Abstract
Alu retroelements, whose retrotransposition requires prior transcription by RNA polymerase III to generate Alu RNAs, represent the most numerous non-coding RNA (ncRNA) gene family in the human genome. Alu transcription is generally kept to extremely low levels by tight epigenetic silencing, but it has been reported to increase under different types of cell perturbation, such as viral infection and cancer. Alu RNAs, being able to act as gene expression modulators, may be directly involved in the mechanisms determining cellular behavior in such perturbed states. To directly address the regulatory potential of Alu RNAs, we generated IMR90 fibroblasts and HeLa cell lines stably overexpressing two slightly different Alu RNAs, and analyzed genome-wide the expression changes of protein-coding genes through RNA-sequencing. Among the genes that were upregulated or downregulated in response to Alu overexpression in IMR90, but not in HeLa cells, we found a highly significant enrichment of pathways involved in cell cycle progression and mitotic entry. Accordingly, Alu overexpression was found to promote transition from G1 to S phase, as revealed by flow cytometry. Therefore, increased Alu RNA may contribute to sustained cell proliferation, which is an important factor of cancer development and progression.
Keywords: Alu retrotransposons, cell cycle, non-coding RNA
1. Introduction
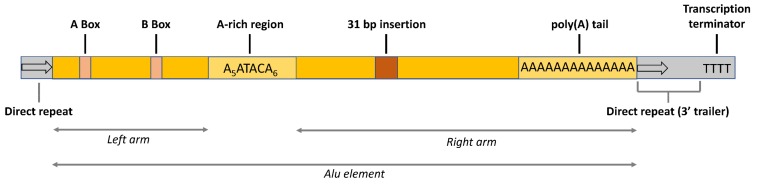
As much as 10% of the human genome is composed of Alu elements, which are highly repetitive retrotransposons belonging to the class of the short interspersed nuclear elements (SINEs), and count for a total of more than one million copies in the whole set of the human chromosomes [1]. It is thought that Alu sequences originated 65 million years ago from the retrotransposition of the 7SL RNA, an event that coincides with the radiation of primates [2,3]. During their amplification, Alu sequences accumulated base substitutions that led to their classification into three subfamilies: the oldest and the intermediate age AluJ and AluS subfamilies, which are no longer retrotranspositionally active, and the youngest AluY subfamily, which is still able to retrotranspose in germ cell lines [4]. Alu retrotransposition depends on non-LTR retroelements LINE-1 (L1)-encoded ORF1p and ORF2p proteins, in order to reintegrate in the genome via a target-primed reverse transcription mechanism. The exact process used by Alu retroelements to target the genome is unknown, but there is strong evidence that Alu retrotransposition is biased towards gene-rich regions [5], both at intergenic loci and at intragenic positions. Possible targets of gene regions are represented by 5′ and 3′ untranslated regions (5′ UTRs and 3′ UTRs) and by introns of protein-coding genes, with a non-random distribution according to gene functional categories [6]. The consensus Alu sequence is about 300 nucleotides in length and is thought to derive from the head to tail fusion of two distinct 7SL RNA genes [7]. The dimeric Alu sequence is composed of a left arm, which harbors the A and B boxes derived from the 7SL RNA polymerase III (Pol III) promoter, and a right arm, which has an additional 31-bp insertion. The left and the right arms are separated by an intermediate A-rich consensus sequence (A5TACA6) and the element ends with a relatively long poly(A) tail (Figure 1). The 3′-trailer region between the Alu poly(A) tail and the first encountered termination signal (a run of at least four Ts or a T-rich non-canonical terminator) is unique to each individual Alu RNA. The potential mutagenic effect that could arise from the frequent insertion of Alu elements during their amplification in primates, Alu’s highly repetitive nature, the lack of a protein-coding potential, and low levels of transcription mainly due to epigenetic silencing, led to Alu elements being referred to as “parasites” of the human genome. However, this hypothesis does not explain the lack of negative selection during evolution, or why Alu elements are maintained at such a high copy number in the human genome. These features instead suggest the possibility that Alus could play important regulatory roles. Indeed, currently there is evidence for the involvement of Alus in a multitude of gene regulatory processes through cis and trans mechanisms. Cis mechanisms rely on (i) the insertion of new transcription factor binding sites that are present in Alu sequences, influencing the expression of genes involved in differentiation and development [8], (ii) the influence of intragenic Alus on pre-mRNA splicing [9], (iii) the evolution of Alu elements into new enhancers, influencing the expression of genes that are far away in the genome [10], and (iv) genomic rearrangements that could arise from Alu insertion, which usually lead to the development of disease [11]. Alu sequences can also influence gene regulation and other processes in trans, due to the ability of Alu transcripts to (i) bind RNA polymerase II (Pol II) and inhibit transcription initiation [12], (ii) regulate mRNA nuclear export via a p54nrb protein (also known as “Nono”) [13,14], (iii) influence translation by binding to the SRP9/14 subunit of the signal recognition particle (SRP) [15], and (iv) activate the NLRP3 inflammasome [16]. Additionally, Alu RNA sequences embedded in longer transcripts may exert other effects, such as the induction of ADAR-dependent RNA editing of mRNAs that carry Alu inverted repeats [17,18], the alteration of translation efficiency by base-pairing of inverted Alu repeats in the 3′ UTR of mRNA genes [19], the stimulation of circRNA biogenesis by backsplicing [20,21], and the control of nuclear localization of long non-coding RNAs [22]. It is known that, in physiological cell conditions, Alu elements are epigenetically silenced [23] and their expression is dramatically increased following different types of cell stress, such as virus infection [24], heat shock [25], cancer progression [26], epithelial to mesenchymal transition [27], and the age-related macular degeneration [28], supporting the hypothesis that Alu RNA may play important roles in both physiological and pathological contexts. However, it is not clear if Alu overexpression functions to overcome the stress condition, or if the increase in Alu RNA is a mere functionally irrelevant consequence of a global change in genome transcription. In spite of the different functions that Alu RNAs may have in gene regulation and other processes, a clear and comprehensive picture of the impact of Alu RNAs on cell growth and proliferation is still lacking. Recently, Di Ruocco et al. [27] found no perturbation of cell cycle distribution of colorectal cancer cells in response to the transient transfection of a synthetic Alu. On the other hand, Sakamoto et al. [29] previously found that HeLa cell growth was inhibited after the transient transfection of endogenous and synthetic Alu sequences, and Baryakin et al. [30] showed that the transient transfection of an Alu-RNA analogue suppresses cell proliferation and induces apoptosis of breast cancer cells. In line with this, Hu et al. [31] reported an inhibition of stem cell proliferation after the microinjection of an AluSx overexpressing vector in Ntera2 cells; Morales-Hernandèz et al. [32] showed that the transient transfection of the NANOG Alu sequence (localized upstream of the NANOG gene) repressed the expression of the pluripotency genes OCT4 and NANOG in Ntera cells; and Castelnuovo et al. [33] reported a differentiation-promoting effect of an Alu-like RNA in neuroblastoma cells. These observations suggest that Alu RNA might exert an inhibitory effect on cell proliferation; it has to be noted, however, that in these studies Alu RNAs were overexpressed through transient transfection and their effects were analyzed in tumor or stem cells, in which the expression of cell cycle genes is already skewed toward proliferation.
Figure 1.
Sequence features of a consensus Alu element. The left arm harbors the internal Pol III promoter, composed of the A Box and B Box. The right arm has a 31-bp insertion. The two arms are separated by an A-rich region and the entire Alu element ends with a poly(A) tail. Grey indicates the genomic repeats that originate from the retrotransposition event and the 3′ trailer sequence between the poly(A) tail and the canonical terminator (at least four Ts).
As an additional step to understand the relationship between Alu expression and cell proliferation, the present study shows that the stable overexpression of two Alu elements of the AluS subfamily shows no effect in HeLa cells, but that it causes a significant upregulation of cell cycle-promoting genes in primary human fibroblasts, suggesting a function for Alu elements in regulating the cell cycle of normal cells.
2. Results
2.1. Alu Sequences, Vectors, and Cell Lines for Overexpression
The main questions we addressed in this study were whether and how an increased level of Alu RNA, derived from transcription of one or more Alu elements, affected the protein-coding transcriptome in human cell lines. HeLa cells were chosen as a model for tumor cells and IMR90 primary fetal lung fibroblast as a model for non-tumor, normal human cells. Alu transcripts are generally extremely similar in their sequence and thus expected to induce very similar functional consequences upon overexpression. We selected two individual Alu elements that we found reproducibly expressed in both tumor and non-tumor cells: AluSq2 and AluSx. Both of these Alus belong to the intermediate-age subfamily AluS, whose members are thought to be inactive for retrotransposition [4]. AluSq2 is antisense to the first intron of the gene NFIA and is expressed in five cell lines (H1-hESC, HeLa-S3, Hep G2, K562, NHEK) according to single-locus Alu expression profiling using ENCODE RNA-Seq data [34]. This Alu element lacks the internal A-rich motif A6TACA5, which is replaced by A3G. AluSx is antisense to the first intron of the AMFR gene and is expressed in NHEK cells [34]. A Control RNA sequence was amplified by PCR from the Escherichia coli LacZ gene, a sequence completely unrelated to any sequence in the human genome, while having a similar GC distribution and content as the two Alus (61% GC in AluSq2, 53% GC in AluSx, 61% GC in the Control sequence). For information on the genomic coordinates of the overexpressed Alu sequences and on the Alus and Control complete nucleotide sequence, see the Supplementary Materials. AluSq2, AluSx, and the Control RNA were inserted into a lentiviral vector under the control of the H1 promoter, an upstream-located (type 3) Pol III promoter that directs the expression of the RNase P RNA (H1 RNA) and is widely used for non-coding RNA (ncRNA) overexpression studies [35] (Figure 2). We chose this strategy because, despite the high expression levels generally observed in vitro for Alu elements carrying canonical A and B boxes, transfected Alu elements with the sole internal promoter usually give barely detectable transcription [36,37], while the H1 promoter was previously shown to drive high Alu transcription in transfected cells [38]. In order to select stable integrants and to directly monitor gene expression from the lentivirus vector, the puromycin resistance and eGFP genes, under the control of the PGK Pol II-dependent promoter, were present in the same vectors. Normal IMR90 human lung fibroblasts and HeLa cells were transformed with the lentivirus constructs carrying AluSq2, AluSx, and the Control RNA sequence, as well as empty vector, and stable integrants were isolated by puromycin selection. No major alteration in cell growth and morphology was observed during cell culture (see Figure S1, Supplementary Materials).
Figure 2.
Schematic representation of the DNA inserted by the lentivirus vector used to generate IMR90 and HeLa cells that stably overexpress Alu sequences. The Pol III H1 promoter (H1p) controls the expression of AluSq2, AluSx, or a Control RNA. An empty vector was used as negative control. The eGFP gene and the gene coding for puromycin resistance are inserted under the control of the PGK gene promoter (PGKp).
2.2. Validation of Alu Overexpression
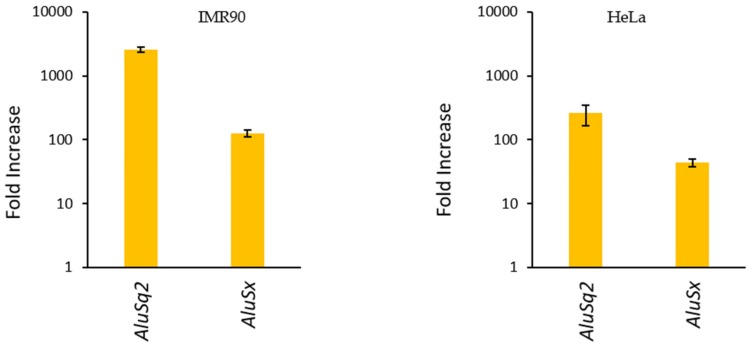
The overexpression of AluSq2, AluSx, and Control RNA in stable integrant IMR90 and HeLa cells was evaluated by RT-qPCR. Since Alu elements are numerically abundant and repetitive, we used primers targeting the unique trailer regions of AluSq2 and AluSx in order to unambiguously detect the transcripts of the transformed elements. As shown in Figure 3, in IMR90 the expression levels of both Alus were increased by 2 (AluSx) or 3 (AluSq2) orders of magnitude with respect to cells transformed with the empty vector, which were used as background control. Overexpression also occurred in HeLa cells, albeit to a lower extent (≈250-fold increase for AluSq2, ≈45-fold increase for AluSx). The lower increase in this case is likely due to higher background expression levels of the two Alus in this tumor cell line. Alu RNA levels were normalized with the expression of the control gene U1 snRNA, which is the most abundant snRNA with ≈1 × 106 copies/cell [39].
Figure 3.
Alu overexpression from lentivirus vector stably inserted in the genome. The bar plots report the fold increase of AluSq2 and AluSx expression in either (left panel) IMR90 or (right panel) HeLa cells that were stably transformed with a lentivirus vector carrying the corresponding Alu. The fold increase was derived from comparisons with cells transformed with the empty vector (background Alu expression). Alu RNA levels were quantified by RT-qPCR analysis, conducted with primers chosen to target unique sequence tracts within the Alu 3′ trailer region. In all measurements, gene expression levels were normalized to U1 gene expression, used as an internal standard.
2.3. Differential Gene Expression Analysis
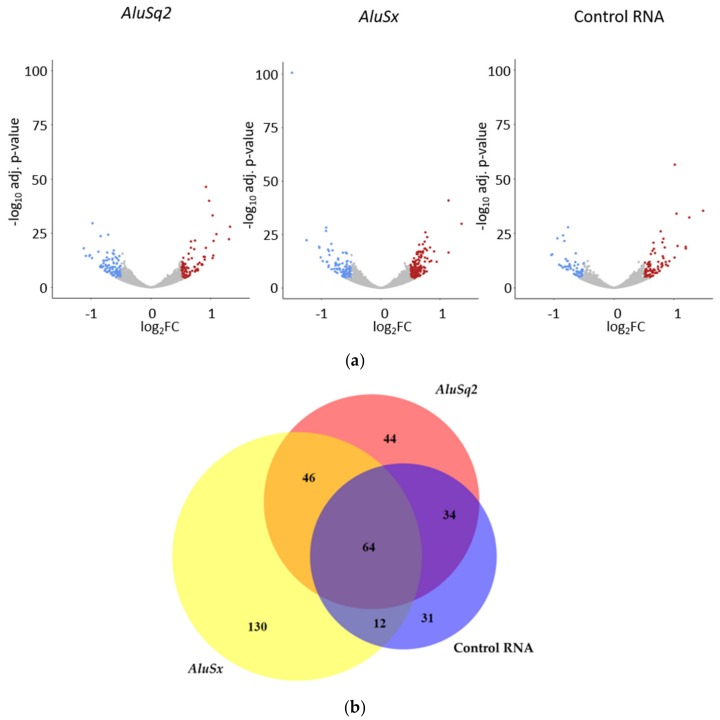
To study Alu-dependent alteration of the protein-coding transcriptome, total RNA was extracted from Alu-overexpressing and Control-transformed IMR90 and HeLa cells and subjected to poly(A) enrichment for RNA-Seq analysis. Differential expression analysis (|log2FC|>0.5, adjusted p-value < 0.001) was then performed on RNA-Seq outputs by comparing cells overexpressing the gene of interest (AluSq2, AluSx, or Control RNA) with cells carrying the empty vector. In the case of IMR90, using DESeq2 [40] a total of 87 upregulated and 101 downregulated genes were found in AluSq2-overexpressing cells, 147 upregulated and 105 downregulated genes in AluSx-overexpressing cells, and 86 upregulated and 55 downregulated genes in cells stably overexpressing the Control RNA. The distribution of up- and downregulated genes in these samples is illustrated by the volcano plots in Figure 4a. The Venn diagram in Figure 4b shows the intersection of genes dysregulated in AluSq2-, AluSx- and Control RNA-overexpressing IMR90 cells. Remarkably, in contrast with IMR90 results, zero and two differentially expressed genes were detected in HeLa cells upon overexpression of AluSq2 or AluSx, respectively (for a complete list of differentially expressed genes in IMR90 and HeLa cells, see Table S1, Supplementary Materials).
Figure 4.
Differentially expressed genes in Alu/Control RNA-overexpressing cells. (a) Volcano plots showing the distribution of the differentially expressed genes in human fibroblasts overexpressing AluSq2, AluSx, or the Control RNA. Blue spots represent downregulated genes, red spots represent upregulated genes. (b) Venn diagram showing the intersection of genes that are differentially expressed in AluSq2, AluSx, and Control RNA-overexpressing cells.
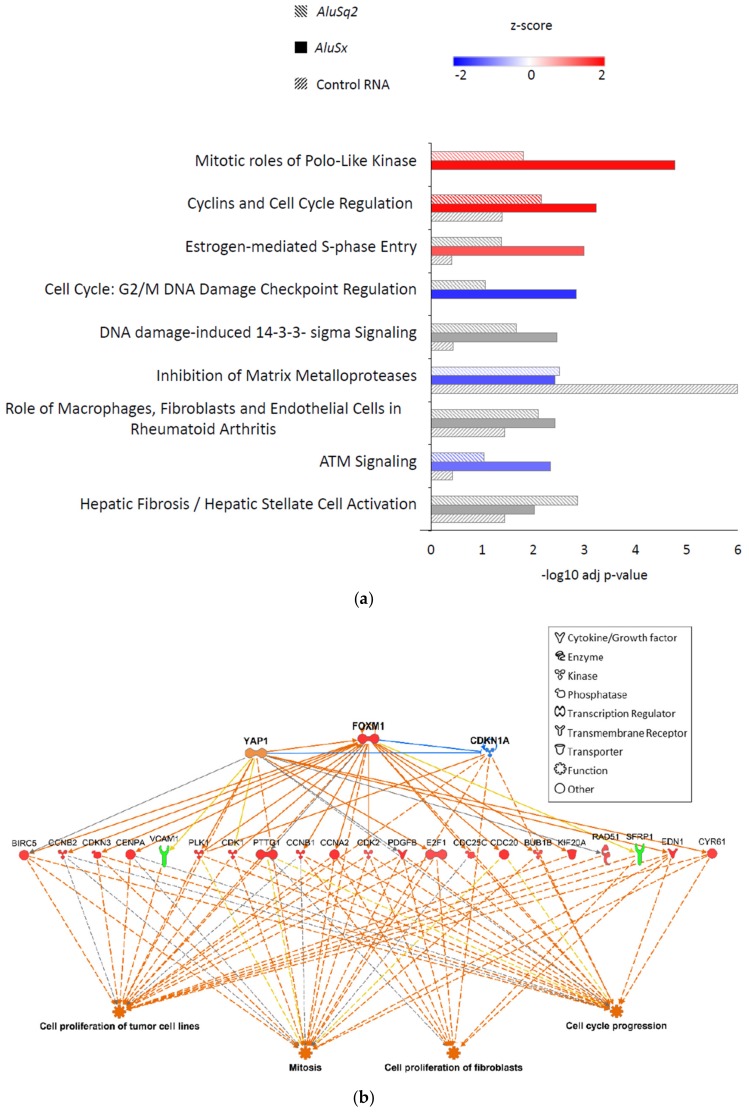
In order to understand which pathways are dysregulated in Alu- or Control-overexpressing IMR90 cells, we performed an enrichment analysis using the Ingenuity Pathway Analysis software (IPA, version number 48207413, Qiagen, Hilden, Germany). As shown in Figure 5a, several pathways related to the cell cycle (mitotic roles of Polo-like kinase, cyclins and cell cycle, estrogen-mediated S phase entry, G2/M DNA damage checkpoint) emerged as significantly enriched in AluSq2- and/or AluSx-overexpressing fibroblasts, with a general trend toward a higher significance in the case of AluSx with respect to AluSq2, but not in Control RNA datasets. In IMR90 cells overexpressing the Control RNA, only the pathway “Inhibition of Matrix Metalloproteases“ resulted in significantly enriched (adjusted p-value < 0.01). This effect was not investigated further. The Alu-specificity of the effect on cell cycle pathways excludes the possibility of gene expression perturbation trivially due to abnormal levels of an exogenous RNA sequence. Interestingly, pathways involved in a positive progression of cell cycle are detected as activated (red bars), whereas pathways that induce an arrest in cell cycle progression are detected as inhibited (blue bars), suggesting a role of Alu RNA in positively regulating cell cycle progression.
Figure 5.
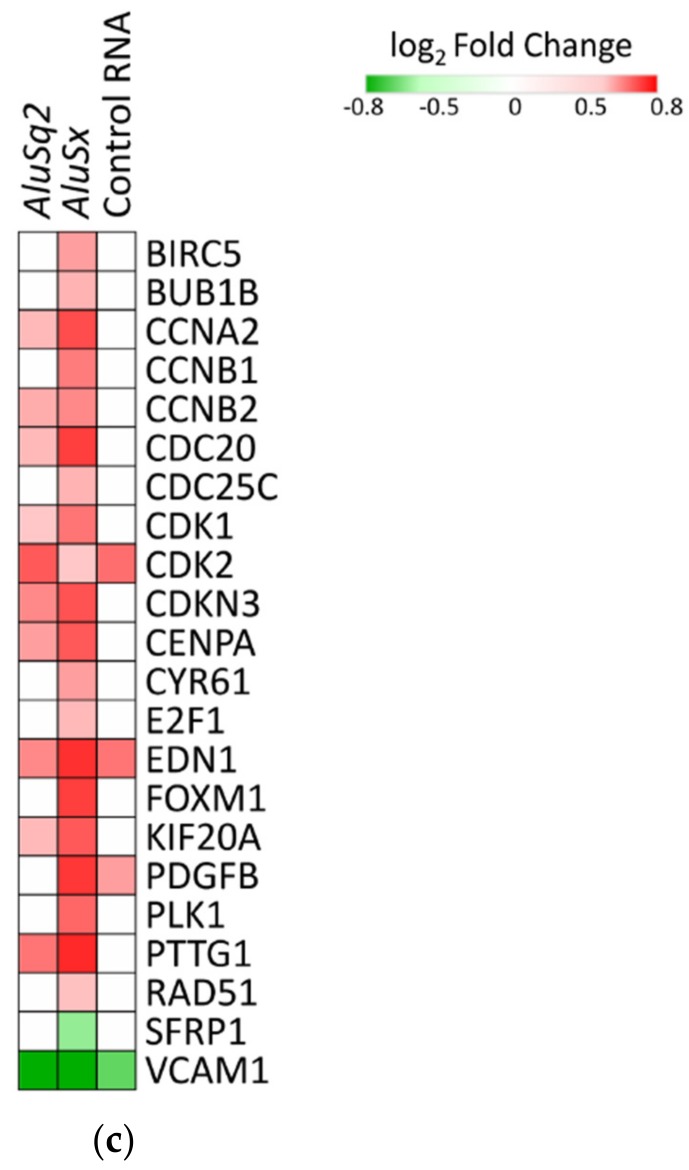
Dysregulation of cell cycle genes in Alu-overexpressing fibroblasts. (a) Pathways enriched in differential transcriptomes of AluSq2, AluSx, and Control RNA transformed fibroblasts as revealed by IPA analysis. Only pathways with p-value (BH correction) <0.01 in at least one comparison are shown and are ordered by AluSx vs. empty vector p-value. Z-score for activated or inhibited pathways is shown in red or blue, respectively. Grey bars: no predictions can be made. (b) Prediction of the upstream regulators that could modulate the expression of differentially expressed genes in AluSx-overexpressing fibroblasts and their effect on cell functions. Dysregulated genes are shown in the middle row as upregulated genes (red symbols) and downregulated genes (green symbols). Regulators are shown in the upper part of the figure. Blue indicates a predicted inhibition of the protein activity, while orange indicates a predicted activation. Blue lines indicate an inhibitory relationship, orange lines show an activating relationship, yellow lines indicate inconsistent relationship, while gray lines stand for no predicted effect. Continuous lines show direct interactions and dashed lines show indirect interactions (less than three passages). (c) Heatmap of differentially expressed genes shown in (b).
In order to predict which molecular species hypothetically influence the dysregulated expression patterns detected in Figure 5a, differentially expressed genes were analyzed for their enrichment in upstream regulators using the IPA software. This analysis is based on literature knowledge about the effects of regulators on gene expression. Therefore, any molecular species that can affect gene expression (i.e., transcription factors, microRNAs, kinases, compounds, or drugs) is taken into account. The literature-documented interaction between the upstream regulator and its target gene may have a direct role (direct interaction) in gene expression or can be involved in a cascade of upstream regulators (indirect interaction). Similarly, the downstream phenotypic effects can be inferred from the differentially expressed genes.
We focused our upstream regulator analysis on transcription factors and regulatory kinase systems known to be involved in the control of mitosis, and applied it to AluSx-dysregulated gene expression profiles. The transcriptional co-activator YAP1, the transcription factor FOXM1, and the cyclin-dependent kinase inhibitor 1A (CDKN1A) were predicted to participate in the control of the expression of dysregulated genes, as revealed by the upstream regulator analysis performed in IPA. Figure 5b shows a network describing the relationship among the most significant upstream regulators, the differentially expressed genes detected in AluSx overexpressing cells, and the inferred effects on cell proliferation of tumor cell lines, mitosis, cell proliferation of fibroblasts, and cell cycle progression. The dysregulated genes belong to CDC proteins, cyclins (CCNB2, CCNB1, CCN1), cyclin-dependent kinases (CDKs), proteins involved in mitotic spindle assembly and chromosome segregation (CENPA, KIF20A, PLK1, BUB1B), and soluble signaling molecules whose dysregulated expression is related to tumorigenesis (CYR61, EDN1, SFRP1, PDGFB). The case of FOXM1 is particularly interesting, since it is detected as an upregulated gene in our dataset as well as predicted as an upstream regulator.
Figure 5c shows the regulation of the differentially expressed genes that are predicted to be controlled by YAP1, FOXM1, and CDKN1A. Of note, eleven of these genes are also found dysregulated in AluSq2; in contrast, we could only detect four differentially expressed genes in the Control RNA-overexpressing cells. Overall, the results of IPA analyses support the hypothesis that AluSx modulates the transcription of cell cycle genes by acting through different mechanisms on key regulators such as YAP1, FOMX1, and CDKN1A.
2.4. Alu RNA Promotes IMR90 Cell Cycle Progression
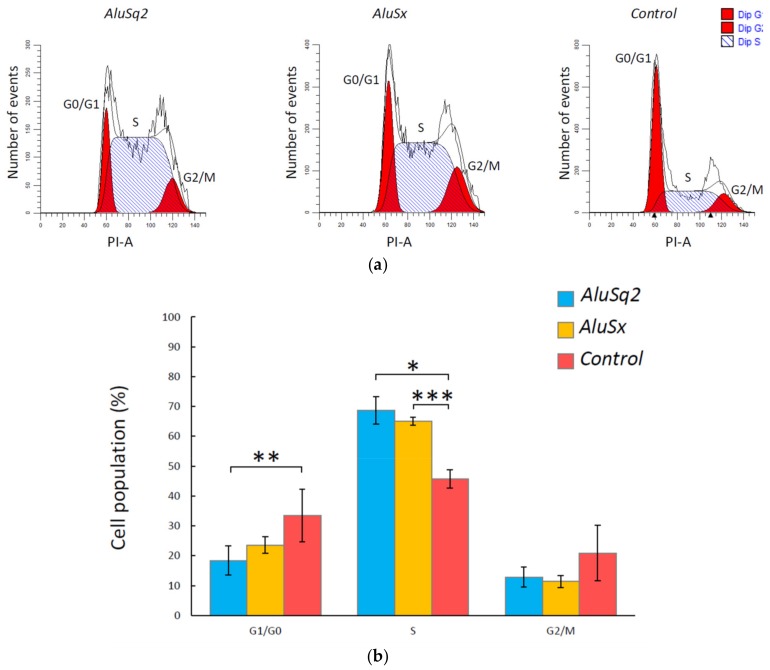
We sought to experimentally assess whether cell cycle progression is affected by Alu RNA-dependent activation of cell cycle pathways. IMR90 cells overexpressing AluSq2, AluSx, or Control RNA were synchronized by serum starvation for 24 h, which resulted in 83 ± 2% of cells in G1 and 8% of cells in S phase, based on propidium iodide (PI) staining. After cell culturing in the starvation medium, serum was re-added and cell cycle distribution analysis was carried out after 24 h by flow cytometry. As shown in Figure 6a, reporting the results from several experiments carried out with two independently transformed IMR90 cell cultures, a significantly higher percentage of Alu-overexpressing cells were in S phase (and correspondingly, less were in G1) with respect to cells overexpressing the Control RNA. This effect was slightly more marked in the case of AluSq2, with 18% and 69% of cells in G1 and S phase, respectively, whereas 33% and 46% of cells in G1 and S phase, respectively, were observed in the Control RNA-overexpressing cells (Figure 6b). The data thus confirm that increased levels of Alu RNA promote cell cycle progression in IMR90 primary fibroblasts.
Figure 6.
AluSq2 and AluSx stimulate cell cycle progression of IMR90 cells. (a) FACS analysis of synchronized IMR90 cells after 24 h of serum re-feeding. ModFit LT analyses revealed an accumulation of AluSq2- and AluSx-overexpressing IMR90 cells in S phase. (b) Bar plot data are derived from at least four independent experiments obtained from two IMR90 cell cultures transformed with a lentivirus vector. * p < 0.05, ** p < 0.01, *** p < 0.0 01 compared to the Control.
2.5. Location Analysis of Alu Elements at Differentially Expressed Loci
As a preliminary attempt to address the mechanism(s) of Alu RNA-dependent alteration of mRNA profiles, we checked for the presence of Alu elements in the promoter (2 kb upstream of the transcription start site (TSS)), in the 5′ UTR and in the 3′ UTR of differentially expressed protein-coding genes. A general trend towards an increase in the number of Alus in the 3′ UTR of downregulated genes and a decrease in the number of Alus in the 3′ UTR of upregulated genes was evident (Table 1). Compared to the total number of protein-coding genes, this effect was barely significant in the case of AluSx, while it was significant at the Fisher exact test for AluSq2-overexpressing IMR90 cells. When the orientation of Alus was taken into account, we could detect an increase of Alu sequences transcribed in an antisense orientation relative to up- and downregulated genes (AluSq2) or only to downregulated ones (AluSx) (Table 1). For a comprehensive list of all the Alu sequences in each differentially expressed gene, see Table S2, Supplementary Materials.
Table 1.
Analysis of the presence of Alu sequences in the 3′ UTR of up- and downregulated genes in IMR90 cells overexpressing AluSq2 or AluSx. Only the Alu sequences that were at least 100-bp long and fully overlapped with the 3′ UTR of target transcripts were considered in the analysis.
| Sample | Genes with Alu in 3′ UTR | Total Number of Analyzed Genes | p-Value (Fisher exact test) | % of Genes with an Alu Element in Their 3′ UTR | % of Anti-sense Alu in Each Gene (Average) |
|---|---|---|---|---|---|
| Genome | 4838 | 19,836 | – | 24.39 | 49.22 |
| AluSq2_Up | 9 | 87 | 0.0005 | 10.34 | 66.67 |
| AluSq2_Down | 37 | 101 | 0.0019 | 36.63 | 55.63 |
| AluSx_Up | 25 | 147 | 0.0082 | 17.01 | 30.00 |
| AluSx_Down | 28 | 105 | 0.0761 | 26.67 | 68.45 |
To verify the possibility of a mechanistic connection of Alu-dependent upregulation with miRNA control, we searched for the presence of miRNA binding sites in AluSq2 and AluSx. We were able to detect only the miRNA binding site for the sequence hsa-mir-619-5p, which was also predicted to target twelve and eleven differentially expressed genes in IMR90 cells overexpressing AluSq2 and AluSx, respectively. In the latter one, the upregulated NCAPD2 is the only gene belonging to the cell cycle (Reactome pathway R-HSA-1640170) (Table S3, Supplementary Materials).
3. Discussion
Our experiments support a positive role of increased Alu expression specifically in the proliferation of primary cells, but not of tumor cells. While the effects of Alu overexpression in primary differentiated cells have never been tested before, its effects on tumor cell proliferation have been addressed by several previous studies, with contrasting results. An early study based on the transient transfection of HeLa cells with Alu-carrying plasmids showed an Alu-dependent inhibition of cell proliferation, yet this effect could not be demonstrated to be caused by Alu RNA [29]. More recently, Alu and 7SL RNA transfection into MCF7 breast adenocarcinoma cells was found to entail a decrease in viability and an induction of pro-apoptotic changes [30]. In another recent study, Alu RNA transfection into the SW480 colorectal cancer cell line was instead found to be without effect on cell viability and cell cycle distribution, yet to induce epithelial-to-mesenchymal transition [27]. In other studies, more subtle effects of the expression of specific Alus were observed in human cells of embryonic origin. In particular, individual or specific subsets of Alu elements were shown to be activated in response to dioxin receptor AHR signaling [32] or to retinoic acid [31], with the production of Alu-derived transcripts inhibiting proliferation and promoting differentiation of human embryonic teratocarcinoma cells (NTera2) or human embryonic stem cells, probably through microRNA-like pathways. Taken together, the results of these studies delineate a complex scenario where cell lineage, differentiation state, and transformation degree are all relevant factors, perhaps together with the peculiar properties of individual Alus, in determining cell behavior in response to Alu overexpression. Within this framework, the most original contribution of our study consists in the use of primary cells (IMR90) and of stable transformation through lentiviral vectors to demonstrate that Alu RNAs can modulate the protein-coding transcriptome so as to promote cell cycling. IMR90 fibroblasts served recently as a good model for oncogenic transformation, also including early events in this process [41,42]. Therefore, the observation that Alu overexpression in these cells reprograms to some extent genome expression towards cell cycling appears as particularly relevant, as it suggests that increased Alu expression may be among the ways through which early oncogenic stimuli exert their effect. Along this line, the lack of effect of Alu overexpression on cell cycling or other pathways in HeLa cells may be due to the transformed state of these cells, with cell cycle and other pathways being already dysregulated as a consequence of this state. With this respect, our observations are in agreement with the recent observation that Alu RNA overexpression does not affect cell proliferation in another carcinoma cell line, SW480 [27]. A recent study reported that Alu transcription in human skin fibroblasts is suppressed in response to serum stimulation, via serum-dependent relocation to Alu of CGGBP1 followed by Pol III dislodging [43]. Since serum stimulation is expected to promote cell cycling, this study establishes a negative correlation between cell proliferation and Alu expression, in contrast with our observation that increased Alu RNA favors cell cycling. The study by Agarwal et al. [43], however, was based on serum starvation/stimulation, known to be complex signal sources producing wide reprogramming of epigenetic and transcriptional profiles, thus making it difficult to unravel the causal relationships between the different phenomena occurring in response to them [44]. Moreover, the causal involvement of Alu RNA in this study is based on the observation that CGGBP1 depletion entails increased Alu RNA levels as measured by RT-qPCR, and this is reflected by higher Alu RNA levels in quiescent cells than in serum-stimulated cells, again as revealed by RT-qPCR. From these observations, the authors infer that CGGBP1 relocation to Alus upon serum stimulation downregulates Alu expression. It should be noted, however (as the authors do), that the Alu RNA detected by this method could in large part be due to Alu sequences that are part of longer, RNA polymerase II-synthesized transcripts. It is thus uncertain whether the phenomena described in this study are causally related to general increase/decrease of genuine, Pol III-dependent Alu transcripts.
Given our experimental protocol, and based on the stable integration of an efficient Alu RNA-expressing cassette into the genome, the observed effects are most likely due to the activity in trans of Alu RNA (even though in cis effects of newly genome-integrated Alu cannot be formally excluded). A key unsolved question put forward by these observations is the mechanisms by which Alu RNAs lead to changes in the levels of specific mRNA subsets in IMR90 cells.
Alu RNA has previously been shown to function as a Pol II-interacting transcriptional repressor during the cellular heat shock response, thus contributing to downregulation of housekeeping genes (but not of other genes) under these conditions [25]. Our results are not inconsistent with the possibility that the overexpressed Alu elements in IMR90 cells could form Alu RNA-Pol II complexes, changing Pol II propensity to act at different subsets of promoters.
As an alternative mechanism, mRNA upregulation could in principle be due to Alu RNA-dependent stabilization of mRNAs. Since regulatory ncRNAs that directly target mRNAs (such as miRNAs) generally cause mRNA destabilization, upregulation by Alu RNA could mechanistically rely on neutralization of mRNA-destabilizing ncRNAs (e.g., by miRNA sponging, [16]). Another possibility is related to the ability of free Alu RNA to interfere with Staufen-mediated mRNA decay, which is known to be favored by Alu sequences present in the 3′ UTR of mRNAs. For example, free Alu RNAs could saturate anti-Alu sequences carried by long non-coding RNAs that normally target mRNA for Staufen-mediated decay [45]. On the other hand, downregulation of protein-coding genes could be caused by RNA-duplexing of free Alu RNA with Alu sequences embedded in the 3′ UTR of mRNAs, inducing mRNA degradation mediated by the RNA-induced silencing complex (RISC), as suggested from our analyses in the case of downregulated genes in AluSx-overexpressing fibroblasts.
As a factor further complicating the landscape of the Alu RNA mechanism of action, it is not known whether the effects that we observe on differentially expressed genes are due to full-length Alu RNA or to shorter processing products, nor whether important interactions of the overexpressed Alu RNA take place in the nucleus or in the cytoplasm. Difficulties in discussing this point derive from the fact that almost nothing is known about the biogenesis and processing pathways of Alu RNAs, except from the proposed role of Dicer1 in Alu RNA processing, which may result in the production of small regulatory RNAs potentially acting as mRNA destabilizers [31].
To what extent the Alu overexpression conditions we established are representative of naturally occurring physiological or pathological states is of course difficult to evaluate. However, increased levels of Alu RNA (or SINE-derived RNA in non-human mammals) are a common feature of cell response to different types of stress, including infection by viruses with the potential to drive cell transformation, such as adenovirus [46] and SV40 [47], and cancer progression [27]. Artificially increased Alu RNA levels in the absence of other oncogenic stimuli thus allow us to distinguish the contribution of these ncRNAs to the cell transformation process. Overall, the results of our analysis suggest that increased Alu RNA levels favor cell cycle progression thus contributing to sustained cell proliferation, which is an important factor for cancer development and progression [48].
4. Materials and Methods
4.1. Cloning of Constructs
The AluSq2 chr1:61057625-61057914 sequence was obtained by PCR amplification on human genomic DNA from saliva using the primer forward 5′-GCCCCAGGTGATCTCTACC-3′ and reverse 5′-GTCCTCGGAGCCGCTAATTT-3′, which anneal 200 nucleotides upstream of the transcription start site (TSS) and 100 nucleotides downstream of the Pol III terminator, respectively. The amplicon was cloned into the pGEM®-T easy vector using the pGEM®-T Easy Vector System kit (Promega, Madison, WI, USA). Subsequently, AluSq2 was amplified from the pGEM®-T easy vector using the primer forward 5′-TAAATATAAAAGATCTGGCCAGGCGCTGTGGCT-3′, which introduces the restriction site BglII, (underlined) and the primer reverse 5′-AATTATTTTACTCGAGAAAAATGGCCACCACCGTTTCC-3′, which introduces the restriction site XhoI (underlined), in order to subclone AluSq2 in the pSUPER.basic vector (OligoEngine™, Seattle, WA, USA). AluSq2 was then subcloned from pSUPER.basic to pSUPER.GFP/neo (OligoEngine™) using the restriction enzymes BglII and XhoI, which allow the insertion of the fragment downstream of the H1 promoter (H1p).
Similarly, the AluSx_chr16:56419511-56419806 sequence was first amplified from human genomic DNA using the primers forward 5′-CCCTTAACTTTTGTACCCTGAGC-3′ and reverse 5′-CACTCTGAACGGGGACAAGTA-3′. The primer forward anneals 183bp upstream of the Alu TSS and the primer reverse 75 bp downstream of the 6T Alu terminator. A second pair of primers (forward 5′-TAAATATAAAAGATCTGGCCAGGCGTGGTGG-3′ and reverse 5′-AATTATTTTACTCGAGAAAAAATGACTTGAAGCTTTGACAGCA-3′) was used to introduce the restriction sites for BglII and XhoI for cloning into the pSUPER.GFP/neo vector downstream of the H1 promoter (H1p).
The Control RNA sequence was amplified by PCR from the LacZ gene (Escherichia coli DH10 genomic DNA), which has the same distribution and GC content as the Alu sequences (61% GC AluSq2, 53% GC AluSx, 61% GC Control RNA sequence). Two rounds of PCR were performed: the first PCR was performed using the primers forward 5′-TAAATATAAAAGATCTGACCAGCGAATACCTGTTCC-3′ and reverse 5′- TTTTTTTTGGGGAGCGTCACACTGAG-3′, obtaining an amplicon that was used as template for a second PCR, which was performed using the same forward primer and the reverse primer 5′-AATTATTTTAAAGCTTTTTTTTTTTTTTTTTTTTGGGGAGCGTCAC-3′. In the last reverse primer, a poly(A) tail typical of Alu sequences was introduced. Moreover, the 3′end contains a HindIII site (underlined) that was used to subclone the Control RNA sequence in AluSx_pSUPER.GFP/neo vector where AluSx was excised using the restriction enzymes BglII and HindIII, in order to exploit the Pol III 6T natural terminator from AluSx.
The sequences H1p-AluSq2, H1p-AluSx, H1p-Control RNA, and H1p-empty present in the pSUPER/GFP.neo expression plasmids were amplified by PCR with XbaI and BamHI restriction sites added at the 5′ and 3′ ends, respectively, of the amplified fragment. Primers that annealed to the H1 sequence and Alu/Control RNA sequence/empty vector were used to amplify the fragment of interest. The primer forward 5′-GAGTTCTAGAGAACGCTGACGTCATCAACCC-3′ was used to start the amplification from the H1 promoter (the XbaI restriction site is underlined), while the primer reverse 5′-CCTCCGGATCCAAAAATGGCCCCACCGTTTCC-3′ annealed to the AluSq2 sequence, 5′-CCTCCGGATCCAAAAAATGACTTGAAGCTTTG-3′ to AluSx, 5′-GAGTGGATCCAAAAAATGACTTGAAGC-3′ to Control RNA, 5′-CCTCCGGATCCGTCGACGGTATCGATAAGCTTAG-3′ to the empty vector (the BamHI restriction site is underlined). The fragments were ligated in a 3rd generation lentiviral vector pRRL-MCS-PGK-GFP-IRES-Puro already digested with XbaI and BamHI. The DNAs were isolated and the inserts sequenced. Clones containing the correct insert were amplified and purified using an Invitrogen PureLink™ HiPure Maxiprep Kit (Carlsbad, CA, USA).
Lentivirus-based vectors encoding AluSq2, AluSx, Control RNA as well as an empty vector were generated by transient cotransfection of 293T cells with a three-plasmid combination, as described previously, with slight modifications [49]. The construct pMD.G was used for the production of the VSV-G viral envelope in combination with the packaging constructs pMDLg/pRRE and pRSV–REV, whereas the pRRL constructs correspond to the different transfer vectors. Briefly, 100 mm dishes of non-confluent 293T cells were co-transfected with the four plasmids, by the CaPi-DNA coprecipitation method [50,51]. Conditioned medium was harvested 48 h later and passed through 0.45 mm filters.
Viral titer was determined by assessing viral p24 antigen concentration by ELISA (the Alliance® HIV-I p24 ELISA Kit, PerkinElmer, Waltham, MA, USA) and hereafter expressed as μg of p24 equivalent units per milliliter.
4.2. Cell Lines and Lentiviral Vector Transduction
IMR90 and HeLa cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were grown in Dulbecco’s Modified Eagle Medium (DMEM, purchased from ThermoFisher Scientific, Waltham, MA, USA) supplemented with 10% FBS (TCB, Tulare, CA, USA) and 100 U/mL penicillin/streptomycin (ThermoFisher Scientific, Waltham, MA, USA) and maintained in a humified atmosphere 5% CO2 at 37 °C. The day before the transduction, IMR90 and HeLa cells in the exponential phase of growth were plated in 6-well plates at 2 × 105 cells per well. The next day, the medium was aspirated and the cells were incubated in 1 ml final volume with virus (0.86 μg/mL AluSq2, 0.75 μg/mL AluSx, 1.2 μg/mL Control RNA, 0.89 μg/mL empty vector) supplemented with 4 μg/mL protamine sulfate. The cells were incubated overnight in a humified atmosphere 5% CO2 at 37 °C and washed with fresh medium the next day. Stable integrants were selected by puromycin selection (2 mg/mL for IMR90 cells and 0.5 µg/mL for HeLa cells) starting 8 days after transformation. After 7 days of growth in selective medium, cells were expanded and grown for another 7–10 days before RNA extraction. All the transformations were performed in duplicate.
Since cells were GFP fluorescent after more than one month of culture growing and we were able to detect the expression (by real-time PCR) of the insert controlled by the promoter H1 after puromycin selection, we could deduce that the lentivirus vector was efficiently inserted in the genome of IMR90 and HeLa cells.
4.3. RNA Extraction and Real-Time PCR
Total RNA was extracted from exponentially growing cells (passage 9 for IMR90 cells) using a Direct-zol™ RNA MiniPrep Plus kit (Zymo Research, Irvine, CA, USA) and the cDNA was synthesized using the SuperScript™ III Reverse Transcriptase (Thermo Fisher, Waltham, MA, USA). Real-Time PCR was performed using the PowerUp™ SYBR® Green Master Mix (Applied Biosystems, Foster City, CA, USA), the Applied Biosystems™ 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA), and a previously optimized pair of primers that anneal to the unique 3′ trailer sequence of Alu elements (primers forward 5′-AAGTGTCACCTCCCCATCTG-3′ and reverse 5′- ACCACCGTTTCCTGAGCTT-3′ for AluSq2, and the primer forward 5′-AATTCAACTATATTAAAACACTTCAGA-3′ and reverse 5′-GACTTGAAGCTTTGACAGCA-3′ for AluSx). The detection of the Control RNA was performed using the same primers employed for cloning. U1 snRNA was used as the internal normalization control.
4.4. RNA-Seq Procedure and Data Analysis
One microgram of total RNA was used to prepare mRNA libraries using the TruSeq stranded mRNA library Preparation kit (Illumina, San Diego, CA, USA). A 50 base-pair single-end stranded sequencing was performed on a HiSeq4000 Sequencer (Illumina, San Diego, CA, USA). Read alignments to the GRCh38 human reference genome were performed using STAR [52]. HTSeq [53] and DESeq2 [40] were used for read counting and differential gene expression, respectively. Genes with |log2FC|≥0.5 and adjusted p-value ≤ 0.001 versus the empty vector were deemed as differentially expressed and visualized using the Volcano Plot workflow on the Galaxy web platform at the public server at https://usegalaxy.eu. Venn diagram was plotted using BioVenn [54] and enrichment analyses were performed with Ingenuity Pathway Analysis (IPA, QIAGEN Inc., Hilden, Germany). Only pathways with overlap p-values (BH-adjusted) < 0.01 were considered significantly enriched. Upstream regulator analysis in IPA was employed to identify the transcriptional regulators that could explain the observed gene expression changes. HeatMaps were visualized with Morpheus software (https://software.broadinstitute.org/morpheus).
4.5. Cell Proliferation Assay
To synchronize IMR90 cells overexpressing Alus/Control sequences, 5 × 105 cells were seeded in 10 cm dishes in growth medium overnight. The next day, the cultures were rinsed with PBS and changed to serum-free medium. After serum starvation for 24 h, the cells were released into cell cycle by addition of 10% serum medium. After 24 h, the cells were trypsinized and fixed with 70% ethanol in PBS for at least 24 h at −20 °C. The day of FACS analysis, the cells were centrifuged and washed with PBS and the cell pellets were resuspended with propidium iodide (PI)-staining solution (100 ug/mL PI, 20 ug/mL RNAse). Samples were incubated in the dark for 15 min at room temperature and then analyzed using a BD FACSCelesta™ flow cytometer (BD Biosciences, San Jose, CA, USA). FACS analyses were performed in at least four replicates deriving from two independently transformed IMR90 cell lines, and the mean of PI fluorescence intensity was obtained from 20,000 cells. Cell cycle distributions were determined with ModFit LT™ software (version 5.0, Verity Software House, Topsham, ME, USA) and the statistical analyses were performed with a Student’s t-test or a Welch’s t-test.
4.6. Bioinformatic Analyses on Promoter, 5′ UTR, 3′ UTR, and miRNA Content of Differentially Expressed Genes
GENCODE v27 (Wellcome Trust Sanger Institute, Hinxton, UK) was used for the analysis of the content of Alu sequences in the promoter, 5′ UTR and 3′ UTR of differentially expressed genes. Only Alu sequences that overlapped for at least 100 bp were taken into account.
The sequences AluSq2 and AluSx were interrogated using the miRBase database for the search of miRNA binding sites. The differentially expressed genes were then interrogated with the miRDB database to search potential binding sites for the sequence has-miR-619-5p.
Acknowledgments
The cloning, virus production, and transduction were done at the UCLA Integrated Molecular Technologies Core which is supported by CURE/P30 DK041301. RNA-sequencing was performed by the Broad Stem Cell Research Center High-throughput Sequencing Core at UCLA. FACS analysis was performed at the Flow Cytometry Core Facility, Broad Stem Cell Research Center (UCLA). The authors wish to thank Arnold J. Berk for providing facilities and for critical reading of the manuscript.
Abbreviations
| ncRNA | non-coding RNA |
| SINEs | short interspersed nuclear elements |
| L1 | non-LTR retroelements LINE-1 |
| UTR | untranslated region |
| FC | fold change |
| IPA | Ingenuity Pathway Analysis |
| H1p | H1 promoter |
| PI | propidium iodide |
Supplementary Materials
Supplementary Materials can be found at https://www.mdpi.com/1422-0067/20/13/3315/s1.
Author Contributions
G.D. and B.M. designed this study. S.C. performed the experiments. D.C. and B.M. performed RNA-seq analysis. G.D., B.M., M.M., and S.C. interpreted the results and supervised this study. A.C. contributed to obtain the preliminary data. M.P. gave experimental support at the University of California, Los Angeles (UCLA). S.C. wrote the manuscript. All authors read and approved the final manuscript.
Funding
This research was funded by the Italian Association for Cancer Research (AIRC), grant number IG 16877 to G.D. AC was supported by a FIRC-AIRC fellowship for Italy.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- 1.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Berger A., Strub K. Multiple Roles of Alu-Related Noncoding RNAs. Prog. Mol. Subcell. Biol. 2011;51:119–146. doi: 10.1007/978-3-642-16502-3_6. [DOI] [PubMed] [Google Scholar]
- 3.Kriegs J.O., Churakov G., Jurka J., Brosius J., Schmitz J. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet. 2007;23:158–161. doi: 10.1016/j.tig.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Comeaux M.S., Roy-Engel A.M., Hedges D.J., Deininger P.L. Diverse cis factors controlling Alu retrotransposition: What causes Alu elements to die? Genome Res. 2009;19:545–555. doi: 10.1101/gr.089789.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Versteeg R., van Schaik B.D., van Batenburg M.F., Roos M., Monajemi R., Caron H., Bussemaker H.J., van Kampen A.H. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 2003;13:1998–2004. doi: 10.1101/gr.1649303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grover D., Majumder P.P., Rao C.B., Brahmachari S.K., Mukerji M. Nonrandom distribution of alu elements in genes of various functional categories: Insight from analysis of human chromosomes 21 and 22. Mol. Biol. Evol. 2003;20:1420–1424. doi: 10.1093/molbev/msg153. [DOI] [PubMed] [Google Scholar]
- 7.Quentin Y. Emergence of master sequences in families of retroposons derived from 7sl RNA. Genetica. 1994;93:203–215. doi: 10.1007/BF01435252. [DOI] [PubMed] [Google Scholar]
- 8.Hamdi H.K., Nishio H., Tavis J., Zielinski R., Dugaiczyk A. Alu-mediated phylogenetic novelties in gene regulation and development. J. Mol. Biol. 2000;299:931–939. doi: 10.1006/jmbi.2000.3795. [DOI] [PubMed] [Google Scholar]
- 9.Lev-Maor G., Sorek R., Shomron N., Ast G. The birth of an alternatively spliced exon: 3’ splice-site selection in Alu exons. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- 10.Su M., Han D., Boyd-Kirkup J., Yu X., Han J.J. Evolution of Alu elements toward enhancers. Cell Rep. 2014;7:376–385. doi: 10.1016/j.celrep.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 11.Deininger P.L., Batzer M.A. Alu repeats and human disease. Mol. Genet. Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- 12.Yakovchuk P., Goodrich J.A., Kugel J.F. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc. Natl. Acad. Sci. USA. 2009;106:5569–5574. doi: 10.1073/pnas.0810738106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L.L., Carmichael G.G. Gene regulation by SINES and inosines: Biological consequences of A-to-I editing of Alu element inverted repeats. Cell Cycle. 2008;7:3294–3301. doi: 10.4161/cc.7.21.6927. [DOI] [PubMed] [Google Scholar]
- 14.Karijolich J., Zhao Y., Alla R., Glaunsinger B. Genome-wide mapping of infection-induced SINE RNAs reveals a role in selective mRNA export. Nucleic Acids Res. 2017;45:6194–6208. doi: 10.1093/nar/gkx180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivanova E., Berger A., Scherrer A., Alkalaeva E., Strub K. Alu RNA regulates the cellular pool of active ribosomes by targeted delivery of SRP9/14 to 40S subunits. Nucleic Acids Res. 2015;43:2874–2887. doi: 10.1093/nar/gkv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tarallo V., Hirano Y., Gelfand B.D., Dridi S., Kerur N., Kim Y., Cho W.G., Kaneko H., Fowler B.J., Bogdanovich S., et al. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell. 2012;149:847–859. doi: 10.1016/j.cell.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daniel C., Silberberg G., Behm M., Ohman M. Alu elements shape the primate transcriptome by cis-regulation of RNA editing. Genome Biol. 2014;15:R28. doi: 10.1186/gb-2014-15-2-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazzari E., Mondala P.K., Santos N.D., Miller A.C., Pineda G., Jiang Q., Leu H., Ali S.A., Ganesan A.P., Wu C.N., et al. Alu-dependent RNA editing of GLI1 promotes malignant regeneration in multiple myeloma. Nat. Commun. 2017;8:1922. doi: 10.1038/s41467-017-01890-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Capshew C.R., Dusenbury K.L., Hundley H.A. Inverted Alu dsRNA structures do not affect localization but can alter translation efficiency of human mRNAs independent of RNA editing. Nucleic Acids Res. 2012;40:8637–8645. doi: 10.1093/nar/gks590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ottesen E.W., Luo D., Seo J., Singh N.N., Singh R.N. Human Survival Motor Neuron genes generate a vast repertoire of circular RNAs. Nucleic Acids Res. 2019;47:2884–2905. doi: 10.1093/nar/gkz034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lubelsky Y., Ulitsky I. Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature. 2018;555:107–111. doi: 10.1038/nature25757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varshney D., Vavrova-Anderson J., Oler A.J., Cowling V.H., Cairns B.R., White R.J. SINE transcription by RNA polymerase III is suppressed by histone methylation but not by DNA methylation. Nat. Commun. 2015;6:6569. doi: 10.1038/ncomms7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panning B., Smiley J.R. Activation of expression of multiple subfamilies of human Alu elements by adenovirus type 5 and herpes simplex virus type 1. J. Mol. Biol. 1995;248:513–524. doi: 10.1006/jmbi.1995.0239. [DOI] [PubMed] [Google Scholar]
- 25.Mariner P.D., Walters R.D., Espinoza C.A., Drullinger L.F., Wagner S.D., Kugel J.F., Goodrich J.A. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol. Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 26.Tang R.B., Wang H.Y., Lu H.Y., Xiong J., Li H.H., Qiu X.H., Liu H.Q. Increased level of polymerase III transcribed Alu RNA in hepatocellular carcinoma tissue. Mol. Carcinog. 2005;42:93–96. doi: 10.1002/mc.20057. [DOI] [PubMed] [Google Scholar]
- 27.Di Ruocco F., Basso V., Rivoire M., Mehlen P., Ambati J., De Falco S., Tarallo V. Alu RNA accumulation induces epithelial-to-mesenchymal transition by modulating miR-566 and is associated with cancer progression. Oncogene. 2018;37:627–637. doi: 10.1038/onc.2017.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaneko H., Dridi S., Tarallo V., Gelfand B.D., Fowler B.J., Cho W.G., Kleinman M.E., Ponicsan S.L., Hauswirth W.W., Chiodo V.A., et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakamoto K., Fordis C.M., Corsico C.D., Howard T.H., Howard B.H. Modulation of HeLa cell growth by transfected 7SL RNA and Alu gene sequences. J. Biol. Chem. 1991;266:3031–3038. [PubMed] [Google Scholar]
- 30.Baryakin D.N., Semenov D.V., Savelyeva A.V., Koval O.A., Rabinov I.V., Kuligina E.V., Richter V.A. Alu- and 7SL RNA Analogues Suppress MCF-7 Cell Viability through Modulating the Transcription of Endoplasmic Reticulum Stress Response Genes. Acta Naturae. 2013;5:83–93. [PMC free article] [PubMed] [Google Scholar]
- 31.Hu Q., Tanasa B., Trabucchi M., Li W., Zhang J., Ohgi K.A., Rose D.W., Glass C.K., Rosenfeld M.G. DICER- and AGO3-dependent generation of retinoic acid-induced DR2 Alu RNAs regulates human stem cell proliferation. Nat. Struct. Mol. Biol. 2012;19:1168–1175. doi: 10.1038/nsmb.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morales-Hernandez A., Gonzalez-Rico F.J., Roman A.C., Rico-Leo E., Alvarez-Barrientos A., Sanchez L., Macia A., Heras S.R., Garcia-Perez J.L., Merino J.M., et al. Alu retrotransposons promote differentiation of human carcinoma cells through the aryl hydrocarbon receptor. Nucleic Acids Res. 2016;44:4665–4683. doi: 10.1093/nar/gkw095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castelnuovo M., Massone S., Tasso R., Fiorino G., Gatti M., Robello M., Gatta E., Berger A., Strub K., Florio T., et al. An Alu-like RNA promotes cell differentiation and reduces malignancy of human neuroblastoma cells. FASEB J. 2010;24:4033–4046. doi: 10.1096/fj.10-157032. [DOI] [PubMed] [Google Scholar]
- 34.Conti A., Carnevali D., Bollati V., Fustinoni S., Pellegrini M., Dieci G. Identification of RNA polymerase III-transcribed Alu loci by computational screening of RNA-Seq data. Nucleic Acids Res. 2015;43:817–835. doi: 10.1093/nar/gku1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brummelkamp T.R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 36.Chu W.M., Liu W.M., Schmid C.W. RNA polymerase III promoter and terminator elements affect Alu RNA expression. Nucleic Acids Res. 1995;23:1750–1757. doi: 10.1093/nar/23.10.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roy A.M., West N.C., Rao A., Adhikari P., Aleman C., Barnes A.P., Deininger P.L. Upstream flanking sequences and transcription of SINEs. J. Mol. Biol. 2000;302:17–25. doi: 10.1006/jmbi.2000.4027. [DOI] [PubMed] [Google Scholar]
- 38.Orioli A. Ph.D. Thesis. University of Parma; Parma, Italy: Mar 12, 2010. Novel insights into human RNA polymerase III transcription: Non canonical termination and biogenesis of potential regulatory RNAs. [Google Scholar]
- 39.Baserga S.J., Steitz J.A. The Diverse World of Small Ribonucleoproteins. In: Gesteland R.F., Atkins J.F., editors. The RNA World. 1st ed. Volume 24. Cold Spring Harbor Monograph Archive; Cold Spring Harbor, NY, USA: 1993. pp. 359–381. [Google Scholar]
- 40.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durrieu-Gaillard S., Dumay-Odelot H., Boldina G., Tourasse N.J., Allard D., Andre F., Macari F., Choquet A., Lagarde P., Drutel G., et al. Regulation of RNA polymerase III transcription during transformation of human IMR90 fibroblasts with defined genetic elements. Cell Cycle. 2018;17:605–615. doi: 10.1080/15384101.2017.1405881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferrari R., Su T., Li B., Bonora G., Oberai A., Chan Y., Sasidharan R., Berk A.J., Pellegrini M., Kurdistani S.K. Reorganization of the host epigenome by a viral oncogene. Genome Res. 2012;22:1212–1221. doi: 10.1101/gr.132308.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Agarwal P., Enroth S., Teichmann M., Jernberg Wiklund H., Smit A., Westermark B., Singh U. Growth signals employ CGGBP1 to suppress transcription of Alu-SINEs. Cell Cycle. 2016;15:1558–1571. doi: 10.4161/15384101.2014.967094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pirkmajer S., Chibalin A.V. Serum starvation: Caveat emptor. Am. J. Physiol. Cell Physiol. 2011;301:C272–C279. doi: 10.1152/ajpcell.00091.2011. [DOI] [PubMed] [Google Scholar]
- 45.Gong C., Maquat L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Panning B., Smiley J.R. Activation of RNA polymerase III transcription of human Alu repetitive elements by adenovirus type 5: Requirement for the E1b 58-kilodalton protein and the products of E4 open reading frames 3 and 6. Mol. Cell Biol. 1993;13:3231–3244. doi: 10.1128/MCB.13.6.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carey M.F., Singh K. Enhanced B2 transcription in simian virus 40-transformed cells is mediated through the formation of RNA polymerase III transcription complexes on previously inactive genes. Proc. Natl. Acad. Sci. USA. 1988;85:7059–7063. doi: 10.1073/pnas.85.19.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 49.Naldini L., Blomer U., Gage F.H., Trono D., Verma I.M. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc. Natl. Acad. Sci. USA. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen C., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell Biol. 1987;7:2745–2752. doi: 10.1128/MCB.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakoda T., Kaibuchi K., Kishi K., Kishida S., Doi K., Hoshino M., Hattori S., Takai Y. smg/rap1/Krev-1 p21s inhibit the signal pathway to the c-fos promoter/enhancer from c-Ki-ras p21 but not from c-raf-1 kinase in NIH3T3 cells. Oncogene. 1992;7:1705–1711. [PubMed] [Google Scholar]
- 52.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anders S., Pyl P.T., Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hulsen T., de Vlieg J., Alkema W. BioVenn—A web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.