Abstract
(1) Background: In a previous study, we found that two phenotypes related to platelet reactivity, measured with the PFA-100 system, were highly heritable. The aim of the present study was to identify genetic determinants that influence the variability of these phenotypes: closure time of collagen-ADP (Col-ADP) and of collagen-epinephrine (Col-Epi). (2) Methods: As part of the GAIT-2 (Genetic Analysis of Idiopathic Thrombophilia (2) Project, 935 individuals from 35 large Spanish families were studied. A genome-wide association study (GWAS) with ≈ 10 M single nucleotide polymorphisms (SNPs) was carried out with Col-ADP and Col-Epi phenotypes. (3) Results: The study yielded significant genetic signals that mapped to the ABO locus. After adjusting both phenotypes for the ABO genotype, these signals disappeared. After adjusting for von Willebrand factor (VWF) or for coagulation factor VIII (FVIII), the significant signals disappeared totally for Col-Epi phenotype but only partially for Col-ADP phenotype. (4) Conclusion: Our results suggest that the ABO locus exerts the main genetic influence on PFA-100 phenotypes. However, while the effect of the ABO locus on Col-Epi phenotype is mediated through VWF and/or FVIII, the effect of the ABO locus on Col-ADP phenotype is partly produced through VWF and/or FVIII, and partly through other mechanisms.
Keywords: platelet reactivity, platelet function test, ABO blood-group system, von Willebrand factor, factor VIII
1. Introduction
Platelet reactivity can be measured using a wide variety of laboratory functional tests. Such tests are classified into two main types [1]: (1) tests based on platelet aggregation activated by agonists, in platelet-rich plasma or in whole blood; (2) tests based on platelet adhesion under shear stress. Among the second group of tests, the PFA-100 system (Siemens Healthcare Diagnostics, Marburg, Germany) measures platelet function by simulating in vitro a vessel wall under shear stress. The vessel wall is simulated by a membrane coated with collagen; it is coated also with ADP (cartridge collagen-ADP) or with epinephrine (cartridge collagen-epinephrine) as platelet agonists. The membrane has a hole through which the anticoagulated blood passes; the closure times (CTs) of this hole are inversely proportional to the functional capacity of platelets. The PFA-100 system was introduced in 1995 with the objective of measuring primary hemostasis in whole blood in vitro as a non-invasive and more accurate method than the bleeding time in vivo [2,3]. Long CTs gave reliable measurements of hemostatic deficiencies due to low levels of von Willebrand factor (VWF) or platelet functional defects [2,4]. The use of PFA-100 was subsequently expanded to the assessment of the effect of platelet hyperreactivity, as indicated by shortened CTs, on arterial and venous thrombotic risk [3,5,6,7,8]. Citrate concentration, platelet count, hematocrit, white blood cell count, circadian rhythm and some dietary elements can influence the CTs [3,9,10]. Age and sex do not have appreciable influence although slight shortening of CTs has been described in neonates, in children and in elderly men [3,10,11]. Obviously, VWF has an important influence on CTs, which correlates inversely with VWF levels [3,4,9,10]. Also, the ABO blood group influences PFA-100 CTs, with O group individuals having longer CTs than those of non-O groups [3,4,9,10,12,13]; this has been interpreted as an effect of the lower levels of VWF in group O individuals [4,9,10]. Some authors have suggested that factor VIII (FVIII) has no impact on the PFA-100 CTs [2,5], while others have found the contrary [14].
The GAIT-2 (Genetic Analysis of Idiopathic Thrombophilia (2) Project is a family-based genetic study designed to identify new genetic markers of thrombotic risk [15]. Using the variance component statistical method, the heritability of intermediate phenotypes that could play a role in thrombotic risk was determined. In the GAIT-2 Project, two PFA-100 phenotypes were included as a measure of platelet reactivity: CTs of collagen-ADP cartridge (Col-ADP) and of collagen-epinephrine cartridge (Col-Epi). Both phenotypes showed a strong genetic component with a heritability of 0.45 for Col-ADP and 0.52 for Col-Epi [16].
The objectives of the present study were: (1) to analyze the genetic correlations between Col-ADP and Col-Epi phenotypes with each other and with other related phenotypes, and (2) to perform a genome-wide association study (GWAS) to identify susceptibility loci for Col-ADP and Col-Epi phenotypes.
2. Results and Discussion
2.1. Genetic Correlations of the PFA-100 Phenotypes
The genetic correlations of Col-ADP and Col-Epi with each other and with the VWF antigen, coagulant FVIII and ABO genotype (considering dominant effect of allele O) are presented in Table 1.
Table 1.
Genetic correlations among PFA phenotypes, von Willebrand factor, coagulation factor VIII and ABO genotype.
| Phenotype | Col-Epi | VWF | FVIII | ABO |
|---|---|---|---|---|
| Col-ADP | ρ = 0.7917 (5.80 × 10−9) | ρ = −0.7002 (1.02 × 10−10) | ρ = −0.6209 (2.66 × 10−8) | ρ = 0.5895 (7.01 × 10−9) |
| Col-Epi | - | ρ = −0.6342 (7.14 × 10−8) | ρ = −0.5947 (3.97 × 10−7) | ρ = 0.4477 (0.0003) |
ρ = genetic correlation and p-value (in brackets); VWF = von Willebrand factor antigen; FVIII = coagulation factor VIII activity; ABO = ABO genotype, considering dominant effect of allele O.
As expected, the genetic correlations of PFA-100 phenotypes with VWF and ABO blood group were statistically significant. In addition, a significant correlation between the PFA-phenotypes and FVIII was also observed. Unlike VWF, which has an important role in primary hemostasis, FVIII is fundamental for coagulation. The mechanism by which the FVIII influences the PFA-100 CTs could be partially explained by its close relationship with the VWF. Both factors circulate together, and their levels are related. Moreover, genetic studies have demonstrated that there is a huge overlap between genetic factors regulating FVIII and VWF [17,18].
2.2. GWAS of the PFA-100 Phenotypes
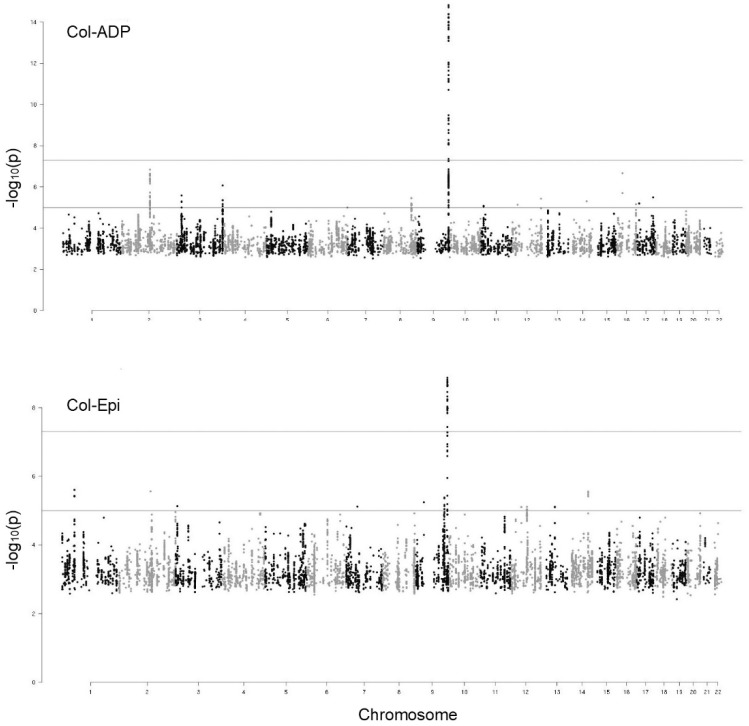
Manhattan plots of the GWAS on Col-ADP and Col-Epi phenotypes are shown in Figure 1.
Figure 1.
Manhattan plots of the GWAS on two PFA-100 phenotypes: collagen-ADP and collagen epinephrine closure times. Dots correspond to SNPs organized by chromosomal order and position and the vertical axis shows the statistical significance expressed as −log10 of the p-values. The horizontal lines mark the 5 × 10−8 p-value threshold of genome-wide significance.
For each phenotype, a signal on chromosome 9 at the ABO locus reached a genome-wide significance level (p-value < 5 × 10−8). Seventy single nucleotide polymorphisms (SNPs) were found to be associated with Col-ADP, with p-values up to 1.50 × 10−15. Of these 70 SNPs, 45 were associated also with Col-Epi but with less statistical significance: p-values up to 1.35 × 10−9. The complete list of these SNPs is described in Supplementary Table S1. For both Col-ADP and Col-Epi phenotypes, all significant SNPs were located within the ABO gene and in its adjacent intergenic region.
To the best of our knowledge, our study is the first GWAS of PFA-100 phenotypes. Previous GWAS reports of platelet reactivity phenotypes were mainly based on aggregometry. Using GWAS and other genetic approaches, several SNPs have been found in association with platelet aggregometry phenotypes. These were located in different genes related to platelet functional receptors, regulators of cytoskeleton and signaling proteins. Among these genes, the following were described: MIR100HG, MME, PIP3-E, GLIS3, LDHAL6A, ANKS1B, PIK3CG, MAGI1, C8orf86, FGFR1, LPAR1, CACNB2, SLC39A12, RPP25, SCAMP5, BMPR1A (revisited in Bunimov et al. [19]), ANKRD26 [20], pannexin [21], ADRA2 [22] and the most relevant PEAR-1 [22,23,24]. Notably, none of these genetic variants was found in the ABO locus. On the other hand, in our study, we did not find any of the SNPs previously reported to be associated with platelet aggregometry. To explain these different results, we should keep in mind that aggregometry, both in platelet-rich plasma and in whole blood, analyzes aggregation, that is, the platelet-platelet binding mediated by fibrinogen. In contrast, the PFA-100 test basically analyzes adhesion under shear stress, which depends mainly on binding with VWF [2,10].
Nineteen of the SNPs that we found in the ABO locus have been previously reported in relation to thrombotic-related conditions (Table 2): Venous thromboembolism [25,26,27,28,29,30], myocardial infarction [31], large-vessel and cardioembolic stroke [32], large-artery arteriosclerosis [33], coronary artery disease [34,35] and coronary artery disease shared with venous thromboembolism [36]. The association of these SNPs with thrombosis and with the PFA-100 phenotypes suggests that this functional platelet test may be useful to estimate thrombotic risk, although further studies are needed to confirm this. Moreover, some of the SNPs have been described also in association with variations in VWF, FVIII, and/or variations in biological factors that may play indirect roles in thrombosis, such as adhesion molecules [17,18,37,38,39].
Table 2.
ABO locus (chromosome 9): SNPs associated with PFA-100 phenotypes which have been previously described in association with thrombosis-related conditions and with variations of biological factors.
| SNP | Position (bp) | Location | MAF | Association with Col-ADP p-Value | Association with Col-Epi p-Value | Association with Thrombosis-Related Conditions [References] | Association with Variations of Biological Factors [References] |
|---|---|---|---|---|---|---|---|
| rs8176719 | 136132908 | coding, 5-UTR, intron | 0.459 | 5.21 × 10−14 | 1.88 × 10−9 | VTE [18,25,26,29] | |
| rs687621 | 136137065 | intron | 0.432 | 1.49 × 10−15 | 3.50 × 10−9 | VTE [18,26], MI [31], LVCES [32] | VWF [17,18,38], ICAM-1 [38] |
| rs687289 | 136137106 | intron | 0.433 | 1.93 × 10−15 | 5.72 × 10−9 | MI [31], LVCES [32] | FVIII [16,17], ICAM-1 [38] |
| rs2519093 | 136141870 | intron | 0.312 | 1.16 × 10−12 | 6.01 × 10−9 | VTE [25], LVCES [32] | - |
| rs514659 | 136142203 | intron | 0.433 | 6.04 × 10−15 | 1.04 × 10−8 | VTE [26], MI [31], LVCES [32], LAA [33] | VWF [38] |
| rs644234 | 136142217 | intron | 0.460 | 1.42 × 10−14 | 2.18 × 10−9 | MI [31], LVCES [32] | E-selectin [38] |
| rs643434 | 136142355 | intron | 0.460 | 1.43 × 10−14 | 2.18 × 10−9 | MI [31], LVCES [32] | - |
| rs545971 | 136143372 | Intron | 0.433 | 6.03 × 10−15 | 1.04 × 10−8 | MI [31], LVCES [32] | - |
| rs612169 | 136143442 | intron | 0.433 | 6.05 × 10−15 | 1.04 × 10−8 | MI [31], LVCES [32] | ICAM-1, E-selectin [38] |
| rs674302 | 136146664 | intron | 0.433 | 6.06 × 10−15 | 1.04 × 10−8 | MI [31], LVCES [32] | - |
| rs500498 | 136148647 | intron | 0.408 | 5.56 × 10−08 | - | VTE [27], LVCES [32] | ICAM-1, E-selectin [38] |
| rs505922 | 136149229 | intron | 0.433 | 9.68 × 10−15 | 1.12 × 10−8 | VTE [27,28], MI [31], LVCES [32] | - |
| rs529565 | 136149500 | intron | 0.434 | 1.86 × 10−14 | 1.01 × 10−8 | VTE [29,30], MI [31], LVCES [32], LAA [33] | - |
| rs630014 | 136149722 | intron | 0.407 | 8.74 × 10−10 | - | VTE [27,28], LVCES [32] | E-selectin [38] |
| rs651007 | 136153875 | intergenic | 0.204 | 8.02 × 10−12 | 2.32 × 10−9 | LVCES [32], LAA [33], CAD [34] | VWF, ICAM-1, E-selectin, cholesterol [38] |
| rs579459 | 136154168 | intergenic | 0.340 | 3.75 × 10−12 | 1.35 × 10−9 | LVCES [32], CAD [35], CAD+VTE [36] | ICAM-1, E- and P-selectin [38] |
| rs649129 | 136154304 | intergenic | 0.338 | 6.88 × 10−12 | 2.19 × 10−9 | LVCES [32] | ICAM-1, LDL-cholesterol [38,39] |
| rs495828 | 136154867 | intergenic | 0.340 | 7.71 × 10−12 | 2.28 × 10−9 | VTE [25,27], LVCES [32] | ACE [38] |
| rs633862 | 136155444 | intergenic | 0.391 | 2.41 × 10−9 | - | LVCES [32] | - |
VTE = venous thromboembolism; MI = myocardial infarction; LVCES = large-vessel and cardioembolic stroke; LAA = large-artery arteriosclerosis; CAD = coronary artery disease; CAD + VTE = coronary artery disease shared with venous thromboembolism; VWF = von Willebrand factor; FVIII = coagulant factor VIII; ACE = angiotensin-converting-enzyme.
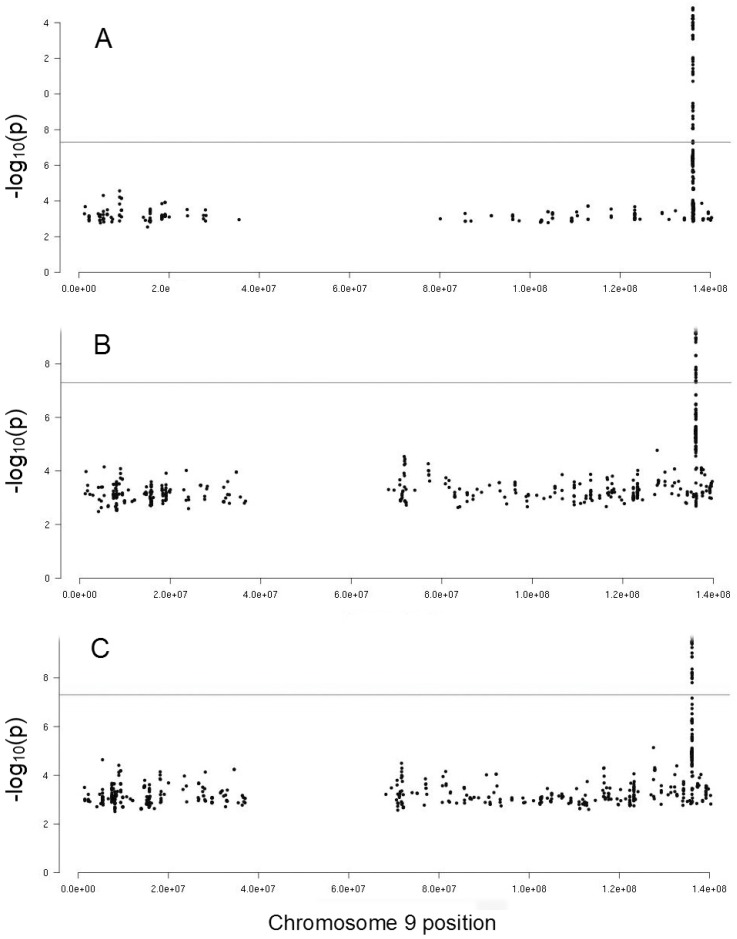
After finding a genetic correlation between the PFA-100 phenotypes and VWF, FVIII and ABO genotype, we adjusted the GWAS results for these factors. As shown in Figure 2 and Supplementary Table S1, after adjusting the Col-ADP results for VWF 50, SNPs remained significant but with lower significance than before adjustment (p-values up to 1.55 × 10−10). After adjusting the Col-ADP, the results for FVIII 46 SNPs remained significant; almost all were the same as for the VWF adjustment, but 6 were different. The remaining SNPs also had a lower significance than before (p-values up to 5.00 × 10−11).
Figure 2.
Manhattan plots of the chromosome 9 region of the GWAS on collagen-ADP closure time phenotype: (A) without adjustments, (B) after adjusting for von Willebrand factor, and (C) after adjusting for factor VIII. Dots correspond to SNPs organized by position and the vertical axis shows the statistical significance expressed as -log10 of the p-values. The horizontal lines mark the 5 × 10−8 p-value threshold of genome-wide significance.
It should be noted that after adjustment of Col-ADP results for ABO genotype, the association signal disappeared completely (data not shown). Regarding Col-Epi, the significant SNPs disappeared with any of the adjustments performed, including VWF, FVIII and ABO genotype.
2.3. Genetic Influence of ABO Locus on PFA-100 Phenotypes
Our GWAS results and adjustments suggested that the ABO gene was the main determinant of variations in PFA-100 CTs since all the significant SNPs associated with PFA-100 phenotypes were at the ABO locus and all of them lost their genome-wide significance when we adjusted for the ABO genotype.
However, there were differences between Col-ADP and Col-Epi phenotypes. With Col-Epi, all significant SNPs also disappeared when adjusted for VWF or FVIII. This suggested that the influence of the ABO gene was related mainly to these factors. Regarding Col-ADP, the adjustment either by VWF or by FVIII reduced the number of significant SNPs in both cases but 50 and 46 respectively did not disappear. This suggested that part of the genetic effect of the ABO gene on the Col-ADP was mediated by VWF and/or FVIII. The relationships among ABO blood group, VWF and FVIII have been previously described. Non-O group individuals have 25% higher levels of VWF and FVIII than O group individuals [37,38]. These differences are attributed to the formation of A and B antigens, catalyzed by specific glucosyltransferases, on the H antigen existing in VWF. They are related to the glycosylation and clearance rate of VWF which is lower in non-O individuals. We previously showed that FVIII and VWF were genetically correlated with thrombotic risk, and demonstrated significant linkage between the ABO locus and plasma levels of VWF and FVIII [40]. More recent GWAS demonstrated that ABO gene is by far the major determinant of VWF and FVIII levels [18]. This can explain the genetic relationship between the PFA-100 phenotypes and the ABO blood group through the VWF and/or FVIII.
According to our results, part of the effect of the ABO locus on the Col-ADP phenotype does not occur due to WVF and/or FVIII. Some studies have suggested alternative mechanisms by which the ABO gene can intervene in platelet reactivity, thrombosis, and cardiovascular diseases. Non-O individuals have higher levels of cholesterol [41]. Also, they have increased amounts of the adhesion molecules and cytokines involved in inflammation that are related to cardiovascular disease [38,39,42]. Ten of the SNPs that we found at the ABO locus, associated with PFA-100 phenotypes, have been described previously in association with adhesion molecules and/or cholesterol (Table 2).
Moreover, the ABO blood group can influence platelet reactivity through various mechanisms related to glycosylation [37,43]. A and B antigens expressed on platelet glycoproteins can modify the activity of the glycoprotein complexes involved in platelet adhesion and the galectin-glycan interactions; galectins from extracellular matrices are potent platelet agonists. A and B antigens, also present on platelet glycosphingolipids, are involved in platelet aggregation and thrombosis by binding cell adhesion molecules.
Glycosylation is essential also for other platelet functions in which no relationship with the ABO blood group has been described so far. An example of this is the glycosylation of the P2Y12 receptor [44]. P2Y12 is a Gi-coupled ADP receptor that contains two potential N-linked glycosylation sites at its extracellular amino-terminus. The lack of glycosylation of this receptor leads to a defective P2Y12-mediated inhibition of the adenylyl cyclase activity resulting in defective platelet reactivity. The fact that the effect of the ABO locus, independent of the VWF and/or FVIII, was demonstrated only for the Col-ADP phenotype gives rise to speculation about the possibility that the ABO locus, among other mechanisms, could influence the glycosylation of the P2Y12 receptor. Further data are required to support this hypothesis.
3. Methods
3.1. Enrollment of Individuals and Families
The enrollment of individuals and families was described in detail in a previous publication [16]. Briefly, 935 individuals from 35 large Spanish families, included in the GAIT-2 Project, were recruited through a proband with idiopathic thrombophilia and the condition of having at least of 10 members in at least 3 generations willing to participate in the study. The exclusion criteria were: Deficiencies of antithrombin, protein S, protein C, heparin cofactor II, or plasminogen, activated protein C resistance, Factor V Leiden, dysfibrinogenemia, lupus anticoagulant and antiphospholipid antibodies. The subjects were questioned about their current medication to confirm that they had not taken antiplatelet drugs in the last two weeks, or other drugs with slight effect on platelet function (such as nonsteroidal anti-inflammatory drugs or serotonin reuptake inhibitor drugs) in the last week.
Among the individuals studied, 465 were male and 470 were female. The mean age was 39.5 (minimum 2.6, maximum 101, SD 21.4), and 197 of them were 18-years of age or younger. There were 86 with venous thrombosis, 47 with arterial thrombosis, and 13 with both venous and arterial thrombosis.
The study was performed according to the Declaration of Helsinki. Written informed consent was obtained from all adult patients and from parents or guardians of children. All procedures were approved by the Institutional Review Board at the Hospital de la Santa Creu i Sant Pau. The GAIT-2 Project was approved on November 23, 2005. At that time, no number was assigned to the approved projects.
3.2. Blood Collection, Laboratory Analyses and DNA Preparation
Whole blood samples were obtained by venipuncture, under basal conditions, after a 12 h overnight fast, and between 9:00 a.m. and 9:30 a.m. to minimize the circadian fluctuation. A 5 mL sample was obtained in EDTA-K3 for determining standard blood cell counts. A 5 mL sample of blood was collected in 3.8% sodium citrate to be analyzed in the PFA-100 device; the phenotypes obtained were: CTs (in seconds) for the cartridge collagen-ADP (Col-ADP phenotype) and for the cartridge collagen-epinephrine (Col-Epi phenotype). To avoid erroneous results due to thrombocytopenia or anemia, 6 individuals with platelet counts down to 100 × 109/L and 8 individuals with hemoglobin down to 110 g/L were excluded for the of PFA-100 measurements. Another sample collected in 3.8% sodium citrate was used to obtain platelet-poor plasma by centrifugation at 2000 g for 20 min at room temperature. This plasma was used for determining VWF antigen by ELISA and coagulant FVIII activity as previously described [40]. VWF antigen and coagulant FVIII were recorded as percentages of an international standard sample. PFA-100 and FVIII activity assays were performed on fresh samples.
DNA was extracted from EDTA blood samples using a standard salting-out procedure [45] or a commercial kit (Wizard, Promega Corp, Madison, WI, USA). The ABO genotype, that distinguishes the A1, A2, B, O1 and O2 alleles, was determined as previously described [46]. The primers used were described in Souto et al. [40].
3.3. Genotyping Filtering and Imputation
We genotyped the samples from 934 individuals, with a combination of HumanOmniExpressExome-8v1.2 (324 individuals and coverage 964,193 variants) and HumanCoreExome-12v1.1 (610 individuals and coverage 542,585 variants). After filtering the datasets by call rate (>98%), Hardy-Weinberg Equilibrium (p-value > 10−6) and minor allele frequency (MAF) (>1%) and deleting the Mendelian errors, we obtained 395,556 SNPs in all of the samples. We estimated haplotypes using SHAPEIT v2 [47] and imputed genotypes to the 1000 genomes phase 1 panel using IMPUTE2 [48]. We imputed 37,985,264 SNPs, of which 10,844,567 remained after filtering again by MAF (>0.5%). The 14 individuals who had thrombocytopenia or anemia were excluded from the association analysis.
3.4. Statistical Correlation Analyses
The correlations among the phenotypes Col-ADP and Col-Epi with each other and, by pairs, with other biologically-related phenotypes, were analyzed by multivariate variance component models, which are an extension of the univariate model [49]. By studying these traits in extended families, we were able to estimate robustly both the genetic (ρg), and the environmental (ρe) correlations between pairs of traits.
3.5. GWAS of PFA-100 Phenotypes
We used two phases for the association analysis of normal transformed quantitative phenotypes with the imputed genotypes. The first was a fast screen of variants with Matrix eQTL [50] using modelLINEAR. The GWAS of each phenotype was calculated adjusting by age. Sex, smoking and and oral contraceptives were not included as covariates, since they did not influence the PFA-100 CTs in the GAIT-2 sample [16]. The “errorCovariance” parameter was used to account for the pedigree effect. Variants with a p-value < 10−3 were recalculated in the second phase with SOLARIUS [51] and SOLAR-Eclipse v 8.1.1 [52], using variance component methods. Only the p-values calculated with SOLAR are reported, since it is considered a gold standard in the field of extended pedigree samples. SOLAR employs the maximum likelihood approach for variance component models with the standard likelihood ratio tests to evaluate the statistical significance of the model’s parameters [53].
4. Conclusions
We report the first GWAS on PFA-100 phenotypes. The results of our study suggest that the ABO locus is the main determinant of these phenotypes and, in the case of Col-Epi, all the ABO influence is mediated by VWF and/or FVIII. However, for Col-ADP, the influence of ABO is only partially mediated by VWF and/or FVIII suggesting that other mechanisms may explain the ABO effect on this phenotype. Much remains to be done to better understand the relationships between ABO blood group and platelet reactivity measured by PFA-100 test, beyond those mediated by VWF and/or FVIII. Unraveling these mechanisms will help to identify novel pathways involved in platelet reactivity.
Acknowledgments
Genotyping was performed by the SNP&SEQ Technology Platform in Uppsala (www.genotyping.se). The facility is part of the National Genomics Infrastructure supported by the Swedish Research Council for Infrastructures and Science for Life Laboratory, Sweden. The SNP&SEQ Technology Platform is also supported by the Knut and Alice Wallenberg Foundation.
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/20/13/3221/s1, Table S1. Description of the 72 significant SNPs associated with PFA-100 phenotypes: position in the ABO gene (chromosome 9), location, and significance levels. For collagen-ADP only, the same parameters after adjusting for von Willebrand factor and for coagulation factor VIII.
Author Contributions
J.C.S., J.M.S., and N.P.-M. designed and coordinated de study. N.P.-M. and A.M.-P. wrote the article. D.L. and N.V. performed and interpreted the PFA-100 test and performed other laboratory analysis and the ABO genotype. A.M.-P. conducted the bioinformatic work related to genotyping and imputation, statistic correlations and GWAS. M.S.-L. and A.H. contributed to data analysis and interpretation of GWAS. All authors critically reviewed and approved the final version of the manuscript.
Funding
This study was supported (in part) by the Spanish Ministerio de Educación y Ciencia, grants SAF 2005-04738 and SAF 2008-08159, and by the Spanish Ministerio de Economía y Competitividad, grants RD12/0042/0032, PI12/00612 and PI15/00269. Maria Sabater-Lleal is a recipient of a Miguel Servet contract from the Spanish Ministerio de Salud – ISCIII, CP17/00142.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Paniccia R., Priora R., Liotta A.A., Abbate R. Platelet Function Tests: A Comparative Review. [(accessed on 17 May 2019)];Vasc Health Risk Manag. 2015 11:133–148. doi: 10.2147/VHRM.S44469. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4340464/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrison P. The role of PFA-100 testing in the investigation and management of haemostatic defects in children and adults. [(accessed on 17 May 2019)];Br. J. Haematol. 2005 130:3–10. doi: 10.1111/j.1365-2141.2005.05511.x. Available online: https://onlinelibrary.wiley.com/doi/epdf/10.1111/j.1365-2141.2005.05511.x. [DOI] [PubMed] [Google Scholar]
- 3.Favaloro E.J. Clinical utility of the PFA-100. [(accessed on 21 June 2019)];Semin. Thromb. Hemost. 2008 34:709–733. doi: 10.1055/s-0029-1145254. Available online: https://www.thieme-connect.com/products/ejournals/pdf/10.1055/s-0029-1145254.pdf. [DOI] [PubMed] [Google Scholar]
- 4.Ardillon L., Ternisien C., Fouassier M., Sigaud M., Lefrançois A., Pacault M., Ribeyrol O., Fressinaud E., Boisseau P., Trossaërt M. Platelet function analyser (PFA-100) results and von Willebrand factor deficiency: A 16-year ‘real-world’ experience. [(accessed on 17 May 2019)];Haemophilia. 2015 5:646–652. doi: 10.1111/hae.12653. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1111/hae.12653. [DOI] [PubMed] [Google Scholar]
- 5.Favaloro E.J. Clinical utility of closure times using the platelet function analyzer-100/200. [(accessed on 17 May 2019)];Am. J. Hematol. 2017 92:398–404. doi: 10.1002/ajh.24620. Available online: https://onlinelibrary.wiley.com/doi/full/10.1002/ajh.24620. [DOI] [PubMed] [Google Scholar]
- 6.Fuchs I., Frossard M., Spiel A., Riedmüller E., Laggner A.N., Jilma B. Platelet function in patients with acute coronary syndrome (ACS) predicts recurrent ACS. [(accessed on 17 May 2019)];J. Thromb. Haemost. 2006 4:2547–2552. doi: 10.1111/j.1538-7836.2006.02239.x. Available online: https://onlinelibrary.wiley.com/doi/full/10.1111/j.1538-7836.2006.02239.x. [DOI] [PubMed] [Google Scholar]
- 7.Reny J.L., De Moerloose P., Dauzat M., Fontana P. Use of the PFA-100 closure time to predict cardiovascular events in aspirin-treated cardiovascular patients: A systematic review and meta-analysis. [(accessed on 17 May 2019)];J. Thromb. Haemost. 2008 6:444–450. doi: 10.1111/j.1538-7836.2008.02897.x. Available online: https://onlinelibrary.wiley.com/doi/full/10.1111/j.1538-7836.2008.02897.x. [DOI] [PubMed] [Google Scholar]
- 8.Vázquez-Santiago M., Vilalta N., Cuevas B., Murillo J., Llobet D., Macho R., Pujol-Moix N., Carrasco M., Mateo J., Fontcuberta J., et al. Short closure time values in PFA-100® are related to venous thrombotic risk. Results from the RETROVE Study. [(accessed on 17 May 2019)];Thromb. Res. 2018 169:57–63. doi: 10.1016/j.thromres.2018.07.012. Available online: https://www.thrombosisresearch.com/article/S0049-3848(18)30420-1/fulltext. [DOI] [PubMed] [Google Scholar]
- 9.Nazarian S.M., Thompson J.B., Gluckman T.J., Laws K., Jani J.T., Kickler T.S., Rade J.J. Clinical and laboratory factors associated with shear-dependent platelet hyper-reactivity in patients on chronic aspirin therapy. [(accessed on 17 May 2019)];Thromb. Res. 2010 126:379–383. doi: 10.1016/j.thromres.2009.04.016. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2975867/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayward C.P., Harrison P., Cattaneo M., Ortel T.L., Rao A.K. Platelet Physiology Subcommittee of the Scientific and Standardization Committee of the International Society on Thrombosis and Haemostasis. Platelet function analyzer (PFA)-100 closure time in the evaluation of platelet disorders and platelet function. [(accessed on 17 May 2019)];J. Thromb. Haemost. 2006 4:312–319. doi: 10.1111/j.1538-7836.2006.01771.x. Available online: https://onlinelibrary.wiley.com/doi/full/10.1111/j.1538-7836.2006.01771.x. [DOI] [PubMed] [Google Scholar]
- 11.Vázquez-Santiago M., Ziyatdinov A., Pujol-Moix N., Brunel H., Morera A., Soria J.M., Souto J.C. Age and gender effects on 15 platelet phenotypes in a Spanish population. [(accessed on 17 May 2019)];Comput. Biol. Med. 2016 69:226–233. doi: 10.1016/j.compbiomed.2015.12.023. Available online: https://www.sciencedirect.com/science/article/pii/S0010482515004163?via%3Dihub. [DOI] [PubMed] [Google Scholar]
- 12.Lippi G., Franchini M. Laboratory screening for abnormalities of primary hemostasis: what’s next? [(accessed on 17 May 2019)];Clin. Chem. 2001 47:2071. Available online: http://clinchem.aaccjnls.org/content/47/11/2071.2.long. [PubMed] [Google Scholar]
- 13.Moeller A., Weippert-Kretschmer M., Prinz H., Kretschmer V. Influence of ABO blood groups on primary hemostasis. [(accessed on 17 May 2019)];Transfusion. 2001 41:56–60. doi: 10.1046/j.1537-2995.2001.41010056.x. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1046/j.1537-2995.2001.41010056.x?sid=nlm%3Apubmed. [DOI] [PubMed] [Google Scholar]
- 14.Favaloro E.J., Thom J., Patterson D., Just S., Baccala M., Dixon T., Meiring M., Koutts J., Rowell J., Baker R. Potential supplementary utility of combined PFA-100 and functional von Willebrand factor testing for the laboratory assessment of desmopressin and factor concentrate therapy in von Willebrand disease. [(accessed on 21 June 2019)];Blood Coagul. Fibrinolysis. 2009 20:475–483. doi: 10.1097/MBC.0b013e32832da1ad. Available online: https://journals.lww.com/bloodcoagulation/Abstract/2009/09000/Potential_supplementary_utility_of_combined.17.aspx. [DOI] [PubMed] [Google Scholar]
- 15.Souto J.C., Almasy L., Borrell M., Blanco-Vaca F., Mateo J., Soria J.M., Coll I., Felices R., Stone W., Fontcuberta J., et al. Genetic susceptibility to thrombosis and its relationship to physiological risk factors: The GAIT study. Genetic Analysis of Idiopathic Thrombophilia. [(accessed on 17 May 2019)];Am. J. Hum. Genet. 2000 67:1452–1459. doi: 10.1086/316903. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1287922/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pujol-Moix N., Vázquez-Santiago M., Morera A., Ziyatdinov A., Remacha A., Nomdedeu J.F., Fontcuberta J., Soria J.M., Souto J.C. Genetic determinants of platelet large-cell ratio, immature platelet fraction, and other platelet-related phenotypes. [(accessed on 17 May 2019)];Thromb. Res. 2015 136:361–366. doi: 10.1016/j.thromres.2015.06.016. Available online: https://www.thrombosisresearch.com/article/S0049-3848(15)30031-1/fulltext. [DOI] [PubMed] [Google Scholar]
- 17.Smith N.L., Chen M.H., Dehghan A., Strachan D.P., Basu S., Soranzo N., Hayward C., Rudan I., Sabater-Lleal M., Bis J.C., et al. Novel associations of multiple genetic loci with plasma levels of factor VII, factor VIII, and von Willebrand factor: The CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium. [(accessed on 17 May 2019)];Circulation. 2010 121:1382–1392. doi: 10.1161/CIRCULATIONAHA.109.869156. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2861278/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sabater-Lleal M., Huffman J.E., de Vries P.S., Marten J., Mastrangelo M.A., Song C., Pankratz N., Ward-Caviness C.K., Yanek L.R., Trompet S., et al. Genome-Wide Association Transethnic Meta-Analyses Identifies Novel Associations Regulating Coagulation Factor VIII and von Willebrand Factor Plasma Levels. [(accessed on 17 May 2019)];Circulation. 2019 5:620–635. doi: 10.1161/CIRCULATIONAHA.118.034532. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6438386/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bunimov N., Fuller N., Hayward C.P. Genetic loci associated with platelet traits and platelet disorders. [(accessed on 17 May 2019)];Semin. Thromb. Hemost. 2013 39:291–305. doi: 10.1055/s-0033-1334466. Available online: https://www.thieme-connect.com/products/ejournals/abstract/10.1055/s-0033-1334466. [DOI] [PubMed] [Google Scholar]
- 20.Chen M.H., Yanek L.R., Backman J.D., Eicher J.D., Huffman J.E., Ben-Shlomo Y., Beswick A.D., Yerges-Armstrong L.M., Shuldiner A.R., O’Connell J.R., et al. Exome-chip meta-analysis identifies association between variation in ANKRD26 and platelet aggregation. [(accessed on 17 May 2019)];Platelets. 2017 28:1–10. doi: 10.1080/09537104.2017.1384538. Available online: https://www.tandfonline.com/doi/abs/10.1080/09537104.2017.1384538?journalCode=iplt20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stierlin F.B., Molica F., Reny J.L., Kwak B.R., Fontana P. Pannexin1 Single Nucleotide Polymorphism and platelet reactivity in a cohort of cardiovascular patients. [(accessed on 17 May 2019)];Cell Commun. Adhes. 2017 23:11–15. doi: 10.1080/15419061.2017.1282469. Available online: https://www.tandfonline.com/doi/full/10.1080/15419061.2017.1282469. [DOI] [PubMed] [Google Scholar]
- 22.Sokol J., Skerenova M., Ivankova J., Simurda T., Stasko J. Association of genetic variability in selected genes in patients with deep vein thrombosis and platelet hyperaggregability. [(accessed on 17 May 2019)];Clin. Appl. Thromb. Hemost. 2018 24:1027–1032. doi: 10.1177/1076029618779136. Available online: https://journals.sagepub.com/doi/full/10.1177/1076029618779136?url_ver=Z39.88-2003&rfr_id=ori%3Arid%3Acrossref.org&rfr_dat=cr_pub%3Dpubmed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson A.D., Yanek L.R., Chen M.H., Faraday N., Larson M.G., Tofler G., Lin S.J., Kraja A.T., Province M.A., Yang Q., et al. Genome-wide metaanalyses identifies seven loci associatedwith platelet aggregation in response to agonists. [(accessed on 17 May 2019)];Nat. Genet. 2010 42:608–613. doi: 10.1038/ng.604. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3057573/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qayyum R., Becker L.C., Becker D.M., Faraday N., Yanek L.R., Leal S.M., Shaw C., Mathias R., Suktitipat B., Bray P.F. Genome-wide association study of platelet aggregation in African Americans. [(accessed on 17 May 2019)];BMC Genet. 2015 6:58. doi: 10.1186/s12863-015-0217-9. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4448541/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heit J.A., Armasu S.M., Asmann Y.W., Cunningham J.M., Matsumoto M.E., Petterson T.M., De Andrade M. A genome-wide association study of venous thromboembolism identifies risk variants in chromosomes 1q24.2 and 9q. [(accessed on 17 May 2019)];J. Thromb. Haemost. 2012 10:1521–1531. doi: 10.1111/j.1538-7836.2012.04810.x. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3419811/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang W., Teichert M., Chasman D.I., Heit J.A., Morange P.E., Li G., Pankratz N., Leebeek F.W., Paré G., de Andrade M., et al. A genome-wide association study for venous thromboembolism: The extended cohorts for heart and aging research in genomic epidemiology (CHARGE) consortium. [(accessed on 17 May 2019)];Genet. Epidemiol. 2013 37:512–521. doi: 10.1002/gepi.21731. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3990406/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Germain M., Saut N., Greliche N., Dina C., Lambert J.C., Perret C., Cohen W., Oudot-Mellakh T., Antoni G., Alessi M.C., et al. Genetics of venous thrombosis: Insights from a new genome wide association study. [(accessed on 17 May 2019)];PLoS ONE. 2011 6:e25581. doi: 10.1371/journal.pone.0025581. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3181335/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trégouët D.A., Heath S., Saut N., Biron-Andreani C., Schved J.F., Pernod G., Galan P., Drouet L., Zelenika D., Juhan-Vague I., et al. Common susceptibility alleles are unlikely to contribute as strongly as the FV and ABO loci to VTE risk: Results from a GWAS approach. [(accessed on 17 May 2019)];Blood. 2009 113:5298–5303. doi: 10.1182/blood-2008-11-190389. Available online: http://www.bloodjournal.org/content/113/21/5298.long?sso-checked=true. [DOI] [PubMed] [Google Scholar]
- 29.Gorski M.M., de Haan H.G., Mancini I., Lotta L.A., Bucciarelli P., Passamonti S.M., Cairo A., Pappalardo E., van Hylckama Vlieg A., Martinelli I., et al. Next-generation DNA sequencing to identify novel genetic risk factors for cerebral vein thrombosis. [(accessed on 17 May 2019)];Thromb. Res. 2018 169:76–81. doi: 10.1016/j.thromres.2018.06.011. Available online: https://www.thrombosisresearch.com/article/S0049-3848(18)30385-2/fulltext. [DOI] [PubMed] [Google Scholar]
- 30.Manco L., Silva C., Fidalgo T., Martinho P., Sarmento A.B., Ribeiro M.L. Venous thromboembolism risk associated with ABO, F11 and FGG loci. [(accessed on 17 May 2019)];Blood Coagul. Fibrinolysis. 2018 6:528–532. doi: 10.1097/MBC.0000000000000753. Available online: https://insights.ovid.com/pubmed?pmid=29995659. [DOI] [PubMed] [Google Scholar]
- 31.Reilly M.P., Li M., He J., Ferguson J.F., Stylianou I.M., Mehta N.N., Burnett M.S., Devaney J.M., Knouff C.W., Thompson J.R., et al. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: Two genome-wide association studies. [(accessed on 17 May 2019)];Lancet. 2011 377:383–392. doi: 10.1016/S0140-6736(10)61996-4. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3297116/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams F.M., Carter A.M., Hysi P.G., Surdulescu G., Hodgkiss D., Soranzo N., Traylor M., Bevan S., Dichgans M., Rothwell P.M., et al. Ischemic stroke is associated with the ABO locus: The EuroCLOT study. [(accessed on 22 Jun 2019)];Ann. Neurol. 2013 73:16–31. doi: 10.1002/ana.23838. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3582024/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ling X., Zheng Y., Tao J., Zheng Z., Chen L. Association study of polymorphisms in the ABO gene with ischemic stroke in the Chinese population. [(accessed on 17 May 2019)];BMC Neurol. 2016 16:146. doi: 10.1186/s12883-016-0671-7. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4992279/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.IBC 50K CAD Consortium Large-scale gene-centric analysis identifies novel variants for coronary artery disease. [(accessed on 17 May 2019)];PLoS Genet. 2011 7:e1002260. doi: 10.1371/journal.pgen.1002260. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3178591/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schunkert H., König I.R., Kathiresan S., Reilly M.P., Assimes T.L., Holm H., Preuss M., Stewart A.F., Barbalic M., Gieger C., et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. [(accessed on 17 May 2019)];Nat. Genet. 2011 43:333–338. doi: 10.1038/ng.784. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3119261/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bruzelius M., Strawbridge R.J., Trégouët D.A., Wiggins K.L., Gertow K., Sabater-Lleal M., Öhrvik J., Bergendal A., Silveira A., Sundström A., et al. Influence of coronary artery disease-associated genetic variants on risk of venous thromboembolism. [(accessed on 17 May 2019)];Thromb. Res. 2014 134:426–432. doi: 10.1016/j.thromres.2014.03.054. Available online: https://www.thrombosisresearch.com/article/S0049-3848(14)00184-4/fulltext. [DOI] [PubMed] [Google Scholar]
- 37.Franchini M., Lippi G. Relative risks of thrombosis and bleeding in different ABO blood groups. [(accessed on 17 May 2019)];Semin. Thromb. Hemost. 2016 42:112–117. doi: 10.1055/s-0035-1564832. Available online: https://www.thieme-connect.com/products/ejournals/abstract/10.1055/s-0035-1564832. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H., Mooney C.J., Reilly M.P. ABO blood groups and cardiovascular diseases. Review Article. [(accessed on 17 May 2019)];Int. J. Vasc. Med. 2012 2012:641917. doi: 10.1155/2012/641917. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3485501/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barbalic M., Dupuis J., Dehghan A., Bis J.C., Hoogeveen R.C., Schnabel R.B., Nambi V., Bretler M., Smith N.L., Peters A., et al. Large-scale genomic studies reveal central role of ABO in sP-selectin and sICAM-1 levels. [(accessed on 17 May 2019)];Hum. Mol. Genet. 2010 19:1863–1872. doi: 10.1093/hmg/ddq061. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2850624/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Souto J.C., Almasy L., Muñiz-Diaz E., Soria J.M., Borrell M., Bayén L., Mateo J., Madoz P., Stone W., Blangero J., et al. Functional effects of the ABO locus polymorphism on plasma levels of von Willebrand factor, factor VIII, and activated partial thromboplastin time. [(accessed on 17 May 2019)];Arterioscler. Thromb. Vasc. Biol. 2000 20:2024–2028. doi: 10.1161/01.ATV.20.8.2024. Available online: https://www.ahajournals.org/action/doSearch?AllField=Souto+JC&publication=atvb. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y., Chen C., Ke X., Xiong L., Shi Y., Li J., Tan X., Ye S. Analysis of circulating cholesterol levels as a mediator of an association between ABO blood group and coronary heart disease. [(accessed on 17 May 2019)];Circ. Cardiovasc. Genet. 2014 7:43–48. doi: 10.1161/CIRCGENETICS.113.000299. Available online: https://www.ahajournals.org/doi/full/10.1161/CIRCGENETICS.113.000299?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%3dpubmed. [DOI] [PubMed] [Google Scholar]
- 42.Paterson A.D., Lopes-Virella M.F., Waggott D., Boright A.P., Hosseini S.M., Carter R.E., Shen E., Mirea L., Bharaj B., Sun L., et al. Genome-wide association identifies the ABO blood group as a major locus associated with serum levels of soluble E-selectin. [(accessed on 17 May 2019)];Arterioscler. Thromb. Vasc. Biol. 2009 29:1958–1967. doi: 10.1161/ATVBAHA.109.192971. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3147250/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong M., Zhang H., Reilly J.P., Chrisitie J.D., Ishihara M., Kumagai T., Azadi P., Reilly M.P. ABO blood group as a model for platelet glycan modification in arterial thrombosis. [(accessed on 17 May 2019)];Arterioscler. Thromb. Vasc. Biol. 2015 35:1570–1578. doi: 10.1161/ATVBAHA.115.305337. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4521621/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong X., Kriz R., Seehra J., Kumar R. N-linked glycosylation of platelet P2Y12 ADP receptor is essential for signal transduction but not for ligand binding or cell surface expression. [(accessed on 17 May 2019)];FEBS Lett. 2004 562:111–117. doi: 10.1016/S0014-5793(04)00191-7. Available online: https://febs.onlinelibrary.wiley.com/doi/full/10.1016/S0014-5793%2804%2900191-7?sid=nlm%3Apubmed. [DOI] [PubMed] [Google Scholar]
- 45.Miller S.A., Dykes D.D., Polesky H.F. A simple salting out procedure for extracting DNA from human nucleated cells. [(accessed on 17 May 2019)];Nucleic Acids Res. 1988 16:1215. doi: 10.1093/nar/16.3.1215. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC334765/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olsson M.L., Chester M.A. A rapid and simple ABO genotype screening method using a novel B/O2 versus A/O2 discriminating nucleotide substitution at the ABO locus. [(accessed on 17 May 2019)];Vox Sang. 1995 69:242–247. doi: 10.1111/j.1423-0410.1995.tb02602.x. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1423-0410.1995.tb02602.x?sid=nlm%3Apubmed. [DOI] [PubMed] [Google Scholar]
- 47.Delaneau O., Zagury J.-F., Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. [(accessed on 17 May 2019)];Nat. Methods. 2013 10:5–6. doi: 10.1038/nmeth.2307. Available online: https://www.nature.com/articles/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 48.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. [(accessed on 17 May 2019)];PLoS Genet. 2009 5:e1000529. doi: 10.1371/journal.pgen.1000529. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2689936/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lynch M., Walsh B. Genetics and Analysis of Quantitative Traits. Sinauer Associates Inc.; Sunderland, MA, USA: 1998. [Google Scholar]
- 50.Shabalin A.A. Matrix eQTL: Ultra fast eQTL analysis via large matrix operations. [(accessed on 17 May 2019)];Bioinformatics. 2012 28:1353–1358. doi: 10.1093/bioinformatics/bts163. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3348564/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ziyatdinov A., Brunel H., Martinez-Perez A., Buil A., Perera A., Soria J.M. Solarius: An R interface to SOLAR for variance component analysis in pedigrees. [(accessed on 17 May 2019)];Bioinformatics. 2016 32:1901–1902. doi: 10.1093/bioinformatics/btw080. Available online: https://academic.oup.com/bioinformatics/article/32/12/1901/1744140. [DOI] [PubMed] [Google Scholar]
- 52.Almasy L., Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. [(accessed on 17 May 2019)];Am. J. Hum. Genet. 1998 62:1198–1211. doi: 10.1086/301844. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1377101/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Self S.G., Liang K.Y. Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. J. Am. Stat. Assoc. 1987;82:605–610. doi: 10.1080/01621459.1987.10478472. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.