Abstract
Orofacial clefting is the most common congenital craniofacial malformation, appearing in approximately 1 in 700 live births. Orofacial clefting includes several distinct anatomic malformations affecting the upper lip and hard and soft palate. The etiology of orofacial clefting is multifactorial, including genetic or environmental factors or their combination. A large body of work has focused on the molecular etiology of cleft lip and clefts of the hard palate, but study of the underlying etiology of soft palate clefts is an emerging field. Recent advances in the understanding of soft palate development suggest that it may be regulated by distinct pathways from those implicated in hard palate development. Soft palate clefting leads to muscle misorientation and oropharyngeal deficiency and adversely affects speech, swallowing, breathing, and hearing. Hence, there is an important need to investigate the regulatory mechanisms of soft palate development. Significantly, the anatomy, function, and development of soft palatal muscles are similar in humans and mice, rendering the mouse an excellent model for investigating molecular and cellular mechanisms of soft palate clefts. Cranial neural crest–derived cells provide important regulatory cues to guide myogenic progenitors to differentiate into muscles in the soft palate. Signals from the palatal epithelium also play key roles via tissue-tissue interactions mediated by Tgf-β, Wnt, Fgf, and Hh signaling molecules. Additionally, mutations in transcription factors, such as Dlx5, Tbx1, and Tbx22, have been associated with soft palate clefting in humans and mice, suggesting that they play important regulatory roles during soft palate development. Finally, we highlight the importance of distinguishing specific types of soft palate defects in patients and developing relevant animal models for each of these types to improve our understanding of the regulatory mechanism of soft palate development. This knowledge will provide a foundation for improving treatment for patients in the future.
Keywords: cleft palate, craniofacial biology/genetics, morphogenesis, muscle biology, signal transduction, anatomy
Introduction
Cleft palate is one of the most common craniofacial deformities in humans and affects crucial physiologic functions, including feeding, swallowing, speech, hearing, middle ear ventilation, and respiration (Goudy et al. 2006; Cooper-Brown et al. 2008). The soft palate is a key component of the oropharyngeal complex in mammals, comprising an array of muscles capable of subtle movements, and performs critical functions during swallowing, respiration, and speech (Lieberman 2011). Previous studies identified a large number of signaling pathways that regulate hard palate development (Chai and Maxson 2006; Bush and Jiang 2012). Recent studies suggested that the regulatory mechanism of soft palate development is distinct from that of the hard palate. Hence, there is an important need for a better understanding of the molecular and cellular regulatory mechanism of soft palate development. Here we review the current state of knowledge of soft palate morphogenesis, focusing on tissue-tissue interactions on the molecular level that guide the formation of a functional soft palate. We highlight how well-defined animal models with specific soft palate clefts that mimic similar conditions in humans can help us gain a better understanding of the regulatory mechanism of soft palate morphogenesis.
Soft Palate Function and Anatomy
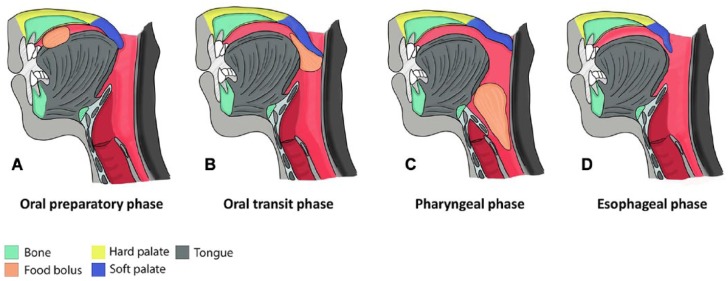
The fleshy soft palate forms approximately the posterior one-third of the roof of the mouth and connects to the bony hard palate to provide physical separation between the oral and nasal cavities (Chai and Maxson 2006). In humans, at the posterior free margin of the soft palate, a conical muscular projection known as the uvula is found at the midline. Unlike the hard palate, the soft palate is mobile and serves as a valve to direct oropharyngeal “traffic” (Keith 1920). The oropharynx is the middle section of the pharynx at the back of the oral cavity between the soft palate and epiglottis, continuous above with the nasopharynx and below with the laryngopharynx (Drake et al. 2005). It serves as the common channel for swallowing and respiration. During food chewing, depression of the soft palate helps close the oropharyngeal isthmus, the gate between the oral cavity and oropharynx, which stops the ordinary swallowing traffic to allow airflow for breathing (Fig. 1A). During swallowing, the soft palate is elevated, and the oropharyngeal isthmus is opened, which stops the breath traffic and directs food into the laryngopharynx (Fig. 1B–D; Lieberman 2011). Therefore, the soft palate is a key player in our most essential activities—breathing and eating, as well as speech and many more.
Figure 1.
Four stages of deglutition (swallowing). (A) During food chewing, depression of the soft palate (dark blue) helps close the oropharyngeal isthmus, the gate between the oral cavity and oropharynx. (B) During swallowing, the soft palate is elevated and touches the posterior pharyngeal wall. At this point, the oropharyngeal isthmus is open. (C) The soft palate stops the breath “traffic” and directs food into the laryngopharynx. (D) Finally, the food enters into esophagus (modified from Lieberman 2011).
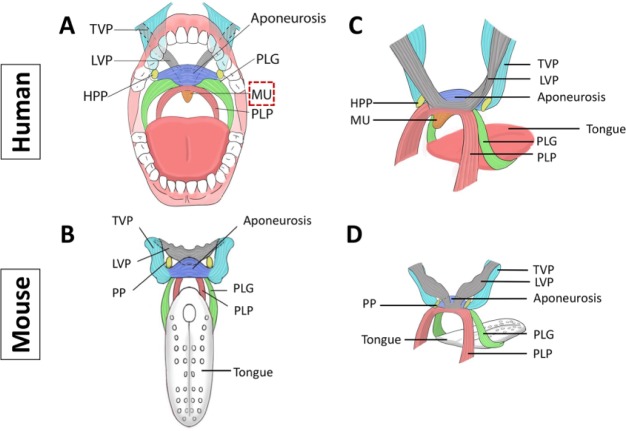
Movement of the soft palate is facilitated by its 5 muscles: the tensor veli palatini (TVP), levator veli palatini (LVP), palatoglossus (PLG), palatopharyngeus (PLP), and musculus uvulae (MU; Fig. 2). Of these, only the MU is uniquely described in humans. Muscles in the soft palate are bilateral, and their fibers interlace at the midline with their partners. All soft palate muscles are innervated by the vagus nerve via the pharyngeal plexus except for TVP, which is innervated by the mandibular branch of the trigeminal nerve (Drake et al. 2005).
Figure 2.
Comparison of soft palate anatomy in humans and mice. Schematic drawings depicting frontal views of the composition and orientation of the soft palate muscles: humans (A) and mice (B). Schematic drawings depicting side views of the composition and orientation of the soft palate muscles: humans (C) and mice (D). Note the tensor veli palatini (TVP) with tendon wrapping around the hamulus of the medial pterygoid palate (HPP). Note also the musculus uvulae (MU), which is present only in humans and is highlighted in panel A by the box with the red dashed line. LVP, levator veli palatini; PLG, palatoglossus; PLP, palatopharyngeus; PP, pterygoid palate. The drawings in panels A to D are based on a human anatomy textbook and 3-dimensional reconstruction of mouse soft palate muscles (Drake et al. 2005; Grimaldi et al. 2015).
The TVP and LVP originate from the soft palate and attach to the base of the skull, whereas the PLG and PLP ascend into the soft palate from the tongue and pharynx, respectively (Drake et al. 2005). These 4 muscles attach to the palatine aponeurosis, which connects the soft palate to the posterior border of the hard palate. This fan-like structure is formed by fibrous horizontal parts of the bilateral TVPs blending along the midline. In the soft palate, the TVP extends laterally and is continuous with the small tendon of the TVP, looping 90° around the hamulus at the inferior border of the medial pterygoid plate and superiorly connecting with the vertical, muscular part of the TVP, which attaches to the pterygoid plate and laterally attaches to the membranous part of the pharyngotympanic tube and the sphenoid bone. The LVP originates from the cartilage of the pharyngotympanic tube and the petrous part of the temporal bone, from which it directly descends and inserts into the palatine aponeurosis. The PLG attaches to the inferior surface of the palatine aponeurosis and inserts into the lateral surface of the tongue. The PLP originates from the superior surface of the palatine aponeurosis and descends into the pharyngeal wall (Drake et al. 2005). The bilateral PLGs and PLPs underlie the palatoglossal arches and palatopharyngeal arches, respectively. The palatoglossal arches define the lateral margins of the oropharyngeal isthmus. The MU originates from the posterior margin of the hard palate and inserts into the connective tissue of the uvula (Drake et al. 2005; Fig. 2A, C).
The functions of soft palatal muscles are defined by their individual courses. The LVP is a strong elevator muscle that aids in closure of the oropharyngeal-nasopharyngeal communication. Both the PLG and the PLP help close the oropharyngeal isthmus during chewing by depressing the soft palate and moving the palatoglossal and palatopharyngeal arches toward the midline. They also assist in elevating the back of the tongue and the pharynx during swallowing. The TVP is responsible for tensing the soft palate, which helps the other muscles work more effectively and achieve a proper seal between the posterior border of the soft palate and the pharyngeal wall (Drake et al. 2005). The TVP also helps open the pharyngotympanic tube during swallowing and yawning to achieve air pressure balance between the middle ear cavity and external ear canal (Drake et al. 2005). Together, these muscles work to accomplish critical physiologic functions of the oropharyngeal complex.
The mouse soft palate has similar tissue components to those of humans; therefore, it is an ideal model for investigation of the molecular and cellular regulatory mechanisms of soft palate morphogenesis. Specifically, the mouse soft palate includes the TVP, LVP, PLG, and PLP muscles. Except for the MU, which is absent in mice, the anatomic features and orientation of soft palate muscles in mice are homologous to those of humans (Fig. 2B, D). Thus, considering these features and its amenability to genetic engineering, the mouse provides an excellent model for investigating soft palate development and defects (Grimaldi et al. 2015).
Soft Palate Development
Soft palate development occurs in the broader context of palatogenesis, which includes formation of the primary and secondary palate. The primary palate is a small portion of the adult hard palate (anterior to the incisive foramen) and is formed through posterior expansion of the frontonasal prominence. The secondary palate constitutes the majority of the hard palate (between the incisive foramen and posterior border of the palatine bone) and the soft palate (posterior to the posterior border of the palatine bone) and is formed through fusion of paired palatal shelves that derive from medial outgrowths of the maxillary prominence. Disruptions to secondary palate development can result in clefting of the hard and soft palate (Yu et al. 2017).
In humans, primordia of the secondary palate initiate early in the sixth week of embryonic development as bilateral outgrowths from the internal aspects of the maxillary prominences to form palatal shelves. Subsequently, the palatal shelves grow inferomedially on either side of the tongue. During the seventh and eighth weeks, the palatal shelves reorient and elevate to their horizontal positions above the dorsum of the tongue as the jaw grows and tongue descends. The palatal shelves complete fusion with the degradation of the medial edge epithelial seam in an anterior-posterior (A-P) direction by the 12th week (Moore and Persaud 2008; Danescu et al. 2015). Soft palate muscles emerge sequentially during the sixth to ninth weeks, with the TVP appearing first and MU last. By the 16th or 17th week, development of the soft palate muscles is complete (Cohen et al. 1993). Despite the aforementioned A-P directionality, fusion of the soft palate is independent from that of the hard palate. In rare cases in humans and mice, the soft palate remains intact despite a cleft of the hard palate (Yu et al. 2005). A recent study demonstrated that there is a difference between the maturation processes in the hard and soft palate, which may correlate with differential gene expression patterns during human palate development. The soft palate fusion process appears to be conserved across mammals (Danescu et al. 2015).
Compared with human palatogenesis, which spans approximately 11 weeks, mouse palatogenesis is normally completed in about 8 days. Secondary palate development begins with formation of paired palatal shelves on embryonic day 11.5 (E11.5), followed by their vertical growth flanking the tongue from E12.5 to E13.5 and elevation to horizontal between E14 and E14.5. Fusion of the palatal shelves starts at E14.5 in an A-P sequence, with hard palate fusion completed by E16.5 (Chai and Maxson 2006; Xu et al. 2006; Iwata et al. 2011). Based on our histologic analysis, only the TVP and LVP regions of the soft palate fuse by E16.5, whereas other soft palatal muscles continue developing until the newborn stage (Grimaldi et al. 2015). Similar to humans, TVP development initiates the earliest, followed by PLG, LVP, and PLP (Grimaldi et al. 2015). Thus, mouse palatogenesis is directly comparable to human palatogenesis, though highly accelerated, making the mouse an excellent model for investigating the regulatory mechanism of palatogenesis.
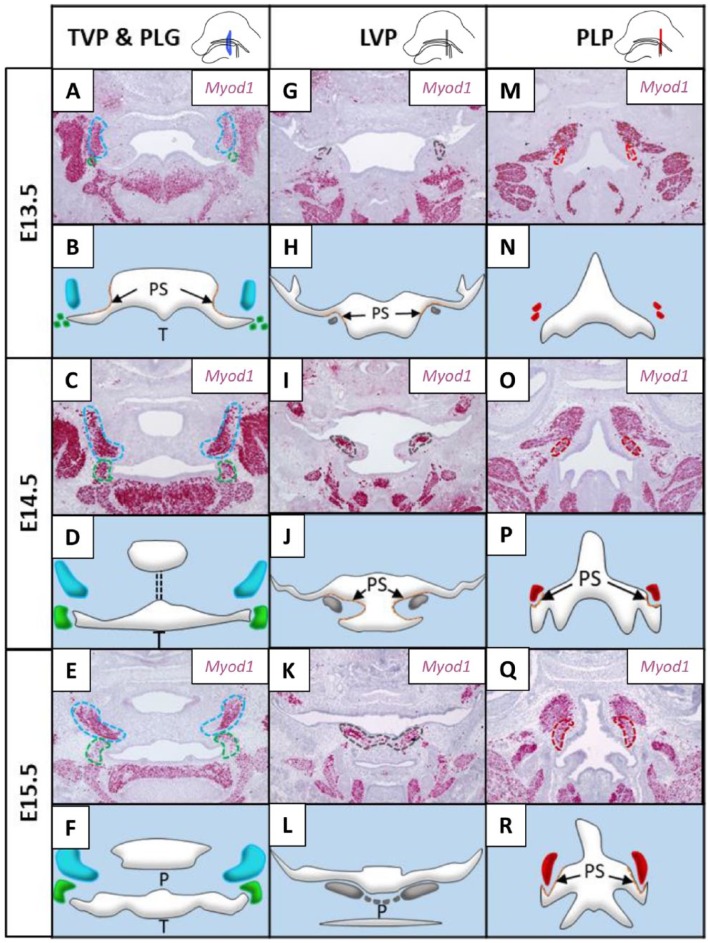
Although the hard and soft palates share some common features and physiologic functions, soft palate development has several unique characteristics. Specifically, cranial neural crest (CNC)–derived mesenchymal cells in the soft palate direct the migration of mesoderm-derived myogenic progenitors into the soft palate in a lateral-to-medial direction following palatal fusion and regulate myogenesis in the soft palate through tissue-tissue interactions (Grimaldi et al. 2015; Fig. 3A–R). For example, CNC-derived mesenchymal cells populate the primordia of palatal shelves in the LVP region very early, while myogenic progenitors are undetectable in the soft palate until E13.5 in the mouse (Fig. 3G–H). As development proceeds, proliferation of CNC-derived mesenchymal cells promotes outgrowth and extension of the palatal shelves toward the midline. In parallel, myogenic progenitors grow medially following the guidance of CNC-derived mesenchymal cells from E13.5 to E15.5 (Fig. 3G–L; Grimaldi et al. 2015). Additionally, A-P axis heterogeneity plays different roles in the hard and soft palate mesenchyme. In the hard palate mesenchyme, several genes show differential expression and regulation along the A-P axis, as evidenced by induction of Msx1 and cell proliferation mediated by Bmp4 exclusively in the anterior region; conversely, Fgf8 specifically induces Pax9 only in the posterior region (Hilliard et al. 2005). In contrast, differential A-P gene expression in the soft palate mesenchyme controls muscle development through tissue-tissue interaction. For example, Dlx5 is expressed in the CNC-derived mesenchyme of the LVP, PLP, and PLG regions. Loss of Dlx5 in the CNC-derived mesenchyme results in defects of the LVP, PLP, and PLG (Lieberman 2011; Sugii et al. 2017).
Figure 3.
Myogenesis of the TVP (tensor veli palatini), LVP (levator veli palatini), PLG (palatoglossus), and PLP (palatopharyngeus) during mouse soft palate development from embryonic day 13.5 (E13.5) to E15.5. (A, C, E, G, I, K, M, O, Q) RNAscope data show the expression of Myod1 during the development of different muscles in the soft palate at E13.5, E14.5, and E15.5. Each muscle primordium is outlined by a dotted line of a color corresponding to the same muscle in the schematic drawings shown below each RNAscope image. (B, D, F, H, J, L, N, P, R) Schematic drawings are based on the expression profile of Myod1 (+) myogenic cells in the primordium of each muscle in the soft palate. P, palate; PS, palatal shelf; T, tongue. The lateral views of the mouse head at the top of the figure show the locations of the sections (Grimaldi et al. 2015).
Soft Palate Defects in Patients and Potential Improvement in Treatment Outcome for Patients
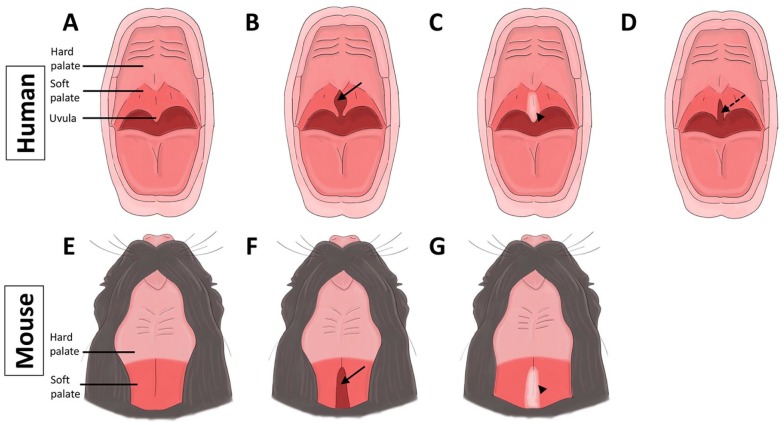
The prevalence of isolated cleft palate is about 6.35 per 10,000 live births, and the prevalence of cleft lip with or without cleft palate is about 10.63 per 10,000 live births (Parker et al. 2010). Approximately 30% of cleft lip and/or palate (CL/P) cases occur with mendelian syndromes, whereas the other 70% are nonsyndromic (Dixon et al. 2011). Genetic or environmental factors or their combination can cause CL/P. Soft palate malformations may appear alone or with cleft hard palate. Thus, it is crucial to investigate the molecular and cellular regulatory mechanisms of soft palate defects in the broader context of CL/P. The Veau classification of cleft palate includes the following: 1) class I, incomplete cleft palate involving soft palate only; 2) class II, complete cleft of the secondary palate; 3) class III, a complete unilateral cleft including lip and palate; and 4) class IV, complete bilateral cleft (Allori et al. 2017). Within class I, soft palate clefts can be further classified as 1) clefts of the soft palate, 2) submucous cleft palate, or 3) bifid uvula (Fig. 4).
Figure 4.
Comparison of soft palate malformations in humans and mice depicting normal palate (A, E), cleft soft palate (B, F; arrows), and submucous cleft palate (C, G; arrowheads) in humans and mice, respectively. (D) Bifid uvula in human is indicated by arrow with dotted line (Xu et al. 2006; Allori et al. 2017).
In different forms of soft palate malformation, muscles are disrupted to different extents. Several properties of the relevant muscles must be considered to achieve effective repair. Each soft palate muscle normally has only 1 skeletal insertion, whereas in patients with cleft soft palate or submucous cleft palate, the muscles may have anomalous attachment with 2 skeletal insertions into the posterior border of the hard palate. For example, the LVP may fail to form a transverse muscular sling, limiting the muscles to isometric contractions and preventing normal soft palatal function (Monroy et al. 2012; Von den Hoff et al. 2018). Moreover, the muscles’ fiber content is abnormal in patients with cleft soft palate. In typically developed individuals, slow- and fast-twitch fibers are present in similar numbers; in individuals with clefts, fast-twitch fibers predominate (Lindman et al. 2001; Hanes et al. 2007). Fast-twitch fibers tire more easily and have a higher activation threshold, whereas slow-twitch fibers are slow to fatigue, with a low activation threshold. As a result, soft palate muscles in cleft patients may fatigue during speech, contributing to velopharyngeal dysfunction (Tachimura et al. 2004; Hanes et al. 2007). Furthermore, fast-twitch fibers are more prone to being damaged during contraction (Macpherson et al. 1997; Rader et al. 2008). Finally, muscles in patients with clefts have reduced blood supply and atrophy due to disorganization at the margin of the cleft and reduced function (Cohen et al. 1994). Currently, surgical intervention is the most common treatment for patients with soft palate malformation. The surgeon dissects the abnormal attachments of the muscles and reconstructs them to seal the cleft in an attempt to restore normal function. However, due to the difficulty of reconstructing the abnormal musculature, 10% to 30% of patients still suffer postoperatively from malfunctions of the soft palate, such as articulation disorders (Marrinan et al. 1998; Von den Hoff et al. 2018). The reduced number and function of satellite cells (the primary muscle stem cells) in cleft soft palate muscles pose a challenge for muscle regeneration (Mozdziak et al. 2001; Von den Hoff et al. 2018). Surgical procedures also often induce fibrosis, which hampers functional muscle regeneration (Von den Hoff et al. 2018).
Clearly, development of new regenerative strategies is necessary to fully restore physiologic functions of the soft palate, increase muscle mass, and prevent fibrosis and fistulae. To regenerate muscle, the significance of tissue-tissue interactions during soft palate development suggests that it will be critical to construct a CNC-like niche to support muscle repair/regeneration. Biological or synthetic scaffolds with directional pores can encourage myofibers to grow in correct alignment. Extrinsic growth factors like those provided by surrounding CNC cells (CNCCs) can promote myogenic precursors to migrate, proliferate, and differentiate properly along the predesigned instructive path within the scaffold (Iwata et al. 2014; Sugii et al. 2017; Von den Hoff et al. 2018). Currently, a few biological and synthetic scaffolds are available for muscle regeneration. There is a need for biomedically engineered scaffolds, which can be synthesized by 3-dimensional bioprinting/curing or electrospinning (Jana et al. 2013; Takeda et al. 2016; Costantini et al. 2017). To prevent muscle fibrosis after surgery, options could be adopted from discoveries concerning fibrosis in other organs, including the lung, kidney, and liver. These options include delivering small molecules, decorin, and microRNAs to prevent fibroblast formation (Yan et al. 2009; Nanthakumar et al. 2015; Lesizza et al. 2017). Several small molecules are available for clinical use, and other promising tools are at preclinical or clinical trial stages. MicroRNAs are a promising therapeutic tool used in cancer treatment (Catela et al. 2017). Recent studies have also suggested microRNAs for treating cleft palate patients (Schoen et al. 2018).
Molecular and Cellular Regulatory Mechanisms of Soft Palate Development
Major signaling pathways, such as Shh, Fgf, Tgf-β, and Wnt, are involved in regulating growth of the palatal shelves through epithelial-mesenchymal interaction. Specifically, our previous studies demonstrated that specific loss of Tgf-β signaling in the palatal epithelium results in cleft soft palate with reduced proliferation of palatal mesenchymal cells (Iwata et al. 2014). Failure of soft palate development in this case is partly caused by disrupted Wnt signaling in the mesenchyme because activation of Wnt signaling can partially rescue the proliferative defect in Tgfbr2 mutant palatal mesenchyme (Iwata et al. 2014). Our findings suggest that Tgf-β signaling in palatal epithelial cells is specifically required for proper activation of Wnt signaling in the soft palate mesenchyme through tissue-tissue interactions (Fig. 5; Iwata et al. 2014). Other members of the Tgf-β family have also been shown to control soft palate development. Constitutive activation of Bmp signaling via Acvr1 leads to submucous cleft palate, highlighting the importance of balanced Bmp/Tgf-β signaling in regulating soft palate development. In the Acvr1 mutant model, there is altered cell proliferation and impaired cell death in the medial edge epithelium, which might interfere with muscle development in the soft palate (Noda et al. 2016). Epithelium-derived Wnt and Shh signaling and transcription factor Tbx1 also play important roles in regulating soft palate morphogenesis via epithelial-mesenchymal interactions. Specifically, downregulation of Shh signaling is required in the medial edge epithelial cells during palatal fusion, whereas constitutive activation of Hedgehog signaling leads to a dysfunctional p63/Irf6 regulatory loop and soft palate cleft (He et al. 2011; Funato et al. 2012; Li et al. 2018).
Figure 5.
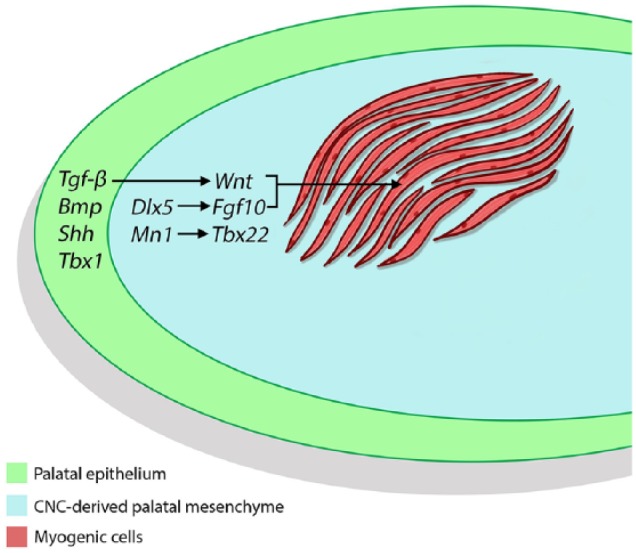
Schematic drawing depicting the mechanism of tissue-tissue interactions between ectoderm-derived palatal epithelial cells, cranial neural crest (CNC)–derived palatal mesenchymal cells, and mesoderm-derived myogenic cells during soft palate development. Signals from the palatal epithelium, such as Tgf-β, regulate Wnt signaling in the CNC-derived palatal mesenchyme, which in turn controls myogenesis (Iwata et al. 2014). Other epithelial signals, such as Bmp, Shh, and Tbx1, are also highlighted here. Transcription factors, such as Dlx5, in CNC-derived cells regulate specific downstream target genes, such as Fgf10, to control myogenesis (Sugii et al. 2017). Transcription factors Mn1 and Tbx22 have been shown to play specific roles in regulating posterior palate development (Liu et al. 2008).
Normal myogenesis is a prerequisite for movement of the soft palate in support of its crucial physiologic functions. Previous studies demonstrated that initiation of myogenesis in vertebrate heads is independent of CNCCs, but migration, patterning, and differentiation of head muscle precursors are regulated by CNCCs (Rinon et al. 2007). CNCCs first migrate into the primordia of the palate, tongue, mandible, and eye, among other structures in the head, then form a scaffold to guide migration and positioning of myogenic progenitors; ultimately, they promote proliferation and differentiation of cranial muscle precursors through cell-cell interactions (Rinon et al. 2007; Hosokawa et al. 2010; Bohnsack et al. 2011; Parada et al. 2012; Han et al. 2014; Tzahor 2015). Differentiated myogenic cells fuse to form myofibers, which are attached to skeletal elements through CNC-derived tendons in a precisely coordinated manner (Chai and Maxson 2006; Han et al. 2012; Tzahor 2015). Ablation of CNCCs leads to abnormalities in differentiation and myofiber organization in the head muscles (Rinon et al. 2007). The tight linkage between CNCCs and cranial muscle precursors indicates crosstalk between the 2 populations via cell-cell interactions.
In the soft palate region, we showed that CNCCs populate the palatal mesenchyme prior to myogenic progenitors migrating into the soft palate (Grimaldi et al. 2015). These myogenic progenitors derive from the pharyngeal mesoderm (Michailovici et al. 2015). Later on, the CNC-derived aponeurosis forms at the level of the TVP (Oka et al. 2012). The tendons and connective tissue surrounding the muscle fibers are derived from CNCCs in the soft palate (Grimaldi et al. 2015). Functional studies showed that, for example, Dlx5-positive cells represent a subset of CNCCs that are adjacent to muscle progenitors in the PLG, LVP, and PLP regions. Loss of Dlx5 leads to a truncated soft palate with missing PLG, LVP, and PLP, due at least in part to decreased secretion of Fgf10 from surrounding CNCCs. Significantly, activation of Fgf10 signaling leads to a rescue of CNCC proliferation and myogenic cell differentiation in Dlx5-/- samples, suggesting that a Dlx5-Fgf10 signaling cascade plays a crucial role in regulating CNC and myogenic cell-cell interaction to control muscle development in the soft palate (Sugii et al. 2017). Other studies showed that transcription factors Mn1 and Tbx22 play crucial roles in regulating soft palate development. Specifically, Mn1 and Tbx22 are expressed in the CNC-derived posterior palatal mesenchyme. Mn1 is required for posterior palatal shelf outgrowth, and loss of Mn1 leads to cleft palate (Liu et al. 2008). Significantly, loss of Mn1 results in downregulation of Tbx22 in the palatal mesenchyme. Loss of Tbx22 results in submucous cleft palate and ankyloglossia in mice, which are similar to the phenotypes of patients with TBX22 mutation (Pauws et al. 2009). Interestingly, the submucous cleft palate in Tbx22 mutant mice is likely due to a defect in the posterior palatine bone, suggesting that defects in hard palate bone formation can have a significant impact on soft palate muscle development. This is likely due to the fact that the CNC-derived aponeurosis, which connects the muscles in the soft palate to the posterior hard palate, may be compromised in Tbx22 mutant mice. CNC-derived cells not only regulate soft palate muscle development (Fig. 5) but also serve as an interface to connect these muscles to the posterior hard palate for them to perform their physiologic functions.
The regulatory mechanism of cell-cell interactions between CNC-derived cells and myogenic cells is well conserved among other craniofacial muscles. TGF-β family members have critical functions in this regard. For instance, deletion of Alk5 in CNC-derived mesenchyme affects the formation of multiple craniofacial muscles, with dramatically reduced total mass in the tongue, eye, and masticatory muscles (Han et al. 2014). Bmp4 and Fgf ligands (Fgf4 and Fgf6), which are targets of Alk5-mediated TGF-β signaling in CNCCs, regulate proliferation and differentiation of myogenic progenitors, respectively (Han et al. 2014). Loss of Tgfbr2 in CNCCs results in microglossia with disorganized, scant muscle cells (Hosokawa et al. 2010). Microglossia in Tgfbr2 mutant mice results from a significant decrease in myogenic cell proliferation, which in turn is due to downregulation of Fgf10 expression. The reduced number of tongue muscle cells in Tgfbr2 mice can be rescued by adding Fgf10, which is expressed only by CNCCs; this suggests a non–cell autonomous mechanism (Hosokawa et al. 2010). Additionally, Bmp signaling represses skeletal muscle differentiation in the head. Myogenic differentiation of cranial paraxial mesoderm initiates upon secretion of Bmp inhibitors, including Noggin and Gremlin from CNCCs (Tzahor et al. 2003). Independent of their role in jaw identity determination, Dlx5/6 plays an important role in regulating masticatory and facial muscle development by repressing Bmp7 and Wnt5a expression, highlighting their role in mediating cell-cell interaction (Heude et al. 2010). Collectively, these findings underscore that better understanding the regulatory mechanism mediated by CNC-derived and myogenic cell-cell interaction will have broad implications for our understanding of craniofacial morphogenesis.
Mouse models have been widely used to investigate the etiology of CL/P (Chai and Maxson 2006; Bush and Jiang 2012), leading to better understanding of molecular and cellular mechanisms and providing valuable knowledge to improve genetic screening for diagnostics and evaluation of recurrence risks (Dixon et al. 2011). However, soft palate malformation–related mouse models are currently limited in number. Genome-wide association studies have shown that mutations to genes in the TGF-β pathway, including TGFβ2, TGFβ3, SMAD3, TGFβR1, and TGFβR2, cause syndromic cleft palate and bifid uvula as part of the autosomal dominant Loeys-Dietz syndrome (Loeys et al. 2005). Significantly, Tgfbr1 and Tgfbr2 mutant mice also exhibit cleft palate, suggesting that Tgf-β signaling pathway is crucial in regulating palatogenesis (Han et al. 2014; Iwata et al. 2014). Recent studies on patients with nonsyndromic orofacial clefts have revealed that differential DNA methylation, epigenetic regulator mutations, and low-frequency genetic variants in noncoding regions may contribute to cleft palate (Alvizi et al. 2017; Shaffer et al. 2019). It is crucial to develop relevant animal models to test how epigenetic factors may control palate development. Furthermore, it will be crucial to integrate the impact of environmental factors into our model of palatogenesis in our effort to gain a better understanding of the etiology of soft palate defects. Finally, a better connection between animal models with specific types of soft palate defects and patients who have similar genotypes/phenotypes will advance our understanding of the regulatory mechanisms of soft palate development and provide a foundation for improving treatment for these patients in the future.
Author Contributions
J. Li, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; G. Rodriguez, X. Han, E. Janečková, contributed to data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; S. Kahng, B. Song, contributed to data acquisition and analysis, critically revised the manuscript; Y. Chai, contributed to conception and design, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Acknowledgments
We thank Bridget Samuels and members of the Chai laboratory for critical reading of the manuscript.
Footnotes
Research studies in the Chai laboratory are supported by the National Institute of Dental and Craniofacial Research of the National Institutes of Health (R37 DE012711 and U01 DE020065 to Y. Chai).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Allori AC, Mulliken JB, Meara JG, Shusterman S, Marcus JR. 2017. Classification of cleft lip/palate: then and now. Cleft Palate Craniofac J. 54(2):175–188. [DOI] [PubMed] [Google Scholar]
- Alvizi L, Ke X, Brito LA, Seselgyte R, Moore GE, Stanier P, Passos-Bueno MR. 2017. Differential methylation is associated with non-syndromic cleft lip and palate and contributes to penetrance effects. Sci Rep. 7(1):2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack BL, Gallina D, Thompson H, Kasprick DS, Lucarelli MJ, Dootz G, Nelson C, McGonnell IM, Kahana A. 2011. Development of extraocular muscles requires early signals from periocular neural crest and the developing eye. Arch Ophthalmol. 129(8):1030–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush JO, Jiang RL. 2012. Palatogenesis: morphogenetic and molecular mechanisms of secondary palate development. Development. 139(2):231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catela Ivkovic T, Voss G, Cornella H, Ceder Y. 2017. MicroRNAs as cancer therapeutics: a step closer to clinical application. Cancer Lett. 407:113–122. [DOI] [PubMed] [Google Scholar]
- Chai Y, Maxson RE. 2006. Recent advances in craniofacial morphogenesis. Dev Dyn. 235(9):2353–2375. [DOI] [PubMed] [Google Scholar]
- Cohen SR, Chen L, Trotman CA, Burdi AR. 1993. Soft-palate myogenesis—a developmental field paradigm. Cleft Palate Craniofac J. 30(5):441–446. [DOI] [PubMed] [Google Scholar]
- Cohen SR, Chen LL, Burdi AR, Trotman CA. 1994. Patterns of abnormal myogenesis in human cleft palates. Cleft Palate Craniofac J. 31(5):345–350. [DOI] [PubMed] [Google Scholar]
- Cooper-Brown L, Copeland S, Dailey S, Downey D, Petersen MC, Stimson C, Van Dyke DC. 2008. Feeding and swallowing dysfunction in genetic syndromes. Dev Disabil Res Rev. 14(2):147–157. [DOI] [PubMed] [Google Scholar]
- Costantini M, Testa S, Mozetic P, Barbetta A, Fuoco C, Fornetti E, Tamiro F, Bernardini S, Jaroszewicz J, Swieszkowski W, et al. 2017. Microfluidic-enhanced 3D bioprinting of aligned myoblast-laden hydrogels leads to functionally organized myofibers in vitro and in vivo. Biomaterials. 131:98–110. [DOI] [PubMed] [Google Scholar]
- Danescu A, Mattson M, Dool C, Diewert VM, Richman JM. 2015. Analysis of human soft palate morphogenesis supports regional regulation of palatal fusion. J Anat. 227(4):474–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MJ, Marazita ML, Beaty TH, Murray JC. 2011. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 12(3):167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake RL, Vogl W, Mitchell AWM. 2005. Gray’s anatomy for students. Philadelphia (PA): Churchill Livingstone. [Google Scholar]
- Funato N, Nakamura M, Richardson JA, Srivastava D, Yanagisawa H. 2012. Tbx1 regulates oral epithelial adhesion and palatal development. Hum Mol Gen. 21(11):2524–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudy S, Lott D, Canady J, Smith RJH. 2006. Conductive hearing loss and otopathology in cleft palate patients. Otolaryngol Head Neck Surg. 134(6):946–948. [DOI] [PubMed] [Google Scholar]
- Grimaldi A, Parada C, Chai Y. 2015. A comprehensive study of soft palate development in mice. PloS One. 10(12):e0145018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han A, Zhao H, Li JY, Pelikan R, Chai Y. 2014. ALK5-mediated transforming growth factor beta signaling in neural crest cells controls craniofacial muscle development via tissue-tissue interactions. Mol Cell Biol. 34(16):3120–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Zhao H, Parada C, Hacia JG, Bringas P, Chai Y. 2012. A TGFβ-Smad4-Fgf6 signaling cascade controls myogenic differentiation and myoblast fusion during tongue development. Development. 139(9):1640–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanes MC, Weinzweig J, Kuzon WM, Panter KE, Buchman SR, Faulkner JA, Yu D, Cederna PS, Larkin LM. 2007. Contractile properties of single permeabilized muscle fibers from congenital cleft palates and normal palates of spanish goats. Plast Reconstr Surg. 119(6):1685–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He FL, Xiong W, Wang Y, Li L, Liu C, Yamagami T, Taketo MM, Zhou CJ, Chen YP. 2011. Epithelial Wnt/beta-catenin signaling regulates palatal shelf fusion through regulation of TGF beta 3 expression. Dev Biol. 350(2):511–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heude E, Bouhali K, Kurihara Y, Kurihara H, Couly G, Janvier P, Levi G. 2010. Jaw muscularization requires Dlx expression by cranial neural crest cells. Proc Natl Acad Sci U S A. 107(25):11441–11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard SA, Yu L, Gu SP, Zhang ZY, Chen YP. 2005. Regional regulation of palatal growth and patterning along the anterior-posterior axis in mice. J Anat. 207(5):655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa R, Oka K, Yamaza T, Iwata J, Urata M, Xu X, Bringas P, Nonaka K, Chai Y. 2010. TGF-beta mediated FGF10 signaling in cranial neural crest cells controls development of myogenic progenitor cells through tissue-tissue interactions during tongue morphogenesis. Dev Biol. 341(1):186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata J, Parada C, Chai Y. 2011. The mechanism of TGF-beta signaling during palate development. Oral Dis. 17(8):733–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata J, Suzuki A, Yokota T, Ho TV, Pelikan R, Urata M, Sanchez-Lara PA, Chai Y. 2014. TGF beta regulates epithelial-mesenchymal interactions through WNT signaling activity to control muscle development in the soft palate. Development. 141(4):909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana S, Cooper A, Zhang MQ. 2013. Chitosan scaffolds with unidirectional microtubular pores for large skeletal myotube generation. Adv Healthc Mater. 2(4):557–561. [DOI] [PubMed] [Google Scholar]
- Keith A. 1920. The engines of the human body: being the substance of Christmas lectures given at the royal institution of Great Britain, Christmas, 1916–1917. Philadelphia (PA): J. B. Lippincott Company. [Google Scholar]
- Lesizza P, Prosdocimo G, Martinelli V, Sinagra G, Zacchigna S, Giacca M. 2017. Single-dose intracardiac injection of pro-regenerative microRNAs improves cardiac function after myocardial infarction. Circ Res. 120(8):1298–1304. [DOI] [PubMed] [Google Scholar]
- Li JY, Yuan Y, He JZ, Feng JF, Han X, Jing JJ, Ho TV, Xu J, Chai Y. 2018. Constitutive activation of hedgehog signaling adversely affects epithelial cell fate during palatal fusion. Dev Biol. 441(1):191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman DE. 2011. The evolution of the human head. Cambridge (MA): Belknap Press of Harvard University Press. [Google Scholar]
- Lindman R, Paulin G, Stal PS. 2001. Morphological characterization of the levator veli palatini muscle in children born with cleft palates. Cleft Palate Craniofac J. 38(5):438–448. [DOI] [PubMed] [Google Scholar]
- Liu W, Lan Y, Pauws E, Meester-Smoor MA, Stanier P, Zwarthoff EC, Jiang R. 2008. The Mn1 transcription factor acts upstream of Tbx22 and preferentially regulates posterior palate growth in mice. Development. 135(23):3959–3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeys BL, Chen J, Neptune ER, Judge DP, Podowski M, Holm T, Meyers J, Leitch CC, Katsanis N, Sharifi N, et al. 2005. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat Genet. 37(3):275–281. [DOI] [PubMed] [Google Scholar]
- Macpherson PC, Dennis RG, Faulkner JA. 1997. Sarcomere dynamics and contraction-induced injury to maximally activated single muscle fibres from soleus muscles of rats. J Physiol. 500(Pt 2):523–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrinan EM, LaBrie RA, Mulliken JB. 1998. Velopharyngeal function in nonsyndromic cleft palate: relevance of surgical technique, age at repair, and cleft type. Cleft Palate Craniofac J. 35(2):95–100. [DOI] [PubMed] [Google Scholar]
- Michailovici I, Eigler T, Tzahor E. 2015. Craniofacial muscle development. Curr Top Dev Biol. 115:3–30. [DOI] [PubMed] [Google Scholar]
- Monroy PLC, Grefte S, Kuijpers-Jagtman AM, Wagener FA, Von den Hoff JW. 2012. Strategies to improve regeneration of the soft palate muscles after cleft palate repair. Tissue Eng Part B Rev. 18(6):468–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KL, Persaud TVN. 2008. The developing human clinically oriented embryology, 8th ed. Philadephia (PA): Saunders. [Google Scholar]
- Mozdziak PE, Pulvermacher PM, Schultz E. 2001. Muscle regeneration during hindlimb unloading results in a reduction in muscle size after reloading. J Appl Physiol. 91(1):183–190. [DOI] [PubMed] [Google Scholar]
- Nanthakumar CB, Hatley RJ, Lemma S, Gauldie J, Marshall RP, Macdonald SJ. 2015. Dissecting fibrosis: therapeutic insights from the small-molecule toolbox. Nat Rev Drug Discov. 14(10):693–720. [DOI] [PubMed] [Google Scholar]
- Noda K, Mishina Y, Komatsu Y. 2016. Constitutively active mutation of ACVR1 in oral epithelium causes submucous cleft palate in mice. Dev Biol. 415(2):306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka K, Honda MJ, Tsuruga E, Hatakeyama Y, Isokawa K, Sawa Y. 2012. Roles of collagen and periostin expression by cranial neural crest cells during soft palate development. J Histochem Cytochem. 60(1):57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada C, Han D, Chai Y. 2012. Molecular and cellular regulatory mechanisms of tongue myogenesis. J Dent Res. 91(6):528–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SE, Mai CT, Canfield MA, Rickard R, Wang Y, Meyer RE, Anderson P, Mason CA, Collins JS, Kirby RS, et al. 2010. Updated national birth prevalence estimates for selected birth defects in the United States, 2004–2006. Birth Defects Res A Clin Mol Teratol. 88(12):1008–1016. [DOI] [PubMed] [Google Scholar]
- Pauws E, Hoshino A, Bentley L, Prajapati S, Keller C, Hammond P, Martinez-Barbera JP, Moore GE, Stanier P. 2009. Tbx22(null) mice have a submucous cleft palate due to reduced palatal bone formation and also display ankyloglossia and choanal atresia phenotypes. Hum Mol Genet. 18(21):4171–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rader EP, Cederna PS, McClellan WT, Caterson SA, Panter KE, Yu D, Buchman SR, Larkin LM, Faulkner JA, Weinzweig J. 2008. Effect of cleft palate repair on the susceptibility to contraction-induced injury of single permeabilized muscle fibers from congenitally-clefted goat palates. Cleft Palate Craniofacial J. 45(2):113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinon A, Lazar S, Marshall H, Buchmann-Moller S, Neufeld A, Elhanany-Tamir H, Taketo MM, Sommer L, Krumlauf R, Tzahor E. 2007. Cranial neural crest cells regulate head muscle patterning and differentiation during vertebrate embryogenesis. Development. 134(17):3065–3075. [DOI] [PubMed] [Google Scholar]
- Schoen C, Glennon JC, Abghari S, Bloemen M, Aschrafi A, Carels CEL, Von den Hoff JW. 2018. Differential microrna expression in cultured palatal fibroblasts from infants with cleft palate and controls. Eur J Orthodont. 40(1):90–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer JR, LeClair J, Carlson JC, Feingold E, Buxo CJ, Christensen K, Deleyiannis FWB, Field LL, Hecht JT, Moreno L, et al. 2019. Association of low-frequency genetic variants in regulatory regions with nonsyndromic orofacial clefts. Am J Med Genet A. 179(3):467–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugii H, Grimaldi A, Li JY, Parada C, Thach VH, Feng JF, Jing JJ, Yuan Y, Guo YX, Maeda H, et al. 2017. The Dlx5-FGF10 signaling cascade controls cranial neural crest and myoblast interaction during oropharyngeal patterning and development. Development. 144(21):4037–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachimura T, Kotani Y, Wada T. 2004. Nasalance scores in wearers of a palatal lift prosthesis in comparison with normative data for Japanese. Cleft Palate Craniofac J. 41(3):315–319. [DOI] [PubMed] [Google Scholar]
- Takeda N, Tamura K, Mineguchi R, Ishikawa Y, Haraguchi Y, Shimizu T, Hara Y. 2016. In situ cross-linked electrospun fiber scaffold of collagen for fabricating cell-dense muscle tissue. J Artif Organs. 19(2):141–148. [DOI] [PubMed] [Google Scholar]
- Tzahor E. 2015. Head muscle development. Results Probl Cell Differ. 56:123–142. [DOI] [PubMed] [Google Scholar]
- Tzahor E, Kempf H, Mootoosamy RC, Poon AC, Abzhanov A, Tabin CJ, Dietrich S, Lassar AB. 2003. Antagonists of Wnt and BMP signaling promote the formation of vertebrate head muscle. Genes Dev. 17(24):3087–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von den Hoff JW, Carvajal Monroy PL, Ongkosuwito EM, van Kuppevelt TH, Daamen WF. 2018. Muscle fibrosis in the soft palate: delivery of cells, growth factors and anti-fibrotics. Adv Drug Deliv Rev [epub ahead of print 11 August 2018]. doi: 10.1016/j.addr.2018.08.002 [DOI] [PubMed] [Google Scholar]
- Xu X, Han J, Ito Y, Bringas P, Urata MM, Chai Y. 2006. Cell autonomous requirement for Tgfbr2 in the disappearance of medial edge epithelium during palatal fusion. Dev Biol. 297(1):238–248. [DOI] [PubMed] [Google Scholar]
- Yan W, Wang PH, Zhao CX, Tang JR, Xiao X, Wang DW. 2009. Decorin gene delivery inhibits cardiac fibrosis in spontaneously hypertensive rats by modulation of transforming growth factor-beta/Smad and p38 mitogen-activated protein kinase signaling pathways. Hum Gene Ther. 20(10):1190–1200. [DOI] [PubMed] [Google Scholar]
- Yu K, Deng M, Naluai-Cecchini T, Glass IA, Cox TC. 2017. Differences in oral structure and tissue interactions during mouse vs. human palatogenesis: implications for the translation of findings from mice. Front Physiol; 8:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Gu SP, Alappat S, Song YQ, Yan MQ, Zhang XY, Zhang GZ, Jiang YP, Zhang ZY, Zhang YD, et al. 2005. Shox2-deficient mice exhibit a rare type of incomplete clefting of the secondary palate. Development. 132(19):4397–4406. [DOI] [PubMed] [Google Scholar]