing HB, Zhao HY, Liu BH, Liang YJ, Liu LY, Cai J, Wan Y, Xing GG (2018) Decreased abundance of TRESK two-pore domain potassium channels in sensory neurons underlies the pain associated with bone metastasis. Sci Signal 11:eaao5150. 10.1126/scisignal.aao5150 [DOI] [PubMed] [Google Scholar]
- Zars T, Fischer M, Schulz R, Heisenberg M (2000) Localization of a short-term memory in Drosophila. Science 288:672–675. 10.1126/science.288.5466.672 [DOI] [PubMed] [Google Scholar]
- Zhong L, Hwang RY, Tracey WD (2010) Pickpocket is a DEG/ENaC protein required for mechanical nociception in Drosophila larvae. Curr Biol 20:429–434. 10.1016/j.cub.2009.12.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Bellemer A, Yan H, Ken H, Jessica R, Hwang RY, Pitt GS, Tracey WD (2012) Thermosensory and non-thermosensory isoforms of Drosophila melanogaster TRPA1 reveal heat sensor domains of a thermoTRP channel. Cell Rep 1:43–55. 10.1016/j.celrep.2011.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Aversive response to noxious mechanical stimulation.

Normal larval locomotion.