Abstract
Columnar structure is a basic unit of the brain, but the mechanism underlying its development remains largely unknown. The medulla, the largest ganglion of the Drosophila melanogaster visual center, provides a unique opportunity to reveal the mechanisms of 3D organization of the columns. In this study, using N-cadherin (Ncad) as a marker, we reveal the donut-like columnar structures along the 2D layer in the larval medulla that evolves to form three distinct layers in pupal development. Column formation is initiated by three core neurons, R8, R7, and Mi1, which establish distinct concentric domains within a column. We demonstrate that Ncad-dependent relative adhesiveness of the core columnar neurons regulates their relative location within a column along a 2D layer in the larval medulla according to the differential adhesion hypothesis. We also propose the presence of mutual interactions among the three layers during formation of the 3D structures of the medulla columns.
SIGNIFICANCE STATEMENT The columnar structure is a basic unit of the brain, but its developmental mechanism remains unknown. The medulla, the largest ganglion of the fly visual center, provides a unique opportunity to reveal the mechanisms of 3D organization of the columns. We reveal that column formation is initiated by three core neurons that establish distinct concentric domains within a column. We demonstrate the in vivo evidence of N-cadherin-dependent differential adhesion among the core columnar neurons within a column along a 2D layer in the larval medulla. The 2D larval columns evolve to form three distinct layers in the pupal medulla. We propose the presence of mutual interactions among the three layers during formation of the 3D structures of the medulla columns.
Keywords: columnar unit, differential adhesion, Drosophila, N-cadherin, optic lobe, visual system
Introduction
The columnar unit is a basic feature of the brain. In the cerebral cortex, microcolumns are cell type-specific radial clusters of neurons that are found in nearly all examined cortical regions (Mountcastle, 1997; Maruoka et al., 2017). However, the developmental mechanisms underlying columnar unit formation remain unclear.
The fly visual center, which receives visual input from the eye, shares structural characteristics with the mammalian cerebral cortex, such as the columnar and layered structures (Sanes and Zipursky, 2010). The retina is the compound eye consisting of 800 ommatidial units, and each ommatidium contains eight photoreceptor neurons, R1–R8. Visual information received in the retina is transmitted to the optic lobe in the brain (Sato et al., 2013; Zhu, 2013; Nériec and Desplan, 2016). R1–R6 project to the first optic ganglion, the lamina, where lamina neurons L1–L5 form a columnar unit called a cartridge. R7, R8, and L1–L5 then project to the second optic ganglion, the medulla, where as many as 100 neurons form a columnar unit. Some of these neurons project through the medulla to the third optic ganglion, the lobula, and lobula plate. In comparison with the simple structures of ommatidia in the retina and cartridges in the lamina, columns in the medulla show complex 3D structures that are analogous to the microcolumns in the mammalian cerebral cortex (see Fig. 1A) (Takemura et al., 2013). Indeed, the developmental mechanisms underlying microcolumns and medulla columns share similarities. A single neuroblast (NB), a neural stem-like cell, produces a group of radially oriented and clonally related neurons in the developing medulla (see Fig. 1A) (Hasegawa et al., 2011; Morante et al., 2011), which is analogous to the radial unit produced from a single neural stem cell and migrates along the same radial fiber in the developing cerebral cortex (Rakic, 1988; Mountcastle, 1997; Noctor et al., 2004; Li et al., 2012; Ohtsuki et al., 2012). These characteristics of the medulla, together with the powerful techniques available in fly genetics, make it an excellent model for studying column formation.
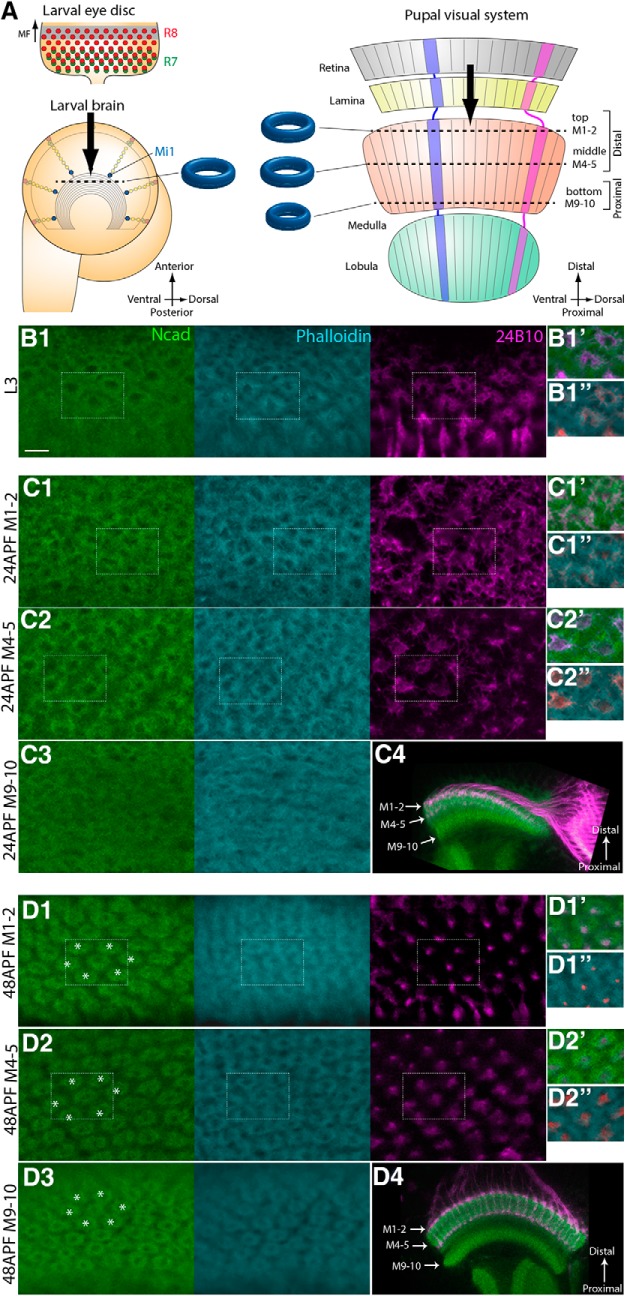
Figure 1.
N-cadherin and phalloidin visualize developing columns. A, Schematics of the developing larval and pupal visual systems. In the larval eye disc, R8 and R7 are sequentially differentiated behind the morphogenetic furrow. The NBs, located on the surface of the larval brain, produce Mi1 medulla neurons. In the pupal visual system, the retina, lamina, medulla, and lobula are topographically connected. Columns are identifiable along the planes indicated by dotted lines in the larval and pupal brains along the three layers (top, M1–M2; middle, M4–M5; bottom, M9–M10). B–D, The donut-like columnar patterns are identifiable in L3 (B1), 24 h APF (C1–C3), and 48 h APF stages (D1–D3) by Ncad (green), phalloidin (blue), and 24B10 (magenta). B1, Single layer of columnar pattern in L3 larval stage. C1–C3, D1–D3, Three layers of columnar patterns in 24 h APF (C1–C3) and 48 h APF (D1–D3) pupal stages. Layers M1–M2 (C1,D1), M4–M5 (C2,D2), and M9-M10 (C3,D3) are shown. Areas indicated by white rectangles are shown at right (B1,C1–C2,D1–D2). In contrast to the clear and regular patterns in layers M1–M2 and M4–M5 (C1,C2), the columnar pattern is irregular in layers M9-M10 at 24 h APF (C3), which become organized by 48 h APF (D3). Distal to the top, dorsal to the right. The following images are oriented in the same manner. C4, D4, Horizontal sections of 24 and 48 h APF brains to visualize the layers M1–M2, M4–M5, and M9-M10. Scale bar: B1, 5 μm.
In this study, we show that the cell adhesion molecule N-cadherin (Ncad) (Iwai et al., 1997) and F-actin-binding molecule phalloidin reveal a donut-like structure of the medulla column. The 2D donut-like pattern in the larval medulla evolves to establish the 3D pattern in the top, middle, and bottom layers during pupal development. We identified three core columnar neurons essential for column formation in larval development. The terminal of R7 occupies the dot-like central region of the larval column. The R8 terminal enwraps the R7 terminal forming a donut-like region that overlaps with the Ncad/phalloidin pattern. The Mi1 terminal occupies a grid-like region outside the R8 terminal. Thus, R7, R8, and Mi1 form the 2D concentric organization composing the three distinct domains in the larval medulla.
Ncad is expressed in R7, R8, L1–L5, and Mi1 neurons (Lee et al., 2001; Ting et al., 2005; Yonekura et al., 2007; Nern et al., 2008; Petrovic and Hummel, 2008; Hasegawa et al., 2013). Ncad-dependent cell adhesion among these neurons may regulate column formation. The differential adhesion hypothesis (DAH) proposes that mixtures of cell populations segregate according to their adhesive properties: the less adhesive cell population is located at the periphery of the cell aggregates and surrounds the more adhesive cells (Steinberg and Takeichi, 1994; Murakawa and Togashi, 2015). Roles of Ncad-dependent differential adhesion have been proposed in lamina cartridge formation (Iwai et al., 2002; Schwabe et al., 2014). In this study, we establish a series of genetic tools that can specifically express fluorescent markers and manipulate Ncad activity in R7, R8, and Mi1 in the medulla. By using these tools, we clearly demonstrate the in vivo evidence of DAH among R7, R8, and Mi1 in the medulla column formation. Additionally, by using a mathematical model based on the differential adhesiveness of these neurons, we also demonstrate that differential adhesion could be the major driving force in establishing the columnar structure.
Materials and Methods
Fly strains.
Fly strains were maintained on standard Drosophila medium at 25°C. For all experiments in this study, both male and female flies were used. The following fly strains were used: sensE2 FRT 80B (Frankfort et al., 2001), sev14 (BDSC5689), bsh1, bsh2 (Hasegawa et al., 2013), Ncad M19FRT40A (Iwai et al., 1997), sensF2-Gal4 (Atkins et al., 2013), sevPM181-Gal4 (Lee et al., 2001), R69B02-Gal4 (L1–L5; BDSC39478), c202-Gal4 (L1) (Rister et al., 2007), R16H03-Gal4 (L2; BDSC48744), R20A03-Gal4 (L3; BDSC48871), R31C06-Gal4 (L4; BL49883), R13E12-Gal4 (Tm3; BDSC48569), R42F06-Gal4 (T4-T5; BDSC41253), drf-Gal4 (Hasegawa et al., 2011), GMR-FRT-stop-FRT-Gal4 (Chen et al., 2014), arm-lacZ FRT80B (BDSC6341), sens-FLPG5 (R8; BDSC55768), 20C11-FLPG5 (R7; BDSC55766), R27G05-FLPG5 (L1–L5; BDSC55765), UAS-bshRNAi (VDRC106634), UAS-NcadRNAi (BDSC27503, VDRC1092), UAS-sevRNAi (BDSC55866), UAS-rpr (BDSC5824), UAS-Ncad7a,13a,18a (Iwai et al., 1997), hs-flp, tub-Gal80 FRT40A, tub-Gal80 FRTG13, and y+ FRT40A. The following fly strains were generated in this work: bshM-Gal4, bshL-Gal4, UAS-myrGFP, UAS>Gal80>myrGFP, R27G05-LexA (L1–L5), sevEnS-LexA, sensF2-LexA, bshM-LexA, LexAop-myrTomato, Ncad-FsF-GFP, and hth-FLP.
Generation of Gal4 and LexA strains.
pGEattB-bshSPGal4 vector was generated by inserting bsh short promoter (bshSP) and Gal4 (IVS-Syn21-Gal4-p10) (Pfeiffer et al., 2012) fragments into pGEattB vector (Huang et al., 2009). Lamina-specific bshL-Gal4 and medulla-specific bshM-Gal4 were generated by inserting bshD fragment together with bshPA and bshPB fragments into pGEattB-bshSPGal4, respectively. pGEattB-LexA vector was generated by inserting LexA (LHV2) (Yagi et al., 2010) fragment into pGEattB. Medulla-specific bshM-LexA was generated by inserting bshD, bshPB, and bshSP fragments into pGEattB-LexA. sevEnS-LexA construct was generated by inserting sev enhancer (sevEnS) and sev promotor (sevPro) fragments into pGEattB-LexA. sensF2-LexA was generated by inserting sens enhancer (sensF2) and sens promoter (sensPro) fragments into pGEattB-LexA. R27G05-LexA was generated by inserting R27G05 and hsp70 promoter fragment into pGEattB-LexA. bshSP (1.4 kb), bshD (2.0 kb), bshPA (552 bp), bshPB (378 bp), sevEnS (1.3 kb), sevPro (1 kb), sensF2 (650 bp), and sensPro (1.5 kb) fragments were amplified using the following PCR primers: bshSP (ACTAGTTAACACAAACCCCATAACCCCC and GTAGCTTCATGGATCCCATTGCCATTGCGCTGGAGGATG), bshD (TATAGATCTGCGGCCGCGTTAGGTTGTTAACCTAAGC and CTGTTTAAGGGAATTCGCCGCTATTGAAGATTACCCAG), bshPA (GAATTCCCTTAAACAGGAACTACAAGTACG and GTTTGTGTTAACTAGTGCCTAATTTCCCATTGTTGC), bshPB (GAATTCCCTTAAACAGGAACTACAAGTACG and GTTTGTGTTAACTAGTGAGCTCCATAGAAGTGGAAAGC), sevEnS (TTCTTGCATGCAATGCGGCCGCTTATTAGCGAGATGCTCGAGGTCTCTC and TTGGTCTGCAGTTGAGTATCCTGGTCTTCCTGCTCGAGATCC), sevPro (CACTCAACTGCAGACCAATTCTGATTGTGTGGGCTAGTTCCC and TTAACGCTTTCATGGCGCGCCTATCTGGATCTGGATGTGCGATTCCCG), sensF2 (AATTCTTGCATGCAATGCGGCCGCGTGGAAAAGTGGCACAGCAAATACTGTCATA and TATCTGAAGCTAGCAACTCGGAAGTTGGCAAAGCTCGAAAGG), and sensPro (GAGTTGCTAGCTTCAGATACGAGAAGATGGGGACTTTATGGCT and CGTTAACGCTTTCATGGCGCGCCGATTCGAACGGATCTTCGATGGGTC).
Generation of UAS>Gal80>myrGFP, UAS-myrGFP, and LexAop-myrTomato strains.
UAS>Gal80>myrGFP was generated by inserting hsp70 promoter-FRT, Syn21-Gal80, STOP-FRT-IVS, and Syn21-myr::GFP into pJFRC164 (Addgene) (Pfeiffer et al., 2012), and was coinjected with PhiC31 integrase into the VK00005 attP landing site. FRT-Gal80-FRT cassette was permanently removed using FLPase to generate UAS-myrGFP fly strain. LexAop-myrTomato was generated by inserting Syn21-myr::Tomato fragment from HM29–37 MCS-tdTomato/pCPCI3 (gift from Hideji Murakoshi) into pJFRC19 and was coinjected with PhiC31 integrase into the ZH-22A attP landing site. hsp70 promoter-FRT, Syn21-Gal80, STOP-FRT-IVS, Syn21-myr::GFP, and Syn21-myr::Tomato fragments were amplified using the following PCR primers: hsp70 promoter-FRT (ACTCTAGCGCTAGCGACGTCG and TTTTTTTTTTAAGTTCTAGCTAGCTTACGTTAGAAG), Syn21-Gal80 (AACTTAAAAAAAAAAATCAAAATGGACTACAACAAGAG and TTGGCTTTAGTCGACTTAACCGGTTTATAAACTATAATGC), STOP-FRT-IVS (GTCGACTAAAGCCAAATAGA and CCTCGAGCCGCGGCCGCCTGA), Syn21-myr::GFP (catGGTACCAACTTAAAAAAAAAAATCAAAATGGGCAACAAATGCTGCAG and TTACTTGTAGAGCTCATCCATG), and Syn21-myr::Tomato (CGGCCGCGGCTCGAGGGTACCAACTTAAAAAAAAAAATCAAAATGGGCAACAAATGCTGCAG and ACGATTCATTTAAGGCCTTTACTTGTACAGCTCGTCC).
Generation of hth-FLP strain.
The third exon of endogenous hth was replaced by the fragment containing attP via the homologous recombination with the homology arms for hth (hthLp and hthRp) (Huang et al., 2009). The hthKnockinFLP-p10 vector was generated by inserting a short bridge fragment containing a part of the second intron and third exon of hth (hth_inEx), FLP, and p10 into pGEattB vector. hthLp (1.8 kb) and hthRp (2.0 kb), hth_inEx, FLP, and p10 fragments were amplified using the following PCR primers: hthLp (CATGCGGCCGCCCAAGTGACCCGAGATGATCTT and CATGGTACCTGGATTGCCTCAAAAACCCAGG), hthRp (CATGGCGCGCCCAGGTAAGAATAATGGCCCCAG and CATCTCGAGCCTTAGCTGCATCATTAGCAGC), hth_inEx (CATCATGCGGCCGCCCTGGGTTTTTGAGGCAATCCA and CATGGCGCGCCGGAATAGCGGATGTCTGCAATGG), FLP (CGCTATTCCGGCGCGCCATGCCACAATTTGATATATTATG and GCTAGCTTATATGCGTCTATTTATGTAG), and p10 (CGCATATAAGCTAGCATGAATCGTTTTTAAAATAAC and TTATACGAAGTTATGGTACCTCGAATCGCTATCCAAGCCAG). The hthKnockinFLP-p10 was coinjected with PhiC31 integrase into the attP site in the hth locus.
Generation of Ncad-FsF-GFP knock-in allele.
Ncad-FsF-GFP knock-in allele was generated by CRISPR/Cas9 technology (Kondo and Ueda, 2013). A knock-in vector containing the homology arms for Ncad (NcadL and NcadR), the flip-out cassette with GFP (FRT-stop-FRT-GFP), and the red fluorescent transformation marker gene (3xP3RFP) was generated by ligating NcadL (555 bp), FRT, stop-FRT, GFP, 3xP3RFP, and NcadR (518 bp) fragments amplified using the following PCR primers: NcadL (TATAGAGCTCACCCATTATGCAGCCGTTTAGTC and TATAGCGGCCGCACAATTCTAGTTCGGTATTGTGGGG), NcadR (TATACTCGAGGCTGGTGGAGCGAGCAGTGA and TATAGGTACCGCGCTCCTAAACTAAACTTTGGTATGC), FRT (TATAGCGGCCGCATTGAAGTTCCTATTCCGAAGTTCC and TATAACTAGTCAAAAGCGCTCTGAAGTTCCTATAC), stop-FRT (TATAACTAGTTAAATCCAGACATGATAAGATACATTGATGAG and TATACTGCAGCAAAAGCGCTCTGAAGTTCCTATAC), GFP (TATACTGCAGATGAGTAAAGGAGAAGAACTTTTC and TATACTCGAGTCTAGTGGATCCAGACATGATAAG), and 3xP3RFP (GCGCCTCGAGAACTTCGTATAGCATACATTA and TATACTCGAGAACGTGTCGGTACCAATTGAGCTC). A gRNA vector (pBFv-U6.2) that recognizes the sequences containing the stop codon of Ncad gene (GAACTAGAATTGTAAGCTGGTGG) was injected to eggs carrying vas-Cas9 (BDSC 55821) together with the knock-in vector. The precise integration of the knock-in vector was verified by PCR and sequencing. The 3xP3RFP flanked by two loxP sites was removed by the heat induction of Cre. We also generated Ncad-GFP allele by permanently removing the FsF cassette from Ncad-FsF-GFP using FLPase. The Ncad-GFP patterns were indistinguishable from the Ncad antibody patterns, suggesting that the Ncad-FsF-GFP allele reliably recapitulates Ncad protein distribution.
Histochemistry.
Immunohistochemistry was performed as described previously (Hasegawa et al., 2011). The following primary antibodies were used: rat anti-Ncad (1:20, Developmental Study Hybridoma Bank), mouse anti-Chaoptin (24B10, 1:20, Developmental Study Hybridoma Bank), guinea pig anti-Bsh (1:500) (Hasegawa et al., 2011), mouse anti-LacZ (1:250, Promega), and chicken anti-LacZ (1:2000, Abcam). The secondary antibodies were used: anti-mouse Cy3, anti-mouse Cy5, anti-guinea pig Cy5, anti-guinea pig FITC, anti-rat Cy5, and anti-chicken Cy3 (Jackson ImmunoResearch Laboratories). Phalloidin staining was performed by TRIC-Phalloidin (1:200, Sigma). Confocal images were obtained by LSM 880 (Carl Zeiss) and a super-resolution unit Airyscan (Carl Zeiss); 40× 1.2 NA water-immersion and 63× 1.4 NA oil-immersion lenses were used for regular confocal and super-resolution imaging, respectively. The default Airyscan filtering mode (filter strength of 6.0) with the 3D reconstruction algorithm was used. Images were processed using LSM image browser ZEN (Carl Zeiss) and Photoshop (Adobe).
Quantification of Ncad expression level.
To quantify the level of Ncad expression, the photon-counting mode of LSM880 confocal microscopy was used. Ncad-FsF-GFP expressing axon terminals in the larval medulla were focused, and numbers of photons emitted from the terminals were quantified in each pixel area to compare the photon density for different neuron types. Sizes of the pixel area were categorized according to the number of photons emitted. Background photons were counted outside the GFP-labeled clones and were subtracted from the number of photons emitted from the axon terminals.
Experimental design and statistical analysis.
The experiments in Figures 6, 7, 8–9 were designed and quantified as described below and statistically tested by t test. p values are indicated in the figure legends.
Figure 6.
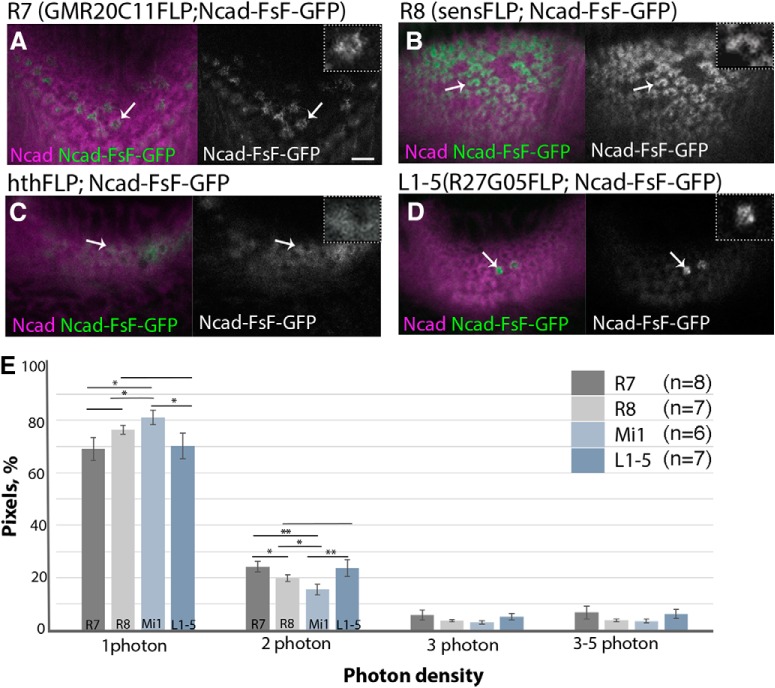
Differential expression of N-cadherin in the columnar neurons. Ncad protein localized at the terminals of columnar neurons are visualized by combining Ncad-FsF-GFP (green or white) and FLPase strains that are specific to R7 (A), R8 (B), Mi1 (C), and L1–L5 (D) in L3 larval medulla. Magenta represents Ncad. A, Ncad-GFP is visualized in R7 under the control of R20C11-FLPG5. Ncad-GFP exists in the central hole area inside the columns (arrows). B, Ncad-GFP is visualized in R8 under the control of sens-FLPG5. C, Ncad-GFP is visualized in Mi1 under the control of hth-FLP. Mi1 was identified by Bsh expression. D, Ncad-GFP is visualized in L1–L5 under the control of R27G05-FLPG5 (arrows). Higher-magnification images are separately visualized in the top right corner of each panel. Scale bar: A, 5 μm. E, Densities of photons that derive from Ncad-GFP at the terminals of columnar neurons are compared in L3 larval brains. Histogram of photon densities at the terminals of R8, R7, Mi1, and L1–L5. Percentages of pixel areas that contain 1, 2, 3, and 3–5 photons are compared for each neuron type. Number of axon terminals examined: R8 (n = 7), R7 (n = 8), Mi1(n = 6), L1–L5 (n = 7). *p < 0.05 (t test). **p < 0.01 (t test).
Figure 7.
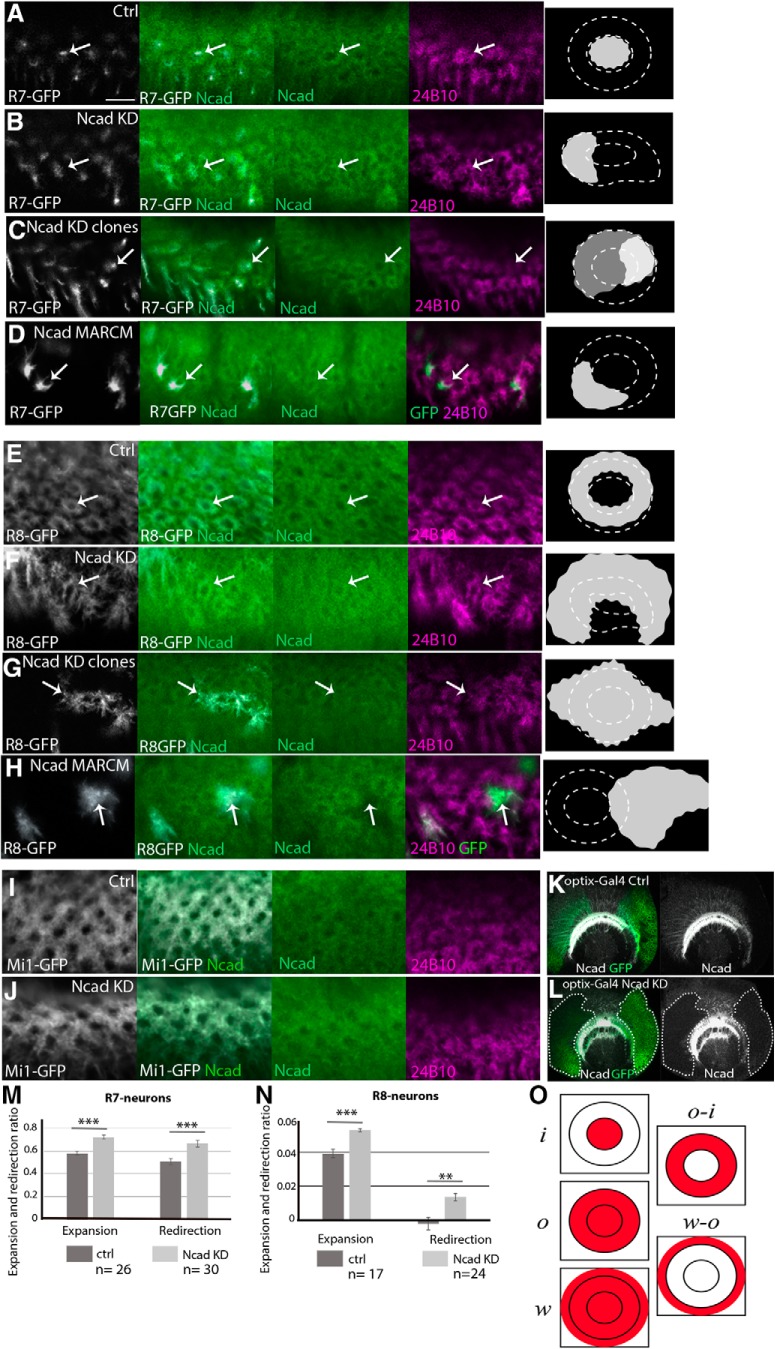
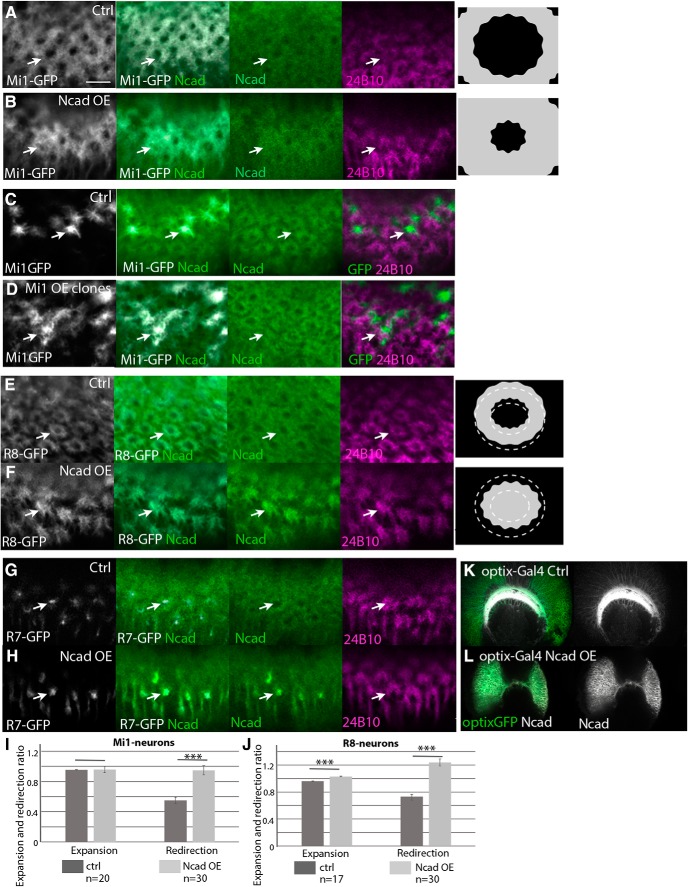
Roles of N-cadherin in columnar organizations. Columnar Ncad pattern (green), and projection patterns of R axons (24B10; magenta) and columnar neurons (UAS-myrGFP; white) in WT and Ncad knockdown backgrounds in L3 larval medulla. A–D, R7 terminals visualized by PM181-Gal4. A, Control. B, Ncad RNAi in R7. R7 terminals are expanded and the columnar Ncad pattern is disorganized (arrows). C, Ncad RNAi in a subset of R7 under the control of R20C11-FLPG5; GMR>Gal80>Gal4. R7 terminals are expanded and redirected to the column periphery (arrows) as quantified in M. D, Ncad mutant MARCM clones visualizing R7 terminals that are expanded and redirected to the periphery of the column (arrows). E–H, R8 terminals visualized by sensF2-Gal4. E, Control. F, Ncad RNAi in R8. R8 terminals are expanded and the columnar Ncad pattern is disorganized (arrows). G, Ncad RNAi in a subset of R8 under the control of sens-FLPG5; GMR>Gal80>Gal4. R8 terminals are expanded and redirected to the column periphery (arrows) as quantified in N. H, Ncad mutant MARCM clones visualizing R8 terinals that are expanded to the column periphery (arrows). Ncad staining is abolished in and around the mutant clones (arrows). I, J, Mi1 terminals visualized by bshM-Gal4. I, Control. J, Ncad RNAi in Mi1. Mi1 terminals and columnar Ncad pattern are not significantly affected. Scale bar: A, 5 μm. K, L, The effect of UAS-Ncad RNAi (BDSC27503) on Ncad protein level was tested by using optix-Gal4 UAS-CD8GFP (green) in larval medulla. Ncad (white). K, Control. L, Ncad signal is significantly downregulated in GFP-positive area indicated by white dotted lines. M, N, Expansion and redirection ratios of R7 (M) and R8 (N) are quantified according to the definition in O. **p < 0.01 (t test). ***p < 0.001 (t test). O, o and i indicate the total GFP amount within the outer and inner rings of Ncad or phalloidin staining, respectively. w indicates the total GFP amount within the entire growth cone of a single R8 axon. Relative GFP density (R7 and Mi1) or amount (R8) between distinct domains was used to calculate the expansion and redirection ratios. Details are described in Materials and Methods.
Figure 8.
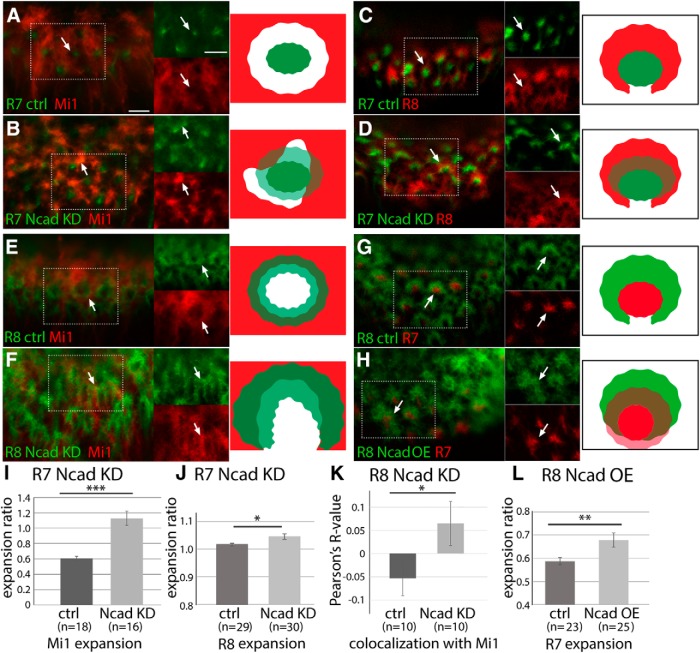
Ncad knockdown reveals differential adhesion between columnar neurons. Influences of Ncad RNAi expression in columnar neurons on other neurons in L3 larval medulla. A–D, R7 terminals are visualized by PM181-Gal4 UAS-myrGFP (green). A, B, Mi1 terminals are visualized by bshM-LexA LexAop-myrTomato (red). A, Control. R7 and Mi1 terminals do not overlap each other (arrows). B, Ncad RNAi in R7. R7 terminals are expanded and Mi1 terminals are disorganized. R7 and Mi1 overlap each other as evident from yellow signals (arrows) as quantified in I. C, D, R8 terminals are visualized by sensF2-LexA LexAop-myrTomato (red). C, Control. R7 terminals are enwrapped by R8 terminals (arrows). D, Ncad RNAi in R7. R7 and R8 overlap each other as evident from yellow signals (arrows) as quantified in J. E–H, R8 terminals are visualized by sensF2-Gal4 UAS-myrGFP (green). E, F, Mi1 terminals are visualized by bshM-LexA LexAop-myrTomato (red). E, Control. R8 and Mi1 terminals are adjacent with each other. F, Ncad RNAi in R8. R8 terminals are expanded and Mi1 terminals are disorganized (arrows). R8 and Mi1 overlap each other as quantified in K. G, H, R7 terminals are visualized by sevEnS-LexA LexAop-myrTomato (red). G, Control. R7 terminals are enwrapped by R8 terminals. H, Ncad overexpression in R8. R7 terminals are expanded toward the column center and overlap with R8 terminals as quantified in L. Rectangles indicated by white dots are separately shown at right (A–H). Scale bars: A, 5 μm. I–L, Expansion of Mi1 (I), R8 (J), and R7 terminals (L), and overlaps between R8 and Mi1 (K) are quantified and statistically tested. *p < 0.05 (t test). **p < 0.01 (t test). ***p < 0.001 (t test).
Figure 9.
Ncad overexpression in the core columnar neurons. Columnar Ncad pattern (green), projection patterns of R axons (24B10; magenta), and columnar neurons (UAS-myrGFP; white) in WT and Ncad overexpression backgrounds in L3 larval medulla. A, B, Mi1 terminals visualized by bshM-Gal4. A, Control. B, Ncad overexpression in Mi1. Mi1 terminals are expanded toward the column center (arrows) as quantified in I. C, D, MARCM clones visualizing Mi1 by bshM-Gal4 UAS-myrGFP. C, Control. D, Ncad overexpression in a subset of Mi1 neurons visualized by GFP. Expansion of Mi1 terminals toward the center of the columns is indicated by arrows. E, F, R8 terminals visualized by sensF2-Gal4. E, Control. F, Ncad overexpression in R8. R8 terminals are redirected toward the column center (arrows) and the central hole area. G, H, R7 terminals visualized by PM181-Gal4. A, Control. B, Ncad overexpression in R7. R7 terminals and columnar Ncad pattern are not significantly affected. Scale bar: A, 5 μm. K, L, The effect of UAS-Ncad on Ncad protein level was tested by using optix-Gal4 UAS-CD8GFP (green) in larval medulla. Ncad (white). K, Control. L, Ncad signal is augmented in GFP-positive medulla neurons. I, J, Expansion and redirection ratios of Mi1 (I) and R8 (J) are quantified according to the definition in Figure 7O and Materials and Methods. ***p < 0.001 (t test).
Quantification of columnar defects.
Fiji was used to quantify the columnar defects (see Figs. 7, 9). Phalloidin was used as a column marker. The outer and inner rings of phalloidin staining were manually outlined by the oval selection tool. Essentially the same results were obtained when Ncad was used as a marker. The total amount of GFP signal and the oval size were measured. Here, o and i indicate the total GFP amount within the outer and inner rings, respectively. o − i indicates the GFP amount within the Ncad+ ring region. The GFP amount was divided by the area size of interest to normalize the size effect. Relative GFP density between distinct domains of the column was calculated as follows. Expansion and redirection ratios of R7 are o/i and (o − i)/i, respectively (see Fig. 7). When we knocked down Ncad in R8, the entire growth cone of a single R8 was outlined by the oval selection tool to measure the total amount of GFP (w). Relative GFP amount between distinct domains of the column was calculated. Expansion and redirection ratios of R8 are w/o and (w − o)/o, respectively (see Fig. 7). When we overexpress Ncad in Mi1 and R8, expansion and redirection ratios are o/(o − i) and i/(o − i), respectively (see Fig. 9).
In Figure 8, individual R7 or R8 terminals were manually outlined by the oval selection tool. The mean intensity of Tomato signal inside the selected area was normalized by the mean intensity of Tomato signal inside a wider area containing multiple columns to obtain expansion ratios of Mi1 or R8. An ROI of 100 × 70 was selected by the rectangle selection tool to crop an area in which both GFP (R8) and Tomato (Mi1) signals are focused. Coloc 2 function of Fiji was used to calculate the Pearson's R value to quantify the colocalization between two neurons.
Mathematics and equations.
We model the neuronal cell segregation by modifying the continuous model of cell–cell adhesion (Murakawa and Togashi, 2015; Carrillo et al., 2019). The densities of the three cell types of neurons at position x ∈ ℝ2 and time t ≥ 0 are denoted by u = u(x, t), v(x, t), w = w(x, t), respectively. Each of the equations in the system represents the evolution in time of the densities u, v, w under a velocity field containing volume exclusion effect and attractive forces. The nondimensional system reads as follows:
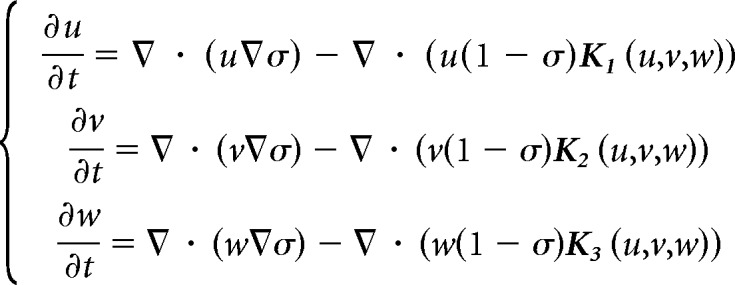 |
where σ = u + v + w is the total population of cells, and the interaction attractive kernels Ki (i = 1, 2, 3) are given by the following:
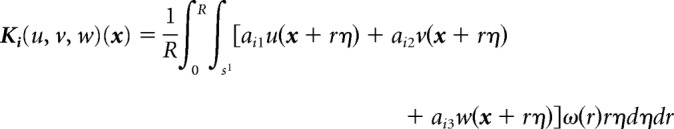 |
Here, R is a positive constant called the sensing radius, which is rescaled to be 1, S1 is the unit circle, aij (i, j = 1, 2, 3) denotes the adhesive strength parameters between the ith and jth species, and ω is a function describing how the adhesion force depends at a distance r from position x, which is defined as ω  1 here. The attractive forces model nonlocal adhesion between the different type of cells characterized by their kernels Ki, the strengths aij, and the shape of the force ω. On the other hand, the repulsive force due to volume constraints is modeled by a local nonlinear diffusivity depending on the total population of cells σ. The main difference with respect to the original model (Murakawa and Togashi, 2015) is the assumption that each cell counts its surrounding linearly to determine the direction of movement and the magnitude of the force decreases as the density at the cell position increases. This modification allows for a more physical balance between the total volume constraint and the adhesion forces terms. The numerical simulations in Figure 10 were performed in a 2D fixed domain of size with the periodic boundary condition. The standard explicit upwind finite volume method was used to discretize the equations, and the nonlocal terms Ki were calculated by a numerical integral (Murakawa and Togashi, 2015; Carrillo et al., 2019). The source codes for the numerical simulations will be deposited to a public repository service.
1 here. The attractive forces model nonlocal adhesion between the different type of cells characterized by their kernels Ki, the strengths aij, and the shape of the force ω. On the other hand, the repulsive force due to volume constraints is modeled by a local nonlinear diffusivity depending on the total population of cells σ. The main difference with respect to the original model (Murakawa and Togashi, 2015) is the assumption that each cell counts its surrounding linearly to determine the direction of movement and the magnitude of the force decreases as the density at the cell position increases. This modification allows for a more physical balance between the total volume constraint and the adhesion forces terms. The numerical simulations in Figure 10 were performed in a 2D fixed domain of size with the periodic boundary condition. The standard explicit upwind finite volume method was used to discretize the equations, and the nonlocal terms Ki were calculated by a numerical integral (Murakawa and Togashi, 2015; Carrillo et al., 2019). The source codes for the numerical simulations will be deposited to a public repository service.
Figure 10.
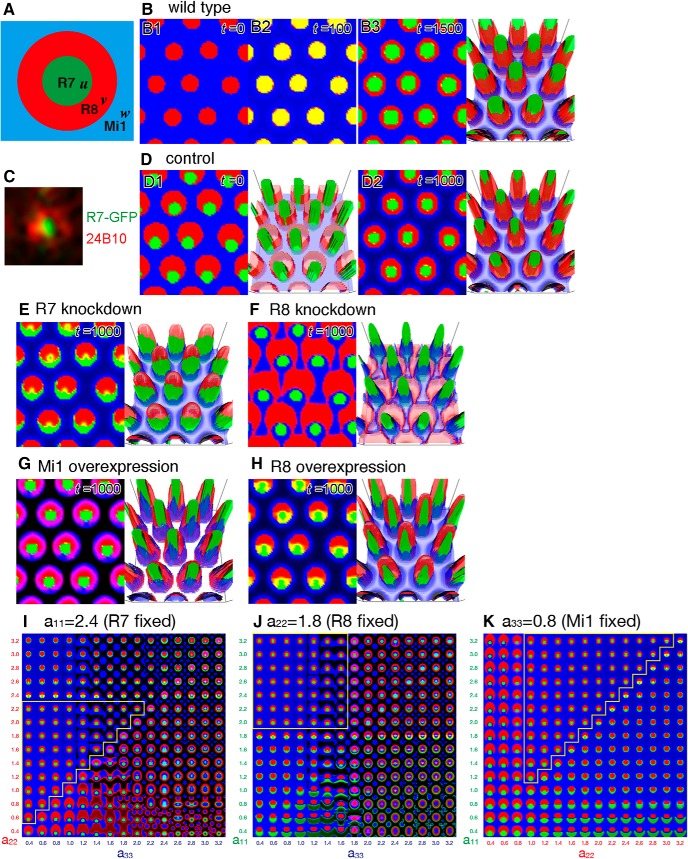
The continuous mathematical model of column formation. A, Schematic of the configuration of a column in the control. The variables u, v, and w are the densities of the terminals of R7 (green), R8 (red), and Mi1 (blue), respectively. B, Numerical results of sequential projections of R8 (B1; t = 0) and R7 (B2; t = 100). Identical initial distributions of R8 and R7 produce clear concentric patterns of R7, R8, and Mi1 (B3; t = 1500). C, Segregated distribution patterns of PM181-Gal4 UAS-myrGFP (green) and 24B10 (red) just below the lamina before the arrival at the medulla, where R8 partially engulfs R7. D–H, Results of numerical simulations based on the initial distribution pattern shown in D1. D1, D2, Control (a11 = 2.4, a22 = 1.8, a33 = 0.8). R7, R8, and Mi1 are clearly segregated (D2; t = 1000). E, R7 knockdown (a11 = 1.7). R7 is excluded from the ring of R8 and partially enwraps R8 (t = 1000). F, R8 knockdown (a22 = 0.6). R8 is expanded and fuses with the neighboring columns (t = 1000). G, Mi1 overexpression (a33 = 1.8). Mi1 penetrates the ring of R8 (t = 1000). H, R8 overexpression (a22 = 2.4). R8 and R7 are expanded toward the column center and column periphery, respectively (t = 1000). B3, D1, D2, E–H, Right panels, 3D views of the results, in which z axis indicates the density of neurites. I–K, The adhesiveness of R7, R8, and Mi1 (a11, a22, and a33) is changed between 0.4 and 3.2 to examine the range of parameter settings by which the normal column pattern can be reproduced. I, a11 is fixed to 2.4. J, a22 is fixed to 1.8. K, a33 is fixed to 0.8. Normal concentric columnar patterns are obtained if the order of adhesiveness is conserved (yellow lines; a11 > a22 > a33).
Results
N-cadherin and phalloidin can visualize developing columns
To visualize the medulla columns, we screened a series of markers expressed in the developing fly brain. We show that the cell adhesion molecule Ncad visualizes a donut-like pattern localized at the boundary of each column in the medulla during the larval and pupal stages (Fig. 1B–D). In contrast to the single layer of this donut-like pattern in the larval brain (Fig. 1B), three layers of similar structures are found in the top (layers M1–M2), middle (layers M4–M5), and bottom (layers M9–M10) of the medulla neuropil at 24 and 48 h after puparium formation (APF) (Fig. 1C,D). The top and middle layers derive from the distal part of the medulla neuropil early in pupal development, whereas the bottom layer is being formed at 24 h APF (Fig. 1C4) and is completely separated from the distal layers at 48 h APF (Fig. 1D4). The irregular arrangement of columns in the bottom layer at 24 h APF becomes well organized by 48 h APF (Fig. 1C3,D3), suggesting that the columns in the bottom layer develop later than those in the top and middle layers. The clear hexagonal arrangement of the columns is observed in all three layers at 48 h APF (Fig. 1D1–D3). The donut-like structure is smaller in the bottom layer than in the top and middle layers.
Additionally, the F-actin-binding molecule phalloidin shows a donut-like pattern similar to that of Ncad, except for the smaller hole area during the larval stage (Fig. 1B–D). Thus, Ncad and phalloidin can be used as markers to visualize the 3D structure of developing medulla columns. As demonstrated later, the arborizations of core columnar neurons overlap with these donut-like structures, suggesting that the above molecular markers precisely visualize the developing medulla columns in larval and pupal stages.
Finally, anti-Chaoptin antibody (24B10), which strongly visualizes the cell membrane of photoreceptor neurons R1–R8 (Van Vactor et al., 1988), also shows a columnar donut-like pattern that overlaps with Ncad and phalloidin in third instar larvae (Fig. 1B1). However, the donut-like patterns marked by 24B10 in layers M1–M2 and M4–M5 are obscured at 24 h APF (Fig. 1C1,C2) and are restricted to the dots inside the donuts by 48 h APF (Fig. 1D1,D2). The single dots of 24B10 are localized in the peripheral parts of the columns in layers M1–M2 (Fig. 1D1) or at the central position within the column in layers M4–M5 at 48 h APF (Fig. 1D2).
Although synapse formation that occurs later in pupal development establishes the neural circuits within and between the columns, we only focused on the larval and early pupal stages in this study to reveal the early steps of column formation before synaptogenesis that have not been studied so far.
Projection patterns of essential neurons within the columns
To identify neurons that contribute to the donut-like patterns, we examined the projection patterns of neurons that project to the medulla columns: the photoreceptor neurons R7-R8, the lamina neurons L1–L5, the medulla neurons Mi1 and Tm3, and lobula plate neurons T4-T5 (Fischbach, 1989).
R8 is the primary neuron that induces the differentiation of other photoreceptor neurons (R1-R7) in the retina (Jarman et al., 1994). Indeed, R8 axons arrive at the medulla earlier than R7 axons (see Fig. 3A1,B2). Interestingly, R8 axon terminals show donut-like shapes that overlap with Ncad from mid-third instar larval stage (Figs. 2A1, 3A3). The R8 terminal is not always circular but occasionally shows a horseshoe-like shape due to a gap (Fig. 2A1, arrows and arrowheads). Super-resolution imaging of the R8 terminals revealed fine filopodia emanating from the bifurcated terminals attached to each other forming a semi-circular shape (Movies 1, 2). In the following text, we call the shape of the R8 terminal “donut-like” for simplicity, except where noted otherwise. After R8, the R7 axons project to the center of the columns (Figs. 2B1, 3B). Both R8 and R7 terminate in the same layer in the larval medulla showing many filopodia, suggesting that the growth cones of R8 and R7 show the donut-like and dot-like structures, respectively (Movie 1). 24B10 preferentially visualizes R8 in the larval stage because the 24B10 signal is undetectable in the central hole area of the column where the R7 axon terminates (Figs. 1B1, 2A1,B1). These patterns gradually change during pupal development. At 48 h APF, R8 axons show semi-donut-like patterns in the superficial part of the M1 layer and faint dot-like patterns in the middle of the column in the M3 layer (Fig. 2A2,A3). The changes in the R8 terminal morphology may be due to the gradual expansion of R8 terminals from the M1 to M3 terminal layers (Özel et al., 2015). The faint signal in the M3 layer is likely due to the immature state of the R8 growth cones and gradually becomes stronger in the later stages of pupal development (Fig. 2G). These developmental processes are regulated by multiple molecules, such as Golden Goal, Flamingo (Tomasi et al., 2008; Hakeda-Suzuki et al., 2011), Ark/Jelly belly (Bazigou et al., 2007), Capricious (Shinza-Kameda et al., 2006), and Sequoia (Petrovic and Hummel, 2008; Kulkarni et al., 2016). Nevertheless, the donut-like morphology of R8 terminals has not been discussed previously. Later in this study, we will show that Ncad is essential for correct column targeting by R8.
Figure 3.
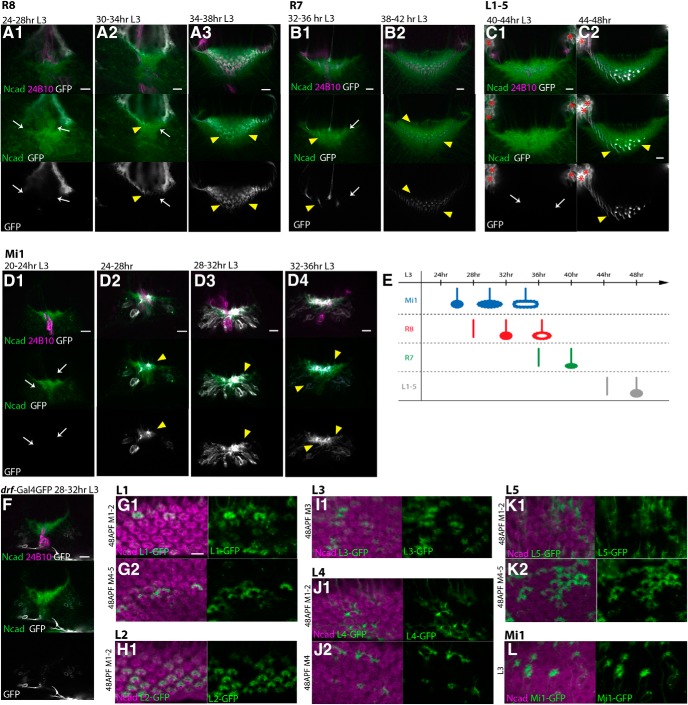
Sequential projections of the columnar neurons. A–E, Sequential projections of R8 (A), R7 (B), L1–L5 (C), and Mi1 (D) are visualized by UAS-myrGFP (white) in L3 larval stages. Magenta represents 24B10. Green represents Ncad. A, R8 terminals are visualized by sensF2-Gal4. R8 terminals are not identifiable before 28 h L3 stage (A1, arrows), start to innervate the medulla neuropil by 32 h, and form the donut-like pattern by 38 h L3 stage (A2,A3, arrowheads). B, R7 terminals are visualized by PM181-Gal4. R7 terminals are not identifiable before 34 h L3 stage (B1, arrows), start to innervate the medulla neuropil after 36 h, and form the dot-like pattern by 42 h L3 stage (B1,B2, arrowheads). C, L1–L5 terminals are visualized by R69B02-Gal4. L1–L5 terminals are not identifiable before 44 h L3 stage (C1, arrows), start to innervate the medulla neuropil, and form the dot-like pattern by 48 h L3 stage (C2, arrowheads). Cell bodies of lamina neurons (asterisks). D, Mi1 terminals are visualized by bshM-Gal4. Mi1 terminals are not identifiable before 24 h L3 stage (D1, arrows), start to innervate the medulla neuropil by 28 h, and form the grid-like pattern by 36 h L3 stage (D2–D4, arrowheads). E, Columnar neurons project to the medulla neuropil in the order of Mi1, R8, R7, and L1–L5. F, Terminals of medulla neurons visualized by drf-Gal4 are not identifiable in the medulla neuropil by 32 h L3 stage. G–K, Projections of individual lamina neurons are visualized by clones expressing UAS-myrGFP (green) in 48 h APF pupal stage in layers M1–M2 (G1,H1,J1,K1), M3 (I1), and M4–M5 (G2,J2,K2). Magenta represents Ncad. L1, L2, L3, L4, and L5 are visualized by clones expressing GFP under the control of c202-Gal4, R16H03-Gal4, R20A03-Gal4, R31C06-Gal4, and bshL-Gal4, respectively. L1 and L2 contribute to layers M1–M2, and L1 and L5 contribute to layers M4–M5. L, Individual Mi1 neurons visualized by clones expressing UAS-myrGFP (green) are projecting to the peripheral region of the columns in L3 stage. Scale bars: A1–G1, 5 μm.
Figure 2.
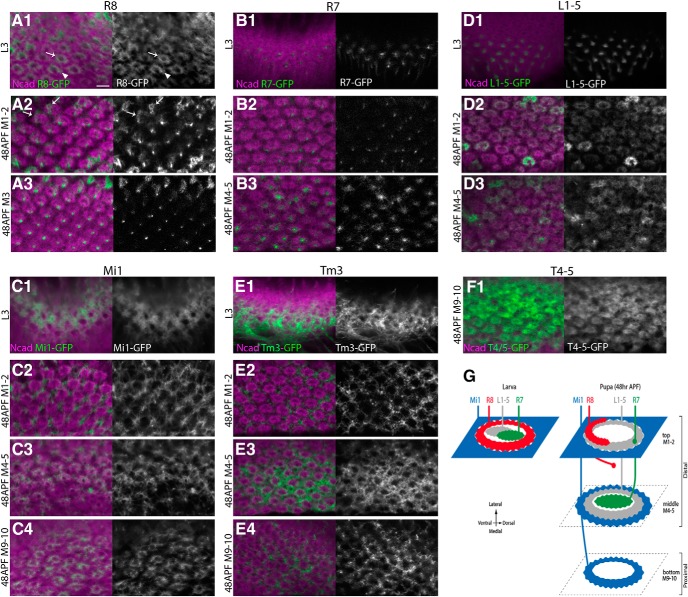
Projection patterns of essential neurons within the columns. Projection patterns of R8, R7, Mi1, L1–L5, Tm3, and T4-T5 are visualized by UAS-myrGFP (green or white) and compared with Ncad pattern (magenta). A1–A3, sensF2-Gal4 visualizes R8 axon terminals showing either donut-like (arrow) or horseshoe-like shape (arrowhead) that overlap with the donut-like Ncad pattern in L3 larval stage (A1). In 48 h APF pupal stage, obscured donut-like (arrows) and faint dot-like patterns are identifiable in layers M1–M2 (A2) and M3 (A3), respectively. B1–B3, PM181-Gal4 visualizes dot-like R7 axon terminals inside the donut-like Ncad pattern in L3 larval (B1) and 48 h APF pupal stages in layers M4–M5 (B3). Faint dot-like patterns of R7 are also identifiable in layers M1–M2 (B2). C1–C3, bshM-Gal4 visualizes grid-like Mi1 axon terminals outside the donut-like Ncad pattern in L3 larval (C1) and 48 h APF pupal stages in layers M1–M2 (C2). Mi1 penetrates inside the donut-like Ncad pattern in layers M4–M5 (C3). In layers M9-M10, Mi1 terminals overlap with the donut-like Ncad pattern (C4). D1–D3, R69B02-Gal4 visualizes dot-like L1–L5 axon terminals inside the donut-like Ncad pattern in L3 larval stage (D1). L1–L5 terminals then overlap with the donut-like Ncad pattern in layers M1–M2 (D2) and M4–M5 (D3) in 48 h APF pupal stage. E1–E3, R13E12-Gal4 visualizes grid-like Tm3 axon terminals outside the donut-like Ncad pattern in L3 larval (E1) and 48 h APF pupal stages in layers M1–M2 (E2), M4–M5 (E3), and M9-M10 (E4). F1, R42F06-Gal4 visualizes T4-T5 axon terminals that overlap with the donut-like Ncad pattern in layers M9-M10 in 48 h APF pupal stage. Scale bar: A1, 5 μm. G, Schematics of the developing columnar units in L3 larval and 48 h APF pupal stages.
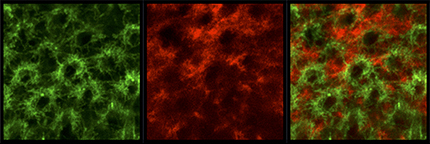
R8 and R7 project to the same layer in the larval medulla. Super-resolution image of sensF2-LexA LexAop-myrTomato (red) and PM181-Gal4 UAS-myrGFP (green) in L3 larval medulla. sensF2-Gal4 UAS-myrGFP (green) and sevEnS-LexA LexAop-myrTomato (red) show the same result.
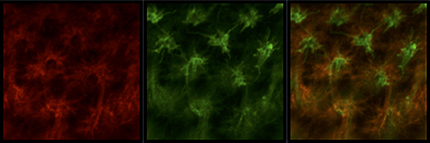
R8 and Mi1 project to the same layer in the larval medulla. Super-resolution image of sensF2-Gal4 UAS-myrGFP (green) and bshM-LexA LexAop-myrTomato (red) in L3 larval medulla. sensF2-LexA LexAop-myrTomato (red) and bshM-Gal4 UAS-myrGFP (green) show the same result.
R7 axons show strong dot-like patterns in the center of the columns in layers M4–M5 (Fig. 2B3), whereas the dot-like pattern of R7 is faint and separate from the column center in layers M1–M2 (Fig. 2B2). These results are consistent with previous observations showing that R7 axons eventually project to the M6 layer, whereas R8 axons initially terminate at M1 and extend to the M3 layer late in pupal development (Ting et al., 2005).
Since Gal4 drivers that are specific to each lamina neuron subtype did not show expression during larval stages, we instead used a pan-lamina Gal4 driver to visualize the axon terminals of all lamina neurons, L1–L5 (Fig. 2D). We found that L1–L5 axons project to the center of columns, most likely overlapping with R7 during the larval stage (Fig. 2B1,D1), but gradually expand early in pupal development, establishing a donut-like shape overlapping with the Ncad pattern in layers M1–M2 and M4–M5 by 48 h APF (Fig. 2D2,D3). The use of Gal4 drivers that are specific to each lamina neuron subtype suggests that the potential sources of the donut-like shape are L1 and L2 in layers M1–M2 and L1 and L5 in layers M4–M5 (Fig. 3G–K).
Mi1 is the primary medulla neuron that is initially produced from medulla NBs and is a unicolumnar neuron because each column contains only one Mi1 (Fischbach, 1989; Hasegawa et al., 2011). The axon terminals of Mi1 were visualized using bshM-Gal4, which is specifically expressed in Mi1 during the larval and early pupal stages. The Mi1 axon terminals were observed in the medulla neuropil at the early third larval instar, before the projections of the R8 and R7 axons (Fig. 3D). The broad pattern of Mi1 terminals is then redirected to the peripheral region outside the columns, showing a grid-like pattern surrounding the R8 terminals (Figs. 2C1, 3D; see Fig. 7I). The terminals of Mi1 are found in the same layer as those of R8 and R7 in the larval medulla (Movies 1, 2). A grid-like pattern of Mi1 terminals was also observed in layers M1–M2 at 48 h APF (Fig. 2C2). However, Mi1 axons partially overlap with the donut-like patterns of Ncad in layers M4–M5 (Fig. 2C3). In layers M9-M10, the Mi1 axon terminal forms a ring shape that clearly overlaps with the donut-like Ncad pattern (Fig. 2C4). Similar to the R8 terminal in larval medulla (Fig. 2A1), the Mi1 terminal in pupal layers M9-M10 also has a gap showing a horseshoe-like shape. This is consistent with the previous observations that the Mi1 axon arborizes in layers M1, M5, and M9-M10 in the pupal and adult brain (Fischbach, 1989; Hasegawa et al., 2011). Although bshM-Gal4 expression is also upregulated in L4 and L5 neurons during pupal development, the use of bshL-Gal4, which is specifically expressed in L4 and L5 throughout development, confirms that Mi1 axons are specifically visualized in the above experiments.
In contrast to Mi1, Tm3 projects to the lobula through the medulla (Fischbach, 1989; Erclik et al., 2017). Tm3 axon terminals show a grid-like pattern surrounding the columns during the larval and 48 h pupal stages, in the top, middle, and bottom layers (Fig. 2E). T4-T5 are columnar lobula plate neurons that show a direction-selective response to moving visual stimuli (Maisak et al., 2013). Although T4-T5 axon terminals were rarely observed in the larval medulla, they show a clear donut-like pattern in layers M9-M10 at the 48 h APF pupal stage (Fig. 2F1). Since the patterns of Ncad/phalloidin staining are unidentifiable in layers M3 (between top and middle) and M6–8 (between middle and bottom) (Fig. 1D4), and the neurons examined above essentially arborize in layers M1–M2, M4–M5, and M9-M10 (Fig. 2G), we mainly focused on these three layers in the following text, except when noted otherwise.
The above results suggest that there are three typical projection patterns of axon terminals: a dot-like pattern at the center of the column, a donut-like pattern that outlines the column, and a grid-like pattern outside the columns. During the larval stage, R7 and L1–L5 show dot-like patterns, R8 shows a donut-like pattern, and Mi1 and Tm3 show grid-like patterns. During pupal development, these patterns change dynamically. At 48 h APF, R7 shows a dot-like pattern, L1–L5 show donut-like patterns, and Mi1 shows grid-like patterns in the distal layers, whereas Mi1 and T4-T5 show donut-like patterns in the proximal layer (Fig. 2G).
Sequential projections of the columnar neurons
Among as many as 100 types of neurons in the medulla column, what are the primary core neurons essential for the formation of columnar units? To address this question, we compared the timing of neural projections during larval development. We found that Mi1 neurons start projecting to the medulla at the mid-third instar larval stage (24–28 h L3), whereas R8 and R7 axons start projecting with short delays of ∼4 and 8 h, respectively, establishing donut-like and dot-like patterns (Figs. 2, 3A–E). Then, the broad projection pattern of Mi1 terminals changes to a grid-like pattern that fills the peripheral region of the columns. L1–L5, T4-T5, and other medulla neurons project to the medulla much later in the larval stage (Fig. 3C,E,F; and data not shown). For example, lamina neurons project to the medulla at the end of the larval stage (44–48 h L3; Fig. 3C). These results suggest that R7, R8, and Mi1 could be the primary neurons for the column formation. Glial processes were not found in the developing columns in larval and 48 h pupal stages (data not shown).
R8, R7, and Mi1 are the core columnar neurons
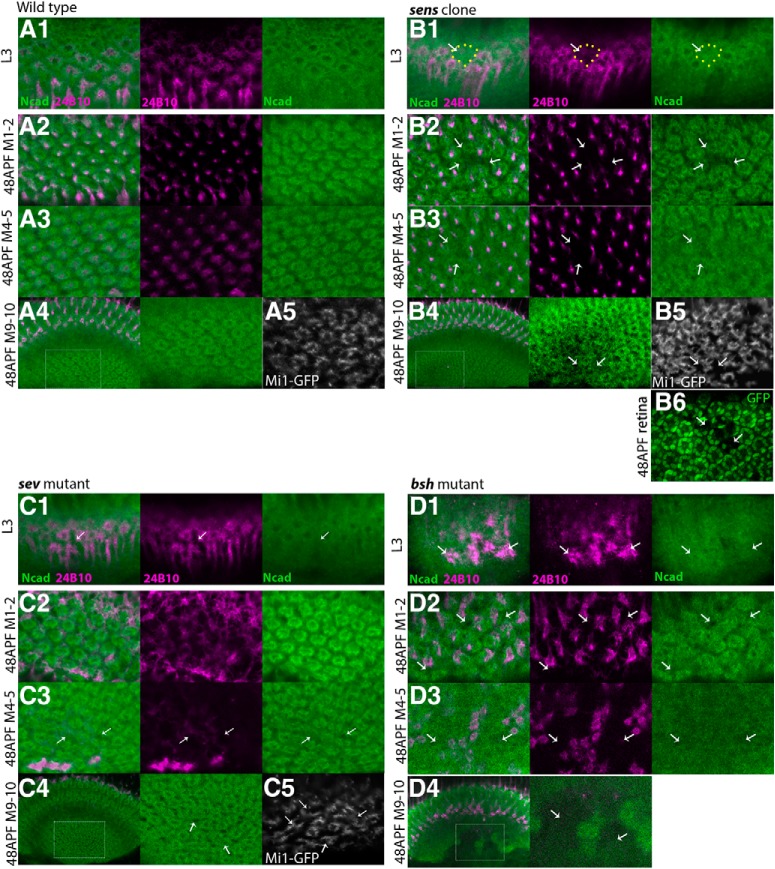
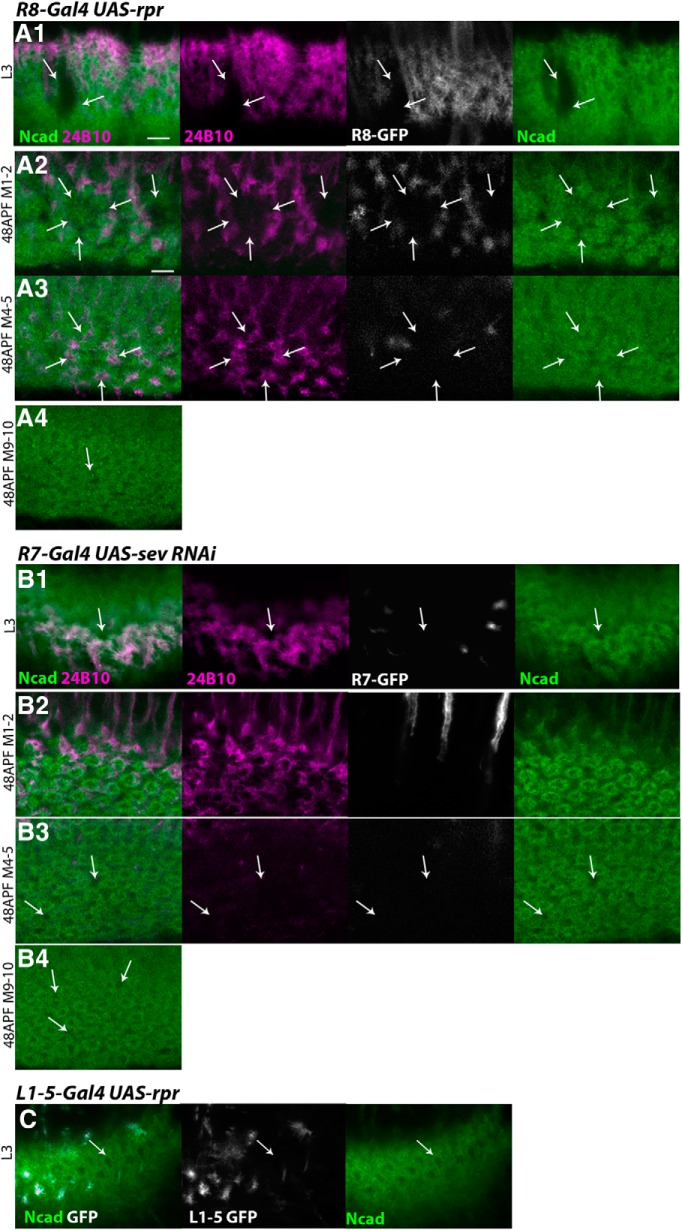
Next, we examined the roles of these core neurons by using mutations in which either R8, R7, or Mi1 was missing. Initially, R8 differentiation was blocked by generating senseless (sens) mutant clones (Frankfort et al., 2001). When sens homozygous clones were generated in the eye, loss of R8 was also revealed by the absence of R8 projection to the medulla visualized by the lack of 24B10. In our experimental condition, the clones are as large as two or three ommatidial units (Fig. 4B6). Indeed, we observed small areas as large as two or three columnar units lacking the 24B10 signal and the donut-like Ncad pattern in the larval medulla (Fig. 4B1; n = 8 of 8) and in layers M1–M2 and M4–M5 in 48 h APF pupal medulla (Fig. 4B2,B3; n = 9 of 9), suggesting that R8 is essential for medulla column formation. In layers M9-M10, we also observed small areas lacking the donut-like Ncad pattern and the ring-like Mi1 axon terminals (Fig. 4A4,A5,B4,B5; n = 4 of 4), suggesting that R8 influences the bottom layer to which R8 does not project. It has been reported that the sens mutation causes the loss of R7 (Frankfort et al., 2001). Therefore, the above defects may be indirectly caused by the loss of R7. To test this possibility, R8 was removed by expressing reaper (rpr), a potent apoptosis inducer. Since the columns were missing when R8 terminals were eliminated while R7 terminals were still present in layers M4–M5, our results suggest that R8 is essential for column formation (Figs. 5A, 1D as a control).
Figure 4.
R8, R7, and Mi1 are the core columnar neurons. Columnar Ncad pattern (green), and projection patterns of R axons (24B10; magenta) and columnar neurons (UAS-myrGFP; white) in WT (A) and in the absence of R8 (B), R7 (C), and Mi1 (D) in L3 larval (A1–D1) and 48 h APF pupal stages (A2–D2, layers M1–M2; A3–D3, layers M4–M5; A4–D4,A5–C5, layers M9-M10). A, In WT, columnar patterns of Ncad are regularly arranged. B, In the presence of sens mutant clones, R axon terminals of 24B10 are lost, and columnar patterns of Ncad are disorganized (arrows) in larval medulla (B1) and in pupal layers M1–M2 (B2), M4–M5 (B3), and M9-M10 (B4). B5, Projection pattern of Mi1 is disorganized in layers M9-M10 due to the loss of R8 in layers M1-M3. B6, sens mutant clone in retina is visualized by the absence of GFP (arrows). C, In sev mutant brains, columnar patterns of Ncad are disorganized (arrows) in larval medulla (C1) and in pupal layers M4–M5 (C3) and M9-M10 (C4). D, In bsh1/bsh2 brains, columnar patterns of Ncad are disorganized or fused with each other (arrows) in larval medulla (D1) and in pupal layers M1–M2 (D2), M4–M5 (D3), and M9-M10 (D4). D4, Neuropil structures visualized by Ncad are missing (arrows). A5–C5, Regularly arranged donut-like Mi1 terminals visualized by bshM-Gal4 in layers M9-M10 are partially lost in the presence of sens mutant clones (B5, arrows) and in sev mutant brains (C5, arrows). Scale bar: A, 5 μm.
Figure 5.
R8 and R7 are essential for medula column formation. A, B, Columnar Ncad pattern (green), and projection patterns of R axons (24B10; magenta) and columnar neurons (UAS-myrGFP; white). A1–A4, R8 is partially eliminated by expressing rpr under the control of sensF2-Gal4 in larval (A1) and 48 h APF pupal stage (A2, layers M1–M2; A3, layers M4–M5; A4, layers M9-M10). Columnar structures are lost when R8 terminals are missing (A1–A4, arrows), whereas R7 terminals are present (A3, arrows). B1–B4, R7 is partially eliminated by expressing sevRNAi under the control of PM181-Gal4 in larval (B1) and 48 h APF pupal stage (B2, layers M1–M2; B3, layers M4–M5; B4, layers M9-M10). R axons and columnar structures are disorganized (B1,B3,B4, arrows). C, Columnar Ncad pattern (green) and projection pattern of L1–L5 neurons (UAS-myrGFP; white). L1–L5 are partially eliminated by expressing rpr under the control of R69B02-Gal4 in larval stage. Columns are not disorganized in the region where L1–L5 are eliminated (arrows). Scale bar: A1, 5 μm.
In the sevenless (sev) whole-body mutant in which R7 is not formed (Simon et al., 1989), we found disorganization of columns as visualized by the donut-like Ncad pattern in the larval medulla and pupal layers M4–M5 and M9-M10 (Fig. 4A,C; n = 14 of 20). The top layer (pupal layers M1–M2) was not significantly affected (Fig. 4C2). In layers M4–M5 and M9-M10, the donut-like column shape is not circular, and there are irregular gaps between the columns (Fig. 4C3,C4, arrows). In the control, the bifurcated axon terminals of Mi1 are close to each other, forming a horseshoe-like shape (Fig. 4A5). In sev mutant, the bifurcated axons terminals are separated and split apart (Fig. 4C5, arrows). Importantly, the abnormal columns and Mi1 axon terminals were observed in the layers to which the R7 axon does not project (Fig. 4A4,A5,C4,C5; n = 8 of 12), suggesting that R7 has nonautonomous effects on column formation in the bottom layer. Since similar columnar defects were observed when sev was specifically knocked down in R7 (Figs. 5B, 1D as a control), the above results confirm the important roles of R7 in the column arrangement in layers M4–M5 and M9-M10.
Finally, we examined the effect of the loss of Mi1 on column formation. Since brain-specific homeobox (bsh) is essential for Mi1 specification in the medulla (Hasegawa et al., 2011, 2013), we examined bsh mutant brains. Strong disorganization of the shape of columns as visualized by the donut-like Ncad pattern was detected in the bsh whole-body mutant in the larval and three layers of the pupal medulla (Fig. 4A,D; n = 20 of 20). A massive disorganization of the neuropil tissue identified by the loss of Ncad signals was observed in layers M9-M10 (Fig. 4A4,D4). These results suggest that Mi1 has important functions in organizing column formation. Since bsh regulates Ncad expression, Ncad expression in Mi1 and/or Mi1 projection itself may be necessary to ensure the integrity of the medulla neuropil (Hasegawa et al., 2013). Bsh is also expressed in L4 and L5 lamina neurons and is required for the specification of L4 and L5 (Hasegawa et al., 2013). However, bsh knockdown specifically in L4–5 using bshL-Gal4 caused no significant defect in column formation (data not shown), suggesting that the above phenotypes were caused by the loss of bsh in Mi1.
Although L1–L5 neurons project to the medulla columns much later than Mi1, R8, and R7, we have examined whether the ablation of L1–L5 can influence the column formation in larval medulla. As expected, the elimination of L1–L5 by the expression of UAS-reaper (rpr) did not have a significant effect on column formation (Figs. 5C, 1B, 2D1). However, it is likely that L1–L5 play important roles in column formation during pupal development.
The above results demonstrate the essential roles of R7, R8, and Mi1 as core columnar neurons in the medulla, although we cannot exclude the possibility that unidentified neurons are also involved in the early steps of column formation. The clearly segregated distributions of R7, R8, and Mi1 in the larval columns remind us of the DAH (Steinberg, 1962). According to this hypothesis, the more adhesive cells contribute to the central part of the cell aggregates surrounded by the less adhesive cell (Steinberg and Takeichi, 1994). Similarly, the dot-like, donut-like, and grid-like patterns of R7, R8, and Mi1, respectively, may reflect the order of their adhesiveness.
Differential expression of Ncad in the core columnar neurons
The mixture of cultured cells expressing different levels of cadherin has been shown to rearrange to form organized cell aggregates, in which more adhesive cells contribute to the internal part enwrapped by the less adhesive external cells (Steinberg and Takeichi, 1994). Therefore, Ncad could be a key molecule that regulates column formation through its differential adhesion. Since R7, R8, and Mi1 terminals form the concentric structure along the same layer in the larval medulla (Movies 1, 2), we focus on the larval medulla as a model system to examine the roles of Ncad in differential adhesion and column formation. As a first step to test this idea, we compared Ncad expression among the core columnar neurons R7, R8, and Mi1, and in L1–L5. We visualized GFP-fused Ncad in these columnar neurons by using an Ncad knock-in allele in which endogenous Ncad is fused with GFP under the control of FLPase (Ncad-FsF-GFP) together with FLP strains specific for columnar neurons. Ncad-GFP expression was clearly detected at the terminals of R7, R8, and L1–L5 (Fig. 6A,B,D). Importantly, the GFP signal was detected at the center of the columns showing a dot-like pattern in larval R7 and L1–L5 (Fig. 6A,D), suggesting that the Ncad protein is present, even within the central hole area of the columns, albeit not visible via Ncad immunostaining. Intriguingly, Ncad-GFP signals were occasionally enriched at the cell membrane of R7 (Fig. 6A), which might be explained by the homophilic adhesion of Ncad-GFP with endogenous Ncad protein expressed in neighboring cells. Thus, the membrane accumulation of Ncad could be the cause of the lack of Ncad immunostaining signal at the central hole area of the columns. We also visualized Ncad-GFP in larval Mi1 neurons using the homothorax (hth)-FLP strain and anti-Bsh antibody, since Mi1 neurons are double-positive for Hth and Bsh (Fig. 6C) (Hasegawa et al., 2011).
According to the DAH, the relative levels of the cell adhesion molecules in a cell type determine the relative location of the cell within a column (Steinberg, 1962). We therefore quantified the relative expression levels of Ncad-GFP at the terminals of R7, R8, and Mi1 by using the photon counting method (Fig. 6E). In this analysis, we modified the laser power and scan speed so that the majority of pixels receive either one or two photons (background photons were subtracted from the number of photons emitted from the axon terminals). Up to 5% pixels received three or more photons. Therefore, we assume that pixels that receive only one photon express lower levels of Ncad-GFP, whereas the pixels that receive two or more photons express higher Ncad-GFP levels. In contrast to the donut-like distribution of Ncad within a column, in which the Ncad signal is lower inside the central hole, the results of photon counting show that the R7 terminals located at the central hole area contain stronger Ncad-GFP signals compared with those of R8 and Mi1. For example, the percentage of pixels emitting one photon was lower for R7 compared with Mi1. In contrast, two-photon pixels were more frequent for R7 compared with R8 and Mi1. Similarly, the R8 terminals expressed Ncad-GFP more strongly than the Mi1 terminals because there were fewer one-photon pixels and more two-photon pixels for R8 compared with Mi1. Although statistically insignificant, pixels emitting three or more photons have also shown a similar tendency in that the axon terminals contain more Ncad-GFP in the order of R7 > R8 > Mi1. Although the relative densities of Ncad-GFP at the L1–L5 terminals were as high as those at the R7 terminals, we cannot draw a clear conclusion about the Ncad-GFP density for each subtype of L-neurons because we do not have reliable subtype-specific FLP strains. Thus, the differential adhesion between R7, R8, and Mi1 can be explained by differential Ncad levels among these columnar neurons.
Roles of N-cadherin in the columnar organizations
Next, we asked whether Ncad-dependent differential adhesion regulates column formation. Here, we define the mutant phenotype of the axon terminal as follows: expanded, when the axon terminal shows ectopic projection and resides in the original location within a column; redirected, when the axon terminal shows ectopic projections while it no longer resides in the original location within a column.
According to the DAH, reduction of adhesiveness would redirect the centrally located neurons toward the peripheral part of the columns. Indeed, the loss of Ncad in R7 expanded its axon terminals toward the peripheral part of the larval column compared with the control, in which R7 occupies the central position, and the shape of the columns as visualized by Ncad and phalloidin was significantly disturbed (Fig. 7A,B; and data not shown). To quantify the expansion and redirection of the R7 terminals, the shape of the columns needs to be preserved. Since the column morphology was largely preserved when Ncad was knocked down in a subset of R7 by the FLP/FRT-mediated clonal analysis (Fig. 7C), we quantified the results in this condition. By measuring the amount of GFP signals detected within a column or within the hole area of the column, we quantified the expansion and redirection ratios of R7, both of which were significantly larger when Ncad was knocked down in R7 compared with the control (Fig. 7M,O; see Materials and Methods). Thus, the R7 terminals are expanded and redirected to the peripheral part of the columns when Ncad is reduced in R7. Similar expansion and redirection phenotypes were observed in Ncad loss-of-function MARCM clones (Fig. 7D; n = 31 of 40).
The results of Ncad knockdown in R8 also support the roles of Ncad-dependent differential adhesion. R8 terminals were deformed and expanded to fuse with neighboring columns upon Ncad knockdown (Fig. 7E,F). By inducing clones of R8 in which Ncad was knocked down, we observed an R8 expansion phenotype (Fig. 7G). By measuring the amount of GFP signals detected within a column and within a single R8 growth cone, we quantified the expansion and redirection ratios of R8 (Fig. 7N,O; see Materials and Methods). We found that the R8 terminals were expanded and redirected to the peripheral part of the columns when Ncad was reduced in R8. We verified the results by generating loss-of-function mutant clones of Ncad using the MARCM technique. As expected, the R8 terminals were expanded (Fig. 7H; n = 20 of 20). However, we could not quantify the expansion and redirection ratios in Ncad mutant MARCM clones because the column morphology was completely disrupted.
According to the DAH, reduction of adhesiveness in neurons that are located at the peripheral part of the column does not significantly affect column formation. To test this idea, we knocked down Ncad in Mi1 by using bshMGal4, which is specifically expressed in Mi1 during the larval stage. Loss of Ncad in Mi1 showed no visible effect on column organization in the larval brain (Fig. 7I,J). Similar results were observed in Ncad loss-of-function MARCM clones for Mi1 (data not shown).
In the above experiments, we knocked down Ncad using two different UAS-Ncad RNAi strains targeting distinct Ncad gene sequences. The effect of Ncad RNAi was confirmed by Ncad staining (Fig. 7K,L). Indeed, during pupal development, R7 terminals frequently failed to stabilize at layers M4–M5 upon Ncad knockdown, which is consistent with the previous results validating the effect of RNAi (Lee et al., 2001).
Since essentially the same results were obtained using phalloidin as a columnar marker instead of Ncad and quantified in Figure 7M, N, the above results are not due to the reduction in Ncad staining induced by RNAi. These results suggest that the core columnar neurons are located within a column according to differential levels of Ncad-dependent adhesion.
Previous extensive studies have shown the importance of Ncad for the proper targeting of R1–R6 in the lamina (Lee et al., 2001) as well as R7 stabilization at the M6 layer (Lee et al., 2001; Clandinin and Zipursky, 2002; Ting et al., 2005; Özel et al., 2015) and L-neuron targeting in the medulla (Nern et al., 2008). However, these studies only focused on the layer targeting events in pupal stages. In this study, we only focused on the earliest step of column formation in the larval stage. The mutual interactions between layer targeting and column formation during pupal development will be studied in the future.
Ncad knockdown reveals differential adhesion between columnar neurons
According to the DAH, reduction of adhesiveness in centrally located neurons would cause a relative increase in the adhesiveness of other neurons and subsequent redirection of these neurons toward the central part of the columns. Since Ncad is a homophilic cell adhesion molecule, Ncad knockdown in one cell type is expected to reduce cell adhesion with others. Therefore, we next examined the influence of Ncad knockdown in R7 and R8 on the projection patterns of the other columnar neurons.
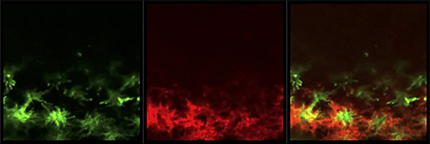
First, we examined the effects of Ncad knockdown in R7 on Mi1 (Fig. 8A,B). We initially focused on the grid-like projection patterns of Mi1 terminals that are separated from the centrally projecting R7 in larval brain. There is a gap between R7 and Mi1 (Fig. 8A, arrows). Upon Ncad knockdown, R7 terminals became diffuse and expanded. At the same time, disorganized and expanded Mi1 terminals overlapped R7 (Fig. 8B, arrows and yellow signals). Since the columnar morphology is significantly disrupted in this condition, we quantified the Mi1 expansion ratio by measuring the Mi1 signal density within the R7 terminal area. The Mi1 expansion was significantly greater when Ncad was knocked down in R7 (Fig. 8I). Similarly, R8 terminals were significantly expanded and overlapped with R7 terminals when Ncad was knocked down in R7 (Fig. 8C,D,J; Movie 3). Although Ncad is thought to regulate layer targeting of R7 terminals during pupal development, we found R7 terminals innervating on the larval medulla neuropil in Ncad RNAi conditions (Figs. 7B,C, 8A–D). These results are consistent with the recent finding that loss of Ncad causes no primary targeting defects but destabilizes R7 growth cones (Clandinin and Zipursky, 2002; Özel et al., 2015).
R7 and R8 terminals significantly overlap upon Ncad knockdown in R7. Super-resolution image of Ncad RNAi in R7. PM181-Gal4 UAS-myrGFP (green) and sensF2-LexA LexAop-myrTomato (red) in L3 larval medulla.
Next, we examined the projections of Mi1 when we induced Ncad RNAi in R8 (Fig. 8E,F). The terminals of R8 overlapping the donut-like pattern were expanded to the column periphery and fused with neighboring R8 terminals in the larval brain (Fig. 8F). The filopodia of R8 were less organized compared with the control. In this condition, the grid-like pattern of Mi1 was disorganized and their terminals were expanded (Fig. 8F). We quantified the defect by measuring the colocalization of R8 and Mi1 signals (Pearson's correlation coefficient). The overlap between R8 and Mi1 was significantly greater when Ncad was knocked down in R8 (Fig. 8K). These results suggest that the relative adhesiveness between two different cell types determines their relative location within a column.
When Ncad was knocked down in R8, R7 terminals were found enwrapped with R8 terminals, and the size of the R7 terminals was 2.8 times larger compared with the control condition (Movie 4; n = 32, p < 0.001 by t test). Since R7 terminals should not be affected according to the DAH, R7 expansion phenotype may be due to Ncad-independent mechanisms. As mentioned later, R7 and R8 are already patterned before their arrival to the medulla by unknown mechanisms (see Fig. 10C). Ncad-independent adhesion between R7 and R8 may be the cause of the expansion of R7 terminals as a result of R8 expansion.
R7 terminals are expanded upon Ncad knockdown in R8. Super-resolution image of Ncad RNAi in R8. sensF2-Gal4 UAS-myrGFP (green) and sevEnS-LexA LexAop-myrTomato (red) in L3 larval medulla.

Overexpression of Ncad in the columnar neurons with different levels of adhesiveness
Since the location of the neurons within the column is determined by their relative adhesiveness according to the DAH, an increase in Ncad levels in neurons that project to the peripheral part of the columns should redirect them toward the central part of the columns. To address this possibility, a UAS-Ncad construct that includes the entire open reading frame of Ncad (Iwai et al., 1997) was used to overexpress Ncad in the core columnar neurons Mi1, R7, and R8.
When we overexpressed Ncad in Mi1 neurons under the control of bshM-Gal4, the Mi1 terminals projecting to the grid-like domain outside the columns in the control were redirected to the column center overlapping with the donut-like Ncad and phalloidin pattern (Fig. 9A,B). The expansion and redirection ratios of Mi1 were quantified by measuring the amount of GFP signals detected within the column and the central hole area (Fig. 9I; see Fig. 7O; see Materials and Methods). Mi1 terminals were significantly redirected to the column center overlapping with R8 terminals. Similar results were observed when Ncad was overexpressed in a subset of Mi1 using the MARCM technique (Fig. 9C,D; n = 11 of 20).
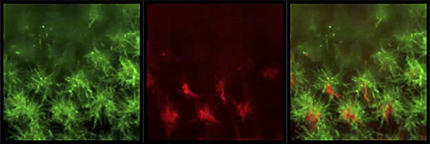
The overexpression of Ncad in R8 neurons led to the expansion and redirection of R8 terminals to the center of the column and the shrinkage of the central hole area (Fig. 9E,F). By measuring the amount of GFP signals detected within the hole area and a single R8 growth cone, we quantified the expansion and redirection ratios of R8 (Fig. 9J; see Fig. 7O; Materials and Methods). Together with the R8 expansion to the column center, R7 terminals were expanded toward the column periphery overlapping with R8 terminals (Fig. 8G,H,L; Movie 5). As we expected, no significant changes were observed in R7 projection after Ncad overexpression in R7 (Fig. 9G,H). These results are consistent with the DAH and demonstrate that the core columnar neurons are located within a column according to the Ncad-dependent differential adhesion.
R8 and R7 terminals significantly overlap upon Ncad overexpression in R8. Super-resolution image of Ncad overexpression in R8. sensF2-Gal4 UAS-myrGFP (green) and sev-LexA LexAop-myrTomato (red) in L3 larval medulla.
The expression level of UAS-Ncad was confirmed by Ncad staining in the medulla neurons (Fig. 9K,L). The flies were cultured at 30°C to enhance the overexpression phenotype. Although we have only tested a single isoform of Ncad (7a, 13a, 18a) for the above overexpression experiments (Iwai et al., 1997), a single Ncad isoform has been shown to rescue the Ncad mutant phenotype, and the biological significance of the diversity of Ncad isoforms has not been demonstrated (Ting et al., 2005).
Differential adhesion of R8, R7, and Mi1 is sufficient to reproduce their columnar distributions
The above results suggest that differential adhesion among R8, R7, and Mi1 plays important roles in establishing their columnar distributions. However, it is unclear whether differential adhesion is sufficient to explain the clear segregation of the terminals of columnar neurons into three distinct domains (Fig. 10A). For example, short-range repulsion between the columnar neurons may also be necessary to establish the distinct domains adjacent to each other. We therefore assessed whether differential adhesion among R8, R7, and Mi1 is sufficient to reproduce the WT and mutant patterns shown above.
The discrete cellular model is commonly used for mathematical modeling of multicellular processes. However, there are tight limitations in the size and shape of the cells in such a cell-based model. Since the terminals of columnar neurons show very irregular shapes that change dynamically, we formulated a continuous model of column formation by modifying the continuous model of cell adhesion (Murakawa and Togashi, 2015; Carrillo et al., 2019). Our model considered the densities of neurites, differential adhesion, and volume exclusion. Volume exclusion is a purely physical phenomenon distinct from active repulsion because it only assumes that a higher cell density causes a pressure to avoid intrusion of surrounding cells. Thus, volume exclusion has an effect similar to passive repulsion. For simplicity, we only focus on R7, R8, and Mi1 during the larval stage. The valuables u, v, and w are the densities of the axon terminals of R7, R8, and Mi1, respectively (Fig. 10A). Initially, the adhesiveness of R7, R8, and Mi1 (a11, a22, and a33) is set to 2.4, 1.8, and 0.8, respectively (a11 > a22 > a33). The adhesiveness between two different cell types was the mean of the two different adhesiveness values (aij = aji = (aii + ajj)/2). According to their sequential projection orders, our simulations are initiated from the regularly spaced projections of R8 into the 2D field uniformly covered with Mi1 terminals (t = 0, v = 0.6, w = 0.2; Figs. 10B1, 3A–E). Small noise is added to the initial distribution of R8. After they reach equilibrium, R7 projects in a manner identical to the initial R8 projection pattern (t = 100, u = 0.6; Fig. 10B2; yellow represents the overlap between R7 and R8). Although there is no spatial bias between the initial distributions of R8 and R7, our model reproduces the distinct and adjacent distribution patterns of R7, R8, and Mi1 showing the dot-like, donut-like, and grid-like patterns, respectively, as found in larval brain (t = 1500; Fig. 2A1–C1).
According to the patterns of R7-GFP and 24B10 just below the lamina before the arrival at the medulla in larval brain, the initial distribution patterns of R8 and R7 are actually biased (Fig. 10C). We assume that the patterning of R7 and R8 occurs during the extension of their axons from the retina to the medulla. R7 axons follow R8 and may be preliminarily paired with R8 by unknown mechanisms. Thus, the prepatterning of R7 and R8 should be important in vivo; however, the results of our mathematical model in Figure 10B show that R7 and R8 can segregate, even in the absence of prepatterning.
24B10 preferentially visualizes R8 in the larval stage (Figs. 1B1, 2A1). Since strong 24B10 signals partially engulf R7, we repeated the numerical simulations based on the initial distributions shown in Figure 10D1. In the control, R7, R8, and Mi1 become clearly segregated, and R7 is completely engulfed by R8 (t = 1000; Fig. 10D2). Qualitatively similar results are obtained by changing the adhesiveness of R7, R8, and Mi1, if the order of adhesiveness is conserved (a11 > a22 > a33; Fig. 10I–K), suggesting that our model reproduces the normal column formation over a wide range of parameter settings. Because our model does not include any active repulsion, the results suggest that differential adhesion is sufficient to explain the clear segregation of these neurons. Of course, the results do not exclude the involvement of repulsive factors in column formation in vivo. In the presence of active repulsion, columnar neurons should be segregated more robustly.
We next asked whether our model also reproduces the results of our genetic experiments. When Ncad was knocked down in R7, R7 terminals were redirected to the peripheral region of the columns (Fig. 7A–C). Similarly, R7 terminals are expanded to the peripheral part partially engulfing R8 when the adhesiveness of R7 is reduced (a11 = 1.7; Fig. 10E). While R7 and Mi1 were adjacent to each other, which is consistent with Figure 8B, R8 terminals were located outside R7 in vivo (Fig. 8D), most likely due to inefficient Ncad knockdown in this condition.
Ncad knockdown in R8 caused expansion of R8 terminals and massive fusion of the columns (Fig. 7E–G). Similarly, R8 terminals are redirected to the peripheral region and fused with the neighboring columns (a22 = 0.6; Fig. 10F). Mi1 are redirected toward the column center, which is consistent with Figure 8F. In contrast to the expansion of R7 terminals together with R8 in vivo (Movie 4), R7 terminals show a dot-like pattern separated from R8 (Fig. 10F). R7 binding with R8 may be regulated by unidentified mechanisms when Ncad is knocked down in R8 in vivo.
When Ncad was overexpressed in Mi1, its terminals intruded inside the columns (Fig. 9A–D). Similar phenomena were observed when the adhesiveness of Mi1 was increased (a33 = 1.8; Fig. 10G; magenta represents the overlap between R8 and Mi1). Similarly to Ncad overexpression in R8 (Fig. 8H), an increased adhesiveness of R8 caused penetration of R8 terminals to the column center and expansion of R7 terminals toward the column periphery (a22 = 2.4, Fig. 10H; yellow represents the overlap between R7 and R8). Since our model only assumes basic physical properties and differential adhesiveness of the cells, differential adhesion could be the major driving force to establish the columnar structure.
Discussion
R8, R7, and Mi1 are essential core neurons for the initial step of column formation
Our results suggest that the photoreceptor neurons R8 and R7 and the medulla neuron Mi1 act as core neurons for the initial step of medulla column formation. At the early third instar larval stage, only Mi1 axons project to the medulla neuropil (Fig. 3D,E). Indeed, Mi1 is a primary medulla neuron initially produced from medulla NBs (Hasegawa et al., 2011). An NB produces Drifter (Drf)-positive neurons after producing Bsh-positive Mi1 neurons. However, axon terminals of Drf-positive neurons were not detectable at the early third instar larval stage (Fig. 3F). At the mid-third instar stage, R8 axons and then R7 axons project to the medulla (Fig. 3A,B,E). The donut-like columnar patterns are then formed by the end of the larval stage, as visualized by Ncad and phalloidin (Fig. 1B1).
Since R8 is the primary photoreceptor neuron that triggers the differentiation of other photoreceptor cells (Jarman et al., 1994), R8 may also play a central role in column formation in the medulla. Indeed, column formation was completely abolished in the absence of R8 axons during the larval and pupal stages (Fig. 4A,B). Thus, R8 is most likely a trigger of column formation that provides a spatiotemporal cue to synchronize the formation of ommatidia in the retina and columns in the medulla. A similar relationship is known to exist between R axons and lamina neurons, with R axons triggering the differentiation of lamina neurons (Huang and Kunes, 1996; Sato et al., 2013).
The column structure was also severely affected in the absence of R7 and Mi1 (Fig. 4C,D). We found that the loss of R7 causes severe defects in column shape and arrangement in layers M4–M5 and M9-M10 (Fig. 4C). The loss of Mi1 caused column distortion in layers M1–M2 and M4–M5 and massive disorganization of the neuropil structure in layers M9-M10 (Fig. 4D). These findings suggest that R8, R7, and Mi1 are the essential core neurons for column formation.
However, projection patterns of these core neurons become diverged in the adult brain. R8 and R7 axons extend and terminate in layers M3 and M6, respectively (Ting et al., 2005). Mi1 axons form the bifurcated hook-like shape, which is distinct from the donut-like shape found in layers M9-M10 at 48 h APF (Fig. 2C4) (Ke et al., 2016). Therefore, the donut-like Ncad structure found in the larval and pupal stages may act as a scaffold to establish synaptic connections in the mature adult columnar structure. In this study, however, we only focused on the early steps of column formation that were largely unknown.
Differential adhesion regulates column formation
Steinberg (Steinberg, 1962; Steinberg and Takeichi, 1994; Foty and Steinberg, 2005) proposed the DAH to explain the self-organizing property of intermixed cells in an aggregate sorting out to form an organized layered structure. The idea of DAH is extensively applied in the field of synthetic biology (Toda et al., 2018). Here, we propose that a similar strategy is used to form the columnar structure in the medulla. A similar idea has been tested in Drosophila lamina cartridges organized by the neurites of R1–R6 and L1–L5 (Schwabe et al., 2014). They demonstrated higher Ncad levels in the lamina dendrites compared with the R axons showing the concentric Ncad distribution in the lamina cartridge. A series of genetic manipulations that invert these relative differences in Ncad levels displace lamina dendrites to the periphery and R axons to the center. However, due to technical limitations, it is difficult to specifically manipulate gene activity and quantify the amount of Ncad in each of the lamina neurons, especially in the early stages. In this study, we identified the core neurons of the medulla column, R8, R7, and Mi1, and developed genetic tools to specifically quantify Ncad levels and manipulate gene activity in each of these neurons. By using these tools, we clearly demonstrated the in vivo evidence of DAH during columnar organization in the larval medulla.
At the beginning of column formation during larval development, R8 overlaps with the donut-like pattern of Ncad and phalloidin, whereas R7 shows a dot-like pattern inside the column, and Mi1 shows a grid-like pattern outside the column (Fig. 2A1–C1). We propose that the relative locations of R7, R8, and Mi1 in a layer can be explained by the DAH. Based on the dot-like, donut-like, and grid-like patterns of R7, R8, and Mi1 during the larval stage, their relative adhesiveness may follow the same order, namely, R7+++, R8++, and Mi1+, respectively, because according to the DAH, the most adhesive cells should be located in the most central position, whereas the least adhesive cells surround the other cells. The concentric domains formed by distinct and adjacent distributions of R7, R8, and Mi1 are clearly explained by our mathematical model in which only volume exclusion and differential adhesion are considered (Fig. 10), suggesting that the differential adhesion among these core neurons could play central roles in column formation. Indeed, Ncad knockdown in R7 and R8 expanded the projections of these neurons toward the column periphery (Fig. 7A–H,M,N). In these conditions, the R8 and Mi1 terminals expanded and overlapped with R7 (Fig. 8A–D), and the Mi1 terminals expanded and overlapped with R8 (Fig. 8E,F). On the other hand, expansion of Mi1 and R8 terminals toward the column center was detected upon overexpression of Ncad in these neurons (Fig. 9A–F,I,J). R7 terminals were expanded toward the column periphery when Ncad was overexpressed in R8 (Fig. 8G,H). Similar results were obtained by numerical simulations based on our mathematical model (Fig. 10E–H). Although L1–L5 projects to the larval column center and is assumed to be the most adhesive (Fig. 2D1), Ncad knockdown in L1–L5 during the larval stage did not show significant defects, most likely because L1–L5 project to the medulla much later than the other neurons (Fig. 3; and data not shown).
The differential adhesion of these cell types may be regulated by the differential expression levels of cell adhesion molecules, such as Ncad. According to the DAH, R7 should be the most adhesive in the larval brain due to the strongest expression or activity of Ncad. Despite the lack of Ncad signals in the center of columns as visualized by Ncad staining (Fig. 2B1), the results of R7-specific Ncad-GFP visualization (Fig. 6A) and Ncad knockdown in R7 (Fig. 7A–C) indicate that R7 indeed produces Ncad. The enrichment of Ncad-GFP at the cell membrane of R7 (Fig. 6A, arrows) or the lower density of R7 neurites compared with the surrounding R8 neurites may explain the loss of Ncad staining within the hole. Indeed, the Ncad level was the highest in R7 among the three core columnar neurons in the larval stage (Fig. 6E). Furthermore, the R8 terminals showed higher Ncad levels than the Mi1 terminals, supporting the Ncad-dependent differential adhesion between R7, R8, and Mi1.
The behavior of R7 upon Ncad knockdown in R8 cannot be explained solely by Ncad function (Movie 4). According to the DAH, R7 should remain in the column center (Fig. 10F). However, R7 terminals expanded together with the R8 terminals. Ncad-independent adhesion between R7 and R8 may be the cause of this phenotype.
Interlayer interactions between the distal and proximal layers
One of the important features of column visualization by using Ncad and phalloidin is that they visualize the columns in three layers of the pupal medulla. The mechanisms of 3D columnar organization across multiple layers would be an important issue in brain development. During the larval stage, there is only one layer in the medulla column. Early in pupal development, the top and middle layers are formed from the distal part of the medulla column. The bottom layers develop later than the top and middle layers at 24 h APF (Fig. 1C4) and are completely separated from the distal layers at 48 h APF (Fig. 1D4).
Our results indicate that the formation of donut-like structures in different layers is tightly coupled with each other. For example, loss of R7 and R8 affected the donut-like columnar structures and Mi1 axon terminals in layers M9-M10, to which they do not project (Fig. 4A4–D4,A5–C5), suggesting intimate interactions across different layers during column formation. The interlayer interactions were also observed when Ncad was knocked down in neurons that only project to the distal layers (data not shown).
Numerous studies have shown that Ncad has a wide range of functions in the developing nervous systems of fly and mammals (Redies et al., 1992; Redies and Takeichi, 1996; Hayashi and Carthew, 2004; Nern et al., 2008). Ncad is essential for the cortical organization of the mouse brain (Kadowaki et al., 2007; Matsunaga et al., 2017). Therefore, Ncad-dependent differential adhesion and interlayer interaction may be the essential mechanism underlying the 3D organization of column formation that is evolutionarily conserved from the fly optic lobe to mammalian brains. Future comparative studies of column formation using fly and mammalian brain systems will reveal key mechanisms of columnar unit formation.
Footnotes
This work was supported by Grant-in-Aid for Japan Society for the Promotion of Science Research Fellow to O.T., Core Research for Evolutional Science and Technology from Japan Science and Technology Agency, Grant-in-Aid for Scientific Research (B) from Ministry of Education, Culture, Sports, Science and Technology, Takeda Science Foundation and Cooperative Research of Network Joint Research Center for Materials and Devices to M.S., and Grant-in-Aid for Scientific Research on Innovative Areas from Ministry of Education, Culture, Sports, Science and Technology to M.S. and T.S. We thank Hugo Bellen, Takashi Hayashi, Aljosha Nern, Takumi Suzuki, and Tetsuo Yasugi for critical comments on the manuscript; Martin Heisenberg, Graeme Mardon, Hideji Murakoshi, Tadashi Uemura, and S. Lawrence Zipursky for reagents and fly strains; Bloomington Stock Center, Vienna Drosophila RNAi Center and DGRC, Kyoto for fly strains; and Developmental Study Hybridoma Bank for antibodies.
The authors declare no competing financial interests.
References
- Atkins M, Jiang Y, Sansores-Garcia L, Jusiak B, Halder G, Mardon G (2013) Dynamic rewiring of the Drosophila retinal determination network switches its function from selector to differentiation. PLoS Genet 9:e1003731. 10.1371/journal.pgen.1003731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazigou E, Apitz H, Johansson J, Lorén CE, Hirst EM, Chen PL, Palmer RH, Salecker I (2007) Anterograde jelly belly and alk receptor tyrosine kinase signaling mediates retinal axon targeting in Drosophila. Cell 128:961–975. 10.1016/j.cell.2007.02.024 [DOI] [PubMed] [Google Scholar]
- Carrillo JA, Murakawa H, Sato M, Togashi H, Trush O (2019) A population dynamics model of cell-cell adhesion incorporating population pressure and density saturation. J Theor Biol 474:14–24. 10.1016/j.jtbi.2019.04.023 [DOI] [PubMed] [Google Scholar]
- Chen Y, Akin O, Nern A, Tsui CY, Pecot MY, Zipursky SL (2014) Cell-type-specific labeling of synapses in vivo through synaptic tagging with recombination. Neuron 81:280–293. 10.1016/j.neuron.2013.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clandinin TR, Zipursky SL (2002) Making connections in the fly visual system. Neuron 35:827–841. 10.1016/S0896-6273(02)00876-0 [DOI] [PubMed] [Google Scholar]
- Erclik T, Li X, Courgeon M, Bertet C, Chen Z, Baumert R, Ng J, Koo C, Arain U, Behnia R, del Valle Rodriguez A, Senderowicz L, Negre N, White KP, Desplan C (2017) Integration of temporal and spatial patterning generates neural diversity. Nature 541:365–370. 10.1038/nature20794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach KF. (1989) The optic lobe of Drosophila melanogaster: I. A Golgi analysis of wild-type structure. Cell Tissue Res 258:441–475. [Google Scholar]
- Foty RA, Steinberg MS (2005) The differential adhesion hypothesis: a direct evaluation. Dev Biol 278:255–263. 10.1016/j.ydbio.2004.11.012 [DOI] [PubMed] [Google Scholar]
- Frankfort BJ, Nolo R, Zhang Z, Bellen H, Mardon G (2001) Senseless repression of rough is required for R8 photoreceptor differentiation in the developing Drosophila eye. Neuron 32:403–414. 10.1016/S0896-6273(01)00480-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakeda-Suzuki S, Berger-Müller S, Tomasi T, Usui T, Horiuchi SY, Uemura T, Suzuki T (2011) Golden goal collaborates with flamingo in conferring synaptic-layer specificity in the visual system. Nat Neurosci 14:314–323. 10.1038/nn.2756 [DOI] [PubMed] [Google Scholar]
- Hasegawa E, Kitada Y, Kaido M, Takayama R, Awasaki T, Tabata T, Sato M (2011) Concentric zones, cell migration and neuronal circuits in the Drosophila visual center. Development 138:983–993. 10.1242/dev.058370 [DOI] [PubMed] [Google Scholar]
- Hasegawa E, Kaido M, Takayama R, Sato M (2013) Brain-specific-homeobox is required for the specification of neuronal types in the Drosophila optic lobe. Dev Biol 377:90–99. 10.1016/j.ydbio.2013.02.012 [DOI] [PubMed] [Google Scholar]
- Hayashi T, Carthew RW (2004) Surface mechanics mediate pattern formation in the developing retina. Nature 431:647–652. 10.1038/nature02952 [DOI] [PubMed] [Google Scholar]
- Huang J, Zhou W, Dong W, Watson AM, Hong Y (2009) From the cover: directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc Natl Acad Sci U S A 106:8284–8289. 10.1073/pnas.0900641106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Kunes S (1996) Hedgehog, transmitted along retinal axons, triggers neurogenesis in the developing visual centers of the Drosophila brain. Cell 86:411–422. 10.1016/S0092-8674(00)80114-2 [DOI] [PubMed] [Google Scholar]
- Iwai Y, Usui T, Hirano S, Steward R, Takeichi M, Uemura T (1997) Axon patterning requires DN-cadherin, a novel neuronal adhesion receptor, in the Drosophila embryonic CNS. Neuron 19:77–89. 10.1016/S0896-6273(00)80349-9 [DOI] [PubMed] [Google Scholar]
- Iwai Y, Hirota Y, Ozaki K, Okano H, Takeichi M, Uemura T (2002) DN-cadherin is required for spatial arrangement of nerve terminals and ultrastructural organization of synapses. Mol Cell Neurosci 19:375–388. 10.1006/mcne.2001.1081 [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grell EH, Ackerman L, Jan LY, Jan YN (1994) Atonal is the proneural gene for Drosophila photoreceptors. Nature 369:398–400. 10.1038/369398a0 [DOI] [PubMed] [Google Scholar]
- Kadowaki M, Nakamura S, Machon O, Krauss S, Radice GL, Takeichi M (2007) N-cadherin mediates cortical organization in the mouse brain. Dev Biol 304:22–33. 10.1016/j.ydbio.2006.12.014 [DOI] [PubMed] [Google Scholar]
- Ke MT, Nakai Y, Fujimoto S, Takayama R, Yoshida S, Kitajima TS, Sato M, Imai T (2016) Super-resolution mapping of neuronal circuitry with an index-optimized clearing agent. Cell Rep 14:2718–2732. 10.1016/j.celrep.2016.02.057 [DOI] [PubMed] [Google Scholar]
- Kondo S, Ueda R (2013) Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195:715–721. 10.1534/genetics.113.156737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni A, Ertekin D, Lee CH, Hummel T (2016) Birth order dependent growth cone segregation determines synaptic layer identity in the Drosophila visual system. Elife 5:e13715. 10.7554/eLife.13715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Herman T, Clandinin TR, Lee R, Zipursky SL (2001) N-cadherin regulates target specificity in the Drosophila visual system. Neuron 30:437–450. 10.1016/S0896-6273(01)00291-4 [DOI] [PubMed] [Google Scholar]
- Li Y, Lu H, Cheng PL, Ge S, Xu H, Shi SH, Dan Y (2012) Clonally related visual cortical neurons show similar stimulus feature selectivity. Nature 486:118–121. 10.1038/nature11110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisak MS, Haag J, Ammer G, Serbe E, Meier M, Leonhardt A, Schilling T, Bahl A, Rubin GM, Nern A, Dickson BJ, Reiff DF, Hopp E, Borst A (2013) A directional tuning map of Drosophila elementary motion detectors. Nature 500:212–216. 10.1038/nature12320 [DOI] [PubMed] [Google Scholar]
- Maruoka H, Nakagawa N, Tsuruno S, Sakai S, Yoneda T, Hosoya T (2017) Lattice system of functionally distinct cell types in the neocortex. Science 358:610–615. 10.1126/science.aam6125 [DOI] [PubMed] [Google Scholar]
- Matsunaga Y, Noda M, Murakawa H, Hayashi K, Nagasaka A, Inoue S, Miyata T, Miura T, Kubo KI, Nakajima K (2017) Reelin transiently promotes N-cadherin-dependent neuronal adhesion during mouse cortical development. Proc Natl Acad Sci U S A 114:2048–2053. 10.1073/pnas.1615215114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morante J, Erclik T, Desplan C (2011) Cell migration in Drosophila optic lobe neurons is controlled by eyeless/Pax6. Development 138:687–693. 10.1242/dev.056069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountcastle VB. (1997) The columnar organization of the neocortex. Brain 120:701–722. 10.1093/brain/120.4.701 [DOI] [PubMed] [Google Scholar]
- Murakawa H, Togashi H (2015) Continuous models for cell-cell adhesion. J Theor Biol 374:1–12. 10.1016/j.jtbi.2015.03.002 [DOI] [PubMed] [Google Scholar]
- Nériec N, Desplan C (2016) From the eye to the brain: development of the Drosophila visual system. Curr Top Dev Biol 116:247–271. 10.1016/bs.ctdb.2015.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Zhu Y, Zipursky SL (2008) Local N-cadherin interactions mediate distinct steps in the targeting of lamina neurons. Neuron 58:34–41. 10.1016/j.neuron.2008.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Martínez-Cerdeño V, Ivic L, Kriegstein AR (2004) Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci 7:136–144. 10.1038/nn1172 [DOI] [PubMed] [Google Scholar]
- Ohtsuki G, Nishiyama M, Yoshida T, Murakami T, Histed M, Lois C, Ohki K (2012) Similarity of visual selectivity among clonally related neurons in visual cortex. Neuron 75:65–72. 10.1016/j.neuron.2012.05.023 [DOI] [PubMed] [Google Scholar]
- Özel MN, Langen M, Hassan BA, Hiesinger PR (2015) Filopodial dynamics and growth cone stabilization in Drosophila visual circuit development. Elife 4:e10721. 10.7554/eLife.10721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic M, Hummel T (2008) Temporal identity in axonal target layer recognition. Nature 456:800–803. 10.1038/nature07407 [DOI] [PubMed] [Google Scholar]
- Pfeiffer BD, Truman JW, Rubin GM (2012) Using translational enhancers to increase transgene expression in Drosophila. Proc Natl Acad Sci U S A 109:6626–6631. 10.1073/pnas.1204520109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. (1988) Specification of cerebral cortical areas. Science 241:170–176. 10.1126/science.3291116 [DOI] [PubMed] [Google Scholar]
- Redies C, Takeichi M (1996) Cadherins in the developing central nervous system: an adhesive code for segmental and functional subdivisions. Dev Biol 180:413–423. 10.1006/dbio.1996.0315 [DOI] [PubMed] [Google Scholar]
- Redies C, Inuzuka H, Takeichi M (1992) Restricted expression of N- and R-cadherin on neurites of the developing chicken CNS. J Neurosci 12:3525–3534. 10.1523/JNEUROSCI.12-09-03525.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rister J, Pauls D, Schnell B, Ting CY, Lee CH, Sinakevitch I, Morante J, Strausfeld NJ, Ito K, Heisenberg M (2007) Dissection of the peripheral motion channel in the visual system of Drosophila melanogaster. Neuron 56:155–170. 10.1016/j.neuron.2007.09.014 [DOI] [PubMed] [Google Scholar]
- Sanes JR, Zipursky SL (2010) Design principles of insect and vertebrate visual systems. Neuron 66:15–36. 10.1016/j.neuron.2010.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Suzuki T, Nakai Y (2013) Waves of differentiation in the fly visual system. Dev Biol 380:1–11. 10.1016/j.ydbio.2013.04.007 [DOI] [PubMed] [Google Scholar]
- Schwabe T, Borycz JA, Meinertzhagen IA, Clandinin TR (2014) Differential adhesion determines the organization of synaptic fascicles in the Drosophila visual system. Curr Biol 24:1304–1313. 10.1016/j.cub.2014.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinza-Kameda M, Takasu E, Sakurai K, Hayashi S, Nose A (2006) Regulation of layer-specific targeting by reciprocal expression of a cell adhesion molecule, capricious. Neuron 49:205–213. 10.1016/j.neuron.2005.11.013 [DOI] [PubMed] [Google Scholar]
- Simon MA, Bowtell DD, Rubin GM (1989) Structure and activity of the sevenless protein: a protein tyrosine kinase receptor required for photoreceptor development in Drosophila. Proc Natl Acad Sci U S A 86:8333–8337. 10.1073/pnas.86.21.8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg MS. (1962) On the mechanism of tissue reconstruction by dissociated cells: I. Population kinetics, differential adhesiveness, and the absence of directed migration. Proc Natl Acad Sci U S A 48:1577–1582. 10.1073/pnas.48.9.1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg MS, Takeichi M (1994) Experimental specification of cell sorting, tissue spreading, and specific spatial patterning by quantitative differences in cadherin expression. Proc Natl Acad Sci U S A 91:206–209. 10.1073/pnas.91.1.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura SY, Bharioke A, Lu Z, Nern A, Vitaladevuni S, Rivlin PK, Katz WT, Olbris DJ, Plaza SM, Winston P, Zhao T, Horne JA, Fetter RD, Takemura S, Blazek K, Chang LA, Ogundeyi O, Saunders MA, Shapiro V, Sigmund C, et al. (2013) A visual motion detection circuit suggested by Drosophila connectomics. Nature 500:175–181. 10.1038/nature12450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CY, Yonekura S, Chung P, Hsu SN, Robertson HM, Chiba A, Lee CH (2005) Drosophila N-cadherin functions in the first stage of the two-stage layer-selection process of R7 photoreceptor afferents. Development 132:953–963. 10.1242/dev.01661 [DOI] [PubMed] [Google Scholar]
- Toda S, Blauch LR, Tang SK, Morsut L, Lim WA (2018) Programming self-organizing multicellular structures with synthetic cell-cell signaling. Science 361:156–162. 10.1126/science.aat0271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi T, Hakeda-Suzuki S, Ohler S, Schleiffer A, Suzuki T (2008) The transmembrane protein golden goal regulates R8 photoreceptor axon-axon and axon-target interactions. Neuron 57:691–704. 10.1016/j.neuron.2008.01.012 [DOI] [PubMed] [Google Scholar]
- Van Vactor D Jr, Krantz DE, Reinke R, Zipursky SL (1988) Analysis of mutants in chaoptin, a photoreceptor cell-specific glycoprotein in Drosophila, reveals its role in cellular morphogenesis. Cell 52:281–290. 10.1016/0092-8674(88)90517-X [DOI] [PubMed] [Google Scholar]
- Yagi R, Mayer F, Basler K (2010) Refined LexA transactivators and their use in combination with the Drosophila Gal4 system. Proc Natl Acad Sci U S A 107:16166–16171. 10.1073/pnas.1005957107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura S, Xu L, Ting CY, Lee CH (2007) Adhesive but not signaling activity of Drosophila N-cadherin is essential for target selection of photoreceptor afferents. Dev Biol 304:759–770. 10.1016/j.ydbio.2007.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. (2013) The Drosophila visual system: from neural circuits to behavior. Cell Adh Migr 7:333–344. 10.4161/cam.25521 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
R8 and R7 project to the same layer in the larval medulla. Super-resolution image of sensF2-LexA LexAop-myrTomato (red) and PM181-Gal4 UAS-myrGFP (green) in L3 larval medulla. sensF2-Gal4 UAS-myrGFP (green) and sevEnS-LexA LexAop-myrTomato (red) show the same result.

R8 and Mi1 project to the same layer in the larval medulla. Super-resolution image of sensF2-Gal4 UAS-myrGFP (green) and bshM-LexA LexAop-myrTomato (red) in L3 larval medulla. sensF2-LexA LexAop-myrTomato (red) and bshM-Gal4 UAS-myrGFP (green) show the same result.

R7 and R8 terminals significantly overlap upon Ncad knockdown in R7. Super-resolution image of Ncad RNAi in R7. PM181-Gal4 UAS-myrGFP (green) and sensF2-LexA LexAop-myrTomato (red) in L3 larval medulla.

R7 terminals are expanded upon Ncad knockdown in R8. Super-resolution image of Ncad RNAi in R8. sensF2-Gal4 UAS-myrGFP (green) and sevEnS-LexA LexAop-myrTomato (red) in L3 larval medulla.

R8 and R7 terminals significantly overlap upon Ncad overexpression in R8. Super-resolution image of Ncad overexpression in R8. sensF2-Gal4 UAS-myrGFP (green) and sev-LexA LexAop-myrTomato (red) in L3 larval medulla.