Abstract
The plant defense responses to pests results in the synchronized change of a complex network of interconnected genes and signaling pathways. An essential part of this process is mediated by the binding of transcription factors to the specific responsive cis-elements within in the promoters of phytophagous-responsive genes. In this work, it is reported the identification and characterization of a bidirectional promoter that simultaneously co-regulate two divergent genes, At5g10300 and At5g10290, upon arthropod feeding. Computational analysis identified the presence of cis-elements within the intergenic region between two loci, mainly from the DOF but also from the AP2/ERF, Golden 2-like and bHLH families. The function of the bidirectional promoter was analyzed using two enhanced variants of the GFP and CherryFP reporter genes, in both orientations, in transient tobacco and stably transformed Arabidopsis plants. Promoter activity was tested in response to feeding of Tetranychus urticae and Pieris brassicae, as well as wounding, flagellin and chitin treatments. Using RT-qPCR assays and confocal microscopy, it was shown that all treatments resulted in the induction of both reporter genes. Furthermore, our findings revealed the asymmetric character of the promoter with stronger activity in the forward than in the reverse orientation. This study provides an example of a bidirectional promoter with a strong potential to be used in plant biotechnology in pest control that requires stacking of the defense genes.
Keywords: bidirectional promoter, cis-elements, defense genes, inducible promoter, reporter gene expression, Tetranychus urticae, Pieris brassicae
Introduction
A bidirectional promoter is the intergenic region between two divergent genes located on complementary strands of the DNA which drives their coordinated transcription in opposite directions. These promoters control of the expression of gene pairs usually involved in the same or in related physiological processes (Mitra et al., 2009; Singh et al., 2009; Oropeza-Aburto et al., 2017). Most of the known bidirectional promoters have been found during the research of single genes, but the development of sequencing technologies and bioinformatics tools discovered that bidirectional nuclear gene promoters are very abundant in eukaryotes, including plants (Wang et al., 2009, 2016; Dhadi et al., 2013). In a first search of bidirectional promoters in the Arabidopsis genome, Wang et al. (2009) identified near 2,500 gene shared intergenic regions, enriched in regulatory elements essential for transcription. The characterization of some of them showed that the majority drove gene pairs within same functional category. Comparative analysis of bidirectional promoters, performed in rice, Arabidopsis and Populus (Dhadi et al., 2009) and in three grass species such as maize, sorghum, and Brachypodium (Krom and Ramakrishna, 2010) provided very useful information about common cis-elements, regulatory mechanisms, co-expression and evolutionary dynamics.
The first characterized bidirectional promoter from plants was identified in maize. Shwarz et al. (1981) found that two genes overlapped by few nucleotides in the maize chloroplast chromosome were transcribed divergently from complementary DNA strands. Since then, bi-directionality of promoters controlling genes in a head-to-head fashion has been described in many species. Some intergenic regions drive an equal expression of the two genes in both directions. One example is the co-expression of two chymotrypsin protease inhibitor encoding genes from rice, OCPI2 and OCPI1, with 1,126 bp distance between their transcription initiation sites in reverse orientations. OCPI2 and OCP1 transcripts were coordinately regulated and induced in an almost identical manner by salt, ABA, low temperature and mechanical wounding treatments in roots and shoot tissues of seedlings (Singh et al., 2009). In other cases, the transcription of some genes under a bidirectional promoter is dependent on the orientation. An example is the tissue-expression profile shown by the two divergent At4g35985 and At4g35987 Arabidopsis genes, encoding a senescence associated protein and a calmodulin methyltransferase, respectively, that shared a 1,258 bp intergenic region. While the genes were highly expressed in apical meristem and shoot and root tips in one orientation, in the opposite strand they were very active in mature tissues (Banerjee et al., 2013). Expression assays of this bidirectional promoter fused to Green Fluorescence Protein (GFP) and β-glucuronidase (GUS) reporter genes in both orientations in stable transformed Arabidopsis and tobacco plants corroborated the tissue specificity, and pointed-out a specific up-regulation under salt and pathogen stresses dependent on the orientation, strength and the exposure time of the stress. Generally, most bidirectional promoters direct the asymmetric transcription of the gene pair under their control, even when they are expressed in the same plant tissue. This is the case of two Arabidopsis genes, At1g71850 (ubiquitin carboxyl-terminal hydrolase) and At1g71860 (tyrosine-specific protein phosphatase) separated by a 347 bp intergenic sequence which exhibited an orientation-dependent expression profile (Liu et al., 2015). Likewise, the intergenic region of the maize defensin-like protein genes Def1 and Def2 determined embryo- and aleurone-specific expression but displayed asymmetric activity (Liu et al., 2016). The deletion of the cis-acting elements contained in this promoter evidenced that the transcription polarity and differential strength were dependent on the presence of specific cis-motifs. Recently, the functional analysis of a bidirectional promoter (1,429 bp) shared by a gene pair from Gossypium hirsutum, Ghrack1 (receptor for activated C kinase1), and Ghhrf1 (E3 ubiquitin-protein ligase), has also showed an orientation dependence (Yang et al., 2018). Using series of expression vectors containing GUS and GFP reporter genes in forward and reverse orientations of the promoter to transform Arabidopsis and tobacco, they found differences on spatial and temporal gene expression controlled by the promoter orientation.
The selection of a promoter is essential for the successful expression for multigene engineering strategies. Bidirectional promoters offer certain advantages including: (i) coordinated expression of two genes; (ii) reduction in time-consuming construction of expression vectors; (iii) prevention of silencing of multigene expression based on the non-repetitive use of the same unidirectional promoter; (iv) stability of the DNA based on the reduced amount of foreign DNA in genetically engineered plants. In this context, to transform a unidirectional promoter into a bidirectional one, or to design artificial bidirectional promoters has been considered as an alternative adapted to transcribe genes under specific physiological and environmental stimuli. Current bioinformatics tools, databases and technologies allow the design and the introduction of cis-elements in bidirectional promoters as an artwork of cis-engineering (Mehrotra et al., 2011). Moreover, changes in the core and flanking sequences of the cis-motifs may lead to alterations in the gene expression in both native and synthetic promoters (Zhang et al., 2019). Xie et al. (2001) created the first artificial plant bidirectional promoter based on the 35S minimal promoter (51 bp from transcription start point) from the cauliflower mosaic virus (CaMV) by fusing the 35S mini-promoter at its 5’ end in the opposite orientation. The same strategy was used by Zheng et al. (2011) to generate a bidirectional methyl jasmonate-inducible promoter derived from poplar. Kumar et al. (2015) combined the construction of bidirectional promoters and a biscistronic approach for coordinated multi-gene expression in maize. They integrated the polyprotein processing T2A ACD sequence from Thosea asigna virus into a synthetic bidirectional promoter derived from the Ubiquitin-1 promoter from Zea mays to improve transgene stacking strategy. Expression analysis of transgenic maize plants confirmed the coordinated expression of four genes conferring insect and herbicide resistance based on this combined approach. More recently, Vogl et al. (2018) generated a library of 168 synthetic bidirectional promoters in the yeast Pichia pastoris by using its natural histone promoters as an engineered template. The rapid screening of expression profiles to optimize gene co-expression suggested the potential of this approach for other eukaryotes, including plants.
In this work, we report the identification and characterization of a bidirectional promoter shared by two divergent genes, At5g10300 and At5g10290, simultaneously transcribed and induced by the two-spotted spider mite feeding. The promoter function was tested by fusing mGFP and mCherryFP reporter genes in both orientations in transiently agroinfiltrated tobacco leaves and in stably transformed Arabidopsis plants. These plants were used to analyze their responses to arthropod feeding and other related defense-treatments by RT-qPCR assays and confocal microscopy.
Materials and Methods
In silico Analysis of the Bifunctional Promoter
The VISTA plots of multiple genomic comparisons of Brassicaceae species different from Arabidopsis thaliana were visualized in the VISTA-Point tool using the pairwise precomputed alignments from the VISTA platform (Frazer et al., 2004). The VISTA plots of pairwise comparisons of putative bifunctional promoter sequences of Arabidopsis lyrata and Arabidopsis halleri species to A. thaliana were generated by the zPicture tool (Ovcharenko et al., 2004) using the sequences retrieved from the Araport11 annotation (Cheng et al., 2017). Multiple sequence alignment was prepared using the MUSCLE program in the SeaView4 platform (Gouy et al., 2010). Transcription factors sites were obtained using the binding site prediction tool in the PlantRegMap and the PlantTFDB 4.0 database (Jin et al., 2017).
Plant Material and Growth Conditions
Arabidopsis thaliana Columbia (Col-0) plants were used as wild-type plants. A. thaliana T-DNA mutants (Salk_022713C and Salk_015889C) were obtained from the Arabidopsis Biological Resource Centre and the European Arabidopsis Stock Centre (NASC).1 For in vitro growth, seeds were surface sterilized with 70% (V/V) ethanol for 2 min, followed by incubation in a solution containing 5% (V/V) sodium hypochlorite and 5% (W/V) SDS for 10 min, and then washed with sterilized distilled H2O. Seeds were plated onto Petri dishes containing 1/2 MS semisolid medium (Sigma-Aldrich), adjusted to pH 5.7 with KOH. For soil growth, a mixture of peat moss and vermiculite (3:2, V/V) was used. Sterilized seeds were stratified in the dark at 4∘C for 5 days. Plates and pots were grown in growth chambers (Sanyo MLR-351-H, Sanyo, Japan) under control conditions (23 ± 1∘C, >70% relative humidity and a 16∘h/8∘h day/night photoperiod).
Nicotiana benthamiana plants were grown in greenhouse under controlled conditions (22 ± 1°C, >70% relative humidity and a 16h/8h day/night photoperiod).
Phytophagous Arthropod Maintenance
A colony of the two-spotted spider mite Tetranychus urticae, London strain (Acari: Tetranychidae), kindly provided by Dr. Miodrag Grbic (Department of Biology, Western Ontario University, Canada), was reared on beans (Phaseolus vulgaris) and maintained in growth chambers (Sanyo MLR-351-H, Sanyo, Japan) at 25 ± 1∘C, >70% relative humidity and a 16∘h/8∘h day/night photoperiod. A Pieris brassicae colony supplied by Prof. Dr. Marcel Dicke and Dr. Pieter Rouweler (Laboratory of Entomology, Wageningen University, Netherlands), was reared on Brussels sprouts (Brassica oleracea L. var. gemmifera) and maintained on growth chambers (Sanyo MLR-350-H, Sanyo, Japan) at 21 ± 1∘C, 50% relative humidity and a 16∘h/8∘h day/night photoperiod.
Construction of Bidirectional Promoter-Reporter Gene Fusions and Plant Transformation
The intergenic region, between At5g10290 and At5g10300 genes (named in figures BP), was amplified using A. thaliana Col-0 genomic DNA as a template following the design schematized in Supplementary Figure 2 and the primers indicated in Supplementary Table 1. The mCherryFP ORF was amplified from the psmRS-mCherryFP plasmid (Barrero-Sicilia et al., 2011). PCRs were performed with Phusion Hot Start II High Fidelity DNA Polymerase according to the manufacturer’s instructions (Thermo Scientific), under the following reaction conditions: 30 s at 98∘C, followed by 30 cycles of 10 s at 98∘C, 20 s at 55∘C for 25 s (bidirectional promoter) or 53∘C for 20 s (mCherryFP) at 72∘C, and 5 min at 72∘C. The target DNA fragments of the forward and reverse orientations of the bidirectional promoter and the mCherryFP gene were ligated into the cloning vector pENTRY3C (Invitrogen). The mCherryFP::BP fusion were inserted in the pmdc107 vector containing the mGFP gene driven by a minimal promoter (Curtis and Grossniklaus, 2003), by homologous recombination using LR Clonase II enzyme mix (Invitrogen). Resultant fusion plasmids mCherryFP::BP::GFP and GFP::BP::mCherryFP, enclosed the two reporter genes driven by the BP in both orientations. The pmdc107, named here pmdc107ø, was used as control vector for all transformation events.
The fusion-constructs were first introduced into the Agrobacterium tumefaciens strain C58C1 Rif (GV3101) by using the freeze-thaw method (Weigel and Glazebrook, 2002). Then, 3-week old N. benthamiana leaves were transiently transformed by Agroinfiltration as described Santamaria et al. (2019), with the fusion-constructs mCherryFP::BP::mGFP, mGFP::BP::mCherryFP and the control pmdc107ø vector. Transient assays were performed after 3 and 4 days of infiltration (dpi). To generate stable transgenic plants, the fusion-constructs and the control vector pmdc107ø were also introduced into A. thaliana Col-0 plants using the Agrobacterium floral dip method (Clough and Bent, 1998). Transformants were selected on MS medium supplemented with 40 mg/L hygromicin. Homozygous lines with one single transgene copy coming from independent transformation events were selected from the T3 generation.
Plant Infestation Assays
To analyse the phenotype of the promoter T-DNA insertion lines in response to spider mites, infestation assays were performed. Three-week-old Arabidopsis T-DNA Salk lines and Col-0 control plants were infested with 20 T. urticae female adults per plant. After 4 days of infestation, leaf damage was assessed by scanning the entire rosette (HP Scanjet 5590 Digital Flatbed Scanner series), according to Cazaux et al. (2014). Leaf damage was calculated in mm2 using Adobe Photoshop CS software. Six replicates were used for each genotype. The experiment was carried out in duplicate. To determine how the bidirectional promoter controls the expression of the two reporter genes in both orientations, feeding assays were performed with T. urticae and P. brassicae. Twenty spider mite female adults or five freshly hatched neonate caterpillar were placed on 3-week old Arabidopsis plants (pmdc107ø control and mCherryFP::BP::mGFP, mGFP::BP::mCherryFP lines), allowing a continuous feeding for 24∘h, in growth chambers (Sanyo MLR-351-H, Sanyo, Japan) at 25 ± 1∘C, >70% relative humidity and a 16∘h/8∘h day/night photoperiod. Six replicates of each plant genotype were performed per treatment and control. From these six plants, three were collected and frozen in liquid N2 for RT-qPCR and the other three plants were observed under the confocal microscopy. All the experiments were repeated three times, as is indicated in Supplementary Figure 3.
Wounding, Flagellin, and Chitin Treatments
Arabidopsis plants (pmdc107ø control and mCherryFP::BP:: mGFP, mGFP::BP::mCherryFP transgenic lines) were also used to analyse the two reporter gene expression in response to mechanical wounding, flagellin (Flg22) and chitin treatments. For wounding, approximately 20% of the leaf area of two leaves, of 3-week old Arabidopsis plants, were pierced with a needle and their leaves were collected 1∘h after. In the case of the Flg22 treatment, plants were sprayed with 200 μl of the flagellin-derived Flg22 peptide (AnaSpec) at a final concentration of 5 μM. Control plants were sprayed with the same amount of water. Plants were collected after 12∘h of treatment. Chitin stock solution were prepared by dissolving chitin from shrimp shells (Sigma-Aldrich) in DMSO at a concentration of 25 mg/mL and subsequently diluted in H2O to a 100 μg/mL concentration for the experiment. The solvent used to solubilize the elicitor (DMSO) was used as control treatment at a final concentration of 0.4% (V/V). Plants were sprayed using 200 μl per plant and leaves were collected after 24∘h of treatment. Growth conditions were the same mentioned above and experimental design and replicates were done according to Supplementary Figure 3.
Nucleic Acid Analysis
Genomic DNA was isolated from Arabidopsis T-DNA insertion lines and Col-0 plants, essentially as described by Sambrook and Russell (2001). The T-DNA homozygous status and gene expression levels of the Salk lines was analyzed by conventional PCR and quantitative PCR (RT-qPCR) analyses (Supplementary Figures 4A–C). Conventional PCR conditions were: 3 min at 94∘C, followed by 30 cycles for 30 s at 94∘C, 40 s at 55∘C, and 1 min 20 s at 72∘C, and finally 7 min at 72∘C.
For RT-qPCR studies, total RNA was isolated from 3-week old rosettes from Arabidopsis T-DNA insertion and Col-0 plants, and from different tissues (seedlings, rosettes, roots and mature seeds) of Arabidopsis plants (pmdc107ø control and mCherryFP::BP::mGFP, mGFP::BP::mCherryFP transgenic lines) after different treatments. Total RNA from seedlings, 3-weel-old-rosettes and roots was extracted by the phenol/chloroform method, followed by precipitation with 8 M LiCl according to Oñate-Sánchez and Vicente-Carbajosa (2008). RNA form mature seeds was isolated by the hot borate method described by Santamaria et al. (2010). Regarding N. benthamiana assays, total RNA was extracted from plants agroinfiltrated with the control vector pmdc107ø, the mGFP::BP::mCherryFP and the mCherryFP::BP::mGFP constructs, by the TRIZOL reagent following manufacturer instructions (Ambion Austin TX). Complementary DNAs (cDNAs) were synthesized from 2 μg of RNA using the Revert AidTM H Minus First Strand cDNA Synthesis Kit (Fermentas) as indicated manufacturer’s instructions. The RT-qPCR conditions used were 40 cycles with 15 s at 95∘C, 1 min at 60∘C and 5 s at 65°C using LightCycler 480 SYBR Green I Master (Roche). RT-qPCR was performed in three samples coming from three independent experiments as previously described by Santamaria et al. (2012), using a SYBR Green Detection System (Roche) and the LightCycler480 Software release 1.5.0 SP4 (Roche). mRNA quantification, expressed as relative expression levels (2–dCt), was normalized to ubiquitin or actin for Arabidopsis and Nicotiana samples, respectively. For mRNA quantification referred as fold change (2–ddCt) in Arabidopsis samples, ubiquitin as housekeeping gene and untreated control sample as internal calibrator were used (Livak and Schmittgen, 2001). Specific primers were designed through PRIMER3.2 Primer sequences are listed in Supplementary Table 1.
Confocal Images
All images were obtained with the TCS-SP8 confocal microscope (Leica). For transient expression experiments, N. benthamiana leaves were observed under the microscope after 3 and 4 days of the agroinfiltration (dpi). Confocal images of Arabidopsis leaves were taken after arthropod feeding assays, mechanical wounding and elicitor treatments in stable transformed Arabidopsis plants. Three independent leaves of each genotype, for control and treatments, were observed. In all experiments, mGFP and mCherryFP signals were sequentially acquired using the following settings: mGFP, excitation 488 nm and emission 491–550 nm; mCherryFP, excitation 561 nm, emission 591–638 nm.
Statistical Analysis
The statistical analysis was performed using GraphPadPrismv6.01 and IBM SPSSv25 statistics. In order to perform the proper statistical analysis, normality and homoscedasticity of the data were previously analyzed. In the case of the foliar damage, data fulfill both assumptions so, one-way ANOVA followed by Tukey’s multiple comparisons test were used. In the case of the gene expression, analysis generalized linear models stats was performed. Wald χ2 followed by LSD test was applied in all the RT-qPCR experiments. In figures, significant differences (P < 0.05) among transgenic lines are reported with different letters.
Results
Structure and Sequence Analysis of an Arabidopsis Intergenic Region Between Two Genes Involved in Defense to T. urticae
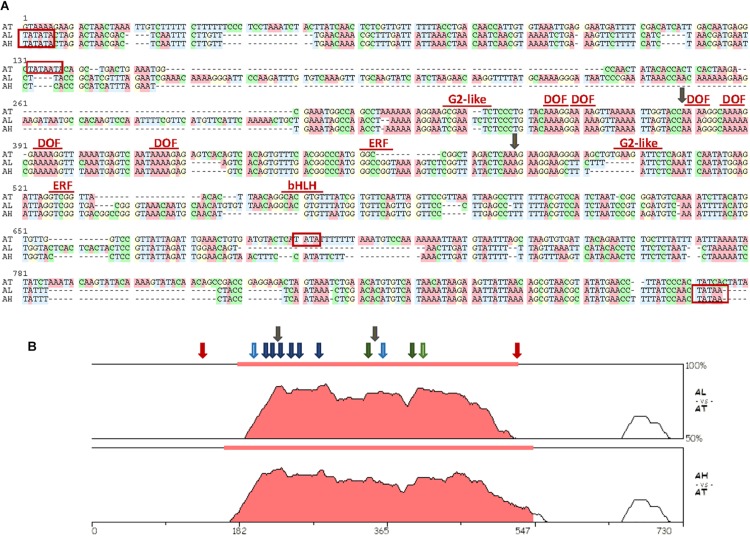
Zhurov et al. (2014) analyzed the transcriptional response to spider mite infestation in resistant Bla-2 and susceptible Kondara Arabidopsis accessions. From this analysis, many differentially expressed genes were identified. Looking at the genomic context of some genes up-regulated in Bla-2 upon mite feeding, we found that one induced gene, At5g10300, shared its potential promoter region with a second gene, At5g10290, annotated in the opposite direction. The At5g10300 and At5g10290 genes encoded a hydroxynitrile lyase and a LRR receptor kinase, respectively. A region of only 645 bp was found between the putative transcription start sites of both genes. To get a first insight on the relevance of this genomic arrangement, a genomic comparison of this region among Brassicaceae species was performed (Supplementary Figure 1). From this comparison, a strong conservation between the genes flanking the promoter was observed (Supplementary Figure 1A). However, conservation in the promoter sequences was mainly restricted to the promoters from Arabidopsis species, with minor similarities with the promoters of Brassicaceae species from other genera (Supplementary Figure 1B). Then, the nucleotide sequences of the promoters from the three Arabidopsis species were aligned to detect conserved functional motifs (Figure 1A). The putative location of TATA boxes was dissimilar between A. thaliana and the other two species. The TATA motifs were located more internally in A. thaliana which reflected in the lower number of bp flanked by the TATA motif in this species (385 bp) than in A. lyrata and A. halleri (764 and 602 bp, respectively). In any case, the strongest conserved region was located in the sequence flanked by the most internal TATA boxes (Figure 1B). Many binding sequences for transcription factors (TFs) were identified in this region. Among them, it is remarkable the presence of several conserved DOF-binding motifs (AAAG/CTTT), as well as some cis-elements potentially recognized by defense-related TFs from the AP2/ERF (GCC box), Golden 2-like (GAATCT) and basic Helix-Loop-Helix, bHLH (CACGTG) TF families.
FIGURE 1.
Sequence characteristics of the region spanning the interval between putative TATA sequences in the bifunctional promoter. (A) Alignment of the nucleotide sequences of the selected interval. Names of transcription factor families mark their putative binding sites. TATA predicted sites are boxed in red. Insertion points for the T-DNA lines used are marked by gray arrows. Transcription factors sites were obtained using the binding site prediction tool in the PlantRegMap and the PlantTFDB 4.0 database. Alignment was prepared using the MUSCLE program in the SeaView4 platform. (B) The VISTA plots of pair-wise comparisons of putative bifunctional promoter sequences of A. lyrata and A. halleri species to A. thaliana as generated by the zPicture tool. Filled portions of the graphs indicate conservation of more than 50% with a width of at least 100 bp. Approximate locations of conserved binding sites for transcription factors are marked by arrows (dark blue, DOF; light blue, G2-like; dark green, ERF; light green, bHLH). TATA predicted sites for A. thaliana are marked by red arrows. Insertion points for the T-DNA lines used are marked by gray arrows.
Expression Analysis of At5g10290 and At5g10300 Genes and Responses to Spider Mite Feeding
To determine whether the bidirectional promoter controls the simultaneous transcription of the gene pair At5g10290 and At5g10300, RT-qPCR assays were carried out in Arabidopsis rosettes of Col-0 plants and two T-DNA insertion lines. Results demonstrated that both genes were transcribed at the same time but the relative transcript abundance of the At5g10290 gene was higher compared to the relative expression of At5g10300 gene in the opposite strand (Supplementary Figure 4D). The alleles generated by the insertion of T-DNA in the promoter (Salk_015889C and Salk_022713C lines) corroborated the promoter bi-directionality. A reduction in the expression levels of the two divergent genes in the mutant lines were observed in comparison to the expression found in Col-0 plants. These data indicated the double knock-down character for At5g10290 and At5g10300 genes of these mutant lines.
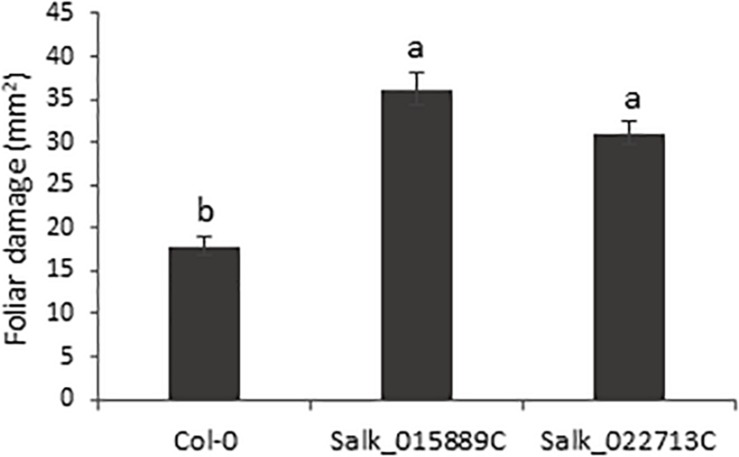
The homozygous T-DNA insertion lines and the Col-0 plants were used to test the involvement of the bidirectional promoter in the plant response to T. urticae feeding. Plants were infested with T. urticae adults and leaf damage was quantified 4 days after infestation. The damage intensity, measured as the chlorotic foliar area, was approximately twofold higher in the knock-down lines than the damage quantified in Col-0 plants after mite infestation (Figure 2). Thus, greater leaf damage parallel to a lower pair gene expression, reflected not only the potential defensive role of the genes located on the opposite strands but also the precise control of the bidirectional promoter.
FIGURE 2.
Plant damage of Arabidopsis Col-0 and T-DNA insertion lines infested with T. urticae. Damaged foliar area was measured after 4 days of spider mite infestation. Data are means ± SE of six replicates. Different letters indicate significant differences (P < 0.05, One-way ANOVA followed by Tukey’s multiple comparisons test).
Functional Analysis of the Bidirectional Promoter in Transiently Transformed N. benthamiana Plants
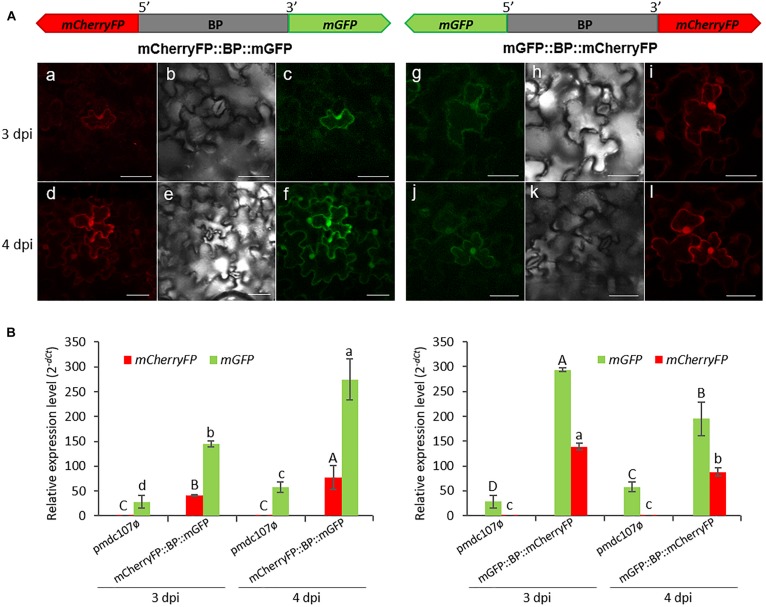
The intergenic region between At5g10290 and At5g10300 genes was used to make transcriptional fusions to mGFP and mCherryFP genes in the opposite orientations. Transient expression assays in agroinfiltrated N. benthamiana leaves revealed that both reporter genes were co-expressed and the emitted fluorescence was localized in the nuclei, membrane and some cytoplasmic areas, independent on the orientation (Figure 3A). No detectable florescence was found under the confocal microscope when the vector pmdc107ø was used (data not shown).
FIGURE 3.
Schematic representation of fusion-constructs and transient expression assays in N. benthamiana leaves. (A) Scheme of mCherryFP and mGFP genes under the bidirectional promoter (BP) and confocal stacks spanning tobacco epidermal cells of transient transformed leaves at 3 and 4 post-infiltration days (dpi). Projections are from mCherryFP (a,d,i,l), mGFP (c,f,g,j) and the corresponding Nomarski snap-shots (b,e,h,k). Scale bars represent 50 μm. (B) Relative expression levels (2–dCt) of mCherryFP and mGFP genes in agroinfiltrated N. benthamiana leaves with the control vector (pmdc107ø) or with the constructs including the bidirectional promoter fused to the two reporter genes. Data are means ± SE of three replicates. Different letters indicate significant differences (P < 0.05 Wald χ2 followed by LSD test). Primers used for this experiment are listed in Supplementary Table 1.
Transcript analysis of the reporter genes performed by RT-qPCR assays, at 3 and 4 dpi, corroborated the simultaneous expression of both genes (Figure 3B). The mGFP gene presented higher expression levels than the mCherryFP gene, independently on the gene orientation. Basal expression levels of the mGFP gene were obtained with the pmdc107ø control vector due to the presence of the minimal promoter contained in the control plasmid. As expected, no detection of the mCherryFP gene was found.
Functional Analysis of the Bidirectional Promoter in Stably Transformed A. thaliana Plants
To investigate the role of the bidirectional promoter in response to different stresses, the same fusion-constructs were used to stably transform A. thaliana plants. The characterization of the homozygous lines with one single transgene copy coming from independent transformation events allowed the selection of lines (1.1 and 2.4 for the mGFP::BP::mCherryFP construct, and 1.3 and 4.6 for the mCherryFP::BP::mGFP construct) for further studies (data not shown). The expression profile of the two reporter genes was studied in T3 plants by using different tissue samples (seedlings, rosettes, roots and mature seeds). Results confirmed the simultaneous transcription of the pair of genes controlled by the bidirectional promoter in all tested plant tissues, independently of the gene location on the complementary strands. Generally, a higher content of mRNAs derived from the mGFP gene than from mCherryFP gene was quantified in all tested tissue samples. Root and seed tissues presented the lowest accumulation of transcripts. As expected, plants transformed with the control pmdc107ø vector had basal expression levels of the mGFP gene but did not express the mCherryFP gene (Supplementary Figure 5).
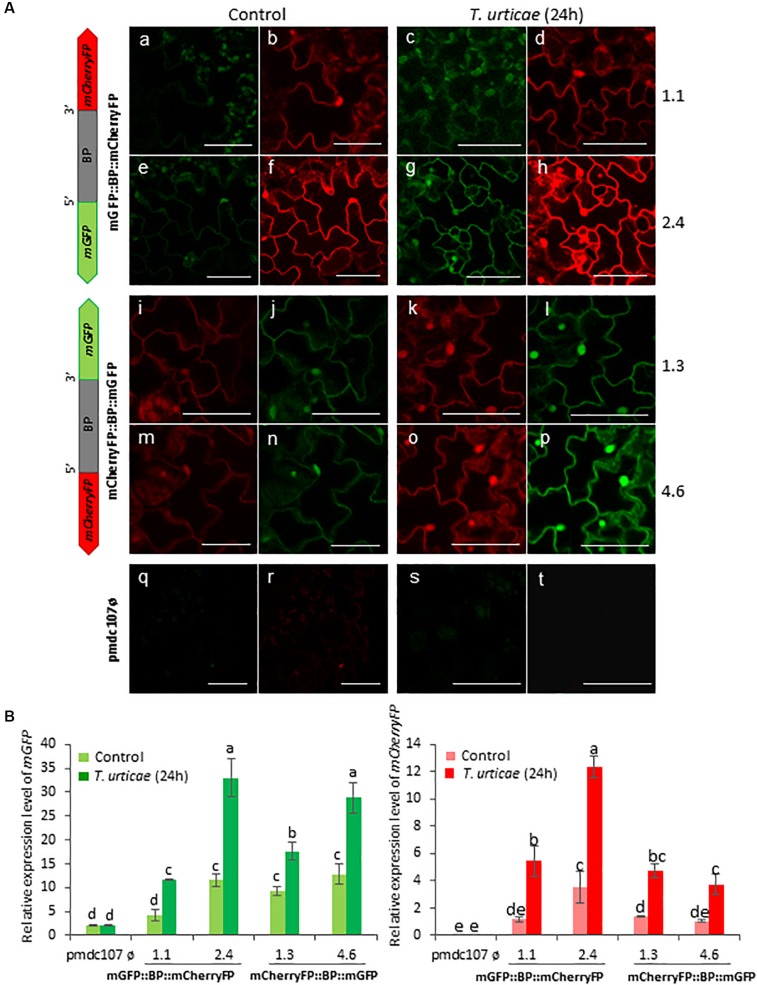
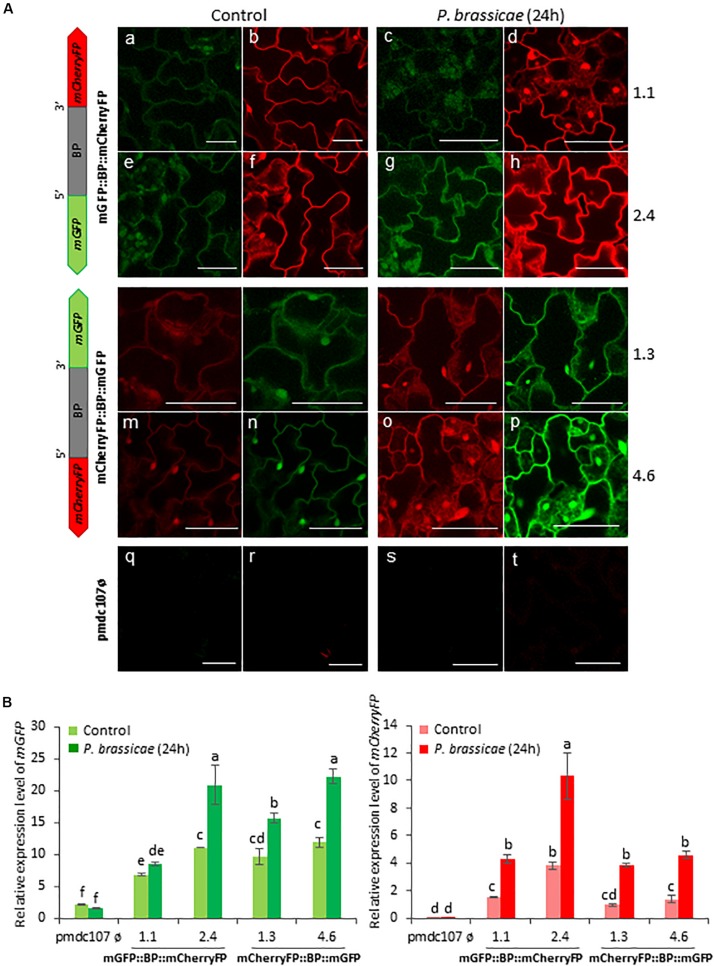
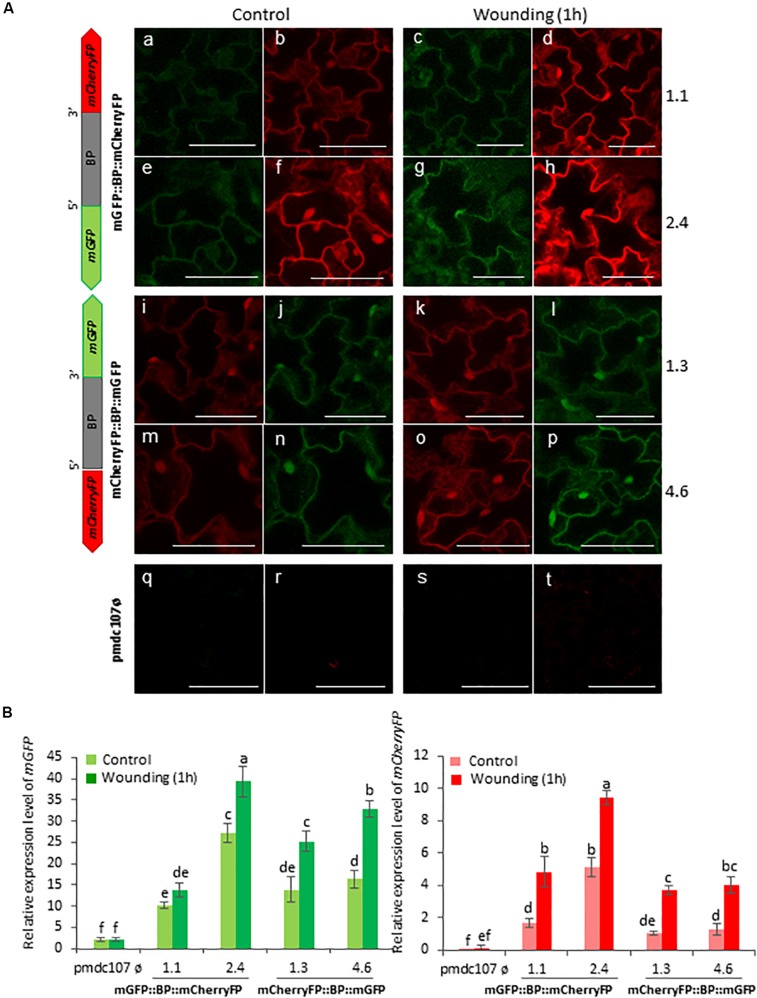
Arabidopsis transformed plants were used to perform feeding bioassays with T. urticae and P. brassicae and the expression of the reporter genes was evaluated under the confocal microscope and/or by RT-qPCR analysis after 24∘h of infestation. Confocal images showed that the two phytophagous arthropods clearly induced the emission of fluorescence of both reporter genes and in both orientations. No detectable fluorescence was found under the confocal microscope in Arabidopsis transformed with the control vector either in non-infested or in infested plants (Figures 4A, 5A). According to the images, the bidirectional promoter played a coordinated control of the expression of the gene pair, although the intensity of the fluorescence was always stronger when the reporter genes were located at the forward orientation in comparison to the location in the reverse orientation. High levels of fluorescence emitted by Arabidopsis leaves in response to spider mite and caterpillar feeding correlated with increased content of reporter transcripts after infestation (Figures 4B, 5B). Once more, the accumulation of mGFP mRNAs was higher than the mCherryFP mRNAs, independently on the gene orientation and on the feeding stress. In addition, similar gene expression patterns and comparable confocal images were observed when transformed Arabidopsis plants were mechanically wounded (Figure 6). Likewise, basal expression levels of the mGFP gene were observed in the transformed control plants (pmdc107ø), but its expression was not up-regulated by biotic or wounding stress.
FIGURE 4.
Schematic representation of fusion-constructs and expression assays in stably transformed Arabidopsis plants in response to T. urticae infestation. (A) Scheme of mCherryFP and mGFP genes under the bidirectional promoter (BD) and confocal stacks spanning Arabidopsis epidermal cells of transformed leaves under control conditions, and after T. urticae infestation. Projections are from mCherryFP (b,d,f,h,i,k,m,o,r,t) and mGFP (a,c,e,g,j,l,n,p,q,s). Bars are 50 μm. (B) Quantitative analyses of mGFP and mCherryFP gene expression in stably transformed Arabidopsis plants in control, and after 24 h of T. urticae infestation. Bars in green and red colors correspond to mGFP and mCherryFP gene expression, respectively. Data are ±SE of three biological replicates. Different letters indicate statistical differences (P < 0.05 Wald χ2 followed by LSD test). Arabidopsis transformed lines for mGFP::BP:mCherryFP construct were 1.1 and 2.4, and for mCherryFP:BP::mGFP were 1.3 and 4.6. Primers used for this experiment are listed in Supplementary Table 1.
FIGURE 5.
Schematic representation of fusion-constructs and expression assays in stably transformed Arabidopsis plants in response to P. brassicae infestation. (A) Scheme of mCherryFP and mGFP genes under the bidirectional promoter (BD) and confocal stacks spanning Arabidopsis epidermal cells of transformed leaves under control conditions, and after P. brassicae infestation. Projections are from mCherryFP (b,d,f,h,i,k,m,o,r,t) and mGFP (a,c,e,g,j,l,n,p,q,s). Bars are 50 μm. (B) Quantitative analyses of mGFP and mCherryFP gene expression in stably transformed Arabidopsis plants in control, and after 24 h of P. brassicae infestation. Bars in green and red colors correspond to mGFP and mCherryFP gene expression, respectively. Data are ±SE of three biological replicates. Different letters indicate statistical differences (P < 0.05 Wald χ2, followed by LSD test). Arabidopsis transformed lines for mGFP::BP:mCherryFP construct were 1.1, 2.4, and for mCherryFP:BP::mGFP were 1.3 and 4.6. Primers used for this experiment are listed in Supplementary Table 1.
FIGURE 6.
Schematic representation of fusion-constructs and expression assays in stably transformed Arabidopsis plants in response to wounding. (A) Scheme of mCherryFP and mGFP genes driven by the bidirectional promoter (BD) and confocal stacks spanning Arabidopsis epidermal cells of transformed plants after 1 h of wounding. Projections are from mCherryFP (b,d,f,h,i,k,m,o,r,t) and mGFP (a,c,e,g,j,l,n,p,q,s). Bars are 50 μm. (B) Quantitative analyses of mGFP and mCherryFP expression by RT-qPCR in Arabidopsis transformed plants after 1 h of wounding. Bars in green and red colors correspond to mGFP and mCherryFP gene expression, respectively. Data are ±SE of three biological replicates. Differential letters indicate statistical differences (P < 0.05 Wald χ2 followed by LSD test). Arabidopsis selected lines for mGP::BBP::mCherryFP construct were 1.1. and 2.4, and for mCherryFP::BD::mGFP were 1.3 and 4.6. Primers used for this experiment are listed in Supplementary Table 1.
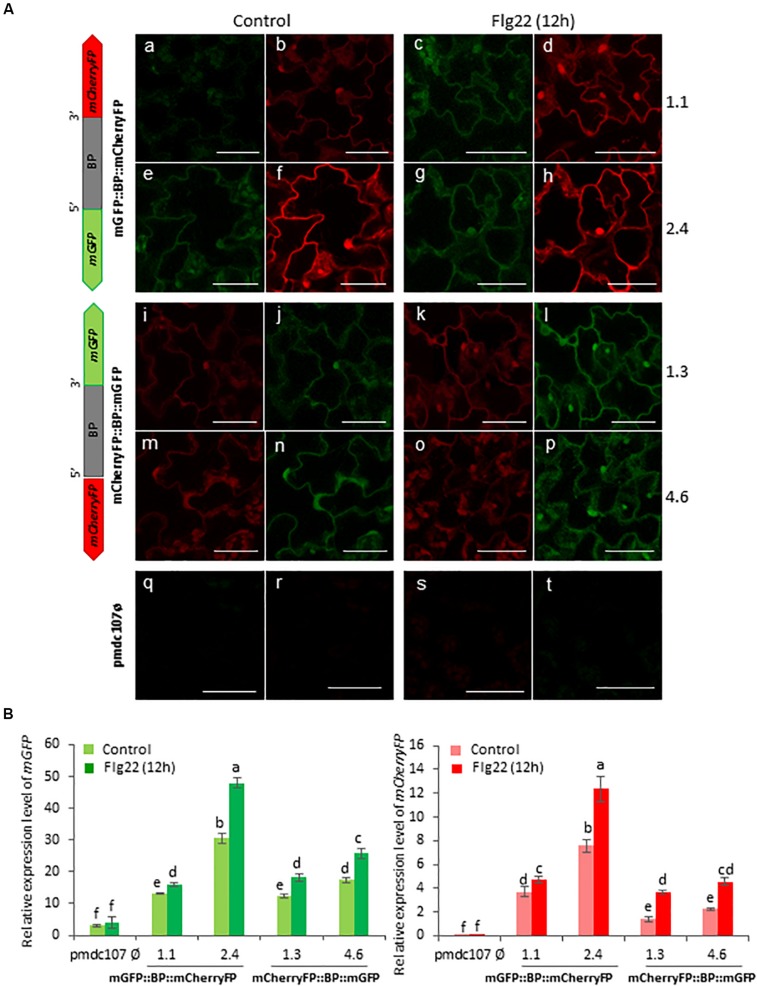
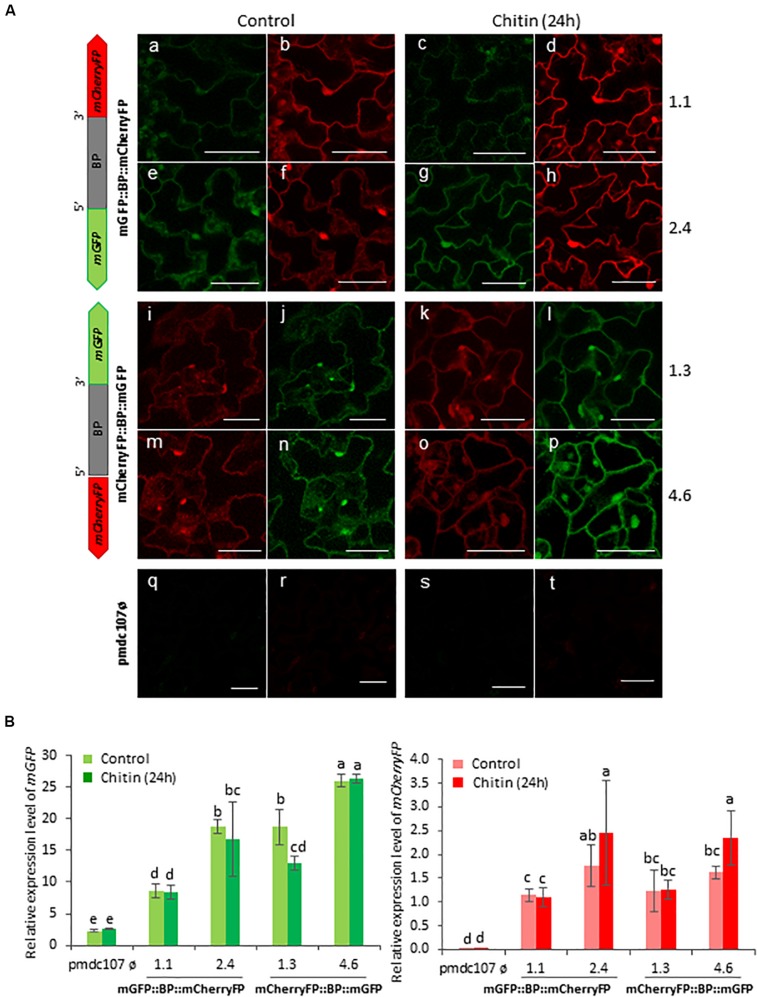
Arabidopsis plants were also treated with two elicitors, flagellin and chitin, and showed a differential response to each one. The Flg22 elicited the expression of the two reporter genes as well as the emission of the fluorescence under confocal microscope as the biotic and wounding treatments did (Figure 7). In contrast, no significant differences were detected after the chitin treatment between treated and non-treated plants (Figure 8). RT-qPCR assays of the two divergent genes, At5g10290 and At5g10300, were also carried out in Arabidopsis rosettes after T. urticae and P. brassicae feeding, as well as under wounding, Flg22 and chitin treatments. Results corroborated that both genes were simultaneously transcribed and induced by all treatments except by chitin (Supplementary Figure 6).
FIGURE 7.
Schematic representation of fusion-constructs and expression assays in stably transformed Arabidopsis plants in response to flagellin treatment. (A) Scheme of mCherryFP and mGFP genes driven by the bidirectional promoter (BD) and confocal stacks spanning Arabidopsis epidermal cells of transformed plants after 1∘h offlagelin22 (Flg22) treatment. Projections are from mCherryFP (b,d,f,h,i,k,m,o,r,t) and mGFP (a,c,e,g,j,l,n,p,q,s). Bars are 50 μm. (B) Quantitative analyses of mGFP and mCherryFP expression by RT-qPCR in Arabidopsis transformed plants after 1 h of Flg22 treatment. Bars in green and red colors correspond to mGFP and mCherryFP gene expression, respectively. Data are ±SE of three biological replicates. Differential letters indicate statistical differences (P < 0.05 Wald χ2 followed by LSD test). Arabidopsis selected lines for mGP::BBP::mCherryFP construct were 1.1. and 2.4, and for mCherryFP::BD::mGFP were 1.3 and 4.6. Primers used for this experiment are listed in Supplementary Table 1.
FIGURE 8.
Schematic representation of fusion-constructs and expression assays in stably transformed Arabidopsis plants in response to Chitin treatment. (A) Scheme of mCherryFP and mGFP genes driven by the bidirectional promoter (BD) and confocal stacks spanning Arabidopsis epidermal cells of transformed plants after 1 h of Chitin treatment. Projections are from mCherryFP (b,d,f,h,i,k,m,o,r,t) and mGFP (a,c,e,g,j,l,n,p,q,s). Bars are 50 μm. (B) Quantitative analyses of mGFP and mCherryFP expression by RT-qPCR in Arabidopsis transformed plants after 1 h of chitin treatment. Bars in green and red colors correspond to mGFP and mCherryFP gene expression, respectively. Data are ±SE of three biological replicates. Differential letters indicate statistical differences (P < 0.05 Wald χ2 followed by LSD test). Arabidopsis selected lines for mGP::BBP::mCherryFP construct were 1.1. and 2.4, and for mCherryFP::BD::mGFP were 1.3 and 4.6. Primers used for this experiment are listed in Supplementary Table 1.
Since both, the reporter genes under the control of the bidirectional promoter and the divergent At5g10290 and At5g10300 genes, were up-regulated in response to pest feeding, wounding and Flg22 treatments but not with chitin, defense signaling pathways in transgenic plants and in T-DNA insertion lines were tested. The expression levels of three genes associated with hormonal pathways such as the plant defensin PDF1.2, the vegetative storage VSP2 and the pathogenesis related PR1, involved in ethylene/jasmonic acid (JA) and salicylic acid (SA) responses, were quantified. Interestingly, the PDF1.2 gene was highly over-expressed in response to spider mite in the Arabidopsis lines transformed with the bidirectional promoter in contrast to the no significant induction quantified in the plants transformed with the control pmdc107ø vector. Besides, mite infestation induced the expression of VSP2 and PR1 genes in all tested plants. In contrast, the induction of VSP2 and PDF1.2 genes mediated by P. brassicae feeding was observed in all Arabidopsis genotypes while the PR1 gene was always down-regulated after this biotic stress (Supplementary Figure 7). Regarding the wounding and elicitor treatments, the PDF1.2 gene was higher induced by Flg22 in plants with the fusion-constructs than in control plants, while was similarly induced by wounding and chitin in all tested plants, independently of the bidirectional promoter presence (Supplementary Figure 8). In addition, the expression profiles of VSP2 and PR1 genes showed a up-regulation mediated by the three treatments, with few differences among all Arabidopsis genotypes (Supplementary Figure 8). Remarkably, the basal transcript content of PDF1.2, VSP2, and PR1 genes in the two Arabidopsis T-DNA insertion lines did not show significant changes in comparison to the expression levels quantified in Col-0 plants (Supplementary Figure 9).
Discussion
Gene pairs oriented in divergent transcriptional configuration and controlled by bidirectional promoters are abundant in plant genomes. In Arabidopsis, more than 13% of its genes are driven by bidirectional promoters (Wang et al., 2009), but the biological relevance of these intergenic regions has been only studied in some cases (Mitra et al., 2009; Kourmpetli et al., 2013; Pratibha et al., 2013; Mishra and Grover, 2014; Liu et al., 2015; Sharma et al., 2017).
Making a search of up-regulated genes in response to spider mite feeding, we identified two over-expressed divergent genes, At5g10290 and At5g10300, separated by an intergenic region. Here, it is proven that the 645 bp intergenic sequence between the transcription start sites of both genes corresponds to a bidirectional promoter. The At5g10290 gene encoded a LRR receptor kinase that could be involved in the perception of molecules derived from phytophagous and/or pathogens initiating the plant transduction defense pathway. The At5g10300 gene encoded a hydroxynitrile lyase that catalyze an enzymatic reaction that release hydrogen cyanide, a toxic compound against herbivores and pathogens (Zhu et al., 2015). The gene pair potentially involved in plant defense, was simultaneously transcribed in the same tissues but the promoter strength was asymmetric as was shown by the differential levels of gene expression quantified by RT-qPCR assays. The asymmetry might be caused by the presence of conserved cis-acting elements in the opposite strands with potential antagonistic or synergistic roles to regulate the transcription (Liu et al., 2015). Co-expression does not necessarily mean identical spatial and temporal expression or equal responses to stresses. A coordinated expression of genes to overcome abiotic and biotic cues is regulated by the cis-motifs contained in the promoter. Thus, T-DNA insertions disturbing the bidirectional promoter architecture presented a negative effect on the gene responses, and subsequently on the damage produced by T. urticae feeding. Since both insertion lines caused the silencing of the opposite genes, the transcriptional regulation most likely involve the whole promoter region. Sequences upstream T-DNA insertion points control both, the nearest and the farthest genes, which reinforces the idea of a strongly coordinated response against a specific stress.
In this context and as many authors previously did, two reporter genes were fused to the promoter in both orientations, for a deep analysis of its function in planta. In previous studies, GUS and GFP genes were the reporters most commonly used but the GUS gene requires an enzymatic reaction to generate the reporter product (Mitra et al., 2009; Wang et al., 2016; Oropeza-Aburto et al., 2017; Yang et al., 2018). This step implies not only a cost and time-consuming but also a time lag between the two reporter quantification assays. To avoid it, enhanced variants of GFP and CherryFP genes encoding fluorescent proteins with different excitation/emission wavelengths were selected here for a rapid gene co-expression analysis in vivo. Transient and stable transformation assays revealed that the two genes coordinately regulated their transcripts and the fluorescent emissions, in terms of constitutive as well as induced expression profiles in response to biotic treatments. The accumulation of mGFP mRNAs was always higher than the mCherryFP mRNA content, independent on the gene orientation, tissue sample and stress treatment, probably due to a major stability of mGFP transcripts. However, gene expression data correlated to the fluorescence observed under the microscope for both reporters.
Interestingly, confocal images and mRNA expression patterns clearly indicated that the bidirectional promoter was inducible by T. urticae and P. brassicae feeding as well as by wounding. Wounding, although is not considered a biotic stress, is an additional effect produced as consequence of the feeding process. These results were in agreement with the up-regulated expression of the pair of genes located on the opposite promoter strands in the resistant Bla-2 Arabidopsis accession, after mite infestation (Zhurov et al., 2014). In addition, the fluorescence emission of both genes in infested Arabidopsis plants corroborated the asymmetrical character of the promoter, with stronger activity in the forward than in the reverse orientation.
There are previous examples of bidirectional promoters whose activity was induced by biotic stresses. Shin et al. (2003) reported the up-regulation of the GUS gene controlled by a bidirectional promoter from hot pepper in response to Tobacco Mosaic Virus infection, in transgenic tobacco plants. Following the same experimental strategy, Banerjee et al. (2013) showed the induction of the GUS staining and the enzymatic GUS activity driven by an Arabidopsis bidirectional promoter when transformed tobacco plants were inoculated with spores from Peronospora tabacina. The main difference between these results and the data presented here is that in the mentioned examples only one GUS-construct with the reporter gene in one orientation was checked. Our findings demonstrated the simultaneous induction of the two reporter genes located in both orientations of the bidirectional promoter mediated by arthropod phytophagous. In addition to the promoter inducibility produced by pests, the pathogen elicitor flagellin also provoked the up-regulation of the reporter genes.
The coordinated function of a bidirectional promoter has to be achieved by the control of the regulatory motifs located in its opposite strands. Computational analysis evidenced the existence of cis-elements potentially recognized by TFs within the intergenic region, mainly from the DOF family but also from the AP2/ERF, Golden 2-like and bHLH families. Most of these motifs are commonly observed in promoters of genes encoding defense responses to pathogen infection and insect attack (Barah et al., 2013; Amorim et al., 2017). Interestingly, putative DOF-binding sequences were particularly abundant in this bidirectional promoter, with six elements close located among them. Although this TF family is not described as the most relevant in defense to biotic stresses, the functional validation of the role played by DOFs in response to pest attack has been shown in the case of the AtDof1.1. This TF, also known as OBP2, was up-regulated by Spodoptera littoralis and methyl jasmonate. It took part of the network involved in the biosynthesis of important defense compounds such as glucosinolates, characteristics of the Brassicaceae family (Skirycz et al., 2006). Likewise, Skirycz et al. (2007) reported that the AtDOF4.2 altered the flavonoid and the hydrocinnamic acid synthesis, which potentially modified the suberin production, an important component of the physical plant barriers with protective roles. In contrast, the importance of the bHLH and AP2/ERF TFs in the regulation of plant immunity to pathogens and insect herbivory is widely reported (Tsuda and Somssich, 2015). Notably, whereas ethylene response factors (ERF) are integrators of hormonal pathways and participate in the transcriptional regulation of JA/ethylene-responsive defense genes (Huang et al., 2016), the basic helix-loop-helix (bHLH) factors are mainly related to the control of different subsets of the JA-dependent transcriptional defense responses (Fernandez-Calvo et al., 2011; Schweizer et al., 2013). Finally, Golden 2-like TFs have been related to the defense against pathogens affecting in some way SA and JA signaling regulatory network (Chen et al., 2016). In consequence, a complex setup of defenses is achieved through the binding to specific responsive elements contained in the promoter sequence, controlled by a hormonal crosstalk with a pivotal role in the orchestration of defenses.
The complexity in the regulation of this network is further supported by the differential over-expression of defense products derived from JA, Et and SA biosynthesis pathways. Whereas VSP2 and PR1 genes were similarly induced by most of the stresses in plants transformed with the fusion-construct or with the control vector, the expression of PDF1.2 after spider mite and Flg22 treatments was significantly higher when the construct harboring the promoter and the reporter genes was present. These alterations in PDF1.2 gene expression were not due to a positional effect of the transgenes since expression patterns were similar between independent transgenic lines. Most likely, the presence of an additional copy of the bidirectional promoter may be in some way modifying the final response associated to the hormonal signaling pathways. This effect cannot be directly attributed to an altered regulation of the endogenous bidirectional promoter caused by the presence of the additional copy. Since the expression of the flanking At5g10300 and At5g10290 genes was similar to that observed in the control line in response to any biotic treatment, hormonal signaling alterations may be probably associated to indirect effects such as the scavenging of transcriptional factors important for a full hormonal response. Interestingly, the basal expression levels of the three hormone-related genes were similar in the two T-DNA insertion lines than in the Col-0 plants, which indicates that the relevance of the bidirectional promoter in the regulation of hormonal signaling pathways relies in the presence of the biotic stress. In the case of the plant response to spider mites, alterations in the expression of PDF1.2 suggest that the ethylene/JA branch-signaling pathway can be involved in the regulation of the Arabidopsis gene pair controlled by the bidirectional promoter to generate defenses. Accordingly, it has been already reported that JA is an essential hormone, but not the unique, for establishing the mite-induced defense responses (Zhurov et al., 2014; Santamaria et al., 2017, 2019).
The relevant finding emerging from this study is that two genes involved in defense against biotic stresses, particularly in the response to spider mite feeding, are packed together, induced by different biotic stresses and simultaneously controlled by a common regulatory mechanism. The bidirectional promoter is the key element in such an efficient defense mechanism and it could be the base to design regulatory modules with the potential to express multigene traits in a coordinately regulated manner.
Data Availability
All datasets for this study are included in the manuscript and the Supplementary Files.
Author Contributions
ID conceived the research. AA and MES performed most of the experimental research. MM performed the bioinformatics analyses. PG-M helped with the microscopic data. All authors participated in the design, acquisition, analysis, and interpretation of the data for the work, and contributed to the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by the project BIO2017-83472-R from the Ministerio de Ciencia y Universidades of Spain. A Ph.D. Studentship from the Universidad Politecnica de Madrid (UPM) financed AA, and a Ramon y Cajal Grant (RYC-2017-21814) financed MES.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00930/full#supplementary-material
References
- Amorim L. L. B., da Fonseca Dos Santos R., Neto J. P. B., Guida-Santos M., Crovella S., Benko-Iseppon A. M. (2017). Transcription factors involved in plant resistance to pathogens. Curr. Protein Pept. Sci. 18 335–351. 10.2174/1389203717666160619185308 [DOI] [PubMed] [Google Scholar]
- Banerjee J., Sahoo D. K., Dey N., Houtz R. L., Maiti I. B. (2013). An intergenic region shared by At4g35985 and At4g35987 in Arabidopsis thaliana is a tissue specific and stress inducible bidirectional promoter analysed in transgenic Arabidopsis and tobacco plants. PloS One 8:e79622 10.1371/journal.pone.0079622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barah P., Winge P., Kusnierczyk A., Tran D. H., Bones A. M. (2013). Molecular signatures in Arabidopsis thaliana in response to insect attack and bacterial infection. PloS One 8:e58987 10.1371/journal.pone.0058987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero-Sicilia C., Hernando-Amado S., Gonzalez-Melendi P., Carbonero P. (2011). Structure, expression profile and subcellular localisation of four different sucrose synthase genes from barley. Planta 234 391–403. 10.1007/s00425-011-1408-x [DOI] [PubMed] [Google Scholar]
- Cazaux M., Navarro M., Bruinsma K. A., Zhurov V., Negrave T., Van Leeuwen T., et al. (2014). Application of two-spotted spider mite Tetranychus urticae for plant-pest interaction studies. J. Vis. Exp. 89:e51738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Wen B., Liu L., Chen X., Gao D., Li L., et al. (2016). GOLDEN 2-like transcription factors of plants. Front. Plant Sci. 7:1509. 10.3389/fpls.2016.01509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C. Y., Krishnakumar V., Chan A. P., Thibaud-Nissen F., Schobel S., Town C. D. (2017). Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89 789–804. 10.1111/tpj.13415 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium- mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.000343.x [DOI] [PubMed] [Google Scholar]
- Curtis M. D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133 462–469. 10.1104/pp.103.027979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhadi S. R., Deshpande A., Driscoll K., Ramakrishna W. (2013). Major cis-regulatory elements for rice bidirectional promoter activity reside in the 5′-untranslated regions. Gene 526 400–410. 10.1016/j.gene.2013.05.060 [DOI] [PubMed] [Google Scholar]
- Dhadi S. R., Krom N., Ramakrishna W. (2009). Genome-wide comparative analysis of putative bidirectional promoters from rice, Arabidopsis and Populus. Gene 429 65–73. 10.1016/j.gene.2008.09.028 [DOI] [PubMed] [Google Scholar]
- Fernandez-Calvo P., Chini A., Fernandez-Barbero G., Chico J. M., Gimenez-Ibañez S., Geerinck J., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23 701–715. 10.1105/tpc.110.080788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K. A., Pachter L., Poliakov A., Rubin E. M., Dubchak I. (2004). VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32 W273–W279. 10.1093/nar/gkh458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O. (2010). Seaview version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27 221–224. 10.1093/molbev/ [DOI] [PubMed] [Google Scholar]
- Huang P. Y., Catinot J., Zimmerli L. (2016). Ethylene response factors in Arabidopsis immunity. J. Exp. Bot. 67 1231–1241. [DOI] [PubMed] [Google Scholar]
- Jin J. P., Tian F., Yang D. C., Meng Y. Q., Kong L., Luo J. C., et al. (2017). PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45 D1040–D1045. 10.1093/nar/gkw98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourmpetli S., Lee K., Hemsley R., Rossignol P., Papageorgiou T., Drea S. (2013). Bidirectional promoters in seed development are related hormone/stress responses. BMC Plant Biol. 13:187. 10.1186/1471-2229-13-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krom N., Ramakrishna W. (2010). Conservation, rearrangement, and deletion of gene pairs during the evolution of four grass genomes. DNA Res. 17 343–352. 10.1093/dnares/dsq022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., AlAbed D., Whitteck J. T., Chen W., Bennett S., Asberry A., et al. (2015). A combinatorial bidirectional and bicistronic approach for coordinate multi-gene expression in corn. Plant Mol. Biol. 87 341–353. 10.1007/s11103-015-0281-6 [DOI] [PubMed] [Google Scholar]
- Liu S. J., Yue Q. J., Zhang W. (2015). Structural and functional analysis of an asymmetric bidirectional promoter in Arabidopsisi thaliana. J. Integr. Plant Biol. 57 162–170. 10.111/jipb.12308 [DOI] [PubMed] [Google Scholar]
- Liu X., Yang W., Li Y., Zhou X., Zhao Q., Fan Y., et al. (2016). The intergenic region of the maize defensin-like protein genes Def1 and Def2 functions as an embryo-specific asymmetric bidirectional promoter. J. Exp. Bot. 67 4403–4412. 10.1093/jxb/erw226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mehrotra R., Gupta G., Sethi R., Bhalothia P., Kumar N., Mehrotra S. (2011). Designer promoter: an artwork of cis engineering. Plant Mol. Biol. 75 527–536. 10.1007/s11103-011-9755-3 [DOI] [PubMed] [Google Scholar]
- Mishra R. C., Grover A. (2014). Intergenic sequence between Arabidopsis caseinolytic protease B-cytoplasmic/heat shock protein100 and choline kinase genes functions as a heat-inducible bidirectional promoter. Plant Physiol. 166 1646–1658. 10.1104/pp.114.250787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra A., Han J., Zhang Z. J., Mitra A. (2009). The intergenic region of Arabidopsis thaliana cab1 and cab2 divergent genes functions as a bidirectional promoter. Planta 229 1015–1022. 10.1007/s00425-008-0859-1 [DOI] [PubMed] [Google Scholar]
- Oñate-Sánchez L., Vicente-Carbajosa J. (2008). DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res. Notes 1:93. 10.1186/1756-0500-1-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oropeza-Aburto A., Cruz-Ramirez A., Mora-Macias J., Herrera-Estrella L. (2017). A phosphate starvation-driven bidirectional promoter as a potential tool for crop improvement and in vitro plant biotechnology. Plant Biotechnol. J. 15 558–657. 10.1111/pbi [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I., Loots G. G., Hardison R. C., Miller W., Stubbs L. (2004). zPicture: dynamic alignment and visualization tool for analyzing conservation profiles. Genome Res. 14 472–477. 10.1101/gr.2129504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratibha P., Singh S. K., Sharma I., Kumar R., Srinivasan R., Bhat S. R., et al. (2013). Characterization of a T-DNA promoter trap line of Arabidopsis thaliana uncovers a cryptic bi-directional promoter. Gene 524 22–27. 10.1016/j.gene.2013.04.031 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W. (2001). Molecular Cloning: A Laboratory Manual, 3rd Edn. New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Santamaria M. E., Cambra I., Martinez M., Pozancos C., Gonzalez-Melendi P., Grbic V., et al. (2012). Gene pyramiding of peptidase inhibitors enhances plant resistance to the spider mite Tetranychus urticae. PloS One 7:e43011. 10.1371/journal.pone.0043011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria M. E., Martinez M., Arnaiz A., Ortego F., Grbic V., Diaz I. (2017). MATi, a novel protein involved in the regulation oh herbivore-associated signalling pathways. Front. Plant Sci. 8:975 10.3389/fpls.2017.00975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria M. E., Martinez M., Arnaiz A., Rioja C., Burow M., Grbic V., et al. (2019). An Arabidopsis TIR-lectin two-domain protein confers defence properties against Tetranychus urticae. Plant Physiol. 179 1298–1314. 10.1104/pp.18.00951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria M. E., Toorop P. E., Rodriguez R., Cañal M. J. (2010). Dormant and non-dormant Castanea sativa Mill. Plant Sci. 178 55–60. 10.1016/j.plantsci.2009.10.002 [DOI] [Google Scholar]
- Schweizer F., Fernandez-Calvo P., Zander M., Diez-Diaz M., Fonseca S., Glauser G., et al. (2013). Arabidopsis basic Helix-Loop-Helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolates biosynthesis, insect performance, and feeding behaviour. Plant Cell 25 3117–3132. 10.1105/tpc.113.115139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., Kumar V., Kumar-Singh S., Thakur S., Siwach P., Sreenivasulu Y., et al. (2017). Promoter trapping and deletion analysis show Arabidopsis thaliana APETALA2 gene promoter is bidirectional and functions as a pollen and ovule-specific promoter in the reverse orientation. Appl. Biochem. Biotechnol. 182 1591–1604. 10.1007/s12010-017-2420-9 [DOI] [PubMed] [Google Scholar]
- Shin R., Kim M. J., Paek K. H. (2003). The CaTin1 (Capsicum annuum TMV-induced Clone 1) and CaTin1-2 genes are linked head-to-head and share a bidirectional promoter. Plant Cell Physiol. 44 549–554. 10.1093/pcp/pcg069 [DOI] [PubMed] [Google Scholar]
- Shwarz Z., Jolly S. O., Steinmetz A. A. (1981). Overlapping divergent genes in maize chloroplast chromosome and in vitro transcription of the gene for tRNAHIS. Proc. Natl. Acad. Sci. U.S.A. 78 3423–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Sahi C., Grover A. (2009). Chymotrypsin protease inhibitor genes family in rice: genomic organization and evidence for the presence of a bidirectional promoter shared between two chymotrypsin protease inhibitor gens. Gene 428 9–19. 10.1016/j.gene.2008.09.028 [DOI] [PubMed] [Google Scholar]
- Skirycz A., Jozefczuk S., Stobiecki M., Muth D., Zanor M. I., Witt I., et al. (2007). Transcription factor AtDOF4;2 affects phenylpropanoid metabolism in Arabidopsis thaliana. New Phytol. 175 425–438. 10.1111/j.1469-8137.2007.02129.x [DOI] [PubMed] [Google Scholar]
- Skirycz A., Reichelt M., Burow M., Birkemeyer C., Rolcik J., Kopka J., et al. (2006). DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 47 10–24. 10.1111/j.1365-313X.2006.02767.x [DOI] [PubMed] [Google Scholar]
- Tsuda K., Somssich I. E. (2015). Transcriptional networks in plant immunity. New Phytol. 206 932–947. 10.1111/nph.13286 [DOI] [PubMed] [Google Scholar]
- Vogl T., Kickenweiz T., Pitzer J., Sturmberger L., Weninger A., Biggs B. W., et al. (2018). Engineered bidirectional promoters enable rapid multi-gene co-expression optimization. Nature Comm. 9:3589 10.1038/s41467-018-0591-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wan L., Li D., Zhu L., Qian M., Deng M. (2009). Searching for bidirectional promoters in Arabidopsis thaliana. BMC Bioinform. 10:S29. 10.1186/1471-2105-10SI-S29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Yan Y., Yang M., Zhou F., Chen H., Lin Y. (2016). Isolation and functional characterization of bidirectional promoters in rice. Front. Plant Sci. 7:766. 10.3389/fpls.2016.00766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., Glazebrook J. (2002). Transformation of Agrobacterium Using the Freeze-thaw Method. New York, NY: Cold Spring Harbor Laboratory Press, 10.1101/pdb.prot4666 [DOI] [PubMed] [Google Scholar]
- Xie M., He Y., Gan S. (2001). Bidirectionalization of polar promoters in plants. Nature Biotechnol. 19 677–679. 10.1038/90296 [DOI] [PubMed] [Google Scholar]
- Yang J., Wang X., Hasi A., Wang Z. (2018). Structural and functional analysis of a bidirectional promoter from Gossypium hirsutum in Arabidopsis. Int. J. Mol. Sci. 19:e3291. 10.3390/ijms19113291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., McHale L. K., Finer J. J. (2019). Changes to the core and flanking sequences of G-box elements lead to increases and decreases in gene expression in both native and synthetic soybean promoters. Plant Biothecnol. J. 17 724–735. 10.1111/pbi.13010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Lei Y., Lin S., Zhang Q., Zhang Z. (2011). Bidirectionalization of methyl jasmonate-inducible plant promoter. Biotechnol. Lett. 33 387–393. 10.1007/s10529-010-0431-5 [DOI] [PubMed] [Google Scholar]
- Zhu W., Liu Y., Zhang R. (2015). A QM/MM study of the reaction mechanism of (R)-hydroxynitrile lyases from Arabidopsis thaliana (AtHNL). Proteins 83 66–77. [DOI] [PubMed] [Google Scholar]
- Zhurov V., Navarro M., Bruinsma K. A., Arbona V., Santamaria M. E., Cazaux M., et al. (2014). Reciprocal responses in the interaction between Arabidopsis and the cell-content-feeding chelicerate herbivore spider mite. Plant Physiol. 164 384–399. 10.1104/pp.113.231555 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets for this study are included in the manuscript and the Supplementary Files.