Abstract
DNA methylation may play a critical role in aging and age-related diseases. DNA methylation phenotypic age (DNAmPhenoAge) is a new aging biomarker and predictor of chronic disease risk. While smoking is a strong risk factor for chronic diseases and influences methylation, its influence on DNAmPhenoAge is unknown. We investigated associations of self-reported and epigenetic smoking indicators with DNAmPhenoAge acceleration in a longitudinal aging study in eastern Massachusetts. DNA methylation was measured in whole blood samples from multiple visits for 692 male participants in the Veterans Affairs Normative Aging Study during 1999–2013. Acceleration was defined using residuals from linear regression of the DNAmPhenoAge on the chronological age. Cumulative smoking (pack-years) was significantly associated with DNAmPhenoAge acceleration, whereas self-reported smoking status was not. We observed significant validated associations between smoking-related loci and DNAmPhenoAge acceleration for 52 CpG sites, where 18 were hypomethylated and 34 were hypermethylated, mapped to 16 genes. The AHRR gene had the most loci (N = 8) among the 16 genes. We generated a smoking aging index based on these 52 loci, which showed positive significant associations with DNAmPhenoAge acceleration. These epigenetic biomarkers may help to predict age-related risks driven by smoking.
Keywords: smoking-related DNA methylation, DNA methylation phenotypic age, aging biomarker, aging acceleration
1. Introduction
DNA methylation is an aging biomarker that has attracted growing attention in recent years. Tobacco smoking is a leading risk factor for many age-related diseases, including cardiopulmonary diseases and various types of cancers [1]. Several studies have revealed, by means of regulating gene expression and genome stability, that DNA methylation is associated with smoking and smoking-related diseases [2]. Smoking-related CpG sites, located in genes such as AHRR, GPR15, and F2RL3, have been reported in an increasing number of epigenome-wide association studies (EWAS) using whole blood samples [3,4]. A recent study generated a smoking index based on 1501 smoking-related loci, suggesting that such methylation indices could be helpful to predict smoking-related health risks [5].
Smoking is a risk factor of disproportional aging that accelerates epigenetic age and decreases life expectancy [6,7]. Aging is characterized by a series of changes at both the cellular and molecular levels, including telomere length shortening, senescence, and variations in gene expression levels. Hannum et al. [8,9] and Horvath (2013) developed the first generation of algorithms to estimate DNA methylation age, which correlates well with chronological age. However, those methods only capture CpG sites that correlate with chronological age and fail to account for an individual’s physiological status [10].
A newly developed epigenetic biomarker, DNA methylation phenotypic age (DNAmPhenoAge), was derived from a series of clinical measurements [11,12,13,14,15] to offer a better biomarker of biological aging as a substitute measurement of “phenotypic aging”. DNAmPhenoAge acceleration is defined as a discrepancy between DNAmPhenoAge and chronological age and has been proposed as an indicator of disproportionate aging.
A recent study associated DNAmPhenoAge with self-reported smoking status [10]. However, no study has investigated the association of smoking-related DNA methylation with DNAmPhenoAge or age acceleration. In the current study, we used a well-established cohort to strengthen and extend prior findings with respect to the association between self-reported and epigenetic smoking indicators and age acceleration in whole blood samples.
2. Materials and Methods
2.1. Study Subjects
This study included 692 participants from the Normative Aging Study (NAS), a closed longitudinal study of aging in men from eastern Massachusetts [16]. A total of 1596 participants free of chronic diseases have been enrolled in NAS since 1984, and they undergo physical examinations every 3–5 years. At each visit, information on demographic factors, lifestyle, and disease history were collected, and physical examinations and laboratory tests were performed. Specifically, we analyzed DNA samples up to 4 visits from 1999–2013. The current study was restricted to the first three visits to observe long-term DNAmPhenoAge acceleration and control for race heterogeneity. Participants provided written informed consent during each visit, and the Veterans Affairs Boston Health Care System Institutional Review Board approved the study (code: AAAQ8739).
2.2. DNA Methylation Data
DNA was extracted from stored blood buffy coats using the QIAamp DNA Blood Kit (Qiagen, Valencia, CA, USA). We treated 500 ng DNA for bisulfite conversion using the EZ-96 DNA Methylation Kit (Zymo Research, Orange, CA, USA). After bisulfite conversion, we hybridized DNA samples to 12-sample Illumina HumanMethylation450 BeadChips per Infinium HD Methylation protocol (Illumina, San Diego, CA, USA). Technical effects resulting from plates/chips were minimized with a two-stage, age-stratified algorithm to randomize the samples. The β-values, ranging from 0–1, were used to represent the methylation status of a specific CpG site, with 0 indicating no methylation and 1 indicating full methylation.
Calculation of DNAmPhenoAge: DNAmPhenoAge was estimated using the method described by Levine et al. [10]. In short, phenotypic age was developed using clinical measurements from the third National Health and Nutrition Examination Survey. A penalized Cox regression model was used to select 10 out of 42 biomarkers. Phenotypic age was then regressed on DNA methylation data from whole blood samples based on an elastic net regression analysis, which led to 513 CpG sites. We calculated DNAmPhenoAge based on these loci for each participant as:
| DNAmPhenoAge = CpG1 × β1 + CpG2 × β2 + CpG3 × β3 +…+ CpG513 × β513 + intercept | (1) |
Values of coefficients and intercepts were extracted from Levine et al. [10]. DNAmPhenoAge acceleration was defined as residuals from a linear regression of DNAmPhenoAge on chronological age.
Selection of smoking-related DNA methylation loci: We selected 151 CpG sites reported from previous smoking EWAS at least twice from a previous systematic review [3]. There was no overlap between these 151 loci and the 513 loci used to predict DNAmPhenoAge.
2.3. Statistical Analysis
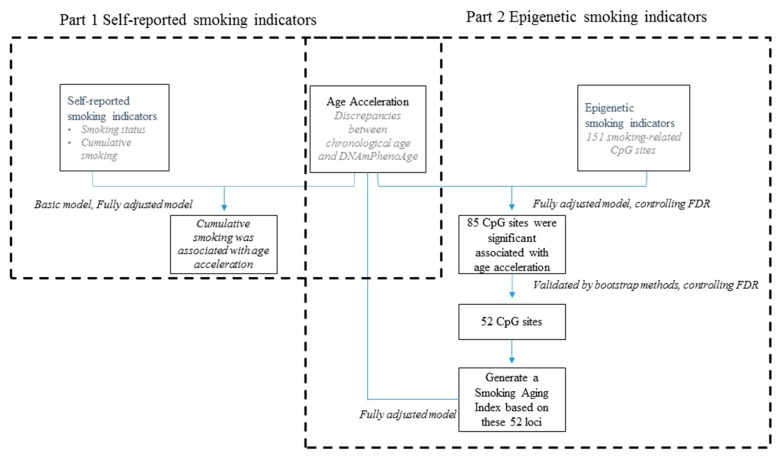
Descriptive statistics were used to present distributions of demographic and clinical variables among all participants. Distributions of these variables across each visit were described as well. Generalized linear mixed models were used to assess the relationship between smoking indicators and DNAmPhenoAge acceleration (Figure 1). We conducted two parts of statistical analysis: first, to explore the association between self-reported smoking indicators and DNAmPhenoAge acceleration; and second, to explore the association between epigenetic smoking indicators and DNAmPhenoAge acceleration.
Figure 1.
Flow chart of the generalized linear mixed model used to assess the relationship between smoking indicators and DNA methylation phenotypic age (DNAmPhenoAge) acceleration of participants in the Veterans Affairs Normative Aging Study in Boston, MA, 1999–2013.
First, we examined the associations between DNAmPhenoAge acceleration and self-reported smoking indicators smoking status (never, current, or former) and cumulative smoking status (pack-years). Model 1 was adjusted for age (years) and leukocyte distribution was estimated using the Houseman et al.’s algorithm [17]. In addition, both visits and random batch effects of methylation measurement were included in the model as random effects. Additionally, Model 2 was adjusted for smoking status, alcohol consumption (abstainer, low, intermediate, or high), body mass index (BMI) (underweight or normal weight BMI <25, overweight BMI ≥25 to <30, or obese BMI ≥30), physical activity (low, metabolic equivalent of task (MET) ≤12 kcal/kg hours/week; median, MET 12–30 kcal/kg hours/week; or high, MET ≥30 kcal/kg hours/week), years of education (≤12 years, 13–16 years, or >16 years), hypertension, stroke, coronary heart disease, and diabetes (yes/no).
Second, we tested if epigenetic indicators were associated with DNAmPhenoAge acceleration. The aforementioned fully adjusted model (Model 2) was used to explore associations between DNAmPhenoAge acceleration and each of the 151 smoking-related loci. False discovery rate (FDR)-adjusted [18] P-values were used to account for multiple testing. The alpha threshold for FDR-adjusted P-value was set at ≤0.05. We performed bootstrap analysis to validate the selected CpGs. We resampled the original dataset to create 1000 simulated datasets for each locus. Then, we ran the model in each dataset and returned the effect estimates for the tested CpG sites. We counted the effect estimates that were greater than or equal to the observed value and divided that count by the number of estimates. We again used FDR-adjusted P-values to select the most robust loci.
Finally, we generated a smoking aging index based on the verified 52 CpG sites [5]. The mean β-value (μc) and standard deviation (SD) (σc) were calculated across the never-smokers for each selected CpG. The smoking aging index was defined as
| (2) |
where, Wc is +1 if the smoking-associated CpG, c, is hypermethylated or −1 if it is hypomethylated in smokers, and βc is the β-value of this CpG in sample s [5]. The smoking aging index for each participant was calculated based on these validated DNAmPhenoAge acceleration-related loci, which were validated by the bootstrap method. Statistical analysis was performed using R 3.5.1 (R core team, 2018).
3. Results
3.1. Characteristics of Participants
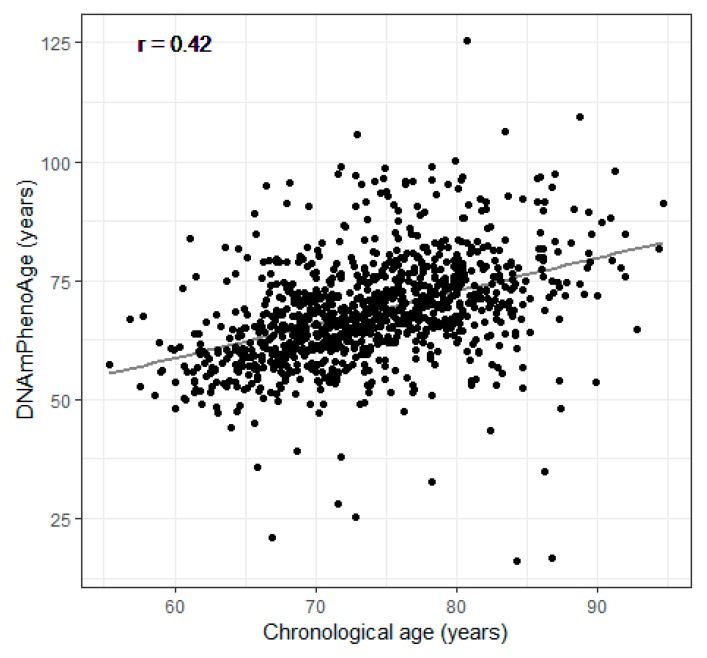
Demographic and clinical characteristics of the participants at all three visits are shown in Table 1. Participants were 55–94 years old with a mean (SD) age of 72.7 (6.7) years at first visit. The mean (SD) of the DNAmPhenoAge was 68.4 (11.9) years at first visit, younger than the chronological age. The chronological age and the DNAmPhenoAge were correlated with each other (r = 0.42, P < 0.001) (Figure 2). The majority of the participants in the cohort were former smokers and never smokers, accounting for 66% and 30% at the first visit, respectively. The percentage for current smokers was only 4% at the first visit. The total years of smoking was 16.9 years on average at the first visit.
Table 1.
Population characteristics of participants in the Veterans Affairs Normative Aging Study in Boston, MA, 1999–2013 a.
| Variable | All Visits | First Visit | Second Visit | Third Visit |
|---|---|---|---|---|
| No. of measures | 1214 | 692 | 418 | 104 |
| Chronological age (years) | 74.3 (7.0) | 72.7 (6.7) | 76.3 (6.8) | 79.8 (6.0) |
| DNAmPhenoAge (years) | 70.5 (12.9) | 68.4 (11.9) | 70.6 (12.2) | 83.6 (13.9) |
| Smoking status | ||||
| Never smoker | 374 (31%) | 207 (30%) | 131 (31%) | 36 (35%) |
| Current smoker | 46 (4%) | 29 (4%) | 15 (4%) | 2 (2%) |
| Former smoker | 794 (65%) | 456 (66%) | 272 (65%) | 66 (63%) |
| Total years of smoking cigarettes (years) | 16.7 (15.8) | 16.9 (15.7) | 16.4 (15.9) | 11.3 (15.5) |
| Alcohol consumption (g/d) | ||||
| Abstainer | 230 (24%) | 146 (24%) | 81 (25%) | 3 (25%) |
| Low (0–40 g/d) | 652 (69%) | 420 (69%) | 223 (70%) | 9 (75%) |
| Intermediate (40–60 g/d) | 37 (4%) | 26 (4%) | 11 (3%) | 0 |
| High (≥60 g/d) | 23(2%) | 18 (3%) | 5 (2%) | 0 |
| Physical activity (MET-hours/week) | ||||
| Low(≤12 kcal/kg hours/week) | 667 (64%) | 408 (63%) | 240 (65%) | 19 (58%) |
| Median (12–30 kcal/kg hours/week) | 227 (22%) | 139 (22%) | 83 (23%) | 5 (15%) |
| High (≥30 kcal/kg hours/week) | 152 (15%) | 98 (15%) | 45 (12%) | 9 (27%) |
| Years of education | ||||
| ≤12 years | 335 (33%) | 215 (33%) | 115 (33%) | 5 (42%) |
| 13–16 years | 477 (47%) | 314 (48%) | 159 (46%) | 4 (33%) |
| >16 years | 209 (20%) | 131 (20%) | 75 (21%) | 3 (25%) |
| Body mass index | ||||
| Underweight or normal weight (<25.0) | 265 (22%) | 139 (20%) | 94 (22%) | 32 (31%) |
| Overweight (≥25.0 to <30.0) | 636 (52%) | 374 (54%) | 214 (51%) | 48 (46%) |
| Obese (≥30.0) | 313 (26%) | 179 (26%) | 110 (26%) | 24 (23%) |
| Chronic diseases | ||||
| Hypertension | 731 (71%) | 464 (70%) | 258 (73%) | 9 (75%) |
| Stroke | 99 (8%) | 53 (8%) | 39 (9%) | 7 (7%) |
| Coronary heart disease | 397 (33%) | 208 (30%) | 146 (35%) | 43 (41%) |
| Diabetes | 193(16%) | 99 (14%) | 73 (17%) | 21 (20%) |
a Mean value (standard deviation) for continuous variables and n (%) for categorical variables.
Figure 2.
Scatter plot of the DNA methylation phenotypic age (DNAmPhenoAge) and the chronological age of participants in the Veterans Affairs Normative Aging Study in Boston, MA, 1999–2013.
3.2. DNAmPhenoAge Acceleration and Smoking Indicators
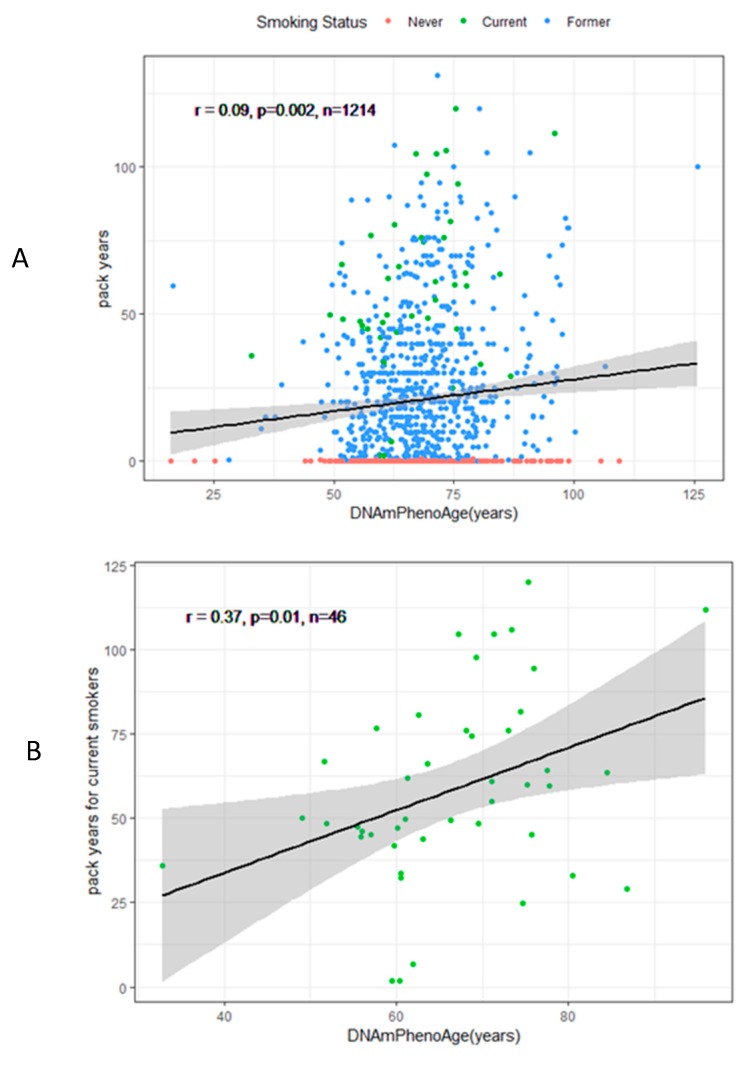
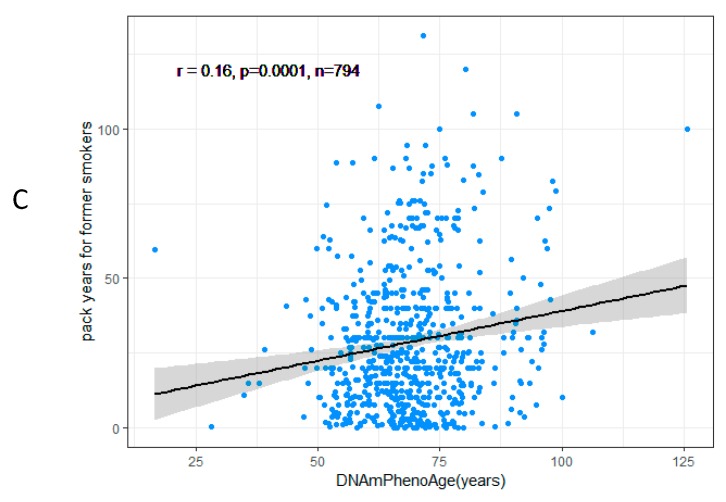
Table 2 illustrates the association of DNAmPhenoAge acceleration with self-reported (smoking status and pack-years) indicators in both the basic model (Model 1) and fully adjusted model (Model 2). Only current smokers had significantly higher DNAmPhenoAge acceleration in Model 1, although not in Model 2. However, cumulative smoking (pack-years) was significantly associated with DNAmPhenoAge acceleration in both models. Figure 3 shows associations between DNAmPhenoAge and pack-years for all participants (r = 0.09, N = 1214, P = 0.002), current smokers (r = 0.37, N = 46, P = 0.01), and former smokers (r = 0.16, N = 794, P = 0.0001).
Table 2.
Association of DNA methylation phenotypic age (DNAmPhenoAge) acceleration with self-reported smoking and epigenetic smoking indicators in the Veterans Affairs Normative Aging Study Cohort in Boston, MA, 1999–2013.
| Smoking Indicator | Model 1 a | Model 2 b | ||||
|---|---|---|---|---|---|---|
| Smoking Status | Estimate # | SE * | P-Value | Estimate # | SE | P-Value |
| Current smoker | 3.41 | 1.83 | 0.06 | 2.69 | 1.86 | 0.15 |
| Former smoker | 0.62 | 0.77 | 0.41 | 0.17 | 0.79 | 0.82 |
| Never smoker | Ref | Ref | ||||
| Cumulative smoking (pack-year) | 0.06 | 0.01 | 3.4 e-5 | 0.04 | 0.01 | 0.003 |
# Effect size of age acceleration per SD (standard deviation) increase of smoking aging index. * SE: standard error. a Model 1: Adjusted for age (years), leukocyte distribution (Houseman et al.’s algorithm), visits, and random batch effect of methylation measurement. b Model 2: Additionally adjusted for smoking status, alcohol consumption (abstainer/low/intermediate/high), body mass index (BMI; underweight or normal weight/overweight/obese), physical activity (metabolic equivalent of task (MET) low ≤12 kcal/kg hours/week, MET median 12–30 kcal/kg hours/week, and MET high ≥30 kcal/kg hours/week), years of education (≤12 years, 13–16 years, >16 years), hypertension, stroke, coronary heart disease, and diabetes (yes/no).
Figure 3.
Scatter plots of the association between pack-years and DNA methylation phenotypic age (DNAmPhenoAge) for (A) all participants, (B) current smokers, and (C) former smokers in the Veterans Affairs Normative Aging Study in Boston, MA, 1999–2013.
Moreover, we assessed associations between the 151 smoking-related CpG candidates and DNAmPhenoAge acceleration using Model 2, with visits and random batch effects of methylation measurement as random effects. A total of 85 of 151 CpG candidates passed the threshold of FDR <0.05, and thus demonstrated significant associations with DNAmPhenoAge acceleration. We attempted to validate theses CpG candidates using bootstrap methods. Validation confirmed that 52 of 85 CpG sites were associated with DNAmPhenoAge acceleration (FDR < 0.05, Table 3). Of these 52 loci, 18 were hypomethylated and 34 were hypermethylated. AHRR had the most related loci (N = 8).
Table 3.
CpG Sites (P < 0.05) associated with DNA methylation phenotypic age (DNAmPhenoAge) acceleration in the Veterans Affairs Normative Aging Study Cohort in Boston, MA, 1999–2013 a.
| Chr | CpG Site | Gene | Effect Size per SD (SE) # | P-Value | FDR- Adjusted P-Value | P-Value for Bootstrap | FDR-Adjusted P-Value |
|---|---|---|---|---|---|---|---|
| 1 | cg25189904 | GNG12 | 1.21 | 6.7E−04 | 2.0E−03 | 1.1E−02 | 2.7E−02 |
| cg04885881 | 1.91 | 3.5E−08 | 4.0E−07 | 2.7E−08 | 3.0E−07 | ||
| cg11314684 | AKT3 | 1.82 | 4.3E−08 | 4.6E−07 | 2.0E−03 | 7.7E−03 | |
| cg09662411 | GFI1 | −1.66 | 2.5E−06 | 1.9E−05 | 5.0E−03 | 1.6E−02 | |
| cg10399789 | GFI1 | 1.58 | 4.5E−06 | 3.0E−05 | 1.7E−02 | 3.3E−02 | |
| cg12547807 | 2.35 | 1.8E−14 | 6.9E−13 | 1.1E−13 | 4.3E−12 | ||
| cg19713429 | CAPZB | 1.89 | 6.6E−09 | 8.3E−08 | 1.0E−03 | 4.5E−03 | |
| cg21140898 | −1.61 | 7.1E−06 | 4.1E−05 | 4.3E−06 | 1.3E−05 | ||
| cg26764244 | GNG12 | 2.01 | 3.1E−09 | 4.4E−08 | 2.0E−03 | 7.7E−03 | |
| cg27537125 | 2.82 | 1.1E−19 | 1.6E−17 | 5.3E−18 | 1.3E−16 | ||
| 2 | cg01940273 | −1.40 | 2.7E−04 | 9.2E−04 | 1.3E−02 | 2.9E−02 | |
| cg03329539 | 1.71 | 2.7E−07 | 2.5E−06 | 1.6E−02 | 3.2E−02 | ||
| cg23079012 | −1.05 | 9.8E−04 | 2.6E−03 | 1.5E−02 | 3.0E−02 | ||
| 3 | cg18642234 | GPX1 | 1.86 | 2.9E−06 | 2.1E−05 | 1.4E−05 | 1.7E−04 |
| cg18754985 | CLDND1 | −1.13 | 1.6E−04 | 5.9E−04 | 3.0E−03 | 1.1E−02 | |
| 4 | cg24556382 | GALNT7 | 1.40 | 3.7E−05 | 1.7E−04 | 7.0E−03 | 2.0E−02 |
| 5 | cg14817490 | AHRR | 1.85 | 2.6E−07 | 2.5E−06 | 1.5E−06 | 3.8E−05 |
| cg05575921 | AHRR | −0.97 | 1.3E−05 | 1.2E−04 | 7.0E−03 | 2.0E−02 | |
| cg25648203 | AHRR | −1.14 | 6.6E−04 | 2.0E−03 | 2.2E−02 | 4.0E−02 | |
| cg26703534 | AHRR | −1.54 | 1.6E−05 | 7.7E−05 | 1.5E−02 | 3.0E−02 | |
| cg01899089 | AHRR | 1.53 | 6.9E−06 | 4.1E−05 | 4.9E−05 | 1.2E−05 | |
| cg01097768 | AHRR | 2.50 | 3.4E−12 | 6.3E−11 | 2.1E−11 | 4.5E−10 | |
| cg11554391 | AHRR | 1.72 | 3.2E−06 | 2.2E−05 | 2.0E−03 | 7.7E−03 | |
| cg17924476 | AHRR | 1.55 | 4.9E−06 | 3.1E−05 | 3.0E−03 | 1.1E−02 | |
| 6 | cg24859433 | −1.44 | 5.8E−05 | 2.4E−04 | 1.5E−02 | 3.0E−02 | |
| cg14753356 | 1.65 | 5.1E−04 | 1.6E−03 | 1.2E−02 | 2.8E−02 | ||
| cg15474579 | CDKN1A | 1.68 | 1.0E−05 | 5.5E−05 | 2.7E−02 | 4.7E−02 | |
| cg20778199 | −3.01 | 5.7E−17 | 4.3E−15 | 3.6E−16 | 2.7E−14 | ||
| 7 | cg11207515 | CNTNAP2 | 1.72 | 5.0E−07 | 4.3E−06 | 5.0E−03 | 1.6E−02 |
| cg25949550 | CNTNAP2 | 2.56 | 8.7E−14 | 2.2E−12 | 6.0E−13 | 1.4E−11 | |
| cg05221370 | LRRN3 | 2.43 | 6.6E−12 | 1.1E−10 | 4.9E−11 | 4.3E−09 | |
| cg07826859 | MYO1G | 1.64 | 4.6E−05 | 2.0E−04 | 2.8E−02 | 4.8E−02 | |
| cg09837977 | LRRN3 | 1.92 | 1.6E−05 | 7.7E−05 | 9.0E−03 | 2.4E−02 | |
| 8 | cg25305703 | 1.39 | 6.7E−05 | 2.7E−04 | 1.3E−02 | 2.9E−02 | |
| 10 | cg25953130 | ARID5B | −1.54 | 1.5E−05 | 7.6E−05 | 7.0E−03 | 2.0E−02 |
| 11 | cg23771366 | PRSS23 | 1.31 | 8.1E−04 | 2.4E−03 | 1.5E−02 | 3.0E−02 |
| cg04039799 | NAV2 | 1.85 | 3.2E−09 | 4.4E−08 | 1.4E−08 | 3.9E−07 | |
| cg16556677 | KCNQ1 | −1.41 | 2.2E−04 | 7.7E−04 | 8.0E−03 | 2.2E−02 | |
| cg16611234 | 1.36 | 1.7E−04 | 5.9E−04 | 7.0E−03 | 2.0E−02 | ||
| 12 | cg02583484 | HNRNPA1 | 1.22 | 3.5E−04 | 1.2E−03 | 2.0E−02 | 3.8E−02 |
| cg04158018 | NFE2 | −1.38 | 1.1E−04 | 4.0E−04 | 4.0E−03 | 1.4E−02 | |
| 13 | cg23681440 | 1.17 | 8.9E−04 | 2.5E−03 | 2.4E−02 | 4.3E−02 | |
| 14 | cg13976502 | C14orf43 | 1.22 | 1.5E−05 | 7.6E−05 | 1.0E−03 | 4.5E−03 |
| cg22851561 | C14orf43 | 2.49 | 1.0E−15 | 5.2E−14 | 3.7E−14 | 6.4E−13 | |
| cg13038618 | 1.20 | 1.1E−03 | 2.9E−03 | 2.9E−02 | 4.8E−02 | ||
| 16 | cg06972908 | ITGAL | 2.64 | 2.5E−14 | 7.7E−13 | 9.3E−12 | 4.6E−10 |
| cg13500388 | CBFB | 1.67 | 1.7E−05 | 7.7E−05 | 1.1E−02 | 2.7E−02 | |
| cg16794579 | XYLT1 | 2.37 | 6.7E−13 | 1.4E−11 | 3.5E−12 | 6.9E−10 | |
| 17 | cg19572487 | RARA | 1.43 | 7.2E−05 | 2.8E−04 | 2.2E−02 | 4.0E−02 |
| cg07465627 | STXBP4 | 1.46 | 1.8E−06 | 1.4E−05 | 6.2E−05 | 7.1E−04 | |
| 19 | cg15187398 | MOBKL2A | 1.84 | 5.1E−07 | 4.3E−06 | 4.2E−06 | 1.8E−05 |
| cg23973524 | CRTC1 | −1.63 | 7.4E−06 | 4.1E−05 | 1.0E−02 | 2.6E−02 |
# SD: Standard deviation; SE: Standard error. a Adjusted for age (years), leukocyte distribution (Houseman et al.’s algorithm), smoking status, alcohol consumption (abstainer/low/intermediate/high), body mass index (BMI; underweight or normal weight/overweight/obese), physical activity (metabolic equivalent of task (MET) low ≤12 kcal/kg hours/week, MET median 12–30 kcal/kg hours/week, and MET high ≥30 kcal/kg hours/week), years of education (≤12 years, 13–16 years, >16 years), hypertension, stroke, coronary heart disease, diabetes, cancer (yes/no), visits, and random batch effect of methylation measurement.
We generated a smoking aging index based on the 52 validated smoking-related loci and compared this new index with AHRR cg05575921, a strong smoking-associated hypomethylated locus. The smoking aging index was highly associated with self-reported smoking indicators in the generalized linear mixed effect model (P < 0.05, data not shown). For cg05575921, negative associations were found in both Model 1 (β = −1.34, P = 0.0001) and Model 2 (β = −0.97, P = 0.007) (Table 3). For the smoking aging index, positive associations were found in both Model 1 (β = 1.92, P < 0.0002) and Model 2 (β = 1.91, P < 0.0001).
4. Discussion
We systematically explored the association between self-reported and epigenetic smoking indicators and DNAmPhenoAge in whole blood samples based on an elderly cohort in eastern Massachusetts. One of the self-reported variables of smoking, cumulative smoking expressed in pack-years, was significantly and strongly associated with DNAmPhenoAge acceleration. For epigenetic indicators, 52 previously confirmed CpG sites (including a robust mono-biomarker of smoking, ch05575921) and smoking aging index derived from these loci were associated with DNAmPhenoAge acceleration.
Tobacco smoking has long been regarded as a leading risk factor for several age-related health outcomes [1,19,20]. A recent study investigated the relationship between DNAmPhenoAge acceleration based on Horvath’s and Hannum et al.’s algorithms and smoking-related DNA methylation [21]. Although our current study assessed DNAmPhenoAge acceleration based on DNAmPhenoAge, our findings are consistent with those of Gao et al., since we both found loci mapping to AHRR, a well-known tumor suppressor gene. One specific AHRR locus, cg05575921, has been regarded as a strong epigenetic smoking indicator in previous smoking EWAS [22,23,24] and was confirmed by our current study [25]. Other consistent loci involve CNTNAP2 (contactin associated protein-like 2) and have been reported associated with several mental diseases [26,27,28], as well as KCNQ1 (voltage-gated KQT-like subfamily Q, member 1) that has been associated with type 2 diabetes [29]. Moreover, our current study also unveiled other genes, such as GNG12 (a negative regulator of inflammation in microglial cells that could impact the neural system) [30], LRRN3 (a member of neuron leucine-rich repeat superfamily that may be associated with neural system development and differentiation processes) [31], and GFI1 (involved in the regulation of hematopoietic stem cells) [32].
There are some inconsistencies between self-reported smoking indicators and epigenetic smoking indicators with respect to their associations with aging. One inconsistency is that the smoking status was not related to either DNAmPhenoAge acceleration in the fully-adjusted model (Table 2) or the smoking aging index (Figure 3). One explanation is that the number of current smokers is far less than former and never smokers, which may diminish statistical power. Another reason might be the susceptibility of different individuals. For instance, genetic polymorphisms may play a role in modifying the health impacts of exposure to environmental health hazards, indicating that some individuals may be more or less likely to be affected by environmental hazards and develop diseases [33,34].
This point leads to our consideration that genetic variation may affect the relationship between epigenetics and aging [35]. A recent study reported findings regarding the heritability of discrepancies between chronological and epigenetic age by examining twins [9]. Moreover, we should also bear in mind that exposure to cigarette smoking is not the only factor that could lead to differences in DNA methylation. Other environmental hazards, such as drinking alcohol, lifestyle or nutritional factors, or interaction effects between these hazards and smoking, might induce similar methylation changes as well [36,37]. This evidence supports the idea that aging rates are influenced by interactions among genetic variation, environmental exposures, and the epigenome.
Our study has the following strengths. We present a longitudinal study to investigate the relationship between smoking-related DNA methylation and aging. Longitudinal studies that investigate aging have advantages over cross-sectional studies, particularly with regard to genetic variations [38]. Furthermore, the cohort longitudinal design allowed us to understand relationships between genetics and other factors that specify inter-subject temporal changes in human DNA methylation profiles.
However, there are also limitations to our study. First, we validated our results using a bootstrap method in the training cohort instead of an external validation cohort. Although it is generally ideal to use another cohort to validate and replicate findings, we could not access another similar phenotypic cohort. Therefore, further studies are needed to validate and replicate our findings. Second, our signature to predict smoking-related aging was derived from elderly males only. Thus, it remains unclear whether the signature would differ in a female population. However, previous EWAS suggest that smoking-associated changes in blood samples are not dependent on sex [39,40]. Nonetheless, future validation in a larger sex-balanced cohort is still needed. Third, measurement inaccuracies may affect the comparability of longitudinal DNAmPhenoAge acceleration. However, we adjusted for batch effect by including it as a random effect in the model and normalized methylation profiles using Horvath’s internal normalization methods. Fourth, loss to follow-up may have biased our findings. However, we used inverse probability weighting to impute missing data as a sensitivity analysis.
5. Conclusions
In summary, our results show that self-reported cumulative smoking and epigenetic smoking indicators were associated with DNAmPhenoAge acceleration. Epigenetic changes at specific smoking-related CpG sites and our combined smoking aging index might be useful biomarkers to predict smoking-associated aging. Further research is warranted to replicate our findings in follow-up studies and to identify the mechanism underlying the effects of smoking on DNAmPhenoAge.
Acknowledgments
We thank all the participants in the Normative Aging Study.
Author Contributions
A.A.B., X.G., and Y.Y. designed this study; Y.Y. did the data analysis and draft the manuscript; E.C. helped with the data analysis; X.G. and C.W. calculated the DNA PhenoAge; L.H. and Y.Z. prepared the DNA methylation data; Y.Y., X.G., A.C.J., E.C., C.W., B.A.C., L.H., Y.Z., P.V., J.S. and, A.A.B. helped revising the paper.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Lee K.W., Pausova Z. Cigarette smoking and DNA methylation. Front. Genet. 2013;4:132. doi: 10.3389/fgene.2013.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakulski K.M., Fallin M.D. Epigenetic epidemiology: promises for public health research. Environ. Mol. Mutagen. 2014;55:171–183. doi: 10.1002/em.21850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao X., Jia M., Zhang Y., Breitling L.P., Brenner H. DNA methylation changes of whole blood cells in response to active smoking exposure in adults: a systematic review of DNA methylation studies. Clin. Epigenetics. 2015;7:113. doi: 10.1186/s13148-015-0148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y., Schöttker B., Florath I., Stock C., Butterbach K., Holleczek B., Mons U., Brenner H. Smoking-associated DNA methylation biomarkers and their predictive value for all-cause and cardiovascular mortality. Environ. Health Perspect. 2015;124:67–74. doi: 10.1289/ehp.1409020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teschendorff A.E., Yang Z., Wong A., Pipinikas C.P., Jiao Y., Jones A., Anjum S., Hardy R., Salvesen H.B., Thirlwell C. Correlation of smoking-associated DNA methylation changes in buccal cells with DNA methylation changes in epithelial cancer. JAMA Oncol. 2015;1:476–485. doi: 10.1001/jamaoncol.2015.1053. [DOI] [PubMed] [Google Scholar]
- 6.Elliott H.R., Tillin T., McArdle W.L., Ho K., Duggirala A., Frayling T.M., Smith G.D., Hughes A.D., Chaturvedi N., Relton C.L. Differences in smoking associated DNA methylation patterns in South Asians and Europeans. Clin. Epigenetics. 2014;6:4. doi: 10.1186/1868-7083-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weidner C.I., Lin Q., Koch C.M., Eisele L., Beier F., Ziegler P., Bauerschlag D.O., Jöckel K.-H., Erbel R., Mühleisen T.W. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Boil. 2014;15:R24. doi: 10.1186/gb-2014-15-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hannum G., Guinney J., Zhao L., Zhang L., Hughes G., Sadda S., Klotzle B., Bibikova M., Fan J.-B., Gao Y. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horvath S. DNA methylation age of human tissues and cell types. Genome Boil. 2013;14:3156. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine M.E., Lu A.T., Quach A., Chen B.H., Assimes T.L., Bandinelli S., Hou L., Baccarelli A.A., Stewart J.D., Li Y. An epigenetic biomarker of aging for lifespan and healthspan. Aging. 2018;10:573. doi: 10.18632/aging.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine M.E. Modeling the rate of senescence: can estimated biological age predict mortality more accurately than chronological age? J. Gerontol. Ser. A Biomed. Sci. Med. Sci. 2012;68:667–674. doi: 10.1093/gerona/gls233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belsky D.W., Caspi A., Houts R., Cohen H.J., Corcoran D.L., Danese A., Harrington H., Israel S., Levine M.E., Schaefer J.D. Quantification of biological aging in young adults. Proc. Natl. Acad. Sci. USA. 2015;112:E4104–E4110. doi: 10.1073/pnas.1506264112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ostro B., Hu J., Goldberg D., Reynolds P., Hertz A., Bernstein L., Kleeman M.J. Associations of mortality with long-term exposures to fine and ultrafine particles, species and sources: results from the California Teachers Study Cohort. Environ. Health Perspect. 2015;123:549–556. doi: 10.1289/ehp.1408565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sebastiani P., Thyagarajan B., Sun F., Schupf N., Newman A.B., Montano M., Perls T.T. Biomarker signatures of aging. Aging Cell. 2017;16:329–338. doi: 10.1111/acel.12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrucci L., Hesdorffer C., Bandinelli S., Simonsick E.M. Frailty as a nexus between the biology of aging, environmental conditions and clinical geriatrics. Public Health Rev. 2010;32:475. doi: 10.1007/BF03391612. [DOI] [Google Scholar]
- 16.Bell B., Rose C.L., Damon A. The Veterans Administration longitudinal study of healthy aging. Gerontologist. 1966;6:179–184. doi: 10.1093/geront/6.4.179. [DOI] [PubMed] [Google Scholar]
- 17.Houseman E.A., Accomando W.P., Koestler D.C., Christensen B.C., Marsit C.J., Nelson H.H., Wiencke J.K., Kelsey K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 19.Breitling L.P., Salzmann K., Rothenbacher D., Burwinkel B., Brenner H. Smoking, F2RL3 methylation, and prognosis in stable coronary heart disease. Eur. Heart J. 2012;33:2841–2848. doi: 10.1093/eurheartj/ehs091. [DOI] [PubMed] [Google Scholar]
- 20.Benayoun B.A., Pollina E.A., Brunet A. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat. Rev. Mol. Cell Boil. 2015;16:593. doi: 10.1038/nrm4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao X., Zhang Y., Breitling L.P., Brenner H. Relationship of tobacco smoking and smoking-related DNA methylation with epigenetic age acceleration. Oncotarget. 2016;7:46878. doi: 10.18632/oncotarget.9795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beach S.R., Dogan M.V., Lei M.K., Cutrona C.E., Gerrard M., Gibbons F.X., Simons R.L., Brody G.H., Philibert R.A. Methylomic aging as a window onto the influence of lifestyle: tobacco and alcohol use alter the rate of biological aging. J. Am. Geriatr. Soc. 2015;63:2519–2525. doi: 10.1111/jgs.13830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Philibert R.A., Beach S.R., Lei M.-K., Brody G.H. Changes in DNA methylation at the aryl hydrocarbon receptor repressor may be a new biomarker for smoking. Clin. Epigenetics. 2013;5:19. doi: 10.1186/1868-7083-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeilinger S., Kühnel B., Klopp N., Baurecht H., Kleinschmidt A., Gieger C., Weidinger S., Lattka E., Adamski J., Peters A. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS ONE. 2013;8:e63812. doi: 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monick M.M., Beach S.R., Plume J., Sears R., Gerrard M., Brody G.H., Philibert R.A. Coordinated changes in AHRR methylation in lymphoblasts and pulmonary macrophages from smokers. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2012;159:141–151. doi: 10.1002/ajmg.b.32021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alarcón M., Abrahams B.S., Stone J.L., Duvall J.A., Perederiy J.V., Bomar J.M., Sebat J., Wigler M., Martin C.L., Ledbetter D.H. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am. J. Hum. Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji W., Li T., Pan Y., Tao H., Ju K., Wen Z., Fu Y., An Z., Zhao Q., Wang T. CNTNAP2 is significantly associated with schizophrenia and major depression in the Han Chinese population. Psychiatry Res. 2013;207:225–228. doi: 10.1016/j.psychres.2012.09.024. [DOI] [PubMed] [Google Scholar]
- 28.Friedman J., Vrijenhoek T., Markx S., Janssen I., Van Der Vliet W., Faas B., Knoers N., Cahn W., Kahn R., Edelmann L. CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol. Psychiatry. 2008;13:261. doi: 10.1038/sj.mp.4002049. [DOI] [PubMed] [Google Scholar]
- 29.Yasuda K., Miyake K., Horikawa Y., Hara K., Osawa H., Furuta H., Hirota Y., Mori H., Jonsson A., Sato Y. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat. Genet. 2008;40:1092. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- 30.Larson K.C., Draper M.P., Lipko M., Dabrowski M. Gng12 is a novel negative regulator of LPS-induced inflammation in the microglial cell line BV-2. Inflamm. Res. 2010;59:15–22. doi: 10.1007/s00011-009-0062-2. [DOI] [PubMed] [Google Scholar]
- 31.Yang J., Wang Y., Cai Y., Yang Z., Wu G., Wen J., Luo X., Huang H. preparation of rabbit anti-rat Lrrn3 polyclonal antibody and study of its expression****⋆. Neural Regen. Res. 2010;5:1030–1034. [Google Scholar]
- 32.Oshima M., Endoh M., Endo T.A., Toyoda T., Nakajima-Takagi Y., Sugiyama F., Koseki H., Kyba M., Iwama A., Osawa M. Genome-wide analysis of target genes regulated by HoxB4 in hematopoietic stem and progenitor cells developing from embryonic stem cells. Blood. 2011;117:e142–e150. doi: 10.1182/blood-2010-12-323212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelada S.N., Eaton D.L., Wang S.S., Rothman N.R., Khoury M.J. The role of genetic polymorphisms in environmental health. Environ. Health Perspect. 2003;111:1055–1064. doi: 10.1289/ehp.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Autrup H. Genetic polymorphisms in human xenobiotica metabolizing enzymes as susceptibility factors in toxic response. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2000;464:65–76. doi: 10.1016/S1383-5718(99)00167-9. [DOI] [PubMed] [Google Scholar]
- 35.Jones M.J., Goodman S.J., Kobor M.S. DNA methylation and healthy human aging. Aging Cell. 2015;14:924–932. doi: 10.1111/acel.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersen A., Dogan M., Beach S., Philibert R. Current and future prospects for epigenetic biomarkers of substance use disorders. Genes. 2015;6:991–1022. doi: 10.3390/genes6040991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ladd-Acosta C., Fallin M.D. The role of epigenetics in genetic and environmental epidemiology. Epigenomics. 2016;8:271–283. doi: 10.2217/epi.15.102. [DOI] [PubMed] [Google Scholar]
- 38.Martino D., Loke Y.J., Gordon L., Ollikainen M., Cruickshank M.N., Saffery R., Craig J.M. Longitudinal, genome-scale analysis of DNA methylation in twins from birth to 18 months of age reveals rapid epigenetic change in early life and pair-specific effects of discordance. Genome Boil. 2013;14:R42. doi: 10.1186/gb-2013-14-5-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shenker N.S., Polidoro S., van Veldhoven K., Sacerdote C., Ricceri F., Birrell M.A., Belvisi M.G., Brown R., Vineis P., Flanagan J.M. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum. Mol. Genet. 2012;22:843–851. doi: 10.1093/hmg/dds488. [DOI] [PubMed] [Google Scholar]
- 40.Besingi W., Johansson Å. Smoke-related DNA methylation changes in the etiology of human disease. Hum. Mol. Genet. 2013;23:2290–2297. doi: 10.1093/hmg/ddt621. [DOI] [PubMed] [Google Scholar]