Abstract
Purpose of review:
Antibiotic stress can evoke considerable genotypic and phenotypic changes in Gram-positive bacteria. Here, we review recent studies describing altered virulence expression in response to cell wall-acting antibiotics and discuss mechanisms that coordinate regulation of the antibiotic response.
Recent findings:
Pleiotropic effects induced by antibiotic exposure include alterations to bacterial metabolism, cell wall structure and antibiotic resistance. In addition, subinhibitory concentrations of cell wall-active antibiotics have increasingly been shown to induce the production of exotoxins and biofilm formation that may influence virulence. Remarkably, phenotypes associated with comparable antibiotic stresses can vary considerably, emphasizing the need to better understand the response to cell wall-active antibiotics. Recent studies support both direct antibiotic recognition and recognition of antibiotic-induced stress to the bacterial cell wall. Specifically, bacterial two-component systems, PASTA kinases and conserved oxidative-stress sensors each contribute to modulating the antibiotic stress response.
Summary:
Bacterial sensory systems and global regulators coordinate signaling in response to cell wall-active antibiotics. Regulation of the antibiotic response is complex and involves integration of signals from multiple response pathways. A better definition of the antibiotic stress response among Gram-positive pathogens may yield novel therapeutic targets to counter antibiotic resistance and virulence factor expression.
Keywords: Antibiotic, cell wall, resistance, stress response
INTRODUCTION
In Gram-positive bacteria, a thick peptidoglycan layer comprises the cell wall and provides structure for the organism. The cell wall also serves a sensory function as bacteria have evolved intricate mechanisms to perceive environmental change including heat, pH, nutrient limitation, and chemical and oxidative stress. Cell wall-active (CWA) antibiotics target biosynthesis of the bacterial cell wall and it is well established that these agents can act as bacterial signaling molecules, evoking diverse biological responses in a concentration-dependent manner (1, 2). Specifically, CWA antibiotics can alter bacterial metabolism, antibiotic resistance and virulence through a coordinated response involving conserved signaling pathways. Considerable heterogeneity exists in the response to antibiotic stress and mechanisms controlling this response remain largely unknown. The current review highlights recent findings of altered virulence potential in response to CWA antibiotics and discusses current insights into signaling pathways that influence the antibiotic stress response.
Altered virulence potential in response to cell wall-active antibiotics
Antibiotics can remarkably influence the production of bacterial toxins. Studies in Staphylococcus aureus have demonstrated that subinhibitory concentrations of CWA antibiotics induce production of potent exotoxins including alpha-hemolysin (3, 4), PVL and TSST-1 (5-7). In Clostridium difficile, subinhibitory CWA antibiotics similarly induce toxin expression (8) including Toxin A and Toxin B (9, 10). Regulation of toxin expression is undoubtedly complex and factors that control toxin expression in response to CWA antibiotics are not completely understood. Recent studies suggest that two-component signaling (TCS) systems, global regulators and certain penicillin-binding protein utilization can influence toxin induction (6, 11). Long noncoding RNA also contributes to the control of antibiotic-induced toxin production in S. aureus (12), indicating an integration of various regulatory pathways in virulence expression.
In addition to toxins, the expression and architecture of bacterial biofilm is modified in response to CWA antibiotics. In S. aureus and Enterococcus faecalis, subinhibitory concentrations of β-lactam and glycopeptide antibiotics induce biofilm formation in a manner dependent upon cell lysis and extracellular DNA (13-16). Recent studies highlight that CWA antibiotics increase bacterial attachment under both static and flow conditions and produce thicker biofilms containing more pillar and channel structures (17). Exposure of existing biofilms to subinhibitory concentrations of antibiotics can similarly lead to biofilm restructuring (18).
Many of the studies assessing the impact of subinhibitory antibiotics on virulence have been conducted in vitro and require validation of antibiotic effects in vivo and in a polymicrobial context. Importantly, strain- and antibiotic-dependent behavior is often observed when monitoring the effects of CWA antibiotics on virulence expression. This points toward a multifaceted response to CWA antibiotics that is dependent upon genetic background, context of the interaction, and multiple interacting signaling pathways, discussed further in the present review.
Two-component signaling systems orchestrate the response to CWA antibiotics
Antibiotic stress can induce an adaptive response in bacteria that comprises the cell wall stress stimulon (CWSS). Notably, the CWSS in Gram-positive organisms is controlled by one or more TCS systems. Although some CWSS TCS systems are triggered by specific classes of antibiotics, others appear to be activated less selectively in response to diverse CWA antibiotic stress (19). Genome-wide transcriptional profiling studies in several Gram-positive pathogens have demonstrated that the CWSS includes a subset of genes that rapidly modulate cell wall metabolism, providing a mechanism of tolerance to antibiotic-induced stress (20-24) (Figure 1A). A systems-wide proteomic analysis in S. aureus similarly found an upregulation of the peptidoglycan biosynthetic pathway following CWA antibiotic stress (25). Activation of CWSS TCS systems results in transcriptional profiles that are strain-dependent (26), and induction kinetics reflect antibiotic- and concentration-dependent growth inhibition (19). CWSS TCS systems provide specificity to the CWA antibiotic response and function in response to disrupted cell wall synthesis, or cell wall hydrolysis, but not toward other external stresses.
Figure 1. An integrated signaling network coordinates the Gram-positive bacterial response to cell wall-active antibiotics.
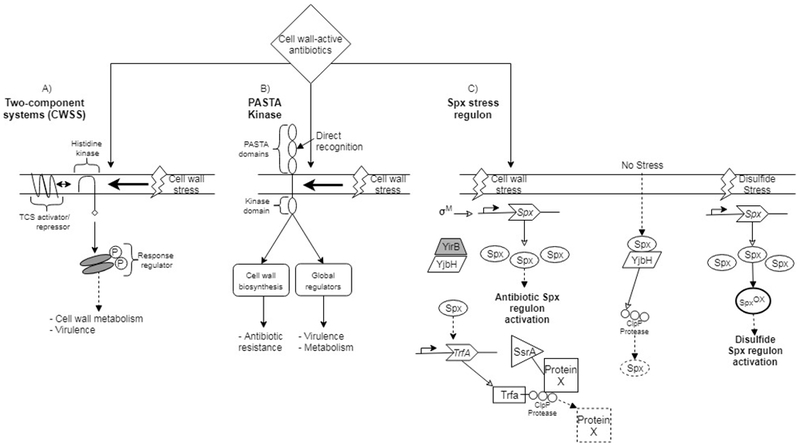
The response to cell wall-active antibiotics involves multiple sensory systems including TCS systems, PASTA kinases and the Spx stress regulon. (A) One of more TCS systems comprise the CWSS. A TCS histidine kinase responds to cell wall stress by autophosphorylation and subsequent transphosphorylation of its response regulator. A third TCS component can act as an activator, or repressor, in regulating TCS signaling. (B) PASTA kinase signaling occurs by direct recognition of antibiotics or, alternatively, by responding to cell wall stress. PASTA kinase activation contributes to modulation of several cellular functions including cell wall biosynthesis, metabolism and virulence by acting on global regulators. (C) The Spx pathway responds to independent stresses. Under no-stress conditions, Spx is bound by YjbH and targeted for degradation. Disulfide stress increases Spx levels and oxidizes Spx to control activation of gene expression. In contrast, antibiotic stress increases Spx, in a manner dependent upon σM, and YirB sequesters YjbH allowing a reduced form of Spx to activate target gene expression. TrfA is induced by Spx during antibiotic stress and serves as a proteasome adapter for recognition of SsrA-tagged substrates. Notably, cross-talk exists among PASTA kinases and TCS systems and between the Spx and TCS systems, coordinating activities of the antibiotic response. Dashed boxes indicated degraded protein.
Modulation of the CWA antibiotic response by PASTA kinases
Recently, serine/threonine kinases, containing extracellular penicillin-binding protein and serine/threonine kinase-associated (PASTA) domains, and their cognate phosphatases have emerged as critical factors regulating the bacterial response to CWA antibiotics. As regulators of cell wall metabolism, PASTA kinases are important in perceiving environmental stress. Genetic deletion of a PASTA kinase sensitizes Gram-positive organisms to CWA antibiotics, supporting an integral role for these kinases in the cell wall stress response and antibiotic resistance (27-32). Likewise, selectively targeting PASTA kinase activity with chemical inhibitors increases susceptibility to β-lactam antibiotics (31, 33-36). Conversely, mutation of an antagonistic Ser/Thr phosphatase in S. aureus contributes to reduced vancomycin susceptibility (37-39) and recapitulates the characteristic thick cell wall phenotype of vancomycin resistance (27).
PASTA kinases coordinate the response to cell wall stress through interactions with bacterial TCS systems and global regulators. For example, S. aureus PASTA kinase signaling targets VraRS, WalKR, GraSR, CcpA, MgrA and SarA (reviewed in (40)) and an E. faecalis PASTA kinase targets CroSR (41), involved in CWA antibiotic resistance. Crosstalk among these independent stress response pathways demonstrates an appreciable level of complexity in the signaling network that underlies the bacterial response to CWA antibiotics. Gene expression studies demonstrate that PASTA kinase activity regulates broad cellular functions including cell wall biosynthesis, metabolism and virulence expression (Figure 1B) (42). Their involvement in toxin expression support that PASTA kinases play a role in the response to subinhibitory concentrations of CWA antibiotics. Remarkably, the contributions of PASTA kinase activity to CWA antibiotic stress are bacteria- and strain-specific. Sequence divergence of PASTA domains (43) and dissimilar interactions with TCS systems among bacteria confound our current understanding of PASTA kinase signaling.
The mechanism by which PASTA kinases recognize cell wall stress remains unclear. Extracellular PASTA domains can recognize the cell wall precursor lipid II and soluble muropeptides (44-47) supporting the notion that cell wall stress could be sensed by altered peptidoglycan precursor pools. Recent evidence also points toward the direct recognition of β-lactam antibiotics by PASTA kinases (45, 46, 48). Intriguingly, a S. aureus isolate that encodes a truncated PASTA kinase, lacking the extracellular PASTA domains, still provides resistance to CWA antibiotics, signifying kinase activation can occur via a mechanism independent of PASTA ligand recognition (32). Further studies are required to clarify the mechanism of PASTA kinase activation, signaling interactions and observed phenotypes in response to CWA antibiotic stress.
Activation of the Spx regulon by CWA antibiotic stress
In addition to TCS systems and PASTA kinases, the Spx stress regulator is highly conserved among Gram-positive bacteria and controls the expression of genes involved in stress tolerance and virulence. Spx is recognized as a transcriptional regulator in the oxidative stress response and spx mutants were recently found to exhibit sensitivity to CWA antibiotic stress (49, 50). Consistent with a role for Spx in the antibiotic stress response, mutation of yjbH, a negative regulator of Spx, reduces bacterial susceptibility to both glycopeptide and β-lactam antibiotics (39, 51).
Induction of the Spx regulon by CWA antibiotics suggests a link between the antibiotic and oxidative stress response. Remarkably, studies in Bacillus subtilus demonstrate that mechanisms of Spx activation during CWA antibiotic stress are distinct from the oxidative stress response (Figure 1C) (50, 52). Specifically, CWA antibiotic stress requires upregulation of spx from a σM promoter, whereas several promoters can activate spx expression during disulfide stress (50). Contrary to oxidized Spx (redox switch) present during disulfide stress, Spx was maintained in a reduced form during antibiotic stress, permitting differential gene regulation in response to different stresses (50). Oxidation of the YjbH regulator was also dispensable for S. aureus β-lactam resistance, but not disulfide stress (51). Thus, Spx is a common regulator in both oxidative and antibiotic stress, but independent signaling interactions provide specificity to each response.
Proteolytic activity regulates adaptation to the CWA antibiotic response by eliminating stress-damaged proteins, controlling cell wall metabolism and regulating virulence (53). In addition to complex transcriptional regulation, Spx is subject to posttranslational oxidation and proteolysis (54). Under non-stress conditions, Spx is targeted to the ClpP protease by YjbH. CWA antibiotic stress induces spx expression but also stabilizes Spx protein levels by the anti-YjbH factor, YirB (52). YirB is induced by the CssRS TCS system in response to CWA antibiotics (52), pointing toward interaction of the Spx pathway with TCS sensory systems. Further, TrfA, a factor contributing to glycopeptide and oxacillin resistance, is induced by Spx in response to CWA antibiotics (55-57). TrfA imparts both specificity and timing to protease-mediated degradation of antitoxins and other factors implicated in the response to antibiotic stress (58). Together, the Spx regulon actively controls both gene expression and protein turnover during the CWA antibiotic stress response.
CONCLUSION
The bacterial response to CWA antibiotics is well appreciated; however, mechanisms directing the outcome of such interactions remain less clear. Multiple conserved sensory systems have now been shown to contribute to perception of CWA antibiotic stress. Alternative sigma factors (20, 59) and the SOS response (60, 61) also play a role in response to antibiotic stress. Interestingly, considerable variation is evident in the global antibiotic response. Strains of the same pathogen can exhibit fundamentally dissimilar responses to the same antibiotic stress (62), highlighting a dependence upon genetic background, the context of the antibiotic interaction and possible epigenetic effects. Phenotypic variations among bacterial subpopulations ultimately contribute to pathogen success and persistence during environmental stress. In the context of genome-wide responses, a strikingly poor correlation between transcriptional and phenotypic fitness suggests that CWA antibiotic stress is quite complex (63). Together, the overall significance of antibiotics toward bacterial evolution and virulence merits the continued investigation of mechanisms controlling CWA antibiotic-induced stress.
KEY POINTS.
Cell wall-active antibiotics can induce the expression of potent exotoxins and alter biofilm formation in Gram-positive bacteria.
Strain-specific responses to cell wall-active antibiotics indicate that complex mechanisms regulate the antibiotic-induced stress response.
Bacterial two-component signaling systems, PASTA kinases and the global Spx stress regulator coordinate a response to cell wall-active antibiotics.
Antibiotic-induced cell wall stress utilizes conserved signaling pathways but independent interactions provide specificity to the response.
ACKNOWLEDGEMENTS
The views expressed here are those of the authors and do not reflect the opinion or policy of the VA or the Government of the United States.
Financial support and sponsorship: This work was supported by the U.S. Department of Veterans Affairs Office of Research and Development Biomedical Laboratory Research Program and the National Institutes of Health Centers of Biomedical Research Excellence (P20GM109007).
ABBREVIATIONS
- CWA
cell wall-active
- CWSS
cell wall stress stimulon
- PASTA
penicillin-binding protein and serine/threonine kinase-associated
- TCS
two-component signaling
Footnotes
Conflicts of interest: none.
REFERENCES AND RECOMMENDED READING
- 1.Bernier SP, Surette MG. Concentration-dependent activity of antibiotics in natural environments. Front Microbiol. 2013;4:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies J, Spiegelman GB, Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol. 2006;9(5):445–53. [DOI] [PubMed] [Google Scholar]
- 3.Kernodle DS, McGraw PA, Barg NL, et al. Growth of Staphylococcus aureus with nafcillin in vitro induces alpha-toxin production and increases the lethal activity of sterile broth filtrates in a murine model. J Infect Dis. 1995;172(2):410–9. [DOI] [PubMed] [Google Scholar]
- 4.Ohlsen K, Ziebuhr W, Koller KP, et al. Effects of subinhibitory concentrations of antibiotics on alpha-toxin (hla) gene expression of methicillin-sensitive and methicillin-resistant Staphylococcus aureus isolates. Antimicrob Agents Chemother. 1998;42(11):2817–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hodille E, Rose W, Diep BA, et al. The Role of Antibiotics in Modulating Virulence in Staphylococcus aureus. Clin Microbiol Rev. 2017;30(4):887–917.** This review article highlights the ability of subinhibitory concentrations of antibiotics to modulate toxin production in S. aureus. It presents a comprehensive survey of the literature regarding effects of antibiotics on S. aureus virulence and the mechanisms underlying altered virulence expression.
- 6.Dumitrescu O, Choudhury P, Boisset S, et al. Beta-lactams interfering with PBP1 induce Panton-Valentine leukocidin expression by triggering sarA and rot global regulators of Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55(7):3261–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevens DL, Ma Y, Salmi DB, et al. Impact of antibiotics on expression of virulence-associated exotoxin genes in methicillin-sensitive and methicillin-resistant Staphylococcus aureus. J Infect Dis. 2007;195(2):202–11. [DOI] [PubMed] [Google Scholar]
- 8.Onderdonk AB, Lowe BR, Bartlett JG. Effect of environmental stress on Clostridium difficile toxin levels during continuous cultivation. Appl Environ Microbiol. 1979;38(4):637–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aldape MJ, Heeney DD, Bryant AE, Stevens DL. Tigecycline suppresses toxin A and B production and sporulation in Clostridium difficile. J Antimicrob Chemother. 2015;70(1):153–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sachdeva M, Leeds JA. Subinhibitory concentrations of LFF571 reduce toxin production by Clostridium difficile. Antimicrob Agents Chemother. 2015;59(2):1252–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuroda H, Kuroda M, Cui L, Hiramatsu K. Subinhibitory concentrations of beta-lactam induce haemolytic activity in Staphylococcus aureus through the SaeRS two-component system. FEMS Microbiol Lett. 2007;268(1):98–105. [DOI] [PubMed] [Google Scholar]
- 12.Horn J, Klepsch M, Manger M, et al. Long Noncoding RNA SSR42 Controls Staphylococcus aureus Alpha-Toxin Transcription in Response to Environmental Stimuli. J Bacteriol. 2018;200(22):e00252–18.* This report shows that subinhibitory concentrations of antibiotics enhance transcription of a regulatory RNA molecule, acting as a mechanism that regulates bacterial toxin production in response to various environmental stimuli.
- 13.Haddadin RN, Saleh S, Al-Adham IS, et al. The effect of subminimal inhibitory concentrations of antibiotics on virulence factors expressed by Staphylococcus aureus biofilms. J Appl Microbiol. 2010;108(4):1281–91. [DOI] [PubMed] [Google Scholar]
- 14.Mirani ZA, Jamil N. Effect of sub-lethal doses of vancomycin and oxacillin on biofilm formation by vancomycin intermediate resistant Staphylococcus aureus. J Basic Microbiol. 2011;51(2):191–5. [DOI] [PubMed] [Google Scholar]
- 15.Kaplan JB, Izano EA, Gopal P, et al. Low levels of beta-lactam antibiotics induce extracellular DNA release and biofilm formation in Staphylococcus aureus. MBio. 2012;3(4):e00198–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu W, Hallinen KM, Wood KB. Interplay between Antibiotic Efficacy and Drug-Induced Lysis Underlies Enhanced Biofilm Formation at Subinhibitory Drug Concentrations. Antimicrob Agents Chemother. 2018;62(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mlynek KD, Callahan MT, Shimkevitch AV, et al. Effects of Low-Dose Amoxicillin on Staphylococcus aureus USA300 Biofilms. Antimicrob Agents Chemother. 2016;60(5):2639–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dale JL, Nilson JL, Barnes AMT, Dunny GM. Restructuring of Enterococcus faecalis biofilm architecture in response to antibiotic-induced stress. NPJ Biofilms Microbiomes. 2017;3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dengler V, Meier PS, Heusser R, et al. Induction kinetics of the Staphylococcus aureus cell wall stress stimulon in response to different cell wall active antibiotics. BMC Microbiol. 2011;11:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mascher T, Margulis NG, Wang T, et al. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol Microbiol. 2003;50(5):1591–604. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen PK, Andersen AZ, Mols M, et al. Genome-wide transcriptional profiling of the cell envelope stress response and the role of LisRK and CesRK in Listeria monocytogenes. Microbiology. 2012;158(Pt 4):963–74. [DOI] [PubMed] [Google Scholar]
- 22.Abranches J, Tijerina P, Aviles-Reyes A, et al. The cell wall-targeting antibiotic stimulon of Enterococcus faecalis. PLoS One. 2014;8(6):e64875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuroda M, Kuroda H, Oshima T, et al. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol. 2003;49(3):807–21. [DOI] [PubMed] [Google Scholar]
- 24.Utaida S, Dunman PM, Macapagal D, et al. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology. 2003;149(Pt 10):2719–32. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Hu Y, Pai PJ, et al. Label-free quantitative proteomics analysis of antibiotic response in Staphylococcus aureus to oxacillin. J Proteome Res. 2014;13(3):1223–33. [DOI] [PubMed] [Google Scholar]
- 26.McCallum N, Spehar G, Bischoff M, Berger-Bachi B. Strain dependence of the cell wall-damage induced stimulon in Staphylococcus aureus. Biochim Biophys Acta. 2006;1760(10):1475–81. [DOI] [PubMed] [Google Scholar]
- 27.Beltramini AM, Mukhopadhyay CD, Pancholi V. Modulation of cell wall structure and antimicrobial susceptibility by a Staphylococcus aureus eukaryote-like serine/threonine kinase and phosphatase. Infect Immun. 2009;77(4):1406–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dias R, Felix D, Canica M, Trombe MC. The highly conserved serine threonine kinase StkP of Streptococcus pneumoniae contributes to penicillin susceptibility independently from genes encoding penicillin-binding proteins. BMC Microbiol. 2009;9:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kristich CJ, Wells CL, Dunny GM. A eukaryotic-type Ser/Thr kinase in Enterococcus faecalis mediates antimicrobial resistance and intestinal persistence. Proc Natl Acad Sci U S A. 2007;104(9):3508–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohlsen K, Donat S. The impact of serine/threonine phosphorylation in Staphylococcus aureus. Int J Med Microbiol. 2010;300(2–3):137–41. [DOI] [PubMed] [Google Scholar]
- 31.Schaenzer AJ, Wlodarchak N, Drewry DH, et al. A screen for kinase inhibitors identifies antimicrobial imidazopyridine aminofurazans as specific inhibitors of the Listeria monocytogenes PASTA kinase PrkA. J Biol Chem. 2017;292(41):17037–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamber S, Schwartzman J, Cheung AL. Role of PknB kinase in antibiotic resistance and virulence in community-acquired methicillin-resistant Staphylococcus aureus strain USA300. Infect Immun. 2010;78(8):3637–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kant S, Asthana S, Missiakas D, Pancholi V. A novel STK1-targeted small-molecule as an “antibiotic resistance breaker” against multidrug-resistant Staphylococcus aureus. Sci Rep. 2017;7(1):5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pensinger DA, Aliota MT, Schaenzer AJ, et al. Selective pharmacologic inhibition of a PASTA kinase increases Listeria monocytogenes susceptibility to beta-lactam antibiotics. Antimicrob Agents Chemother. 2014;58(8):4486–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaenzer AJ, Wlodarchak N, Drewry DH, et al. GW779439X and Its Pyrazolopyridazine Derivatives Inhibit the Serine/Threonine Kinase Stk1 and Act As Antibiotic Adjuvants against beta-Lactam-Resistant Staphylococcus aureus. ACS Infect Dis. 2018;4(10):1508–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vornhagen J, Burnside K, Whidbey C, et al. Kinase Inhibitors that Increase the Sensitivity of Methicillin Resistant Staphylococcus aureus to beta-Lactam Antibiotics. Pathogens. 2015;4(4):708–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cameron DR, Ward DV, Kostoulias X, et al. Serine/threonine phosphatase Stp1 contributes to reduced susceptibility to vancomycin and virulence in Staphylococcus aureus. J Infect Dis. 2012;205(11):1677–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Passalacqua KD, Satola SW, Crispell EK, Read TD. A mutation in the PP2C phosphatase gene in a Staphylococcus aureus USA300 clinical isolate with reduced susceptibility to vancomycin and daptomycin. Antimicrob Agents Chemother. 2012;56(10):5212–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renzoni A, Andrey DO, Jousselin A, et al. Whole genome sequencing and complete genetic analysis reveals novel pathways to glycopeptide resistance in Staphylococcus aureus. PLoS One. 2011;6(6):e21577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wright DP, Ulijasz AT. Regulation of transcription by eukaryotic-like serine-threonine kinases and phosphatases in Gram-positive bacterial pathogens. Virulence. 2014;5(8):863–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kellogg SL, Kristich CJ. Convergence of PASTA kinase and two-component signaling in response to cell wall stress in Enterococcus faecalis. J Bacteriol. 2018;200(12):e00086–18.** This reports demonstrates cross-communication between the E. faecalis CroRS two-component signaling pathway and IreK PASTA kinase signaling pathway in response to cell wall-active antibiotics. These findings support complex interactions among antibiotic-responsive signaling networks.
- 42.Donat S, Streker K, Schirmeister T, et al. Transcriptome and functional analysis of the eukaryotic-type serine/threonine kinase PknB in Staphylococcus aureus. J Bacteriol. 2009;191(13):4056–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones G, Dyson P. Evolution of transmembrane protein kinases implicated in coordinating remodeling of gram-positive peptidoglycan: inside versus outside. J Bacteriol. 2006;188(21):7470–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hardt P, Engels I, Rausch M, et al. The cell wall precursor lipid II acts as a molecular signal for the Ser/Thr kinase PknB of Staphylococcus aureus. Int J Med Microbiol. 2017;307(1):1–10. [DOI] [PubMed] [Google Scholar]
- 45.Maestro B, Novakova L, Hesek D, et al. Recognition of peptidoglycan and beta-lactam antibiotics by the extracellular domain of the Ser/Thr protein kinase StkP from Streptococcus pneumoniae. FEBS Lett. 2011;585(2):357–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mir M, Asong J, Li X, et al. The extracytoplasmic domain of the Mycobacterium tuberculosis Ser/Thr kinase PknB binds specific muropeptides and is required for PknB localization. PLoS Pathog. 2011;7(7):e1002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shah IM, Laaberki MH, Popham DL, Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135(3):486–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turapov O, Loraine J, Jenkins CH, et al. The external PASTA domain of the essential serine/threonine protein kinase PknB regulates mycobacterial growth. Open Biol. 2015;5(7):150025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kajfasz JK, Mendoza JE, Gaca AO, et al. The Spx regulator modulates stress responses and virulence in Enterococcus faecalis. Infect Immun. 2012;80(7):2265–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rojas-Tapias DF, Helmann JD. Induction of the Spx regulon by cell wall stress reveals novel regulatory mechanisms in Bacillus subtilis. Mol Microbiol. 2018;107(5):659–74.** This report describes several key differences in the mechanism of activation of the Spx regulon in response to cell wall stress, when compared to disulfide stress.
- 51.Gohring N, Fedtke I, Xia G, et al. New role of the disulfide stress effector YjbH in beta-lactam susceptibility of Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55(12):5452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rojas-Tapias DF, Helmann JD. Stabilization of Bacillus subtilis Spx under cell wall stress requires the anti-adaptor protein YirB. PLoS Genet. 2018;14(7):e1007531.** This reports demonstrates interactions between a B. subtilus two-component signaling pathway and an Spx regulatory factor, highlighting the complexity of the response to cell wall stress.
- 53.Frees D, Gerth U, Ingmer H. Clp chaperones and proteases are central in stress survival, virulence and antibiotic resistance of Staphylococcus aureus. Int J Med Microbiol. 2014;304(2):142–9. [DOI] [PubMed] [Google Scholar]
- 54.Zuber P Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J Bacteriol. 2004;186(7):1911–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jousselin A, Kelley WL, Barras C, et al. The Staphylococcus aureus thiol/oxidative stress global regulator Spx controls trfA, a gene implicated in cell wall antibiotic resistance. Antimicrob Agents Chemother. 2013;57(7):3283–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Renzoni A, Kelley WL, Barras C, et al. Identification by genomic and genetic analysis of two new genes playing a key role in intermediate glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53(3):903–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roch M, Clair P, Renzoni A, et al. Exposure of Staphylococcus aureus to subinhibitory concentrations of beta-lactam antibiotics induces heterogeneous vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother. 2014;58(9):5306–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Donegan NP, Marvin JS, Cheung AL. Role of adaptor TrfA and ClpPC in controlling levels of SsrA-tagged proteins and antitoxins in Staphylococcus aureus. J Bacteriol. 2014;196(23):4140–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller HK, Carroll RK, Burda WN, et al. The extracytoplasmic function sigma factor sigmaS protects against both intracellular and extracytoplasmic stresses in Staphylococcus aureus. J Bacteriol. 2012;194(16):4342–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maiques E, Ubeda C, Campoy S, et al. beta-lactam antibiotics induce the SOS response and horizontal transfer of virulence factors in Staphylococcus aureus. J Bacteriol. 2006;188(7):2726–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Plata KB, Riosa S, Singh CR, et al. Targeting of PBP1 by beta-lactams determines recA/SOS response activation in heterogeneous MRSA clinical strains. PLoS One. 2013;8(4):e61083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jensen PA, Zhu Z, van Opijnen T. Antibiotics Disrupt Coordination between Transcriptional and Phenotypic Stress Responses in Pathogenic Bacteria. Cell Rep. 2017;20(7):1705–16.** This report combines genome-wide RNA-seq and Tn-seq profiling approaches to compare the network of transcriptionally- and phenotypicaly-important genes during stress. A coordinated response was evident during metabolic stress, whereas the response to antibiotic stress was largely uncoordinated.
- 63.van Opijnen T, Dedrick S, Bento J. Strain Dependent Genetic Networks for Antibiotic-Sensitivity in a Bacterial Pathogen with a Large Pan-Genome. PLoS Pathog. 2016;12(9):e1005869. [DOI] [PMC free article] [PubMed] [Google Scholar]


