Abstract
Background
Prognostic markers specific to a particular cancer type can assist in the evaluation of survival probability of patients and help clinicians to assess the available treatment modalities.
Methods
Gene expression data was analyzed from three independent colon cancer microarray gene expression data sets (N = 1052). Survival analysis was performed for the three data sets, stratified by the expression level of the LINE-1 type transposase domain containing 1 (L1TD1). Correlation analysis was performed to investigate the role of the interactome of L1TD1 in colon cancer patients.
Results
We found L1TD1 as a novel positive prognostic marker for colon cancer. Increased expression of L1TD1 associated with longer disease-free survival in all the three data sets. Our results were in contrast to a previous study on medulloblastoma, where high expression of L1TD1 was linked with poor prognosis. Notably, in medulloblastoma L1TD1 was co-expressed with its interaction partners, whereas our analysis revealed lack of co-expression of L1TD1 with its interaction partners in colon cancer.
Conclusions
Our results identify increased expression of L1TD1 as a prognostic marker predicting longer disease-free survival in colon cancer patients.
Electronic supplementary material
The online version of this article (10.1186/s12885-019-5952-2) contains supplementary material, which is available to authorized users.
Keywords: Human L1TD1 gene, Colon cancer, Prognostic factors, Biomarkers, Survival analysis
Background
Colon cancer is the third leading cancer, both in terms of newly diagnosed cases and mortality [1]. Despite the fact that chemotherapeutic agents, such as oxaliplatin and irinotecan, have markedly improved the survival rate in colon cancer [2], identification of patients likely to respond well to chemotherapy could increase the survival rate. Our study identifies LINE-1 type transposase domain containing 1 (L1TD1) as a novel positive prognostic marker for colon cancer.
Stem cell-like gene signatures have been detected in various cancers [3, 4], and embryonic stem cell factors have been associated with enhanced tumorigenesis and poor prognosis [5–7]. L1TD1 is an RNA-binding protein required for self-renewal of undifferentiated embryonic stem cells [8]. Recently, L1TD1 protein was shown to form a core interaction network with the canonical pluripotency factors OCT4, NANOG, LIN28, and SOX2 in human embryonic stem cells (hESCs) [9], and L1TD1 depletion resulted in downregulation of the pluripotency markers OCT4, NANOG, and LIN28 in hESCs [10]. L1TD1 has previously been shown to be essential for self-renewal of embryonal carcinoma cells [10] and to support the growth of seminoma cells [10].
We studied L1TD1 immunoexpression in colon adenocarcinoma tissue sections and analyzed three independent gene expression microarray data sets of colon cancer patients to assess the prognostic significance of L1TD1 in colon cancer [11–13]. Our findings suggest that L1TD1 is a promising prognostic marker for colon cancer.
Methods
Microarray data sets
Raw microarray data sets (Table 1) were downloaded from Gene Expression Omnibus (GEO) [17]. Three colon cancer gene expression microarray data sets comprising a total of 1052 clinical samples were analyzed [11–13]. Either due to non-tumoral origin (i.e. normal tissue) or due to missing associated survival information, 124 samples had to be excluded from the survival analysis (928 samples remained). Additionally, two seminoma [14, 15] and one stem cell [16] gene expression microarray data sets were analyzed to assess the co-expression of L1TD1 and its interaction partners (Additional file 2: Table S1). The stem cell data set was composed of samples from ten hESCs, 49 induced pluripotent stem cells (iPSCs), five cancer cell lines, and six non-cancerous somatic cell lines. A summary of the data sets used is presented in Table 1.
Table 1.
Summary of the data sets used in the study. The GEO accession numbers (GEO ID) are listed together with alias names used to refer to the individual data sets, the microarray platform, the total number of samples, and the number of samples used in the survival analysis
| GEO ID | Total Samples | Survival Analysis | Platform | Alias |
|---|---|---|---|---|
| GSE14333 [11] | 290 | 226 | Affymetrix HG-U133Plus2 | colon1 |
| GSE17536 [12] | 177 | 145 | Affymetrix HG-U133Plus2 | colon2 |
| GSE39582 [13] | 585 | 557 | Affymetrix HG-U133Plus2 | colon3 |
| GSE3218 [14] | 107 | Not used | Affymetrix HG-U133A | seminoma1 |
| GSE10783 [15] | 34 | Not used | Affymetrix HG-U133A | seminoma2 |
| GSE42445 [16] | 70 | Not used | Agilent-028004 SurePrint G3 Human GE 8x60K | hESC1 |
Gene expression analysis
The CEL files, containing the probe intensity measurements of the Affymetrix probes were normalized using the Universal exPression Code (UPC) normalization method from the Bioconductor package “SCAN.UPC” [18] and the Robust Multiarray Average (RMA) normalization method from the Bioconductor package “affy” [19, 20]. The UPC normalization method provides a score between 0.0 and 1.0, which represents the probability of a particular gene being expressed in a particular sample [18]. The UPC scores were used to categorize the samples in all data sets based on their L1TD1 expression status as L1TD1 high (UPC > =0.60) and L1TD1 low (UPC < 0.60). The UPC threshold of 0.6 was determined by calculating a weighted mean (by sample size) of the local minima between the two peaks in the bimodal distributions of UPC scores for L1TD1 over the three colon cancer data sets (Additional file 1: Fig. S1). RMA provides normalized log2 intensity values. RMA normalized gene expression values were used to calculate pairwise correlations between genes. To correct for multiple testing, the false discovery rate (FDR) was controlled using the Benjamini-Hochberg procedure [21]. The probe “219955_at” was chosen as the primary probe for the quantification of L1TD1 because it was present in both of the Affymetrix platforms used in this study (HG-U133Plus2 and HG-U133A).
Gene list descriptions
Interaction partners
The 311 interaction partners of L1TD1 were determined using mass spectrometry and co-immunoprecipitation in our earlier study [9]. 306 interaction partners of L1TD1 were identified by performing a mass spectrometry analysis on co-immunoprecipitated proteins with two different anti-L1TD1 antibodies (recognizing different epitopes on L1TD1). In addition, for 5 proteins (NANOG, OCT4 (POU5F1), SOX2, DNMT3B, and TRIM28) that were challenging to detect using mass spectrometry, the interactions were shown using immunoprecipitation and Western blotting. Out of the 311 interaction partners, 285 corresponded to genes that had probes associated to them in the microarray platforms used in this study.
Top 20 interaction partners
The top 20 interaction partners of L1TD1 were determined on the basis of their co-expression with L1TD1 in the seminoma and stem cell data sets. First, the interaction partners were ranked in descending order of their Spearman rank correlation value with L1TD1 in these data sets. Then, the maximum rank over the data sets was selected as a representative statistic for each interaction partner. The list was ordered (ascending) based on this maximum rank and 20 interaction partners were selected from the top of the list.
Top 20 co-expressed genes with L1TD1 in colon cancer
Out of all the genes in the microarray data sets (27213 unique probe-gene mappings), top 20 genes were selected based on their co-expression with L1TD1 in the colon cancer data sets. First, all the genes in the microarray data sets were ranked in descending order of their Spearman rank correlation value with L1TD1 separately for each colon cancer data set. Then, the maximum rank over these data sets was selected as a representative statistic for each gene. The list was ordered (ascending) based on this maximum rank and 20 genes were selected from the top of the list.
Survival analysis of microarray data
Disease-free survival was analyzed in each data set with the Kaplan-Meier method as implemented in the R package “survival” [22, 23] and survival curves were plotted using the R package “survminer” [24]. The log-rank test was used to compare survival rates between the two L1TD1 groups (L1TD1 high and L1TD1 low).
Association between L1TD1 expression and clinicopathological variables
We investigated the association of age and sex and other publicly accessible clinicopathological variables to the L1TD1 gene expression in the three gene expression data sets. The variables included cancer stage [11–13], prior-therapy received by the patients [11–13], tumor location [11–13], chromosomal instability [13], CpG island methylation status [13], DNA mismatch repair proficiency [13], mutation status of BRAF (B-Raf proto-oncogene, serine/threonine kinase), mutation status of KRAS (KRAS proto-oncogene, GTPase), and mutation status of TP53 (Tumor Protein p53) [13]. For variables with only two categories, Wilcoxon rank sum test [25] was used for the analysis of statistical significance. For variables with more than two categories, Kruskal-Wallis test [26] was used. Association of L1TD1 expression with age was investigated using Pearson correlation [27].
Analysis of TCGA Colon adenocarcinoma RNA-seq data set
RNA-seq data from The Cancer Genome Atlas Colon Adenocarcinoma [28] (TCGA-COAD) data set was acquired from Genomic Data Commons (portal.gdc.cancer.gov). The FPKM-UQ normalized (Fragments Per Kilobase of transcript per Million mapped reads Upper Quartile) RNA-seq counts from the primary tumor samples (N = 521) were used to validate the correlation analyses performed using the microarray data sets. Due to lack of an evident choice of the intensity threshold to designate samples into high and low L1TD1 expression groups, we fitted a mixture of two Gaussian distributions and evaluated two different thresholds (Additional file 1: Figure S2): FPKM-UQ value where the ratio of the two Gaussian distributions was equal, and FPKM-UQ value where the ratio of the two Gaussian distributions was 10%. These two thresholds were then used to perform survival analysis using disease-free survival with Kaplan-Meier method.
Results
High expression of L1TD1 associates with longer disease-free survival
Across the three colon cancer microarray data sets, 26.7% of the colon cancer patients were categorized to have high L1TD1 expression (Table 2, Additional file 1: Figure S3). The proportion was lower than that observed in seminoma (48.6 and 50%) and stem cell (88.6%) data sets (Table 2, Additional file 1: Figure S3).
Table 2.
Proportion of samples with high expression of L1TD1. The samples were categorized based on their L1TD1 expression level (high L1TD1+ or low L1TD1-) in the different data sets used in this study. For the colon cancer data sets, only tumor samples with complete survival information were considered
| Data set | L1TD1 + | L1TD1 - | Total | Percentage of L1TD1 + |
|---|---|---|---|---|
| colon1 | 64 | 162 | 226 | 28.3% |
| colon2 | 44 | 101 | 145 | 30.3% |
| colon3 | 140 | 417 | 557 | 25.1% |
| Total (Colon Cancer) | 248 | 680 | 928 | 26.7% |
| seminoma1 | 52 | 55 | 107 | 48.6% |
| seminoma2 | 17 | 17 | 34 | 50.0% |
| hESC1 | 62 | 8 | 70 | 88.6% |
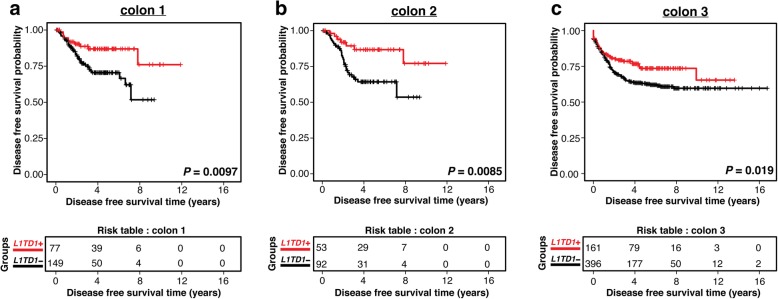
Kaplan-Meier analysis of 928 samples from the three colon cancer data sets revealed that the colon cancer samples with high L1TD1 expression had longer disease-free survival as compared to those with no/low L1TD1 expression (Fig. 1). The difference was statistically significant in all the three data sets (log-rank test P < 0.05).
Fig. 1.
Survival curves for colon cancer. Kaplan-Meier curves showing disease-free survival for the three colon cancer data sets (a-c). The curves present survival data for the two groups of colon cancer patients based on L1TD1 expression level (high or low). The red curve corresponds to the patients with high L1TD1 expression and the black curve corresponds to the patients with low L1TD1 expression. The x-axis shows disease-free survival time in years and the y-axis shows the probability of disease-free survival. The risk table shows the number of patients at risk at the given time point
L1TD1 expression was higher in the samples from early cancer stages as compared to those from later stages in all the three data sets (P < 0.05), whereas differences between the later stages were typically not statistically significant (Additional file 1: Figure S4A-C). In the dataset colon3, L1TD1 expression was high for samples with mutated KRAS (P < 0.0001), wild-type TP53 (P < 0.0001), and negative chromosomal instability marker (P < 0.0001) (Additional file 1: Figure S4D-F). Additionally, significant associations were observed between L1TD1 expression and tumor location or tumor differentiation status (P < 0.0001) (Additional file 1: Figure S4G-I). Age, sex, prior therapy (chemo-, radio- or adjuvant therapy), BRAF mutation status, CpG island methylation status, or DNA mismatch repair proficiency did not show statistically significant associations with the L1TD1 expression (Additional file 1: Figure S5).
Interactome of L1TD1 is not co-expressed in colon cancer
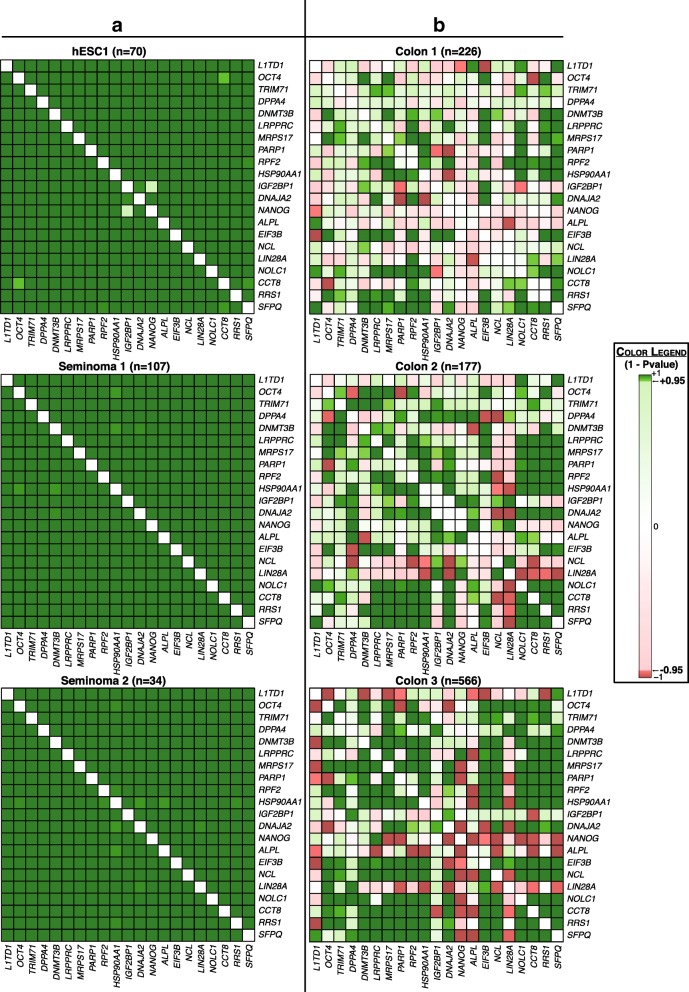
To examine the potential role of the previously identified interaction partners of L1TD1 [9] (Additional file 2: Table S1) in its prognostic performance in colon cancer, Spearman rank correlation matrices were calculated between the expression levels of L1TD1 and its interaction partners [9]. Interestingly, the high positive correlation observed among L1TD1 and its top 20 interaction partners in seminoma and stem cell data sets (P < 0.0001, Fig. 2a) was absent in all three colon cancer data sets (Fig. 2b). However, the interaction partners did not consistently improve the predictive prognostic power obtained with L1TD1 alone (Additional file 2: Table S2).
Fig. 2.
Co-expression of interaction partners of L1TD1. Heatmaps showing signed P-value of Spearman rank correlation for the 20 most significantly co-expressed interaction partners of L1TD1 determined on the basis of the seminoma and stem cell data sets. Co-expression in (a) seminoma and stem cell data sets, and (b) colon cancer data sets. The signed P-value of Spearman rank correlation was defined as 1 - P-value of Spearman rank correlation multiplied by the sign of the correlation
Genes co-expressed with L1TD1 in colon cancer
We identified genes that were co-expressed with L1TD1 in colon cancer patients using Spearman rank correlation (Table 3, Additional file 2: Table S3). Although none of the top 20 co-expressed genes outperformed L1TD1 as independent prognostic marker for colon cancer in all the three data sets, five genes had statistically significant (P < 0.05) impact on survival in at least two out of the three colon cancer data sets (Table 4): Serine peptidase inhibitor Kazal type 4 (SPINK4), Resistin-like beta (RETNLB), Asparaginase-like 1 Protein (ASRGL1), Chloride channel accessory 1 (CLCA1), and Fc fragment of IgG binding protein (FCGBP) (Additional file 1: Figure S6).
Table 3.
Top 20 co-expressed genes with L1TD1 in colon cancer. The Spearman rank correlation values (rs) with L1TD1 are shown together with their false discovery rates (FDR) separately for each colon cancer data set
| colon 1 | colon 2 | colon 3 | |||||
|---|---|---|---|---|---|---|---|
| Rank | Gene Name | r s | FDR | r s | FDR | r s | FDR |
| 1 | RETNLB | 0.47 | 9.26E-13 | 0.53 | 3.69E-10 | 0.45 | 0.00 |
| 2 | CLCA1 | 0.45 | 5.65E-12 | 0.43 | 1.10E-05 | 0.45 | 0.00 |
| 3 | HEPACAM2 | 0.43 | 1.05E-10 | 0.41 | 3.98E-05 | 0.46 | 0.00 |
| 4 | FOXA3 | 0.41 | 1.14E-09 | 0.43 | 1.06E-05 | 0.43 | 0.00 |
| 5 | FCGBP | 0.41 | 1.14E-09 | 0.39 | 2.15E-04 | 0.47 | 0.00 |
| 6 | ST6GALNAC1 | 0.40 | 4.55E-09 | 0.39 | 1.87E-04 | 0.43 | 2.57E-24 |
| 7 | SPINK4 | 0.44 | 2.99E-11 | 0.38 | 3.91E-04 | 0.43 | 5.06E-25 |
| 8 | KIAA1324 | 0.40 | 4.60E-09 | 0.44 | 7.71E-06 | 0.39 | 0.00 |
| 9 | KLF4 | 0.40 | 4.60E-09 | 0.37 | 4.61E-04 | 0.41 | 0.00 |
| 10 | GMDS | 0.46 | 1.50E-12 | 0.40 | 9.95E-05 | 0.38 | 0.00 |
| 11 | SLITRK6 | 0.43 | 5.87E-11 | 0.36 | 1.14E-03 | 0.46 | 0.00 |
| 12 | SERPINA1 | 0.42 | 1.35E-10 | 0.38 | 3.84E-04 | 0.35 | 1.26E-16 |
| 13 | LINC00261 | 0.34 | 1.45E-06 | 0.35 | 2.09E-03 | 0.48 | 0.00 |
| 14 | ITLN1 | 0.35 | 4.43E-07 | 0.33 | 3.97E-03 | 0.42 | 0.00 |
| 15 | MUC2 | 0.39 | 8.64E-09 | 0.33 | 4.90E-03 | 0.38 | 0.00 |
| 16 | DEFA5 | 0.37 | 5.72E-08 | 0.35 | 1.78E-03 | 0.33 | 6.77E-14 |
| 17 | ASRGL1 | 0.40 | 4.55E-09 | 0.32 | 6.22E-03 | 0.41 | 0.00 |
| 18 | SLC27A2 | 0.36 | 2.17E-07 | 0.36 | 9.05E-04 | 0.33 | 2.44E-13 |
| 19 | RNF186 | 0.32 | 8.44E-06 | 0.36 | 1.30E-03 | 0.34 | 1.89E-14 |
| 20 | PCCA | 0.37 | 1.05E-07 | 0.37 | 7.52E-04 | 0.33 | 2.95E-13 |
Table 4.
Prognostic assessment of genes that co-express with L1TD1 in colon cancer
| Gene | colon1 | colon2 | colon3 |
|---|---|---|---|
| L1TD1 | 0.009729 | 0.008520 | 0.018607 |
| SPINK4 | 0.007148 | 0.001854 | 0.880992 |
| RETNLB | 0.325642 | 0.012519 | 0.009064 |
| ASRGL1 | 0.015986 | 0.521116 | 0.016293 |
| CLCA1 | 0.030053 | 0.006496 | 0.710961 |
| FCGBP | 0.028617 | 0.047080 | 0.292182 |
| ITLN1 | 0.088225 | 0.043802 | 0.844453 |
| FOXA3 | 0.077752 | 0.609721 | 0.093598 |
| PCCA | 0.064797 | 0.601176 | 0.107992 |
| DEFA5 | 0.136904 | 0.157008 | 0.737800 |
| GMDS | 0.318171 | 0.170255 | 0.000919 |
| HEPACAM2 | 0.368837 | 0.687066 | 0.098125 |
| SERPINA1 | 0.000008 | 0.493419 | 0.911649 |
| RNF186 | 0.700045 | 0.541107 | 0.010793 |
| KLF4 | 0.938136 | NA | 0.220231 |
| ST6GALNAC1 | 0.593332 | 0.880638 | 0.030027 |
| MUC2 | 0.624983 | 0.505661 | 0.842770 |
| KIAA1324 | 0.220079 | 0.969530 | 0.730810 |
| SLITRK6 | 0.750696 | 0.894483 | 0.085490 |
| LINC00261 | 0.823520 | 0.823442 | 0.269044 |
| SLC27A2 | 0.883481 | 0.975288 | 0.002906 |
Statistical significance of the top 20 co-expressed genes in the survival analysis of colon cancer patients in the three data sets. Genes with statistically significant association with disease-free survival (log-rank test P < 0.05) in at least two colon cancer data sets are underlined
Validation in TCGA Colon adenocarcinoma RNA-seq data set
To further validate our findings from the three colon cancer microarray data sets, we analyzed the TCGA Colon Adenocarcinoma [28] (TCGA-COAD) RNA-seq data set containing 521 patient samples. When the samples were stratified for L1TD1 expression using the threshold where the ratio of the two Gaussian distributions was 10%, Kaplan-Meier analysis supported that the colon cancer samples with high L1TD1 expression had longer disease-free survival as compared to those with no/low L1TD1 expression (P = 0.038, Additional file 1: Figure S2C). Additionally, we were able to reproduce the findings from the correlation analyses, indicating a lack of correlation between L1TD1 and its top 20 interaction partners (Additional file 1: Figure S2D) and confirming significant correlations between L1TD1 and genes that were co-expressed with L1TD1 in the colon cancer microarray data sets (Additional file 1: Figure S2E).
Discussion
In this study, we examined the prognostic value of L1TD1 in colon cancer patients. We found compelling evidence of L1TD1 being a positive prognostic marker for colon cancer (Fig. 1). We demonstrated this by survival analysis of 928 samples from three independent gene expression data sets of colon cancer patients and further confirmed the results in the TCGA Colon Adenocarcinoma RNA-seq data set of 521 colon cancer patients.
Expression of L1TD1 has earlier been reported to be highly specific to embryonic stem cells [10], brain [29], and colon (Additional file 1: Figure S7). Besides these healthy tissues, L1TD1 expression has also been reported in seminoma [10], embryonic carcinomas [10], medulloblastoma [30], and colon adenocarcinoma (Additional file 1: Figures S3 and S7). Expression of L1TD1 at high levels in colon cancer cells led us to hypothesize that high expression of L1TD1 in colon cancer might be associated with prognosis. Earlier reports have demonstrated the association of stem cell pluripotency factors with poor prognosis in different cancer types, including medulloblastoma [30] and seminoma [15]. Interestingly, our results were in contrast with previous studies, suggesting that in colon cancer, high expression of L1TD1 is linked to better prognosis. In the three colon cancer data sets, expression of L1TD1 was associated with samples of low clinical cancer stage (Additional file 1: Figure S4A-C), which can perhaps be a reason for its prognostic significance.
In an attempt to understand the distinctive role of L1TD1 in different cancers, we investigated the co-expression of L1TD1 with its currently known interaction partners. We discovered that, unlike in hESCs and seminomas, L1TD1 was not co-expressed with its interaction partners in colon cancer (Fig. 2). This points to the potential participation of L1TD1’s interaction partners in the contrasting prognostic outcome. This was further supported by a recent study in medulloblastoma, showing an association of high L1TD1 expression with poor clinical outcome and significant co-expression between L1TD1 and its interaction partner OCT4 [30]. Together, these findings suggest that the co-expression of L1TD1 with its interaction partners might be required for manifesting an aggressive and detrimental phenotype. This is the first time that an embryonic stem cell factor has been shown to lead to contrasting outcomes in cancer, taking into consideration to the presence or absence of strong co-expression with its interaction partners.
We also investigated genes that were co-expressed with L1TD1 in colon cancer. Among the top 20 co-expressed genes, six had previously been linked to colon cancer. Chloride Channel Accessory 1 (CLCA1) is a tumor suppressor protein that regulates differentiation and proliferation of colorectal cancer cells. Its low expression has been associated with tumorigenesis, metastasis, and chromosomal instability, as well as poor prognosis in colorectal cancer [31]. Kruppel Like Factor 4 (KLF4) is a target of the tumor suppressor gene Adenomatous Polyposis Coli (APC) and its overexpression reduces cell migration and invasion in vitro and tumorigenicity of colon cancer cells in vivo [32]. GDP-mannose-4,6-dehydratase (GMDS) has been shown to have exon deletions linked to progression of colorectal cancer [33]. Also, an in vitro study found that GMDS deficiency in colon cancer cells made them resistant to receptor-mediated apoptosis [34]. High expression of Mucin 2 (MUC2) has been associated with longer disease-free survival in colorectal cancer patients [35]. Frameshift mutations resulting in premature termination of translation of Propionyl-CoA Carboxylase Alpha Subunit (PCCA) have been reported in colon and gastric cancer [36]. Investigation of the potential role of Alpha-1-antitrypsin (SERPINA1) expression in cancers provides controversial results; it has been associated with good prognosis in breast and colon cancer on protein atlas [37] (https://www.proteinatlas.org/ENSG00000197249-SERPINA1/pathology), but there are also reports that associate it with poor prognosis in colon cancer [38], gastric cancer [39] and cutaneous squamous cell carcinoma [40].
Several of the co-expressed genes have been linked to various other cancers. Down-regulation of Fc fragment of IgG binding protein (FCGBP) has been associated with decreased overall survival in gallbladder adenocarcinoma [41] and with progression of prostate cancer in Transgenic adenocarcinoma Mouse Prostate (TRAMP) [42]. Upregulation of ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (ST6GALNAC1) has been associated with good prognosis in breast cancer [43]. Additionally, siRNA-mediated silencing of ST6GALNAC1 has been shown to lead to reduced growth, migration and invasion of gastric cancer cells in vitro [44]. Estrogen-Induced Gene 121 Protein (KIAA1324), Long Intergenic Non-Protein Coding RNA 261 (LINC00261), and Intelectin 1 (ITLN1) have been shown to function as tumor suppressors in gastric cancer, with decreased expression associated with poor prognosis [45–47]. Low expression of Asparaginase-Like 1 Protein (ASRGL1) has been suggested as a marker for poor prognosis in endometrial carcinoma [48], whereas reduced levels of Solute carrier family 27 member 2 (SLC27A2) have been associated with poor survival in lung cancer [49]. SLIT and NTRK- like protein 6 (SLITRK6) is a known bladder tumor antigen, and is currently under investigation in clinical trials as a target for antibody-drug conjugate therapy [50]. HEPACAM family member 2 (HEPACAM2) is a paralog of Hepatocyte Cell Adhesion Molecule (HEPACAM), which is known to act as a tumor suppressor by promoting differentiation [51]. HEPACAM2, however, is a relatively newly-identified molecule and is not well-studied.
Conclusion
Our study of gene expression data from four clinical colon cancer data sets produced promising evidence in support of L1TD1 as a marker for good prognosis in colon cancer. Our results emphasize the need for further investigation and validation of L1TD1 as a potential prognostic marker in larger cohorts of colon cancer. Finally, this work also underscores the potential merits of investigating co-expressed genes to markers of interest.
Additional files
Figure S1. Density distributions of UPC scores for L1TD1 in the three colon cancer microarray data sets. A dashed black line indicates the UPC threshold of 0.6, which was used to stratify the samples into L1TD1+ and L1TD1- groups in the three data sets. Figure S2. Analysis of the primary tumor samples in The Cancer Genome Atlas Colon Adenocarcinoma (TCGA-COAD) data set. (A) Estimation of FPKM-UQ normalized RNA-seq counts by fitting Gaussian distributions. (B-C) Kaplan-Meier curves using the two thresholds for designating L1TD1 high and low samples (grey and red dashed lines, respectively). (D) Heatmaps showing signed P-value of Spearman rank correlation for the 20 most significantly co-expressed interaction partners of L1TD1. (E) The Spearman rank correlation values (rs) between L1TD1 and its top 20 co-expressed genes (Table 3). The correlations in the TCGA-COAD data set are shown with their false discovery rate (FDR). Figure S3. Heatmaps showing expression level of L1TD1 and its top 20 interaction partners in the samples of (A) colon cancer data sets, and (B) seminoma and stem cell data sets. Figure S4. Boxplots of UPC scores of L1TD1 stratified based on the indicated clinicopathological parameters in the different colon cancer microarray data sets. Figure S5. Boxplots of UPC scores of L1TD1 stratified based on the indicated clinicopathological parameters in the different colon cancer microarray data sets. Figure S6. Kaplan-Meier curves showing disease-free survival for the three colon cancer data sets (columns). The curves present survival data for the two groups of colon cancer patients based on gene expression level (high or low) of SPINK4, RETNLB, ASRGL1, CLCA1, and FCGBP (rows). Grey = high gene expression, Black = low gene expression. Figure S7. Formalin-fixed and paraffin-embedded tissue microarray blocks were stained with immunohistochemistry using anti-L1TD1 (Atlas Antibodies, HPA028501). (A) Normal colon tissue, (B) colorectal adenocarcinoma sample. (PDF 2132 kb)
Table S1. 311 Interaction partners of L1TD1 were determined using Mass spectrometry and co-immunoprecipitation in our earlier publication (Emani, Närvä, 2015, Stem cell reports). 306 Interaction partners of L1TD1 were identified by performing a Mass spectrometry analysis on co-immunoprecipitated proteins with two different anti-L1TD1 antibodies (recognizing different epitopes on L1TD1). In addition, we included 5 more proteins (NANOG, OCT4 (POU5F1), SOX2, DNMT3B, and TRIM28) that were challenging to detect using Mass spectrometry but the interactions were shown using Immunoprecipitation and Western Blotting. This makes a total of 311 proteins that are referred to in this work as “Interaction partners” of L1TD1. Table S2. Colon cancer samples with a high L1TD1 expression and a concomitant lack of expression of the listed interaction partner were compared to colon cancer samples with a low L1TD1 expression in the three data sets, this table lists the P-values (log-rank test) for these comparisons. P-value less (more significant) than the one obtained by comparing L1TD1 high and low sample groups are highlighted. Table S3. Table lists the 20 genes that had a positive correlation with L1TD1 in the colon cancer data sets. The table lists their UNIPROT ID and UNIRPOT protein name. (PDF 211 kb)
Acknowledgements
We would like to thank the funding agencies supporting the study. We would also like to thank Dr. Aidan McGlinchey for proofreading the manuscript.
Abbreviations
- APC
Adenomatous Polyposis Coli
- ASRGL1
Asparaginase-like 1 Protein
- CLCA1
Chloride channel accessory 1
- DNMT3B
DNA (cytosine-5)-methyltransferase 3B
- FCGBP
Fc fragment of IgG binding protein
- FDR
False discovery rate
- GEO
Gene Expression Omnibus
- GMDS
GDP-mannose-4,6-dehydratase
- HEPACAM
Hepatocyte Cell Adhesion Molecule
- HEPACAM2
HEPACAM family member 2
- hESC
Human embryonic stem cell
- iPSCs
Induced pluripotent stem cell
- ITLN1
Intelectin 1
- KIAA1324
Estrogen-Induced Gene 121 Protein
- KLF4
Kruppel Like Factor 4
- L1TD1
LINE-1 type transposase domain containing 1
- LIN28
Protein lin-28 homolog A
- LINC00261
Long Intergenic Non-Protein Coding RNA 261
- MUC2
Mucin 2
- NANOG
Homeobox protein NANOG
- OCT4
POU domain, class 5, transcription factor 1
- PCCA
Propionyl-CoA Carboxylase Alpha Subunit
- RETNLB
Resistin-like beta
- RMA
Robust Multiarray Average
- SERPINA1
Alpha-1-antitrypsin
- SLC27A2
Solute carrier family 27 member 2
- SLITRK6
SLIT and NTRK- like protein 6
- SOX2
Transcription factor SOX-2
- SPINK4
Serine peptidase inhibitor Kazal type 4
- ST6GALNAC1
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 1
- TRAMP
Transgenic adenocarcinoma Mouse Prostate
- TRIM28
Transcription intermediary factor 1-beta
- UPC
Universal exPression Code
Author contributions
Conceptualization: MRE, CH, AR, RL, LLE; Methodology: DC, MRE, RK, CB, JH, CH, AR, RL, LLE; Investigation: DC, RK, CB, JH; Formal analysis: DC, RK, CB; Visualization: DC, RK; Writing – original draft: DC; Writing – review and editing: DC, RK, CB, CH, LLE; Supervision: RL, LLE; Funding Acquisition: MRE, CH, AR, RL, LLE; All authors have read and approved the manuscript.
Funding
DC has been supported by University of Turku and University of Turku Graduate School.
MRE has been supported by the Academy of Finland (265723). CH and AR have received financial support from the Sigrid Jusélius Foundation, the Finnish Cancer Foundation, Helsinki University Central Hospital Research Funds, and Finska Läkaresällskapet. RL has been supported by the Academy of Finland, AoF, Centre of Excellence in Molecular Systems Immunology and Physiology Research (2012–2017) grant 250114; by the AoF grants 292335, 294337, 319280, 292482, 31444, and by grants from the Sigrid Jusélius Foundation (SJF) and the Finnish Cancer Foundation. LLE reports grants from the European Research Council ERC (677943), European Union’s Horizon 2020 research and innovation programme (675395), Academy of Finland (296801, 304995, 310561 and 313343), Juvenile Diabetes Research Foundation JDRF (2–2013-32), Tekes – the Finnish Funding Agency for Innovation (1877/31/2016), and Sigrid Juselius Foundation, during the conduct of the study. The funding bodies had no role in the study design, data collection and analysis, interpretation of the data and results, or in writing the manuscript.
Availability of data and materials
All requests for access to data and material are to be addressed jointly to Riitta Lahesmaa and Laura L. Elo. Publicly available datasets can be accessed at Gene Expression Omnibus (GEO IDs listed in Table 1).
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
There are no financial or non-financial competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Deepankar Chakroborty, Maheswara Reddy Emani, and Riku Klén should be considered equal first authors.
Riitta Lahesmaa and Laura L. Elo should be considered equal senior authors.
Contributor Information
Deepankar Chakroborty, Email: deepankar.chakroborty@utu.fi.
Maheswara Reddy Emani, Email: memani@broadinstitute.org.
Riku Klén, Email: riku.klen@utu.fi.
Camilla Böckelman, Email: camilla.bockelman@helsinki.fi.
Jaana Hagström, Email: jaana.hagstrom@hus.fi.
Caj Haglund, Email: Caj.Haglund@hus.fi.
Ari Ristimäki, Email: ari.ristimaki@helsinki.fi.
Riitta Lahesmaa, Phone: +358-(0)40 718 4813, Email: riitta.lahesmaa@utu.fi.
Laura L. Elo, Phone: +358 (0)29 450 3796, Email: laura.elo@utu.fi
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Wilson PM, Ladner RD, Lenz H-J. Predictive and prognostic markers in colorectal cancer. Gastrointest Cancer Res. 2007;1:237–246. [PMC free article] [PubMed] [Google Scholar]
- 3.Atlasi Y, Mowla SJ, Ziaee SAM, Bahrami A-R. OCT-4, an embryonic stem cell marker, is highly expressed in bladder cancer. Int J Cancer. 2007;120:1598–1602. doi: 10.1002/ijc.22508. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiou SH, Yu CC, Huang CY, Lin SC, Liu CJ, Tsai TH, et al. Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin Cancer Res. 2008;14:4085–4095. doi: 10.1158/1078-0432.CCR-07-4404. [DOI] [PubMed] [Google Scholar]
- 6.Meng H-M, Zheng P, Wang X-Y, Liu C, Sui H-M, Wu S-J, et al. Over-expression of Nanog predicts tumor progression and poor prognosis in colorectal cancer. Cancer Biol Ther. 2010;9:295–302. doi: 10.4161/cbt.9.4.10666. [DOI] [PubMed] [Google Scholar]
- 7.Lu X, Mazur SJ, Lin T, Appella E, Xu Y. The pluripotency factor nanog promotes breast cancer tumorigenesis and metastasis. Oncogene. 2014;33:2655–2664. doi: 10.1038/onc.2013.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong RC-B, Ibrahim A, Fong H, Thompson N, Lock LF, Donovan PJ. L1TD1 is a marker for undifferentiated human embryonic stem cells. PLoS One. 2011;6:e19355. doi: 10.1371/journal.pone.0019355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emani MRR, Närvä E, Stubb A, Chakroborty D, Viitala M, Rokka A, et al. The L1TD1 protein interactome reveals the importance of post-transcriptional regulation in human pluripotency. Stem Cell Reports. 2015;4:519–528. doi: 10.1016/j.stemcr.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Närvä E, Rahkonen N, Emani MR, Lund R, Pursiheimo JP, Nästi J, et al. RNA-binding protein L1TD1 interacts with LIN28 via RNA and is required for human embryonic stem cell self-renewal and cancer cell proliferation. Stem Cells. 2012;30:452–460. doi: 10.1002/stem.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jorissen RN, Gibbs P, Christie M, Prakash S, Lipton L, Desai J, et al. Metastasis-associated gene expression changes predict poor outcomes in patients with dukes stage B and C colorectal cancer. Clin Cancer Res. 2009;15:7642–7651. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with Colon Cancer. Gastroenterology. 2010;138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marisa L, de Reyniès A, Duval A, Selves J, Gaub MP, Vescovo L, et al. Gene expression classification of Colon Cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korkola JEJ, Houldsworth J, Chadalavada RSV, Olshen AB, Dobrzynski D, Reuter VE, et al. Down-regulation of stem cell genes, including those in a 200-kb gene cluster at 12p13. 31, is associated with in vivo differentiation of human male germ cell tumors. Cancer Res. 2006;66:820–827. doi: 10.1158/0008-5472.CAN-05-2445. [DOI] [PubMed] [Google Scholar]
- 15.Korkola JE, Houldsworth J, Feldman DR, Olshen AB, Qin L-X, Patil S, et al. Identification and validation of a gene expression signature that predicts outcome in adult men with germ cell tumors. J Clin Oncol. 2009;27:5240–5247. doi: 10.1200/JCO.2008.20.0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koyanagi-Aoi M, Ohnuki M, Takahashi K, Okita K, Noma H, Sawamura Y, et al. Differentiation-defective phenotypes revealed by large-scale analyses of human pluripotent stem cells. Proc Natl Acad Sci U S A. 2013;110:20569–20574. doi: 10.1073/pnas.1319061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edgar R. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piccolo S, Withers M. Multiplatform single-sample estimates of transcriptional activation. Proc Natl Acad Sci U S A. 2013;110:17778–17783. doi: 10.1073/pnas.1305823110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irizarry R, Hobbs B, Collin F. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 20.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 22.Therneau TM, Grambsch PM. Modeling survival data: extending the cox model. New York: Springer; 2000. [Google Scholar]
- 23.Therneau TM. A package for survival analysis in S. 2015. [Google Scholar]
- 24.Kassambara A, Kosinski M. survminer: Drawing Survival Curves using “ggplot2.” 2016.
- 25.Wilcoxon F. Individual comparisons by ranking methods. Biom Bull. 1945;1:80. doi: 10.2307/3001968. [DOI] [Google Scholar]
- 26.Kruskal WH, Wallis WA. Use of ranks in one-criterion variance analysis. J Am Stat Assoc. 1952;47:583–621. doi: 10.1080/01621459.1952.10483441. [DOI] [Google Scholar]
- 27.Pearson K. Notes on regression and inheritance in the case of two parents. Proc R Soc London. 1895;58:240–242. doi: 10.1098/rspl.1895.0041. [DOI] [Google Scholar]
- 28.Muzny DM, Bainbridge MN, Chang K, Dinh HH, Drummond JA, Fowler G, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwabuchi K a, Yamakawa T, Sato Y, Ichisaka T, Takahashi K, Okita K, et al. ECAT11/L1td1 is enriched in ESCs and rapidly activated during iPSC generation, but it is dispensable for the maintenance and induction of pluripotency. PLoS One. 2011;6:e20461. doi: 10.1371/journal.pone.0020461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santos MCT, Silva PBG, Rodini CO, Furukawa G, Marco Antonio DS, Zanotto-Filho A, et al. Embryonic stem cell-related protein L1TD1 is required for cell viability, Neurosphere formation, and Chemoresistance in Medulloblastoma. Stem Cells Dev. 2015;24:150810085436001. doi: 10.1089/scd.2015.0052. [DOI] [PubMed] [Google Scholar]
- 31.Yang B, Cao L, Liu J, Xu Y, Milne G, Chan W, et al. Low expression of chloride channel accessory 1 predicts a poor prognosis in colorectal cancer. Cancer. 2015;121:1570–1580. doi: 10.1002/cncr.29235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dang DT, Chen X, Feng J, Torbenson M, Dang LH, Yang VW. Overexpression of Krüppel-like factor 4 in the human colon cancer cell line RKO leads to reduced tumorigenecity. Oncogene. 2003;22:3424–3430. doi: 10.1038/sj.onc.1206413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakayama K, Moriwaki K, Imai T, Shinzaki S, Kamada Y, Murata K, et al. Mutation of GDP-mannose-4,6-dehydratase in colorectal cancer metastasis. PLoS One. 2013;8:e70298. doi: 10.1371/journal.pone.0070298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moriwaki Kenta, Shinzaki Shinichiro, Miyoshi Eiji. GDP-mannose-4,6-dehydratase (GMDS) Deficiency Renders Colon Cancer Cells Resistant to Tumor Necrosis Factor-related Apoptosis-inducing Ligand (TRAIL) Receptor- and CD95-mediated Apoptosis by Inhibiting Complex II Formation. Journal of Biological Chemistry. 2011;286(50):43123–43133. doi: 10.1074/jbc.M111.262741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu H-P, Lai M-D, Lee J-C, Yen M-C, Weng T-Y, Chen W-C, et al. Mucin 2 silencing promotes colon cancer metastasis through interleukin-6 signaling. Sci Rep. 2017;7:5823. doi: 10.1038/s41598-017-04952-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.JO Y. S., OH H. R., KIM M. S., YOO N. J., LEE S. H. Frameshift mutations of OGDH, PPAT and PCCA genes in gastric and colorectal cancers. Neoplasma. 2016;63(05):681–686. doi: 10.4149/neo_2016_504. [DOI] [PubMed] [Google Scholar]
- 37.Uhlén Mathias, Björling Erik, Agaton Charlotta, Szigyarto Cristina Al-Khalili, Amini Bahram, Andersen Elisabet, Andersson Ann-Catrin, Angelidou Pia, Asplund Anna, Asplund Caroline, Berglund Lisa, Bergström Kristina, Brumer Harry, Cerjan Dijana, Ekström Marica, Elobeid Adila, Eriksson Cecilia, Fagerberg Linn, Falk Ronny, Fall Jenny, Forsberg Mattias, Björklund Marcus Gry, Gumbel Kristoffer, Halimi Asif, Hallin Inga, Hamsten Carl, Hansson Marianne, Hedhammar My, Hercules Görel, Kampf Caroline, Larsson Karin, Lindskog Mats, Lodewyckx Wald, Lund Jan, Lundeberg Joakim, Magnusson Kristina, Malm Erik, Nilsson Peter, Ödling Jenny, Oksvold Per, Olsson Ingmarie, Öster Emma, Ottosson Jenny, Paavilainen Linda, Persson Anja, Rimini Rebecca, Rockberg Johan, Runeson Marcus, Sivertsson Åsa, Sköllermo Anna, Steen Johanna, Stenvall Maria, Sterky Fredrik, Strömberg Sara, Sundberg Mårten, Tegel Hanna, Tourle Samuel, Wahlund Eva, Waldén Annelie, Wan Jinghong, Wernérus Henrik, Westberg Joakim, Wester Kenneth, Wrethagen Ulla, Xu Lan Lan, Hober Sophia, Pontén Fredrik. A Human Protein Atlas for Normal and Cancer Tissues Based on Antibody Proteomics. Molecular & Cellular Proteomics. 2005;4(12):1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Kwon CH, Park HJ, Choi JH, Lee JR, Kim HK. Snail and serpinA1 promote tumor progression and predict prognosis in colorectal cancer. Oncotarget. 2014;6. [DOI] [PMC free article] [PubMed]
- 39.Kwon CH, Park HJ, Lee JR, Kim HK, Jeon TY, Jo H-J, et al. Serpin peptidase inhibitor clade a member 1 is a biomarker of poor prognosis in gastric cancer. Br J Cancer. 2014;111:1993–2002. doi: 10.1038/bjc.2014.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farshchian M, Kivisaari A, Ala-aho R, Riihilä P, Kallajoki M, Grénman R, et al. Serpin peptidase inhibitor clade a member 1 (SerpinA1) is a novel biomarker for progression of cutaneous squamous cell carcinoma. Am J Pathol. 2011;179:1110–1119. doi: 10.1016/j.ajpath.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiong L, Wen Y, Miao X, Yang Z. NT5E and FcGBP as key regulators of TGF-1-induced epithelial-mesenchymal transition (EMT) are associated with tumor progression and survival of patients with gallbladder cancer. Cell Tissue Res. 2014;355:365–374. doi: 10.1007/s00441-013-1752-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gazi MH, He M, Cheville JC, Young CYF. Downregulation of IgG fc binding protein (fc gammaBP) in prostate cancer. Cancer Biol Ther. 2008;7:70–75. doi: 10.4161/cbt.7.1.5131. [DOI] [PubMed] [Google Scholar]
- 43.Patani N, Jiang W, Mokbel K. Prognostic utility of glycosyltransferase expression in breast cancer. Cancer Genomics Proteomics. 2008;5:333–340. [PubMed] [Google Scholar]
- 44.Tamura F, Sato Y, Hirakawa M, Yoshida M, Ono M, Osuga T, et al. RNAi-mediated gene silencing of ST6GalNAc I suppresses the metastatic potential in gastric cancer cells. Gastric Cancer. 2016;19:85–97. doi: 10.1007/s10120-014-0454-z. [DOI] [PubMed] [Google Scholar]
- 45.Kang JM, Park S, Kim SJ, Kim H, Lee B, Kim J, et al. KIAA1324 suppresses gastric Cancer progression by inhibiting the Oncoprotein GRP78. Cancer Res. 2015;75:3087–3097. doi: 10.1158/0008-5472.CAN-14-3751. [DOI] [PubMed] [Google Scholar]
- 46.Fan Y, Wang Y-F, Su H-F, Fang N, Zou C, Li W-F, et al. Decreased expression of the long noncoding RNA LINC00261 indicate poor prognosis in gastric cancer and suppress gastric cancer metastasis by affecting the epithelial–mesenchymal transition. J Hematol Oncol. 2016;9:57. doi: 10.1186/s13045-016-0288-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Li D, Zhao X, Xiao Y, Mei H, Pu J, Xiang X, et al. Intelectin 1 suppresses tumor progression and is associated with improved survival in gastric cancer. Oncotarget. 2015;6:16168–16182. doi: 10.18632/oncotarget.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edqvist P-HD, Huvila J, Forsström B, Talve L, Carpén O, Salvesen HB, et al. Loss of ASRGL1 expression is an independent biomarker for disease-specific survival in endometrioid endometrial carcinoma. Gynecol Oncol. 2015;137:529–537. doi: 10.1016/j.ygyno.2015.03.055. [DOI] [PubMed] [Google Scholar]
- 49.Su J, Wu S, Tang W, Qian H, Zhou H, Guo T. Reduced SLC27A2 induces cisplatin resistance in lung cancer stem cells by negatively regulating Bmi1-ABCG2 signaling. Mol Carcinog. 2016;55:1822–1832. doi: 10.1002/mc.22430. [DOI] [PubMed] [Google Scholar]
- 50.Morrison Kendall, Challita-Eid Pia M., Raitano Arthur, An Zili, Yang Peng, Abad Joseph D., Liu Wendy, Lortie Dawn Ratay, Snyder Josh T., Capo Linnette, Verlinsky Alla, Aviña Hector, Doñate Fernando, Joseph Ingrid B.J., Pereira Daniel S., Morrison Karen, Stover David R. Development of ASG-15ME, a Novel Antibody–Drug Conjugate Targeting SLITRK6 , a New Urothelial Cancer Biomarker. Molecular Cancer Therapeutics. 2016;15(6):1301–1310. doi: 10.1158/1535-7163.MCT-15-0570. [DOI] [PubMed] [Google Scholar]
- 51.Moh MC, Shen S. The roles of cell adhesion molecules in tumor suppression and cell migration: a new paradox. Cell Adhes Migr. 2009;3:334–336. doi: 10.4161/cam.3.4.9246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Density distributions of UPC scores for L1TD1 in the three colon cancer microarray data sets. A dashed black line indicates the UPC threshold of 0.6, which was used to stratify the samples into L1TD1+ and L1TD1- groups in the three data sets. Figure S2. Analysis of the primary tumor samples in The Cancer Genome Atlas Colon Adenocarcinoma (TCGA-COAD) data set. (A) Estimation of FPKM-UQ normalized RNA-seq counts by fitting Gaussian distributions. (B-C) Kaplan-Meier curves using the two thresholds for designating L1TD1 high and low samples (grey and red dashed lines, respectively). (D) Heatmaps showing signed P-value of Spearman rank correlation for the 20 most significantly co-expressed interaction partners of L1TD1. (E) The Spearman rank correlation values (rs) between L1TD1 and its top 20 co-expressed genes (Table 3). The correlations in the TCGA-COAD data set are shown with their false discovery rate (FDR). Figure S3. Heatmaps showing expression level of L1TD1 and its top 20 interaction partners in the samples of (A) colon cancer data sets, and (B) seminoma and stem cell data sets. Figure S4. Boxplots of UPC scores of L1TD1 stratified based on the indicated clinicopathological parameters in the different colon cancer microarray data sets. Figure S5. Boxplots of UPC scores of L1TD1 stratified based on the indicated clinicopathological parameters in the different colon cancer microarray data sets. Figure S6. Kaplan-Meier curves showing disease-free survival for the three colon cancer data sets (columns). The curves present survival data for the two groups of colon cancer patients based on gene expression level (high or low) of SPINK4, RETNLB, ASRGL1, CLCA1, and FCGBP (rows). Grey = high gene expression, Black = low gene expression. Figure S7. Formalin-fixed and paraffin-embedded tissue microarray blocks were stained with immunohistochemistry using anti-L1TD1 (Atlas Antibodies, HPA028501). (A) Normal colon tissue, (B) colorectal adenocarcinoma sample. (PDF 2132 kb)
Table S1. 311 Interaction partners of L1TD1 were determined using Mass spectrometry and co-immunoprecipitation in our earlier publication (Emani, Närvä, 2015, Stem cell reports). 306 Interaction partners of L1TD1 were identified by performing a Mass spectrometry analysis on co-immunoprecipitated proteins with two different anti-L1TD1 antibodies (recognizing different epitopes on L1TD1). In addition, we included 5 more proteins (NANOG, OCT4 (POU5F1), SOX2, DNMT3B, and TRIM28) that were challenging to detect using Mass spectrometry but the interactions were shown using Immunoprecipitation and Western Blotting. This makes a total of 311 proteins that are referred to in this work as “Interaction partners” of L1TD1. Table S2. Colon cancer samples with a high L1TD1 expression and a concomitant lack of expression of the listed interaction partner were compared to colon cancer samples with a low L1TD1 expression in the three data sets, this table lists the P-values (log-rank test) for these comparisons. P-value less (more significant) than the one obtained by comparing L1TD1 high and low sample groups are highlighted. Table S3. Table lists the 20 genes that had a positive correlation with L1TD1 in the colon cancer data sets. The table lists their UNIPROT ID and UNIRPOT protein name. (PDF 211 kb)
Data Availability Statement
All requests for access to data and material are to be addressed jointly to Riitta Lahesmaa and Laura L. Elo. Publicly available datasets can be accessed at Gene Expression Omnibus (GEO IDs listed in Table 1).