Abstract
Facioscapulohumeral muscular dystrophy (FSHD) is characterized by misexpression of the double homeobox 4 (DUX4) developmental transcription factor in mature skeletal muscle, where it is responsible for muscle degeneration. Preventing expression of DUX4 mRNA is a disease-modifying therapeutic strategy with the potential to halt or reverse the course of disease. We previously reported that agonists of the β-2 adrenergic receptor suppress DUX4 expression by activating adenylate cyclase to increase cAMP levels. Efforts to further explore this signaling pathway led to the identification of p38 mitogen-activated protein kinase as a major regulator of DUX4 expression. In vitro experiments demonstrate that clinically advanced p38 inhibitors suppress DUX4 expression in FSHD type 1 and 2 myoblasts and differentiating myocytes in vitro with exquisite potency. Individual small interfering RNA–mediated knockdown of either p38α or p38β suppresses DUX4 expression, demonstrating that each kinase isoform plays a distinct requisite role in activating DUX4. Finally, p38 inhibitors effectively suppress DUX4 expression in a mouse xenograft model of human FSHD gene regulation. These data support the repurposing of existing clinical p38 inhibitors as potential therapeutics for FSHD. The surprise finding that p38α and p38β isoforms each independently contribute to DUX4 expression offers a unique opportunity to explore the utility of p38 isoform-selective inhibitors to balance efficacy and safety in skeletal muscle. We propose p38 inhibition as a disease-modifying therapeutic strategy for FSHD.
SIGNIFICANCE STATEMENT
Facioscapulohumeral muscular dystrophy (FSHD) currently has no treatment options. This work provides evidence that repurposing a clinically advanced p38 inhibitor may provide the first disease-modifying drug for FSHD by suppressing toxic DUX4 expression, the root cause of muscle degeneration in this disease.
Introduction
Facioscapulohumeral muscular dystrophy (FSHD) is one of the most prevalent muscular dystrophies (Padberg et al., 1995; Deenen et al., 2014), yet there is currently no treatment available and few clinical trials of promising therapies are ongoing. Thus, there is a desperate need to identify drug targets to stop or reverse progression of the disease. As described in its name, FSHD typically presents as facial, shoulder, and upper arm weakness, which eventually progresses to involve nearly all skeletal muscle groups (Tawil et al., 2014). Although most individuals develop symptoms during their second or third decade of life, a small number have a more acute pediatric onset (Goselink et al., 2017). FSHD is caused by misexpression of the double homeobox 4 (DUX4) transcription factor in skeletal muscle. DUX4 is encoded by a retrogene located within each unit of the D4Z4 macrosatellite repeat array on chromosome 4q35 and is normally expressed in preimplantation embryos, where it activates an early developmental program that marks the cleavage stage of embryogenesis (Tawil et al., 2014; De Iaco et al., 2017; Hendrickson et al., 2017; Whiddon et al., 2017).
In normal skeletal muscle, DUX4 is silenced, likely through repeat-mediated epigenetic repression (van Overveld et al., 2003; Zeng et al., 2009; Snider et al., 2010; Daxinger et al., 2015; Das and Chadwick, 2016). In patients with FSHD, deletion of a subset of D4Z4 repeats [FSHD type 1 (FSHD1)] or damaging variants in epigenetic regulators of the D4Z4 array [FSHD type 2 (FSHD2)] (Lemmers et al., 2010, 2012; van den Boogaard et al., 2016) leads to inefficient D4Z4 repression in somatic cells; when combined with a permissive chromosome 4 haplotype that provides a polymorphic polyadenylation site this results in the ectopic expression of DUX4 in muscle cells (Lemmers et al., 2010; Snider et al., 2010; Tawil et al., 2014). DUX4 misexpression in skeletal muscle causes many cellular phenotypes, ultimately leading to cell death (Winokur et al., 2003; Kowaljow et al., 2007; Bosnakovski et al., 2008; Snider et al., 2009; Wallace et al., 2011; Geng et al., 2012; Young et al., 2013; Feng et al., 2015; Rickard et al., 2015; Shadle et al., 2017).
Because of its causative role in FSHD, targeting DUX4 is an obvious therapeutic approach to halt or reverse disease progression (Himeda et al., 2015). Blocking transcription of DUX4 mRNA with small molecule drugs is an attractive option because all downstream pathologic mechanisms are inherently disrupted and small molecules are generally amenable to oral dosing, avoiding delivery complications associated with RNA interference or gene editing technologies. However, subsequent to chromatin decompaction associated with FSHD, the mechanisms responsible for bursts of DUX4 mRNA synthesis are still poorly understood and limited small molecule drug targets have been identified (Block et al., 2013; Campbell et al., 2017; Teveroni et al., 2017; Cruz et al., 2018).
Previously, we reported on the discovery of β-2 adrenergic receptor agonists as modulators of DUX4 expression (Campbell et al., 2017). Since β2 adrenergic signaling has been extensively characterized in skeletal muscle, we began systematically testing small molecule inhibitors that could potentially interfere with the pathway (for a review, see Joassard et al., 2013). One example of kinases that are activated by β2 adrenergic signaling in a protein kinase A–dependent manner is the set of p38 mitogen-activated protein kinases (MAPKs) (Moule and Denton, 1998; Zheng et al., 2000; Aggeli et al., 2002; McAlees and Sanders, 2009). The p38 MAPKs are classically involved in the cellular response to stressful stimuli, including inflammatory cytokines, and have been heavily pursued by pharmaceutical companies for diseases with inflammatory components, such as rheumatoid arthritis, resulting in an abundance of chemical tools from p38α selective to pan-p38 inhibitors (Xing, 2015). In exploring the hypothesis that activation of p38 plays a role in β2-agonist repression of DUX4, we determined that rather than blocking the ability of β2-agonists to suppress DUX4, p38 inhibitors instead potently inhibit the expression of DUX4 in the absence of β2 agonism. Here, we identify p38α and p38β MAPKs as having important roles in the pathogenic expression of DUX4 in FSHD and demonstrate that clinically advanced p38 inhibitors decrease expression of DUX4 and DUX4 target genes in FSHD patient–derived muscle cells at inhibitor levels that do not negatively affect muscle differentiation. In a mouse pharmacology model of FSHD gene regulation, these inhibitors suppress DUX4 at blood levels that are efficacious in published preclinical models of inflammatory diseases and that have been routinely achieved in human clinical trials. These findings suggest that existing clinical p38 inhibitors are repurposing candidate drugs for FSHD therapeutic development.
Materials and Methods
Ethics Statement.
This research used preexisting deidentified human cell lines from approved repositories. These studies were determined to not be human subject research by the Saint Louis University Institutional Review Board. Primary human myoblast cell lines were obtained from the Fields Center at the University of Rochester (http://www.urmc.rochester.edu/fields-center.aspx) and immortalized by retroviral transduction of cyclin-dependent kinase 4 and human telomerase reverse transcriptase (MB200, FSHD2) (Snider et al, 2010; Stadler et al., 2011). An immortalized FSHD1 cell line (54-2) and isogenic control line from a mosaic male patient (54-6, non-FSHD) were also used (Krom et al., 2012).
Cell Culture.
Immortalized myoblasts were grown in Ham’s F-10 Nutrient Mix (Gibco, Waltham, MA) supplemented with 20% Corning USDA–approved source FBS (Corning, Corning, NY), 100 U/100 μg penicillin/streptomycin (Gibco), 10 ng/ml recombinant human fibroblast growth factor (Promega Corporation, Madison, WI), and 1 μM dexamethasone (Sigma-Aldrich, St. Louis, MO). Differentiation of myoblasts into myotubes was achieved by switching confluent myoblast monolayers into Dulbecco’s modified Eagle’s medium/F-12 Nutrient Mix (1:1; Gibco) supplemented with 2% knockout serum replacement (Gibco), 100 U/100 μg penicillin/streptomycin, 10 μg/ml insulin, and 10 μg/ml transferrin (knockout serum replacement media) for 40 hours.
Compounds.
Individual compounds were purchased from Sigma-Aldrich, Tocris Bioscience (A-485 catalog #6387, BIRB 796 catalog #5989, SB203580 catalog #1202, SCIO 469 catalog #3528, PF-3644022 catalog #4279; Bio-Techne Corporation, Minneapolis, MN), MedChemExpress (PH-797804 catalog #HY-10403, losmapimod catalog #10402, acumapimod catalog #HY-16715, MK@-IN-1 catalog #HY-12834; Monmouth Junction, NJ), or Selleck Chemicals (pexmetinib catalog #S7799, BMS-582949 catalog #S8124, neflamapimod catalog #S1458, pamapimod catalog #S8125, ralimetinib catalog #S1494, SB202190 catalog #S1077, TAK-715 catalog #S2928, VX-702 catalog #S6005, eFT-508 catalog #S8275; Houston, TX), dissolved in 100% DMSO as concentrated stocks and stored at −20°C until use. For in vitro experiments, concentrated DMSO stocks were first diluted in 100% DMSO to 2000-fold the final concentration, and then diluted 2000-fold into culture media before addition to cells. Experiments were performed in triplicate with S.D. represented. For determination of the 50% inhibitory concentration (IC50) for each compound, 11-point concentration response curves were generated by first creating 3-fold serial dilutions of compounds from concentrated stocks in 100% DMSO in 96-well plates. Serial dilutions were then further diluted 2000-fold into culture medium before addition to cells. The IC50 values were determined by nonlinear regression using a four-parameter logistic equation (http://www.graphpad.com; GraphPad Prism Software Inc., San Diego, CA). Data are presented as IC50 values with two significant digits.
DUX4 Activity Assay.
DUX4 activity assay was performed as detailed in our previous studies (Campbell et al., 2017). Briefly, control 54-6 (non-FSHD) myoblasts in one well of a six-well plate were cotransfected with 75 ng of the DUX4 expression vector pCS2-DUX4 (Geng et al., 2012) and 2.925 µg of pGL3-promoter vector (Promega) using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA) following the manufacturer’s instructions. Three hours after transfection, cells were trypsinized and distributed to 42 wells of a 96-well plate. Two hours later (a time at which there is a low but detectable level of DUX4 target gene expression), six wells were harvested to represent the baseline gene expression state, while DMSO control or compounds were added to the remaining wells as indicated in the figures. Twenty hours later, the remaining wells were harvested to represent the endpoint gene expression state. DUX4 activity was determined by normalizing DUX4 target gene mRNA levels at the 25-hour endpoint to the levels at the 5-hour baseline and setting that value to 100 in the absence of drug.
mRNA Expression Analyses.
For screening of compounds in 96-well plates for IC50 determinations and DUX4 activity assay, cell lysates were prepared using Cells-to-Ct Bulk Lysis Reagents (Invitrogen, Carlsbad, CA). For other gene expression analyses, total RNA was extracted from whole cells using the E.Z.N.A. Total RNA Kit or xenograft tissues using the E.Z.N.A. Tissue RNA Kit (OMEGA Bio-Tek, Norcrass, GA) and RNA/DNA was isolated from xenograft tissue using the E.Z.NA. DNA/RNA Kit (OMEGA Bio-Tek) according to the manufacturer’s instructions. Quantitative real-time polymerase chain reaction (PCR) was carried out on a QuantStudio 5 (Applied Biosystems, Foster City, CA). For all gene detection except for DUX4, TaqMan Gene Expression Assays (Applied Biosystems) and TaqMan Fast Virus 1-Step Master Mix (Invitrogen) were used. For DUX4 expression, isolated RNA was treated with DNase I (Thermo Fisher Scientific) and reverse transcribed into cDNA using Superscript IV (Thermo Fisher Scientific) and Oligo (dT) Primer (Invitrogen) following the manufacturer’s protocol. Quantitative real-time PCR was performed using a custom Taqman primer/probe set and TaqMan Gene Expression Master Mix (Applied Biosystems). The relative expression levels of target genes were normalized to that of the reference gene ribosomal protein L30 (RPL30), which was included in multiplex (two gene) PCR reactions, using the ΔΔCt method (Livak and Schmittgen, 2001) after confirming equivalent amplification efficiencies of reference and target molecules.
Small Interfering RNA Transfections.
Duplex small interfering RNAs (siRNAs) were obtained from Thermo Fisher Scientific (Silencer Select). Transfections of siRNAs into myoblasts were carried out, in triplicate, using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. Briefly, cells were seeded at 1 × 105 cells/well in 12-well plates and transfected ∼20 hours later with 2 μl Lipofectamine RNAiMAX and 10 pmol of either gene-specific siRNAs or a scrambled nonsilencing control siRNA diluted in 100 μl Opti-MEM Reduced Serum Medium. For myoblast knockdowns, cells were harvested for RNA analysis 72 hours later. For myotube experiments, 24–48 hours following transfection, cells were switched to differentiation medium and harvested for RNA analysis 40 hours later. Where noted, a double transfection protocol was followed to ensure efficient depletion of preexisting proteins. Cells were transfected a second time 24 hours after the first transfection, switched to differentiation medium 24 hours later, and harvested for RNA analysis 40 hours after switching to differentiation medium.
Western Blotting.
Samples from siRNA knockdown experiments were obtained by lysing cells by addition of SDS-PAGE loading buffer. Reduced and boiled samples were run on NuPage 4–12% Bis-Tris precast polyacrylamide gels (Life Technologies, Carlsbad, CA) and transferred to Immobilon-FL polyvinylidene difluoride membrane (Millipore, Burlington, MA). After blocking in 0.2% I-Block (Thermo Fisher Scientific) in Tris-buffered saline/Tween 20 for 1 hour at room temperature, membranes were incubated with appropriate primary antibodies diluted at 1:1000 in 1X Tris-buffered saline/Tween 20 overnight at 4°C. Membranes were then incubated with Li-Cor near-infrared fluorescently labeled secondary antibodies diluted at 1:15,000 in 1X Tris-buffered saline/Tween 20 for 1 hour at room temperature. Blots were scanned and analyzed with the Li-Cor Odyssey CLx Imaging System (Li-Cor Biosciences, Lincoln, NE). Quantitation of bands was normalized to α-tubulin using Image Studio software (Li-Cor Biosciences).
Antibodies.
The following antibodies were used: α-tubulin mouse mAb (926-42213; Li-Cor Biosciences); p38α MAPK polyclonal Rabbit Ab (9218S; Cell Signaling Technology, Danvers, MA); p38β MAPK (C28C2) Rabbit mAb (2339; Cell Signaling Technology); IRDye 680 goat anti-mouse secondary (926-68070; Li-Cor Biosciences); and IRDye 800 goat anti-rabbit secondary (926-32211; Li-Cor Biosciences).
Animals.
Male NOD-Rag immunodeficient mice (strain #007799 NOD.CgRag1tm1Mom Il2rgtm1Wjl/SzJ; Jackson Laboratories) were used for the xenograft model of FSHD because the absence of T, B, and NK cells in this strain makes them suitable for xenograft transplantation (Silva-Barbosa et al., 2005). All protocols were approved by the Institutional Animal Care and Use Committee of Saint Louis University.
Plasma Drug Levels.
Terminal plasma samples were diluted with control naive mouse plasma as appropriate to bring samples into the standard curve range (1–1000 ng/ml) or were run undiluted. Enalapril was used as an internal standard prior to extraction at a 100 ng/ml final concentration. Samples were vortexed for 5 minutes to fully incorporate the internal standard. Afterward, 180 ml of acetonitrile was added, vortexed for 5 minutes, and centrifuged for 5 minutes at 4°C and greater than 2100g. The supernatant was transferred to a 96-well sample plate and heat sealed for liquid chromatography–tandem mass spectrometry analysis using a system consisting of an LC-20AD pump (Shimadzu, Kyoto, Japan), an HTC PAL autosampler (Leap Technologies, Carrboro, NC), and a Sciex API-4000 mass spectrometer in electrospray ionization mode (AB Sciex, Foster City, CA). The multiple reaction monitoring transitions for losmapimod were m/z: 383 > 267. An Amour C18 reverse-phase column (2.1 × 30 mm, 5 μm; Analytical Sales and Services, Pompton Plains, NJ) was used for chromatographic separation. Mobile phases were 0.1% formic acid (aqueous) and 0.1% formic acid in acetonitrile (organic) with a flow rate of 0.35 ml/min. The starting phase was 10% acetonitrile for 0.9 minutes, increased to 90% acetonitrile over 0.4 minutes, maintained for an additional 0.2 minutes, returned to 10% acetonitrile over 0.4 minutes, and then held for 2 minutes. Peak areas were integrated using Analyst 1.5.1 (AB Sciex).
Cell and Barium Chloride Preparations for Injection.
Barium chloride was dissolved in 0.9% saline to a concentration of 2.4% w/v. The barium chloride solution was then sterile filtered (0.2 μm filter; Thermo Scientific). Growing FSHD myoblasts were detached with 0.25% trypsin 2.21 mM EDTA (Corning) and resuspended in growth media for enumeration. Cells were then centrifuged at 900g for 5 minutes and suspended in 30 ml of sterile saline. This saline rinse was repeated one more time before cells were suspended in saline at ∼6.67 × 107 cells/ml. The cells were stored on ice and injected within 1 to 2 hours of preparation. Just prior to injection, cells were combined with an equal volume of barium chloride solution yielding 1.2% w/v barium chloride solution with 3.33 × 107 cells/ml.
Barium Chloride Xenograft Pharmacology Model of FSHD.
NOD-Rag immunodeficient mice were anesthetized with 3%–3.5% isoflurane to effect and the injection site was shaved and cleaned with betadine. Barium chloride cell suspensions were injected using a 26-gauge needle into three sites along the tibialis anterior muscle. Injections were 10 μl/site in volume amounting to 30 μl and 1 × 106 cells injected in total. Mice recovered for 1 to 2 hours prior to administration of test compounds. PH-797804 and losmapimod were dissolved in DMSO prior to addition of the remaining vehicle components. PH-797804 was administered subcutaneously twice a day at 5 ml/kg in 10% DMSO, 40% polyethylene glycol, and 50% saline vehicle. Losmapimod was administered by mouth twice a day at 10 ml/kg in 10% DMSO and 90% 0.5% methylcellulose. At the termination of the study, mice were euthanized via CO2 asphyxiation and blood and tissue samples were collected for bioanalytical or quantitative PCR analysis. For FSHD endpoints, the entire xenograft muscle was harvested, weighed, and homogenized in lysis buffer as described in mRNA Expression Analyses. Lysis buffer was then transferred such that 30 mg of tissue was used in the RNA isolation procedure and 10 mg of tissue was used in the DNA/RNA procedure to prevent column clogging.
Taqman Gene Expression Assay Identification Numbers.
Taqman assays were purchased from Applied Biosystems (Thermo Fisher Scientific): MBD3L2, Hs00544743_m1; LEUTX, Hs01028718_m1; MYH2, Hs00430042_m1; MAPK11, Hs00177101_m1; MAPK14, Hs01051152_m1; MYOG, Hs01072232_m1; RPL30, Hs00265497_m1; ZSCAN4, Hs00537549_m1; and DUX4, primers GCCGGCCCAGGTACCA and CAGCGAGCTCCCTTGCA with probe 6FAM-CAGTGCGCACCCCG-MGBNFQ.
siRNA Assay Identifications and Target Sequences.
All siRNAs were purchased with Silencer Select Chemistry (Ambion; Life Technologies): MAPK14, s3585, CCTAAAACCTAGTAATCT; MAPK11, s11155, GCGACTACATTGACCAGCT; and negative control: Silencer Select Negative Control #1.
Statistical Analysis.
All statistics were performed using GraphPad 4 software. One-way ANOVA was used for multigroup comparisons; Dunnett’s post test was used to compare individual groups to the control. An unpaired two-tailed t test was used for two sample comparison. The P values and statistical comparison used are listed in each figure caption.
Results
p38 Inhibitors Suppress DUX4 in FSHD Myotubes and Myoblasts.
We have previously screened several small molecule compound libraries to identify bromodomain and extra-terminal motif inhibitors and agonists of the β-2 adrenergic receptor as inhibitors of DUX4 expression in FSHD muscle cells (Campbell et al., 2017). Efforts to further explore the signaling pathways regulating DUX4 expression in FSHD led us to screen p38 MAPK inhibitors in a high throughput assay for DUX4. Detection of DUX4 mRNA in FSHD muscle cells for drug screening purposes is challenging for many reasons (Snider et al., 2009, 2010; Geng et al., 2012; Feng et al., 2015). Therefore, DUX4 was measured indirectly by quantitating mRNA levels for the DUX4-regulated gene MBD3L2, which is a sensitive and highly specific marker of DUX4 expression since it is not expressed in normal muscle cultures or tissue (Geng et al., 2012; Yao et al., 2014; Campbell et al., 2017). Differentiating cultures of MB200 (FSHD2) myoblasts that express elevated levels of DUX4 and DUX4-target gene mRNA upon differentiation into multinucleated myotubes (myocytes) were used. Figure 1A shows the concentration-response curve for MBD3L2 RNA levels in MB200 (FSHD2) myotubes differentiated in the presence of the clinical p38α/β inhibitor PH-797804 for 40 hours. The levels of MBD3L2 RNA induced during differentiation are suppressed >95% in a concentration-dependent manner with a potent IC50 value of 0.41 nM, in line with the reported IC50 value of 2.3 nM for PH-797804 on p38α enzymatic activity (Selness et al., 2011). Since the p38 family of kinases is known to play important roles in the myogenic program (Wu et al., 2000), it was important to distinguish effects on DUX4 expression from effects on myocyte differentiation. We noted that only at high concentrations of PH-797804 (>100 nM) was there a noticeable delay in myotube formation. Key markers of myocyte differentiation (myogenin, MYOG, and myosin heavy chain, MYH2) were likewise only inhibited at concentrations greater than 100 nM, and even at 1–10 μM the inhibition was incomplete, reaching a maximum of ∼60% inhibition (Fig. 1A). To further demonstrate that the effects of p38 inhibition on DUX4 target gene expression were independent of myocyte differentiation, cultures of proliferating FSHD myoblasts were treated with PH-797804. Figure 1B shows that multiple DUX4 targets are suppressed with treatment in both 54-2 (FSHD1) and MB200 (FSHD2) proliferating myoblast cultures, consistent with suppression of DUX4 expression being independent of the differentiation state of the cells. To confirm that p38 inhibition by PH-797804 had a direct effect on DUX4, we performed experiments in both FSHD1 and FSHD2 myocytes treated with several drug concentrations for 40 hours during differentiation. Figure 1C (54-2, FSHD1) and Fig. 1D (MB200, FSHD2) demonstrate that p38 inhibitor PH-797804 treatment reduces RNA levels for DUX4 and DUX4 targets (MBD3L2, ZSCAN4, and LEUTX) at drug concentrations that have negligible effects on markers of differentiation MYOG and MYH2. These data confirm that p38 inhibition directly suppresses DUX4 expression, and consequently the expression of DUX4 target genes is similarly reduced.
Fig. 1.
p38 Inhibitor PH-797804 reduces DUX4 and DUX4 target gene expression in FSHD patient-derived proliferating myoblasts and differentiating myotubes. (A) PH-797804 concentration-response curve. Differentiating MB200 (FSHD2) cells were treated with PH-797804 in an 11-point dilution series for 40 hours. RNA levels for DUX4 target MBD3L2 and differentiation markers MYOG and MYH2 were determined in cell lysates by quantitative real-time (qRT) PCR. (B) PH-797804 in FSHD myoblasts. Proliferating cultures of 54-2 (FSHD1) and MB200 (FSHD2) myoblasts were treated with 100 nM PH-797804 for 72 hours. RNA was isolated and analyzed for DUX4 targets MBD3L2, ZSCAN4, and LEUTX. (C and D) Differentiating cultures of 54-2 (FSHD1) and MB200 (FSHD2) muscle cells were treated with varying concentrations of PH-797804, as indicated, for 40 hours and the cultures were analyzed for DUX4, DUX4 target (MBD3L2, ZSCAN4, and LEUTX), and differentiation marker (MYOG and MYH2) RNA levels by qRT-PCR. Data are expressed as relative expression (mean with S.D.) with the expression in absence of inhibitor set to 1. *P < 0.01 vs. control (unpaired, two-tailed t test). **P < 0.01 vs. control (one-way ANOVA with Dunnett’s post test).
To determine if PH-797804 was representative of the class of p38 inhibitors, we screened 12 commercially available p38 inhibitors that had reached clinical testing and two commonly cited chemical probes in differentiating FSHD myocytes. All p38 inhibitors were active with concentration-response curves similar to those of PH-797804, exhibiting an approximate 3-fold order of magnitude difference in their ability to suppress DUX4 target MBD3L2 expression versus having an effect on differentiation markers (Supplemental Fig. 1). Inhibitors were similarly active in 54-2 (FSHD1) and MB200 (FSHD2) myocytes with IC50 values reported in Table 1, suggesting that DUX4 expression is exquisitely sensitive to p38 inhibition in both genetic backgrounds. Interestingly, inhibitors of the p38 inflammatory pathway target MAPK-activated protein kinase 2 (MAPKAPK2, MK2) (Mourey et al., 2010) were not active at suppressing DUX4, nor were inhibitors of MAPK-interacting serine/threonine kinase 1/2 (MKNK1/2, MNK1/2) (Reich et al., 2018) (Table 1).
TABLE 1.
Potency of Inhibitors in differentiating cultures of FSHD1 and FHD2 muscle cells
| Inhibitor | Mechanism/Selectivity | FSHD1 EC50 | FSHD2 EC50 | Clinical Advancement |
|---|---|---|---|---|
| nM | nM | |||
| Acumapimod (BCT197) | p38α/β | 13 | 6.3 | Phase II, active |
| ARRY-614 (Pexmetinib) | p38/Tie2 | 0.33 | 0.18 | Phase I, discontinued |
| BMS-582949 | p38α, 5× selective over p38β | 17 | 4.2 | Phase II, discontinued |
| Doramapimod (BIRB796) | p38α/β/γ/δ | 0.83 | 4.6 | Phase II, discontinued |
| Losmapimod (GW856553) | p38α/β | 14 | 2.4 | Phase III, discontinued |
| Neflamapimod (VX-745) | p38α, 22X selective over p38β | 12 | 13 | PhasePhs II, active |
| Ralimetinib (LY2228820) | p38α/β JNK2, JNK3 > JNK1 | 4.0 | 1.6 | Phase II, discontinued |
| Pamapimod (RO4402257) | P38α, 34X selective over p38β | 5.5 | 2.2 | Phase II, discontinued |
| PH-797804 | p38α, 4X selective over p38β | 0.15 | 0.41 | Phase II, discontinued |
| SB202190 | p38α/β | 6.0 | 5.2 | Tool compound |
| SB203580 | p38α/β | 17 | 9.5 | Tool compound |
| TAK-715 | p38α, 28X selective over p38β | 4.4 | 12 | Phase II, discontinued |
| Talmapimod (SCIO 469) | p38α, 10X selective over p38β | 1.6 | 5.0 | Phase II, discontinued |
| VX-702 | p38α, 14X selective over p38β | 4.1 | 14 | Phase II, discontinued |
| Tomivosertib (eFT-508) | MKNK1/2 (MNK1/2) | >10,000 | >10,000 | Phase II, active |
| PF-3644022 | MAPKAPK2/3 (MK2/3) | >10,000 | >10,000 | Tool compound |
| MK2-IN-1 | MAPKAPK2 (MK2) | >10,000 | >10,000 | Tool compound |
DUX4 Activity.
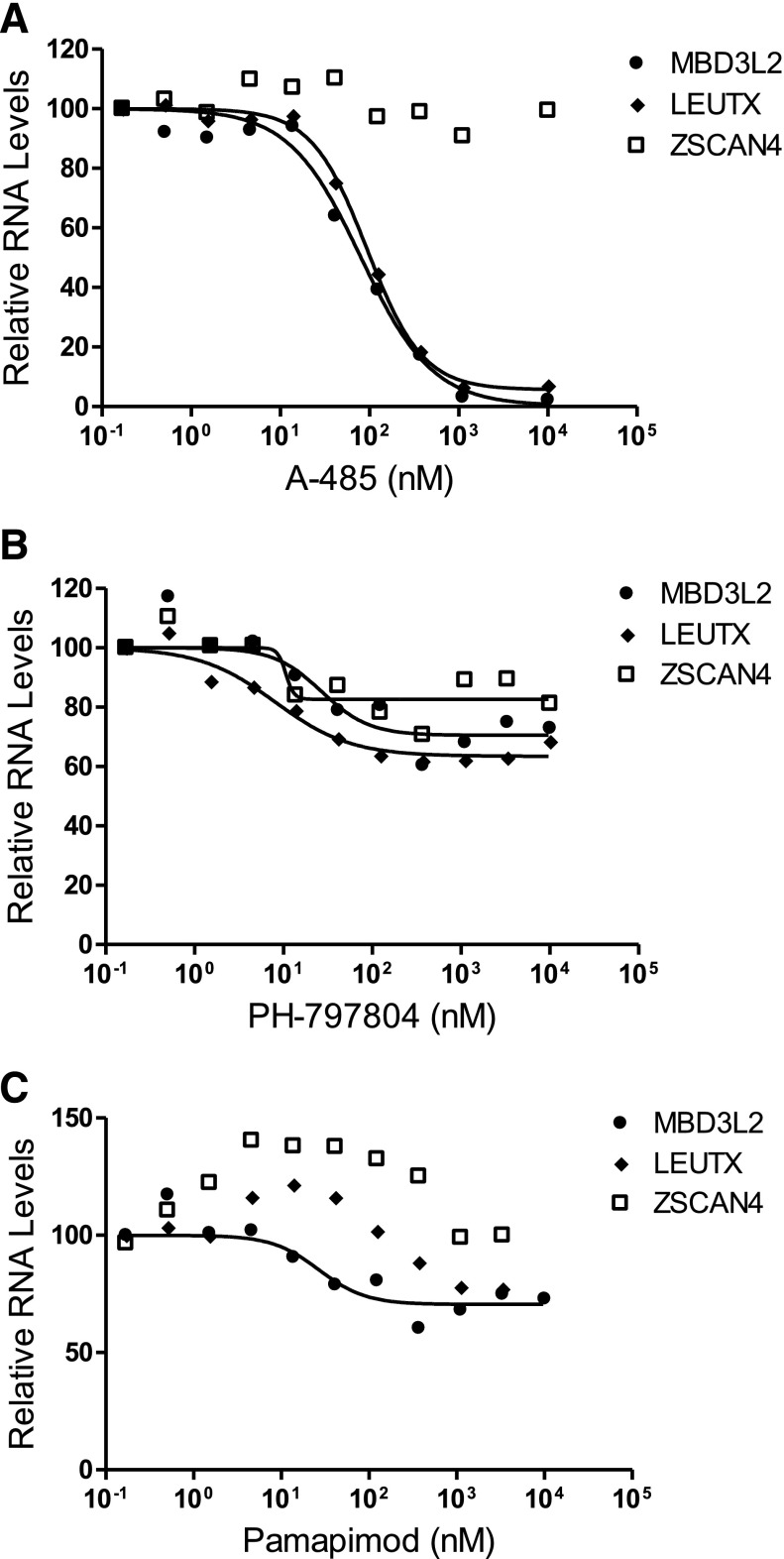
DUX4 protein is likely a target for post-translational modifications, including acetylation and phosphorylation, which could affect its function. It was possible that p38 inhibition not only suppressed the transcription of DUX4 mRNA as shown previously, but also affected the transcriptional activation function of DUX4 by altering its post-translational modifications. To test this possibility, we used forced expression of DUX4 in normal skeletal muscle myoblasts to determine if p38 inhibition would alter the ability of transiently expressed DUX4 to induce expression of its target genes. The 54-6 (non-FSHD) myoblasts were transfected with the pCS2-DUX4 plasmid (Geng et al., 2012), which drives DUX4 expression from the cytomegalovirus promoter, and then treated with various concentrations of two different p38 inhibitors for 20 hours. Since DUX4 protein interacts with the transcriptional coactivator p300 and utilizes its acetyltransferase activity to induce expression of many DUX4 target genes (Choi et al., 2016), we used the p300 acetyltransferase activity inhibitor A-485 (Lasko et al., 2017) as a control. Figure 2A shows that DUX4-induced increases in targets MBD3L2 and LEUTX are reduced by >95% in a concentration-dependent manner by A-485, demonstrating the utility of this assay in measuring DUX4 activity. Interestingly, DUX4 target ZSCAN4 mRNA levels were not affected by A-485 treatment (Fig. 2A), suggesting that DUX4 activates some target genes independent of p300 acetyltransferase activity. When transfected cells are exposed to PH-797804 during this time frame, there is a slight decrease in DUX4 targets (∼35%–40% maximal inhibition); however, significant transcriptional activation activity remains that is insensitive to p38 inhibition (Fig. 2B). Additionally, the p38α-biased inhibitor pamapimod (Hill et al., 2008) did not block DUX4-induced target gene expression (Fig. 2C). These data indicate that p38 inhibition has a primary activity of suppressing DUX4 expression and potentially minor activity of partially reducing DUX4 transactivation function.
Fig. 2.
DUX4 activity: p38 inhibitors do not block DUX4 transcriptional activation function. Non-DUX4 expressing 54-6 (normal) myoblasts were transfected with a DUX4 expression plasmid and treated with varying concentrations of control p300 inhibitor A-485 (A), p38 inhibitor PH-797804 (B) or p38 inhibitor pamapimod (C), as indicated, for 20 hours. Cultures were analyzed for DUX4 targets (MBD3L2, ZSCAN4, and LEUTX) RNA levels by quantitative real-time PCR. Data are expressed as relative expression with the expression in absence of inhibitor set to 100.
siRNA Targeting p38α and p38β.
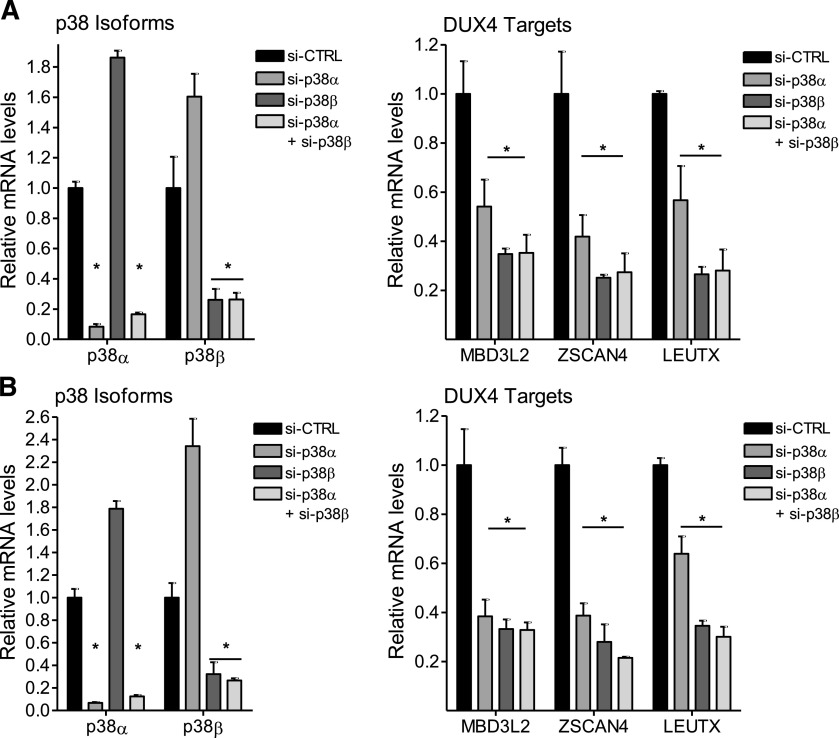
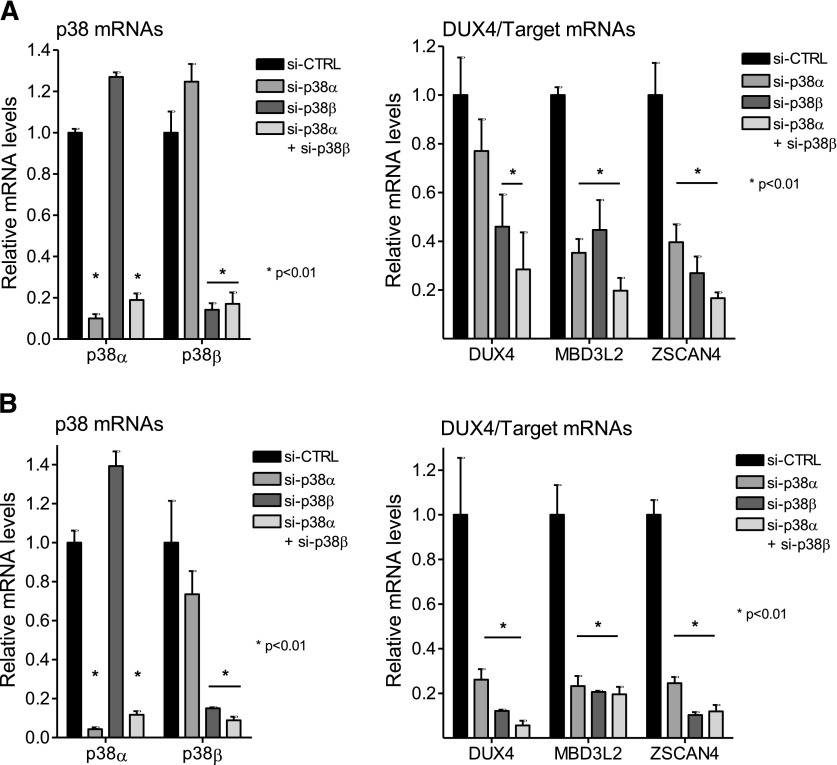
The majority of p38 inhibitors to enter clinical testing are p38α/p38β isoform-selective inhibitors with several having bias toward p38α (for reviews, see Goldstein and Gabriel, 2005; Yong et al., 2009; Norman, 2015). An exception is BIRB796, which inhibits the α, β, γ, and δ isoforms (Kuma et al., 2005). The data presented previously suggested that either p38α, p38β, or both isoforms play a role in promoting DUX4 expression in FSHD myoblasts and differentiating myocytes. The consistency of inhibition of DUX4 target genes (Table 1) appeared to track with the potency of the drugs toward the p38α isoform. To determine the relative roles of each isoform, we used siRNAs to knock down individual isoforms in FSHD1 and FSHD2 myoblasts and myotubes. Figure 3A shows that in proliferating 54-2 (FSHD1) myoblast cultures, siRNAs targeting p38α or p38β, individually or in combination, effectively and selectively decrease levels of their respective target mRNAs (left panel). Interestingly, individual knockdown of p38α mRNA resulted in increased levels of p38β mRNA and individual knockdown of p38β mRNA resulted in increased levels of p38α mRNA (Fig. 3A, left panel). Since DUX4 expression in myoblasts is difficult to measure, we looked at the effects of p38 knockdown on DUX4 target gene expression. Individual knockdown of each isoform or combined knockdown of both resulted in decreased expression of multiple DUX4 target genes (Fig. 3A, right panel), suggesting that both isoforms are individually required to support DUX4 mRNA expression. Similar results were obtained using MB200 (FSHD2) myoblasts (Fig. 3B, left and right panels). These results are surprising because instead of having redundant roles, p38α and p38β appear to have independent roles in supporting DUX4 mRNA synthesis. We extended these experiments to differentiating FSHD myotubes where DUX4 expression is increased during the differentiation process (Snider et al., 2010; Jones et al., 2012; Krom et al., 2012; Tassin et al., 2013). For these experiments, confluent monolayers of myoblasts were transfected with siRNAs 1 or 2 days prior to differentiation to allow for efficient depletion of target proteins. Differentiation was then induced for 40 hours until multinucleated myotubes predominated in the cultures. Similar to results in myoblasts, siRNAs targeting p38α or p38β, individually or in combination, effectively decrease levels of their respective target mRNA in 54-2 (FSHD1) and MB200 (FSHD2) myotubes (Fig. 4, A and B, respectively, left panels). Western analysis confirmed that p38α protein levels were decreased only when siRNA targeting p38α was used alone or in combination with siRNA targeting p38β (Supplemental Fig. 2). Similarly, p38β protein levels were decreased only when siRNA targeting p38β was used alone or in combination with siRNA targeting p38α (Supplemental Fig. 2). In myotubes, knockdown of p38β increased the expression of p38α at the mRNA and protein levels, while knockdown of p38α did not significantly affect p38β levels. Inspection of quantitative PCR cycle times indicated that p38β is expressed much less abundantly than p38α in our FSHD myotube cultures at the mRNA level. Nonetheless, individual knockdown of each p38 isoform alone or combined knockdown of both resulted in decreased expression of DUX4 and multiple DUX4 target genes in FSHD1 and FSHD2 myotubes (Fig. 4, A and B, respectively, right panels), although the decrease in DUX4 did not reach significance with individual knockdown of p38α in 54-2 (FSHD1) myotubes. It is worth noting that knockdown of p38β decreases DUX4 expression even as p38α levels are increased and that combined knockdown of both isoforms does not decrease DUX4 expression below the level generated with knockdown of p38β alone. Additionally, knockdown of the p38γ isoform did not affect DUX4 expression (Supplemental Fig. 3). These combined data in FSHD myoblasts and myotubes suggest that p38α and p38β are both necessary and play nonredundant roles in supporting pathogenic DUX4 expression in FSHD muscle cells.
Fig. 3.
p38 siRNAs suppress DUX4 in FSHD myoblasts. 54-2 (FSHD) (A) or MB200 (FSHD2) (B) myoblasts were transfected with siRNAs targeting p38α (si-p38α), p38β (si-p38β), or control siRNAs (si-CTRL) twice starting 72 hours before harvest and again 24 hours before harvest to ensure efficient knockdown. Myoblasts were maintained in growth media before RNA was isolated and analyzed for p38 (left panels) and DUX4 target [MBD3L2, ZSCAN4, and LEUTX (right panels)] expression levels. Data are expressed as relative expression (mean with S.D.) with the expression in the presence of si-CTRL set to one. *P < 0.01 vs. si-CTRL (one-way ANOVA with Dunnett’s post test).
Fig. 4.
p38 siRNAs suppress DUX4 in FSHD myotubes. (A) 54-2 (FSHD1) myoblasts were transfected with siRNA targeting p38α (si-p38α), p38β (si-p38β), or control (si-CTRL) siRNAs, as indicated, 24 hours prior to induction of differentiation. Cells were induced to differentiate and harvested 40 hours later. RNA was isolated and analyzed for p38 (left panel) and DUX4, MBD3L2, ZSCAN4, and LEUTX (right panel) expression levels. (B) MB200 (FSHD2) myoblasts were transfected with the indicated siRNAs on two consecutive days prior to inducing differentiation. Twenty-four hours after the second transfection, cells were induced to differentiate and RNA was isolated 40 hours later and analyzed as in (A). Data are expressed as relative expression (mean with S.D.) with the expression in the presence of si-CTRL set to 1. *P < 0.01 vs. si-CTRL (one-way ANOVA with Dunnett’s post test).
p38 Inhibitors Are Active in a Xenograft Model of FSHD.
To test the ability of clinically advanced p38 inhibitors to suppress DUX4 expression in vivo, we developed a pharmacology model of human FSHD gene regulation. The primate-specific organization of the DUX4 gene imbedded within D4Z4 repeats (Clapp et al., 2007; Leidenroth et al., 2012) and the loss of epigenetic repression of DUX4 within the context of human genetic mutations represent barriers to the development of a transgenic mouse model that recapitulates DUX4 misexpression in FSHD. To most accurately model FSHD epigenetic dysregulation of DUX4, we used a mouse xenograft approach in immunodeficient mice in which tibialis anterior muscles are injured by barium chloride injection to induce a regenerative response (Hardy et al., 2016). The regenerating muscle environment is conducive to engraftment of transplanted human myoblasts that respond to cues from the microenvironment, express appropriate markers of differentiation, and become part of mature myofibers (Silva-Barbosa et al., 2005). Variations of this model have been used to demonstrate engraftment and differentiation of human FSHD patient–derived myoblasts or muscle biopsies (Krom et al., 2012; Chen et al., 2016; Sakellariou et al., 2016). To treat FSHD xenograft mice with p38 inhibitors to suppress DUX4, it was necessary to determine the time frame of DUX4 expression after xenotransplantation. Since it is well established that DUX4 is induced upon differentiation of myoblasts into multinucleated myotubes in vitro in the time frame of 1.5 to 6 days (Snider et al., 2010; Jones et al., 2012; Krom et al., 2012; Tassin et al., 2013; Balog et al., 2015) and that the corresponding elevated levels of DUX4 protein are sufficient to cause myotube death (Block et al., 2013; Rickard et al., 2015), we profiled DUX4 expression during the first week after xenotransplantation of MB200 (FSHD2) myoblasts. Total RNA from xenograft mice tibialis anterior muscles was analyzed using human-specific primers and Taqman probes for DUX4 and DUX4 target gene RNA levels by quantitative real-time PCR. Supplemental Fig. 4 demonstrates that DUX4 mRNA levels increase over the first few days and peak at about the fourth day after xenotransplantation (left panel). RNA levels for representative DUX4 target gene MBD3L2 peak at days 5 to 6 and begin to decline by day 7 (Supplemental Fig. 2, right panel). These data suggest that DUX4 expression is induced as transplanted FSHD myoblasts begin to differentiate in vivo and that this process peaks within the first week.
We reasoned that treating animals for 4 days, starting immediately after the xenograft procedure through the peak of DUX4 expression, was appropriate to evaluate agents intended to suppress DUX4 expression in FSHD. We treated xenograft mice systemically with two different inhibitors: PH-797804 and losmapimod. Figure 5A shows that treatment of xenograft mice with PH-797804 by subcutaneous injection twice daily resulted in dose-dependent decreases in mRNA levels for DUX4 (left panel) and DUX4 targets MBD3L2, ZSCAN4, and LEUTX (right three panels, respectively). The highest dose of PH-797804 (5 mg/kg b.wt.) elicited a decrease of approximately 80% for DUX4 and its targets compared with vehicle. Similarly, treatment of xenograft mice with losmapimod orally twice daily resulted in dose-dependent decreases in DUX4 and DUX4 target mRNA levels (Fig. 5B). In this case, mRNA levels for DUX4 and its targets were decreased by approximately 80% with losmapimod dosed at 6 mg/kg twice daily and by greater than 90% at a dose of 18 mg/kg twice daily. In preclinical models of inflammation, losmapimod has been efficacious at a dose of 12 mg/kg per day in food (Willette et al., 2009; Yeung et al., 2018). To understand how suppression of DUX4 was related to plasma drug levels, we measured trough levels of losmapimod in plasma at sacrifice (∼14 hours after last dose). Supplemental Fig. 6 shows that terminal (trough) plasma levels of losmapimod in the 6 mg/kg dosing group were still above the in vitro IC50 values of losmapimod (average of 16 nM in plasma vs. an IC50 of 2.4 nM in MB200 cells) (see Table 1).
Fig. 5.
p38 Inhibitors suppress DUX4 expression in a mouse xenograft pharmacology model of FSHD gene regulation. (A) PH-797804 was administered to xenograft mice twice daily by subcutaneous injections at the indicated doses for 4 days. RNA was isolated from excised tibialis anterior (TA) muscles and analyze for DUX4, MBD3L2, ZSCAN4, and LEUTX RNA levels. (B) Losmapimod was administered orally to xenograft mice twice daily at the indicated doses for 4 days. RNA was isolated and analyzed as in (A). (C) Losmapimod was administered orally to xenograft mice twice daily at 6 mg/kg for 14 days. RNA and DNA were isolated from excised TA muscles and analyzed for differentiation marker MYH2 RNA levels and two-copy gene (hTERT and mTfrc) DNA levels. Data are expressed as relative expression (mean with S.E.) with the expression in the vehicle groups set to 1. For MYH2 analysis, the 14-day levels were compared with levels in 4-day animals (day 4) not treated with drugs, which was set to 1. *P < 0.01 vs. vehicle (one-way ANOVA with Dunnett’s post test). #P = 0.030 vs. vehicle (unpaired two-tailed t test).
To further assess the potential utility of losmapimod in FSHD and the effects of p38 inhibitor treatment on the differentiation of transplanted FSHD myoblasts, xenograft mice were treated for 14 days with losmapimod dosed at 6 mg/kg twice daily. Muscle RNA was analyzed for the late differentiation marker myosin heavy chain (MYH2) while terminal plasma samples were used to measure trough drug levels that are reported in Supplemental Fig. 6B. Figure 5C shows that compared with the low level present 4 days after xenotransplantation, levels of MYH2 RNA increase dramatically in mice treated with vehicle or losmapimod over the period during which fully formed mouse myofibers develop, between 4 and 14 days after xenotransplantation (left panel) (Hardy et al., 2016). These data indicate treatment of xenograft mice with losmapimod at a level that represses DUX4 by 80% is compatible with robust differentiation of the introduced human FSHD myoblasts. Since DUX4 expression during the differentiation process is sufficient to cause cell death (Block et al., 2013; Rickard et al., 2015), we also looked at the survival of human FSHD cells by measuring the human cell DNA copy number relative to mouse cell DNA after 14 days of losmapimod treatment. Fig. 5C (right panel) shows that xenograft mice treated with losmapimod had an approximately 2-fold increase in human cell copy number relative to mouse. Analysis of the PCR cycle times for the human TERT gene suggests an even greater absolute increase in human cell number per xenograft (5.5-fold) (Supplemental Fig. 5) with a 2.7-fold increase in mouse cell number per xenograft, although this analysis is semiquantitative. These data suggest that p38 treatment suppresses DUX4 expression and promotes the survival and incorporation of differentiating human FSHD cells in regenerating mouse muscle.
Discussion
FSHD is one of the most prevalent muscular dystrophies, yet development of disease-modifying therapies lags far behind that for other dystrophies. The consensus identification of DUX4 protein expression in skeletal muscle as causing the disease has given a clear therapeutic target. Approaches include targeting the DUX4 mRNA with antisense oligonucleotide technology and targeting DUX4 transcriptional activity or the activity of gene products downstream from DUX4 (Himeda et al., 2015; Banerji et al., 2018). These approaches have challenges to overcome, such as muscle delivery for antisense oligonucleotide–based therapeutics. Furthermore, there is incomplete understanding of the pathophysiology of FSHD in relation to the downstream gene targets of DUX4 that mediate its toxic effects. We have focused instead on identifying small molecule drugs capable of suppressing the transcription of DUX4 mRNA such that no DUX4 protein is produced, an approach that bypasses many of these challenges.
In this study, we demonstrated that DUX4 mRNA synthesis in FSHD myoblasts and myotubes is exquisitely sensitive to p38α/β inhibitors. The IC50 vaues shown in Table I are in line with published activities determined for isolated p38α or p38β enzymes (Xing, 2015), strongly suggesting that DUX4 expression is positively regulated by p38α or p38 α/β kinase activity. One desirable characteristic of a drug intended for degenerative muscular dystrophy is that the treatment does not negatively affect the ability of muscle cells to participate in the muscle regenerative process. The suppression of DUX4 by p38 inhibitors is independent of myocyte differentiation, evidenced by the activity of inhibitors in proliferating myoblasts. Furthermore, in differentiating myotubes DUX4 is suppressed at drug concentrations that do not affect markers of differentiation. In fact, decreases in markers of muscle differentiation (MYOG and MHY2) require three orders of magnitude higher drug concentrations than for DUX4 suppression. Even at high drug concentrations, MYOG and MHY2 decreases are not complete and may be an artifact of in vitro differentiation. Systemic administration of losmapimod was able to decrease DUX4 expression and its downstream targets without decreasing maturation markers after chronic dosing over 14 days.
The sensitivity of DUX4 in differentiating muscle cells to p38 inhibition at drug concentrations that do not prevent differentiation is surprising given the literature describing the roles of p38 in skeletal muscle. Four p38 isoforms exist in mammals: p38α, p38β, p38γ, and p38δ (Cuenda and Rousseau, 2007; Cuadrado and Nebreda, 2010). The α, β, and γ isoforms are expressed in skeletal muscle (Wang et al., 2008), and p38α involvement in regulating the skeletal muscle differentiation process is well documented. Activation of p38α/β has been shown to be important in regulating muscle progenitor satellite cell activation from quiescence and reestablishment of the satellite cell pool (Troy et al., 2012). The latter mechanism led to the proposal of using p38 inhibition to help reestablish the muscle stem cell pool in situations where this pool is depleted, such as in aged skeletal muscle (Bernet et al., 2014). Also, p38 has been implicated in key temporally ordered events involving myogenic regulatory transcription factors and the establishment of muscle-specific gene expression (Zetser et al., 1999; Wu et al., 2000; Bergstrom et al., 2002; Penn et al., 2004). Importantly, p38 is involved in several epigenetic mechanisms including recruitment of methyltransferase and nucleosome remodeling complexes to muscle-specific gene promoters (Simone et al., 2004; Rampalli et al., 2007) and inactivation of methyltransferases at specific genes (Chatterjee et al., 2016). In fact, p38α binds to and regulates many promoters during myogenesis (Segalés et al., 2016).
How can inhibition of p38α/β selectively reduce DUX4 without disrupting muscle differentiation? Several observations suggest this is possible. First, DUX4 expression is exquisitely sensitive to p38 inhibition, with much less inhibitor required than in the case of blocking differentiation. For example, SB203580 (Kumar et al., 1997) inhibited DUX4 in differentiating FSHD myocytes with IC50 values of 17 nM (FSHD1) and 9.8 nM (FSHD2), while muscle differentiation gene markers were partially inhibited at IC50 values >1500 nM (Supplemental Fig. 1). Second, mice with muscle-specific deletion of p38α grow and function normally, albeit with smaller muscles and delayed myofiber growth and maturation (Brien et al., 2013). Additionally, muscle-specific p38α knockout mice exhibit an increase in the muscle satellite cell pool, consistent with the previously mentioned role of p38 in regulating satellite cell activation and restricting excess myoblast proliferation (Troy et al., 2012) and the proposal that p38 inhibition may be a therapeutic option in muscle degenerative disorders in which the stem cell niche appears to be diminished (Bernet et al., 2014; Dumont and Rudnicki, 2016). Notably, when we treated xenograft mice for 14 days with the p38 inhibitor losmapimod, the total number of both human and mouse cells increased compared with nontreated animals (Fig. 5; Supplemental Fig. 5), consistent with p38 inhibition promoting myoblast proliferation even as the expression of a late differentiation marker (MYH2) was induced and maintained.
The molecular mechanisms that tie DUX4 expression to p38 activity remain to be elucidated. A surprise finding from these studies is that DUX4 expression is dependent on both α and β p38 isoforms (Figs. 3 and 4). The result that simultaneously knocking down both isoforms does not suppress DUX4 more than solely knocking down p38β implies that each kinase plays a distinct mechanistic role such that when p38β is knocked down, DUX4 is suppressed even when p38α levels are increased. It should be noted that p38β is generally expressed at lower levels than the other isoforms (Cuadrado and Nebreda, 2010) and appears to be dispensable for muscle differentiation (Perdiguero et al., 2007; Ruiz-Bonilla et al., 2008). This offers an opportunity to explore the utility of p38 isoform-selective inhibitors to balance efficacy and safety. Due to its role in inflammation (Allen et al., 2000), previous efforts have focused on developing p38α inhibitors for various inflammatory diseases (Kumar et al., 2003). Most of these inhibitors also inhibit p38β due to the high sequence similarity between the closely related proteins (Kondoh et al., 2016), and no p38β-selective inhibitors have been described. Given the roles of p38α in muscle differentiation as well as its role in inflammation (Page et al., 2010) it is worth pursuing the development of p38β-selective inhibitors. Interestingly, p38β inhibition has independently been suggested as a therapeutic strategy for muscle cachexia (Ding et al., 2017).
Since DUX4 is a transcription factor that is likely regulated by post-translational modifications, it was important to determine if p38 inhibition had any effect on the transcriptional activation function of DUX4. We used the p300 acetyltransferase activity inhibitor A-485 as a control since DUX4 has been reported to interact with p300 to activate target genes (Choi et al., 2016). Indeed, A-485 inhibited induction of DUX4 targets MBD3L2 and LEUTX. Interestingly, ZSCAN4 RNA levels were not decreased by A-485, suggesting that this DUX4 target is induced independent of p300 acetyltransferase activity, even though DUX4 has been shown to induce p300-dependent acetylation of histones in the vicinity of the ZSCAN4 DUX4 binding sites (Choi et al., 2016). The significance of this finding remains to be explored. In the case of p38 inhibitors, there are minor and inconsistent decreases in some DUX4 targets by p38 inhibitors. It is difficult to conclude from these data if p38 plays any role in DUX4 activity or if the small decreases are assay artifacts due to potential interference of cytomegalovirus promoter by p38 inhibitors (Bruening et al., 1998). Nonetheless, high concentrations of p38 inhibitors do not block DUX4 activity and the primary effect of p38 inhibition in FSHD cells is to suppress the synthesis of DUX4 mRNA.
The p38 inhibitors suppress the expression of DUX4 and its targets at clinically relevant doses. It is clear from in vivo testing of two clinically advanced inhibitors that p38 inhibition effectively reduces the peak levels of DUX4 expression in a dose-dependent manner (Fig. 5). The doses of 5 mg/kg twice a day for PH-797804 and 6 mg/kg twice a day for losmapimod reduce DUX4 and DUX4 target RNA levels by approximately 80%. These dosing levels have been used in translational animal models (Willette et al., 2009; Xing et al., 2012). Losmapimod has been extensively characterized in humans. Efficacy and safety studies have used repeated dosing with losmapimod at 7.5 mg, twice daily (O’Donoghue et al., 2016; Fisk et al., 2018); a detailed study of drug exposure demonstrated a stable plasma trough concentration of around 16 ng/ml (39 nM) (Ino et al., 2015). Losmapimod was efficacious in our xenograft mouse model with a terminal drug concentration in the plasma of 16 nM. These data suggest there is sufficient muscle exposure by dosing losmapimod at 7.5 mg twice a day to substantially reduce DUX4 in FSHD patient muscle. A more detailed analysis of pharmacokinetic and pharmacodynamic relationships, specifically in muscle tissue, will enable accurate prediction of human dose requirements.
To conclude, the data presented here support the repurposing of existing p38α or dual p38α/β inhibitors as potential therapeutics to suppress DUX4 expression in FSHD and suggests the development of p38β-selective inhibitors to perhaps more specifically target DUX4. These studies uncover the exciting possibility of a disease-modifying treatment for FSHD by targeting the root cause of disease through p38 MAPK inhibition.
Acknowledgments
We thank Silvère M. van der Maarel for critical reading of the manuscript, Nikolaos Atkinson for technical assistance and screening of compounds, and Mary Campbell and Stacey Arnett for PK analysis and dose selection for PH-797804.
Abbreviations
- DUX4
double homeobox 4
- FSHD
facioscapulohumeral muscular dystrophy
- FSHD1
facioscapulohumeral muscular dystrophy type 1
- FSHD2
facioscapulohumeral muscular dystrophy type 2
- MAPK
mitogen-activated protein kinase
- PCR
polymerase chain reaction
- siRNA
small interfering RNA
Authorship Contributions
Participated in research design: Oliva, Galasinski, Campbell, Meyers, Tapscott, Sverdrup.
Conducted experiments: Oliva, Richey, Campbell, Modi, Zhong, Sverdrup.
Contributed new reagents or analytic tools: Tawil.
Performed data analysis: Oliva, Richey, Campbell, Sverdrup.
Wrote or contributed to the writing of the manuscript: Oliva, Galasinski, Campbell, Tapscott, Sverdrup.
Footnotes
This work was supported by the National Institutes of Health National Institute of Neurologic Disorders and Stroke [Grant NS069539]; the Muscular Dystrophy Association [Grant 576054]; and Ultragenyx Pharmaceutical, Inc.
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Aggeli IK, Gaitanaki C, Lazou A, Beis I. (2002) α1- and β-adrenoceptor stimulation differentially activate p38-MAPK and atrial natriuretic peptide production in the perfused amphibian heart. J Exp Biol 205:2387–2397. [DOI] [PubMed] [Google Scholar]
- Allen M, Svensson L, Roach M, Hambor J, McNeish J, Gabel CA. (2000) Deficiency of the stress kinase p38α results in embryonic lethality: characterization of the kinase dependence of stress responses of enzyme-deficient embryonic stem cells. J Exp Med 191:859–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balog J, Thijssen PE, Shadle S, Straasheijm KR, van der Vliet PJ, Krom YD, van den Boogaard ML, de Jong A, F Lemmers RJ, Tawil R, et al. (2015) Increased DUX4 expression during muscle differentiation correlates with decreased SMCHD1 protein levels at D4Z4 [published correction appears in Epigenetics (2016) 11:175]. Epigenetics 10:1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji CRS, Panamarova M, Pruller J, Figeac N, Hebaishi H, Fidanis E, Saxena A, Contet J, Sacconi S, Severini S, et al. (2018) Dynamic transcriptomic analysis reveals suppression of PGC1α/ERRα drives perturbed myogenesis in facioscapulohumeral muscular dystrophy. Hum Mol Genet 28:1244–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom DA, Penn BH, Strand A, Perry RL, Rudnicki MA, Tapscott SJ. (2002) Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol Cell 9:587–600. [DOI] [PubMed] [Google Scholar]
- Bernet JD, Doles JD, Hall JK, Kelly Tanaka K, Carter TA, Olwin BB. (2014) p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nat Med 20:265–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block GJ, Narayanan D, Amell AM, Petek LM, Davidson KC, Bird TD, Tawil R, Moon RT, Miller DG. (2013) Wnt/β-catenin signaling suppresses DUX4 expression and prevents apoptosis of FSHD muscle cells. Hum Mol Genet 22:4661–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosnakovski D, Xu Z, Gang EJ, Galindo CL, Liu M, Simsek T, Garner HR, Agha-Mohammadi S, Tassin A, Coppée F, et al. (2008) An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J 27:2766–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brien P, Pugazhendhi D, Woodhouse S, Oxley D, Pell JM. (2013) p38α MAPK regulates adult muscle stem cell fate by restricting progenitor proliferation during postnatal growth and repair. Stem Cells 31:1597–1610. [DOI] [PubMed] [Google Scholar]
- Bruening W, Giasson B, Mushynski W, Durham HD. (1998) Activation of stress-activated MAP protein kinases up-regulates expression of transgenes driven by the cytomegalovirus immediate/early promoter. Nucleic Acids Res 26:486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell AE, Oliva J, Yates MP, Zhong JW, Shadle SC, Snider L, Singh N, Tai S, Hiramuki Y, Tawil R, et al. (2017) BET bromodomain inhibitors and agonists of the beta-2 adrenergic receptor identified in screens for compounds that inhibit DUX4 expression in FSHD muscle cells. Skelet Muscle 7:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee B, Wolff DW, Jothi M, Mal M, Mal AK. (2016) p38α MAPK disables KMT1A-mediated repression of myogenic differentiation program. Skelet Muscle 6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, King OD, Zhang Y, Clayton NP, Spencer C, Wentworth BM, Emerson CP, Jr, Wagner KR. (2016) Morpholino-mediated knockdown of DUX4 toward facioscapulohumeral muscular dystrophy therapeutics. Mol Ther 24:1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Gearhart MD, Cui Z, Bosnakovski D, Kim M, Schennum N, Kyba M. (2016) DUX4 recruits p300/CBP through its C-terminus and induces global H3K27 acetylation changes. Nucleic Acids Res 44:5161–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapp J, Mitchell LM, Bolland DJ, Fantes J, Corcoran AE, Scotting PJ, Armour JA, Hewitt JE. (2007) Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. Am J Hum Genet 81:264–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz JM, Hupper N, Wilson LS, Concannon JB, Wang Y, Oberhauser B, Patora-Komisarska K, Zhang Y, Glass DJ, Trendelenburg AU, et al. (2018) Protein kinase A activation inhibits DUX4 gene expression in myotubes from patients with facioscapulohumeral muscular dystrophy. J Biol Chem 293:11837–11849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado A, Nebreda AR. (2010) Mechanisms and functions of p38 MAPK signalling. Biochem J 429:403–417. [DOI] [PubMed] [Google Scholar]
- Cuenda A, Rousseau S. (2007) p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim Biophys Acta 1773:1358–1375. [DOI] [PubMed] [Google Scholar]
- Das S, Chadwick BP. (2016) Influence of repressive histone and DNA methylation upon D4Z4 transcription in non-myogenic cells. PLoS One 11:e0160022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxinger L, Tapscott SJ, van der Maarel SM. (2015) Genetic and epigenetic contributors to FSHD. Curr Opin Genet Dev 33:56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deenen JC, Arnts H, van der Maarel SM, Padberg GW, Verschuuren JJ, Bakker E, Weinreich SS, Verbeek AL, van Engelen BG. (2014) Population-based incidence and prevalence of facioscapulohumeral dystrophy. Neurology 83:1056–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A, Planet E, Coluccio A, Verp S, Duc J, Trono D. (2017) DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet 49:941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Zhang G, Sin KW, Liu Z, Lin RK, Li M, Li YP. (2017) Activin A induces skeletal muscle catabolism via p38β mitogen-activated protein kinase. J Cachexia Sarcopenia Muscle 8:202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont NA, Rudnicki MA. (2016) Targeting muscle stem cell intrinsic defects to treat Duchenne muscular dystrophy. NPJ Regen Med 1:16006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Snider L, Jagannathan S, Tawil R, van der Maarel SM, Tapscott SJ, Bradley RK. (2015) A feedback loop between nonsense-mediated decay and the retrogene DUX4 in facioscapulohumeral muscular dystrophy. eLife 4:e04996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk M, Cheriyan J, Mohan D, Forman J, Mäki-Petäjä KM, McEniery CM, Fuld J, Rudd JHF, Hopkinson NS, Lomas DA, et al. (2018) The p38 mitogen activated protein kinase inhibitor losmapimod in chronic obstructive pulmonary disease patients with systemic inflammation, stratified by fibrinogen: a randomised double-blind placebo-controlled trial. PLoS One 13:e0194197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng LN, Yao Z, Snider L, Fong AP, Cech JN, Young JM, van der Maarel SM, Ruzzo WL, Gentleman RC, Tawil R, et al. (2012) DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev Cell 22:38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein DM, Gabriel T. (2005) Pathway to the clinic: inhibition of P38 MAP kinase. A review of ten chemotypes selected for development. Curr Top Med Chem 5:1017–1029. [DOI] [PubMed] [Google Scholar]
- Goselink RJM, Voermans NC, Okkersen K, Brouwer OF, Padberg GW, Nikolic A, Tupler R, Dorobek M, Mah JK, van Engelen BGM, et al. (2017) Early onset facioscapulohumeral dystrophy - a systematic review using individual patient data. Neuromuscul Disord 27 (12):1077–1083, doi: 10.1016/j.nmd.2017.09.007 . [DOI] [PubMed] [Google Scholar]
- Hardy D, Besnard A, Latil M, Jouvion G, Briand D, Thépenier C, Pascal Q, Guguin A, Gayraud-Morel B, Cavaillon JM, et al. (2016) Comparative study of injury models for studying muscle regeneration in mice. PLoS One 11:e0147198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson PG, Doráis JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, et al. (2017) Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet 49:925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RJ, Dabbagh K, Phippard D, Li C, Suttmann RT, Welch M, Papp E, Song KW, Chang KC, Leaffer D, et al. (2008) Pamapimod, a novel p38 mitogen-activated protein kinase inhibitor: preclinical analysis of efficacy and selectivity. J Pharmacol Exp Ther 327:610–619. [DOI] [PubMed] [Google Scholar]
- Himeda CL, Jones TI, Jones PL. (2015) Facioscapulohumeral muscular dystrophy as a model for epigenetic regulation and disease. Antioxid Redox Signal 22:1463–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ino H, Takahashi N, Terao T, Igarashi H, Sarai N. (2015) Safety, tolerability, pharmacokinetics, and pharmacodynamics of losmapimod in healthy Japanese volunteers. Clin Pharmacol Drug Dev 4:262–269. [DOI] [PubMed] [Google Scholar]
- Joassard OR, Durieux AC, Freyssenet DG. (2013) β2-Adrenergic agonists and the treatment of skeletal muscle wasting disorders. Int J Biochem Cell Biol 45:2309–2321. [DOI] [PubMed] [Google Scholar]
- Jones TI, Chen JC, Rahimov F, Homma S, Arashiro P, Beermann ML, King OD, Miller JB, Kunkel LM, Emerson CP, Jr, et al. (2012) Facioscapulohumeral muscular dystrophy family studies of DUX4 expression: evidence for disease modifiers and a quantitative model of pathogenesis. Hum Mol Genet 21:4419–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh Y, Honda K, Hiranuma S, Hayashi T, Shimizu T, Watanabe N, Osada H. (2016) Comparative chemical array screening for p38γ/δ MAPK inhibitors using a single gatekeeper residue difference between p38α/β and p38γ/δ. Sci Rep 6:29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowaljow V, Marcowycz A, Ansseau E, Conde CB, Sauvage S, Mattéotti C, Arias C, Corona ED, Nuñez NG, Leo O, et al. (2007) The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul Disord 17:611–623. [DOI] [PubMed] [Google Scholar]
- Krom YD, Dumonceaux J, Mamchaoui K, den Hamer B, Mariot V, Negroni E, Geng LN, Martin N, Tawil R, Tapscott SJ, et al. (2012) Generation of isogenic D4Z4 contracted and noncontracted immortal muscle cell clones from a mosaic patient: a cellular model for FSHD. Am J Pathol 181:1387–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma Y, Sabio G, Bain J, Shpiro N, Márquez R, Cuenda A. (2005) BIRB796 inhibits all p38 MAPK isoforms in vitro and in vivo. J Biol Chem 280:19472–19479. [DOI] [PubMed] [Google Scholar]
- Kumar S, Boehm J, Lee JC. (2003) p38 MAP kinases: key signalling molecules as therapeutic targets for inflammatory diseases. Nat Rev Drug Discov 2:717–726. [DOI] [PubMed] [Google Scholar]
- Kumar S, McDonnell PC, Gum RJ, Hand AT, Lee JC, Young PR. (1997) Novel homologues of CSBP/p38 MAP kinase: activation, substrate specificity and sensitivity to inhibition by pyridinyl imidazoles. Biochem Biophys Res Commun 235:533–538. [DOI] [PubMed] [Google Scholar]
- Lasko LM, Jakob CG, Edalji RP, Qiu W, Montgomery D, Digiammarino EL, Hansen TM, Risi RM, Frey R, Manaves V, et al. (2017) Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 550:128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidenroth A, Clapp J, Mitchell LM, Coneyworth D, Dearden FL, Iannuzzi L, Hewitt JE. (2012) Evolution of DUX gene macrosatellites in placental mammals. Chromosoma 121:489–497. [DOI] [PubMed] [Google Scholar]
- Lemmers RJ, Tawil R, Petek LM, Balog J, Block GJ, Santen GW, Amell AM, van der Vliet PJ, Almomani R, Straasheijm KR, et al. (2012) Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet 44:1370–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camaño P, Dauwerse JG, Snider L, Straasheijm KR, van Ommen GJ, Padberg GW, et al. (2010) A unifying genetic model for facioscapulohumeral muscular dystrophy. Science 329:1650–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. [DOI] [PubMed] [Google Scholar]
- McAlees JW, Sanders VM. (2009) Hematopoietic protein tyrosine phosphatase mediates β2-adrenergic receptor-induced regulation of p38 mitogen-activated protein kinase in B lymphocytes. Mol Cell Biol 29:675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moule SK, Denton RM. (1998) The activation of p38 MAPK by the β-adrenergic agonist isoproterenol in rat epididymal fat cells. FEBS Lett 439:287–290. [DOI] [PubMed] [Google Scholar]
- Mourey RJ, Burnette BL, Brustkern SJ, Daniels JS, Hirsch JL, Hood WF, Meyers MJ, Mnich SJ, Pierce BS, Saabye MJ, et al. (2010) A benzothiophene inhibitor of mitogen-activated protein kinase-activated protein kinase 2 inhibits tumor necrosis factor α production and has oral anti-inflammatory efficacy in acute and chronic models of inflammation. J Pharmacol Exp Ther 333:797–807. [DOI] [PubMed] [Google Scholar]
- Norman P. (2015) Investigational p38 inhibitors for the treatment of chronic obstructive pulmonary disease. Expert Opin Investig Drugs 24:383–392. [DOI] [PubMed] [Google Scholar]
- O’Donoghue ML, Glaser R, Cavender MA, Aylward PE, Bonaca MP, Budaj A, Davies RY, Dellborg M, Fox KA, Gutierrez JA, et al. LATITUDE-TIMI 60 Investigators (2016) Effect of losmapimod on cardiovascular outcomes in patients hospitalized with acute myocardial infarction: a randomized clinical trial. JAMA 315:1591–1599. [DOI] [PubMed] [Google Scholar]
- Padberg GW, Frants RR, Brouwer OF, Wijmenga C, Bakker E, Sandkuijl LA. (1995) Facioscapulohumeral muscular dystrophy in the Dutch population. Muscle Nerve Suppl 2:S81–S84. [PubMed] [Google Scholar]
- Page TH, Brown A, Timms EM, Foxwell BM, Ray KP. (2010) Inhibitors of p38 suppress cytokine production in rheumatoid arthritis synovial membranes: does variable inhibition of interleukin-6 production limit effectiveness in vivo? Arthritis Rheum 62:3221–3231. [DOI] [PubMed] [Google Scholar]
- Penn BH, Bergstrom DA, Dilworth FJ, Bengal E, Tapscott SJ. (2004) A MyoD-generated feed-forward circuit temporally patterns gene expression during skeletal muscle differentiation. Genes Dev 18:2348–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdiguero E, Ruiz-Bonilla V, Gresh L, Hui L, Ballestar E, Sousa-Victor P, Baeza-Raja B, Jardí M, Bosch-Comas A, Esteller M, et al. (2007) Genetic analysis of p38 MAP kinases in myogenesis: fundamental role of p38α in abrogating myoblast proliferation. EMBO J 26:1245–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampalli S, Li L, Mak E, Ge K, Brand M, Tapscott SJ, Dilworth FJ. (2007) p38 MAPK signaling regulates recruitment of Ash2L-containing methyltransferase complexes to specific genes during differentiation. Nat Struct Mol Biol 14:1150–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich SH, Sprengeler PA, Chiang GG, Appleman JR, Chen J, Clarine J, Eam B, Ernst JT, Han Q, Goel VK, et al. (2018) Structure-based design of pyridone–aminal eFT508 targeting dysregulated translation by selective mitogen-activated protein kinase interacting kinases 1 and 2 (MNK1/2) inhibition. J Med Chem 61:3516–3540. [DOI] [PubMed] [Google Scholar]
- Rickard AM, Petek LM, Miller DG. (2015) Endogenous DUX4 expression in FSHD myotubes is sufficient to cause cell death and disrupts RNA splicing and cell migration pathways. Hum Mol Genet 24:5901–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Bonilla V, Perdiguero E, Gresh L, Serrano AL, Zamora M, Sousa-Victor P, Jardí M, Wagner EF, Muñoz-Cánoves P. (2008) Efficient adult skeletal muscle regeneration in mice deficient in p38β, p38γ and p38δ MAP kinases. Cell Cycle 7:2208–2214. [DOI] [PubMed] [Google Scholar]
- Sakellariou P, O’Neill A, Mueller AL, Stadler G, Wright WE, Roche JA, Bloch RJ. (2016) Neuromuscular electrical stimulation promotes development in mice of mature human muscle from immortalized human myoblasts. Skelet Muscle 6:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segalés J, Islam AB, Kumar R, Liu QC, Sousa-Victor P, Dilworth FJ, Ballestar E, Perdiguero E, Muñoz-Cánoves P. (2016) Chromatin-wide and transcriptome profiling integration uncovers p38α MAPK as a global regulator of skeletal muscle differentiation. Skelet Muscle 6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selness SR, Devraj RV, Devadas B, Walker JK, Boehm TL, Durley RC, Shieh H, Xing L, Rucker PV, Jerome KD, et al. (2011) Discovery of PH-797804, a highly selective and potent inhibitor of p38 MAP kinase. Bioorg Med Chem Lett 21:4066–4071. [DOI] [PubMed] [Google Scholar]
- Shadle SC, Zhong JW, Campbell AE, Conerly ML, Jagannathan S, Wong CJ, Morello TD, van der Maarel SM, Tapscott SJ. (2017) DUX4-induced dsRNA and MYC mRNA stabilization activate apoptotic pathways in human cell models of facioscapulohumeral dystrophy. PLoS Genet 13:e1006658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Barbosa SD, Butler-Browne GS, Di Santo JP, Mouly V. (2005) Comparative analysis of genetically engineered immunodeficient mouse strains as recipients for human myoblast transplantation. Cell Transplant 14:457–467. [DOI] [PubMed] [Google Scholar]
- Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. (2004) p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet 36:738–743. [DOI] [PubMed] [Google Scholar]
- Snider L, Asawachaicharn A, Tyler AE, Geng LN, Petek LM, Maves L, Miller DG, Lemmers RJ, Winokur ST, Tawil R, et al. (2009) RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum Mol Genet 18:2414–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider L, Geng LN, Lemmers RJ, Kyba M, Ware CB, Nelson AM, Tawil R, Filippova GN, van der Maarel SM, Tapscott SJ, et al. (2010) Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet 6:e1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler G, Chen JC, Wagner K, Robin JD, Shay JW, Emerson CP, Jr, Wright WE. (2011) Establishment of clonal myogenic cell lines from severely affected dystrophic muscles - CDK4 maintains the myogenic population. Skelet Muscle 1:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin A, Laoudj-Chenivesse D, Vanderplanck C, Barro M, Charron S, Ansseau E, Chen YW, Mercier J, Coppée F, Belayew A. (2013) DUX4 expression in FSHD muscle cells: how could such a rare protein cause a myopathy? J Cell Mol Med 17:76–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawil R, van der Maarel SM, Tapscott SJ. (2014) Facioscapulohumeral dystrophy: the path to consensus on pathophysiology. Skelet Muscle 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teveroni E, Pellegrino M, Sacconi S, Calandra P, Cascino I, Farioli-Vecchioli S, Puma A, Garibaldi M, Morosetti R, Tasca G, et al. (2017) Estrogens enhance myoblast differentiation in facioscapulohumeral muscular dystrophy by antagonizing DUX4 activity. J Clin Invest 127:1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy A, Cadwallader AB, Fedorov Y, Tyner K, Tanaka KK, Olwin BB. (2012) Coordination of satellite cell activation and self-renewal by Par-complex-dependent asymmetric activation of p38α/β MAPK. Cell Stem Cell 11:541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boogaard ML, Lemmers RJLF, Balog J, Wohlgemuth M, Auranen M, Mitsuhashi S, van der Vliet PJ, Straasheijm KR, van den Akker RFP, Kriek M, et al. (2016) Mutations in DNMT3B modify epigenetic repression of the D4Z4 repeat and the penetrance of facioscapulohumeral dystrophy. Am J Hum Genet 98:1020–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Overveld PG, Lemmers RJ, Sandkuijl LA, Enthoven L, Winokur ST, Bakels F, Padberg GW, van Ommen GJ, Frants RR, van der Maarel SM. (2003) Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet 35:315–317. [DOI] [PubMed] [Google Scholar]
- Wallace LM, Garwick SE, Mei W, Belayew A, Coppee F, Ladner KJ, Guttridge D, Yang J, Harper SQ. (2011) DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann Neurol 69:540–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Xu Q, Xiao F, Jiang Y, Wu Z. (2008) Involvement of the p38 mitogen-activated protein kinase α, β, and γ isoforms in myogenic differentiation. Mol Biol Cell 19:1519–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiddon JL, Langford AT, Wong CJ, Zhong JW, Tapscott SJ. (2017) Conservation and innovation in the DUX4-family gene network. Nat Genet 49:935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willette RN, Eybye ME, Olzinski AR, Behm DJ, Aiyar N, Maniscalco K, Bentley RG, Coatney RW, Zhao S, Westfall TD, et al. (2009) Differential effects of p38 mitogen-activated protein kinase and cyclooxygenase 2 inhibitors in a model of cardiovascular disease. J Pharmacol Exp Ther 330:964–970. [DOI] [PubMed] [Google Scholar]
- Winokur ST, Chen YW, Masny PS, Martin JH, Ehmsen JT, Tapscott SJ, van der Maarel SM, Hayashi Y, Flanigan KM. (2003) Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum Mol Genet 12:2895–2907. [DOI] [PubMed] [Google Scholar]
- Wu Z, Woodring PJ, Bhakta KS, Tamura K, Wen F, Feramisco JR, Karin M, Wang JY, Puri PL. (2000) p38 and extracellular signal-regulated kinases regulate the myogenic program at multiple steps. Mol Cell Biol 20:3951–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L. (2015) Clinical candidates of small molecule p38 MAPK inhibitors for inflammatory diseases. MAP Kinase 4:24–30. [Google Scholar]
- Xing L, Devadas B, Devraj RV, Selness SR, Shieh H, Walker JK, Mao M, Messing D, Samas B, Yang JZ, et al. (2012) Discovery and characterization of atropisomer PH-797804, a p38 MAP kinase inhibitor, as a clinical drug candidate. ChemMedChem 7:273–280. [DOI] [PubMed] [Google Scholar]
- Yao Z, Snider L, Balog J, Lemmers RJ, Van Der Maarel SM, Tawil R, Tapscott SJ. (2014) DUX4-induced gene expression is the major molecular signature in FSHD skeletal muscle. Hum Mol Genet 23:5342–5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung YT, Yin S, Lu B, Fan S, Yang R, Bai R, Zhang C, Bode AM, Liu K, Dong Z. (2018) Losmapimod overcomes gefitinib resistance in non-small cell lung cancer by preventing tetraploidization. EBioMedicine 28:51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong HY, Koh MS, Moon A. (2009) The p38 MAPK inhibitors for the treatment of inflammatory diseases and cancer. Expert Opin Investig Drugs 18:1893–1905. [DOI] [PubMed] [Google Scholar]
- Young JM, Whiddon JL, Yao Z, Kasinathan B, Snider L, Geng LN, Balog J, Tawil R, van der Maarel SM, Tapscott SJ. (2013) DUX4 binding to retroelements creates promoters that are active in FSHD muscle and testis. PLoS Genet 9:e1003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, de Greef JC, Chen YY, Chien R, Kong X, Gregson HC, Winokur ST, Pyle A, Robertson KD, Schmiesing JA, et al. (2009) Specific loss of histone H3 lysine 9 trimethylation and HP1γ/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD). PLoS Genet 5:e1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetser A, Gredinger E, Bengal E. (1999) p38 mitogen-activated protein kinase pathway promotes skeletal muscle differentiation. Participation of the MEF2C transcription factor. J Biol Chem 274:5193–5200. [DOI] [PubMed] [Google Scholar]
- Zheng M, Zhang SJ, Zhu WZ, Ziman B, Kobilka BK, Xiao RP. (2000) β2-Adrenergic receptor-induced p38 MAPK activation is mediated by protein kinase A rather than by Gi or Gβγ in adult mouse cardiomyocytes. J Biol Chem 275:40635–40640. [DOI] [PubMed] [Google Scholar]